Abstract
The reaction centre light-harvesting 1 (RC–LH1) complex is the core functional component of bacterial photosynthesis. We determined the cryo-electron microscopy (cryo-EM) structure of the RC–LH1 complex from Rhodospirillum rubrum at 2.5 Å resolution, which reveals a unique monomeric bacteriochlorophyll with a phospholipid ligand in the gap between the RC and LH1 complexes. The LH1 complex comprises a circular array of 16 αβ-polypeptide subunits that completely surrounds the RC, with a preferential binding site for a quinone, designated QP, on the inner face of the encircling LH1 complex. Quinols, initially generated at the RC QB site, are proposed to transiently occupy the QP site prior to traversing the LH1 barrier and diffusing to the cytochrome bc1 complex. Thus, the QP site, which is analogous to other such sites in recent cryo-EM structures of RC–LH1 complexes, likely reflects a general mechanism for exporting quinols from the RC–LH1 complex.
Keywords: carotenoids, cryo-electron microscopy, light-harvesting, photosynthesis, quinone, reaction centre
Introduction
Reaction centre-light harvesting complex 1 (RC–LH1) complexes are the central functional units of photosynthesis in purple phototrophic bacteria. Solar energy absorbed by a circular LH1 assembly migrates to an enclosed membrane-bound RC [1], where a succession of charge separation and protonation events produces a quinol that leaves the RC–LH1 complex, carrying protons and electrons to a cytochrome (cyt) bc1 complex [2,3]. The LH1 complex is formed from an oligomeric assembly of transmembrane αβ heterodimers, which bind bacteriochlorophyll (BChl) and carotenoid pigments and curve round the central RC complex. In some phototrophic bacteria, the ring of LH1 subunits is incomplete and the ‘missing’ LH1 αβ subunits create a gap through which quinones and quinols can enter and leave the RC–LH1 complex to sustain repeated turnovers of the photosystem. In such complexes one or more transmembrane polypeptides forms part of this interrupted LH1 assembly [4–7]. In other RC–LH1 complexes the RC is fully encircled by LH1 and there is no obvious entry and exit point for quinone traffic [8–10]. Structures of these complexes have revealed small pores in the LH1 antenna that are apparently sufficient to allow the passage of quinones across the LH1 barrier. One such complex, from the phototrophic bacterium Rhodospirillum (Rsp.) rubrum, has been particularly well-studied over many years, with the lack of detailed structural information for this RC–LH1 complex at odds with its important roles in studies of energy transfer [11–13], carotenoid function [14–16] and in vitro LH1 assembly [17–19]. Furthermore, more detailed structural data are required to augment earlier NMR work [20] as well as structural information from cryo-electron microscopy (cryo-EM) and atomic force microscopy studies of 2-D crystals [21–24]. A high-resolution structure would also address some intriguing and unsolved aspects of the Rsp. rubrum RC–LH1 complex, which include the use of geranylgeraniol (GG) and phytol to esterify the BChl and bacteriopheophytin (BPhe) pigments, respectively [25,26], the shape of the complex that likely imparts curvature on the intracytoplasmic membrane [27,28], and the potentially problematic, continuous LH1 barrier round the Rsp. rubrum RC that nevertheless allows rapid quinone traffic between the RC and cyt bc1 complexes [29].
Here, we used cryo-EM to determine the structure of the RC–LH1 complex from Rsp. rubrum at 2.5 Å resolution, which shows the detailed organization of all protein and cofactor components, a preferential site for quinone diffusion across the fully circular LH1 complex, and a unique monomeric BChl, with a phospholipid ligand, in the gap between the RC and LH1 complexes.
Materials and methods
Cell culture and protein purification
Wild type cells of Rsp. rubrum strain S1 were cultured photosynthetically in M22+ medium under illumination (100 μmol of photons m−2 s−1) at 30°C in 20 L screw-capped vessels. When the culture reached an absorbance at 680 nm of 1.6 cells were harvested by centrifugation at 3290g for 30 min. The harvested cells were washed using working buffer (20 mM HEPES, pH 7.8). Washed cells were suspended in the working buffer with a few grains of DNase and MgCl2 and broken by three passages through a French Press at 18 000 psi. The broken cell suspension was applied to a two-step sucrose gradient (15% and 40% (w/w) in a SW32 ultracentrifuge tube), and spun for 4 h at 100 000g. Photosynthetic membranes were collected at the 15%–40% sucrose interface and pelleted. After re-suspension in working buffer, the absorbance of the membrane solution was adjusted to ∼100 at 880 nm. For solubilization of the core complexes, the absorbance at 880 nm of the photosynthetic membrane was adjusted to 60 (1 cm pathlength), and n-Dodecyl-β-d-Maltoside (β-DDM) was added to a final concentration of 3% (w/w). This mixture was then stirred in the dark at 4°C for 30 min. Unsolubilized material was removed by centrifugation for 1 h at 211 000g. The clarified supernatant was loaded onto an DEAE-Sepharose ion exchange column pre-equilibrated with running buffer (working buffer solution containing 0.03% β-DDM). The column was washed using 2 column volumes of running buffer followed by stepwise washing to 120 mM NaCl. A 100 ml gradient of 120–300 mM NaCl then was used to elute the complexes from the column. Fractions were monitored using an A880/A820 absorption ratio, and fractions with a ratio >2.0 were pooled, and concentrated for the next purification step on a Superdex 200 gel filtration column. Fractions with an A880/A820 absorbance ratio >2.2 were pooled and used for cryo-EM data collection.
Cryo-EM data collection
The protein concentration was adjusted to an absorbance of 100 at 880 nm. 3.0 µl protein solution was applied to a glow-discharged holey carbon grid (Quantifoil grid R1.2/1.3, 300 mesh Cu). The grid was plunged into liquid ethane cooled by liquid nitrogen using a FEI Vitrobot 4. Parameters were set as following: blotting time 2.5 s, sample chamber humidity 99%, sample chamber temperature 4°C. The frozen grid was stored in liquid nitrogen before use. Data were recorded at the Cambridge Pharmaceutical Cryo-EM Consortium on a ThermoFisher Scientific Titan Krios G3i Cryo-Transmission Electron Microscope (Cryo-TEM) equipped with a Falcon 4 direct electron detector. The microscope was operated at 300 kV accelerating voltage, at a nominal magnification of 120 000×, corresponding to a pixel size of 0.65 Å at the specimen level. The detector was operated in counting mode. A total dose of 45 electrons per Å2 was divided between 42 frames within 12.21 s exposure time, resulting in an electron dose of 1.07 e−/Å2/frame. In total, 9 024 movies were collected with defocus values varied from 0.8 to 2.2 µm. A typical cryo-EM image, averaged from motion corrected movie frames, is shown in Supplementary Figure S1A.
