Abstract
Hepatitis B virus (HBV), which was discovered in 1965, is a threat to global public health. HBV infects human hepatocytes and leads to acute and chronic liver diseases, and there is no cure. In cells infected by HBV, viral DNA can be integrated into the cellular genome. HBV DNA integration is a complicated process during the HBV life cycle. Although HBV integration normally results in replication-incompetent transcripts, it can still act as a template for viral protein expression. Of note, it is a primary driver of hepatocellular carcinoma (HCC). Recently, with the development of detection methods and research models, the molecular biology and the pathogenicity of HBV DNA integration have been better revealed. Here, we review the advances in the research of HBV DNA integration, including molecular mechanisms, detection methods, research models, the effects on host and viral gene expression, the role of HBV integrations in the pathogenesis of HCC, and potential treatment strategies. Finally, we discuss possible future research prospects of HBV DNA integration.
Key words: hepatitis B virus, integration, non-homologous end joining, insertional mutagenesis, hepatocellular carcinoma
Graphical Abstract
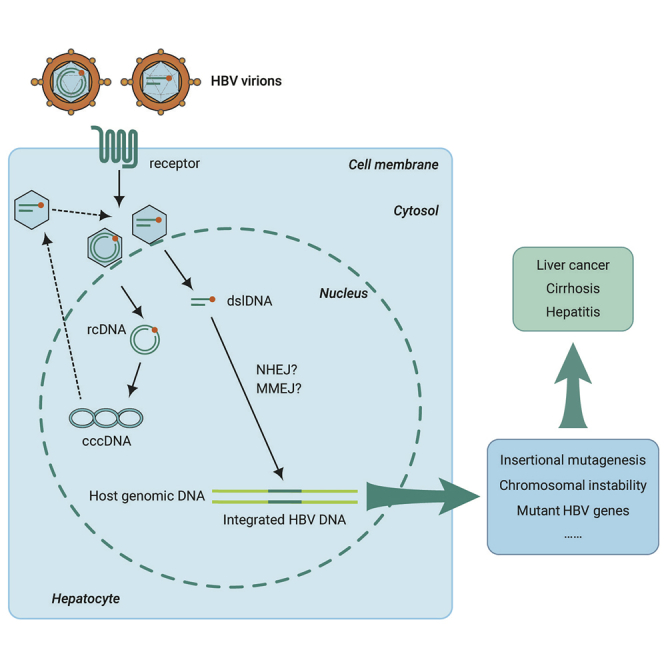
Public Summary
-
•
HBV DNA integration is associated with hepatocarcinogenesis via multiple mechanisms
-
•
HBV double-stranded linear DNA (dslDNA) is the dominant substrate for integration into the host genome
-
•
The insertion sites of HBV DNA integration occur throughout the whole host genome using the NHEJ or MMEJ DNA repair pathway
-
•
HBV DNA integration should be used as a clinical indicator for disease monitoring and treatment of patients with HBV infection
Main Text
Introduction
In 1965, Dr. Baruch Blumberg made the discovery of the “Australia antigen,” subsequently identified as the hepatitis B virus (HBV) surface antigen (HBsAg).1 Because of this work, Dr. Blumberg was awarded the Nobel Prize in Physiology or Medicine in 1976. HBV infection chronically infects about 250 million individuals worldwide and is a major global health burden.2,3 Chronic HBV infection is recognized as a high risk factor for developing liver cirrhosis and hepatocellular carcinoma (HCC).4 After infection, the viral genome is established as a minichromosome, called covalently closed circular DNA (cccDNA). The cccDNA is essential to establishing persistent HBV infection as a stable intracellular HBV replication intermediate, which is localized to the nucleus of infected cells as an episomal plasmid-like molecule that can produce progeny virus.5 Stably integrated HBV DNA in the host genome is another stable form of viral DNA. Although no progeny virus is produced, integrated HBV DNA can produce viral RNAs and proteins.6, 7, 8, 9 HBV DNA integration was first reported in 1980 in human HCC.10,11 Since then, technologies used in HBV integration research include Southern blots, in situ hybridization, polymerase chain reaction (PCR), and next-generation sequencing (NGS).7,12, 13, 14, 15, 16 HBV DNA integration events occur at a rate of ~1 integration per 103–104 infected cells in animal models17, 18, 19 and ~1 integration per 104 infected cells in the early viral life cycle in an in vitro infection model.20 HBV DNA integration has a close relationship with HCC and many studies have been done to explore its role in the development of HCC. In the genome of hepatic cancer cells, HBV DNA integration occurs more often (86.4%) than in normal liver tissues (30.7%).21 Observed mechanisms leading to tumorigenesis include insertional mutagenesis acting in cis of key HCC-associated genes, induction of chromosomal instability, and the expression of mutant HBV genes from integrated HBV DNA.21, 22, 23, 24 Here, we review and discuss current research advances in HBV DNA integration, including molecular mechanisms, detection methods, research models, and roles in disease and potential treatment strategies.
HBV Life Cycle
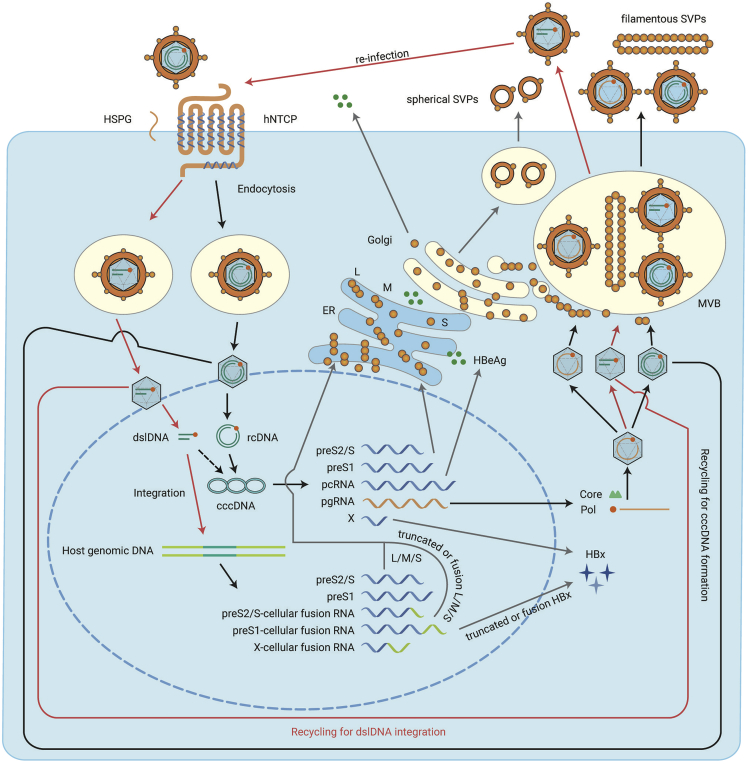
HBV belongs to the Hepadnaviridae, a family of small enveloped hepatotropic DNA viruses. Unlike other DNA viruses, HBV replicates its DNA through reverse transcription, which also determines the complexity of the HBV life cycle. The HBV life cycle is complex, including processes, such as viral entry, cccDNA formation, transcription, replication, assembly, secretion, and integration (Figure 1). HBV enters hepatocytes from the basolateral (sinusoidal) membrane, where the viral receptor, sodium taurocholate cotransporting polypeptide (NTCP), is localized.25 This process can be subdivided into adsorption and entry. Adsorption is mediated by glycosaminoglycans, such as heparan sulfate proteoglycans, that can recruit HBV to the cell surface through non-covalent binding.26,27 As the functional receptor for HBV, NTCP interacts with the preS1 domain of HBV large surface protein (L) to complete the HBV entry process.28 Mediated by NTCP, the relaxed circular DNA (rcDNA) containing nucleocapsids enter the cytoplasm and are released in the nucleus,29 where the rcDNA is repaired to form cccDNA with the assistance of host factors, including TDP2, DNA polymerase (POL) κ, POLα, DNA ligase 1 and 3, and flap endonuclease 1.5,30, 31, 32, 33, 34, 35, 36 As a template for HBV transcription, cccDNA transcribes five kinds of HBV RNAs (0.7 kb, 2.1 kb, 2.4 kb, longer and shorter 3.5 kb RNAs) under the action of host RNA polymerase II.37 Transcription of cccDNA is controlled by four promoters (the Core, preS1, preS2, and X promoters) and two enhancers (enhancers I and II).38,39 The 0.7-kb RNA can be translated to HBV X protein (HBx) which acts as a transcriptional regulator. The 2.1-kb RNA can be translated to HBV small surface protein (S) and middle surface protein (M). The 2.4-kb RNA can be translated to HBV large surface protein (L). L, M, and S can self-assemble to form empty subviral particles (SVPs) (including spherical SVPs and filamentous SVPs) that are secreted,40, 41, 42, 43 with only filamentous SVPs and virions containing significant amounts of L protein. The spherical SVPs are secreted through the constitutive secretory pathway. The filamentous SVPs are secreted by the endosomal sorting complex required for transport (ESCRT) machinery through multivesicular bodies (MVB).43 The longer 3.5-kb RNA is termed pre-Core RNA (pcRNA) and can be translated to pre-Core protein, better known as HBV e antigen (HBeAg).44, 45, 46 The shorter 3.5-kb RNA is pre-genomic RNA (pgRNA) that has two roles, as the translation template for HBV polymerase (Pol) and Core proteins and as the replication template for intra-capsid (formed by Core protein polymerization) reverse transcription by Pol to form HBV rcDNA.45, 46, 47, 48 These nucleocapsids can then be enveloped by HBV surface proteins (L, M, and S) to form mature virions and secrete through the ESCRT/MVB pathway.40 Alternatively, these nucleocapsids can also be transported to the nucleus to form cccDNA.
Figure 1.
HBV Life Cycle.
The HBV life cycle, including viral entry, cccDNA formation, transcription, replication, assembly, secretion, and dslDNA integration is shown. The red arrows indicate the routes of dslDNA integration from both the de-novo-infected nucleocapsids and the cytoplasm-produced nucleocapsids. The black dotted arrow indicates the formation of cccDNA-like molecules from dslDNA. See text for detailed description. ER, endoplasmic reticulum; MVB, multivesicular body.
