Abstract
Transferases have emerged as among the best catalysts for enzyme-mediated bioorthogonal functional group installation to advance innovative in vitro, cell-based and in vivo chemical biology applications. This review introduces the key considerations for selecting enzyme catalysts and chemoselective reactions most amenable to bioorthogonal platform development and highlights relevant key technology development and applications for one ubiquitous transferase subclass – methyltransferases (MTs). Within this context, recent advances in MT-enabled bioorthogonal labeling/conjugation relevant to DNA, RNA, protein, and natural products (i.e., complex small molecule metabolites) are highlighted.
Keywords: transferase; S-adenosyl-L-methionine (AdoMet, SAM); glycosyltransferase; natural product
INTRODUCTION
A range of innovative chemoselective bioorthogonal/biocompatible conjugation strategies have been developed to enable in situ conjugation of reporters and affinity ligands for molecular tracking and mechanistic studies in cells and even in live animals [1–3] (Figure 1). Ideally, the chemoselective functional groups employed in such applications must: afford exquisite reactivity/efficiency under physiological conditions; provide notable selectivity/compatibility within the context of living cells and in vivo; lead to conjugated products that are metabolically stable and non-toxic; and not infringe on the native biology to be studied. While, many bioorthogonal/biocompatible conjugation strategies rely on metabolic incorporation of non-native biomolecules synthetically-modified to display a chemoselective group, the use of enzyme-mediated strategies to install the requisite chemoselective functional groups is on the rise [3–5]. This review briefly introduces the catalysts and chemistries most amenable to enzyme-mediated chemoselective functional group installation and highlights recent key technology development and applications of methyltransferase-mediated bioorthogonal/biocompatible conjugation.
Figure 1. Overview of bioorthogonal strategies used in chemical biology research.
The center of the figure reflects key steps in native biological systems. Specifically, in the biomolecular assembly step metabolic building blocks (e.g., nucleotides, amino acids, acetate, isoprenes, etc.; represented by grey cylinders) are commonly used to generate functional biopolymers (e.g., RNA, DNA, proteins, lipids, etc.). In the ligand binding step, certain biopolymers (e.g., proteins, RNA or DNA complexes) are able to selectively bind ligands (e.g., hormones, small molecules, metabolies, drugs, etc.; highlighted in light blue). The upper panel (yellow) reflects an application of a ligand analogue (light blue) bearing a bioorthogonal chemoselective ‘tag’ (purple puzzle shape). This ligand can be tracked in situ via a chemoselective bioorthogonal reaction with a partner reagent (dark blue puzzle piece) commonly appended with a fluorescent reporter or affinity capture ligand (yellow burst). The lower panel (green) reflects an application of a metabolite building block analogue bearing a bioorthogonal chemoselective functional group (purple puzzle shape). This building block can be tracked in situ via a similar chemoselective labeling strategy using a partner reagent (dark blue puzzle piece) commonly appended with a fluorescent reporter or affinity capture ligand (yellow burst). Transferases are able to introduce bioorthogonal chemoselective functional groups (purple puzzle shape) at many different stages including: modification of the ligand prior to (a) or after (b) binding to it’s molecular target as well as modification of metabolic building blocks (c) or biopolymers prior to (d) or after (e) ligand binding. Transferase reactions (grey panels) typically utilize a co-substrate comprised of the transfer group (e.g., alkyl, glycosyl, acyl, etc.) and ‘activating group’ (e.g., S-adenosylhomocysteine/AdoHcy or SAH, nucleotide diphosphate/NDP, coenzyme A/CoA, etc.; represented by the green cube).
DESIRED CATALYST PROPERTIES: THE CASE FOR METHYLTRANSFERASES
There are three key considerations to selecting a catalyst for chemoselective functional group installation – substrate scope, catalyst efficiency, and catalyst abundance. Specifically, an ideal catalyst requires a uniquely balanced substrate scope that encompasses target (probe or biomolecule) selectivity with permissivity toward non-native chemoselective group installation. For intracellular applications, such non-native substrates must also be membrane permeable or transported via active/passive transport mechanisms. Catalytic efficiency is an essential requirement to enable subsequent rapid reporter conjugation and detection. For in situ applications catalytic turnover ultimately contributes to both probe/assay sensitivity and temporal resolution. Finally, catalyst abundance refers to natural/engineered distribution (cellular, tissue and organism) and abundance (protein levels), all of which ultimately influence the range of potential applications. Considering these factors, transferases (Enzyme Commission class 2; EC2) that catalyze macromolecule/metabolite ‘tailoring’ reactions have typically been favored for chemoselective functional group installation, catalyst development and applications.
