Abstract
Cartilaginous fish, or Chondrichthyes, are the oldest extant vertebrates to possess the MHC and the Ig superfamily–based Ag receptors, the defining genes of the gnathostome adaptive immune system. In this work, we have identified a novel MHC lineage, UEA, a complex multigene nonclassical class I family found in sharks (division Selachii) but not detected in chimaeras (subclass Holocephali) or rays (division Batoidea). This new lineage is distantly related to the previously reported nonclassical class I lineage UCA, which appears to be present only in dogfish sharks (order Squaliformes). UEA lacks conservation of the nine invariant residues in the peptide (ligand)–binding regions (PBR) that bind to the N and C termini of bound peptide in most vertebrate classical class I proteins, which are replaced by relatively hydrophobic residues compared with the classical UAA. In fact, UEA and UCA proteins have the most hydrophobic-predicted PBR of all identified chondrichthyan class I molecules. UEA genes detected in the whale shark and bamboo shark genome projects are MHC linked. Consistent with UEA comprising a very large gene family, we detected weak expression in different tissues of the nurse shark via Northern blotting and RNA sequencing. UEA genes fall into three sublineages with unique characteristics in the PBR. UEA shares structural and genetic features with certain nonclassical class I genes in other vertebrates, such as the highly complex XNC nonclassical class I genes in Xenopus, and we anticipate that each shark gene, or at least each sublineage, will have a unique function, perhaps in bacterial defense.
The Chondrichthyes or cartilaginous fish (chimaeras, rays, and sharks) are the oldest living animals with the MHC-based adaptive immune system, which is present in all jawed vertebrates or gnathostomes (1, 2). Studies of cartilaginous fish MHC genes have shown them to be multifaceted, with different class I/II lineages and high polymorphism of the classical class I (UAA) and class II genes (2–7). However, this group of early-branching vertebrates also preserves several ancestral features of the immune system that were lost in some or all “higher” vertebrates, such as the cluster-type organization of Ig genes (8), close genetic linkage of MHC class I processing and presenting genes (9, 10), somatic hypermutation of TCR genes (11), and the MHC linkage of β2-microglobulin (β2m) (12).
Classical MHC class I molecules (class Ia) are ubiquitously expressed and bind peptidic Ags in the peptide-binding region (PBR), comprised of the class I α1 and α2 domains, and present the peptides to CD8-positive T cells. Class Ia molecules also have an α3 domain belonging to the C1-type of the Ig superfamily (IgSF), a connecting piece (CP), a transmembrane (TM) domain, and a cytoplasmic tail (CYT) (13, 14). The class I α-chain, made up of three domains, forms a heterodimer with the IgSF C1 member β2m. Nonclassical class I molecules (class Ib), although generally having the same basic structure as class Ia, can be distinguished from class Ia molecules by their low levels of polymorphism, tissue-specific expression, and/or lack of conservation of 9 aa residues that bind to the N and C termini of the bound peptide in the PBR (2, 15–18). Unlike class Ia molecules, class Ib molecules like CD1 and MR1 present nonpeptidic ligands to innate T cells (NKT, MAIT, and γδ cells), and others like FcRn have a “closed” PBR, bind no Ag in the groove at all, and serve other functions besides Ag presentation (19). Class Ib genes can either be MHC linked or map to other chromosomal regions [for example, see (1, 15)].
Four class I lineages have been identified so far and characterized in Chondrichthyes, namely the classical UAA (2) found in all gnathostomes and the nonclassical UBA (2), UCA (4), and UDA (5). Previous work showed that UBA is multigenic and was derived from a UAA ancestor (2, 5), whereas UDA is an ancient (as old as UAA), MHC-linked, single or low-copy lineage found in all cartilaginous fish (5). UCA was detected as a highly divergent class I gene in the spiny dogfish Squalus acanthias (4) but was not studied further.
In this study, we have identified a novel shark-specific nonclassical class I lineage, UEA, distantly related to UCA in the PBR and present in all shark species examined (division Selachii), which emerged ~350 million y ago (MYA) (20). Unlike UAA and UDA, UEA is found in a large multigene family with three distinct sublineages. In this study, we describe the fundamental characteristics of this new, complex class Ib gene family and speculate on its relationship to the previously described UCA and many other vertebrate class I lineages.
Materials and Methods
Database searches
Several sequences representative of all MHC class I lineages described in chondrichthyan taxa (sharks, rays, and chimaeras) were used as templates to search for additional MHC class I–like sequences in this taxonomic group. Searches were performed on Sequence Read Archive (SRA), Transcriptome Shotgun Assembly (TSA) and Whole Genome Sequence (WGS) databases publicly accessible on the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov, as of November 1, 2019) using the following amino acid sequences as queries: MHC class Ia UAA: nurse shark (Ginglymostoma cirratum), Gici UAA: AAF66110; clearnose skate (Raja eglanteria), Raeg UAA: AGH32767; and elephant shark (Callorhinchus milii), Cami UAA: AFP04519; MHC class Ib UBA: nurse shark, Gici UBA: AAC60347; horn shark (Heterodontus francisci), Hefr UBA: AAC60348; whale shark (Rhincodon typus), Rhty UBA: XP_020392556; MHC class Ib UCA spiny dogfish (S. acanthias), Sqac UCA: AAN78091; MHC class Ib UDA: nurse shark, Gici UDA: MN339476; little skate (Leucoraja erinacea), Leer UDA: LS-transcriptB2-ctg40765 (skatebase.org), and the elephant shark, Cami UDA: AFP03335. Some SRA datasets from RNA sequencing (RNAseq) bioprojects on Selachii taxa were excluded from the analysis when the data referred to species whose genera had multiple datasets and included nonimmunity-related tissues (e.g., retina, pectoral fin bud, or ampullary receptor cells). In the case of Batoid (rays and skates) and Holocephalan taxa (chimaeras and ratfish), all datasets were included in the analysis because of their underrepresentation in public databases. A total of 34 species covering the main chondrichthyan orders were included in the analysis (more details in Supplemental Table I). This comprised 1 species of Holocephalans, 23 species of Selachians, and 9 species of Batoids. All but one order of Batoids (sensu Ref. 21) and five out of the nine orders of Selachians (sensu Ref. 22) are represented.
Bioinformatic filtering
Depending on the database used (i.e., TSA, SRA, or WGS) (Supplemental Table I), we applied different protocols to extract MHC class I sequences. Briefly, for TSA, we downloaded the available transcripts and used blastx to search for matching sequences against the set of MHC class I queries (see information above). We only retained transcripts with blast e-values less than 0.001 and covering more than 50% of the query sequence. For SRA, raw reads were downloaded, cleaned using Trimmomatic (23), and then assembled using SPAdes (24) to obtain the final transcripts. Yielded transcripts were subjected to the same blastx procedure as described for TSA. For the WGS dataset, a direct search on the contigs for every species was performed using as the query a set of known proteins belonging to the MHC class I (the same query sequences used above) and performing a tblastn to locate the exact positions in the contigs using very restrictive parameters (e-value 10−7). After a manual refinement to avoid overlaps or redundancy, the corresponding gene was extracted from the contigs, including a flanking region of 200 nts, to ensure the whole gene sequence was included. The sequences obtained were translated to amino acids, and only those having basic features of MHC class I structure, namely the PBR (α1 and α2 domains) and the IgSF α3 domain, were further analyzed. To confirm the main characteristics of the MHC molecules, the ScanProsite tool was also used (https://prosite.expasy.org/scanprosite). We excluded from further analyses all of the nearly identical sequences (differing by 3 aa or less) to be conservative and reduce methodological errors.
Sequence alignments and phylogenetic trees
Deduced amino acid sequences were aligned using the ClustalX program in Geneious Prime v2.1 (25) with manual adjustments (representative alignment in Fig. 1; Supplemental Fig. 1, Supplemental Table II). The neighbor-joining (NJ) phylogenetic trees of the class I peptide-binding domains (α1 and α2) (Fig. 2A) and IgSF (α3 domain) (Fig. 2B) were constructed in MEGA 6.06 (26) using the p-distance model with uniform rates among sites and pairwise deletions for the gaps/missing data. Branch support was evaluated using 1000 bootstrap replicates of the data. Human, chicken, and Chinese alligator CD1 molecules were used as outgroups because they are among the most divergent vertebrate class I lineages (17). To infer the relationships of chondrichthyan MHC class I genes with those of other vertebrates, we also included some representative lineages from other vertebrates (Fig. 2A, Supplemental Table II).
FIGURE 1.
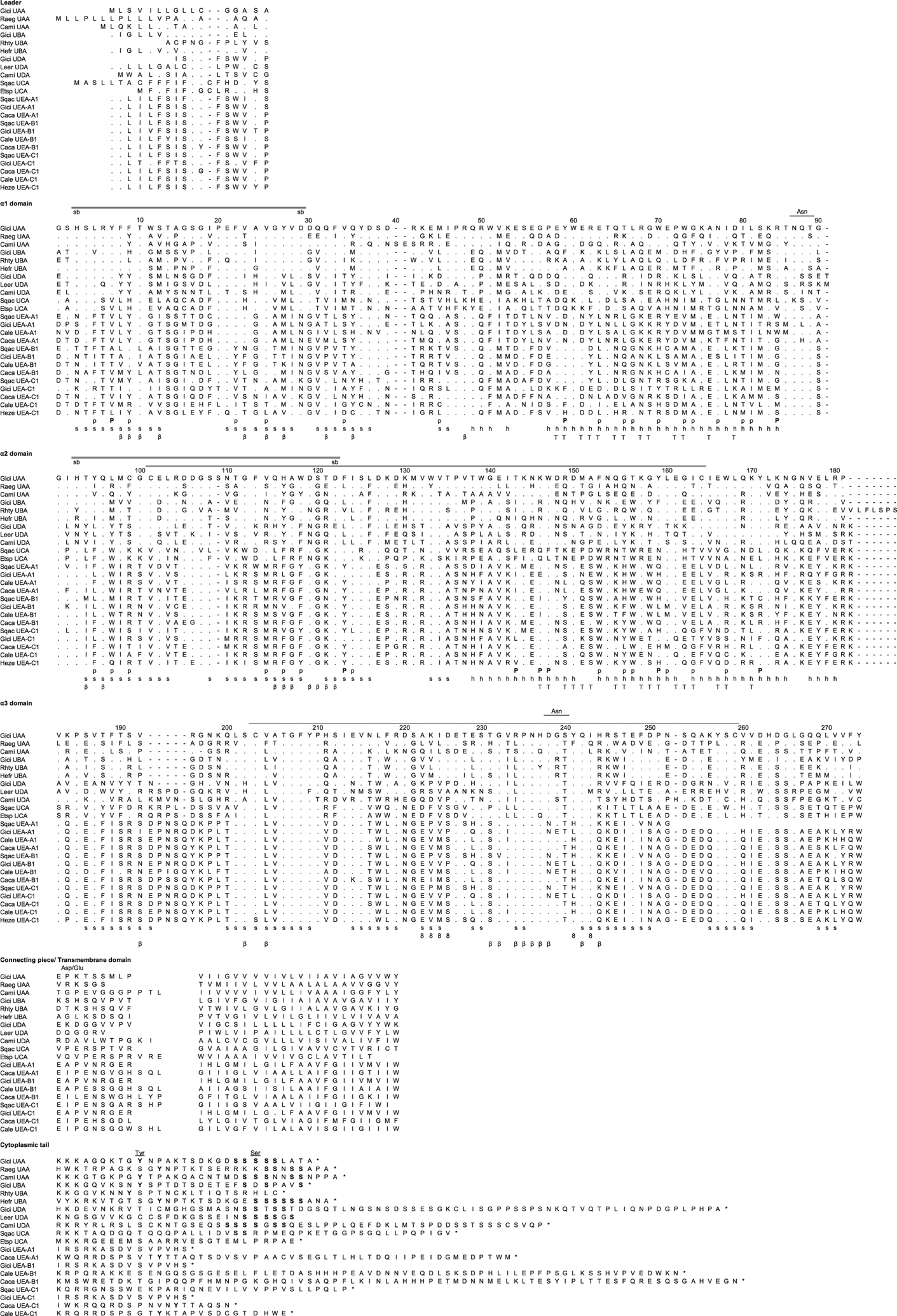
Amino acid alignment of the representative MHC class I lineages in cartilaginous fish, UAA, UBA, UCA, UDA, and the new multicopy lineage UEA. GenBank accession numbers are listed in Supplemental Table II. Dashes indicate gaps, and asterisks indicate the stop codon. s and h indicate the β strands and α helices, and the line connecting the two Cys (C) in the α2 and α3 domain indicates the class I canonical disulfide bridge. The double line between the amino acids His (H) and Asp (D) indicates the possible salt bridge, whereas eight β and T indicate the residues that are potential binding with CD8, β2m, and TCR, respectively. P marks the invariant residues that bind to the N and C termini of the bound peptide in the classical class I molecules, and p indicates the other 28 potential conserved peptide binding residues. The Asn (N) marks the asparagine-linked glycosylation site, and Asp/Glu (D/E) indicates the typical aspartic acid and glutamic acid residues found in the CP (light shade). The underlined Asn above the a3 domain in Fig. 1 denotes a potential glycosylation site for some UEA sequences. Because of the high diversity in the CYT, the alignment would be merely speculative. The numbering of amino acid positions is based on human HLA-A2.