Other enveloped nucleocapsids containing double-stranded linear DNA (dslDNA), HBV RNA, or the enveloped empty capsid can also form in the HBV life cycle.49, 50, 51, 52 These particles are secreted out of the cells after which they can conduct viral entry. In both de-novo-infected nucleocapsids and in cytoplasm-produced nucleocapsids dslDNA can be circularized by the non-homologous end joining (NHEJ) DNA repair pathway into cccDNA-like molecules.5,53 As NHEJ is error-prone, many of these molecules are functionally defective.5 Furthermore, dslDNA is the dominant substrate for integration into the host genome.18 A study performed in an in vitro infection model demonstrated that the dslDNA in de-novo-infected nucleocapsids was the main contributor of HBV DNA integration.20 However, some NGS studies report that a few virus-cell junctions are not correlated to the dslDNA termini, suggesting a place for other HBV DNA forms.21,54,55 Cohesive end linear DNA is another HBV DNA form thought to occur caused by denaturation at the 5′ cohesive overlap region of HBV rcDNA and subsequent extension of the recessed 3′ ends to create terminal redundancies between direct repeat 1 (DR1) and DR2 which may play this role in integration.18,53,56 HBV RNA in the nucleocapsids may be an additional candidate for viral integration along with HBV spliced variants, as HBV spliced variants lose the cis-acting signal for circular DNA and thus support dslDNA.20,57 The integrated HBV DNA can transcribe viral mRNAs or viral-cellular fusion mRNAs, which is discussed below.
Molecular Mechanisms of HBV DNA Integration
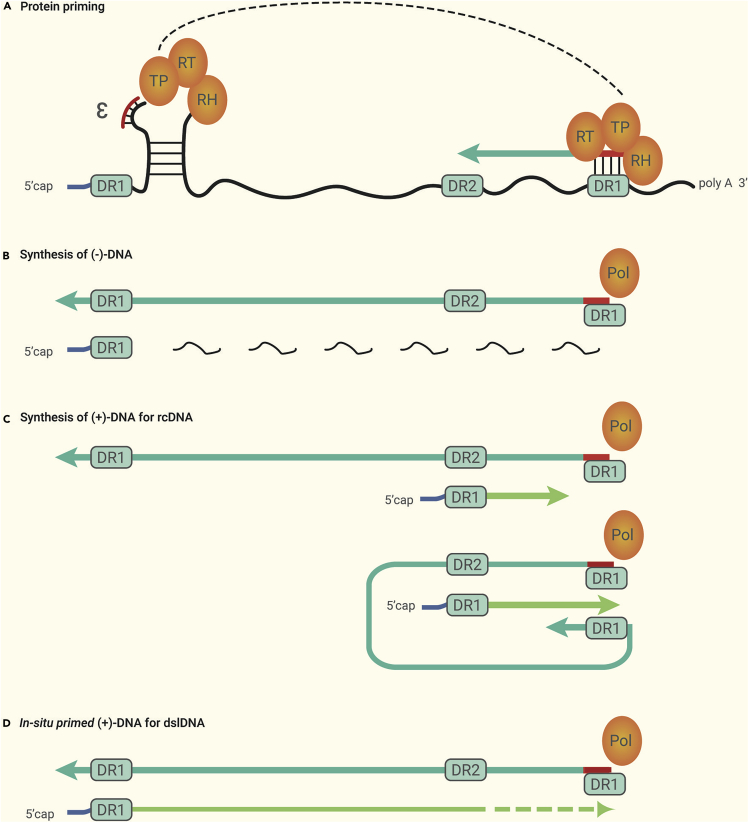
The dslDNA is synthesized from pgRNA like the rcDNA (Figure 2). First, a short DNA oligo templated by the sequence of the ϵ packaging signal internal bulge, which is phosphodiester-linked to the terminal protein (TP) domain of the Pol protein, is generated by protein priming at the 5′ ϵ of pgRNA mediated by Pol protein.58,59 The DNA oligo then is transferred to proximal DR1 at the 3′ terminal of the pgRNA, which subsequently primes the synthesis of the negative strand DNA ((−)-DNA) mediated by the reverse transcriptase domain of the Pol protein. During (−)-DNA extension, most of the pgRNA is degraded by the RNase H domain of the Pol protein, except for the capped 5′ end, and the undegraded RNA oligo serves as the positive strand DNA ((+)-DNA) primer. Subsequently, the RNA primer is transferred to DR2 of (−)-DNA, which primes the synthesis of (+)-DNA and generates rcDNA. However, in 5%–20% of cases, the RNA primer performs the direct (in situ) extension yielding dslDNA, also known as the in-situ-primed linear DNA.5,52,60
Figure 2.
Synthesis of rcDNA and dslDNA.
As the template, pgRNA is preformed for protein priming (A) mediated by the TP domain of Pol, and the (−)-DNA is subsequently synthesized (B). For the synthesis of rcDNA (C), the RNA primer is transferred to DR2 of (−)-DNA and primes the synthesis of (+)-DNA. For the synthesis of dslDNA (D), the RNA primer performs the direct (in situ) extension yielding (+)-DNA. See text for detailed description.
HBV dslDNA is ~10 nucleotides (nt) longer than the genome length (e.g., genotype A, serotype adw2, GenBank accession number X02763.1)61,62 and contains several features (1) a capped RNA oligomer at the 5′ end of (+)-DNA, (2) Pol protein, which is covalently linked to the 5′ end of the (−)-DNA, (3) DR1 at 1,826–1,836 nt and DR2 at 1,592–1,602 nt which have identical sequences (ttcacctctgc), and (4) DR1 at the 5′ end of (−)-DNA, which has only four nucleotides (tgaa) and the nucleotide “t,” which is covalently linked to the Pol protein. Like rcDNA, the plus strand of dslDNA is incomplete. HBV integration occurs early in the viral life cycle as demonstrated in in vitro infection models20,63 and appears to have the same timeline as the formation of cccDNA, indicating a similar process between the two events. Thus, the first step of HBV integration is likely to remove or repair these structures to remove the RNA oligomer and Pol protein and repair the incomplete plus strand.
The second step of HBV integration is to integrate HBV DNA into the host genome. Although some aspects of this process remain unknown, much progress has been made. As a member of the Hepadnaviridae, the biological characteristics of duck HBV (DHBV) are very similar to HBV. Most present knowledge of HBV is derived from DHBV models. A study using recombinant restriction enzyme-induced host genomic double-stranded breaks in an in vitro DHBV infection model showed that integration preferentially occurs at these sites.56 A series of studies on HBV or DHBV reported that the integration breakpoints in the viral DNA frequently occur near the DR1 or DR2 sites (1,600–1,800 nt in HBV genome).18,21,22,54,55,63, 64, 65, 66, 67 The sizes of integrated viral DNA fragments range from 28 base pairs (bp) to the full-length viral DNA sequence,68 with deletions at the termini of up to 200 bp from the integrated viral DNA being common.18,56,64,65
The NHEJ DNA repair pathway is typically associated with deletions and sometimes insertions of sequences at the termini.69 As there is almost no sequence homology between the viral DNA and the cellular DNA in the majority of virus-cell junctions,20,55,56 NHEJ is proposed as a mechanism used for HBV DNA integration. However, a few integration junctions have short homologous sequences between the viral DNA and the cellular DNA,20,54,70,71 suggesting that the microhomology-mediated end joining (MMEJ) (also known as non-classical or alternative NHEJ) DNA repair pathway72 is the mechanism for these cases.
The insertion sites of HBV DNA integration occur throughout the whole host genome.21,22,54,55,65,66,68 Earlier studies using PCR-based techniques suggested a random process with no sites of preferential integration.73 More robust high-throughput sequencing studies have found various recurrent sites of insertion in the genes encoding human telomerase reverse transcriptase (hTERT),21,74 mixed-lineage leukemia 4 (MLL4),21,75 cyclin e1 (CCNE1),21 extra spindle pole bodies like 1 (ESPL1),76 and in structures such as telomeres and CpG islands.54 Recurrent sites of integration are preferentially observed in tumor cells and greater enrichment of integration into specific genes or features has been detected in HCC tissue compared with non-tumor tissues.21,22,54,77,78 A plausible suggested mechanism is that, after random integration of viral DNA occurs, those hepatocytes carrying recurrent sites of integration experience clonal expansion as they have a selective advantage during tumorigenesis.24 Thus, in a recent study more virus-integrated hepatocytes are observed in HCC tumors when compared with matched control tissues.55 However, recurrent sites of integration, such as the retrotransposon sequences long interspersed nuclear elements 1 and 2 have been detected in HepaRG cells, a human hepatocyte stem cell line and an in vitro HBV infection model, which has no clonal expansion during HBV infection.63 A similar study in a non-clonally expanding in vitro HBV infection model via NTCP-dependent uptake of HBV found no obvious genomic recurrent sites of integration, although HBV DNA integration occurred across the whole genome.20
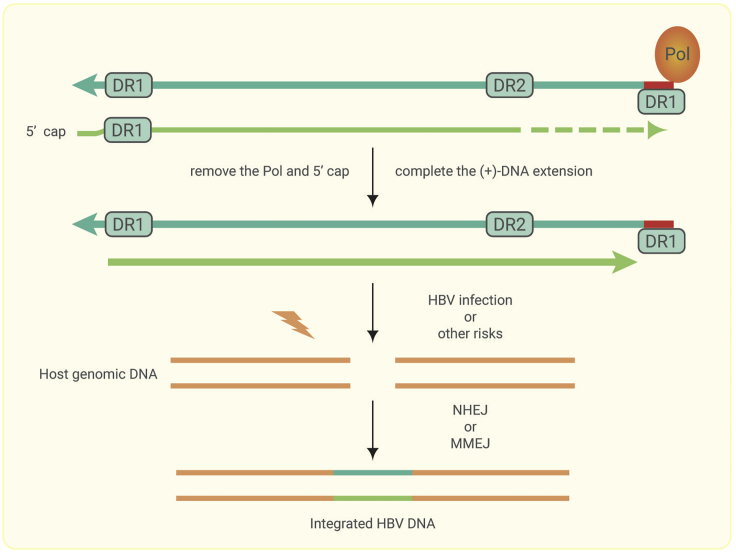
A conceptual model is thus proposed to describe the process of HBV integration during HBV infection (Figure 3): (1) host chromosomal DNA double-stranded breaks are formed by HBV infection79 or endogenous metabolism.80 (2) The dslDNA in de-novo-infected nucleocapsids or in cytoplasm-produced nucleocapsids is repaired by viral proteins or host proteins. (3) The mature dslDNA is randomly integrated into the host genome. The use of the NHEJ or MMEJ DNA repair pathways depends on the sequence characteristics at the ends of the viral DNA and the host double-stranded break DNA. (4) During hepatocarcinogenesis, recurrent sites of integration are a result of positive selection and clonal expansion.
Figure 3.
Conceptual Model for HBV DNA Integration.
The dslDNA is first repaired by removing the Pol protein and the structure of the 5′ cap, after which the (+)-DNA extension is completed. Host genomic DNA double-stranded breaks can result from HBV infection or other factors. Finally, the mature dslDNA is randomly integrated into the host genome by NHEJ or MMEJ.
Detection Methods for HBV DNA Integration
HBV DNA integration has been studied for 40 years since the first reports in 1980.10,11 Knowledge of HBV DNA integration has grown rapidly, largely limited by detection capabilities. From the earliest Southern blot hybridization to the current high-throughput sequencing technologies, each method has unique capabilities and limitations to further the study of HBV DNA integration.
