Abstract
This review summarizes over a decade of investigations into how membrane-binding proteins from the HIV-1 virus interact with lipid membrane mimics various HIV and host T-cell membranes. The goal of the work was to characterize at the molecular level both the elastic and structural changes that occur due to HIV protein/membrane interactions, which could lead to new drugs to thwart the HIV virus. The main technique used to study these interactions is diffuse X-ray scattering, which yields the bending modulus, KC, as well as structural parameters such as membrane thickness, area/lipid and position of HIV peptides (parts of HIV proteins) in the membrane. Our methods also yield information about lipid chain order or disorder caused by the peptides. This review focuses on three stages of the HIV-1 life cycle: 1) infection, 2) Tat membrane transport, and 3) budding. In the infection stage, our lab studied three different parts of HIV-1 gp41 (glycoprotein 41 fusion protein): 1) FP23, the N-terminal 23 amino acids that interact non-specifically with the T-cell host membrane to cause fusion of two membranes, and its trimer version, 2) CRAC (cholesterol recognition amino acid consensus sequence), on the MPER (membrane proximal external region) near the membrane-spanning domain, and 3) LLP2 (lentiviral lytic peptide 2) on the CTT (cytoplasmic C-terminal tail). For Tat transport, we used membrane mimics of the T-cell nuclear membrane as well as simpler models that varied charge and negative curvature. For membrane budding, we varied the myristoylation of the MA31 peptide as well as the negatively charged lipid. These studies show that HIV peptides with different roles in the HIV life cycle affect differently the relevant membrane mimics. In addition, the membrane lipid composition plays an important role in the peptides’ effects.
INTRODUCTION
1. Methods
The goal of this review article is to present an overview of several years’ investigations from the Tristram-Nagle/Nagle lab of HIV-1 peptide/lipid interactions, studied by the method of X-ray diffuse scattering. This method obtains both elastic and structural parameters of membranes. The samples are well-oriented stacks of ~2000 bilayers on highly polished silicon wafers, hydrated through the vapor. Diffuse scattering is caused by the thermal fluctuations which occur when lipid bilayers are fully hydrated [3], which in turn causes the exiting X-ray scattering vectors to be deflected outside of the usual lamellar Bragg orders. There is actually much more information in the diffuse scatter than in the Bragg peaks, where the “lobes” of off-specular diffuse scatter yield elastic parameters as well as bilayer structure. The analysis derives from equilibrium statistical mechanics and uses a height-height pair correlation in the out-of-plane direction, where we simultaneously solve for the Caillé parameter, η, and the in-plane correlation length, ξ [3, 4]. The original Caillé theory [7] employed a continuum model of smectic liquid crystals which has gradually been replaced by the following, more realistic, discrete free energy functional [8–12]
where N is the number of bilayers in the Z (vertical) direction, Lr is the domain size in the r (horizontal) direction, KC is the bending modulus, un is the vertical membrane displacement, and B is the compressibility modulus. From this analysis we are able to obtain KC and B independently, where KC describes the bending of a single bilayer, such as in cell membranes. While the bilayer stack is fairly thick (10 μm), it behaves as a single bilayer since a single lamellar D-spacing is measured even with inclusions, indicating that the inclusions and water have penetrated to all layers.
In 2000, Pabst et al. published the use of X-rays to study diffuse data from fluctuating lipid bilayers in capillaries, but their method relies upon isotropic samples where the elastic constants KC and B are unable to be separated [14]. Moreover, the resolution of the electron density profiles obtained (yielding bilayer thickness) is rough, due to the use of a very simple model of the bilayer and low signal-to-noise ratio in the data. In 2001, our lab was the first to publish the X-ray diffuse scattering method to obtain both elastic moduli, KC and B, as well as high-resolution structure from fluctuating oriented lipid membranes [3], although Salditt’s lab in Germany published a similar method using specular X-ray scattering from fluctuating, oriented lipid bilayers soon thereafter in 2002 [19], and in 2006, Li’s lab published an X-ray study of DOPC using oriented stacks of fully hydrated bilayers, yielding KC and B separately [20]. More details of our low-angle X-ray scattering (LAXS) methods are given in Supplementary Material.
2. Focus on HIV-1 protein/lipid interactions
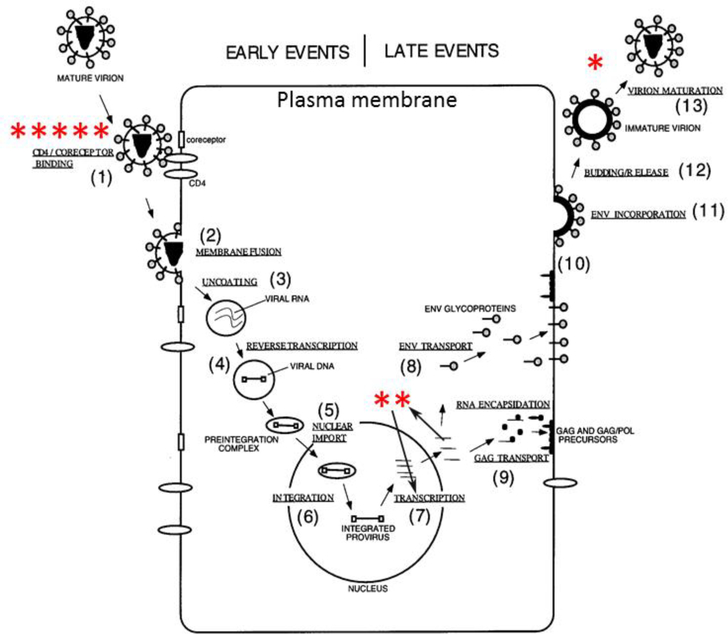
With such a powerful, probe-free method to study lipid/protein interactions, we looked for research areas where this method could yield fruitful, biologically-relevant results. Although we have published in several areas [21–27], we were attracted to the area of human immunodeficiency (HIV) due to its biological relevance and also due to its strong support at the NIH. HIV remains a health threat worldwide. Every year there are 2 million new cases, 1.5 million deaths and 37 million people living with HIV. In developed countries, the death rate from HIV/acquired immunodeficiency syndrome (AIDS) has decreased since the advent of highly active anti-retroviral therapy (HAART) in 1996. However, in the U.S., every year there are 50,000 new cases, 17,000 deaths and 1.2 million people living with HIV, so it remains a serious health problem. Therefore, any information concerning interactions of proteins, or parts of proteins, where a membrane interaction is crucial for the success of the protein, could become a future drug target. Fig. 1 summarizes the HIV life cycle, where the asterisks represent our published papers. This review article will sequentially present results obtained regarding 1) HIV viral entry into the host T-cell, 2) translocation of Tat through the nuclear membrane and 3) budding out of new HIV virions at the T-cell plasma membrane.