FIGURE 2.
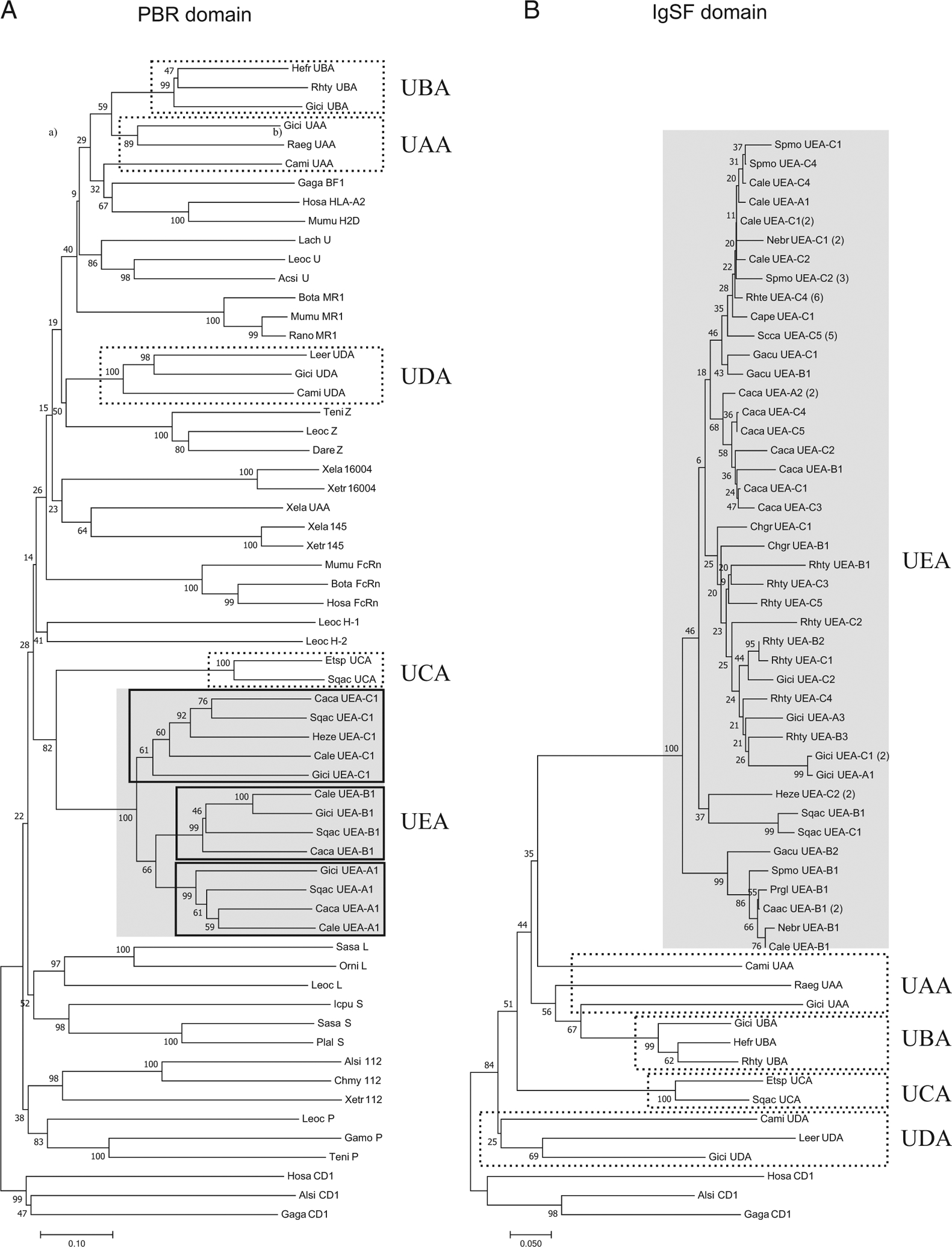
(A) Phylogenetic tree of representative MHC class I lineages in cartilaginous fish and other selected vertebrate class I molecules with the α1 and α2 domains (PBR); (B) phylogenetic tree of cartilaginous fish class I lineages with the IgSF α3 domains. These trees were constructed using the NJ method and rooted with CD1. Bootstrap support values are shown as percentages on the branches. GenBank accession numbers for all the sequences are listed in Supplemental Table II, and the corresponding alignment is found in Supplemental Fig. 1. Common names are shown, followed by the abbreviation of the scientific name (two letters of the genus and two letters of species). The sublineages are represented by the letters A, B, and C, followed by the number corresponding to a specific sequence. The scale bar indicates the number of amino acid differences per sequence.
Northern blotting
To compare the expression between the new lineage (UEA) and the class Ia UAA, we performed Northern blotting. Ten micrograms of total RNA from various nurse shark tissues (brain, epigonal, gill, gonad, liver, muscle, pancreas, spiral valve, spleen, stomach, thymus, and WBCs) were electrophoresed on denaturing 1% agarose gel. The RNA was then transferred onto nitrocellulose membranes as previously described (2). Hybridization was done using a [32P]-labeled probe encoding the α3 domains of UAA and UEA as well as the loading control nucleoside diphosphate kinase probe (27) under high-stringency conditions (50% formamide, 6× SSC, 5× Denhardt solution, 10 mM EDTA, 5% SDS, and 100 μg/ml sheared salmon sperm DNA) (2). The membrane was air dried and exposed to x-ray film until the desired signal strength was obtained.
RNAseq
Total RNA samples from all tissues mentioned above (except spiral valve because of an insufficient amount of RNA left) were used in transcriptome sequencing (RNAseq) as a complementary and quantitative approach to assess the relative gene expression of the UEA lineage. Tissue-specific mRNA libraries were obtained using poly-A enrichment and sequenced on a NovaSeq platform aiming at 30 million reads per tissue (150PE). Read quality was checked using the FastQC software, but no further filters were needed given the high quality of the data. Raw reads from the 11 libraries were used to assemble the nurse shark transcriptome through SPAdes 3.13 (24) with specific parameters for RNAseq (-rna), using k-mer = 73 as the best k-mer tested. The obtained transcripts were blasted against UEA and UAA sequences, retrieved as indicated in the “Bioinformatic filtering” section, and from G. cirratum and the close relative Rhincodon typus, to select all variants for these genes in the new nurse shark transcriptome.
Gene expression quantification was performed for UEA and UAA selected transcripts (as in the Northern blot) using bioinformatic mapping of the paired reads with RSEM (28). We used α3 domain sequences as reference for the mapping to allow direct comparison with the Northern blot results and because this domain is more conserved within lineages and may serve as a better proxy for lineage-wide expression in multicopy gene lineages such as UEA and UAA.
Southern blotting
Southern blots were performed to estimate the presence/absence and number of UEA genes in various chondrichthyan species (including species without genome and transcriptome data, i.e., spotted ratfish Hydrolagus colliei, cownose ray Rhinoptera bonasus, thornback ray R. clavata, and horned shark H. francisci) and to confirm the data obtained with the bioinformatic searches (in species with genome and transcriptome data, i.e., little skate L. erinacea, nurse shark G. cirratum, bamboo shark Chiloscyllium punctatum, sand-tiger shark Carcharias taurus, lesser spotted dogfish Scyliorhinus canicula, lemon shark Negaprion brevirostris, spiny dogfish S. acanthias, and blue shark Prionace glauca). Ten micrograms of genomic DNA (gDNA) extracted from shark erythrocytes were digested with BamHI for 48 h, followed by electrophoresis in a 0.8% agarose gel. The digested gDNA was transferred onto a nitrocellulose membrane via capillary transfer, and a [32P]-labeled probe encoding the α3 domain of nurse shark UEA was used for hybridization under low-stringency conditions (30% formamide, 6× SSC, 5× Denhardt solution, 0.5% SDS, and 100 μg/ml sheared salmon sperm DNA) (2). The membrane was exposed to x-ray film for different periods to obtain the optimal signal strength.
Results
Identification of the UEA lineage in all shark species tested (Selachii) and its general biochemical properties
Taking advantage of the new wealth of data available for transcriptomes and genomes in cartilaginous fish (Supplemental Table I), we searched for MHC class I genes in an extended taxonomic representation of sharks, rays, and chimaeras. Our searches (representative sequences in Fig. 1; the entire list of sequences in Supplemental Table II, Supplemental Fig. 1; and available at https://doi.org/10.6084/m9.figshare.14932545.v2) yielded a novel group of sequences clearly distinct from the previously reported cartilaginous fish MHC class I lineages. These sequences were found in all shark species with transcriptome and/or genome data but not in any of the chimaera and ray taxa studied. Following MHC nomenclature for nonmammalian vertebrates (29) and taking into consideration the previously reported cartilaginous fish lineages, we christened this new lineage UEA.
The nine invariant residues that bind to the N and C termini of bound peptides in almost all classical class I PBR are not conserved in UEA sequences (Table I), one of the features that defines UEA as a nonclassical molecule (16, 18). The UEA α3 domain sequences have the typical disulfide bridge found in nearly all IgSF domains (Fig. 1; Supplemental Fig. 1). Of the two conserved salt bridges in the α1 (H3 and D29) and α2 domains (H93 and D122) found in most class I molecules (including all lineages in chondrichthyan fish), only the UEA α2 domain bridge is present. Unexpectedly, the α2 domain disulfide bridge present in all other vertebrate class I molecules (and β1 domain of class II), an important element for its secondary and tertiary structure (30), is absent in this new lineage. The canonical α2 domain cysteines are replaced with hydrophobic residues in UEA, similar to the few cases in which the disulfide bond is lost in IgSF domains (13). All UEA sequences have the typical class I Asn-linked glycosylation site at the C terminus of the α1 domain (N87), an Asp/Glu-containing CP, and a hydrophobic TM region (Fig. 1; Supplemental Fig. 1). A second putative Asn-linked glycosylation site (N238) is detected in some UEA IgSF domains between the C and D β strands, and there is an insertion of 5 aa between the A and B IgSF β strands. The length of the CYT in different UEA sequences (Fig. 1) varies greatly when compared with the conserved size of the UAA tail. Although the alignment of the CYT could not be achieved because of its high sequence divergence, the typical UAA tyrosine (Y320), which regulates endocytosis and sorting (31), is not present in most of the UEA sequences. Additionally, the serine content of the CYT is lower than in the other cartilaginous fish MHC class I lineages. Considering the residues that interact with CD8 (positions 222, 223, 224, 225, 241; and 243; Fig. 1; for examples, see Refs. 17, 32, and 33), most are conservatively changed (i.e., different amino acids but with the same properties: charge and polarity) with the exception of E225 (Q/L in UEA). The same conservation is found for the 30 residues that interact with β2m (Fig. 1) (32, 34). Overall, the UEA PBR has less than 30% amino acid identity (on average) to the PBRs of UAA, UBA, UCA, and UDA, and the UEA IgSF domain has 35–41% amino acid identity to the IgSF domains of these four lineages (Table II).
Table I.