Data processing
Image processing was performed within RELION 3.1. Beam-induced movement of dose fractionated images were corrected using RELION's built-in motioncorr2 [30] on 5 × 5 patches. CTF parameters were determined using CTFFIND4 [31]. In total, 1 519 688 particles were picked based on the particle coordinates calculated from cisTEM [32] with a box size of 380 × 380, corresponding to a 24.7 nm square. These particles were subjected to reference-free two-dimensional classification. 1 128 646 (74%) particles from good 2D classes were selected for 3D classification. The resulting good 2D classes were subjected to an initial 3D model calculation using EMAN2 [33] for maximum-likelihood-based 3D classification. The best 3D class out of four, containing 519 005 particles (34%), was selected for high resolution 3D reconstruction and refinement, resulting in a 3.2 Å resolution 3D map. After CTF refinement, including anisotropic magnification, beam-tilt, trefoil, fourth order aberration, per particle defocus and per-image astigmatism estimation, Bayesian polishing was performed with the default parameters provided by RELION, improving the resolution of the 3D map to 2.7 Å. The selected particles for the 3D refinement were re-extracted using a 512 × 512 box size for a final CTF refinement and Bayesian polishing, producing a 2.5 Å resolution map.
Modeling and refinement
Initially, the crystal structure of the reaction centre from Rhodobacter (Rba.) sphaeroides (PDB 3I4D) was fitted to the cryo-EM map as a rigid body using the fit in map function of Chimera [34]. The polypeptides of the RC-H, RC-M and RC-L subunits were mutated according to those from Rsp. rubrum using COOT [35]. They were then manually adjusted and real space refined for both polypeptides and cofactors. For LH1, a single subunit of the LH1 complex, αβBChl2Car, was built based on structural similarity compared with the LH1 of Thermochromatium (Tch.) tepidum [36]. It was docked into the density map so that the two BChl a molecules in the model fitted comfortably into their density in the map. Amino acid sequences then were replaced with those from Rps. rubrum, and real space refined in COOT. This subunit was copied to the other 15 αβ subunits, forming a closed LH1 ring. Real space refinement was performed on individual αβ subunits. All-trans spirilloxanthin and ubiquinone-10 molecules (QB and QP) were also fitted into the density map independently using COOT.
Analysis of pigment composition shows that the major carotenoid in the core complex is spirilloxanthin [37]; BChl a is esterified by a GG tail [38], whereas BPhe is esterified by phytol. Rsp. rubrum contains two types of quinone, ubiquinone-10 and rhodoquinone-10 [39]. All-trans spirilloxanthin and 15-cis spirilloxanthin fitted well into the LH1 and RC, respectively. BChl aGG was used for the LH1 BChl a pair, RC special pair and RC accessory BChl a. Based on the 2.5 Å resolution map, RC QA and QB and QP were assigned to UQ-10, whereas QF was assigned to RQ-10. In addition, two tetramyristoyl cardiolipin and two phosphatidylglycerol lipids were modeled into the map. Restraints for novel ligands were generated using eLBOW [40] with the ideal values for bonds and angles obtained from Mogul [41,42]. All of the LH1 subunits and ligands underwent real-space refinement using ISOLDE [43]. The final model was subjected to global refinement and minimization using PHENIX [44]. The final refinement statistics are summarized in Supplementary Table S1. The quality of fit for the structural model within the electron density map was validated using EMRinger [45].
Results and discussion
Overall structure of the RC–LH1 complex
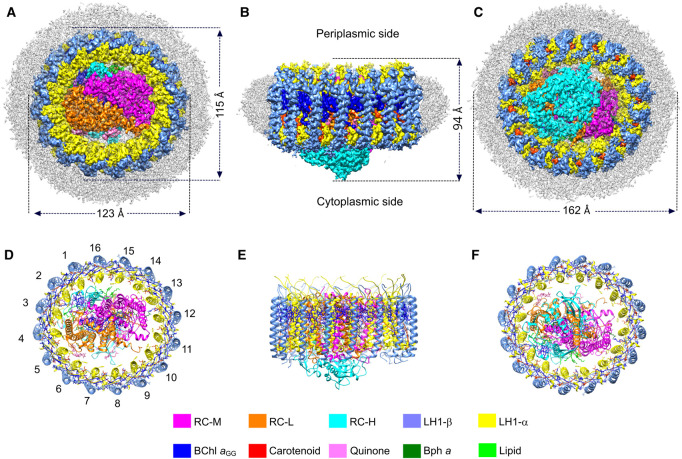
In this study, we report the structure of Rsp. rubrum RC–LH1 complex at 2.5 Å, determined by cryo-EM. In total, 9,024 cryo-EM movies were recorded, from which 1 519 688 particles were picked for further data processing yielding a final resolution of 2.5 Å (Figure 1, Supplementary Figures S1, S2, Supplementary Table S1). The density map (Figure 1A–C) shows the overall shape of the complex, which is slightly elliptical in projection with short and long axes of 115 and 123 Å (Figure 1A), and 94 Å in height (Figure 1B). As Figure 1B also shows, the shape of the complex in the plane of the membrane is cylindrical, yet the chromatophore membranes of Rsp. rubrum, in which RC–LH1 complexes are the dominant complex [46], are curved with a ∼50–100 nm diameter [27,28]. Only a few degrees of conical angle are required to account for this level of curvature, and we conclude that in the native membrane a ring of lipid molecules binds to the outer face of the LH1-β polypeptide on the cytoplasmic side of the complex. Inspection of the density map shows disordered detergent molecules in the position that lipids are suggested to occupy, and lipids bound tightly in these positions round the LH1 ring would confer a slight conical shape on the complex. There are precedents for bound, and structurally resolved, rings of lipids and detergents in RC–LH1 complexes from Rhodopseudomonas (Rps.) palustris (RC–LH114-W; Protein Data Bank (PDB) 6Z5S, and RC–LH116, 6Z5R) [6], Rba. veldkampii (7DDQ) [7], Tch. tepidum (5Y5S) [9] and Thiorhodovibrio (Trv.) strain 970 (7C9R) [10]. Ribbon models of the complex are shown in Figure 1D–F. Although the RC is completely surrounded by the LH1 complex only one H-bond connects the RC and the LH1 ring, between LH1-α1 Ser34 and RC-H Trp9 (3.1 Å); we suggest that this point of contact is a potential site for initiating the assembly of a curved array of LH1 αβ subunits that culminates in a fully encircled RC, so it is assigned as αβ(1). Each of the 16 pairs of transmembrane α and β polypeptides, numbered in Figure 1D, binds two BChl aGG molecules creating a ring of 32 closely spaced and paired BChl a pigments. In this respect, the Rsp. rubrum LH1 antenna is very similar to the 16-subunit ring of Tch. tepidum [9] and to the RC–LH116 complex of Rps. palustris [6].
Figure 1. Cryo-EM structure of the RC–LH1 core complex from Rsp. rubrum.