Southern Blot Hybridization
Southern blot hybridization was first used to report the presence of HBV DNA integration both in tumor tissue and in HCC cell lines.10,11 Total DNA extracted from the cells or tissues infected by HBV is digested with a restriction enzyme, after which the 32P-labeled HBV DNA used as the hybridization probe binds to the digested DNAs, which are separated through gel electrophoresis. The autoradiograph is subsequently presented on film, from which the HBV DNA integration can be judged.10,11,81 However, this method cannot detect cells below a detection limit of 103–105 copies of the HBV genome, thus biasing it toward hepatocytes that have undergone clonal expansion.7 In addition, the integrated sequence information cannot be determined.
In Situ Hybridization
In situ hybridization can identify the location of genes in cells and has been used to investigate the chromosomal integration sites of HBV DNA.82 Cellular DNA fragments are isolated from the DNA of the integrant clones and then radiolabeled with [3H]dTTP for use as a probe for in situ hybridization. The target metaphase chromosomes are then extracted and hybridization is performed. Like Southern blot hybridization, the sensitivity of this technique is low, but improved in situ hybridization techniques with higher sensitivity, such as fluorescence in situ hybridization (FISH), have been used for the detection of HBV DNA integration in the chromosomes.83 However, FISH faces drawbacks, such as a high off-target rate resulting in a high noise-to-signal ratio thus necessitating high specificity and careful design of the probe.
Cloning and Nucleotide Sequencing
Cloning and nucleotide sequencing was the earliest technique used to identify the sequences of integrated HBV DNA and the adjacent cellular DNA.84, 85, 86 Although the detailed integrated sequence information can be identified, this method is laborious and inefficient. A cellular genomic library must be constructed through a specific restriction enzyme digest and then screened with the HBV probe. The positive clones are subsequently sequenced.84 During this process, many integrated sequences may be lost due to the choice of restriction enzyme and the amplification of the genomic library. This method is not suitable to screen a large number of unknown virus-cell junctions.
PCR is a molecular biology technique used to amplify specific DNA fragments. PCR can greatly increase the amount of target DNA making it a suitable method to detect HBV integration from samples that are always obtained in small amounts. Common PCR strategies used for HBV integration are Alu-PCR and inverse-nested PCR (invPCR).
Alu-PCR
Alu-PCR is a “DNA fingerprinting” technique based on using Alu elements as target loci, Alu-repeated sequences being one of the most abundant repetitive elements interspersed in the human genome at a mean interval of about 4 kb and accounting for more than 10% of the human genome.13,87, 88, 89, 90 Alu elements are frequently mapped within or adjacent to HBV integration fragments.84 The primers specific for HBV DNA and Alu-repetitive sequences are used to amplify virus-cell DNA junctions13 and many HBV DNA integrations in patients with HBV infection have been detected.91, 92, 93 Alu-PCR is a very sensitive method for the detection of HBV DNA integration. However, this method can only effectively detect the integration near Alu sequences, thus losing other integration information.
invPCR
invPCR is used to amplify the unknown cellular DNA adjacent to an integrated sequence. Restriction enzymes are used to digest DNA, after which circularization of cleavage products through self-ligation and amplification using specific outward facing primers based on the HBV sequence is done.90,94 Normally, a restriction enzyme that has only one cutting site in the HBV sequence is chosen. In this situation, the left, right, or both ends of the virus-cell junction can be detected because the cleavage site in the genomic DNA can be either upstream or downstream of the integrated HBV DNA or both.65,66,95,96 Nevertheless, in one study two restriction enzymes were used to specially detect the right end of the virus-cell junction.20 This method is highly sensitive; some studies suggest that single copy virus-cell junctions can be detected.20,97 However, the use of restriction enzymes may limit the detection of some integration junctions due to the deficiency of the corresponding cutting sites upstream and downstream of the virus-cell DNA junctions.
Some uncommon PCR technologies also used to detect HBV DNA integration are fluorescence in situ PCR15 and adaptor-ligation/suppression-PCR.75
High-Throughput Sequencing Technology
High-throughput sequencing technology, also known as NGS technology can sequence millions of reads in a single run and does not require previous knowledge of the target sequences.98 Depending on the strategy and target, NGS can be used for whole-genome sequencing (WGS), whole-exome sequencing, RNA sequencing (RNA-seq), single-cell sequencing, and captured/enriched NGS among others. Many of these technologies have been used for the detection of HBV DNA integration.16,21,22,54,55,68,70,99, 100, 101, 102 WGS and RNA-seq were first used to detect HBV integration in HCC patients in 201216,21,55 and are both sensitive and comprehensive at the identification and quantification of viral integrants across the human genome and transcriptome. RNA-seq allows for analysis of differential gene expression and can be more sensitive in reporting sub-clonal HBV integrations than WGS due to a greater sequencing coverage, especially if the sub-clonal genomic integration greatly alters the produced transcript.103 However, those integrations resulting in no RNA transcription cannot be detected by RNA-seq.
WGS is a powerful method to detect HBV integration in HCC genomes limited by the high cost of performing population-scale studies. Thus, high-throughput viral integration detection (HIVID), a sensitive low cost method, was created99 and has been used widely.22,54,68,100,101 HIVID uses a set of HBV probes to enrich HBV sequence fragments, which are then sequenced using a high-throughput platform. Compared with WGS, HIVID is cost-effective while still providing high specificity and sensitivity to detect viral integration throughout the human genome.99
Research Models for HBV DNA Integration
Currently, most knowledge about HBV DNA integration is acquired from liver tissues of patients infected with HBV65,96,104 or non-primate animal models of other hepadnaviruses, such as woodchuck infected with woodchuck hepatitis virus and duck infected with DHBV.18,64,65 Studies of HBV DNA integration in HBV models have been stymied due to the lack of models for HBV infection. With the discovery of the HBV receptor NTCP, a series of HBV in vitro infection models have been established,105 which is of use in the study of HBV DNA integration. HBV DNA integration occurs in the early stages of infection,20,63 so here we introduce models that support HBV infection.
HBV in vitro infection models consists of primary human hepatocytes, HepaRG/HepaRG-human NTCP (hNTCP) cells, and hNTCP-expressing hepatoma cells (such as HepG2-hNTCP cells and Huh7-hNTCP cells). Studies using these models show that HBV DNA is integrated quickly after infection with a variety of host sequences which often consist of tandemly repeating non-coding DNAs; the HBV genotypes and various cell lines may affect the detection of HBV DNA integration.20,63
HBV in vivo infection models consists of chimpanzee, macaque expressing hNTCP, Tupaia and human liver chimeric mice.105 Chimpanzees remain the only immunocompetent non-human hosts fully susceptible to infection with HBV.106 In the livers of chimpanzees with chronic HBV infection, clonally expanded hepatocytes carried integrated viral DNA.66 Human liver chimeric mice harbor primary human hepatocytes incorporated into the murine liver parenchyma and can support HBV infection for extended periods of time.19,105 Despite HBV DNA integration was detected in this model, integrated viral DNA was not observed to contribute to antigen production.19 Macaques expressing hNTCP are a non-human primate that can support HBV infection,107 while Tupaia are the only non-primate animals experimentally susceptible to HBV infection.108,109 Although infection of adult Tupaia results in acute self-limited infection, neonate Tupaia can be infected with HBV for chronic infection of the liver with moderate levels of viremia and histopathological changes, such as mild fibrosis and HCC development.108,109 The Tupaia model may be a suitable animal model for studying the relationship between HBV DNA integration and HCC, but at the time of writing the literature is silent on HBV DNA integration using either macaque expressing hNTCP or Tupaia.
The Roles of HBV DNA Integration in Disease
HBV DNA integration is a common phenomenon during the HBV life cycle and has been widely reported, from cell lines infected with HBV,20,63 chimpanzees chronically infected with HBV,8,66 and patients chronically infected with HBV with and without HCC.54,65 HBV DNA integration is an early event in HBV infection,20,63 which can run through all stages of chronic HBV infection. Chronic HBV infection can cause a series of liver diseases, including chronic hepatitis, cirrhosis, and HCC, with HBV DNA integration playing a role in the progression of these diseases.
HBV-related liver diseases are mainly driven by sustained inflammation110 modulated by immune cells. As an important general phenomenon in liver diseases, HBV DNA integration appears to have some relationship with the immune response in the liver microenvironment. Chronic inflammation associated with HBV infection can increase the exposure of hepatocytes to oxidative stress, leading to greater levels of double-stranded DNA breaks which form the substrate for HBV DNA integration.56,111 It has been shown that HBV DNA integration is increased under oxidative stress.79 Furthermore, the immune system in the liver microenvironment may facilitate HBV DNA integration through clonal hepatocyte expansion. HBV-specific T cells, HBV DNA integration, and clonal hepatocyte expansion has been detected in chronic hepatitis B patients considered immune tolerant.96 HBV-specific T cells selectively kill hepatocytes undergoing HBV virus replication, leading to clonal expansion of HBV DNA-integrated hepatocytes as they bypass the HBV-specific T cell immune response. Meanwhile, HBV DNA integration also affects the immune response and leads to inflammation in the liver microenvironment. High levels of HBsAg can lead to T cell exhaustion, resulting in limited or weak T cell responses.112,113 Integrated HBV DNA has been reported as another source of HBsAg apart from cccDNA8 and may serve as a persistent factor impairing the HBV-specific T cell response. In addition, hepatic CD205+ macrophages from HBsAg-transgenic mice exhibited an activated phenotype and expressed higher levels of inflammatory cytokine, chemokine, and phagocytosis-related genes,110 suggesting that HBsAg produced from integrated HBV DNA may induce inflammation through the immune response in the liver microenvironment. The relationship between HBV DNA integration and the immune system in the liver microenvironment is complex and lacks direct evidence, thus perhaps serving as an important future direction of study.