Peptide-binding residues in MHC class I molecules from different lineages
| Position of peptide-binding residues* | |||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| α1 domain | α2 domain | ||||||||||||||||||||||||||||||||||||
| Sample | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||||||||||
| 2 | 2 | 3 | 4 | 5 | 6 | 6 | 6 | 7 | 7 | 7 | 7 | 8 | 8 | 8 | 9 | 9 | 9 | 1 | 1 | 1 | 2 | 2 | 4 | 4 | 4 | 5 | 5 | 5 | 5 | 6 | 6 | 6 | 7 | ||||
| 5 | 7 | 9 | 2 | 4 | 4 | 5 | 9 | 3 | 6 | 7 | 0 | 3 | 4 | 7 | 0 | 1 | 4 | 5 | 7 | 9 | 4 | 6 | 8 | 3 | 4 | 3 | 6 | 7 | 2 | 5 | 6 | 9 | 0 | 3 | 7 | 1 | |
| nurse shark, Gici UAA | L | Y | F | F | A | V | M | Y | E | T | L | W | W | G | N | I | L | R | Y | L | C | V | H | W | F | I | T | K | W | F | G | T | Y | L | I | W | Y |
| clearnose skate, Raeg UAA | L | Y | Y | F | A | V | L | Y | E | K | L | W | G | F | N | T | L | R | I | R | F | S | Y | W | L | I | T | K | W | I | Q | N | Y | L | T | W | Y |
| elephant shark, Cami UAA | L | Y | Y | F | I | V | M | Y | Q | T | A | W | T | Y | N | T | V | R | V | Q | Y | I | Y | Y | F | I | T | K | W | L | Q | E | Y | L | I | W | Y |
| horn shark, Hefr UBA | L | Y | F | F | G | V | L | A | K | L | A | R | T | F | R | I | L | R | I | M | T | N | Y | W | F | L | I | N | W | G | G | W | Y | L | E | W | Y |
| whale shark, Rhty UBA | L | Y | L | F | I | I | L | A | K | L | A | Q | D | F | F | R | I | R | Y | M | T | N | Y | W | V | L | I | R | W | G | Q | W | Y | L | E | W | Y |
| nurse shark, Gici UBA | V | Y | H | F | I | V | L | F | K | L | A | M | H | F | Y | I | F | R | Y | M | V | N | F | W | L | I | I | R | W | N | E | W | F | L | E | W | Y |
| chicken, Gaga BF1 | L | Y | H | Y | D | V | Y | Y | Q | I | G | N | S | V | S | T | L | R | V | W | F | R | V | Y | F | I | T | K | W | V | G | W | Y | L | T | W | Y |
| mouse, Mumu H2D | . | Y | E | Y | S | V | Y | Y | E | K | A | Q | W | F | S | N | L | Y | L | Q | S | L | F | Y | Y | I | T | K | W | A | H | Y | Y | L | E | W | Y |
| human, Hosa HLA-A2 | M | Y | F | F | A | V | M | Y | E | K | V | H | T | H | D | T | L | Y | L | M | F | H | Y | Y | Y | I | T | K | W | V | Q | L | Y | L | T | W | Y |
| coelacanth, Lach U | - | - | - | - | - | N | Y | S | E | T | R | P | Y | G | F | I | A | R | V | R | Y | F | D | Y | F | I | T | K | W | M | Q | W | Y | L | I | W | Y |
| chinese sturgeon, Acsi U | L | Y | Y | F | A | I | A | Y | N | N | L | A | A | F | N | I | A | R | A | T | Y | R | E | Y | Y | L | T | K | W | F | Q | R | Y | L | T | W | Y |
| spotted gar, Leoc U | Y | Y | Y | F | V | N | M | F | G | I | L | W | T | F | G | I | L | R | W | L | Y | F | F | Y | Y | I | T | K | W | D | F | Q | Y | L | T | W | Y |
| cow, Bota MR1 | L | Y | R | F | S | T | K | H | Y | L | L | W | A | F | E | Q | L | H | Y | R | I | L | Y | Y | F | L | I | V | W | E | Y | Q | W | L | E | W | F |
| mouse, Mumu MR1 | L | Y | R | F | S | T | K | H | Y | L | L | W | T | F | E | H | L | H | Y | R | I | L | Y | Y | F | I | T | A | W | E | Y | Q | W | L | E | W | F |
| rat, Rano MR1 | L | Y | R | F | S | T | K | H | Y | L | L | W | T | F | E | H | L | H | Y | R | I | L | Y | Y | F | I | T | A | W | E | Y | Q | W | L | E | W | F |
| spotted gar, Leoc Z | L | Y | Y | I | E | R | K | Y | G | S | R | K | W | F | N | I | L | R | L | W | H | D | Y | Y | F | L | T | K | W | L | Y | T | Y | L | E | W | F |
| tetraodon, Teni Z | L | Y | Y | I | E | H | K | Y | G | S | R | K | W | F | N | I | L | R | L | W | H | D | Y | Y | F | L | T | K | W | L | Y | T | Y | L | E | W | F |
| zebra fish, Dare Z | L | Y | Y | I | E | R | K | Y | G | S | R | K | W | F | N | I | L | R | L | W | H | D | Y | Y | F | L | T | K | W | L | Y | T | Y | L | E | W | F |
| elephant shark, Cami UDA | L | Y | Y | F | H | T | N | E | E | K | L | R | L | Y | N | I | A | R | I | Y | S | R | Y | F | L | L | T | K | W | E | Y | K | Y | S | I | W | Y |
| little skate, Leer UDA | L | Y | Y | F | H | T | D | Y | E | R | L | R | L | Y | N | I | A | R | L | Y | S | R | Y | F | L | I | T | K | W | N | Y | K | Y | T | I | W | Y |
| nurse shark, Gici UDA | L | Y | Y | F | H | T | D | Y | E | R | L | R | L | S | N | I | A | R | L | Y | S | R | Y | F | L | I | T | K | W | D | Y | K | Y | T | I | W | Y |
| african clawed frog, Xela 16004 | L | Y | F | Y | I | G | V | H | M | W | G | Y | C | H | D | F | F | R | Y | V | F | E | F | Y | F | M | V | V | W | S | K | H | H | M | D | W | Y |
| western clawed frog, Xetr 16004 | L | Y | F | Y | V | G | V | H | M | W | G | Y | C | H | D | F | F | R | Y | V | F | E | F | F | F | M | I | V | W | S | K | H | H | M | D | W | Y |
| african clawed frog, Xela 145 | Y | Y | Y | F | A | G | F | L | Y | I | F | W | E | F | G | K | M | H | L | W | Y | D | D | Y | F | M | I | K | W | E | K | M | Y | L | Q | T | Y |
| western clawed frog, Xetr 145 | Y | Y | Y | F | A | G | F | L | Y | I | F | W | D | F | G | K | M | H | L | W | Y | D | D | Y | F | M | I | K | W | E | K | M | Y | L | Q | T | Y |
| african clawed frog, Xela UAA | L | Y | Y | F | T | E | D | Y | E | K | S | I | T | F | N | V | A | R | V | W | Y | E | H | Y | F | F | T | K | W | A | R | N | Y | L | I | G | Y |
| mouse, Mumu FcRn | L | Y | L | F | A | L | A | Y | E | D | L | K | L | F | A | T | L | I | L | G | L | A | F | L | F | M | V | L | W | A | K | E | F | L | S | R | H |
| cow, Bota FcRn | L | Y | F | F | V | L | A | Y | E | D | L | Q | L | F | A | A | L | E | M | G | L | A | F | L | F | M | V | K | W | A | K | E | F | L | S | R | H |
| human, Hosa FcRn | L | Y | L | F | V | L | A | Y | E | D | L | K | L | F | A | A | L | K | L | G | L | A | F | L | F | M | I | R | W | A | K | E | F | L | S | R | H |
| spotted gar, Leoc H-1 | L | Y | G | V | V | S | V | G | N | V | L | F | I | Y | A | E | R | P | V | L | C | C | L | H | Y | L | A | G | W | F | T | V | F | L | Y | W | F |
| spotted gar, Leoc H | L | Y | Y | F | A | L | I | F | E | R | N | Q | V | F | T | L | M | S | Y | R | R | D | F | Y | Y | L | V | K | W | R | E | C | Y | L | E | V | L |
| channel catfish, Icpu S | F | L | L | Y | Q | Y | T | L | A | L | F | N | N | M | A | T | A | Q | Y | A | S | L | H | F | F | L | Y | I | R | W | I | V | Y | Y | R | H | F |
| atlantic salmon, Sasa S | L | R | C | N | Q | Y | E | F | V | N | L | S | V | M | A | V | T | H | Y | A | G | M | H | Y | F | V | Y | K | R | D | R | L | H | Y | G | W | L |
| ayu, Plal S | L | R | C | H | Q | Y | G | F | F | N | L | N | V | M | A | L | T | H | Y | A | G | M | H | Y | F | I | Y | S | R | D | R | I | H | Y | E | W | L |
| spotted gar, Leoc L | M | W | F | F | V | E | V | N | K | A | A | T | S | M | K | L | A | H | Y | R | A | A | D | Y | L | L | D | - | L | D | E | Q | T | Y | L | V | Y |
| tilapia, Orni L | L | A | A | F | V | G | V | L | P | V | L | I | S | M | R | L | V | R | Q | R | T | F | D | S | S | L | W | L | K | F | L | Y | I | Y | F | T | F |
| atlantic salmon, Sasa L | L | A | A | F | V | A | F | E | G | V | F | M | H | M | R | H | L | H | Q | R | A | T | N | F | A | I | W | M | R | E | L | Y | V | L | L | T | I |
| spiny dogfish, Sqac UCA | L | V | H | F | A | M | V | Q | E | S | A | A | I | M | G | N | T | L | F | W | K | V | F | F | F | I | L | Q | F | W | T | W | N | L | T | G | L |
| velvet belly, Etsp UCA | L | V | H | F | A | M | V | Q | E | S | A | A | I | M | G | N | A | V | F | W | K | V | F | F | F | I | F | Q | L | W | T | W | N | L | T | A | L |
| nurse shark, Gici UEA-A1 | F | V | Y | F | G | S | M | N | L | L | G | R | V | M | T | T | I | S | L | W | R | M | L | F | F | I | I | E | W | W | H | W | H | L | E | R | R |
| bull shark, Cale UEA-A1 | F | V | Y | F | G | S | M | D | L | L | G | R | V | M | T | T | I | W | F | W | R | M | F | F | Y | I | T | K | W | W | H | W | Q | L | E | L | Y |
| great white shark, Caca UEA-A1 | F | V | Y | F | G | S | M | N | L | L | G | R | V | M | T | T | I | G | L | W | R | M | F | F | Y | I | I | K | W | W | H | W | Q | L | E | L | Y |
| spiny dogfish, Sqac UEA-A1 | F | V | Y | F | G | S | M | D | L | L | G | R | V | M | N | T | I | W | F | W | R | M | F | Y | Y | I | T | K | W | W | H | W | Q | L | E | L | Y |
| nurse shark, Gici UEA-B1 | I | T | F | Y | G | T | T | Y | L | Q | A | L | M | A | S | T | I | G | L | W | R | M | V | F | Y | I | T | K | W | W | F | W | L | M | E | R | R |
| bull shark, Cale UEA-B1 | I | T | F | Y | G | T | T | Y | L | Q | G | L | M | A | N | T | I | G | L | W | R | M | V | F | Y | I | T | I | W | W | F | W | L | M | E | R | R |
| great white shark, Caca UEA-B1 | F | V | Y | Y | G | A | I | Y | L | R | G | H | I | A | N | M | I | N | F | W | R | M | F | F | Y | I | T | K | W | W | Y | W | Q | L | E | R | R |
| spiny dogfish, Sqac UEA-B1 | F | A | F | Y | G | T | T | Y | L | Q | G | H | M | A | N | T | I | G | L | M | R | M | V | F | Y | I | T | I | W | W | H | W | H | L | E | H | C |
| nurse shark, Gici UEA-C1 | R | T | F | V | A | A | S | F | D | L | S | Y | L | L | N | A | I | M | L | W | R | M | F | F | F | I | L | K | W | W | Y | W | Y | L | T | S | F |
| bull shark, Cale UEA-C1 | F | V | R | I | S | G | C | F | I | L | A | H | M | A | N | T | V | M | F | W | A | M | F | F | Y | I | T | K | W | W | Y | W | Y | L | E | C | Y |
| great white shark, Caca UEA-C1 | L | V | Y | V | N | N | M | Y | L | V | G | K | I | A | N | A | M | S | F | W | T | M | F | F | Y | I | T | K | W | W | L | W | H | M | G | H | Y |
| spiny dogfish, Sqac UEA-C1 | L | V | Y | V | N | N | M | Y | L | L | G | R | K | A | N | T | I | G | F | W | S | M | V | Y | Y | L | T | K | W | W | Y | W | H | L | G | D | Y |
| atlantic cod, Gamo P | L | F | S | F | Q | S | A | V | L | C | R | H | L | K | H | F | Q | T | Q | R | G | E | W | V | F | V | I | R | W | R | V | F | F | V | H | M | L |
| tetraodon, Teni P | L | F | S | F | Q | S | E | I | E | Q | Y | S | L | R | S | I | N | V | Q | R | G | E | W | V | F | I | V | S | W | R | V | F | F | L | Q | M | I |
| spotted gar, Leoc P | L | Y | S | L | I | S | E | D | E | E | T | R | L | - | I | K | R | M | L | R | R | D | Y | F | F | L | T | D | W | K | Y | T | F | L | D | T | F |
| western clawed frog, Xetr 112 | F | W | K | N | V | G | F | S | Q | H | C | C | L | R | D | D | L | D | L | F | K | T | Y | L | F | M | I | I | W | - | I | Y | I | S | E | R | N |
| chinese alligator, Alsi 112 | Y | R | Q | T | N | T | L | R | G | E | C | W | W | I | E | A | L | A | M | V | H | T | L | L | I | L | L | K | W | K | G | M | P | S | I | L | T |
| green sea turtle, Chmy 112 | L | H | Q | L | H | S | L | F | G | A | C | W | W | V | K | A | L | E | M | I | K | T | Y | L | V | L | V | R | W | T | V | W | I | Q | H | F | T |
| human, Hosa CD1 | F | V | Q | T | G | H | A | E | L | I | F | Y | G | F | E | D | F | D | I | G | A | L | G | L | F | L | F | L | I | I | T | V | L | L | T | Y | V |
| chinese alligator, Alsi CD1 | L | L | V | T | G | H | I | Q | L | A | I | Y | L | I | T | T | V | G | I | I | M | Y | S | R | L | V | V | S | F | T | R | L | L | L | T | E | F |
| chicken, Gaga CD1 | L | L | H | V | G | G | I | D | I | S | I | Y | D | F | L | M | Y | V | F | S | I | F | I | Y | F | L | E | M | A | L | V | I | L | L | T | I | F |
The nine invariant residues that bind to the N- (light gray) and C-termini (dark gray) of bound peptides found in the classical class I and 28 other predicted peptide-binding residues are shown. In bold are the classic lineages. Distinct class I lineages are separated by dashed lines.
