Abstract
We describe a new chromogenic agar medium, ABC medium (αβ-chromogenic medium), which includes two substrates, 3,4-cyclohexenoesculetin-β-d-galactoside and 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside, to facilitate the selective isolation of Salmonella spp. This medium exploits the fact that Salmonella spp. may be distinguished from other members of the family Enterobacteriaceae by the presence of α-galactosidase activity in the absence of β-galactosidase activity. A total of 1,022 strains of Salmonella spp. and 300 other gram-negative strains were inoculated onto this medium. Of these, 1,019 (99.7%) strains of Salmonella spp. produced a characteristic green colony, whereas only 1 strain (0.33%) of non-Salmonella produced a green colony. A total of 283 stool samples were cultured onto desoxycholate citrate (DC) agar and ABC medium by direct inoculation and after selective enrichment in selenite broth. Overall, the sensitivity and specificity were superior for ABC medium (100 and 90.5%, respectively) than for DC agar (88 and 26.9%, respectively). We conclude that ABC medium offers a high degree of specificity for the detection of Salmonella spp. in stool samples.
Members of the genus Salmonella constitute the most important causes of food poisoning in the United Kingdom, and isolation by culture remains the most reliable method for their detection. A wide variety of selective media has been developed for this purpose. These media traditionally rely on the visualization of simple biochemical features such as the production of hydrogen sulfide or the nonfermentation of lactose. While confirmation of suspect colonies with such media are successful, most are highly nonspecific and consequently place a heavy burden on the laboratory in terms of serological and biochemical confirmation of suspect colonies (1, 8).
In recent years chromogenic media have been developed for the detection of Salmonella spp. Such media detect a combination of biochemical characteristics and are consequently highly specific. Rambach agar was the first medium of this type. It uses a chromogenic substrate for β-galactosidase in conjunction with propylene glycol, which is fermented by Salmonella spp. to generate acid (7). Although highly specific, Rambach agar has the disadvantage that it does not detect S. typhi. SM-ID agar is another chromogenic agar that is based on principles similar to those for Rambach agar and that incorporates β-galactosidase and glucuronic acid (6). This medium allows the detection of a wider range of serotypes including S. typhi and S. paratyphi (4, 5).
We describe a new agar, ABC medium (αβ-chromogenic medium), which incorporates two chromogenic substrates. The first substrate, 3,4-cyclohexenoesculetin-β-d-galactoside (CHE-GAL), is incorporated to visualize β-galactosidase-producing organisms as black colonies in the presence of iron. The second substrate, 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside (X-α-Gal), is hydrolyzed by strains of Salmonella spp., which are visualized as green colonies.
MATERIALS AND METHODS
X-α-Gal and isopropyl-β-d-thiogalactopyranoside (IPTG) were obtained from Melford Laboratories Ltd., Ipswich, United Kingdom. Ferric ammonium citrate was obtained from the Sigma Chemical Company, Poole, United Kingdom. CHE-GAL was kindly synthesized by L. Armstrong of the University of Northumbria, Newcastle-upon-Tyre, United Kingdom, by a previously described method (2).
Modified DCA Hynes Base was kindly supplied by Lab M Ltd., Bury, United Kingdom. This comprised the following (per liter): beef extract (5 g), peptone (15 g), sodium citrate (8.5 g), sodium desoxycholate (5 g), and agar (12 g). This was supplemented with the following (per liter): X-α-Gal (80 mg), CHE-GAL (0.3 g), ferric ammonium citrate (0.5 g), and IPTG (30 mg). The agar was prepared in 1-liter batches and was autoclaved at 116°C for 3 min before it was poured into plates.
A total of 1,022 strains of Salmonella spp. consecutively isolated from enteric samples were collected in our laboratories over a 3-year period. These strains had been isolated with a combination of desoxycholate citrate (DC) agar (Lab M) and mannitol-lysine-crystal violet-brilliant green agar (Lab M) following both direct culture and enrichment with selenite broth. Serotyping of all of these strains was performed by the Central Public Health Laboratory, Colindale, United Kingdom. Three hundred strains of other gram-negative bacteria isolated from stool samples with DC agar were also collected. These were identified with the API 20E system (bioMérieux, La Balme-les-Grottes, France). All 1,322 strains were freshly subcultured onto Columbia agar (Lab M) before they were streaked onto ABC medium. The cultures were incubated at 37°C for 18 hours, and the colonies were examined for color production.
Over a 3-month period 283 consecutive diarrheal stool samples from both hospitalized and nonhospitalized patients were cultured onto both ABC medium and DC agar both directly and following enrichment in selenite broth. This was performed quantitatively by inoculating a 50-μl sample of the stool onto each agar and into selenite broth with a Gilson pipette. Following incubation a 50-μl sample of selenite broth was inoculated onto each plate. All agar media were used within 1 week of preparation. Cultures were incubated for 18 h and examined for the presence of characteristic colonies.
