Main Text
The human brain is considered to be the most complex and mysterious organ of our body. Due to technological and ethical considerations, we have not been able to fully map the multi-scale structures and understand the organizing principles. As the origin of human consciousness and intelligence, it inspires awe and marvel. Since the beginning of the 21st century, brain science has increasingly become popular around the whole world, and numerous large-scale brain initiatives have been launched to explore the structure and function of the human brain, to decode the nature of consciousness, and to address the problem of neuropsychiatric diseases.1
Exploring the human brain is just like exploring a new continent, as one would need a brain map for navigation. Therefore, brain atlases have always been an important approach to the study of brain structure and function. Compared with understanding the external world, our acquisition of the knowledge of our brain has gone through a longer and more complicated journey. Beginning with the detailed descriptions of brain structure by a 16th-century Flemish anatomist Andreas Vesalius (1514–1564), a large number of neuroanatomical descriptions, pictures, and images have emerged. It is generally accepted that brain function depends largely on its structure, and brain regions with different architectures are likely to perform different functions. Therefore, it is essential to identify the distinct regions for a better understanding of brain functions. For more than 100 years, the study of the human brain has been directed in the important area of brain parcellation, from the classic Brodmann map to the Jülich-Brain proposed by Amunts et al.,2 and to the recently published human Brainnetome atlas3 and Human Connectome Project (HCP) multimodal parcellation4 using in vivo MRI data.
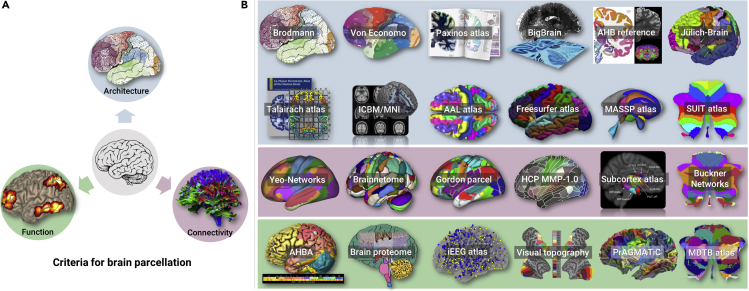
A retrospective look at the history of the human brain atlases can reveal the trends and tell us about future directions. Three main different structural and functional criteria or features, namely architecture, functional activities, and anatomical and functional connectivity (Figure 1A), have been applied in human brain parcellation, and there are many milestones in the history of mapping the human brain (Figure 1B). It is generally accepted that the story of human brain mapping began in 1909 when the German neuroanatomist Korbinian Brodmann (1868–1918) published a book in which he processed postmortem human and non-primate brains with Nissen's stain and parcellated each brain into more than 50 regions based on the size, shape, and density of neuronal cells (i.e., cytoarchitecture). This is the well-known Brodmann atlas, which was the classic map of the human brain and is still widely used today. During the same period, Vogt et al. used myelin information as a feature (i.e., myelin architecture) to divide the cerebral cortex into more than 200 regions, the major ones of which were similar to those based on cytoarchitecture. Later, Von Economo and Koskinas developed cytoarchitectonic techniques, but, using refined protocols, obtained a whole-brain map in 1925, and identified more cortical areas. In terms of location and size, the cortical areas vary between different brains. These early brain atlases were mainly based on limited postmortem brains, which were inevitably affected by individual differences, and resulted in large discrepancies when mapped to individual brains in applications. To address these questions, the Jülich research center in Germany has been dedicated to creating the Jülich-Brain for decades, which is a probabilistic map of cellular structures synthesized from 10 brain specimens, reflecting structural variations across individual brains. In a previous study by the same group, they collaborated with a research team from McGill University in Canada to obtain the BigBrain template with a resolution of 20μm using tissue sections and 3D image reconstruction, which greatly improved the understanding of the microscopic architecture of the human brain. These microstructure brain maps can provide reliable histological information, serving as a reference and validation for other types of brain atlases. However, all of these above-mentioned human brain atlases are still based on information collected from postmortem brains, and the histology-based cytoarchitectonic mapping procedures are extremely laborious and time consuming.
Figure 1.