The position of peptide-binding residues is based on the human HLA-A2 sequence. Sequence information is listed in Supplemental Table II.
Table II.
Percentage amino acid identity between and within class I lineages and sublineages in cartilaginous fish
| Between Lineages | ||||
|---|---|---|---|---|
| UAA | UBA | UCA | UDA | |
| PBR (α1 + α2) | ||||
| UEA | 29.5 | 27.7 | 27.4 | 27.2 |
| UEA-A | 27.5 | 27.9 | 31.0 | 26.3 |
| UEA-B | 29.5 | 27.0 | 26.3 | 27.5 |
| UEA-C | 30.1 | 27.9 | 26.8 | 27.4 |
| UAA | UBA | UCA | UDA | |
| IgSF (α3) | ||||
| UEA | 39.7 | 44.3 | 41.0 | 35.2 |
| UEA-A | 38.3 | 40.9 | 36.8 | 32.1 |
| UEA-B | 38.6 | 45.9 | 41.7 | 36.4 |
| UEA-C | 40.6 | 44.6 | 42.0 | 35.6 |
| UAA | UBA | UCA | UDA | |
| PBR + IgSF (α1 + α2 + α3) | ||||
| UEA | 32.8 | 33.3 | 31.9 | 30.1 |
| UEA-A | 29.4 | 29.0 | 32.7 | 28.2 |
| UEA-B | 31.6 | 29.5 | 31.0 | 29.6 |
| UEA-C | 32.2 | 29.4 | 32.0 | 29.7 |
| Within Lineage (UEA)a | ||||
| α1 | α2 | IgSF | PBR | |
| UEA | 48.8 | 69.3 | 86.4 | 59.3 |
| UEA-A | 75.2 | 72.9 | 87.3 | 74.0 |
| UEA-B | 75.9 | 79.4 | 82.9 | 77.7 |
| UEA-C | 56.0 | 71.5 | 89.2 | 63.9 |
Sequence information listed in Supplemental Table II.
UEA falls into three distinct sublineages
As has been observed in other classical and nonclassical class I lineages (32), UEA is more diverse in the PBR α1 compared with the α2 domains, and the IgSF is extremely well conserved (average amino acid identity: 48.8% for α1, 69.3% for α2, and 86.4% for IgSF) (note the relative size of the branch lengths in the phylogenetic tree in Fig. 2; Table II). UEA can be clearly subdivided into three sublineages, which we designated as UEA-A, UEA-B, and UEA-C, considering the level of amino acid sequence identity in the PBR (Table II) and the residues present in each subgroup (Fig. 1, Supplemental Fig. 1). The sublineage UEA-C (average 56.0% identity) is the most diverse in the α1 domain when compared withUEA-A (average 75.2% identity) and UEA-B (average 75.9% identity). In contrast, the α2 and IgSF domain show similar levels of amino acid sequence identity among sublineages (average values for UEA-A, -B, and -C: 72.9%, 79.4%, and 71.5% for the α2 domain and 86.4%, 87.3%, and 89.2% for the IgSF domain, respectively). Overall, the amino acid alignment (Fig. 1) shows that sublineages UEA-A and -B are distinguished by many “fixed” positions (overall) when compared with the UEA-C sublineage, which is equally similar to both UEA-A and UEA-B. This characteristic is clearly visible when the phylogenetic trees are separated by domains (see Fig. 2, Supplemental Fig. 2, Discussion). Although it is possible to detect some distinctive positions (e.g., position 59 in which UEA-B and C have Tyr, His, or Phe, whereas UEA-A have Asn or Asp; position 70 in which UEA-A and -C share the Lys or Arg residues, whereas lineage B has Leu or His residues), the majority of the sequences is highly variable (Fig. 1, Supplemental Fig. 1). There are some conserved residues in the different sublineages that potentially could bind to the TCR (T in Fig. 1, based on Ref. 35). For example, three residues in the α2 domain of UEA-A at 161–163 (all Glu) are well conserved within the sublineage and could interact with a conserved region of TCRs.
Evolutionary relationship among MHC class I lineages, sublineages, and domains
To understand the evolutionary relationship among all chondrichthyan MHC class I amino acid sequences, NJ phylogenetic trees (Fig. 2) were constructed using either the PBR (α1 and α2 domains, Fig. 2A) or the IgSF (α3 domains, Fig. 2B), rooted with the divergent nonclassical class I molecule CD1. In both trees, UEA formed a single, well-supported clade (100% bootstrap value). Unlike UBA, UEA is clearly not derived from any class I lineage (Fig. 2) but has an old, divergent relationship with UCA in the PBR (Fig. 2A), especially in the α2 domain (Supplemental Fig. 2B).
Three UEA sublineages are clearly distinguished in the PBR tree. UEA-A and -B are each well-supported sublineages exhibiting some fixed positions (overall) when compared with the UEA-C sublineage. The UEA-C appears as a sister group of both UEA-A and UEA-B, which are somewhat more closely related. Domain-wise, the sublineages group precisely in the α1 domain tree, whereas some sequences are mixed in the α2 tree. Note that the UEA-A and -B sublineages have fewer sequences than UEA-C (11, 15, and 37 sequences per sublineage UEA-A, -B, and -C, respectively; Supplemental Table II), but they share a similar taxonomic representation, at least when comparing UEA-B and UEA-C (7, 12, and 13 species per sublineage UEA-A, -B, and -C, respectively). In the cartilaginous fish IgSF α3 domain tree (Fig. 2B), the lineages of class I genes within the cartilaginous fish were generally well supported but not the distinction of the UEA sublineages. Rather, the IgSF UEA sequences group according to taxon (species, family, or order) regardless of sublineage, suggesting within-species homogenization of this exon among the three ancient UEA sublineages, perhaps via gene conversion [concerted evolution (36)]. Indeed, the percentage of identity of IgSF nucleotide sequences among sublineages at the within-species level range from 88 to 100% (average range: 92–100%). This feature of differential evolutionary histories of the PBR and IgSF domain has been documented previously for many vertebrate class I families (see Discussion).
UEA is an MHC-linked, multigenic, likely nonpolymorphic lineage
We tested for UEA’s presence and gene number in different cartilaginous fish species via Southern blotting under low-stringency conditions with a UEA probe encoding the conserved IgSF α3 domain (“Chondroblot” in Fig. 3). Consistent with our bioinformatic searches, UEA genes were found in all shark species tested (division Selachii) but not detected in Holocephalans (represented by ratfish H. colliei) or Batoids (represented by cownose ray R. bonasus, little skate L. erinacea, thornback ray R. clavata, and marbled electric ray Torpedo marmorata). Also consistent with multiple sequences retrieved per species with the bioinformatic searches, we detected a large number of bands in all shark species, indicative of a large multigene family.
FIGURE 3.
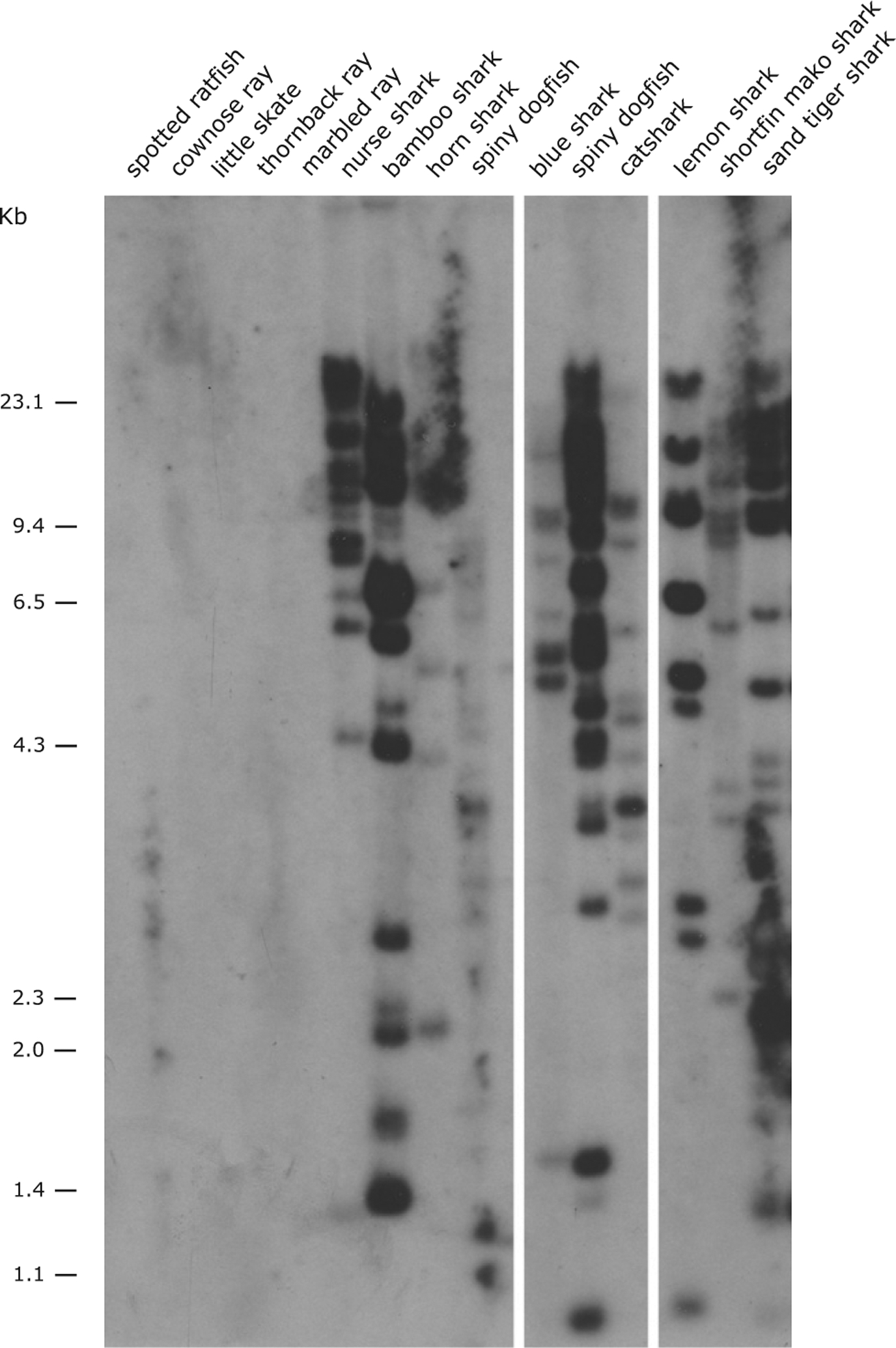
Presence of the UEA lineage in Chondrichthyan species via Southern blotting. BamHI-digested DNA from different species was hybridized with the nurse shark MHC class I UEA α3 domain probe under low-stringency conditions. The chondrichthyan species used were one chimaera (spotted ratfish H. colliei), four rays (cownose ray R. bonasus, little skate L. erinacea, thornback ray R. clavata, and marbled electric ray T. marmorata), and eight sharks (nurse shark G. cirratum, bamboo shark C. punctatum, horn shark H.s francisci, sand-tiger shark C. taurus, lesser spotted dogfish S. canicula, lemon shark N. brevirostris, spiny dogfish S. acanthias, and blue shark P. glauca). The marker size (kb) is shown on the left.