Figure 1.
HIV life cycle (figure adapted from Ref. [1]). Asterisks represent works published by the Tristram-Nagle/Nagle lab at 3 steps in the HIV life cycle: HIV entry *****[2], [5], [6], [13], [15], Tat translocation **[16], [17], and budding of new HIV virions *[18].
HIV-1 fusion protein, gp41
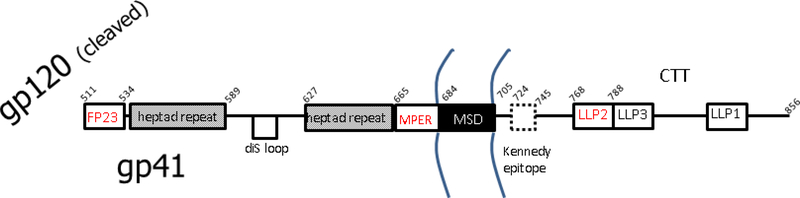
We begin with the HIV-1 infection step, shown in Fig. 1, which requires the fusion protein, glycoprotein41 (gp41). Fig. 2 shows a linear arrangement of well-recognized domains of gp41, where those in red type were studied by our X-ray methods. After FP23, we focus on the CRAC motif in the membrane proximal external region (MPER) of gp41, followed by the lentivirus lytic peptide2 (LLP2) on the cytoplasmic C-terminal tail (CTT) of gp41. The common thread is that all three of these domains are thought to interact with membranes during the crucial infectivity step involving fusion of the viral membrane with the target T-cell membrane.
Figure 2.
Linear sequence of recognized epitopes in gp41, adapted from Refs. [28–30]. Abbreviations: disulfide loop (diS loop), membrane proximal external region (MPER), membrane spanning domain (MSD), lentivirus lytic peptide (LLP). The region to the right of TM is the cytoplasmic C-terminal tail (CTT) [30]. Dotted line for the Kennedy epitope indicates that it sometimes is exposed extracellularly [30]. Curved lines indicate the membrane.
3. FP23 in gp41
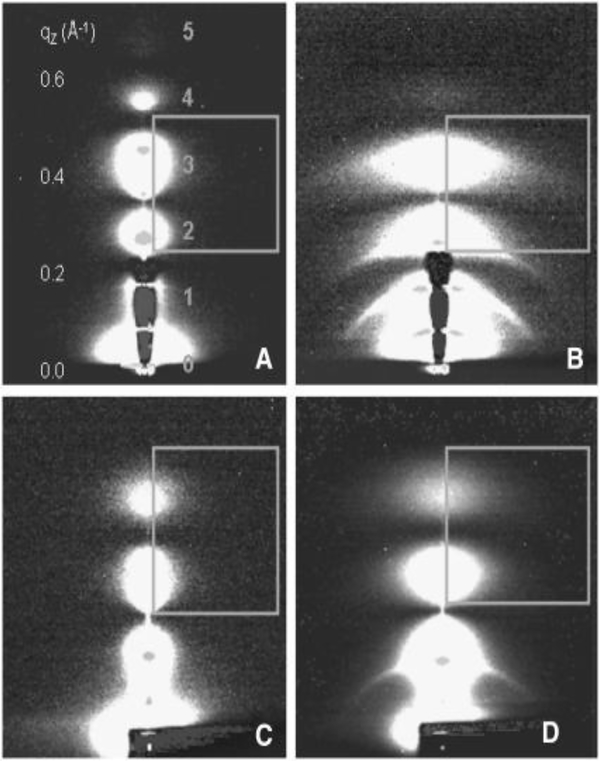
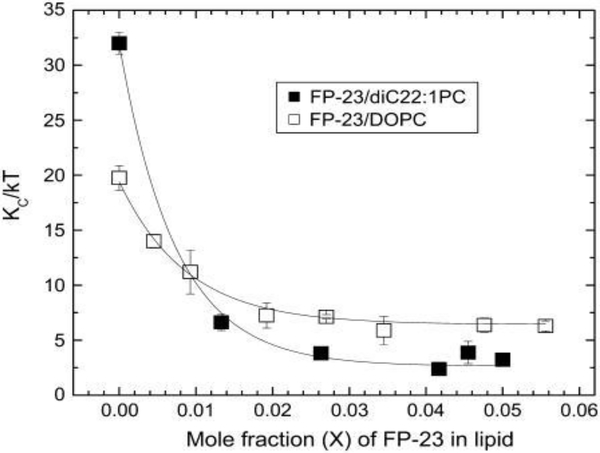
In step 1 of the 1.5 day life cycle (Fig. 1), the HIV-1 virion membrane interacts both specifically and non-specifically with the T-cell membrane. Fusion of the membrane of enveloped viruses such as HIV-1 with the target T-cell is required for infection [31]. The HIV-1 envelope glycoprotein gp160 contains two noncovalently bound subunits, gp120 and the fusion protein gp41 [32], in a trimer arrangement (“lollipops” on HIV virion in Fig. 1); the trimer is the naturally occurring form during HIV-1 infection. Numbers on gp41 in Fig. 2 indicate amino acid sequence from gp160 before cleavage into gp120 and gp41. After the initial docking step in which sites on gp120 interact with the CD4 and chemokine receptors on the target membrane [33, 34], the N-terminal hydrophobic region of the viral gp41 envelope protein is thought to provide the crucial perturbation of the target membrane that induces fusion of the viral and T-cell membranes, thus allowing the viral RNA to be injected into the host cell [35, 36]. Thus the N-terminus of gp41 (FP23, 23 amino acids) is the subject of many investigations, including three involving our lab [2, 6, 37]. Fig. 3 shows the low-angle X-ray scattering we obtained by adding FP23 to DOPC (3D) and to a thicker lipid diC22:1PC (3B) at ~16:1 lipid:peptide molar ratio. As shown, the lobes of diffuse scattering (numbered) are broader in 3B and 3D than those in Fig. 3A and 3C (controls). This increase in lateral extension was partially due to an increase in mosaic spread, or degree of misorientation, but this was not the major factor. The effect of adding FP23 to these two simple membranes was to increase fluctuations, resulting in enlarged diffuse scatter. When the decrease in white lobe intensity in the horizontal direction is analyzed in the fitting boxes shown, KC and B are obtained using liquid crystal theory as we have described previously [3, 4]. Fig. 4 compares the fit of the liquid crystal theory to the data in Fig. 3C and 3D. Fig. 5 shows an exponential decrease in KC with increase in FP23 in both DOPC and diC22:1PC.