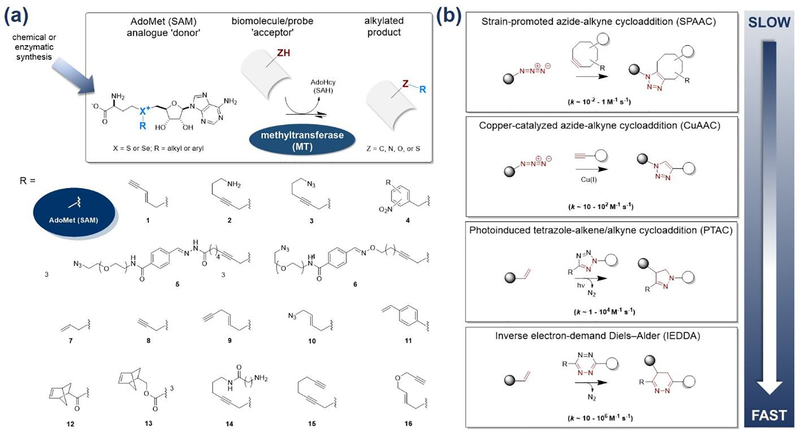
Of EC2 transferases, methyltransferases (MTs) are currently the catalysts most favored for chemoselective functional group installation. MTs catalyze the transfer of a methyl group from S-adenosyl-L-methionine (SAM or AdoMet; the ‘donor’) to a substrate nucleophile (carbon, nitrogen, oxygen or sulfur; the ‘acceptor’) [6–8] (Figure 2a). The substrate scope of MTs is exceptionally broad and includes macromolecular substrates (DNA, RNA, and proteins) and small molecules (primary and secondary metabolites). MTs are also able to use non-native S/Se -alkyl-substituted AdoMet donors to afford non-native alkylation, including S/Se-alkyl substituents bearing chemoselective functional groups [4,7,8]. While AdoMet analogues are unsuitable for cell-based studies due to poor uptake and chemical stability, these reagents can be generated in cells from corresponding non-native methionine analogues and ATP via methionine adenosyltransferases (MAT) [9–12]. Alternative enzyme-based approaches for non-native AdoMet analogue synthesis [13,14] and new chemically-stable AdoMet isosteric substrates [15] have also been recently reported. MT natural abundance and distribution is high and MTs are critical to all walks of life [4,6–8]. Thus, MT-based platforms are anticipated to offer inroads to rich biology and support a vast array of impactful applications. MT catalytic efficiency is the perhaps the greatest liability in the context of chemoselective applications. For example, the catalytic efficiencies for MTs discussed in this review range from ~10 to 7,200 M−1 s−1 and the corresponding range for AdoMet-producing enzymes is ~145 – 340 M−1 s−1.
Figure 2.
(a) The native function of a methyltransferase (MT) is to transfer a methyl group from an S-activated AdoMet donor to an acceptor C-, N-, O- or S-nucleophile (upper panel). As described in this review, MTs are capable of also using non-native S- or Se-activated alkyl or aryl (light blue) AdoMet analogues and thereby catalyze differential alkylation of target biomolecules. The lower panel highlights the structure of the S/Se-substituent for representative examples highlighted in this review. (b) Chemoselective reactions that fit criteria for transferase-mediated platforms.
CHEMOSELECTIVE REACTION PRIORITIZATION
Chemoselective reaction efficiency and selectivity are a cornerstone of effective bioorthogonal conjugation platforms [1–3]. Specifically, to be effective, such reactions must proceed at high reaction rates (efficiency) in cellular environments (selectivity). The molecular properties (e.g, size, stability, polarity/charge and/or hydrophobicity/hydrophilicity) of the selected non-native chemoselective functional groups are also key to selecting chemistries for transferase-mediated platforms. Specifically, these features influence an enzyme’s substrate recognition, turnover and lifetime must align with the selected transferase’s substrate scope. Based on these key parameters, the following chemoselective reactions are considered most amenable to transferase-mediated strategies (Figure 2b).
Modified Huisgen 1,3-dipolar cycloaddition reactions (CuAAC and SPAAC) [16].
Poor reaction rates limited the synthetic utility of Huisgen 1,3-dipolar cycloaddition prior to the advent of the copper-catalyzed azide-alkyne cycloaddition (CuAAC) reaction by Sharpless and co-workers (often referred as the first conceptual example of ‘click’ chemistry) [17]. Despite favorable reaction rates (10–100 M−1 s−1), chemoselectivity and stability of reaction products, the metal dependence of the CuACC reaction limited use in living cells and tissues. To circumvent this liability, Bertozzi and co-workers advanced copper-free strain-promoted azide-alkyne cycloaddition (SPAAC) reactions driven by the ring strain of cyclooctyne reactants such difluorooctyne (DIFO, 0.08 M−1 s−1) [18]. Alternative constrained reactants and electronic activation strategies led to SPAAC reaction rate improvements of nearly two orders in magnitude [19,20].
Photoinduced tetrazole-alkene/alkyne cycloaddition reactions (PTAC).
This ‘photoclick’ reaction, first reported by Lin and co-workers, was also inspired by the work of Huisgen [21]. Photo-induced tetrazole cycloreversion to generate short-lived reactive nitrile imine intermediates serves the basis for this reaction. These intermediates rapidly (>50 M−1 s−1) undergo 1,3-dipolar cycloaddition reactions with alkynes or alkenes to give fluorescent cyclic pyrazolines. Conceptual advances include optimizing ring strain and electronics, leading to rate improvements of nearly two orders in magnitude [22,23]. While this method offers exceptional utility for spatial and temporal control in cell/tissue-based applications, the dependence on light may limit in vivo applications.
Inverse electron-demand Diels–Alder reactions (IEDDA).
Unlike a classical Diels–Alder reaction in which an electron-rich diene reacts with an electron-poor dienophile, IEDDA reaction exploit an electron-rich dienophile (alkenes/alkynes) and an electron-poor diene (typically tetrazines) [24]. Early IEDDA proof of concept was reported by Fox and co-workers using trans-cyclooctene and tetrazine (>103 M−1 s−1). Continued development has focused on tuning reactivity via electronic and steric perturbation to afford rate improvements of over two orders in magnitude [24,25]. Photoinduction (photo-IEDDA) and orthogonality, both among exclusive IEDDA reactions and between IEDDA reactions and other chemoselective reactions (e.g., SPAAC and CuAAC), have also been reported [26,27].
MT-ENABLED BIOORTHOGONAL APPLICATIONS
Initial work in the field was based on MT-mediated single turnover reactions using aziridine-based AdoMet analogues to give fused AdoMet analogue-acceptor adducts [28,29]. Weinhold and Klimasăuskas were first to demonstrate MT-catalyzed transfer of non-native groups from catalytically-competent AdoMet analogues [30] which set the stage for what they subsequently described as the MT-directed “Transfer of Activated Groups” (mTAG) platform for chemoselective conjugation. This section is limited to recent applications of MTs in combination with catalytically-competent AdoMet analogues bearing non-native S/Se-alkyl groups with chemoselective functionality.