As we have done in several other studies to examine MHC linkage in nurse shark families (5, 7, 37), we attempted to uncover restriction fragment length polymorphisms (RFLP) to use for segregation analyses (Fig. 4). Unfortunately, despite the multigenic nature of UEA, five different restriction enzymes (BamHI, EcoRI, HaeIII, MindIII, and RsaI) yielded no RFLP, suggesting that this family is relatively nonpolymorphic. Bioinformatic searches using the genome of the whale shark (Rhincodon typus) (38) revealed at least 17 UEA genes located on 13 scaffolds (NW_018043287; NW_018046740; NW_018051575; NW_018034418; NW_018030408; NW_018035489; NW_0180603390; NW_018029627; NW_018034418; NW_018052231; NW_018052231; NW_018046740; and NW_018051575). One gene model (XM_020510578) in the sublineage UEA-C is in the same scaffold (NW_018030408) as the UDA gene (XM_020510579). Thus, based on our previous work showing linkage between UDA and UAA genes in nurse sharks (5), it is highly likely that UEA-C genes are also MHC linked. In two other scaffolds (NW_018060336 and NW_018034418), we found three and two UEA-C genes, respectively, suggesting that the genes belonging to the sublineage UEA-C are distributed in tandem across the MHC. The search performed in the whitespotted bamboo shark (C. plagiosum) genome revealed the presence of MHC class I genes on chromosome 37 (accession number CM012992) as previously reported by Zhang et al. (39). On this chromosome, we found two UAA loci, one UDA locus, and six UEA loci (four UEA-B and two UEA-C) (data not shown). The gene identification was performed with the α1 domain because it allows the best identification of the sublineages (Supplemental Fig. 2). Unfortunately, in the remaining species (Supplemental Table I), the available genome assemblies remain highly fragmented. In summary, the data support MHC linkage of at least some of the UEA genes.
FIGURE 4.
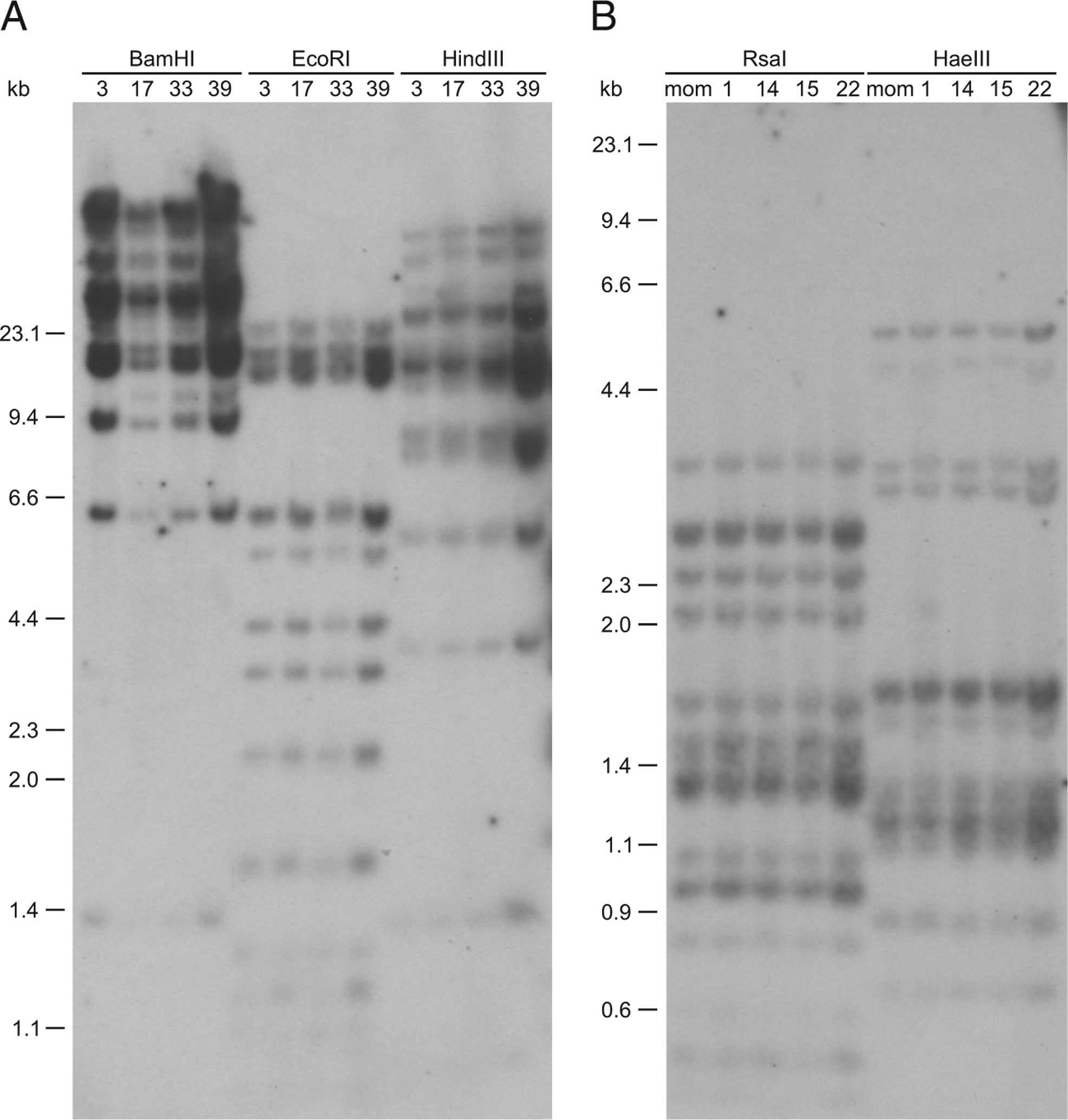
UEA lineage is relatively nonpolymorphic in nurse shark individuals. The Southern blot was performed with gDNA from different unrelated and related nurse shark individuals using five restriction enzymes [(A) BamHI, EcoRI, and HindIII; (B) RsaI and HaeIII]. The restriction fragments were hybridized with nurse shark α3 domain of UEA probe, and no RFLP are detected. Marker size (kb) is shown on the left.
Expression of the multicopy UEA lineage
The expression of the UEA lineage was analyzed in different nurse shark tissues and compared with the classical MHC class I UAA via Northern blotting (Fig. 5) and RNAseq (Fig. 6). A UEA IgSF α3 probe was used to detect the expression of all sublineages simultaneously because this domain is the most conserved among all sublineages (Table III). Distinctive banding patterns were detected in each tissue, consistent with the expression of multiple genes in this large class I family, in addition to tissue-specific expression patterns (Fig. 5). A band at ~1.9 kbp was detected in several tissues, which may be a major transcript or group of transcripts. The Northern blot also shows other bands between 3.6 and 5.0 kbp in the epigonal and thymus and above 6.6 kbp in epigonal, spleen, and WBC.
FIGURE 5.
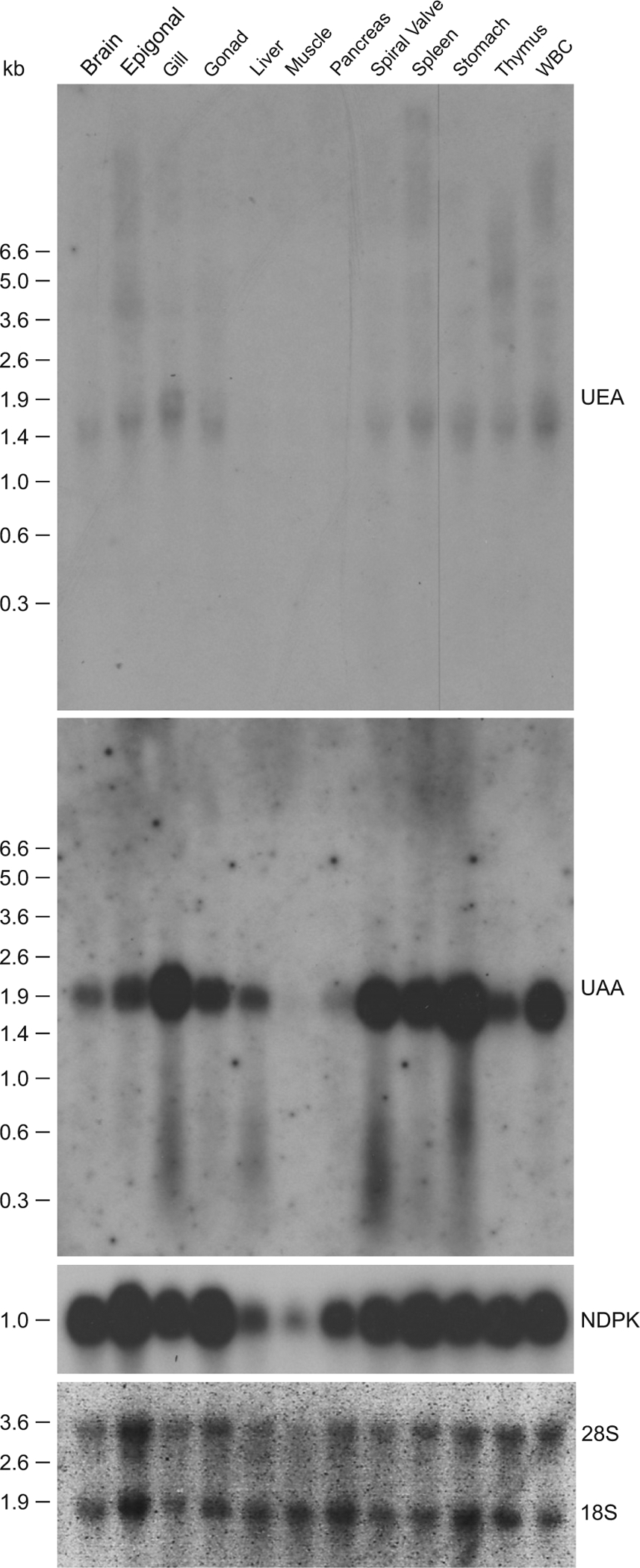
Unique pattern of expression of the UEA lineage in nurse shark compared with UAA via Northern blotting. Nucleoside diphosphate kinase was used as a loading control (ubiquitous expression), although we found it to have lower expression in muscle and pancreas in the presence of similar amounts of 28S and 18S rRNA across samples. The tissues analyzed are shown in the top, and RNA marker size (kb) on the left of the blot.
FIGURE 6.