Parcellation Criteria and Typical Human Brain Atlases
Criteria for human brain parcellation (A), including architecture, function, and connectivity, and the collections of available human brain atlases (B). As one of the criteria for brain parcellation, function contains diverse and complex contents, so here we list information including genetic and protein expressions in this category. The available human brain atlases are listed alphabetically as follows:
AAL atlas: automated anatomical labeling atlas, https://www.gin.cnrs.fr/en/tools/aal/
AHBA: Allen Human Brain Atlas integrating anatomic and genomic information https://human.brain-map.org
AHB reference: Allen Human Brain reference atlas, a 2D coronal cellular resolution anatomical reference atlas, http://atlas.brain-map.org
BigBrain: microscopic 3D model with maps of cytoarchitectonic structures, https://bigbrainproject.org
Brain proteome: protein-coding genes are classified based on RNA expression in the brain https://www.proteinatlas.org/humanproteome/brain
Brainnetome: human Brainnetome atlas based on connectional architecture http://atlas.brainnetome.org
Buckner-networks: cerebellar parcellation estimated by intrinsic functional connectivity http://surfer.nmr.mgh.harvard.edu/fswiki/CerebellumParcellation_Buckner2011
Caret surface: http://brainvis.wustl.edu/wiki/index.php/Caret:About
Freesurfer atlas: Desikan-Killiany Atlas and Destrieux Atlas https://surfer.nmr.mgh.harvard.edu/fswiki/CorticalParcellation
Gordon parcel: resting-state functional connectivity-boundary map-derived parcels https://sites.wustl.edu/petersenschlaggarlab/resources/
HCP MMP-1.0: a multimodal parcellation of the human cerebral cortex, https://balsa.wustl.edu/study/show/RVVG
ICBM/MNI: an unbiased standard MRI brain template, http://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin2009
iEEG Atlas: Montreal Neurological Institute (MNI) Open Intracranial EEG Atlas, https://mni-open-ieegatlas.research.mcgill.ca
Jülich-Brain: a 3D probabilistic atlas of the human brain's cytoarchitecture, https://jubrain.fz-juelich.de/apps/cytoviewer2/cytoviewer.php##mitte
https://ebrains.eu/services/atlases/brain-atlases
MDTB atlas: multi-domain task battery maps http://www.diedrichsenlab.org/imaging/mdtb.htm
Paxinos atlas: the fourth edition of Atlas of the Human Brain, http://atlas.thehumanbrain.info
PrAGMATiC: A Probabilistic and Generative Model of Areas Tiling the Cortex, https://gallantlab.org/huth2016
Subcortex parcel: Melbourne subcortex atlas, https://github.com/yetianmed/subcortex
SUIT atlas: a spatially unbiased atlas template of the human cerebellum, http://www.diedrichsenlab.org/imaging/suit.htm
Talairach atlas: a 3D coordinate system of Talairach space, http://www.talairach.org
Visual topography: probabilistic maps of visual topography in human cortex https://scholar.princeton.edu/napl/resources
Yeo-Networks: cortical parcellation estimated by intrinsic functional connectivity, https://surfer.nmr.mgh.harvard.edu/fswiki/CorticalParcellation_Yeo2011
With the advancement of neuroimaging technology, especially in the past 40 years, non-invasive neuroimaging methods represented by MRI have made great progress. In the early days, the Tournoux-Talairach 3D coordinate system opened the era of digitalization of brain mapping. To overcome the shortcomings of the Talairach template based on data from a single subject (a 60-year-old woman), Evans and his group from Canada established the MNI (Montreal Neurological Institute) brain template using a large sample of normal human brain imaging data starting in 1995. The anatomical reference space of MNI was also defined to facilitate comparisons between individuals. Since then, several probabilistic brain atlases have been developed in this de facto standard reference space, which was mapped by MRI using the macro-scale anatomical features of the human brain, such as the automated anatomical labeling atlas, the Harvard-Oxford atlas, the Desikan Kiliany atlas in FreeSurfer, and cerebellar SUIT (spatially unbiased template) probability atlas. However, for most of the existing MRI-based brain atlases using structural magnetic resonance images, the brain regions were defined mainly according to the topological distribution of sulci and gyri, with coarse boundaries and even obvious errors, making it difficult to correspond to the functional anatomy.
In recent years, it has been possible to non-invasively delineate the structural and functional areas of the human brain and map its connectivity patterns using multimodal brain imaging, which provides a new approach to the development of the next generation of human brain atlases. However, there are still some problems that need further study to resolve.