Figure 3.
2D CCD scattering intensity images obtained at CHESS at 30 °C of (A) diC22:1PC, (B) FP23/diC22:1PC (1:15) (C) DOPC, and (D) FP23/DOPC (1:17). The white regions define diffuse scattering lobes that are numbered 1–5 in A. The lamellar Bragg orders appear as narrow, horizontal lines, showing through the semi-transparent beam-stop in A and B, or as dots on the lobes in all images. The gray boxes are the fitting boxes within which the diffuse scattering theory is used to fit to the data to obtain KC and B. The samples are oriented stacks of ~2000 bilayers on highly polished silicon wafers prepared by evaporation of organic solvent using the rock and roll method [37]. The dried lipid/peptide films are hydrated through the vapor in a thick-walled hydration chamber [24] to full hydration. The flat samples are rotated between −3° and 7° during 30–60 second dezingered exposures. Figure from Ref. [2].
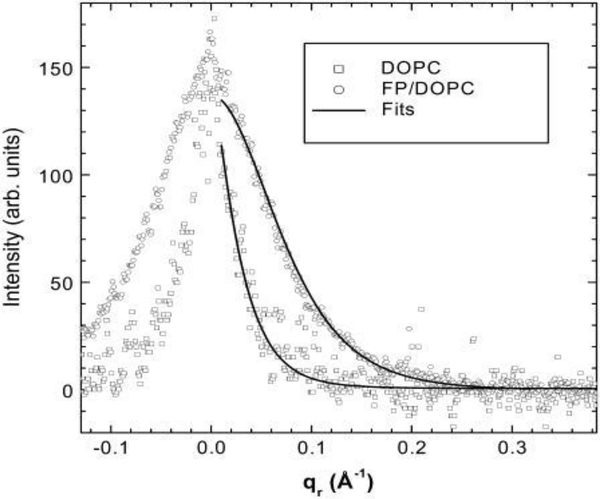
Figure 4.
(left). Normalized and background subtracted DOPC and FP23/DOPC (1:17) diffuse scattering data as a function of qr at the position of the highly diffuse 5th order peak are shown by open symbols. The corresponding fits from the diffuse scattering theory are shown by black lines. Figure from Ref. [2].
Figure 5.
(right). Effect on bending modulus KC, in units of thermal energy kT, when adding FP23 to diC22:1PC and to DOPC. The lines are exponential fits to the data. Figure from Ref. [2].
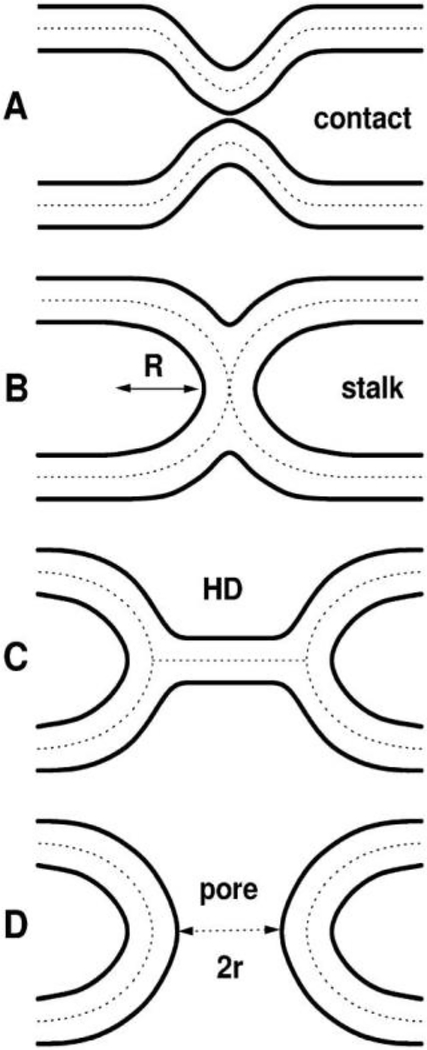
As shown in Fig. 5, FP23 dramatically reduces KC as a function of increasing concentration, lowering the energy required to bend a membrane by a factor of ~13 for the thicker diC22:1PC and ~3 for DOPC. In Fig. 6 are shown the well-known hypothetical fusion intermediates. The energy required to bend a membrane depends on KC and also on the spontaneous membrane curvature (CS), as E=KC/2(C-CS)2, so lowering KC would lower this energy. Our calculations in Ref. [2] indicate that our concentrations are physiologically relevant, and that the estimated effective radius R of the stalk can be as large as 45 Å and still attain the concentration Xe (~0.008) of FP23, where Xe is from the exponent in the exponential fit to the data in Fig. 5. However, it is possible that each FP23 might reside primarily in one monolayer, or in a staggered arrangement in both monolayers, which could induce a permanent local spontaneous curvature in the bilayer. If so, the main result that the bending modulus decreases could be construed as FP23 inducing a local spontaneous curvature that might also reduce the energy of curved fusion intermediates. However, a staggered, regular pattern would manifest as in-plane scattering that we do not observe.
Figure 6.
Well-known hypothetical fusion intermediates. Lipid bilayer surfaces are indicated by solid lines. Dotted lines in the hydrocarbon interior divide the bilayers into monolayers. A. Contact of the virus and target membranes, B. stalk that allows lipid mixing, C. hemifusion diaphragm (HD), D. pore. Figure from Ref. [2].
Since our methods using X-rays rely on the 1–10 nm scale, we attempted to verify if the more traditional methods of obtaining KC, such as micromechanical manipulation (pipette aspiration) (MM) and shape or fluctuation analysis (SA), which both use giant unilamellar vesicles (GUVs) at the micron length scale, would confirm our dramatic results for FP23. We resolve this issue in the next paragraph, but first here is some background. For pure lipids, the absolute values of KC from our X-ray methods are in good agreement with MM [38], while SA results give larger KC values by a factor of 1.5–2 [39]. To try to understand this difference between X-ray and SA, we tested whether the sugars that are required for both these methods alter KC, but we found no effect of glucose or sucrose [40], so that cannot be the reason for the disparity. Recently, the Nagle lab has included a new elastic modulus, molecular tilt, which was initially shown to increase the absolute value of KC by ~20% [41]. However, newer data with many pure lipids [42] or as a function of temperature [43] yield a KC with tilt analysis that is ~1.5 times higher than without considering tilt, thus removing the disparity between X-ray and some SA studies. All of the measurements discussed in this review were carried out without the newer molecular tilt field analysis since we are not so concerned with absolute values of KC, rather with the relative effect of peptides on the pure lipid KC’s. In general, the effect of HIV peptides on KC is larger than a factor two.