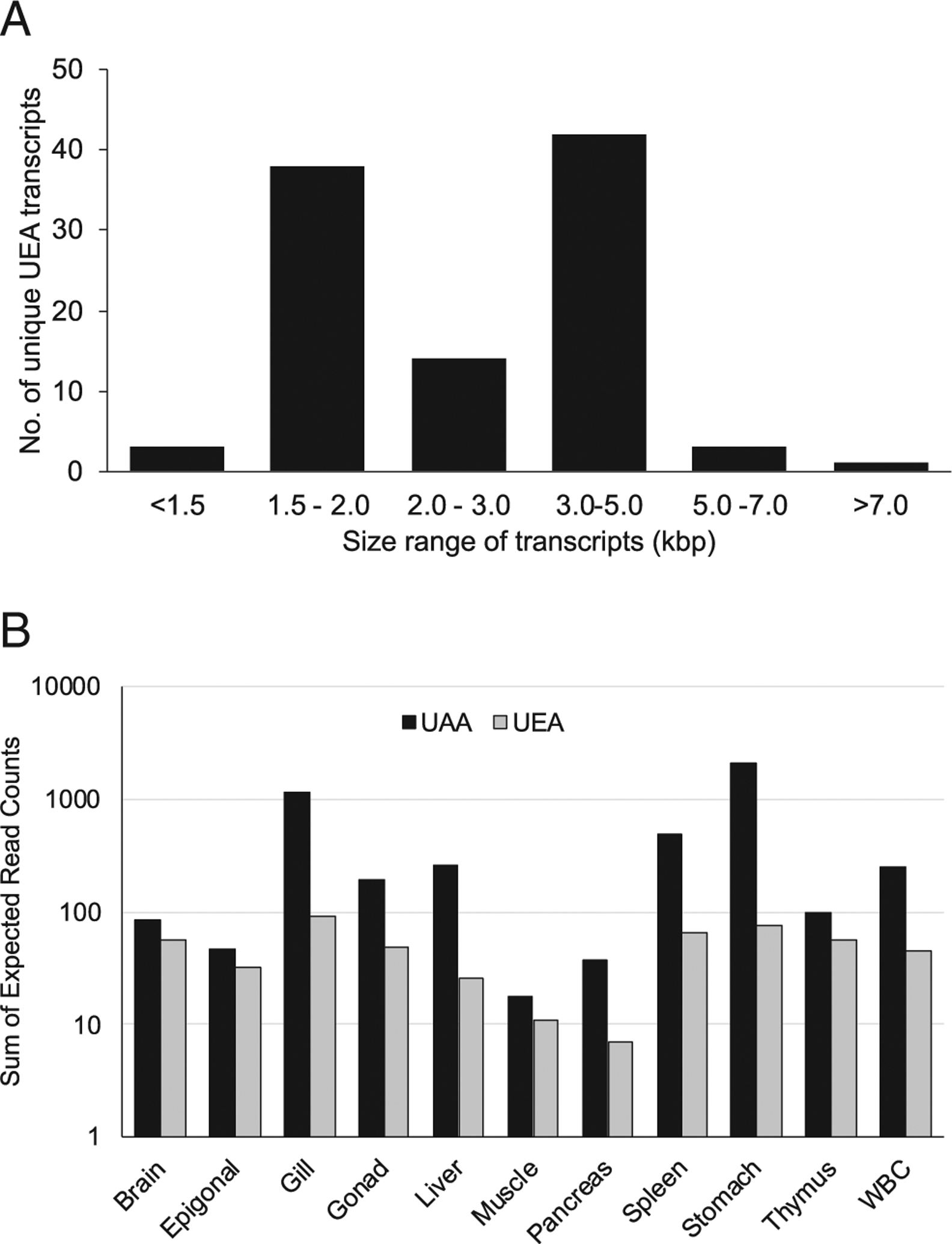
Expression of UEA and UAA genes in the nurse shark G. cirratum based on RNAseq data from multiple tissues. (A) UEA transcript size distribution for a total of 101 unique transcripts retrieved across all tissues. (B) Expression levels (measured as the expected read count in each transcript) of the UEA and UAA genes in each of 11 tissues. Note that the y-axis is in logarithmic scale.
Table III.
Percentage hydrophobicity between and within class I lineages and sublineages in cartilaginous fish and CD1
| PBR | UAA | UBA | UCA | UDA | UEA | UEA-A | UEA-B | UEA-C | CD1 |
|---|---|---|---|---|---|---|---|---|---|
| 192 aa (α1 and α2) | 42.1 | 48.6 | 45.0 | 38.8 | 42.6 | 43.8 | 41.7 | 42.3 | 50.1 |
| 9 aaa | 22.2 | 48.1 | 66.7 | 22.2 | 37.1 | 39.8 | 34.1 | 37.6 | 80.8 |
| 37 aab | 53.2 | 63.1 | 71.6 | 35.1 | 60.3 | 63.1 | 58.6 | 59.3 | 69.1 |
| Number of sequencesc | 3 | 3 | 2 | 3 | 63 | 11 | 15 | 37 | 3 |
The CD1 lineage is used for comparison because it is the most studied molecule with a hydrophobicity groove (61).
The nine invariant residues that bind to the N and C termini of bound peptides found in the classical class I.
The previous nine invariant residues and the 28 other predicted peptide-binding residues.
Sequence information listed in Supplemental Table II. Percentages were obtained using Geneious Prime v2.1 (25).
The RNAseq data corroborated the Northern blot results, retrieving multiple UEA transcripts (n = 101) with lengths ranging between 1111 and 7688 bp. Transcript size showed a bimodal distribution at 1.5–2.0 kbp and 3.0–5.0 kbp (Fig. 6A), consistent with the Northern blot. Transcript size variation was due to a combination of variable intron sizes retained in many of the transcripts and to variable 5’ and 3’ untranslated regions. Expression levels across tissues based on unique α3 query sequences (available at https://doi.org/10.6084/m9.figshare.14932545.v2) was higher for the single copy UAA compared with the multicopy UEA (up to 10× higher in some tissues), with the former exhibiting marked variability among tissues, whereas the latter showed more even expression levels (Fig. 6B). Overall, UAA and UEA had the highest expression in gill and stomach and the lowest in muscle and pancreas (Figs. 5, 6B).
UCA is distantly related to UEA and is detected only in order Squaliformes
The UCA lineage was identified in 2003 by Wang et al. (4) in the spiny dogfish S. acanthias as a highly divergent class I gene. Despite our targeted searches for UCA sequences in all chondrichthyan species available in the genome and transcriptome datasets, we only detected UCA in another squaloid shark species, the velvet-belly lanternshark Etmopterus spinax. Both species are Squaliform sharks, and thus, the extant UCA lineage appears to be exclusive to this order of cartilaginous fish (Supplemental Table I). UCA is likely represented by a single locus because all sequences we detected were identical; of 84 MHC class I sequences from the spiny dogfish transcriptomic data, six were UCA with 100% identity. Only a single sequence of UCA was detected in the velvet-belly lanternshark transcriptome from a total of eight MHC class I sequences. Phylogenetic trees of the PBR show that UCA and UEA indeed share an early common ancestor (Fig. 2, Supplemental Fig. 2), but both lineages are found in the Squaliform sharks; note that the lack of the canonical disulfide bridge in the α2 domain is shared by UCA and UEA. The potential phylogenetic scenario for emergence and maintenance of the UCA and UEA lineages is addressed in the Discussion (Fig. 7). Interestingly, UCA shows the highest level of hydrophobicity for chondrichthyan MHC class I lineages in the conserved “peptide”-binding sites, comparable to the highly hydrophobic groove of the CD1 lineage (Table III). Taking in consideration the nine UAA invariant residues that bind to the N (light gray) and C termini (dark gray) of bound peptides, the percentage of hydrophobic residues range from 22.2% (UDA) to 66.7% (UCA) and 80.8% (CD1). All potential binding residues (37 aa) have the percentage of hydrophobic residues ranging from 35.1% (UDA) to 71.6% (UCA) and 69.1% (CD1). These results contrast with the percentage of hydrophobic residues obtained for the entire PBR (~180 residues), in which all lineages are similar (38.8% UDA, 48.6% UCA, and 50% CD1).
FIGURE 7.
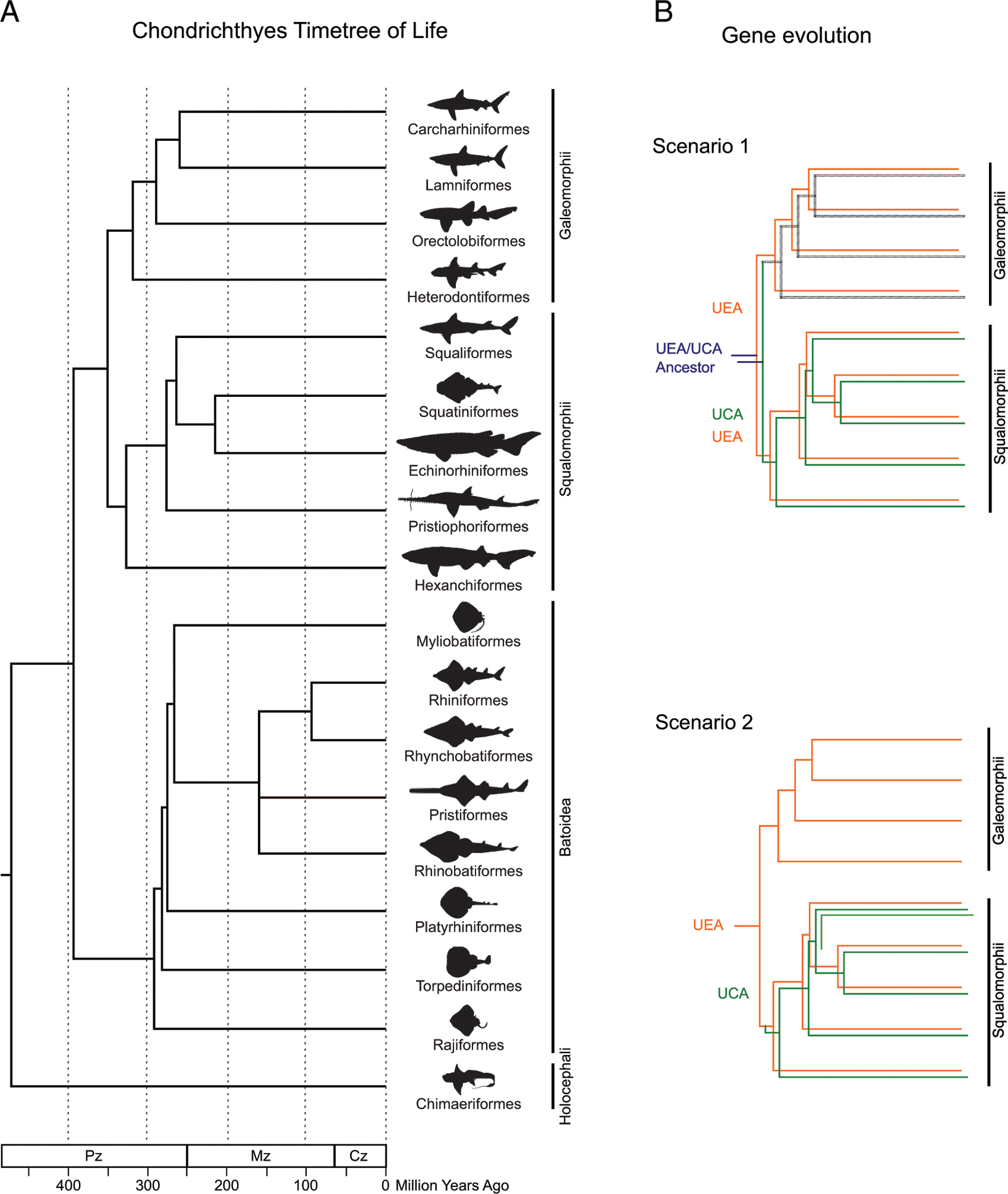
(A) Time tree of life for chondrichthyan orders. The divergence times are show in million y (MYA) in the branches. The division Selachii include the Galeomorphii and Squalomorphii; Elasmobranchs include Selachii and Batoidea. (B) Schematic representation of possible scenarios for the evolutionary relationship between UCA and UEA. Scenario 1: UEA (orange) and UCA (green) emerged from a common ancestor (blue) in sharks, but UCA was lost (black) in superorder Galeomorphii. Scenario 2: UCA emerged from a UEA ancestor within Squalomorphii (or within order Squaliformes only, highlighted as a single lighter green branch). Superorder Galeomorphii and Squalomorphii comprise the division Selachii. Approximate node age estimation was based on Ref. 20. C, Carboniferous; CZ, Cenozoic; D, Devonian; J, Jurassic; K, Cretaceous; Mz, Mesozoic; O, Ordovician; P, Permian; Pg, Paleogene; Pz, Paleozoic; S, Silurian; Tr, Triassic.