Firstly, most of these human brain atlases, including the current version of the human Brainnetome atlas, are still population-based atlases. Therefore, to construct individualized brain atlases for specific individuals remains a challenging topic due to the large variations in brain size, connectivity, and function across subjects. To meet the requirements in clinical applications, a precise individual brain atlas is needed to reflect the individual characteristics of the brain architecture, as well as demonstrate high repeatability and consistency across different MRI scanners and brain states. Recently, several approaches utilizing machine learning for integrating connectivity information for brain atlas individualization have been proposed. For example, Tian, et al.5 have trained machine learning classifiers to recognize functional boundaries within the human subcortex for each individual. The promising results obtained suggest that advanced machine learning approaches deserve to be considered as a powerful tool for mapping the human brain.
Secondly, it is conceptually and technically challenging to integrate the information of different aspects of the human brain, including the micro- and macro-structural segregation, the regional specialization of function, and connectivity, as well as temporal dynamics. To understand human brain organization, it is inevitable that these different aspects of brain mapping need to be integrated into a multimodal human brain atlas. Traditional neuroanatomical investigations have been complemented by emerging methods for identifying distinct brain areas and functional imaging for visualizing response properties of the brain to experimentally purported tasks and brain connectivity. The connectivity-based approach (i.e., the delineation of cortical areas based on locally distinct connection patterns), in turn, may be pursued using structural (white matter fiber tracts) and functional (coherent fluctuations in brain activity signals) connectivity as well as co-activation patterns during neuroimaging experiments. While these methods present unprecedented tools for mapping the human brain, the relationship between the ensuing structurally, functionally, and connectionally defined modules, the convergence and discrepancies among approaches, and finally the synthesis toward a deeper understanding of brain organization is yet to be investigated.
Thirdly, it needs to combine invasive neuroscience techniques and using non-human primate models to verify the biological substrates of the imaging-based atlas. Non-human primates have the closest evolutionary relationship with humans, especially in terms of the structure and function of the nervous system, and they are more closely related to humans than other experimental animals. At present, mapping of the non-human primate brains is incomplete and relatively preliminary, which has limited our understanding of brain structure and function. Therefore, comparative studies among primate species will help to further clarify the similarities and differences in the structure and function of the brain between non-human primates and humans. It is not only important to understand the unique cognitive functions of the human brain but also essential for the establishment of non-human primate models of major brain diseases and the development of new techniques for diagnosis and therapy.
In summary, numerous forms of brain atlases have been developed to map the human brain. The trends in human brain mapping are from the early printed 2D brain atlases to the current digital 3D and 4D brain atlases; from cross-sectional data based on specimens to atlases based on in vivo multimodal neuroimaging; from single modal atlases with only anatomical brain information to multimodal brain atlases integrating population-based anatomical and functional information. In the future, with the advancement of new technologies and tools for brain research, the human brain atlas will go local instead of global and dynamic instead of static, which will be consistent with other biological information, such as the gene and protein expression patterns, cell types, wiring patterns, and the spatiotemporal dynamic changes during normal development and the aging process, or in different disease states. The next generation of human brain atlases will provide basic tools for probing the mysteries of the most complex organ, as well as providing solutions for biomarker discoveries for early diagnosis and therapy of various brain diseases.
Published Online: December 16, 2020
References
- 1.Adams A., Albin S., Amunts K., et al. International brain initiative: an innovative framework for coordinated global brain research efforts. Neuron. 2020;105:212–216. doi: 10.1016/j.neuron.2020.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Amunts K., Mohlberg H., Bludau S., et al. Julich-Brain: a 3D probabilistic atlas of the human brain's cytoarchitecture. Science. 2020;369:988–992. doi: 10.1126/science.abb4588. [DOI] [PubMed] [Google Scholar]
- 3.Fan L., Li H., Zhuo J., et al. The human brainnetome atlas: a new brain atlas based on connectional architecture. Cereb. Cortex. 2016;26:3508–3526. doi: 10.1093/cercor/bhw157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glasser M.F., Coalson T.S., Robinson E.C., et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian Y., Margulies D.S., Breakspear M., et al. Topographic organization of the human subcortex unveiled with functional connectivity gradients. Nat. Neurosci. 2020;23:1421–1432. doi: 10.1038/s41593-020-00711-6. [DOI] [PubMed] [Google Scholar]