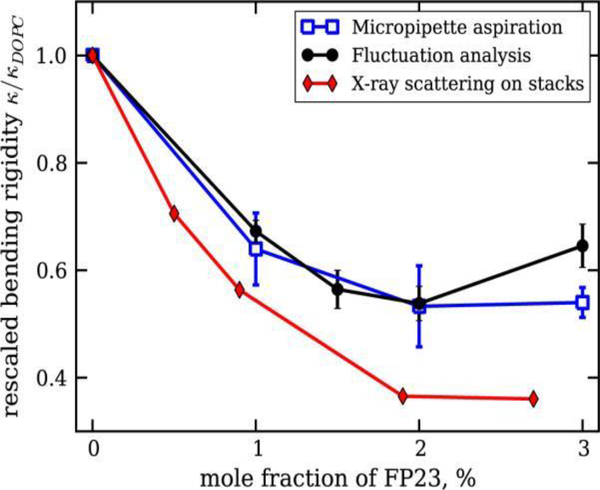
In order to compare our FP23 results with SA and MM, Dr. Tristram-Nagle teamed with Dr. Rumiana Dimova’s lab where postdoc Pavlo Shchelokovskyy carried out data collection and analysis. Fig. 7 shows the effect of FP23 in DOPC on bending modulus (rigidity) Kc (κ) using MM, SA and X-ray scattering, normalized to 1 to avoid differences in absolute values. As shown, there was a general decrease in KC due to added FP23 in all cases, but slightly less for MM and SA than for X-ray. The small discrepancy of MM and SA compared to X-ray could be due to the different length scales in these methods. This decrease in the bending rigidity is similar to that observed with co-surfactants [44] and is theoretically expected for transmembrane inclusions [45–47].
Figure 7.
Relative changes of the bending rigidity as a function of the FP23 concentration in DOPC bilayers. See legend for details. Figure from Ref. [37].
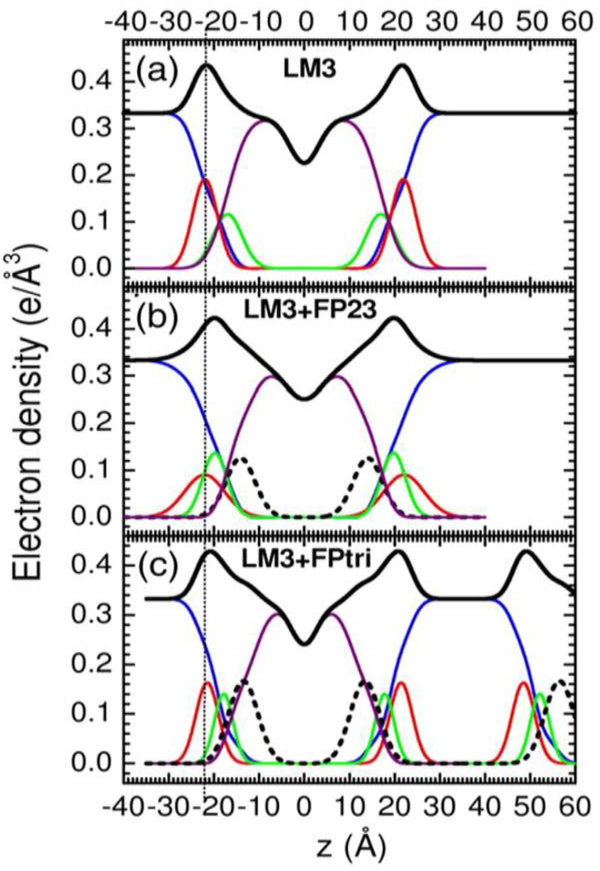
In a separate collaboration with the Weliky and Kooijman labs, we investigated spontaneous curvature as well as KC when FP23 was added to a more heterogeneous membrane mimic [6]. LM3 is a T-cell plasma membrane mimic consisting of a mixture of six lipids: POPC:POPE:POPS:SoyPI:Egg SM:chol in a 10:5:2:1:2:10 mole ratio. For this study, we were fortunate to obtain the trimer version of FP23 due to the efforts of Wei Chang, a graduate student at Michigan State. As shown in Table 1, we found that KC was also dramatically decreased in LM3 due to the effects of FP23 and of FP-trimer (FPtri). We also tested a water-soluble monomeric form of FP23, FPwsm. Even though this peptide was initially co-solubilized in organic solvent with the lipid, it partitioned into the aqueous phase and had little effect on LM3. When electron density profiles were constructed, we found that both FP23 and FPtri localized near the interfacial region, as shown in Fig. 8 (dotted lines). At this location, FP23 can act as a wedge to induce curvature in a flat bilayer, thus facilitating the formation of fusion intermediates.
Table1.
Bending modulus KC
| Lipid | x Chol | Peptide | x FP | KC/kT |
|---|---|---|---|---|
| LM3 | 0.33 | 0 | 32±2 | |
| LM3 | 0.33 | FP23 | 0.048 | 5.3±0.3 |
| LM3 | 0.33 | FP23 | 0.091 | 4.4±0.3 |
| LM3 | 0.33 | FPtri | 0.016 | 6.6±1.3 |
| LM3 | 0.33 | FPwsm | 0.048 | 25±2 |
| DOPC | 0 | 19±0.9 | ||
| DOPC | FP23 | 0.063 | 6.2±0.4 | |
| DOPC | 0.3 | 0 | 18±0.4 | |
| DOPC | 0.3 | FP23 | 0.063 | 5.8±0.7 |
| diC22:1PC | 0 | 31±3 | ||
| diC22:1PC | FP23 | 0.063 | 2.4±0.8 | |
| diC22:1PC | 0.3 | 0 | 30±3 | |
| diC22:1PCl | 0.3 | FP23 | 0.063 | 2.0±0.8 |
Adapted from Ref. [15].
Figure 8.
(right). Electron density vs. distance (z) from the bilayer center for samples of (a) LM3, (b) .048 mole fraction FP23/LM3, and (c) .016 mole fraction FPtri/LM3. Line colors: Black is total electron density, blue is water, red is phosphate plus average headgroup moiety, green is glycerol-carbonyl, purple is hydrocarbon region including cholesterol, black dotted line is peptide. (c) shows part of a second adjacent bilayer at large values of z, and the presence of a large water spacing between adjacent bilayer is emphasized in all panels. Adapted from Ref. [6].