HBV DNA integration is more common in HBV-related HCC than in the non-tumor tissue,21,67 and is associated with hepatocarcinogenesis via multiple mechanisms, including insertional mutagenesis acting in cis of HCC-associated genes, induction of chromosomal instability, and the expression of mutant HBV genes from the stably integrated HBV DNA.21, 22, 23, 24
Insertional Mutagenesis
Insertional mutagenesis caused by HBV DNA integration is common as the insertion sites of HBV DNA integration occur throughout the whole host genome.21,22,54,55,65,66,68 In HCC patients with HBV infection, many recurrent integration sites that may cause mutation of HCC-associated genes have been identified. The first gene that was reported to be a recurrent HBV integration site was hTERT in two HCC tumor samples.114,115 The telomerase reverse transcriptase encoded by hTERT plays an important role in overriding cellular senescence and its dysregulation in somatic cells is linked to carcinogenesis.116 The activation of telomerase and its role in maintenance of telomere length are key events to drive hepatocarcinogenesis.117, 118, 119 A recent study reported that the E74-like ETS transcription factor 4 (ELF4), which normally drives HBV gene transcription, can bind to the chimeric HBV enhancer I at the TERT promoter, resulting in the activation of telomerase.74 A histone methyltransferase encoded by MLL4 plays an important role in epigenetics and gene expression in cancer cells75 and is a part of the ASC-2 complex implicated in the p53 tumor suppressor pathway.120 Other members of the MLL family are also frequently mutated in solid tumors.121,122 Viral integration within the MLL4 gene results in a >20-fold increase in the transcript level of MLL4.55 As the key cell-cycle regulators in eukaryotic cells, Cyclins are the major targets for oncogenic signals. Cyclin E1 is encoded by CCNE1 and is required for the cell-cycle G1/S phase transition. The expression of CCNE1 was increased by approximately 30-fold in tumors with HBV integration compared with normal controls.21 HBV integration at CCNE1 suggests a mechanism of dysregulation of the G1/S phase transition leading to HCC;123 however, a study of overexpressed CCNE1 in transgenic mice found chromosomal instability in addition to dysregulated cell-cycle control through accelerated S phase entry in CCNE1 overexpressed mice liver tissues.124 HBV integration in the ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 (SERCA1) gene in HCC tissue was reported to result in an HBx/SERCA1 fusion protein, which drove oncogenesis by inducing apoptosis.119
Chromosomal Instability
Chromosomal instability, a hallmark of cancer cells, is another possible consequence of HBV DNA integration and may contribute to hepatocarcinogenesis.125, 126, 127, 128, 129 It has been hypothesized that viral DNA integration may cause chromosomal instability, which is recognized as one of the factors leading to tumorigenesis.130 This hypothesis was subsequently tested by a WGS study, in which the somatic copy number variations (CNVs) near the 344 HBV integration sites in tumors were examined. The results indicated that somatic CNVs were positively correlated with the number of HBV integration reads, suggesting that HBV integration might impair chromosomal stability and cause the copy number to change.21 A similar study reported the same result.55
Telomeres are repetitive DNA regions at the ends of chromosomes and maintain chromosomal stability by protecting them from degradation and fusion events. HBV integration sites are markedly enriched in the vicinity of telomeres in HCC compared with matched control tissue,54 suggesting a mechanism involving chromosomal stability. Furthermore, as discussed above, HBV integrations into hTERT and long non-coding RNAs involved in telomere maintenance and chromosome localization have been identified.131,132
Mutant HBV Genes
Mutant HBV genes are also the products of HBV DNA integration and can stably express mutant HBV proteins, which may promote hepatocarcinogenesis. The sequence of dslDNA determines that the integrated HBV DNA can at most transcribe one intact 2.4/2.1-kb HBV mRNA (coding HBsAg), with integrated HBV DNA reported as another source of HBsAg apart from cccDNA.8,67 Because deletions at the termini of up to 200 bp from the integrated viral DNA are common,18,56,64,65 truncated HBsAg can also be produced. Intact HBsAg and some truncated HBsAg are associated with endoplasmic reticulum stress responses, thereby increasing the risk of HCC.23,133,134 Moreover, the HBsAg derived from integrated HBV DNA can participate in the assembly and release of hepatitis delta virus (HDV), a satellite virus of HBV that requires HBsAg for production of new virions135 in vitro.136 Chronic co-infection of HBV and HDV in patients often causes severe necroinflammation and is associated with a higher risk of cirrhosis and HCC development compared with patients with only HBV infection.137, 138, 139
Furthermore, as enhancer I is recognized to be active in the integrated form, the incomplete HBx open reading frame (ORF) can be transcribed to produce truncated HBx,140,141 which is functional in transcriptional transactivation.142 Overexpression of the truncated HBx can induce stem cell-like properties,143 transformation and inhibition of apoptosis,144,145 and tumor invasion.146,147 Meanwhile, HBx cellular transcripts can be produced due to the lack of a stop codon in the 3′ end of the HBx ORF.70,75,148,149
HBV DNA transcription and stability are further regulated by epigenetic effects, along with both viral and host factors.150, 151, 152 Thus, integrated HBV DNA in the host genome may also recruit viral or host factors which perform epigenetic modifications, affecting either integrated HBV DNA expression or the host genome. Indeed, as described above, E74-like ELF4 can bind to the chimeric HBV enhancer I at the TERT promoter, resulting in telomerase activation.74 Clarification of the influence of integrated HBV DNA regulated by transcription factors or epigenetic modifications may be an important issue.
Despite the hepatocarcinogenic effects of HBV DNA integration, in some cases it can be utilized for HCC treatment. A study which aimed to detect and use the HBV-host-integrated DNAs in plasma samples as potential circulating biomarkers to detect the tumor load in HBV-related HCC patients may help to monitor the residual tumor and recurrence clonality after tumor resection.101
The current goal of HBV antiviral therapy is to achieve a “functional cure” consisting of persistently undetectable HBV DNA in serum, loss of HBsAg, preferably with seroconversion (from HBsAg to antibody to HBsAg, anti-HBs), and normal liver enzymes and histology after stopping treatment.39,153 Persistently undetectable HBV DNA in serum can commonly be achieved through current antiviral therapies (using nucleos(t)ide analogs or PegIFNα), but the loss of HBsAg is rare,154 perhaps because current treatments cannot clear cccDNA, the main source of HBsAg. Furthermore, integrated HBV DNA can express HBsAg, making the loss of HBsAg more difficult.8,154,155 In this way, HBV DNA integration will seriously hinder the achievement of a functional cure.
Potential Therapeutic Strategies for HBV DNA Integration
Many therapeutic approaches for HBV infection have been developed and used to cure HBV infection.39,156,157 Currently, there are no treatment strategies targeting the integrated HBV DNA. This may be due to the complexity of HBV DNA integration, as the HBV DNA sequence integrated host genome is variable, consists of many different integration sites, which are mostly unable to produce RNAs. Potential treatment strategies to target integrated HBV DNA may be through gene therapies or RNA interference (RNAi)-based therapeutic targeting transcripts derived from integrated HBV DNA. In a human clinical trial with an RNAi-based therapeutic targeting HBV transcripts, ARC-520 strongly reduced HBsAg levels in treatment-naïve patients positive for HBeAg but the reduction in HBsAg levels was significantly less in patients who were HBeAg-negative, indicating that ARC-520 could not target HBsAg mRNA transcribed from integrated HBV DNA.8,158 Improved RNAi therapies that can target viral transcripts regardless of origin could be a promising treatment strategy.
Gene therapies mainly depend on gene-editing tools, such as CRISPR-Cas9, zinc-finger nucleases, and transcription activator-like effector nucleases, which have been used to target HBV cccDNA.36,157,159, 160, 161, 162, 163, 164 Gene therapy is still in its early stages of exploration and has many issues to be solved before clinical application is viable. The most basic requirement of gene editing in clinical use is to ensure both accuracy and safety, with off-target effects being troubling. The sequence and site diversity of HBV DNA integration will be problematic in the application of gene therapies. It should be noted that most of the impact of HBV DNA integration comes from the destruction of the continuity of the host genome by insertion, and it is difficult for current gene-editing techniques to remove the inserted HBV DNA sequence. Overall, the use of gene therapies to target integrated HBV DNA is still in its early stages.
Possible Future Research Prospects of HBV DNA Integration
HBV cccDNA is the primary reason that HBV continues to lack a functional cure. Therefore, many studies are devoted to removing or inactivating cccDNA. However, HBV DNA integration also plays an important role in HBV-related diseases, especially HCC. Although research on HBV DNA integration has made great strides, problems remain.
The Role of HBV Itself in HBV DNA Integration
HBV DNA integration occurs early in the viral life cycle20,63 and integration sites are at double-stranded breaks of the host genome.56 Can HBV infection cause double-stranded breaks in the host genome? The role, if any, that viral protein plays in the integration process needs to be clarified.
Generation of Mature HBV DNA Integrated Fragment dslDNA
Similar to cccDNA, HBV dslDNA contains several features: (1) a capped RNA oligomer at the 5′ end of (+)-DNA , (2) Pol protein, which is covalently linked to the 5′ end of the (−)-DNA , and (3) incomplete plus-strand dslDNA. Does HBV dslDNA need to be repaired before integration? Is the mechanism similar to the repair mechanism of HBV rcDNA? What virus or host proteins are involved in this process?
The Integration Mechanisms of HBV DNA
The NHEJ or MMEJ DNA repair pathways are used to perform HBV DNA integration. Why are these two cellular processes hijacked? What is the mechanism of integration, and are there other ways of integration? Moreover, a determination of whether it is random integration into the host genome needs to be investigated.
Appropriate Animal Research Models
The role of HBV DNA integration in HBV-related liver diseases is complex. Appropriate animal models that can simulate the pathogenesis of HBV will help us understand more clearly the role of HBV DNA integration in disease and assist related drug development.
The Relationship between HBV DNA Integration and the Immune Response in the Liver Microenvironment
This relationship is closely related to the occurrence and development of HBV-related diseases. Understanding this will prove useful for the monitoring and treatment of the disease.
Mapping HBV DNA Integration in the Host Genome
This map should include HBV DNA integration sites, integration sequences, transcripts, protein expression, and related pathological changes. This will enable the development of the full picture that HBV DNA integration plays in the occurrence of disease and provide more precise targets for targeted gene therapies. Furthermore, this will support the location of HBV DNA integration sites that cause disease through analysis of HBV DNA integration in individual patients, allowing for precision medicine.
Summary
During the past four decades, great strides have been made in the HBV DNA integration field, although some key aspects remain muddled, such as the molecular mechanism of integration and the role of HBV integration in HCC initiation and development. It remains hopeful that continual progress in the development of detection methods and research models, such as NTCP-expressing hepatoma cell lines and animals will result in a comprehensive and detailed understanding of HBV DNA integration. As a part of the cellular genome, integrated HBV DNA is more stable than cccDNA. While many direct-acting antiviral and host-targeting agents aimed at cccDNA destruction or silencing have been developed, they do not address the problem of integrated HBV DNA. It should be recognized that, in addition to cccDNA, integrated HBV DNA plays an important role in HBV-related diseases, especially HCC. HBV DNA integration should be used as a clinical indicator for disease monitoring and treatment of patients with HBV infection. Even if cccDNA can be cleared in the future, a functional cure may not be feasible because of the fundamental challenge integrated HBV DNA will continue to pose. Thus, the exact role of integrated HBV DNA in disease remains a question of interest in the development of curative therapeutics and to gain a clearer picture of HBV biology.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (project no. 82041004), the Fundamental Research Funds for the Central Universities and Gilead Sciences Research Scholars Program in Liver Disease Asia.
Declaration of interests
The authors declare no competing interests.