Representative recent DNA applications [31].
Building on the mTAG precedent, Neely and collaborators reported DNA adenine-N6 MT M.TaqI-mediated DNA CuAAC using 1 (Figure 2a) for DNA mapping [32] and a subsequent comparative study of M. TaqI-mediated N-hydroxysuccinimidyl (NHS) ester amide coupling or CuAAC/SPAAC using amine 2 or azide 3, respectively [33]. These studies revealed SPAAC to outperform CuAAC and amide coupling and also noted DNA decomposition/damage in the CuAAC reactions. Weinhold and co-workers also recently reported the use of a similar M. TaqI-driven strategy with 3 and SPAAC to introduce fluorophores and affinity labels as probes to study DNA origami nanostructure folding [34]. Innovative new MT-enabled methods for DNA photocaging open the door to ‘reversible’ selective modification of DNA as exemplified by the M.TaqI-catalyzed introduction of photocleavable protecting groups (Figure 2a; 4) [35,36]. This work demonstrated a feasible platform for photoregulation using simple in vitro transcription/translation systems where subtle changes in the photoprotecting group substitution pattern led to modulation of both MT turnover and/or photoreaction efficiency. Rentmeister and co-workers recently extended this platform via the development of new MATs (Cryptosporidium hominus MAT and an engineered Methanocaldococcus jannaschii thermostable MAT variant) to produce photocaged AdoMet analogues in situ [12]. Neely, Fernandez-Trillo and team also recently put forth tools for an innovative in vitro DNA ‘write, remove, rewrite’ approach [37]. Specifically, they demonstrated M.TaqI-mediated and the cytosine-C5 MT M.MpeI-catalyzed installation of a bifunctional DNA tag comprised of a hydrazone-linked 5 or oxime-linked terminal azide 6. This modification allowed for SPAAC-mediated conjugation and, in the case of the hydrazone linker, selective hydrolysis in the presence of NH2OH and chemoselective reinstallation of modifiers via the exposed hydrazide.
Representative recent RNA applications [31,38].
Rentmeister and colleagues have reported a range of mTAG RNA-based applications in recent years using three fundamental MT model systems – variants of the 5’-cap mRNA adenine-N2 MT trimethylguanosine synthase2 (GlaTgs2) [39–41,44], 5’-cap mRNA gaunine-N7 MT Ecm1 [42–44] and mRNA adenine-N6 MTs and METTL3–14 and METTL16 [45,46]. Chemoselective reactions employed in these studies include CuAAC [41,45,46], SPAAC [40–44,46], PTAC and IEDDA [39,43,44] using non-native AdoMet analogues (Figure 2a; 1, 7-13). Many of these studies evaluated the impact of mRNA non-native alkylation on RNA processing (primarily reverse transcription) and corresponding MT-catalyzed reactions were conducted in increasingly complex reaction environments (in vitro [41,44,46], cell lysates [39,40,43] and live cells [45]). Four specific recent advances relating to MT-mediated RNA applications are particularly noteworthy. First, reminiscent to previously reported MT-enabled DNA photocaging (Figure 2a; 4), conceptually similar RNA studies were recently reported [46]. Rentmeister and co-workers also demonstrated proof of concept of novel norbornene-based AdoMet co-substrates (e.g., 12,13) for RNA MTs to facilitate downstream IEDDA conjugation reactions [43]. Third, this same team published an innovative study that revealed the ability to use adenine-N2 MT (GlaTgs2 variant) and gaunine-N7 MT (Ecm1) in tandem, two distinct non-native AdoMet co-substrates 10 and 11, and subsequent SPAAC and IEDDA to afford selective differential mRNA labeling and FRET [44]. Finally, these researchers also reported a cell-based platform to identify METTL3-METTL14 target sites where the resulting termination of reverse transcription and bioconjugation method faciltiated RNA fragment capture and next generation sequencing [45]. Klimašauskas, Vilkaitis and colleagues recently extended these tools to the small-RNA 2’-O-MT HEN1 from Arabidopsis thaliana, a duplex-driven MT [47]. Specifically, this work employed amines 2 and 14 (NHS ester amide coupling), azide 3 (CuAAC) and alkyne 15 (CuAAC) as tools to map the sequence/context specificity of HEN1 in vitro.
Representative recent protein applications.
A range of diverse strategies have been developed for bioorthgonal chemoselective protein modification [48]. Within this context, the Weinhold group were the first to demonstrate MT-mediated chemoselective protein modification via CuAAC using the Neurospora crassa histone 3 lysine 9 N-MT (H3K9) Dim-5 and 1 (Figure 2a) [49]. Luo and co-workers were among the first to advance similar tools for proteomics using oncogenic H3K9 N-MTs EuHMT1 (GLP1) and EuHMT2 (G9a) as models. Coined ‘bioorthogonal profiling of protein methylation’ (BPPM), their proof of concept utilized EuHMT1/2 variants engineered for improved turnover with 10 followed by SPAAC to identify non-histone EuHMT1/2 substrates in cell lysates [50]. The same group used a similar strategy to identify substrates for protein arginine N-MT PRMT3 using a PRMT3 variant (M233G) and 16 [51]. This team extended the concept to living cells via the inclusion of a MAT (I117A) and H3K9 N-MT (EuHMT1-Y1211A or EuHMT2-Y1154A) engineered to favor 1 followed by CuAAC-based labeling. Referred to as ‘clickable chromatin enrichment with parallel DNA sequencing’ (CliEn-seq) [52], this proof of concept study highlighted MAT-catalyzed intracellular production of 1 from cell-permeable methionine analogues, in situ chromatin modification by engineered EuHMT1/2 and subsequent enrichment of uniquely modified chromatins via CuAAC-enabled capture for sequencing. In vitro functional annotation of two putative lysine N-MTs METTL21A and METTL10 employed similar tactics. Specifically, the use of 8 in cell lysates followed by CuAAC confirmed METTL21A to function as a histone N-MT and revealed METTL10 to EF1A1 lysine 318 [53]. Zumbusch and colleagues recently reported an alternative method for conceptually similar cell-based proteomics [54]. In this study, HEK293T and HeLa S3 cells engineered to produce enhanced green fluorescent protein (eGFP)-fusions with protein targets of interest (e.g., p53, Akt1, GAPDH, histones H2B, H3, H4; microtubule-associated protein RP/EB family member EB1; Foxo1; and heat shock proteins HSPA1 and HSPA8; valosin-containing protein VCP; and nucleolin) were electroporated with 8. Subsequent CuAAC-mediated fluorophore conjugation enabled intracellular fluorescence lifetime imaging (FLIM) FRET of each target protein’s alkylation state and localization.