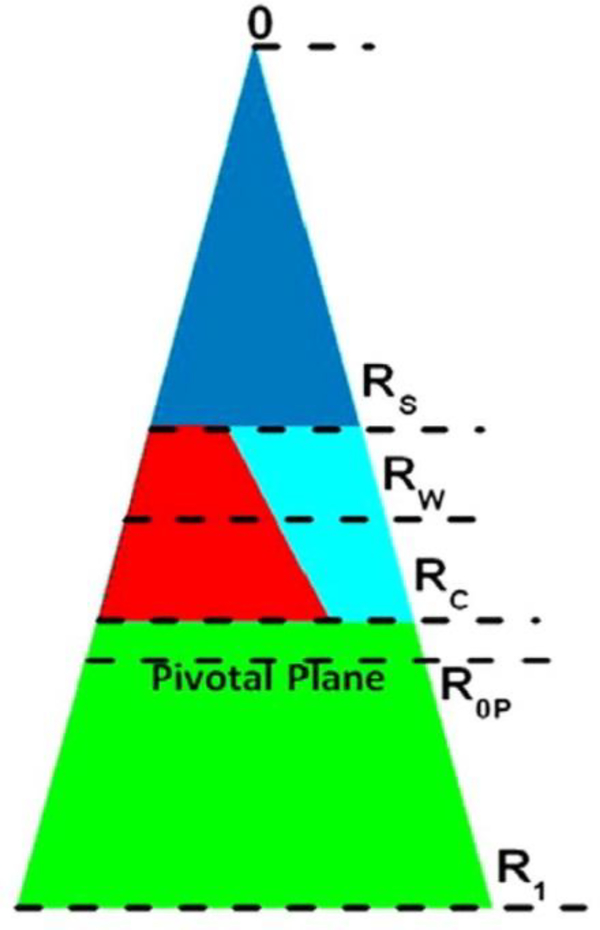
While the work reviewed so far used diffuse scattering, a related standard method to study spontaneous curvature employs X-ray scattering from lipids such as DOPE that form hexagonal II phases [48, 49]. Using this method it is possible to quantify curvature as the inverse of the radius of the pivotal plane, defined as that radius at which the area does not change as the monolayer bends upon hydration. Fig. 9 shows the location of the pivotal plane (R0P) as well as other locations of interest, which include RW, the radius of the HII water cylinder. In addition, RS is the radius from the center of the HII tube to the lipid steric thickness, which bisects the headgroup region with associated water; RC is the radius to the start of the hydrocarbon region; R1 is the effective lipid radius, including void region. Using these radii and the equations of Leikin et al. [50], we calculate three areas: AW, the area per lipid molecule at the Luzzati plane (RW), AP, the area per unit cell at the pivotal plane (R0P), and A1, the area corresponding to R1. Table 2 gives numerical values for these quantities. The main result is that FP23 decreases R0P, thereby increasing the spontaneous curvature of the mixture beyond that of the already highly curved HII DOPE lipid. Since 1/R0P = the spontaneous curvature (CS), then the energy for forming fusion intermediates is further reduced beyond the KC reduction, since E=KC/2(C-CS)2.
Figure 9.
Schematic for the packing of lipid and water in the hexagonal II phase. The cylindrical water tubes extend in the direction perpendicular to the page, with the tube’s center at 0. The triangle extending from 0 to R1 represents the piece-of-pie space occupied by a monolayer of lipid and peptide and its associated water. The dark blue triangle is occupied by water. The lower green trapezoid is the hydrophobic volume. The region between RS and RC represents the interfacial region, which is occupied by water (light blue) and lipid headgroup (red). FP23 is located partially in the red region and partially in the green region. The pivotal plane is at R0P. Figure from Ref. [6].
Table 2.
Radial and areal results for DOPE and 2 mol% FP23/DOPE
| Sample | ϕW | D hex | R W | R 0P | R S | R C | R 1 | A W | A P | A 1 |
|---|---|---|---|---|---|---|---|---|---|---|
| DOPE Ref.[50] | 0.30 | 63.8 | 21.1 | 28.5 | — | — | — | 49.6 | 64.2 | — |
| DOPE | 0.30 | 63.3 | 21.0 | 27.3 | 16.9 | 25.6 | 38.5 | 50.0 | 65.0 | 91.6 |
| DOPE/FP23 | 0.28 | 62.3 | 20.0 | 25.6 | 15.8 | 24.5 | 37.8 | 50.1 | 64.2 | 94.5 |
Units are in the appropriate powers of Angstroms. Table from Ref. [6].
5. CRAC motif on MPER in gp41
The membrane proximal external region (MPER) region is in close proximity to the transmembrane region of the HIV-1 membrane and this region contains a highly-conserved consensus sequence having the pattern L/V-X(1–5)-Y-X(1–5)-R/K in which X(1–5) represents 1–5 residues of any amino acid. In HIV-1 this sequence is LWYIK, which has been shown to bind to cholesterol; mutations in the CRAC sequence reduce HIV infectivity [51]. In a collaboration with the Epand lab [5], LWYIK was compared to IWYIK, a non-CRAC motif peptide, in the absence and presence of cholesterol. Previous calorimetric work using SOPC/cholesterol (1:1 molar ratio) mixtures suggested that LWYIK binds directly to cholesterol, while IWYIK binds to SOPC [52, 53]. In Ref. [5] in addition to obtaining KC, we obtained Sxray, a chain order parameter. This method was developed in our lab due to the efforts of Thalia Mills, our postdoc [54, 55]; it analyzes diffuse X-ray scattering in the wide-angle region due to chain-chain correlations. The analysis uses a Mauer-Saupe distribution of tilt angles to model the chain disorder to yield Sxray, which was shown to parallel SCD from NMR [54]. Table 3 shows the KC and Sxray results when LWYIK and IWYIK were added to SOPC or SOPC/cholesterol (30 mole percent). As shown, both LWYIK and IWYIK at 9:1 lipid:peptide mole ratio decreased KC, thus softening SOPC or SOPC/cholesterol, where this effect was slightly greater with IWYIK. In addition, both peptides disordered lipid chains as evidenced by a decrease in Sxray. More details of our wide-angle X-ray scattering (WAXS) methods are given in Supplementary Material. Table 3 also shows structural results that are obtained using form factors and electron density profiles as described below.
Table 3.