Discussion
Unique features of the UEA class I family
This new MHC class I lineage, UEA, is present in all shark species (Selachii) but not in rays (Batoidea) or chimaeras (Holocephali). UEA has most of the typical features of MHC class I lineages such as leader peptide, α1, α2, and α3 domains, and CP/TM/CYT regions. These molecules also show the N-glycosylation site in the α1 domain, the salt bridge in the α2 domain, and the disulfide bridge in the α3 domain. However, at least two important elements, the salt bridge in the α1 domain and, especially, the disulfide bridge in the α2 domain, are not conserved in UEA (Fig. 1, Supplemental Fig. 1). The absence of the salt bridge was also reported for CD1 and PROCR (17), lineages that diverged from classical class I at least 350 MYA. However, CD1/PROCR also lost the second salt bridge in the α2 domain, and they have preserved the disulfide bridge in the α2 domain (17). This disulfide bridge, formed between the thiol groups in two α2 domain Cys residues, is important for the secondary and tertiary structure of class I proteins (30). This feature has been lost in both UEA and UCA, so far the only two MHC class I lineages or genes with this condition. Further studies are needed to predict the consequences of this absence on the structure of the protein, but disruption of this disulfide bridge in HLA-A class I proteins results in a delay in the maturation of the H chain during biosynthesis, decrease in the total amount of mature H chains within the cells, and lower levels of class I surface expression (40). A novel potential N-glycosylation site is found in some UEA sequences in the α3 domain, mostly from nurse shark and spiny dogfish (Fig. 1, Supplemental Fig. 1). The role of this potential N-glycosylation site is unknown. However, the well-conserved Asn78 N-linked glycan is frequently associated with class I protein structure and quality control events in the endoplasmic reticulum (41–43). One of the features that remains unclear is UEA’s associations with β2m, CD8, and/or TCR, like the classical class I molecules (Fig. 1, Supplemental Fig. 1); although most of the potential interacting residues are conservatively changed (maintaining the basic amino acid properties), there are several residues that completely change the charge or polarity, making the association with these molecules unclear.
UEA immunogenetics
As is seen both by the presence of multiple sequences from the same species, and the Southern blotting data (Fig. 3), the UEA lineage is a large, multigene family. UEA is subdivided in three sublineages, UEA-A, -B, and -C, based on differences in the PBR (Figs. 1, 2A, Supplemental Figs. 1, 2). The three sublineages were found in transcriptome datasets from different shark taxa, and thus, all are expressed. The UEA-C sublineage has the most divergent sequences in the α1 domain and the most conserved sequences in the IgSF α3 domain (Table II). UEA-A and -B are each well-supported sublineages exhibiting some fixed positions (overall) when compared with the UEA-C sublineage. Although bootstrap support values are not high, the UEA-C appears as a sister group to another cluster bearing UEA-A and UEA-B as closer relatives. Preliminary data from Southern blotting in nurse sharks suggests that the UEA family is relatively nonpolymorphic, as we detected no UEA RFLP (Fig. 4), whereas UAA RFLP can be readily detected (7). Low polymorphism is generally expected for nonclassical class I genes, and this will be a topic of future studies.
Data mined from the emerging cartilaginous fish genome projects show that at least some of the UEA genes are MHC linked. UEA genes were found in tandem in the genomes of whale shark (38) and whitespotted bamboo shark (Y. Zhang, H. Gao, H. Li, J. Guo, M. Wang, Q. Xu, J. Wang, M. Lv, X. Guo, Q. Liu, et al, manuscript posted on bioRxiv), and thus, it is likely that homogenization of the IgSF α3 domain exon may occur via gene conversion of these closely linked genes. With such a complex gene family, it is likely that numerous gene conversions and recombinational events take place within this lineage. Gene segmental exchange has been widely reported for MHC genes in other species (44–46), and tandem class I gene organization is common in gnathostomes (32, 47–50) (see below).
Relationship between UCA and UEA
The PBR phylogenetic tree suggests that both UCA and UEA are independent and evolutionary old lineages not derived from any previously reported class I lineage, unlike the UBA lineage that is clearly derived from UAA (Fig. 2, Supplemental Fig. 2). In turn, UEA and UCA are ancestrally related in the PBR, as seen in the phylogenetic trees, and both lineages lack the canonical disulfide bridge in the α2 domain (Fig. 1, Supplemental Fig. 1). However, the exon encoding the UCA and UEA IgSF domains is not homogenized, suggesting a long and independent evolutionary history (Fig. 2). Based on the above features and the presence of both lineages only in Selachii, we hypothesize two evolutionary scenarios (Fig. 7B): 1) UEA emerged in the ancestor of sharks at the same time as UCA, but UCA was lost in superorder Galeomorphii; or 2) UCA emerged from a UEA ancestor within superorder Squalomorphii or possibly within only one of its orders (i.e., Squaliformes). The UCA gene was (so far) only found in two species of the order Squaliformes that are also the only representatives of the superorder Squalomorphii in our dataset; thus, UCA is found in one order but maybe is present in all Squalomorphii orders. In scenario 1, both UEA and UCA would have originated ~350 MYA in the ancestor of sharks, whereas in scenario 2, UCA would be not older than ~327 million y (20) (Fig. 7A). UCA’s long divergence time could explain its distinctive features from UEA, in addition to a possibly different function (and selective pressure) consistent with its highly hydrophobic binding site. It should be reiterated that the squaloid sharks have retained both the UCA and UEA lineages, suggesting that the UEA sublineages emerged independently and not from UCA.
Where does UEA fit into the big class I picture?
The main features of UEA lineage are like certain nonclassical class I genes in other vertebrates, namely the XNC lineage in amphibian Xenopus (32, 51, 52), nonclassical class I genes in the amphibian Ambystoma (axolotl) (49), and UT in marsupials and monotremes (50). The XNC is a highly complex class I family with several sublineages, much like UEA. XNC and UEA genes have very high variability in PBR, with the α1 domain being more diverse and gene specific than the α2 domain. Only one residue of the nine conserved classical class I peptide-binding residues that bind the N and C termini of bound peptides are conserved: Y171 for Xenopus XNC and W174 for UEA. The α3 IgSF domains are very similar among the different members of the family for both XNC and UEA, suggestive of concerted evolution by gene conversion. Such homogenization of the exon encoding the α3 domain has been widely reported for other vertebrate species from humans to fishes (53–57), and the XNC and UEA data typify the fundamentally differential selection pressures on the PBR (even differences between the α1 and α2 domains) and the IgSF domains. The XNC genes are minimally polymorphic (32), and our preliminary data suggest the UEA genes also have limited polymorphism.
Note that there are also differences between XNC and UEA. Although the gene conversion of the α3 domain is specific to UEA and does not extend to any of the other shark class I lineages (even the related UCA), there seems to be concerted evolution of XNC with the classical class I gene (32), which is a half chromosome away from XNC (58), and even with Xenopus class I genes on other chromosomes (59). It will be important to perform fine mapping of the cartilaginous fish class I genes on the MHC chromosomal region; during the 400 million y since the emergence of the five class I lineages, interesting chromosomal “blocks” may have arisen to safeguard the integrity of particular lineages.
As expected for a multigene family, UEA shows a very complicated expression pattern (Figs. 4, 5), much like the XNC genes. In addition to the expression of multiple transcripts of different sizes consistently shown by the Northern blot and RNAseq data, the Northern blot also suggests a tissue-specific expression with the presence of different major bands in various tissues. This pattern is in part supported by the RNAseq data; we found that only half of the UEA α3 transcripts were expressed in seven or more tissues (data not shown). Likewise, different XNC genes or sublineages have a tissue-specific expression (51), and two XNC genes have been shown to activate NKT cells (52) for defense against tumors and intracellular bacteria. NKT cells have been best studied in mammals for their recognition of the CD1 and MR1 nonclassical class I molecules and also for recognition of tumors and bacteria. During T cell development, mammalian NKT cells are not positively selected by the thymic epithelium but rather by nonclassical class I molecules expressed by cortical thymocytes (19), and indeed, many of the XNC genes are expressed by cortical thymocytes (60). UEA shows a complex expression pattern in the thymus (Fig. 5), and we speculate, based on precedent, low levels of polymorphism and its relatively hydrophobic binding site, that UEA (and UCA) functions to stimulate innate T cells for antibacterial defense. The elasmobranch UBA lineage, also multicopy but with a relatively hydrophilic PBR, is also a candidate for ligands stimulating NKT cells (2).
Regardless of UEA’s function, the discovery of this multigenic class I family, in combination with the other four chondrichthyan class I lineages, further reveals the complexity of adaptive, and likely innate, immunity in the cartilaginous fish. This work demonstrates a previously unrecognized “Big Bang” of class I lineages at the origins of gnathostome adaptive immunity as no MHC molecules have been detected in jawless fish (61). Three other class I lineages are present in the cartilaginous fish (Almeida et al., manuscript in preparation), suggesting that when class I genes first emerged in the vertebrates, there was a rapid emergence of lineages, more complex than in most gnathostome classes. Delving into the functions of these lineages may reveal primordial functions of the class I/T cell system.
Supplementary Material
Acknowledgments
This work was supported by Portuguese Foundation for Science and Technology Award PD/BD/114542/2016 (T.A.), Contract IF/00376/2015 (P.J.E.), Contract DL57/2016 (A.V.), and Project Grant PTDC/ASP-PES/28053/2017, FEDER Funds through Operational Competitiveness Factors Program-COMPETE Contract POCI-01-0145-FEDER-022184 (A.M.M.), and National Institutes of Health Grants AI140326-26 (Y.O.) and AI02877 (M.F.F.).