References
- 1.Blumberg B.S., Alter H.J., Visnich S. A "new" antigen in leukemia sera. JAMA. 1965;191:541–546. doi: 10.1001/jama.1965.03080070025007. [DOI] [PubMed] [Google Scholar]
- 2.WHO Hepatitis B Fact Sheet No. 204. 2017. http://www.who.int/mediacentre/factsheets/fs204/en/
- 3.Schweitzer A., Horn J., Mikolajczyk R.T., Krause G., Ott J.J. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546–1555. doi: 10.1016/S0140-6736(15)61412-X. [DOI] [PubMed] [Google Scholar]
- 4.Polaris Observatory C. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol. Hepatol. 2018;3:383–403. doi: 10.1016/S2468-1253(18)30056-6. [DOI] [PubMed] [Google Scholar]
- 5.Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972–1984. doi: 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 6.Tu T., Budzinska M.A., Shackel N.A., Urban S. HBV DNA integration: molecular mechanisms and clinical implications. Viruses. 2017;9:75. doi: 10.3390/v9040075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budzinska M.A., Shackel N.A., Urban S., Tu T. Cellular genomic sites of hepatitis B virus DNA integration. Genes. 2018;9:365. doi: 10.3390/genes9070365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wooddell C.I., Yuen M.F., Chan H.L., Gish R.G., Locarnini S.A., Chavez D., Ferrari C., Given B.D., Hamilton J., Kanner S.B., et al. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci. Transl. Med. 2017;9:eaan0241. doi: 10.1126/scitranslmed.aan0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu B., Wang R., Fu J., Su M., Du M., Liu Y., Li H., Wang H., Lu F., Jiang J. Integration of hepatitis B virus S gene impacts on hepatitis B surface antigen levels in patients with antiviral therapy. J. Gastroenterol. Hepatol. 2018;33:1389–1396. doi: 10.1111/jgh.14075. [DOI] [PubMed] [Google Scholar]
- 10.Chakraborty P.R., Ruiz-Opazo N., Shouval D., Shafritz D.A. Identification of integrated hepatitis B virus DNA and expression of viral RNA in an HBsAg-producing human hepatocellular carcinoma cell line. Nature. 1980;286:531–533. doi: 10.1038/286531a0. [DOI] [PubMed] [Google Scholar]
- 11.Brechot C., Pourcel C., Louise A., Rain B., Tiollais P. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature. 1980;286:533–535. doi: 10.1038/286533a0. [DOI] [PubMed] [Google Scholar]
- 12.Kawai S., Yokosuka O., Imazeki F., Maru Y., Saisho H. State of HBV DNA in HBsAg-negative, anti-HCV-positive hepatocellular carcinoma: existence of HBV DNA possibly as nonintegrated form with analysis by Alu-HBV DNA PCR and conventional HBV PCR. J. Med. Virol. 2001;64:410–418. doi: 10.1002/jmv.1066. [DOI] [PubMed] [Google Scholar]
- 13.Minami M., Poussin K., Brechot C., Paterlini P. A novel PCR technique using Alu-specific primers to identify unknown flanking sequences from the human genome. Genomics. 1995;29:403–408. doi: 10.1006/geno.1995.9004. [DOI] [PubMed] [Google Scholar]
- 14.Georgi-Geisberger P., Berns H., Loncarevic I.F., Yu Z.Y., Tang Z.Y., Zentgraf H., Schroder C.H. Mutations on free and integrated hepatitis B virus DNA in a hepatocellular carcinoma: footprints of homologous recombination. Oncology. 1992;49:386–395. doi: 10.1159/000227078. [DOI] [PubMed] [Google Scholar]
- 15.Matsui H., Shiba R., Matsuzaki Y., Asaoka H., Hosoi S., Doi M., Ohno T., Tanaka N., Muto H. Direct detection of hepatitis B virus gene integrated in the Alexander cell using fluorescence in situ polymerase chain reaction. Cancer Lett. 1997;116:259–264. doi: 10.1016/s0304-3835(97)00199-7. [DOI] [PubMed] [Google Scholar]
- 16.Fujimoto A., Totoki Y., Abe T., Boroevich K.A., Hosoda F., Nguyen H.H., Aoki M., Hosono N., Kubo M., Miya F., et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat. Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 17.Summers J., Jilbert A.R., Yang W., Aldrich C.E., Saputelli J., Litwin S., Toll E., Mason W.S. Hepatocyte turnover during resolution of a transient hepadnaviral infection. Proc. Natl. Acad. Sci. U S A. 2003;100:11652–11659. doi: 10.1073/pnas.1635109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang W., Summers J. Integration of hepadnavirus DNA in infected liver: evidence for a linear precursor. J. Virol. 1999;73:9710–9717. doi: 10.1128/jvi.73.12.9710-9717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allweiss L., Volz T., Giersch K., Kah J., Raffa G., Petersen J., Lohse A.W., Beninati C., Pollicino T., Urban S., et al. Proliferation of primary human hepatocytes and prevention of hepatitis B virus reinfection efficiently deplete nuclear cccDNA in vivo. Gut. 2018;67:542–552. doi: 10.1136/gutjnl-2016-312162. [DOI] [PubMed] [Google Scholar]
- 20.Tu T., Budzinska M.A., Vondran F.W.R., Shackel N.A., Urban S. Hepatitis B virus DNA integration occurs early in the viral life cycle in an in vitro infection model via sodium taurocholate cotransporting polypeptide-dependent uptake of enveloped virus particles. J. Virol. 2018;92:e02007-17. doi: 10.1128/JVI.02007-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sung W.K., Zheng H., Li S., Chen R., Liu X., Li Y., Lee N.P., Lee W.H., Ariyaratne P.N., Tennakoon C., et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat. Genet. 2012;44:765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 22.Yan H., Yang Y., Zhang L., Tang G., Wang Y., Xue G., Zhou W., Sun S. Characterization of the genotype and integration patterns of hepatitis B virus in early- and late-onset hepatocellular carcinoma. Hepatology. 2015;61:1821–1831. doi: 10.1002/hep.27722. [DOI] [PubMed] [Google Scholar]
- 23.Wang H.C., Huang W., Lai M.D., Su I.J. Hepatitis B virus pre-S mutants, endoplasmic reticulum stress and hepatocarcinogenesis. Cancer Sci. 2006;97:683–688. doi: 10.1111/j.1349-7006.2006.00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu T., Budzinska M.A., Shackel N.A., Jilbert A.R. Conceptual models for the initiation of hepatitis B virus-associated hepatocellular carcinoma. Liver Int. 2015;35:1786–1800. doi: 10.1111/liv.12773. [DOI] [PubMed] [Google Scholar]
- 25.Meier P.J., Stieger B. Bile salt transporters. Annu. Rev. Physiol. 2002;64:635–661. doi: 10.1146/annurev.physiol.64.082201.100300. [DOI] [PubMed] [Google Scholar]
- 26.Schulze A., Gripon P., Urban S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology. 2007;46:1759–1768. doi: 10.1002/hep.21896. [DOI] [PubMed] [Google Scholar]
- 27.Leistner C.M., Gruen-Bernhard S., Glebe D. Role of glycosaminoglycans for binding and infection of hepatitis B virus. Cell. Microbiol. 2008;10:122–133. doi: 10.1111/j.1462-5822.2007.01023.x. [DOI] [PubMed] [Google Scholar]
- 28.Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z., Huang Y., Qi Y., Peng B., Wang H., et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blondot M.L., Bruss V., Kann M. Intracellular transport and egress of hepatitis B virus. J. Hepatol. 2016;64:S49–S59. doi: 10.1016/j.jhep.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Koniger C., Wingert I., Marsmann M., Rosler C., Beck J., Nassal M. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc. Natl. Acad. Sci. U S A. 2014;111:E4244–E4253. doi: 10.1073/pnas.1409986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi Y., Gao Z., Xu G., Peng B., Liu C., Yan H., Yao Q., Sun G., Liu Y., Tang D., et al. DNA polymerase kappa is a key cellular factor for the formation of covalently closed circular DNA of hepatitis B virus. PLoS Pathog. 2016;12:e1005893. doi: 10.1371/journal.ppat.1005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long Q., Yan R., Hu J., Cai D., Mitra B., Kim E.S., Marchetti A., Zhang H., Wang S., Liu Y., et al. The role of host DNA ligases in hepadnavirus covalently closed circular DNA formation. PLoS Pathog. 2017;13:e1006784. doi: 10.1371/journal.ppat.1006784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui X., McAllister R., Boregowda R., Sohn J.A., Cortes Ledesma F., Caldecott K.W., Seeger C., Hu J. Does tyrosyl DNA phosphodiesterase-2 play a role in hepatitis B virus genome repair? PLoS One. 2015;10:e0128401. doi: 10.1371/journal.pone.0128401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitamura K., Que L., Shimadu M., Koura M., Ishihara Y., Wakae K., Nakamura T., Watashi K., Wakita T., Muramatsu M. Flap endonuclease 1 is involved in cccDNA formation in the hepatitis B virus. PLoS Pathog. 2018;14:e1007124. doi: 10.1371/journal.ppat.1007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang L., Sheraz M., McGrane M., Chang J., Guo J.T. DNA polymerase alpha is essential for intracellular amplification of hepatitis B virus covalently closed circular DNA. PLoS Pathog. 2019;15:e1007742. doi: 10.1371/journal.ppat.1007742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia Y., Guo H. Hepatitis B virus cccDNA: formation, regulation and therapeutic potential. Antiviral Res. 2020;180:104824. doi: 10.1016/j.antiviral.2020.104824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rall L.B., Standring D.N., Laub O., Rutter W.J. Transcription of hepatitis B virus by RNA polymerase II. Mol. Cell. Biol. 1983;3:1766–1773. doi: 10.1128/mcb.3.10.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doitsh G., Shaul Y. Enhancer I predominance in hepatitis B virus gene expression. Mol. Cell. Biol. 2004;24:1799–1808. doi: 10.1128/MCB.24.4.1799-1808.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia Y., Liang T.J. Development of direct-acting antiviral and host-targeting agents for treatment of hepatitis B virus infection. Gastroenterology. 2019;156:311–324. doi: 10.1053/j.gastro.2018.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patient R., Hourioux C., Roingeard P. Morphogenesis of hepatitis B virus and its subviral envelope particles. Cell. Microbiol. 2009;11:1561–1570. doi: 10.1111/j.1462-5822.2009.01363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerlich W.H., Heermann K.H., Lu X. Functions of hepatitis B surface proteins. Arch. Virol. Suppl. 1992;4:129–132. doi: 10.1007/978-3-7091-5633-9_28. [DOI] [PubMed] [Google Scholar]
- 42.Patient R., Hourioux C., Sizaret P.Y., Trassard S., Sureau C., Roingeard P. Hepatitis B virus subviral envelope particle morphogenesis and intracellular trafficking. J. Virol. 2007;81:3842–3851. doi: 10.1128/JVI.02741-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang B., Himmelsbach K., Ren H., Boller K., Hildt E. Subviral hepatitis B virus filaments, like infectious viral particles, are released via multivesicular bodies. J. Virol. 2015;90:3330–3341. doi: 10.1128/JVI.03109-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jean-Jean O., Levrero M., Will H., Perricaudet M., Rossignol J.M. Expression mechanism of the hepatitis B virus (HBV) C gene and biosynthesis of HBe antigen. Virology. 1989;170:99–106. doi: 10.1016/0042-6822(89)90356-5. [DOI] [PubMed] [Google Scholar]