(A–C) Views of the RC–LH1 density map, colored as in the key at the bottom of the figure. Detergent and other disordered molecules are in grey. (A) View of the slightly elliptical LH1 ring from the periplasmic side of the membrane, showing the diameters of the long and short axes. (B) View in the plane of the membrane showing the height of the complex. (C) Perpendicular view from the cytoplasmic side. (D–F) Ribbon models corresponding to (A–C), made using Chimera [58]; the LH1 subunits are numbered in (D).
Structure of the LH1 complex
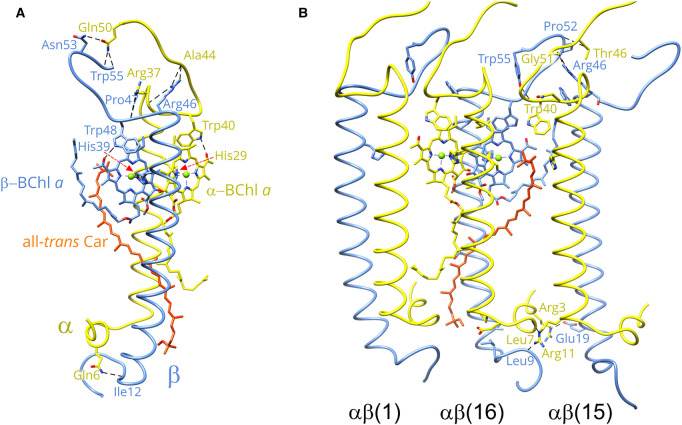
The LH1 αβ subunits are the building blocks of the LH1 ring; Figure 2A shows the structure of a single αβ subunit, which is stabilized by a series of protein–protein, BChl–protein, BChl–BChl, carotenoid–BChl and carotenoid–protein contacts. LH1α consists of a short N-terminal helix of nine residues that lies parallel to the surface of the membrane on the cytoplasmic side, followed by a transmembrane domain and a 14-residue C-terminal region that contains a loop structure. The LH1 β-polypeptide has the same topology as LH1α. Hydrogen bonds between C- and N-terminal residues link the α and β polypeptides and contribute to the formation of LH1 αβ subunits: α-Gln50—β-Asn53, 2.9 Å; α-Gln50—β-Trp55, 2.9 Å; α-Ala44—β-Arg46, 2.9 Å; α-Arg37—β-Arg46, 2.8 Å; α-Arg37—β-Pro47, 2.8 Å and α-Gln6—β-Ile12, 2.8 Å. However, the binding of BChl and carotenoid pigments, and the ensuing pigment-pigment interactions, are the major driving forces that create the LH1αβ(BChl)2 units, which behave in a modular fashion that can form either open or closed rings [4–10], or, in some RC-minus mutants, ellipses, and spirals of variable size [47]. LH1α His29 and LH1β His39 supply the ligands (2.6 and 2.5 Å, respectively) for binding a pair of opposing, excitonically coupled BChls. These pigments are modeled with GG ‘tails’ (BChl aGG) in the well-resolved density map (Supplementary Figure S2). As with other light-harvesting LH1 complexes, hydrogen bonds from C-terminal residues (LH1α Trp40 and LH1β Trp48) to BChl C3 acetyl groups (2.6 and 2.5 Å, respectively) tune the absorption properties of the bound BChls [48,49]. A combination of excitonic coupling within and between BChl dimers, together with hydrogen bonding to LH1-α and LH1-β aromatic side chains, redshifts the absorption from ∼770 nm for monomeric BChl in solvent to 880 nm in the RC–LH1 complex. The progressive red-shifting of absorption, as more LH1αβ(BChl)2 units associate, has been examined extensively using in vitro reconstitution approaches [17–19]. Some of these studies employed a carotenoid-free mixture of polypeptides and BChls, but the native complex necessarily binds carotenoids, both for harvesting light in the 450–550 nm region of the spectrum and for photoprotection. Many studies of excited state dynamics have focused on the Rsp. rubrum LH1 complex, for example [11–14], but the location and conformation of the carotenoid, spirilloxanthin, have remained unknown. Here, the structure of the RC–LH1 complex shows that the spirilloxanthins in the LH1 ring are in the all-trans configuration, making contacts with the N-terminal helical region (α3-RIWQLF) of the n − 1 α-polypeptide and the transmembrane region (α26-LLIHFILL) of the n + 1 α-polypeptide. The central region of the spirilloxanthin is in close contact (4.2 Å) with the tail of the α-BChl aGG (Figure 2B). The extent of these contacts reveals a third, stabilizing, role for the carotenoid, analogous to the same role for these pigments in LH2 complexes [50,51]. Thus, carotenoids stabilize individual LH1αβ subunits, and interactions between spirilloxanthins and n + 1, n, n − 1 polypeptides interlock LH1 αβ subunits and promote the formation of a curved, oligomeric assembly. A subset of this interlocking ring structure is shown in Figure 2B, in which hydrogen-bonds help to stabilize associations between adjacent LH1αβ subunits. Focusing on subunits 15 and 16, at the periplasmic (C-terminal) side of the complex these bonds are between β(15)-Gly51 and β(16)-Arg46, 3.0 Å; β(15)-Pro52 and α(16)-Thr46, 3.3 Å; β(15)-Tyr55 and α(16)-Trp40, 2.6 Å. On the N-terminal side there are hydrogen bonds between α(15)-Glu19 and β(16)-Arg3, 2.8 Å; α(15)-Leu7 and α(16)-Arg11, 2.8 Å; β(15)-Leu 9 and α(16)-Arg 11, 2.8 Å.
Figure 2. Protein–protein and protein–pigment interactions in the Rsp. rubrum RC–LH1 core complex.
(A) A single LH1 αβ subunit, containing one α-polypeptide (yellow), one β-polypeptide (cornflower blue), two BChl aGG molecules colored according to their cognate polypeptide, and one all-trans spirilloxanthin (red-orange). All residues involved in H-bonds are labelled. (B) Inter-subunit interactions between three adjoining LH1 αβ subunits. Pigments are only shown in the middle subunit for clarity. Only residues involved in inter-subunit H-bonds are labelled.