Representative recent natural product and small molecule applications (Figure 3).
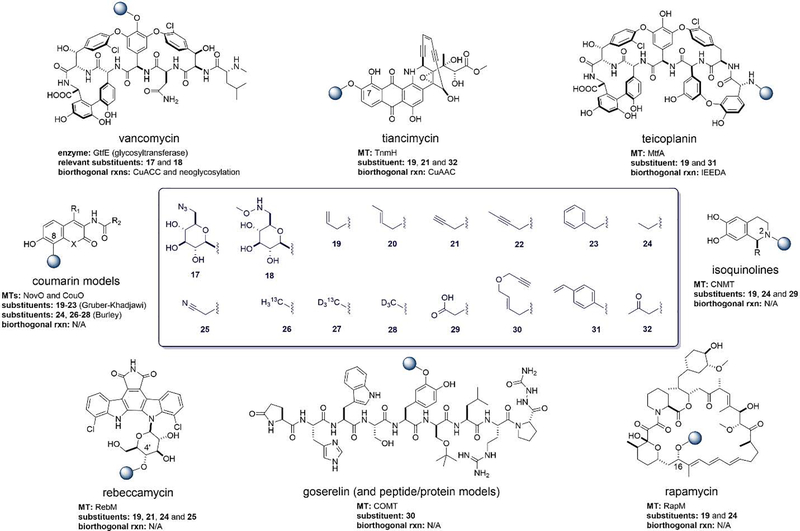
Figure 3.
Natural products for which transferase-enable bioorthogonal chemoselective modification and/or MT-enable non-native alkylation has been reported. The regiochemistry of non-native modification is highlight by the blue ball with corresponding non-native substituents illustrated in the center box.
Transferase-mediated bioactive natural product (NP) chemoselective modification was first demonstrated using the vancomycin glycosyltransferase GtfE and non-native sugars bearing azides 17 and alkoxyamines 18 to afford CuAAC [55] or alkoxyamine-based ‘neoglycosylation’ [56], respectively. Proof-of concept MT-catalyzed non-native NP alkylation was first established by Gruber-Khadjawi and colleagues using the novobiocin and coumermycin 8-C-MTs NovO and CouO [57]. Using synthetic non-native AdoMet analogues 19–23 and simple coumarin models, this study demonstrated NovO/CouO-catalyzed five non-native 8-C-alkyl groups and also highlighted some permissivity toward alternative acceptors. To circumvent limitations associated with synthetic AdoMets within this context, Singh, Thorson and colleagues were the first to develop MAT-MT coupled systems for natural product ‘alkylrandomization’ using the indolocarbazole rebeccamycin sugar 4’-O-MT RebM as a model [9]. This work highlighted the survey of five diverse MATs for non-native AdoMet production, use of hMAT2 to produce AdoMet analogues bearing 18 non-native S-substitutions and the application of coupled hMAT2-RebM reactions to generate 4 rebeccamycin analogues bearing 4’-O non-native alkyl groups (Figure 3; Reb-19, 21, 24 and 25). Similar NP biosynthetic MT alkyl permissivity and corresponding coupled multi-enzyme platforms have since been reported for a range of NPs including: rapamycin (16-O-MT RapM: coupled hMAT-RapM system to produce 16-O-alkyl rapalogs Rap-19 and 24 [58]); coumarins (novobiocin 8-C-MT NovO: coupled SalL-NovO system to produce model coumarins including 8-C-isotopically-labeled coumarin-26–28 derivatives [59,60]) and alkaloids/phenolics (coclaurine N-MT CNMT: coupled hMAT-CNMT system to produce 2-N-alkyl isoquinolone-19 and isoquinolone-24 [61]; carboxy-S-adenosyl-L-methionine synthase in conjunction with CNMT or catechol-O-methyltransferase COMT to produce N- and O-carboxymethylated isoquinoline-29 molecules [62]). Micklefield and colleagues also introduced an innovative coupled fungal tyrosinase-mammalian catechol-O-MT (COMT) system for selective peptide O-alkylation with non-native alkyl groups (Figure 3; peptide-30) [63]. Brieke and collaborators reported the first proof of concept for MT-mediated chemoselective conjugation. This study employed two glycopeptide α-N-MTs (A40926 MtfAdbv and pekiskomycin MtfApek) and led to the production of teicoplanin aglycon-19 and teicoplanin aglycon-31 (Figure 3), the latter of which was successfully used in IEDDA conjugation reactions to tether an affinity ligand or fluorophore [64]. Enediyne MT-mediated chemoselective conjugation was also recently demonstrated using the permissive tiancimycin 7-O-MT TnmH [65]. This study highlighted TnmH to turnover a range of anthraquinone acceptors and, in the presence of non-native AdoMet donors (19,21, and 32), catalyze production of tiancimycin 7-O-alkyl analogues. The corresponding tiancimycin-21 (Figure 3) was subsequently used in CuAAC reactions to introduce tethers for putative antibody-conjugation. This work set the stage for future production of tiancimycin-antibody conjugates and subsequent preclinical evaluation studies.