Abbreviations used in this article:
- class Ia
classical MHC class I molecule
- class Ib
nonclassical class I molecule
- CP
connecting piece
- CYT
cytoplasmic tail
- gDNA
genomic DNA
- IgSF
Ig superfamily
- β2m
β2-microglobulin
- MYA
million y ago
- NJ
neighbor-joining
- PBR
peptide-binding region
- RFLP
restriction fragment length polymorphism
- RNAseq
RNA sequencing
- SRA
Sequence Read Archive
- TM
transmembrane
- TSA
Transcriptome Shotgun Assembly
- WGS
Whole Genome Sequence
Footnotes
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Kasahara M, Kandil E, Salter-Cid L, and Flajnik MF. 1996. Origin and evolution of the class I gene family: why are some of the mammalian class I genes encoded outside the major histocompatibility complex? Res. Immunol 147: 278–284, discussion 284–285. [DOI] [PubMed] [Google Scholar]
- 2.Bartl S, Baish MA, Flajnik MF, and Ohta Y. 1997. Identification of class I genes in cartilaginous fish, the most ancient group of vertebrates displaying an adaptive immune response. J. Immunol 159: 6097–6104. [PubMed] [Google Scholar]
- 3.Okamura K, Ototake M, Nakanishi T, Kurosawa Y, and Hashimoto K. 1997. The most primitive vertebrates with jaws possess highly polymorphic MHC class I genes comparable to those of humans. Immunity 7: 777–790. [DOI] [PubMed] [Google Scholar]
- 4.Wang C, Perera TV, Ford HL, and Dascher CC. 2003. Characterization of a divergent non-classical MHC class I gene in sharks. Immunogenetics 55: 57–61. [DOI] [PubMed] [Google Scholar]
- 5.Almeida T, Esteves PJ, Flajnik MF, Ohta Y, and Veríssimo A. 2020. An ancient, MHC-linked, nonclassical class I lineage in cartilaginous fish. J. Immunol 204: 892–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almeida T, Gaigher A, Muñoz-Mérida A, Neves F, Castro LFC, Flajnik MF, Ohta Y, Esteves PJ, and Veríssimo A. 2020. Cartilaginous fish class II genes reveal unprecedented old allelic lineages and confirm the late evolutionary emergence of DM. Mol. Immunol 128: 125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohta Y, Okamura K, McKinney EC, Bartl S, Hashimoto K, and Flajnik MF. 2000. Primitive synteny of vertebrate major histocompatibility complex class I and class II genes. Proc. Natl. Acad. Sci. USA 97: 4712–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hinds KR, and Litman GW. 1986. Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature 320: 546–549. [DOI] [PubMed] [Google Scholar]
- 9.Ohta Y, McKinney EC, Criscitiello MF, and Flajnik MF. 2002. Proteasome, transporter associated with antigen processing, and class I genes in the nurse shark Ginglymostoma cirratum: evidence for a stable class I region and MHC haplotype lineages. J. Immunol 168: 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohta Y, Powis SJ, Lohr RL, Nonaka M, Pasquier LD, and Flajnik MF. 2003. Two highly divergent ancient allelic lineages of the transporter associated with antigen processing (TAP) gene in Xenopus: further evidence for co-evolution among MHC class I region genes. Eur. J. Immunol 33: 3017–3027. [DOI] [PubMed] [Google Scholar]
- 11.Ott JA, Castro CD, Deiss TC, Ohta Y, Flajnik MF, and Criscitiello MF. 2018. Somatic hypermutation of T cell receptor α chain contributes to selection in nurse shark thymus. Elife 7: e28477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohta Y, Shiina T, Lohr RL, Hosomichi K, Pollin TI, Heist EJ, Suzuki S, Inoko H, and Flajnik MF. 2011. Primordial linkage of β2-microglobulin to the MHC. J. Immunol 186: 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams AF, and Barclay AN. 1988. The immunoglobulin superfamily–domains for cell surface recognition. Annu. Rev. Immunol 6: 381–405. [DOI] [PubMed] [Google Scholar]
- 14.Halaby DM, and Mornon JPE. 1998. The immunoglobulin superfamily: an insight on its tissular, species, and functional diversity. J. Mol. Evol 46: 389–400. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto K, Okamura K, Yamaguchi H, Ototake M, Nakanishi T, and Kurosawa Y. 1999. Conservation and diversification of MHC class I and its related molecules in vertebrates. Immunol. Rev 167: 81–100. [DOI] [PubMed] [Google Scholar]
- 16.Madden DR, Gorga JC, Strominger JL, and Wiley DC. 1991. The structure of HLA-B27 reveals nonamer self-peptides bound in an extended conformation. Nature 353: 321–325. [DOI] [PubMed] [Google Scholar]
- 17.Dijkstra JM, Yamaguchi T, and Grimholt U. 2018. Conservation of sequence motifs suggests that the nonclassical MHC class I lineages CD1/PROCR and UT were established before the emergence of tetrapod species. Immunogenetics 70: 459–476. [DOI] [PubMed] [Google Scholar]
- 18.Adams EJ, and Luoma AM. 2013. The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annu. Rev. Immunol 31: 529–561. [DOI] [PubMed] [Google Scholar]
- 19.Salio M, Silk JD, Jones EY, and Cerundolo V. 2014. Biology of CD1- and MR1-restricted T cells. Annu. Rev. Immunol 32: 323–366. [DOI] [PubMed] [Google Scholar]
- 20.Heinicke MP, Naylor GJP, and Hedges SB. 2009. Cartilaginous fishes (Chondrichthyes) Hedges SB, and Kumar S, eds. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 21.Aschliman NC 2011. The Batoid Tree of Life: Recovering the Patterns and Timing of the Evolution of Skate, Rays and Allies (Chondrichthyes : Batoidea) Doctoral dissertation, Florida State University, Tallahassee, FL. [Google Scholar]
- 22.Naylor GJP, Caira JN, Jensen K, Rosana KAM, Straube N, and Lakner C. 2012. Elasmobranch phylogeny: a mitochondrial estimate based on 595 species. In Biology of Sharks and Their Relatives Carrier JC, Musick JA, and Heithaus MR, eds. CRC Press, Boca Raton, FL, p. 31–56. [Google Scholar]
- 23.Bolger AM, Lohse M, and Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol 19: 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Stecher G, Peterson D, Filipski A, and Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasahara M, Vazquez M, Sato K, McKinney EC, and Flajnik MF. 1992. Evolution of the major histocompatibility complex: isolation of class II A cDNA clones from the cartilaginous fish. Proc. Natl. Acad. Sci. USA 89: 6688–6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, and Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ballingall KT, Bontrop RE, Ellis SA, Grimholt U, Hammond JA, Ho C-S, Kaufman J, Kennedy LJ, Maccari G, Miller D, et al. 2018. Comparative MHC nomenclature: report from the ISAG/IUIS-VIC committee 2018. Immunogenetics 70: 625–632. [DOI] [PubMed] [Google Scholar]
- 30.Kastin A, ed. 2013. Handbook of Biologically Active Peptides Academic Press, Cambridge, MA. [Google Scholar]
- 31.Roeth JF, Williams M, Kasper MR, Filzen TM, and Collins KL. 2004. HIV-1 Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail. J. Cell Biol 167: 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flajnik MF, Kasahara M, Shum BP, Salter-Cid L, Taylor E, and Du Pasquier L. 1993. A novel type of class I gene organization in vertebrates: a large family of non-MHC-linked class I genes is expressed at the RNA level in the amphibian Xenopus. EMBO J 12: 4385–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao GF, Tormo J, Gerth UC, Wyer JR, McMichael AJ, Stuart DI, Bell JI, Jones EY, and Jakobsen BK. 1997. Crystal structure of the complex between human CD8α(α) and HLA-A2. Nature 387: 630–634. [DOI] [PubMed] [Google Scholar]
- 34.Shields MJ, Assefi N, Hodgson W, Kim EJ, and Ribaudo RK. 1998. Characterization of the interactions between MHC class I subunits: a systematic approach for the engineering of higher affinity variants of β 2-microglobulin. J. Immunol 160: 2297–2307. [PubMed] [Google Scholar]
- 35.Reche PA, and Reinherz EL. 2003. Sequence variability analysis of human class I and class II MHC molecules: functional and structural correlates of amino acid polymorphisms. J. Mol. Biol 331: 623–641. [DOI] [PubMed] [Google Scholar]
- 36.Nei M, and Rooney AP. 2005. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet 39: 121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Criscitiello MF, Ohta Y, Graham MD, Eubanks JO, Chen PL, and Flajnik MF. 2012. Shark class II invariant chain reveals ancient conserved relationships with cathepsins and MHC class II. Dev. Comp. Immunol 36: 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Read TD, Petit III RA, Joseph SJ, Alam MT, Weil MR, Ahmad M, Bhimani R, Vuong JS, Haase CP, Webb DH, et al. 2017. Draft sequencing and assembly of the genome of the world’s largest fish, the whale shark: Rhincodon typus Smith 1828. [Published erratum appears in 2017 BMC Genomics. 18: 755.]BMC Genomics 18: 532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Gao H, Li H, Guo J, Ouyang B, Wang M, Xu Q, Wang J, Lv M, Guo X, et al. 2020. The white-spotted bamboo shark genome reveals chromosome rearrangements and fast-evolving immune genes of cartilaginous fish. Iscience 23: 101754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warburton RJ, Matsui M, Rowland-Jones SL, Gammon MC, Katzenstein GE, Wei T, Edidin M, Zweerink HJ, McMichael AJ, and Frelinger JA. 1994. Mutation of the α 2 domain disulfide bridge of the class I molecule HLA-A*0201. Effect on maturation and peptide presentation. Hum. Immunol 39: 261–271. [DOI] [PubMed] [Google Scholar]
- 41.Blees A, Januliene D, Hofmann T, Koller N, Schmidt C, Trowitzsch S, Moeller A, and Tampé R. 2017. Structure of the human MHC-I peptide-loading complex. Nature 551: 525–528. [DOI] [PubMed] [Google Scholar]
- 42.Ryan SO, and Cobb BA. 2012. Roles for major histocompatibility complex glycosylation in immune function. Semin. Immunopathol 34: 425–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neerincx A, and Boyle LH. 2019. Preferential interaction of MHC class I with TAPBPR in the absence of glycosylation. Mol. Immunol 113: 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Promerová M, Králová T, Bryjová A, Albrecht T, and Bryja J. 2013. MHC class IIB exon 2 polymorphism in the Grey partridge (Perdix perdix) is shaped by selection, recombination and gene conversion. PLoS One 8: e69135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richman AD, Herrera LG, Nash D, and Schierup MH. 2003. Relative roles of mutation and recombination in generating allelic polymorphism at an MHC class II locus in Peromyscus maniculatus. Genet. Res 82: 89–99. [DOI] [PubMed] [Google Scholar]
- 46.Hosomichi K, Miller MM, Goto RM, Wang Y, Suzuki S, Kulski JK, Nishibori M, Inoko H, Hanzawa K, and Shiina T. 2008. Contribution of mutation, recombination, and gene conversion to chicken MHC-B haplotype diversity. J. Immunol 181: 3393–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hansen JD, Strassburger P, Thorgaard GH, Young WP, and Du Pasquier L. 1999. Expression, linkage, and polymorphism of MHC-related genes in rainbow trout, Oncorhynchus mykiss. J. Immunol 163: 774–786. [PubMed] [Google Scholar]
- 48.Ohta Y, Goetz W, Hossain MZ, Nonaka M, and Flajnik MF. 2006. Ancestral organization of the MHC revealed in the amphibian Xenopus. J. Immunol 176: 3674–3685. [DOI] [PubMed] [Google Scholar]
- 49.Sammut B, Du Pasquier L, Ducoroy P, Laurens V, Marcuz A, and Tournefier A. 1999. Axolotl MHC architecture and polymorphism. Eur. J. Immunol 29: 2897–2907. [DOI] [PubMed] [Google Scholar]
- 50.Papenfuss AT, Feng ZP, Krasnec K, Deakin JE, Baker ML, and Miller RD. 2015. Marsupials and monotremes possess a novel family of MHC class I genes that is lost from the eutherian lineage. BMC Genomics 16: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salter-Cid L, Nonaka M, and Flajnik MF. 1998. Expression of MHC class Ia and class Ib during ontogeny: high expression in epithelia and coregulation of class Ia and lmp7 genes. J. Immunol 160: 2853–2861. [PubMed] [Google Scholar]
- 52.Banach M, Edholm E-S, and Robert J. 2017. Exploring the functions of non-classical MHC class Ib genes in Xenopus laevis by the CRISPR/Cas9 system. Dev. Biol 426: 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joly E, and Rouillon V. 2006. The orthology of HLA-E and H2-Qa1 is hidden by their concerted evolution with other MHC class I molecules. Biol. Direct 1: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinto RD, Randelli E, Buonocore F, Pereira PJB, and dos Santos NMS. 2013. Molecular cloning and characterization of sea bass (Dicentrarchus labrax, L.) MHC class I heavy chain and β2-microglobulin. Dev. Comp. Immunol 39: 234–254. [DOI] [PubMed] [Google Scholar]
- 55.Nonaka MI, Aizawa K, Mitani H, Bannai HP, and Nonaka M. 2011. Retained orthologous relationships of the MHC Class I genes during euteleost evolution. Mol. Biol. Evol 28: 3099–3112. [DOI] [PubMed] [Google Scholar]
- 56.Geliebter J, and Nathenson SG. 1987. Recombination and the concerted evolution of the murine MHC. Trends Genet 3: 107–112. [Google Scholar]
- 57.Alcaide M, Edwards SV, and Negro JJ. 2007. Characterization, polymorphism, and evolution of MHC class II B genes in birds of prey. J. Mol. Evol 65: 541–554. [DOI] [PubMed] [Google Scholar]
- 58.Courtet M, Flajnik M, and Du Pasquier L. 2001. Major histocompatibility complex and immunoglobulin loci visualized by in situ hybridization on Xenopus chromosomes. Dev. Comp. Immunol 25: 149–157. [DOI] [PubMed] [Google Scholar]
- 59.Ohta Y, Kasahara M, O’Connor TD, and Flajnik MF. 2019. Inferring the “primordial immune complex”: origins of MHC class I and antigen receptors revealed by comparative genomics. J. Immunol 203: 1882–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goyos A, and Robert J. 2009. Tumorigenesis and anti-tumor immune responses in Xenopus. Front. Biosci 14: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flajnik MF 2018. A cold-blooded view of adaptive immunity. Nat. Rev. Immunol 18: 438–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