The reaction centre, and the space between this complex and LH1
The RC consists of H, M and L subunits (Figure 1, Supplementary Figure S2), and the internal arrangement of BChl, bacteriopheophytin (BPhe), carotenoid (Car), quinone and Fe cofactors is shown in Figure 3A. Supplementary Figure S2 shows the quality of fits of atomic models for all the cofactors and protein components to their respective densities within the 2.5 Å resolution map. The unusual presence of both GG and phytol ‘tails’ of the various RC pigments in Rsp. rubrum has been known for many years [25]. BChl a esterified with a GG tail (Supplementary Figure S3), BChl aGG, was used in our structural model for the RC special pair and accessory BChl pigments; the BPhe pigments are esterified with phytol. Surprisingly, we identified an extra BChl aGG, localized between the second LH1αβ subunit and the RC special pair (Figure 3A). This pigment is not bound to a protein, and instead it is coordinated by an oxygen within the headgroup of a neighboring phosphatidylglycerol molecule. This type of coordination has not been observed before in RC–LH1 complexes, but in the RC from the green sulfur bacterium Chlorobaculum tepidum [52] it has been proposed that the headgroup of a phosphatidylglycerol molecule coordinates a water molecule forming the axial ligand of the A0 Chl a [53]. One of the acyl chains of the RC-LH1 lipid is close to the GG tail of a BChl bound to the α-polypeptide of LH1αβ(2) and to a spirilloxanthin molecule (Supplementary Figure S4). Presumably the interactions between LH1, a lipid, and the extra BChl are sufficient to position this pigment consistently, so it becomes a defined feature in the density map.
Figure 3. Arrangement of pigments and cofactors in the Rsp. rubrum RC–LH1 core complex.
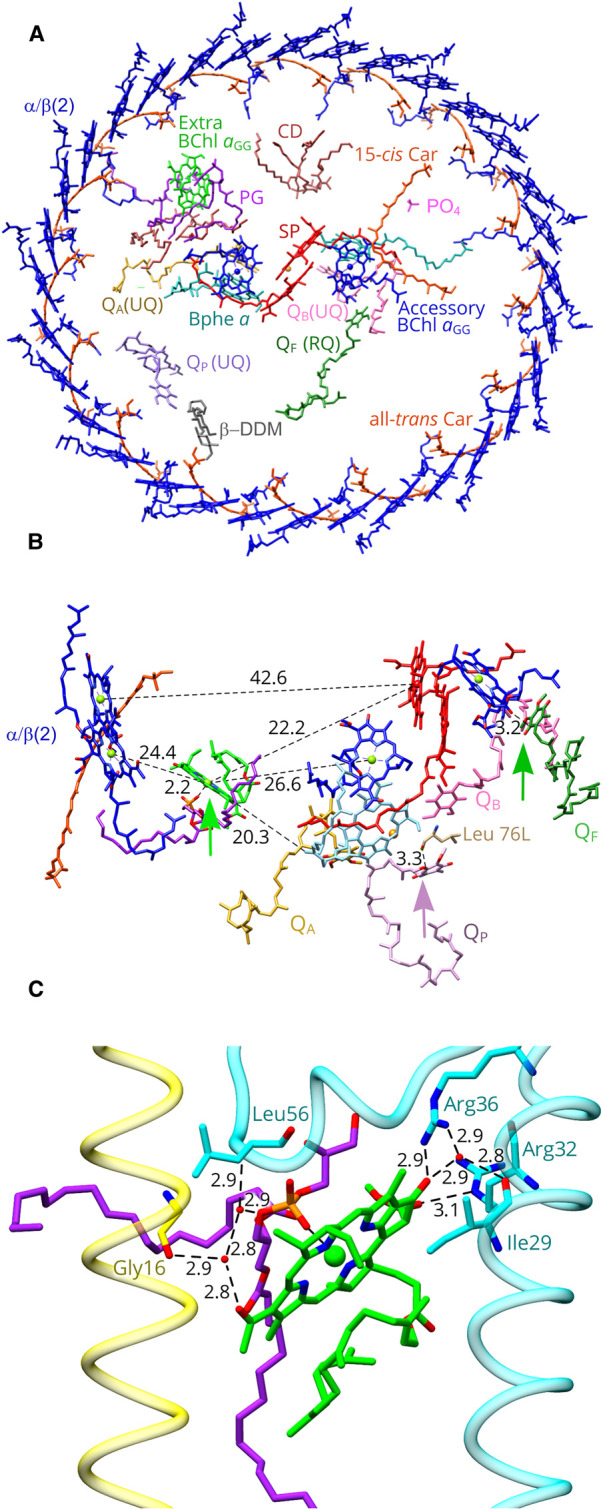
(A) Perpendicular view from the periplasmic side of the membrane. Water molecules were omitted for clarity. PG, Phosphatidyl glycerol; CD, cardiolipin; SP, the reaction centre special pair of BChl aGG molecules. (B) Location and pigment environment of the extra BChl aGG (bright green; colours as in panel (A)). The central Mg ion of this BChl aGG is coordinated by the nearby PG lipid molecule, indicated by a bright green arrow. A lone pair π interaction between QP and RC-L Leu 76 is indicated with a mauve arrow, and a mid-green arrow points to a lone pair π interaction between non-protein bound rhodoquinone RQ and the accessory BChl aGG on the B-branch of the RC. (C) Detailed view of the bonding environment for the extra BChl aGG (green). Three water molecules are shown as small red spheres, with hydrogen bonds shown as dashed lines with the distances in Ångstroms indicated. LH1 α is in yellow, The RC-H subunit is in cyan, and the PG is in magenta.
The function of this BChl aGG pigment, unique in currently determined RC–LH1 structures, is unknown. Figure 3 displays the distances to neighboring pigments, showing that the extra BChl aGG is located approximately midway between the BChl aGG pair attached to LH1αβ(2) and the RC special pair of BChl aGG pigments, a distance of 42.6 Å, with its macrocycle tilted ∼60° from the special pair. The distance from the special pair is 22.2 Å, and 26.6 Å separates the extra BChl aGG from the accessory BChl aGG on the active (A branch) of the bifurcated network of RC cofactors. In the context of the inverse sixth power distance dependence for Förster energy transfer [54], halving the gap between the LH1 donor and RC acceptor pigments could exert a significant effect on LH1 to RC transfer. However, the absorption maximum of this pigment is also a factor to consider. Figure 3C shows the environment of this extra BChl aGG which, in addition to its ligand with the nearby phosphatidyl glycerol (magenta), forms a series of hydrogen bonds with the RC H-subunit and with the second LH1α subunit. The C132 ester of this BChl aGG hydrogen-bonds to RCH-Arg32 NE, and the C131 keto bonds to RCH-Arg36. In addition, three water molecules (small red spheres in Figure 3C) form a hydrogen bond network between the C3-acetyl of BChl aGG, the backbone oxygen of LH1α–Gly16, the lipid and the backbone N of RCH–Leu56. Finally, there is also a water bridging to the RCH–Ile29 OH. As a monomeric pigment, the absorption maximum of this extra BChl aGG is likely to be ∼800 nm by analogy with, for example, LH2 complexes [55,56]. A protein engineering study was conducted on the β-Arg30 residue near to the monomeric B800 BChl in the LH2 complex of Rba. sphaeroides. Raman spectroscopy of β-Arg30 mutants showed that the loss of a hydrogen bond to the C3-acetyl group, and it was concluded that the absorption red-shift attributable solely to the H-bond is ∼10 nm [55]. Thus the absorption maximum of a monomeric BChl, ∼780 nm in solvent, would likely absorb no further to the red than 800 nm and this is proposed to apply to the BChl aGG in the Rsp. rubrum RC–LH1 complex. If this is the case, energy transfer from 880 nm LH1 BChls to this new BChl would be ‘uphill’; onward excitation transfer to the RC special pair absorbing at 865 nm would be correspondingly ‘downhill’, though, and energetically level to the accessory RC BChl absorbing near 800 nm. Overall, the extra BChl aGG could act as a conduit for energy transfer to the RC, but early picosecond transient absorption experiments showed no difference between Rsp. rubrum and Rba. sphaeroides RC–LH1 complexes, with rates of energy transfer in the 30–40 ps range [12].