CONCLUSIONS AND PROSPECTUS
This review points to notable advances in bioorthogonal chemoselective reaction and enzyme reagent (enzymes and non-native substrates) development to support emerging transferase-mediated bioorthogonal platforms. While limited proof of concept examples for cell-based applications of MT-enabled bioorthogonal labeling/conjugation exist, three key barriers to broader use remain. First, unlike many of the commercially available kits/reagents for bioorthogonal chemoselective conjugation, the reagents (enzymes and non-native substrates) for MT-enabled platforms are highly specialized and largely inaccessible to the broader research community. Improved access to non-native AdoMet analogues via commercial sources and/or user-friendly production methods is expected to present new opportunities and unlock new discoveries. Second, the stability, PK, biodistribution and cellular uptake of non-native AdoMet analogues remain substantive barriers to cell-based and in vivo applications. Strategies to stabilize and/or ‘deliver’ AdoMet analogues as well as improved methods for in situ (cell and/or tissue-specific) production are anticipated to help circumvent these roadblocks. Finally, MT reaction rates and bioorthogonality (i.e., selectivity for non-native substrates) fall far short of the corresponding rates or selectivities of the best bioorthogonal chemoselective reactions. The ongoing discovery and evolution/engineering of improved catalysts is expected to continue to narrow this gap. Cumulatively, such platform improvements are expected to usher in broader application and impact including, but not limited to, lead discovery or development efforts.
ACKNOWLEDGEMENTS
We gratefully acknowledge the support of R37 AI52218, R01 GM115261, the Center of Biomedical Research Excellence (COBRE) in Pharmaceutical Research and Innovation (CPRI, NIH P20 GM130456), the University of Kentucky College of Pharmacy and the National Center for Advancing Translational Sciences (UL1TR000117 and UL1TR001998).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare the following competing financial interest: J.S.T. is a co-founder of Centrose (Madison, WI, USA).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1]. Nguyen SS, Prescher JA: Developing bioorthogonal probes to span a spectrum of reactivities. Nat Rev Chem 2020, 4:476–489. ● This is an excellent comprehensive review of bioorthogonal reaction development and applications.
- [2]. Smeenk MLWJ, Agramunt J, Bonger KM: Recent developments in bioorthogonal chemistry and the orthogonality within. Curr Opin Chem Biol 2020, 60:79–88. ● This is a terrific brief, timely overview of concepts and reactions relating to bioorthogonality.
- [3].Porte K, Riberaud M, Châtre R, Audisio D, Papot S, Taran F: Bioorthogonal reactions in animals. Chembiochem 2021, 22:100–113. [DOI] [PubMed] [Google Scholar]
- [4].Tomkuvienė M, Mickutė M, Vilkaitis G, Klimašauskas S: Repurposing enzymatic transferase reactions for targeted labeling and analysis of DNA and RNA. Curr Opin Biotechnol 2019, 55:114–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Romero E, Jones BS, Hogg BN, Rué Casamajo A, Hayes MA, Flitsch SL, Turner NJ, Schnepel C: Enzymatic late-stage modifications: Better late than never. Angew Chem Int Ed Engl 2021, doi: 10.1002/anie.202014931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sun Q, Huang M, Wei Y: Diversity of the reaction mechanisms of SAM-dependent enzymes. Acta Pharm Sin B 2020, doi: 10.1016/j.apsb.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Huber TD, Johnson BR, Zhang J, Thorson JS: AdoMet analog synthesis and utilization: Current state of the art. Curr Opin Biotechnol 2016, 42:189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bennett MR, Shepherd SA, Cronin VA, Micklefield J: Recent advances in methyltransferase biocatalysis. Curr Opin Chem Biol 2017, 37:97–106. [DOI] [PubMed] [Google Scholar]
- [9]. Singh S, Zhang J, Huber TD, Sunkara M, Hurley K, Goff RD, Wang G, Zhang W, Liu C, Rohr J, et al. : Facile chemoenzymatic strategies for the synthesis and utilization of S-adenosyl-(L)-methionine analogues. Angew Chem Int Ed Engl 2014, 53:3965–3969. ● This is the first study to describe a coupled MAT-MT-mediated approach to differential alkylation of a natural product. In this work, five diverse MATs were assessed for non-native AdoMet production, hMAT2 production of AdoMet analogues bearing 18 non-native S-substitutions was demonstrated and proof of concept for coupled hMAT2-RebM-catalyzed reactions to produce novel indolocarbazoles (rebeccamycins) was highlighted.
- [10].Bothwell IR, Luo M: Large-scale, protection-free synthesis of Se-adenosyl-l-selenomethionine analogues and their application as cofactor surrogates of methyltransferases. Org Lett 2014, 16:3056–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Huber TD, Clinger JA, Liu Y, Xu W, Miller MD, Phillips GN Jr, Thorson JS: Methionine adenosyltransferase engineering to enable bioorthogonal platforms for AdoMet-utilizing enzymes. ACS Chem Biol 2020, 15:695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Michailidou F, Klöcker N, Cornelissen NV, Singh RK, Peters A, Ovcharenko A, Kümmel D, Rentmeister A: Engineered SAM synthetases for enzymatic generation of AdoMet analogs with photocaging groups and reversible DNA modification in cascade reactions. Angew Chem Int Ed Engl 2021, 60:480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Davis TD, Kunakom S, Burkart MD, Eustaquio AS: Preparation, assay, and application of chlorinase SalL for the chemoenzymatic synthesis of S-adenosyl-L-methionine and analogs. Methods Enzymol 2018, 604:367–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Tang Q, Grathwol CW, Aslan-Üzel AS, Wu S, Link A, Pavlidis IV, Badenhorst CPS, Bornscheuer UT: Directed evolution of a halide methyltransferase enables biocatalytic synthesis of diverse SAM analogs. Angew Chem Int Ed Engl 2021, 60:1524–1527. ● The authors describe the application of an iodide-based assay for the directed evolution of a halide MT (HMT) from Arabidopsis thaliana for improved production of S-alkyl AdoMet analogues. Single vessel HMT-MT coupled systems for alkylation of small molecule models (luteolin and 3,4-dihydroxybenzaldehyde) was also demonstrated.