RESULTS
Of 1,022 strains of Salmonella spp. tested, 1,019 (99.7%) produced a characteristic green colony on ABC medium. These included the following Salmonella serotypes: aberdeen (n = 1), agama (n = 1), agona (n = 2), albany (n = 1), anatum (n = 3), arechavaleta (n = 1), bareilly (n = 1), blockley (n = 2), bonn (n = 1), braenderup (n = 5), brandenburg (n = 3), bredeney (n = 1), concord (n = 3), derby (n = 2), durban (n = 1), emek (n = 1), enteritidis (n = 556), galiema (n = 2), goldcoast (n = 6), grumpensis (n = 1), hadar (n = 13), heidelberg (n = 16), hvittingfoss (n = 1), idikan (n = 1), indiana (n = 7), infantis (n = 8), java (n = 5), lanka (n = 1), lexington (n = 1), manhattan (n = 1), mbandaka (n = 5), molade (n = 2), montevideo (n = 7), muenchen (n = 1), napoli (n = 1), newington (n = 2), newport (n = 9), oranienburg (n = 1), panama (n = 3), poona (n = 4), senftenberg (n = 1), tennessee (n = 1), thompson (n = 3), typhi (n = 2), typhimurium (n = 236), virchow (n = 76), wangata (n = 4), weltevreden (n = 7), zanzibar (n = 1), and unnamed Salmonella serotypes (n = 5).
Commonly occurring serotypes were further examined by the Central Public Health Laboratory by phage typing. The strains of S. enteritidis comprised 22 distinct phage types, of which phage type 4 was by far the commonest (439 strains). Others which were routinely phage typed included S. typhimurium (18 distinct types), S. virchow (10 distinct types), and S. hadar (6 distinct types).
Three strains of Salmonella spp. produced atypical colonies on ABC medium and would have been interpreted as nonpathogens. These included one strain of S. arizonae which produced a black colony due to β-galactosidase production and two strains, S. braenderup and S. saintpaul, which failed to produce α-galactosidase and which remained colorless.
Of the 300 strains of other bacteria tested, 299 strains produced either clear or black colonies. These included Acinetobacter species (n = 15), Aeromonas caviae (n = 5), Aeromonas hydrophila (n = 5), Aeromonas sobria (n = 5), Citrobacter diversus (n = 10), Citrobacter freundii (n = 20), Enterobacter aerogenes (n = 5), Enterobacter agglomerans (n = 5), Enterobacter cloacae (n = 20), Escherichia coli (n = 39), Escherichia hermannii (n = 2), Hafnia alvei (n = 10), Klebsiella oxytoca (n = 20), Klebsiella pneumoniae (n = 20), Morganella morganii (n = 10), Proteus mirabilis (n = 20), Proteus penneri (n = 5), Proteus vulgaris (n = 10), Providencia rettgeri (n = 5), Providencia stuartii (n = 10), Pseudomonas aeruginosa (n = 10), Serratia liquefaciens (n = 10), Serratia marcescens (n = 10), Shigella dysenteriae (n = 2), Shigella flexneri (n = 5), Shigella sonnei (n = 10), Stenotrophomonas maltophilia (n = 6), and Yersinia enterocolitica (n = 5). Only one strain (0.33%) produced a green colony characteristic of Salmonella spp. This was a highly atypical strain of E. coli which failed to produce β-galactosidase.
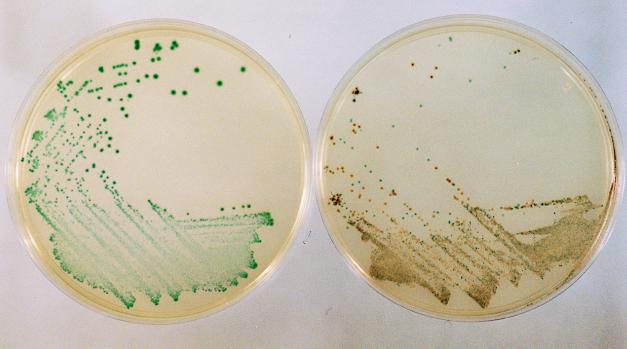
From 283 stool samples a total of 25 strains of Salmonella spp. were recovered with a combination of DC agar and ABC medium. All of these strains were isolated as typical green colonies on ABC medium (13 from direct culture and a further 12 following enrichment), whereas only 22 strains were recovered with DC agar alone (13 from direct culture and a further 9 following enrichment). The sensitivity of ABC medium (100%) therefore compared favorably with that of DC agar (88%). A total of 130 distinct non-lactose-fermenting colonies requiring further investigation were isolated on DC agar, and of these, 35 were eventually confirmed to be Salmonella spp. In contrast, a total of 42 green colonies were isolated on ABC medium, and 38 of these were confirmed to be Salmonella spp. (these comprised 25 distinct strains, some of which were isolated on both direct culture and after enrichment culture). The specificity of ABC medium (90.5%) was therefore clearly superior to that of DC agar (26.9%). The four non-Salmonella strains which produced green colonies were confirmed with the API 20E system to be E. coli (two strains), C. freundii (one strain), and E. aerogenes (one strain). All four of these were biochemically atypical since they failed to produce β-galactosidase. The characteristic appearance of Salmonella spp. on ABC medium is shown in Fig. 1.