Four quinone molecules were identified: QA, QB and Qp are assigned as ubiquinone-10, based on their well-resolved densities (Supplementary Figure S2). Both the RC QA and QB sites are occupied, and there is a clear density for another quinone that sits between the RC and the inner face of the LH1 complex. Despite the disorder presumed to exist in the space between the RC and LH1 complexes, we could assign a rhodoquinone-10 at a position designated QF (‘free’ quinone) based on the match between its chemical structure and the density shape (Figure 3A, Supplementary Figures S2, S3). ‘Free’ quinones, with no surrounding protein to provide a defined binding pocket, were also found in Rps. palustris, Tch. tepidum and Rba. veldkampii RC–LH1 complexes [6,7,9]. There is a lone pair π interaction between the head of QF and the C131 acetyl group of the accessory BChl aGG on the B-branch.
In the Rsp. rubrum structure, there is one spirilloxanthin molecule per LH1αβ(BChl)2 unit, and this stoichiometry has been proposed to create a series of pores round the LH1 ring that could allow passage of quinone across the LH1 ring by means of ‘breathing motions’ [29,55]. This view of quinone diffusion does not assign any particular location or set of LH1 subunits for traversing the LH1 ring. However, the Rsp. rubrum RC–LH1 structure adds to the lengthening list of complexes [5–10] that have a conserved QP quinone (Figure 4A). The head group of the QP quinone in the Rsp. rubrum LH1 makes lone pair π interactions with the backbone oxygen of RC-L Leu 76, and the isoprenyl tail makes contacts with LH1α4 and 5. As with other such QP sites, we suggest that transient docking into this binding pocket prepares the quinone for passage through an adjacent pore between LH1αβ(BChl)2 units. This does not rule out migration paths across other parts of the LH1 ring, although these routes may be less probable. In particular, we note the structurally resolved β-DDM detergent molecule (Figure 3A, grey), which fits well into the density near to QP (Supplementary Figure S5). This detergent molecule could have displaced a lipid, or a quinone, or it could sit in a pore in the LH1 ring between subunits 6 and 7, indicating another possible point of exit for quinol produced at the RC QB site.
Figure 4. Location of a quinone/quinol channel adjacent to the QP site in the LH1 ring.
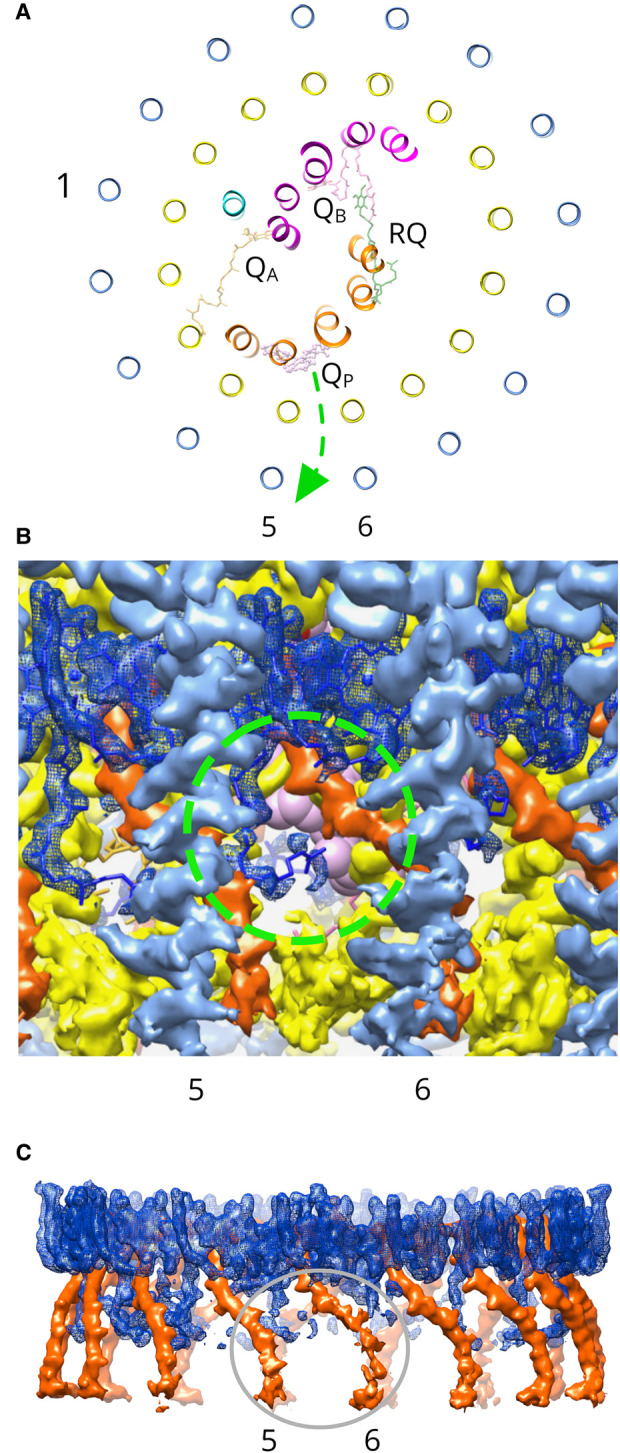
(A) The RC–LH1 complex viewed from the periplasmic side, with α-helices represented as ribbons and appearing as circles in the case of LH1 polypeptides. Only quinone cofactors are shown. Subunits are colored as in Figure 1. LH1-αβ subunits 1, 5 and 6 are labeled. A green arrow indicates the proposed path taken by a quinol though the pore. (B) View of the LH1 complex in the plane of the membrane, from outside the complex. The LH1 α-polypeptide is in yellow, the β-polypeptide in cornflower blue, BChl aGG molecules are blue, and the all-trans spirilloxanthin is in red-orange. The green dashed circle shows the pore, with the weak density of this particular GG tail allowing a view of the QP quinone (pink) visible in the background. (C) Densities of pigments adjacent to the LH1 pore. The grey ellipse delineates the weaker densities for the GG tail and the carotenoid at these positions in the LH1 ring.