Structure and interaction results
| Sample | KC/kT | 2DC (Å) | A+ (Å2) | Zpep | Sxray |
|---|---|---|---|---|---|
| SOPC | 21.5±2.8 | 29.2±0.4 | 67.0±0.9 | --- | 0.33 ± 0.03 |
| SOPC/I (9:1) | 5.5±0.5 | 26.1 | 75.7 | 17.3 | 0.22 ± 0.02 |
| SOPC/L (9:1) | 10.0±1.0 | 26.2 | 75.8 | 17.0 | 0.24 ± 0.02 |
| SOPC/Chol (7:3) | 32.3±1.2* | 32.9* | 73.8±1.0* | --- | 0.65 ± 0.06 |
| SOPC/Chol (7:3)/ I (9:1) | 5.7±3.1 | 29.1±1.5** | 109±5** | 10±0** | 0.39 ± 0.04 |
| SOPC/Chol (7:3)/ L (9:1) | 9.1±1.2 | 29.8±2.8** | 111±8** | 10.5±0.3** | 0.37 ± 0.04 |
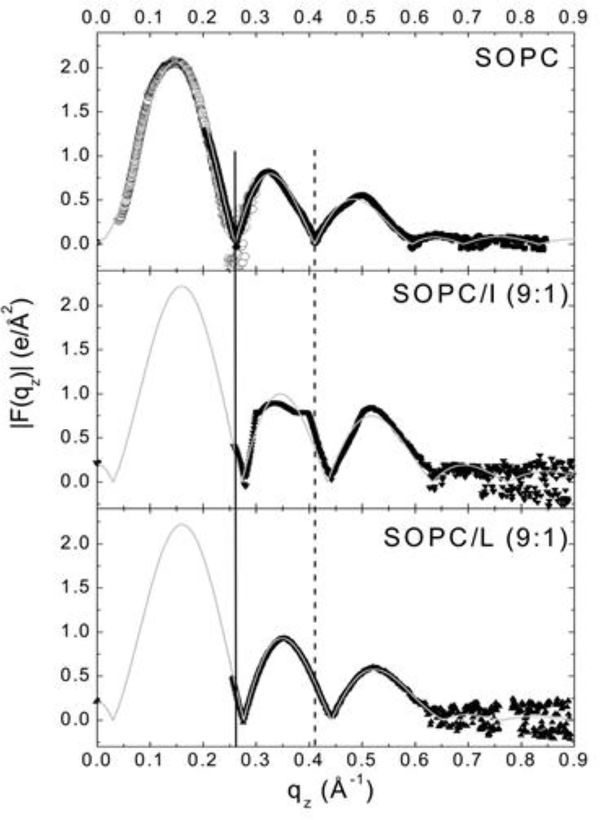
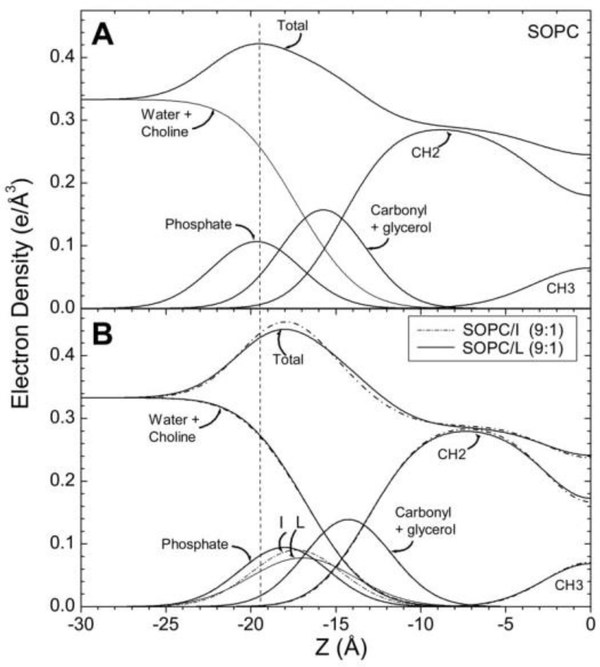
In order to obtain electron density profiles, form factor data are collected from the intensity patterns, similar to those shown in Fig. 3, as described previously [3, 4]. Form factor data are shown in Fig. 10 for SOPC containing LWYIK (L) or IWYIK (I). Fig. 10 shows a shift of the form factor data of SOPC containing either LWYIK or IWYIK to higher qz, indicating a thinning of the SOPC bilayer. When the form factor data are fit to a model of the bilayer, electron density profiles are constructed [57] (shown in Fig. 11), and structural changes are quantitated. Thinning of the hydrocarbon thickness (2DC) by ~3 Å was caused by the addition of either LWYIK or IWYIK, and both peptides located in the headgroup region near the phosphate group in pure SOPC. When 30% cholesterol was added to SOPC, then both peptides located in the hydrocarbon region, ~10 Å from the bilayer center (previously unpublished). Therefore, these structural, elastic and chain order results surprisingly did not exhibit large differences between LWYIK and IWYIK.
Figure 10.
(left). |F(qz)| data for SOPC, SOPC/IWYIK (9:1) and SOPC/LWYIK (9:1). Solid symbols represent data from oriented samples. Open symbols for only SOPC were obtained from unilamellar vesicles (ULV). The solid and dotted vertical lines show shifts in the qz values for the zeros compared to SOPC. The thin solid curves show the fits of the model to the data. Figure from Ref. [5].
Figure 11.
(right). Electron density profiles of SOPC (A) and SOPC/peptides (B) obtained using the H2 model with a Gaussian added for the peptides. Component groups are labeled and the thinning of the bilayer caused by either peptide is emphasized by the dotted vertical line. Figure from Ref. [5].
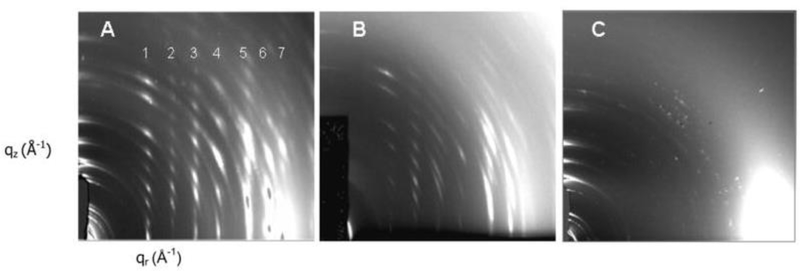
However, when the concentration of cholesterol was increased to 50 mole percent, which is closer to the 45 mole percent in the HIV viral membrane [58], dramatic differences were observed in the ability of LWYIK vs. IWYIK to crystallize out cholesterol. Either peptide induced the initial formation of oriented, 3D crystals during the rock and roll thin film procedure [37], which produced a highly crystalline oriented pattern as shown in Fig. 12A and B, as compared with a few unoriented crystalline reflections in the wide-angle pattern of dried SOPC/50% cholesterol membranes (12C). The crystalline pattern in Fig. 12A caused by IWYIK had stronger, sharper reflections in several samples than did the LWYIK patterns, where in both patterns the spacings are consistent with the ~17 Å dimension of the cholesterol molecule [5]. When these samples were hydrated, the crystalline patterns dissipated, but more rapidly for the crystals formed by LWYIK. We interpret these patterns to indicate that LWYIK binds directly to cholesterol, thus interfering with its ability to form highly crystalline arrays, while IWYIK binds to the nearby SOPC lipid, thus sequestering cholesterol away from the lipid, allowing it to form long-range, highly crystalline arrays. This could be an indication that in vivo, LWYIK binds directly to cholesterol and aids in the fusion process of the viral and host cell membranes.