FIG. 1.
Typical colonies produced by S. typhi ATCC 19430 on ABC medium (left) and a wild strain of S. napoli isolated in mixed culture from a stool sample (right).
DISCUSSION
From the retrospective survey of 1,022 strains of salmonellae, 3 strains of Salmonella spp. which did not produce a typical reaction on ABC medium were identified. It is clear that no single medium will reliably detect all known Salmonella serotypes. For this reason it is recommended by various regulatory bodies that a combination of complementary media be used for the reliable detection of all Salmonella spp. (8). ABC medium is no exception to this, and it is clear that some less common serotypes, in particular, those which produce β-galactosidase (3, 5), may not be detected.
Given the fact that both media use the same selective base, it was notable to find that three strains (12%) of Salmonella spp. were isolated only on ABC medium. Because of this apparent discrepancy, it was decided to perform an intensive retrospective examination of the relevant enrichment cultures on DC agar. To do this we took as many individual non-lactose-fermenting colonies as were available and spotted them onto ABC medium. We also took a sweep of the confluent growth and respread it onto ABC medium. An examination of these subcultures revealed that small numbers of Salmonella spp. were indeed present on DC medium but were mixed with a predominant growth of commensal organisms, some of which were indistinguishable from Salmonella spp. in their colonial appearance.
Incorporation of chromogenic substrates into selective agars has a significant impact on cost. However, we believe that this is offset by the substantially reduced labor time and consumable costs associated with the detection of Salmonella spp. on traditional agars.
In conclusion, we believe that ABC medium offers a potentially valuable addition to the array of media available for the isolation of Salmonella spp. Further studies are required to evaluate the reactions of many more serotypes, and the medium needs to be evaluated against a wide selection of the best media currently available.
REFERENCES
- 1.Dusch H, Altwegg M. Evaluation of five new plating media for isolation of Salmonella species. J Clin Microbiol. 1995;33:802–804. doi: 10.1128/jcm.33.4.802-804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.James A L, Perry J D, Ford M, Armstrong L, Gould F K. Evaluation of cyclohexenoesculetin-β-d-galactoside and 8-hydroxyquinoline-β-d-galactoside as substrates for the detection of β-galactosidase. Appl Environ Microbiol. 1996;62:3868–3870. doi: 10.1128/aem.62.10.3868-3870.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kühn H, Wonde B, Rabsch W, Reissbrodt R. Evaluation of Rambach agar for detection of Salmonella subspecies I to VI. Appl Environ Microbiol. 1994;60:749–751. doi: 10.1128/aem.60.2.749-751.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monnery I, Freydiere A M, Baron C, Rousset A M, Tigaud S, Boude-Chevalier M, de Montclos H, Gille Y. Evaluation of two new chromogenic media for detection of Salmonella in stools. Eur J Clin Microbiol Infect Dis. 1994;13:257–261. doi: 10.1007/BF01974547. [DOI] [PubMed] [Google Scholar]
- 5.Pignato S, Giammanco G, Giammanco G. Rambach agar and SM-ID medium sensitivity for presumptive identification of Salmonella subspecies I-VI. J Med Microbiol. 1995;43:68–71. doi: 10.1099/00222615-43-1-68. [DOI] [PubMed] [Google Scholar]
- 6.Poupart M C, Mounier M, Denis F, Sirot J, Couturier C, Villeval F. Abstracts of the 5th European Congress of Clinical Microbiology and Infectious Diseases. 1991. A new chromogenic ready-to-use medium for Salmonella detection, abstr. 1254. [Google Scholar]
- 7.Rambach A. New plate medium for facilitated differentiation of Salmonella spp. from Proteus spp. and other enteric bacteria. Appl Environ Microbiol. 1990;56:301–303. doi: 10.1128/aem.56.1.301-303.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warburton D W, Bowen B, Konkle A, Crawford C, Durzi S, Foster R, Fox C, Gour L, Krohn G, LaCasse P, Lamontagne G, McDonagh S, Arling V, Mackenzie J, Todd E C D, Oggel J, Plante R, Shaw S, Tiwari N P, Trottier Y, Wheeler B D. A comparison of six different plating media used in the isolation of Salmonella. Int J Food Microbiol. 1994;22:277–289. doi: 10.1016/0168-1605(94)90179-1. [DOI] [PubMed] [Google Scholar]