The QP pore is circled in green in Figure 4B, and the QP quinone molecule can be seen by virtue of the weaker density for the GG tail of the BChl, relative to other pigments in the ring (Figure 4C). Similarly, the density for the cytoplasmic end of the spirilloxanthin in LH1αβ (6) is also weaker than for neighboring carotenoids. Counterparts of the QP quinone have been found in several RC–LH1 structures [5–10], and in each case QP associates with the inner face of the LH1 complex adjacent to a pore that would allow quinone movement across the LH1 ring. In the case of the Blastochloris viridis RC–LH1 [8], QP is adjacent to a region of relatively weak density for the BChl phytol and the associated carotenoid, which was proposed to reflect conformational flexibility leading to local disorder where quinones diffuse across the complex. We propose that transient occupation of the QP site increases the probability that the quinol will pass through the adjacent pore in the RC–LH1 complex of Rsp. rubrum.
Acknowledgements
We acknowledge the European Synchrotron Radiation Facility for provision of initial beam time on CM01 (MX-2117) and we would like to thank Dr. Michael Hons for his assistance.
Abbreviations
- β-DDM
n-Dodecyl-β-d-Maltoside
- BChl aGG
BChl a esterified with a GG tail
- BChl
Bacteriochlorophyll a
- BPhe
bacteriopheophytin
- cryo-EM
cryo-electron microscopy
- GG
geranylgeraniol
- Rba.
Rhodobacter
- RC–LH1
reaction centre light-harvesting 1 complex
- Rps.
Rhodopseudomonas
- Rsp.
Rhodospirillum
- Tch.
Thermochromatium
- Trv.
Thiorhodovibrio
Data Availability
The cryo-EM density map has been deposited in the World Wide Protein Data Bank (wwPDB) under accession code EMD-13110 and the coordinates have been deposited in the Protein Data Bank (PDB) under accession number 7OY8.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
P.Q., D.J.K.S. and C.N.H. were supported by the Biotechnology and Biological Sciences Research Council (BBSRC) UK, award number BB/M000265/1, and C.N.H. and D.J.K.S. are supported by European Research Council Synergy Award 854126. T.I.C. acknowledges Wellcome Trust grant 209407/Z/17/Z.
Open Access Statement
Open access for this article was enabled by the participation of University of Sheffield in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with JISC.
CRediT Author Contribution
C. Neil Hunter: Conceptualization, Supervision, Funding acquisition, Writing — original draft, Project administration, Writing — review and editing. Pu Qian: Conceptualization, Data curation, Formal analysis, Supervision, Validation, Investigation, Visualization, Methodology, Writing — original draft. Tristan Ian Croll: Software, Formal analysis, Validation, Methodology. David JK Swainsbury: Visualization, Writing — review and editing. Pablo Castro-Hartmann: Investigation. Nigel W. Moriarty: Formal analysis. Kasim Sader: Investigation.
Supplementary Material
References
- 1.Gardiner, A.T., Nguyen-Phan, T.C. and Cogdell, R.J. (2020) A comparative look at structural variation among RC-LH1 ‘Core’ complexes present in anoxygenic phototrophic bacteria. Photosynth. Res. 145, 83–96 10.1007/s11120-020-00758-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sener, M., Strumpfer, J., Singharoy, A., Hunter, C.N. and Schulten, K. (2016) Overall energy conversion efficiency of a photosynthetic vesicle. eLife 5, e09541 10.7554/eLife.09541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singharoy, A., Maffeo, C., Delgado-Magnero, K.H., Swainsbury, D.J.K., Sener, M., Kleinekathofer, U.et al. (2019) Atoms to phenotypes: molecular design principles of cellular energy metabolism. Cell 179, 1098–1111.e23 10.1016/j.cell.2019.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qian, P., Papiz, M.Z., Jackson, P.J., Brindley, A.A., Ng, I., Olsen, J.D.et al. (2013) The 3-D structure of the Rhodobacter sphaeroides RC-LH1-PufX complex: dimerization and quinone channels promoted by PufX. Biochemistry 52, 7575–7585 10.1021/bi4011946 [DOI] [PubMed] [Google Scholar]
- 5.Xin, Y., Shi, Y., Niu, T., Wang, Q., Niu, W., Huang, X.et al. (2018) Cryo-EM structure of the RC-LH core complex from an early branching photosynthetic prokaryote. Nat. Commun. 9, 1568 10.1038/s41467-018-03881-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swainsbury, D.J.K., Qian, P., Jackson, P.J., Faries, K.M., Niedzwiedzki, D., Martin, E.C.et al. (2021) Structures of Rhodopseudomonas palustris RC-LH1 complexes with open or closed quinone channels. Sci. Adv. 7, eabe2631 10.1126/sciadv.abe2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bracun, L., Yamagata, A., Christianson, B.M., Terada, T., Canniffe, D.P., Shirouzu, M.et al. (2021) Cryo-EM structure of the photosynthetic RC-LH1-PufX supercomplex at 2.8-Å resolution. Sci. Adv. 7, eabf8864 10.1126/sciadv.abf8864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qian, P., Siebert, C.A., Wang, P.Y., Canniffe, D.P. and Hunter, C.N.C. (2018) Cryo-EM structure of the Blastochloris viridis LH1-RC complex at 2.9 Å. Nature 556, 203–208 10.1038/s41586-018-0014-5 [DOI] [PubMed] [Google Scholar]
- 9.Yu, L.J., Suga, M., Wang-Otomo, Z.Y. and Shen, J.R. (2018) Structure of photosynthetic LH1-RC supercomplex at 1.9 Å resolution. Nature 556, 209–213 10.1038/s41586-018-0002-9 [DOI] [PubMed] [Google Scholar]
- 10.Tani, K., Kanno, R., Makino, Y., Hall, M., Takenouchi, M., Imanishi, M.et al. (2020) Cryo-EM structure of a Ca2+-bound photosynthetic LH1-RC complex containing multiple αβ-polypeptides. Nat. Commun. 11, 4955 10.1038/s41467-020-18748-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valkunas, L., Akesson, E., Pullerits, T. and Sundström, V. (1996) Energy migration in the light-harvesting antenna of the photosynthetic bacterium Rhodospirillum rubrum studied by time-resolved excitation annihilation at 77K. Biophys. J. 70, 2373–2379 10.1016/S0006-3495(96)79804-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visscher, K.J., Bergström, H., Sundström, V., Hunter, C.N. and van Grondelle, R. (1989) Temperature dependence of energy transfer from the long wavelength antenna bchl-896 to the reaction center in Rhodospirillum rubrum, Rhodobacter sphaeroides (w.t. and M21 mutant) from 77 to 177 K, studied by picosecond absorption spectroscopy. Photosynth. Res. 22, 211–217 10.1007/BF00048300 [DOI] [PubMed] [Google Scholar]