- 45.Cattaneo R., Will H., Schaller H. Hepatitis B virus transcription in the infected liver. EMBO J. 1984;3:2191–2196. doi: 10.1002/j.1460-2075.1984.tb02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enders G.H., Ganem D., Varmus H. Mapping the major transcripts of ground squirrel hepatitis virus: the presumptive template for reverse transcriptase is terminally redundant. Cell. 1985;42:297–308. doi: 10.1016/s0092-8674(85)80125-2. [DOI] [PubMed] [Google Scholar]
- 47.Hirsch R.C., Lavine J.E., Chang L.J., Varmus H.E., Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature. 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 48.Bartenschlager R., Junker-Niepmann M., Schaller H. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J. Virol. 1990;64:5324–5332. doi: 10.1128/jvi.64.11.5324-5332.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Bommel F., Bartens A., Mysickova A., Hofmann J., Kruger D.H., Berg T., Edelmann A. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology. 2015;61:66–76. doi: 10.1002/hep.27381. [DOI] [PubMed] [Google Scholar]
- 50.Wang J., Shen T., Huang X., Kumar G.R., Chen X., Zeng Z., Zhang R., Chen R., Li T., Zhang T., et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J. Hepatol. 2016;65:700–710. doi: 10.1016/j.jhep.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 51.Luckenbaugh L., Kitrinos K.M., Delaney W.E.t., Hu J. Genome-free hepatitis B virion levels in patient sera as a potential marker to monitor response to antiviral therapy. J. Viral Hepat. 2015;22:561–570. doi: 10.1111/jvh.12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Staprans S., Loeb D.D., Ganem D. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J. Virol. 1991;65:1255–1262. doi: 10.1128/jvi.65.3.1255-1262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang W., Mason W.S., Summers J. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J. Virol. 1996;70:4567–4575. doi: 10.1128/jvi.70.7.4567-4575.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao L.H., Liu X., Yan H.X., Li W.Y., Zeng X., Yang Y., Zhao J., Liu S.P., Zhuang X.H., Lin C., et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat. Commun. 2016;7:12992. doi: 10.1038/ncomms12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang Z., Jhunjhunwala S., Liu J., Haverty P.M., Kennemer M.I., Guan Y., Lee W., Carnevali P., Stinson J., Johnson S., et al. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res. 2012;22:593–601. doi: 10.1101/gr.133926.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bill C.A., Summers J. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc. Natl. Acad. Sci. U S A. 2004;101:11135–11140. doi: 10.1073/pnas.0403925101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abraham T.M., Lewellyn E.B., Haines K.M., Loeb D.D. Characterization of the contribution of spliced RNAs of hepatitis B virus to DNA synthesis in transfected cultures of Huh7 and HepG2 cells. Virology. 2008;379:30–37. doi: 10.1016/j.virol.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu J., Boyer M. Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J. Virol. 2006;80:2141–2150. doi: 10.1128/JVI.80.5.2141-2150.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones S.A., Boregowda R., Spratt T.E., Hu J. In vitro epsilon RNA-dependent protein priming activity of human hepatitis B virus polymerase. J. Virol. 2012;86:5134–5150. doi: 10.1128/JVI.07137-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao X.L., Yang J.R., Lin S.Z., Ma H., Guo F., Yang R.F., Zhang H.H., Han J.C., Wei L., Pan X.B. Serum viral duplex-linear DNA proportion increases with the progression of liver disease in patients infected with HBV. Gut. 2016;65:502–511. doi: 10.1136/gutjnl-2014-308989. [DOI] [PubMed] [Google Scholar]
- 61.Valenzuela P., Gray P., Quiroga M., Zaldivar J., Goodman H.M., Rutter W.J. Nucleotide sequence of the gene coding for the major protein of hepatitis B virus surface antigen. Nature. 1979;280:815–819. doi: 10.1038/280815a0. [DOI] [PubMed] [Google Scholar]
- 62.Nassal M. Hepatitis B viruses: reverse transcription a different way. Virus Res. 2008;134:235–249. doi: 10.1016/j.virusres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 63.Chauhan R., Churchill N.D., Mulrooney-Cousins P.M., Michalak T.I. Initial sites of hepadnavirus integration into host genome in human hepatocytes and in the woodchuck model of hepatitis B-associated hepatocellular carcinoma. Oncogenesis. 2017;6:e317. doi: 10.1038/oncsis.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Summers J., Mason W.S. Residual integrated viral DNA after hepadnavirus clearance by nucleoside analog therapy. Proc. Natl. Acad. Sci. U S A. 2004;101:638–640. doi: 10.1073/pnas.0307422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mason W.S., Liu C., Aldrich C.E., Litwin S., Yeh M.M. Clonal expansion of normal-appearing human hepatocytes during chronic hepatitis B virus infection. J. Virol. 2010;84:8308–8315. doi: 10.1128/JVI.00833-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mason W.S., Low H.C., Xu C., Aldrich C.E., Scougall C.A., Grosse A., Clouston A., Chavez D., Litwin S., Peri S., et al. Detection of clonally expanded hepatocytes in chimpanzees with chronic hepatitis B virus infection. J. Virol. 2009;83:8396–8408. doi: 10.1128/JVI.00700-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen W., Zhang K., Dong P., Fanning G., Tao C., Zhang H., Guo S., Wang Z., Hong Y., Yang X., et al. Noninvasive chimeric DNA profiling identifies tumor-originated HBV integrants contributing to viral antigen expression in liver cancer. Hepatol. Int. 2020;14:326–337. doi: 10.1007/s12072-020-10016-2. [DOI] [PubMed] [Google Scholar]
- 68.Yang L., Ye S., Zhao X., Ji L., Zhang Y., Zhou P., Sun J., Guan Y., Han Y., Ni C., et al. Molecular characterization of HBV DNA integration in patients with hepatitis and hepatocellular carcinoma. J. Cancer. 2018;9:3225–3235. doi: 10.7150/jca.26052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mao Z., Bozzella M., Seluanov A., Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lau C.C., Sun T., Ching A.K., He M., Li J.W., Wong A.M., Co N.N., Chan A.W., Li P.S., Lung R.W., et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer cell. 2014;25:335–349. doi: 10.1016/j.ccr.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 71.Budzinska M.A., Shackel N.A., Urban S., Tu T. Sequence analysis of integrated hepatitis B virus DNA during HBeAg-seroconversion. Emerg. Microbes Infect. 2018;7:142. doi: 10.1038/s41426-018-0145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McVey M., Lee S.E. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genetics. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matsubara K., Tokino T. Integration of hepatitis B virus DNA and its implications for hepatocarcinogenesis. Mol. Biol. Med. 1990;7:243–260. [PubMed] [Google Scholar]
- 74.Sze K.M., Ho D.W., Chiu Y.T., Tsui Y.M., Chan L.K., Lee J.M., Chok K.S., Chan A.C., Tang C.N., Tang V.W., et al. HBV-TERT promoter integration harnesses host ELF4 resulting in TERT gene transcription in hepatocellular carcinoma. Hepatology. 2020 doi: 10.1002/hep.31231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saigo K., Yoshida K., Ikeda R., Sakamoto Y., Murakami Y., Urashima T., Asano T., Kenmochi T., Inoue I. Integration of hepatitis B virus DNA into the myeloid/lymphoid or mixed-lineage leukemia (MLL4) gene and rearrangements of MLL4 in human hepatocellular carcinoma. Hum. Mutat. 2008;29:703–708. doi: 10.1002/humu.20701. [DOI] [PubMed] [Google Scholar]
- 76.Hu B., Huang W., Wang R., Zang W., Su M., Li H., Wang H., Cao B., Deng D., Li Q.Q., et al. High rate of detection of human ESPL1-HBV S fusion gene in patients with HBV-related liver cancer: a Chinese case-control study. Anticancer Res. 2020;40:245–252. doi: 10.21873/anticanres.13946. [DOI] [PubMed] [Google Scholar]
- 77.Cao S., Wendl M.C., Wyczalkowski M.A., Wylie K., Ye K., Jayasinghe R., Xie M., Wu S., Niu B., Grubb R., et al. Divergent viral presentation among human tumors and adjacent normal tissues. Sci. Rep. 2016;6:28294. doi: 10.1038/srep28294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li X., Zhang J., Yang Z., Kang J., Jiang S., Zhang T., Chen T., Li M., Lv Q., Chen X., et al. The function of targeted host genes determines the oncogenicity of HBV integration in hepatocellular carcinoma. J. Hepatol. 2014;60:975–984. doi: 10.1016/j.jhep.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 79.Dandri M., Burda M.R., Burkle A., Zuckerman D.M., Will H., Rogler C.E., Greten H., Petersen J. Increase in de novo HBV DNA integrations in response to oxidative DNA damage or inhibition of poly(ADP-ribosyl)ation. Hepatology. 2002;35:217–223. doi: 10.1053/jhep.2002.30203. [DOI] [PubMed] [Google Scholar]
- 80.Vilenchik M.M., Knudson A.G. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. U S A. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shafritz D.A., Shouval D., Sherman H.I., Hadziyannis S.J., Kew M.C. Integration of hepatitis B virus DNA into the genome of liver cells in chronic liver disease and hepatocellular carcinoma. Studies in percutaneous liver biopsies and post-mortem tissue specimens. N. Engl. J. Med. 1981;305:1067–1073. doi: 10.1056/NEJM198110293051807. [DOI] [PubMed] [Google Scholar]
- 82.Tokino T., Matsubara K. Chromosomal sites for hepatitis B virus integration in human hepatocellular carcinoma. J. Virol. 1991;65:6761–6764. doi: 10.1128/jvi.65.12.6761-6764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang T.H., Zhang Q.J., Xie Q.D., Zeng L.P., Zeng X.F. Presence and integration of HBV DNA in mouse oocytes. World J. Gastroenterol. 2005;11:2869–2873. doi: 10.3748/wjg.v11.i19.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choo K.B., Liu M.S., Chang P.C., Wu S.M., Su M.W., Pan C.C., Han S.H. Analysis of six distinct integrated hepatitis B virus sequences cloned from the cellular DNA of a human hepatocellular carcinoma. Virology. 1986;154:405–408. doi: 10.1016/0042-6822(86)90467-8. [DOI] [PubMed] [Google Scholar]