- [15].Huber TD, Wang F, Singh S, Johnson BR, Zhang J, Sunkara M, Van Lanen SG, Morris AJ, Phillips GN Jr, Thorson JS: Functional AdoMet isosteres resistant to classical AdoMet degradation pathways. ACS Chem Biol 2016, 11:2484–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Breugst M, Reissig HU: The Huisgen reaction: Milestones of the 1,3-dipolar cycloaddition. Angew Chem Int Ed Engl 2020, 59:12293–12307. ● This article provides a comprehensive overview of important advances relevant to Huisgen 1,3-dipolar cycloaddition-inspired bioorthogonal reaction development and synthetic applications.
- [17].Neumann S, Biewend M, Rana S, Binder WH: The CuAAC: Principles, homogeneous and heterogeneous catalysts, and novel developments and applications. Macromol Rapid Commun 2020, 41:e1900359. [DOI] [PubMed] [Google Scholar]
- [18].Dommerholt J, Rutjes FPJT, van Delft FL: Strain-promoted 1,3-dipolar cycloaddition of cycloalkynes and organic azides. Top Curr Chem (Cham) 2016, 374:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Svatunek D, Houszka N, Hamlin TA, Bickelhaupt FM, Mikula H: Chemoselectivity of tertiary azides in strain-promoted alkyne-azide cycloadditions. Chemistry 2019, 25:754–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Burke EG, Gold B, Hoang TT, Raines RT, Schomaker JM: Fine-tuning strain and electronic activation of strain-promoted 1,3-dipolar cycloadditions with endocyclic sulfamates in SNO-OCTs. J Am Chem Soc 2017, 139:8029–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Kumar GS, Lin Q: Light-triggered click chemistry. Chem Rev 2020, doi: 10.1021/acs.chemrev.0c00799. ● This review presents a recent comprehensive perspective on light-triggered chemistries including, but not limited to, the photoinduced tetrazole-alkene/alkyne cycloaddition reactions.
- [22].Jiang S, Wu X, Liu H, Deng J, Zhang X, Yao Z, Zheng Y, Li B, Yu Z: Ring-strain-promoted ultrafast diaryltetrazole–alkyne photoclick reactions triggered by visible light. ChemPhotoChem 2020, 4:327–331 [Google Scholar]
- [23].Li J, Kong H, Zhu C, Zhang Y: Photo-controllable bioorthogonal chemistry for spatiotemporal control of bio-targets in living systems. Chem Sci 2020, 11:3390–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Oliveira BL, Guo Z, Bernardes GJL: Inverse electron demand Diels-Alder reactions in chemical biology. Chem Soc Rev 2017, 46:4895–4950. ● This article provides a comprehensive review on IEDDA development and the available toolbox reagents.
- [25].Wu H, Devaraj NK: Advances in tetrazine bioorthogonal chemistry driven by the synthesis of novel tetrazines and dienophiles. Acc Chem Res 2018, 51:1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Reisacher U, Ploschik D, Rönicke F, Cserép GB, Kele P, Wagenknecht HA: Copper-free dual labeling of DNA by triazines and cyclopropenes as minimal orthogonal and bioorthogonal functions. Chem Sci 2019, 10:4032–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Macias-Contreras M, He H, Little KN, Lee JP, Campbell RP, Royzen M, Zhu L: SNAP/CLIP-tags and strain-promoted azide-alkyne cycloaddition (SPAAC)/inverse electron demand Diels-Alder (IEDDA) for intracellular orthogonal/bioorthogonal labeling. Bioconjug Chem 2020, 31:1370–1381. [DOI] [PubMed] [Google Scholar]
- [28].Pljevaljcic G, Pignot M, Weinhold E: Design of a new fluorescent cofactor for DNA methyltransferases and sequence-specific labeling of DNA. J Am Chem Soc 2003, 125:3486–3492. [DOI] [PubMed] [Google Scholar]
- [29].Zhang C, Weller RL, Thorson JS, Rajski SR: Natural product diversification using a non-natural cofactor analogue of S-adenosyl-L-methionine. J Am Chem Soc 2006, 128:2760–2761. [DOI] [PubMed] [Google Scholar]
- [30]. Dalhoff C, Lukinavicius G, Klimasăuskas S, Weinhold E: Direct transfer of extended groups from synthetic cofactors by DNA methyltransferases. Nat Chem Biol 2006, 2:31–32. ●● The authors describe the pioneering discovery that non-native S-alkyl AdoMet cosubstrates could be used for DNA MTs. This article provides the conceptual foundation upon which all MT-enabled bioorthogonal strategies have been built.
- [31]. Klöcker N, Weissenboeck FP, Rentmeister A: Covalent labeling of nucleic acids. Chem Soc Rev. 2020, 49:8749–8773. ● This review presents an excellent recent comprehensive review of methods for DNA and RNA labeling including, but not limited to, MT-mediated chemoselective modification.