Figure 12.
2D CCD images of crystalline X-ray wide-angle diffraction patterns in the dried state of (A) SOPC/chol (5:5)/I (9:1), (B) SOPC/chol (5:5)/L (9:1), (C) SOPC/chol (5:5). Figure from Ref. 5.
Our result that LWYIK resides in the hydrocarbon interior in SOPC/Chol (7:3) membranes, but in the headgroup region in SOPC membranes, disagrees with the result of the Brown lab [59], who found the CRAC motif bacterial leukotoxin to remain at the bilayer surface of ULV containing 33% cholesterol using fluorescence and MD simulation [60], and also disagrees with the molecular simulation of Epand’s lab that found LWYIK to reside at the interface in cholesterol-containing membranes [61]. However, in another investigation from the Epand lab, a nearest neighbor recognition (NNR) study demonstrated that LWYIK is sensitive to the packing of the bilayer, suggesting that the peptide must at least partially penetrate into the membrane [52]. The fact that LWYIK caused the Sxray value to decrease by a factor of 2 in SOPC/Chol (7:3) membranes, while only slightly in SOPC membranes, supports the differential location of the CRAC motif found in the electron density profiles in our study. In another study, fluorescence measurements of the tryptophan moiety showed that either LWYIK or KIYWL tethered to cholesterol or to a phospholipid favor the bilayer interface region in liquid-disordered lipids [62]. This agrees with our location of free LWYIK in pure SOPC membranes.
6. LLP2 on CTT in gp41
The motivation for this study was to determine how a moiety on the cytoplasmic C-terminal tail (CTT) of the HIV-1 virion (see Fig. 2) is able to affect the fusion process that occurs externally. The biochemical question is: How does a signal get transmitted to the other side of the membrane? The physics question is: How does the membrane carry information from one side to the other? Three conserved amphipathic, primarily α-helical segments in the CTT referred to as lentiviral lytic peptides (LLP1, LLP2 and LLP3) have been shown to bind and perturb membranes [63–67]. To understand the function of the CTT, deletions of the entire CTT have been carried out with different results depending upon cell type. For so-called permissive cell types, deletion has little effect on Env (gp120 and gp41) incorporation into virions [68, 69]. However, the majority of T-cell lines are non-permissive in that CTT deletion results in a 10-fold reduction in Env incorporation and a complete blockage of viral replication [70]. Murakami and Freed found the block to be at the early stage of Env incorporation [68]. Less drastic changes in the CTT have been carried out using site-directed mutations. Kalia et al. [71] produced a mutant MX2 of LLP2 where two highly conserved arginine residues were replaced with negatively charged glutamic acid, thus decreasing the net charge from +3 to −1. In HIV virions, MX2 mutation showed no loss of Env expression or infectivity, while when MX2 was in the T-cell membrane, both viral-initiated cell death and syncytium formation (cell-cell fusion) were greatly decreased [71], which was similar in permissive and non-permissive cell types. In Ref. [15] we collaborated with the Montelaro lab to employ membrane mimics of both HIV-1 virion and T-cell membranes, and added WT LLP2 and the MX2 mutated form of LLP2. The lipid compositions were based on [58, 72] and are shown in Table 4. In addition to the HIV, HIV minus negative charge and T-cell membrane mimics, an HIV mimic devoid of cholesterol was also attempted, but this membrane phase separated into fluid and gel phases and was thus not analyzable. A lipid extract from attenuated HIV-1 virions provided another membrane mimic.
Table 4.
Lipid composition of membrane mimics
| Lipid | Mole percentage | ||
|---|---|---|---|
| HIV | HIV-neg | T-cell | |
| C 16 Ceramide | 0.8 | 1.0 | 1.1 |
| Soy Phosphoinositol (PI) | 1.3 | 0 | 6.3 |
| Brain PI(4)P | 1.3 | 0 | 0.9 |
| Brain PI(4,5)P2 | 1.3 | 0 | 1.1 |
| 12:0 diHydroSphingomyelin (diHSM) | 1.7 | 1.8 | 2.7 |
| PO-Phosphoethanolamine (POPE) | 4.2 | 4.7 | 6.3 |
| PO-Phosphoserine (POPS) | 7.5 | 0 | 9.1 |
| Plasmalogen PE | 7.5 | 8.5 | 10.9 |
| Egg Sphingomyelin (Egg SM) | 14.2 | 16.0 | 16.3 |
| PO-PhosphoCholine (POPC) | 15.1 | 17.0 | 15.4 |
| Cholesterol (Chol) | 45.2 | 51.0 | 29.9 |
When the diffuse X-ray scattering data were analyzed as detailed above, the results in Table 5 were obtained. Both WT LLP2 and MX2 caused membrane softening as indicated by a lowered KC, which was even lower for MX2, in T-cell membrane mimics. There was little change in Sxray order parameter, indicating that the lipid chains were not the cause of the membrane softening. In the HIV and HIV-neg mimics, the KC value was quite low for the control lipid, indicating a soft membrane even before peptides were added. This was also true for the membranes created from lipids extracted from HIV virions. These lower KC values (6–8.2/kT) are surprising because of the high cholesterol content (45 mole %) of these membranes [56]. The KC values and Sxray order parameters were little affected by the addition of either peptide to HIV, HIV-neg or HIV extracted membranes. Therefore, this study showed that very realistic mimics of the T-cell and HIV-1 virion membranes have quite different elastic moduli. Since both WT and MX2 lowered KC, as we saw previously with FP23 and with the CRAC motif, this cannot be the source of the difference between their mechanisms of action. To explore this further, we obtained electron density profiles.
Table 5.