- 13.Kosumi, D., Maruta, S., Horibe, T., Fujii, R., Sugisaki, M., Cogdell, R.J.et al. (2011) Ultrafast energy-transfer pathway in a purple-bacterial photosynthetic core antenna, as revealed by femtosecond time-resolved spectroscopy. Angew. Chem. Int. Ed. Engl. 50, 1097–1100 10.1002/anie.201003771 [DOI] [PubMed] [Google Scholar]
- 14.Gradinaru, C.C., Kennis, J.T., Papagiannakis, E., van Stokkum, I.H., Cogdell, R.J., Fleming, G.R.et al. (2001) An unusual pathway of excitation energy deactivation in carotenoids: singlet-to-triplet conversion on an ultrafast timescale in a photosynthetic antenna. Proc. Natl Acad. Sci. U.S.A. 98, 2364–2369 10.1073/pnas.051501298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akahane, J., Rondonuwu, F.S., Fiedor, L., Watanabe, Y. and Koyama, Y. (2004) Dependence of singlet energy transfer on the conjugation length of carotenoids reconstituted into the LH1 complex from Rhodospirillum rubrum G9. Chem. Phys. Lett. 393, 184–191 10.1016/j.cplett.2004.06.021 [DOI] [Google Scholar]
- 16.Nakagawa, K., Suzuki, S., Fujii, R., Gardiner, A.T., Cogdell, R.J., Nango, M.et al. (2008) Probing the effect of the binding site on the electrostatic behavior of a series of carotenoids reconstituted into the light-harvesting 1 complex from purple photosynthetic bacterium Rhodospirillum rubrum detected by Stark spectroscopy. J. Phys. Chem. B 112, 9467–9475 10.1021/jp801773j [DOI] [PubMed] [Google Scholar]
- 17.Parkes-Loach, P.S., Sprinkle, J.R. and Loach, P.A. (1988) Reconstitution of the B873 light-harvesting complex of Rhodospirillum rubrum from the separately isolated α- and β-polypeptides and bacteriochlorophyll a. Biochemistry 27, 2718–2727 10.1021/bi00408a011 [DOI] [PubMed] [Google Scholar]
- 18.Davis, C.M., Bustamante, P.L. and Loach, P.A. (1995) Reconstitution of the bacterial core light-harvesting complexes of Rhodobacter sphaeroides and Rhodospirillum rubrum with isolated α- and β-polypeptides, bacteriochlorophyll a and carotenoid. J. Biol. Chem. 270, 5793–5804 10.1074/jbc.270.11.5793 [DOI] [PubMed] [Google Scholar]
- 19.Fiedor, L., Akahane, J. and Koyama, Y. (2004) Carotenoid-induced cooperative formation of bacterial photosynthetic LH1 complex. Biochemistry 43, 16487–16496 10.1021/bi0481287 [DOI] [PubMed] [Google Scholar]
- 20.Wang, Z.Y., Gokan, K., Kobayashi, M. and Nozawa, T. (2005) Solution structures of the core light-harvesting alpha and beta polypeptides from Rhodospirillum rubrum: implications for the pigment-protein and protein-protein interactions. J. Mol. Biol. 347, 465–477 10.1016/j.jmb.2005.01.017 [DOI] [PubMed] [Google Scholar]
- 21.Karrasch, S., Bullough, P.A. and Ghosh, R. (1995) The 8.5 Å projection map of the light-harvesting complex I from Rhodospirillum rubrum reveals a ring composed of 16 subunits. EMBO J. 14, 631–638 10.1002/j.1460-2075.1995.tb07041.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jamieson, S.J., Wang, P., Qian, P., Kirkland, J.Y., Conroy, M.J., Hunter, C.N.et al. (2002) Projection Structure of the photosynthetic reaction centre-antenna complex of Rhodospirillum rubrum at 8.5 Å resolution. EMBO J 21, 3927–3935 10.1093/emboj/cdf410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian, P., Addlesee, H.A., Ruban, A.V., Wang, P., Bullough, P.A. and Hunter, C.N. (2003) A reaction center – light harvesting 1 complex from a Rhodospirillum rubrum mutant with altered esterifying pigments: characterization by optical spectroscopy and cryo-electron microscopy. J. Biol. Chem. 278, 23678–23685 10.1074/jbc.M302753200 [DOI] [PubMed] [Google Scholar]
- 24.Fotiadis, D., Qian, P., Philippsen, A., Bullough, P.A., Engel, A. and Hunter, C.N. (2004) Structural analysis of the RC-LH1 photosynthetic core complex of Rhodospirillum rubrum using atomic force microscopy. J. Biol. Chem. 279, 2063–2068 10.1074/jbc.M310382200 [DOI] [PubMed] [Google Scholar]
- 25.Francke, C. and Amesz, J. (1995) The size of the photosynthetic unit in purple bacteria. Photosynth. Res. 46, 347–352 10.1007/BF00020450 [DOI] [PubMed] [Google Scholar]
- 26.Addlesee, H.A. and Hunter, C.N. (2002) Rhodospirillum rubrum possesses a variant of the bchP gene, encoding geranylgeranyl-bacteriopheophytin reductase. J. Bacteriol. 184, 1578–1586 10.1128/JB.184.6.1578-1586.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vatter, A.E. and Wolfe, R.S. (1958) The structure of photosynthetic bacteria. J. Bacteriol. 75, 480–488 10.1128/jb.75.4.480-488.1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holt, S.C. and Marr, A.G. (1965) Isolation and purification of the intracytoplasmic membranes of Rhodospirillum rubrum. J. Bacteriol. 89, 1413–1420 10.1128/jb.89.5.1413-1420.1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aird, A., Wrachtrup, J., Schulten, K. and Tietz, C. (2007) Possible pathway for ubiquinone shuttling in Rhodospirillum rubrum revealed by molecular dynamics simulation. Biophys. J. 92, 23–33 10.1529/biophysj.106.084715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zivanov, J., Nakane, T., Forsberg, B.O., Kimanius, D., Hagen, W.J.H., Lindahl, E.et al. (2018) New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 10.7554/eLife.42166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohou, A. and Grigorieff, N. (2015) CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant, T., Rohou, A. and Grigorieff, N. (2018) cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 10.7554/eLife.35383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang, G., Peng, L., Baldwin, P.R., Mann, D.S., Jiang, W., Rees, I.et al. (2007) EMAN2: an extensible image processing suite for electron microscopy. J Struct. Biol. 157, 38–46 10.1016/j.jsb.2006.05.009 [DOI] [PubMed] [Google Scholar]
- 34.Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C.et al. (2004) UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 35.Emsley, P. and Cowtan, K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Struct. Biol. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 36.Niwa, S., Yu, L.-J., Takeda, K., Hirano, Y., Kawakami, T., Wang-Otomo, Z.-Y.et al. (2014) Structure of the LH1–RC complex from Thermochromatium tepidum at 3.0 Å. Nature 508, 228–232 10.1038/nature13197 [DOI] [PubMed] [Google Scholar]