- [32].Vranken C, Deen J, Dirix L, Stakenborg T, Dehaen W, Leen V, Hofkens J, Neely RK: Super-resolution optical DNA Mapping via DNA methyltransferase-directed click chemistry. Nucleic Acids Res 2014, 42:e50–e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lauer MH, Vranken C, Deen J, Frederickx W, Vanderlinden W, Wand N, Leen V, Gehlen MH, Hofkens J, Neely RK: Methyltransferase-directed covalent coupling of fluorophores to DNA. Chem Sci 2017, 8:3804–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Heck C, Torchinsky D, Nifker G, Gularek F, Michaeli Y, Weinhold E, Ebenstein Y: Label as you fold: Methyltransferase-assisted functionalization of DNA nanostructures. Nanoscale 2020. 12:20287–20291. [DOI] [PubMed] [Google Scholar]
- [35]. Anhäuser L, Muttac F, Rentmeister A: Reversible modification of DNA by methyltransferase-catalyzed transfer and light-triggered removal of photo-caging groups. Chem Commun 2018, 54:449–451. ● This report highlights the first example of MT-enabled strategies to incorporate removable protecting groups on biomolecules. Specifically, the authors describe the application of a DNA MT and novel S-nitrobenyzl AdoMet analogues for DNA photocaging.
- [36].Heimes M, Kolmar L, Brieke C. Efficient cosubstrate enzyme pairs for sequence-specific methyltransferase-directed photolabile caging of DNA. Chem Commun (Camb) 2018. 54:12718–12721. [DOI] [PubMed] [Google Scholar]
- [37]. Wilkinson AA, Jagu E, Ubych K, Coulthard S, Rushton AE, Kennefick J, Su Q, Neely RK, Fernandez-Trillo P: Site-selective and rewritable labeling of DNA through enzymatic, reversible, and click chemistries. ACS Cent Sci 2020, 6:525–534. ● This paper describes the development and application of an innovative AdoMet-based reagent set to write, remove, and rewrite biomolecule modifications. Specifically, MT-mediated bifunctional DNA modifiers were comprised of a hydrazone-linked terminal azide for SPAAC reactions (write), the hydrazone-linker of which could be selectively hydrolyzed to the hydrazide (remove) and then selectively reacted with aldehydes (rewrite).
- [38]. Muthmann N, Hartstock K, Rentmeister A: Chemo-enzymatic treatment of RNA to facilitate analyses. Wiley Interdiscip Rev RNA 2020, 11:e1561. ● This review presents a terrific recent comprehensive review of methods for RNA labeling including, but not limited to, MT-mediated chemoselective modification.
- [39].Holstein JM, Stummer D, Rentmeister A: Enzymatic modification of 5′-capped RNA with a 4-vinylbenzyl group provides a platform for photoclick and inverse electron-demand Diels-Alder reaction. Chem Sci 2015, 6:1362–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Holstein JM, Schulz D, Rentmeister A: Bioorthogonal site-specific labeling of the 5′-cap structure in eukaryotic mRNAs. Chem Commun 2014, 50:4478–4481. [DOI] [PubMed] [Google Scholar]
- [41].Muttach F, Rentmeister A: A Biocatalytic Cascade for Versatile One-Pot Modification of mRNA Starting from Methionine Analogues. Angew Chemie - Int Ed 2016, 55:1917–1920. [DOI] [PubMed] [Google Scholar]
- [42].Holstein JM, Anhäuser L, Rentmeister A: Modifying the 5′-Cap for Click Reactions of Eukaryotic mRNA and To Tune Translation Efficiency in Living Cells. Angew Chemie - Int Ed 2016, 55:10899–10903. [DOI] [PubMed] [Google Scholar]
- [43]. Muttach F, Muthmann N, Reichert D, Anhäuser L, Rentmeister A: A benzylic linker promotes methyltransferase catalyzed norbornene transfer for rapid bioorthogonal tetrazine ligation. Chem Sci 2017, 8:7947–7953. ● This paper presents the first example of MT-enabled strategies to incorporate norbornene-based substitutions.
- [44]. Holstein JM, Muttach F, Schiefelbein SHH, Rentmeister A: Dual 5′ Cap Labeling Based on Regioselective RNA Methyltransferases and Bioorthogonal Reactions. Chem - A Eur J 2017, 23:6165–6173. ● This paper describes reactions that combine two MTs (an adenine-N2 MT and gaunine-N7 MT) in tandem, two distinct non-native AdoMet co-substrates, and two discrete chemoselective reactions (SPAAC and IEDDA) to afford selective differential mRNA labeling and FRET.
- [45]. Hartstock K, Nilges BS, Ovcharenko A, Cornelissen NV, Püllen N, Lawrence-Dörner AM, Leidel SA, Rentmeister A: Enzymatic or in vivo installation of propargyl groups in combination with click chemistry for the enrichment and detection of methyltransferase target sites in RNA. Angew Chem Int Ed 2018, 57:6342–6346. ● This report highlights a cell-based strategy to identify RNA MT target sites based by termination of reverse transcription due to non-native alkylation. Corresponding CuACC faciltiated RNA fragment capture and next generation sequencing.
- [46].Ovcharenko A, Weissenboeck FP, Rentmeister A: Tag-free internal RNA labeling and photocaging based on mRNA methyltransferases. Angew Chemie Int Ed 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Osipenko A, Plotnikova A, Nainytė M, Masevičius V, Klimašauskas S, Vilkaitis G: Oligonucleotide-addressed covalent 3’-terminal derivatization of small RNA strands for enrichment and visualization. Angew Chem Int Ed Engl. 2017, 56:6507–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Braun AC, Gutmann M, Lühmann T, Meinel L: Bioorthogonal strategies for site-directed decoration of biomaterials with therapeutic proteins. J Control Release 2018, 273:68–85. [DOI] [PubMed] [Google Scholar]
- [49].Peters W, Willnow S, Duisken M, Kleine H, Macherey T, Duncan KE, Litchfield DW, Lüscher B, Weinhold E: Enzymatic site-specific functionalization of protein methyltransferase substrates with alkynes for click labeling. Angew Chem Int Ed Engl 2010, 49:5170–5173. [DOI] [PubMed] [Google Scholar]
- [50]. Islam K, Bothwell I, Chen Y, Sengelaub C, Wang R, Deng H, Luo M: Bioorthogonal profiling of protein methylation using azido derivative of S-adenosyl-L-methionine. J Am Chem Soc 2012, 134:5909–5915. ● This paper presents the first example advancing MT-mediated technologies for proteomics applications. Specifically, proof of concept application of ‘Bioorthogonal Profiling of Protein Methylation’ (BPPM) technology led to the identification of the nonhistone targets of MTs EuHMT1/2.