- 85.Yaginuma K., Kobayashi M., Yoshida E., Koike K. Hepatitis B virus integration in hepatocellular carcinoma DNA: duplication of cellular flanking sequences at the integration site. Proc. Natl. Acad. Sci. U S A. 1985;82:4458–4462. doi: 10.1073/pnas.82.13.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takada S., Gotoh Y., Hayashi S., Yoshida M., Koike K. Structural rearrangement of integrated hepatitis B virus DNA as well as cellular flanking DNA is present in chronically infected hepatic tissues. J. Virol. 1990;64:822–828. doi: 10.1128/jvi.64.2.822-828.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jelinek W.R., Schmid C.W. Repetitive sequences in eukaryotic DNA and their expression. Annu. Rev. Biochem. 1982;51:813–844. doi: 10.1146/annurev.bi.51.070182.004121. [DOI] [PubMed] [Google Scholar]
- 88.Kariya Y., Kato K., Hayashizaki Y., Himeno S., Tarui S., Matsubara K. Revision of consensus sequence of human Alu repeats—a review. Gene. 1987;53:1–10. doi: 10.1016/0378-1119(87)90087-4. [DOI] [PubMed] [Google Scholar]
- 89.Batzer M.A., Deininger P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 90.Hui E.K., Wang P.C., Lo S.J. Strategies for cloning unknown cellular flanking DNA sequences from foreign integrants. Cell. Mol. Life Sci. 1998;54:1403–1411. doi: 10.1007/s000180050262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Murakami Y., Saigo K., Takashima H., Minami M., Okanoue T., Brechot C., Paterlini-Brechot P. Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut. 2005;54:1162–1168. doi: 10.1136/gut.2004.054452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saitta C., Tripodi G., Barbera A., Bertuccio A., Smedile A., Ciancio A., Raffa G., Sangiovanni A., Navarra G., Raimondo G., et al. Hepatitis B virus (HBV) DNA integration in patients with occult HBV infection and hepatocellular carcinoma. Liver Int. 2015;35:2311–2317. doi: 10.1111/liv.12807. [DOI] [PubMed] [Google Scholar]
- 93.Rendon J.C., Cortes-Mancera F., Restrepo-Gutierrez J.C., Hoyos S., Navas M.C. Molecular characterization of occult hepatitis B virus infection in patients with end-stage liver disease in Colombia. PLoS one. 2017;12:e0180447. doi: 10.1371/journal.pone.0180447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tsuei D.J., Chen P.J., Lai M.Y., Chen D.S., Yang C.S., Chen J.Y., Hsu T.Y. Inverse polymerase chain reaction for cloning cellular sequences adjacent to integrated hepatitis B virus DNA in hepatocellular carcinomas. J. Virol. Methods. 1994;49:269–284. doi: 10.1016/0166-0934(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 95.Ruan P., Dai X., Sun J., He C., Huang C., Zhou R., Chemin I. Integration of hepatitis B virus DNA into p21-activated kinase 3 (PAK3) gene in HepG2.2.15 cells. Virus Genes. 2020;56:168–173. doi: 10.1007/s11262-019-01725-4. [DOI] [PubMed] [Google Scholar]
- 96.Mason W.S., Gill U.S., Litwin S., Zhou Y., Peri S., Pop O., Hong M.L., Naik S., Quaglia A., Bertoletti A., et al. HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology. 2016;151:986–998.e4. doi: 10.1053/j.gastro.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tu T., Jilbert A.R. Detection of hepatocyte clones containing integrated hepatitis B virus DNA using inverse nested PCR. Methods Mol. Biol. 2017;1540:97–118. doi: 10.1007/978-1-4939-6700-1_9. [DOI] [PubMed] [Google Scholar]
- 98.Meyerson M., Gabriel S., Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 99.Li W., Zeng X., Lee N.P., Liu X., Chen S., Guo B., Yi S., Zhuang X., Chen F., Wang G., et al. HIVID: an efficient method to detect HBV integration using low coverage sequencing. Genomics. 2013;102:338–344. doi: 10.1016/j.ygeno.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 100.Chen X.P., Long X., Jia W.L., Wu H.J., Zhao J., Liang H.F., Laurence A., Zhu J., Dong D., Chen Y., et al. Viral integration drives multifocal HCC during the occult HBV infection. J. Exp. Clin. Cancer Res. 2019;38:261. doi: 10.1186/s13046-019-1273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li C.L., Ho M.C., Lin Y.Y., Tzeng S.T., Chen Y.J., Pai H.Y., Wang Y.C., Chen C.L., Lee Y.H., Chen D.S., et al. Cell-free virus-host chimera DNA from hepatitis B virus integration sites as a circulating biomarker of hepatocellular cancer. Hepatology. 2020 doi: 10.1002/hep.31230. [DOI] [PubMed] [Google Scholar]
- 102.Fujimoto A. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Ann. Oncol. 2017;28(Suppl 9):ix31. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 103.Shiraishi Y., Fujimoto A., Furuta M., Tanaka H., Chiba K., Boroevich K.A., Abe T., Kawakami Y., Ueno M., Gotoh K., et al. Integrated analysis of whole genome and transcriptome sequencing reveals diverse transcriptomic aberrations driven by somatic genomic changes in liver cancers. PLoS one. 2014;9:e114263. doi: 10.1371/journal.pone.0114263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tu T., Mason W.S., Clouston A.D., Shackel N.A., McCaughan G.W., Yeh M.M., Schiff E.R., Ruszkiewicz A.R., Chen J.W., Harley H.A., et al. Clonal expansion of hepatocytes with a selective advantage occurs during all stages of chronic hepatitis B virus infection. J. Viral Hepat. 2015;22:737–753. doi: 10.1111/jvh.12380. [DOI] [PubMed] [Google Scholar]
- 105.Allweiss L., Dandri M. Experimental in vitro and in vivo models for the study of human hepatitis B virus infection. J. Hepatol. 2016;64:S17–S31. doi: 10.1016/j.jhep.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 106.Maynard J.E., Berquist K.R., Krushak D.H., Purcell R.H. Experimental infection of chimpanzees with the virus of hepatitis B. Nature. 1972;237:514–515. doi: 10.1038/237514a0. [DOI] [PubMed] [Google Scholar]
- 107.Burwitz B.J., Wettengel J.M., Muck-Hausl M.A., Ringelhan M., Ko C., Festag M.M., Hammond K.B., Northrup M., Bimber B.N., Jacob T., et al. Hepatocytic expression of human sodium-taurocholate cotransporting polypeptide enables hepatitis B virus infection of macaques. Nat. Commun. 2017;8:2146. doi: 10.1038/s41467-017-01953-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Walter E., Keist R., Niederost B., Pult I., Blum H.E. Hepatitis B virus infection of Tupaia hepatocytes in vitro and in vivo. Hepatology. 1996;24:1–5. doi: 10.1002/hep.510240101. [DOI] [PubMed] [Google Scholar]
- 109.Yang C., Ruan P., Ou C., Su J., Cao J., Luo C., Tang Y., Wang Q., Qin H., Sun W., et al. Chronic hepatitis B virus infection and occurrence of hepatocellular carcinoma in tree shrews (Tupaia belangeri chinensis) Virol. J. 2015;12:26. doi: 10.1186/s12985-015-0256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yong L., Li M., Gao Y., Deng Y., Liu W., Huang D., Ren C., Liu M., Shen J., Hou X. Identification of pro-inflammatory CD205(+) macrophages in livers of hepatitis B virus transgenic mice and patients with chronic hepatitis B. Sci. Rep. 2017;7:46765. doi: 10.1038/srep46765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Barash H., E R.G., Edrei Y., Ella E., Israel A., Cohen I., Corchia N., Ben-Moshe T., Pappo O., Pikarsky E., et al. Accelerated carcinogenesis following liver regeneration is associated with chronic inflammation-induced double-strand DNA breaks. Proc. Natl. Acad. Sci. U S A. 2010;107:2207–2212. doi: 10.1073/pnas.0908867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Boni C., Fisicaro P., Valdatta C., Amadei B., Di Vincenzo P., Giuberti T., Laccabue D., Zerbini A., Cavalli A., Missale G., et al. Characterization of hepatitis B virus (HBV)-specific T-cell dysfunction in chronic HBV infection. J. Virol. 2007;81:4215–4225. doi: 10.1128/JVI.02844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bertoletti A., Ferrari C. Innate and adaptive immune responses in chronic hepatitis B virus infections: towards restoration of immune control of viral infection. Gut. 2012;61:1754–1764. doi: 10.1136/gutjnl-2011-301073. [DOI] [PubMed] [Google Scholar]
- 114.Paterlini-Brechot P., Saigo K., Murakami Y., Chami M., Gozuacik D., Mugnier C., Lagorce D., Brechot C. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene. 2003;22:3911–3916. doi: 10.1038/sj.onc.1206492. [DOI] [PubMed] [Google Scholar]
- 115.Gozuacik D., Murakami Y., Saigo K., Chami M., Mugnier C., Lagorce D., Okanoue T., Urashima T., Brechot C., Paterlini-Brechot P. Identification of human cancer-related genes by naturally occurring hepatitis B virus DNA tagging. Oncogene. 2001;20:6233–6240. doi: 10.1038/sj.onc.1204835. [DOI] [PubMed] [Google Scholar]
- 116.Cao Y., Bryan T.M., Reddel R.R. Increased copy number of the TERT and TERC telomerase subunit genes in cancer cells. Cancer Sci. 2008;99:1092–1099. doi: 10.1111/j.1349-7006.2008.00815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nault J.C., Mallet M., Pilati C., Calderaro J., Bioulac-Sage P., Laurent C., Laurent A., Cherqui D., Balabaud C., Zucman-Rossi J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 2013;4:2218. doi: 10.1038/ncomms3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Borah S., Xi L., Zaug A.J., Powell N.M., Dancik G.M., Cohen S.B., Costello J.C., Theodorescu D., Cech T.R. Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science. 2015;347:1006–1010. doi: 10.1126/science.1260200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Barthel F.P., Wei W., Tang M., Martinez-Ledesma E., Hu X., Amin S.B., Akdemir K.C., Seth S., Song X., Wang Q., et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 2017;49:349–357. doi: 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee J., Kim D.H., Lee S., Yang Q.H., Lee D.K., Lee S.K., Roeder R.G., Lee J.W. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc. Natl. Acad. Sci. U S A. 2009;106:8513–8518. doi: 10.1073/pnas.0902873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lee S., Lee J., Lee S.K., Lee J.W. Activating signal cointegrator-2 is an essential adaptor to recruit histone H3 lysine 4 methyltransferases MLL3 and MLL4 to the liver X receptors. Mol. Endocrinol. 2008;22:1312–1319. doi: 10.1210/me.2008-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Natarajan T.G., Kallakury B.V., Sheehan C.E., Bartlett M.B., Ganesan N., Preet A., Ross J.S., FitzGerald K.T. Epigenetic regulator MLL2 shows altered expression in cancer cell lines and tumors from human breast and colon. Cancer Cell Int. 2010;10:13. doi: 10.1186/1475-2867-10-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hai H., Tamori A., Kawada N. Role of hepatitis B virus DNA integration in human hepatocarcinogenesis. World J. Gastroenterol. 2014;20:6236–6243. doi: 10.3748/wjg.v20.i20.