- 37.Evans, M.B., Cogdell, R.J. and Britton, G. (1988) Determination of the bacteriochlorophyll:Carotenoid ratios of the B890 antenna complex of Rhodospirillum rubrum and the B800–850 complex of Rhodobacter sphaeroides. Biochim. Biophys. Acta Bioenergetics 935, 292–298 10.1016/0005-2728(88)90224-1 [DOI] [Google Scholar]
- 38.Katz, J.J., Strain, H.H., Harkness, A.L., Studier, M.H., Svec, W.A., Janson, T.R.et al. (1972) Esterifying alcohols in the chlorophylls of purple photosynthetic bacteria. A new chlorophyll, bacteriochlorophyll (gg), all-trans-geranylgeranyl bacteriochlorophyllide a. J. Am. Chem. Soc. 94, 7938–7939 10.1021/ja00777a054 [DOI] [PubMed] [Google Scholar]
- 39.Hiraishi, A. and Hoshino, Y. (1984) Distribution of rhodoquinone in Rhodospirillaceae and its taxonomic implications. J. Gen. Appl. Microbiol. 30, 435–448 10.2323/jgam.30.435 [DOI] [Google Scholar]
- 40.Moriarty, N.W., Grosse-Kunstleve, R.W. and Adams, P.D. (2009) Electronic ligand builder and optimization workbench (eLBOW): A tool for ligand coordinate and restraint generation. Acta Crystallogr. D Struct. Biol. 65, 1074–1080 10.1107/S0907444909029436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bruno, I.J., Cole, J.C., Kessler, M., Luo, J., Motherwell, W.D.S., Purkis, L.H.et al. (2004) Retrieval of crystallographically-derived molecular geometry information. J. Chem. Inf. Comput. Sci. 44, 2133–2144 10.1021/ci049780b [DOI] [PubMed] [Google Scholar]
- 42.Bruno, I.J., Cole, J.C., Edgington, P.R., Kessler, M., Macrae, C.F., McCabe, P.et al. (2002) New software for searching the Cambridge Structural Database and visualizing crystal structures. Acta Crystallogr. B 58, 389–397 10.1107/S0108768102003324 [DOI] [PubMed] [Google Scholar]
- 43.Croll, T. (2018) ISOLDE: a physically realistic environment formodel building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 74, 519–530 10.1107/S2059798318002425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim, D.N. and Sanbonmatsu, K. (2019) CryoFIT: user-friendly fitting of high-resolution cryo-EM reconstructions in PHENIX. Abstracts of Papers of the American Chemical Society 257
- 45.Barad, B.A., Echols, N., Wang, R.Y.-R., Cheng, Y., Dimaio, F., Adams, P.D.et al. (2015) EMRinger: side chain directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 10.1038/nmeth.3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Picorel, R., Bélanger, G. and Gingras, G. (1983) Antenna holochrome B880 of Rhodospirillum rubrum SI. Pigment, phospholipid, and polypeptide composition. Biochemistry 22, 2491–2497 10.1021/bi00279a028 [DOI] [Google Scholar]
- 47.Olsen, J.D., Adams, P.G. and Hunter, C.N. (2014) Aberrant assembly intermediates of the RC-LH1-PufX core complex of Rhodobacter sphaeroides imaged by atomic force microscopy. J. Biol. Chem. 289, 29927–29936 10.1074/jbc.M114.596585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olsen, J.D., Sockalingum, G.D., Robert, B. and Hunter, C.N. (1994) Modification of a hydrogen bond to a bacteriochlorophyll a molecule in the light-harvesting 1 antenna of Rhodobacter sphaeroides. Proc. Natl Acad. Sci. U.S.A. 91, 7124–7128 10.1073/pnas.91.15.7124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sturgis, J., Olsen, C.N., Robert, B. and Hunter, C.N. (1997) The functions of conserved tryptophan residues of the core light harvesting complex of Rhodobacter sphaeroides. Biochemistry 36, 2772–2778 10.1021/bi962524a [DOI] [PubMed] [Google Scholar]
- 50.Lang, H.P., Cogdell, R.J., Takaichi, S. and Hunter, C.N. (1995) Complete DNA sequence, specific Tn5 insertion map, and gene assignment of the carotenoid biosynthesis pathway of Rhodobacter sphaeroides. J. Bacteriol. 177, 2064–2073 10.1128/jb.177.8.2064-2073.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chi, S.C., Mothersole, D.J., Dilbeck, P., Niedzwiedzki, D.M., Zhang, H., Qian, P.et al. (2015) Assembly of functional photosystem complexes in Rhodobacter sphaeroides incorporating carotenoids from the spirilloxanthin pathway. Biochim. Biophys. Acta 1847, 189–201 10.1016/j.bbabio.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen, J.-H., Wu, H., Xu, C., Liu, X.-C., Huang, Z., Chang, S.et al. (2020) Architecture of the photosynthetic complex from a green sulfur bacterium. Science 370, eabb6350 10.1126/science.abb6350 [DOI] [PubMed] [Google Scholar]
- 53.Gisriel, C.J., Azai, C. and Cardona, T. (2021) Recent advances in the structural diversity of reaction centers. Photosynth. Res. 10.1007/s11120-021-00857-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Şener, M.K., Strümpfer, J., Hsin, J., Chandler, D., Scheuring, S., Hunter, C.N.et al. (2011) Förster energy transfer theory as reflected in the structures of photosynthetic light harvesting systems. ChemPhysChem 12, 518–531 10.1002/cphc.201000944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gall, A., Sturgis, J.D., Fowler, G.J.S., Hunter, C.N. and Robert, B. (1997) The influence of the protein binding site on the absorption properties of the monomeric bacteriochlorophyll in Rhodobacter sphaeroides LH2 complex. Biochemistry 36, 16282–16287 10.1021/bi9717237 [DOI] [PubMed] [Google Scholar]
- 56.Swainsbury, D.J.K., Faries, K.M., Niedzwiedzki, D., Martin, E.C., Flinders, A.J., Canniffe, D.P.et al. (2019) Engineering of B800 bacteriochlorophyll binding site specificity in Rhodobacter sphaeroides LH2. Biochim. Biophys. Acta Bioenergetics 1860, 209–223 10.1016/j.bbabio.2018.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walz, T. and Ghosh, R. (1997) Two-dimensional crystallization of the light-harvesting I–reaction centre photounit from Rhodospirillum rubrum. J. Mol. Biol. 265, 107–111 10.1006/jmbi.1996.0714 [DOI] [PubMed] [Google Scholar]
- 58.Pettersen, E.F., Goddard, T.D., Huang, C.C., Meng, E.C., Couch, G.S., Croll, T.I.et al. (2021) UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 10.1002/pro.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density map has been deposited in the World Wide Protein Data Bank (wwPDB) under accession code EMD-13110 and the coordinates have been deposited in the Protein Data Bank (PDB) under accession number 7OY8.