- [51].Guo H, Wang R, Zheng W, Chen Y, Blum G, Deng H, Luo M. Profiling substrates of protein arginine N-methyltransferase 3 with S-adenosyl-L-methionine analogues. ACS Chem Biol 2014, 9:476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wang R, Islam K, Liu Y, Zheng W, Tang H, Lailler N, Blum G, Deng H, Luo M: Profiling genome-wide chromatin methylation with engineered posttranslation apparatus within living cells. J Am Chem Soc 2013, 135:1048–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shimazu T, Barjau J, Sohtome Y, Sodeoka M, Shinkai Y: Selenium-based S-adenosylmethionine analog reveals the mammalian seven-beta-strand methyltransferase METTL10 to be an EF1A1 lysine methyltransferase. PLoS One 2014, 9:105394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Doll F, Steimbach RR, Zumbusch A: Direct imaging of protein-specific methylation in mammalian cells. ChemBioChem 2019, 20:1315–1325. [DOI] [PubMed] [Google Scholar]
- [55]. Fu X, Albermann C, Jiang J, Liao J, Zhang C, Thorson JS: Antibiotic optimization via in vitro glycorandomization. Nat Biotechnol 2003, 21:1467–1469. ●● The authors describe the pioneering application of non-native sugar nucleotides for complex natural product (vancomycin) differential glycosylation (glycorandomization), including the use of sugars bearing azides and subsequent CuAAC for downstream diversification. This article provides the conceptual foundation transferase-enabled bioorthogonal strategies.
- [56].Peltier-Pain P, Marchillo K, Zhou M, Andes DR, Thorson JS: Natural product disaccharide engineering through tandem glycosyltransferase catalysis reversibility and neoglycosylation. Org Lett 2012, 14:5086–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Stecher H, Tengg M, Ueberbacher BJ, Remler P, Schwab H, Griengl H, Gruber-Khadjawi M: Biocatalytic Friedel-Crafts alkylation using non-natural cofactors. Angew Chem Int Ed Engl 2009, 48:9546–9548. ● ● This is the first study to describe MT-catalyzed differential alkylation of a natural product model and the first to describe the use of non-native non-native S-alkyl AdoMet analogues with C-MTs. This work enabled the production of a number novel C-alkyl coumarins and included the incorporation of alkyl groups with chemoselective functionality (allyl, alkynes).
- [58].Law BJC, Struck AW, Bennett MR, Wilkinson B, Micklefield J: Site-specific bioalkylation of rapamycin by the RapM 16-O-methyltransferase. Chem Sci 2015, 6:2885–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sadler JC, Humphreys LD, Snajdrova R, Burley GA: A tandem enzymatic sp2-C-methylation process: Coupling in situ S-adenosyl-L-methionine formation with methyl transfer. Chembiochem 2017, 18:992–995. [DOI] [PubMed] [Google Scholar]
- [60].McKean IJW, Sadler JC, Cuetos A, Frese A, Humphreys LD, Grogan G, Hoskisson PA, Burley GA: S-adenosyl methionine cofactor modifications enhance the biocatalytic repertoire of small molecule C-alkylation. Angew Chem Int Ed Engl 2019, 58:17583–17588. [DOI] [PubMed] [Google Scholar]
- [61].Bennett MR, Thompson ML, Shepherd SA, Dunstan MS, Herbert AJ, Smith DRM, Cronin VA, Menon BRK, Levy C, Micklefield J: Structure and biocatalytic scope of coclaurine N-methyltransferase. Angew Chem Int Ed Engl 2018, 57:10600–10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Herbert AJ, Shepherd SA, Cronin VA, Bennett MR, Sung R, Micklefield J: Engineering orthogonal methyltransferases to create alternative bioalkylation pathways. Angew Chem Int Ed Engl 2020, 59:14950–14956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Struck AW, Bennett MR, Shepherd SA, Law BJ, Zhuo Y, Wong LS, Micklefield J: An enzyme cascade for selective modification of tyrosine residues in structurally diverse peptides and proteins. J Am Chem Soc 2016. 138:3038–3045. [DOI] [PubMed] [Google Scholar]
- [64]. Brieke C, Yim G, Peschke M, Wright GD, Cryle MJ: Catalytic promiscuity of glycopeptide N-methyltransferases enables bio-orthogonal labeling of biosynthetic intermediates. Chem Commun (Camb) 2016, 52:13679–13682. ● This is the first study to describe MT-mediated chemoselective (IEDDA) modification of an NP. Chemoenzymatic synthesis of α-N-allyl and α-N-benzylvinyl teichoplanin aglycon analogues were reported, the latter of which was successfully used in proof of concept IEDDA conjugation reactions.
- [65]. Adhikari A, Teijaro CN, Yan X, Chang CY, Gui C, Liu YC, Crnovcic I, Yang D, Annaval T, Rader C, et al. : Characterization of TnmH as an O-methyltransferase revealing insights into tiancimycin biosynthesis and enabling a biocatalytic strategy to prepare antibody-tiancimycin conjugates. J Med Chem 2020, 63:8432–8441. ● TnmH in the presence of enediyne precursor and non-native AdoMet donors enabled production of tiancimycin 7-O-allyl, 7-O-propanone and 7-O-propargyl analogues, the latter of which was successfully employed in CuAAC reactions. This study is the first to describe use of MT-mediated strategies to introduce tethers applicable to the production of preclinical antibody-drug conjugate (ADC) leads.