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Aziz K., Limzerwala J.F., Sturmlechner I., Hurley E., Zhang C., Jeganathan K.B., Nelson G., Bronk S., Fierro Velasco R.O., van Deursen E.J., et al. Ccne1 overexpression causes chromosome instability in liver cells and liver tumor development in mice. Gastroenterology. 2019;157:210–226.e12. doi: 10.1053/j.gastro.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bakhoum S.F., Cantley L.C. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell. 2018;174:1347–1360. doi: 10.1016/j.cell.2018.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nigg E.A. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 127.D'Assoro A.B., Lingle W.L., Salisbury J.L. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- 128.Lengauer C., Kinzler K.W., Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 129.Gao C., Furge K., Koeman J., Dykema K., Su Y., Cutler M.L., Werts A., Haak P., Vande Woude G.F. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc. Natl. Acad. Sci. U S A. 2007;104:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Neuveut C., Wei Y., Buendia M.A. Mechanisms of HBV-related hepatocarcinogenesis. J. Hepatol. 2010;52:594–604. doi: 10.1016/j.jhep.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 131.Yang X.B., Wu L.C., Lin J.Z., Wang A.Q., Wan X.S., Wu Y., Robson S.C., Sang X.T., Zhao H.T. Distinct hepatitis B virus integration patterns in hepatocellular carcinoma and adjacent normal liver tissue. Int. J. Cancer. 2017;140:1324–1330. doi: 10.1002/ijc.30547. [DOI] [PubMed] [Google Scholar]
- 132.Ding D., Lou X., Hua D., Yu W., Li L., Wang J., Gao F., Zhao N., Ren G., Li L., et al. Recurrent targeted genes of hepatitis B virus in the liver cancer genomes identified by a next-generation sequencing-based approach. PLoS Genet. 2012;8:e1003065. doi: 10.1371/journal.pgen.1003065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liu S., Zhang H., Gu C., Yin J., He Y., Xie J., Cao G. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J. Natl. Cancer Inst. 2009;101:1066–1082. doi: 10.1093/jnci/djp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang H.C., Wu H.C., Chen C.F., Fausto N., Lei H.Y., Su I.J. Different types of ground glass hepatocytes in chronic hepatitis B virus infection contain specific pre-S mutants that may induce endoplasmic reticulum stress. Am. J. Pathol. 2003;163:2441–2449. doi: 10.1016/S0002-9440(10)63599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bonino F., Heermann K.H., Rizzetto M., Gerlich W.H. Hepatitis delta virus: protein composition of delta antigen and its hepatitis B virus-derived envelope. J. Virol. 1986;58:945–950. doi: 10.1128/jvi.58.3.945-950.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Freitas N., Cunha C., Menne S., Gudima S.O. Envelope proteins derived from naturally integrated hepatitis B virus DNA support assembly and release of infectious hepatitis delta virus particles. J. Virol. 2014;88:5742–5754. doi: 10.1128/JVI.00430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fattovich G., Bortolotti F., Donato F. Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J. Hepatol. 2008;48:335–352. doi: 10.1016/j.jhep.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 138.Fattovich G., Giustina G., Christensen E., Pantalena M., Zagni I., Realdi G., Schalm S.W. Influence of hepatitis delta virus infection on morbidity and mortality in compensated cirrhosis type B. The European Concerted Action on Viral Hepatitis (Eurohep) Gut. 2000;46:420–426. doi: 10.1136/gut.46.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mariscotti C., Vigano P., Costa R., Gubertini G., Cargnel A. Histological and immunohistochemical features in chronic delta hepatitis. Basic Appl. Histochem. 1988;32:247–254. [PubMed] [Google Scholar]
- 140.Shamay M., Agami R., Shaul Y. HBV integrants of hepatocellular carcinoma cell lines contain an active enhancer. Oncogene. 2001;20:6811–6819. doi: 10.1038/sj.onc.1204879. [DOI] [PubMed] [Google Scholar]
- 141.Schluter V., Meyer M., Hofschneider P.H., Koshy R., Caselmann W.H. Integrated hepatitis B virus X and 3' truncated preS/S sequences derived from human hepatomas encode functionally active transactivators. Oncogene. 1994;9:3335–3344. [PubMed] [Google Scholar]
- 142.Kumar V., Jayasuryan N., Kumar R. A truncated mutant (residues 58–140) of the hepatitis B virus X protein retains transactivation function. Proc. Natl. Acad. Sci. U S A. 1996;93:5647–5652. doi: 10.1073/pnas.93.11.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ng K.Y., Chai S., Tong M., Guan X.Y., Lin C.H., Ching Y.P., Xie D., Cheng A.S., Ma S. C-Terminal truncated hepatitis B virus X protein promotes hepatocellular carcinogenesis through induction of cancer and stem cell-like properties. Oncotarget. 2016;7:24005–24017. doi: 10.18632/oncotarget.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tu H., Bonura C., Giannini C., Mouly H., Soussan P., Kew M., Paterlini-Brechot P., Brechot C., Kremsdorf D. Biological impact of natural COOH-terminal deletions of hepatitis B virus X protein in hepatocellular carcinoma tissues. Cancer Res. 2001;61:7803–7810. [PubMed] [Google Scholar]
- 145.Ma N.F., Lau S.H., Hu L., Xie D., Wu J., Yang J., Wang Y., Wu M.C., Fung J., Bai X., et al. COOH-terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin. Cancer Res. 2008;14:5061–5068. doi: 10.1158/1078-0432.CCR-07-5082. [DOI] [PubMed] [Google Scholar]
- 146.Wang Q., Zhang W., Liu Q., Zhang X., Lv N., Ye L., Zhang X. A mutant of hepatitis B virus X protein (HBxDelta127) promotes cell growth through a positive feedback loop involving 5-lipoxygenase and fatty acid synthase. Neoplasia. 2010;12:103–115. doi: 10.1593/neo.91298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sze K.M., Chu G.K., Lee J.M., Ng I.O. C-Terminal truncated hepatitis B virus x protein is associated with metastasis and enhances invasiveness by C-Jun/matrix metalloproteinase protein 10 activation in hepatocellular carcinoma. Hepatology. 2013;57:131–139. doi: 10.1002/hep.25979. [DOI] [PubMed] [Google Scholar]
- 148.Chami M., Gozuacik D., Saigo K., Capiod T., Falson P., Lecoeur H., Urashima T., Beckmann J., Gougeon M.L., Claret M., et al. Hepatitis B virus-related insertional mutagenesis implicates SERCA1 gene in the control of apoptosis. Oncogene. 2000;19:2877–2886. doi: 10.1038/sj.onc.1203605. [DOI] [PubMed] [Google Scholar]
- 149.Yuan Y., Zhao K., Yao Y., Liu C., Chen Y., Li J., Wang Y., Pei R., Chen J., Hu X., et al. HDAC11 restricts HBV replication through epigenetic repression of cccDNA transcription. Antiviral Res. 2019;172:104619. doi: 10.1016/j.antiviral.2019.104619. [DOI] [PubMed] [Google Scholar]
- 150.Quasdorff M., Protzer U. Control of hepatitis B virus at the level of transcription. J. Viral Hepat. 2010;17:527–536. doi: 10.1111/j.1365-2893.2010.01315.x. [DOI] [PubMed] [Google Scholar]
- 151.Dandri M. Epigenetic modulation in chronic hepatitis B virus infection. Semin. Immunopathol. 2020;42:173–185. doi: 10.1007/s00281-020-00780-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang Y., Li C., Zhang Y., Zhu H., Kang Y., Liu H., Wang J., Qin Y., Mao R., Xie Y., et al. Comparative analysis of CpG islands among HBV genotypes. PLoS one. 2013;8:e56711. doi: 10.1371/journal.pone.0056711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liang T.J., Block T.M., McMahon B.J., Ghany M.G., Urban S., Guo J.T., Locarnini S., Zoulim F., Chang K.M., Lok A.S. Present and future therapies of hepatitis B: from discovery to cure. Hepatology. 2015;62:1893–1908. doi: 10.1002/hep.28025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu. and European Association for the Study of the Liver EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 155.Zoulim F., Testoni B., Lebosse F. Kinetics of intrahepatic covalently closed circular DNA and serum hepatitis B surface antigen during antiviral therapy for chronic hepatitis B: lessons from experimental and clinical studies. Clin. Gastroenterol. Hepatol. 2013;11:1011–1013. doi: 10.1016/j.cgh.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 156.Gehring A.J., Protzer U. Targeting innate and adaptive immune responses to cure chronic HBV infection. Gastroenterology. 2019;156:325–337. doi: 10.1053/j.gastro.2018.10.032. [DOI] [PubMed] [Google Scholar]
- 157.Fanning G.C., Zoulim F., Hou J., Bertoletti A. Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat. Rev. Drug Discov. 2019;18:827–844. doi: 10.1038/s41573-019-0037-0. [DOI] [PubMed] [Google Scholar]
- 158.Yuen M.F., Schiefke I., Yoon J.H., Ahn S.H., Heo J., Kim J.H., Lik Yuen Chan H., Yoon K.T., Klinker H., Manns M., et al. RNA interference therapy with ARC-520 results in prolonged hepatitis B surface antigen response in patients with chronic hepatitis B infection. Hepatology. 2020;72:19–31. doi: 10.1002/hep.31008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Weber N.D., Stone D., Sedlak R.H., De Silva Feelixge H.S., Roychoudhury P., Schiffer J.T., Aubert M., Jerome K.R. AAV-mediated delivery of zinc finger nucleases targeting hepatitis B virus inhibits active replication. PLoS One. 2014;9:e97579. doi: 10.1371/journal.pone.0097579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cradick T.J., Keck K., Bradshaw S., Jamieson A.C., McCaffrey A.P. Zinc-finger nucleases as a novel therapeutic strategy for targeting hepatitis B virus DNAs. Mol. Ther. 2010;18:947–954. doi: 10.1038/mt.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Maepa M.B., Roelofse I., Ely A., Arbuthnot P. Progress and prospects of anti-HBV gene therapy development. Int. J. Mol. Sci. 2015;16:17589–17610. doi: 10.3390/ijms160817589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Seeger C., Sohn J.A. Complete spectrum of CRISPR/Cas9-induced mutations on HBV cccDNA. Mol. Ther. 2016;24:1258–1266. doi: 10.1038/mt.2016.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Weber N.D., Stone D., Jerome K.R. TALENs targeting HBV: designer endonuclease therapies for viral infections. Mol. Ther. 2013;21:1819–1820. doi: 10.1038/mt.2013.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dreyer T., Nicholson S., Ely A., Arbuthnot P., Bloom K. Improved antiviral efficacy using TALEN-mediated homology directed recombination to introduce artificial primary miRNAs into DNA of hepatitis B virus. Biochem. Biophys. Res. Commun. 2016;478:1563–1568. doi: 10.1016/j.bbrc.2016.08.152. [DOI] [PubMed] [Google Scholar]