Bending and chain order results
| Sample | T-cell mimic | HIV mimic | HIV-neg mimic | HIV extracted | |||
|---|---|---|---|---|---|---|---|
| KC | Sxray | KC | Sxray | KC | Sxray | KC | |
| Membrane | 16.6 | 0.71 | 8.2 | 0.80 | 7.0 | 0.80 | 6.1 |
| /WT(40:1) | 12.6 | 0.72 | 8.2 | 0.79 | 7.0 | 0.78 | 6.8 |
| /WT(20:1) | 7.7 | 0.73 | 5.4 | 0.78 | – | 0.81 | – |
| /MX2(40:1) | 5.3 | 0.67 | 6.8 | 0.77 | 6.8 | 0.79 | * |
| /MX2(20:1) | 4.1 | 0.72 | 8.9 | 0.80 | 6.5 | 0.77 | – |
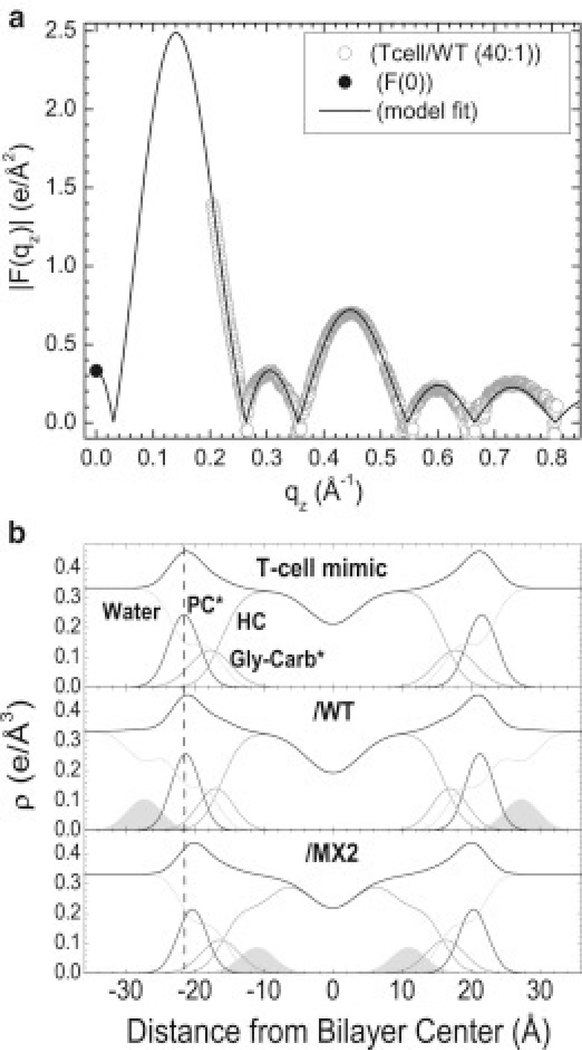
Fig.13A shows a typical form factor and fit to the Scattering Density Profile (SDP) model program [73] from the T-cell X-ray data with WT, 40:1 lipid:peptide molar ratio [15]. When X-ray data from HIV membrane mimics were analyzed, there was no difference in the form factors when peptides were added compared to the control, indicating that the peptides had not penetrated the HIV membranes. This is the reason that there was also no change in KC or Sxray. Fig. 13B shows the electron density profiles for T-cell membranes and with 40:1 lipid:peptide mole ratio. As shown, there was slight thinning in membrane thickness when WT LLP2 was added, while there was a greater thinning when MX2 was added to the T-cell mimic. In addition, there was a deeper penetration of MX2 into the hydrocarbon core, while WT LLP2 remained outside of the phosphate headgroup. A theoretical challenge is to understand how the cytoplasmic CTT influences the extracellular domain of the gp41 Env protein. A potential biophysical mechanism involves the lateral pressure profile of the membrane [74], which would be affected locally by embedded LLP2 and which would then affect other nearby parts of gp41, such as the membrane spanning domain (MSD). Perturbation of the MSD of gp41 could affect the conformational changes necessary for fusion as several studies with modified MSDs have demonstrated inhibition of fusion [75–77]. With regard to the T-cell membrane, the deeper embedding of MX2 would alter the lateral pressure profile compared to WT and that would affect the TMD of the gp41 Env protein differently, correlating well to the inhibition of cell-cell fusion upon mutation. With regard to the HIV membrane mimic, the very weak interaction of LLP2, either WT or MX2, predicts that MX2 would have little effect on Env incorporation or infectivity, as observed experimentally. The cartoon in Fig. 14 summarizes the interaction of LLP2 WT and MX2 with the two membrane mimics.
Figure 13.
(a) Model fit to |F(qz)| obtained from X-ray data and F(0) obtained from volumetric data for T-cell/WT. (b) Electron density profiles for T-cell mimic control with WT and MX2 at 40:1 lipid:peptide molar ratio. The component groups are water, PC* (phosphocholine*), Gly-carb* (glycerol plus carbonyls*), HC (lipid hydrocarbon chains plus cholesterol), and LLP2 peptides (shaded). * indicates a weighted average group. Vertical dotted line facilitates comparison of membrane thickness. Figure from Ref. [15].
Figure 14.
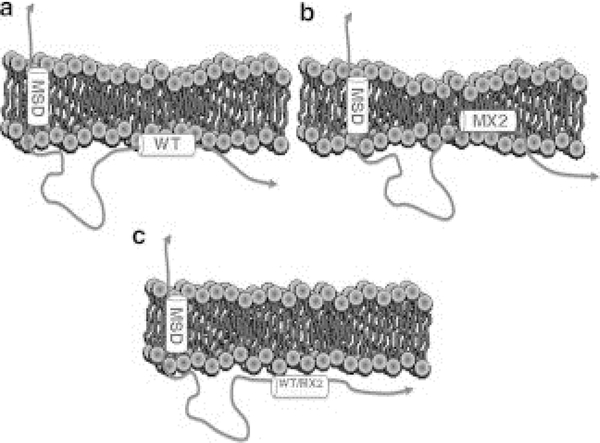
Cartoon showing differences in the interaction of LLP2 with T-cell and HIV membranes. a. WT LLP2 binds to the outer headgroup region and thins the T-cell membrane. b. MX2 binds more deeply into the hydrocarbon interior and thins the T-cell membrane to a greater extent. c. WT and MX2 LLP2 only weakly bind to the HIV virion membrane. MSD, membrane spanning domain. Figure from Ref. [15].
7. Tat47–57 peptide
As shown in Fig. 1, Tat translocates through the T-cell nuclear membrane to enhance transcription of DNA into RNA. Two investigations from our lab [16, 17] focused on this transactivator of transcription, Tat, which plays a role in AIDS progression. Tat peptide Y47GRKKRRQRRR57 is part of a larger 86-amino acid protein that transactivates a gene on the HIV-1 promoter [78, 79], and is thought to be responsible for cellular uptake [80]. In order to do this, Tat must translocate from the cytoplasm into the nucleus across the nuclear membrane. Tat peptide47–57 is particularly rich in basic amino acids; deletion of three out of eight positive charges in this region caused loss of its ability to translocate [80]. Many cell penetrating peptides (CPPs), including those derived from Tat, are capable of carrying cargo into live cells [81, 82]. It is controversial, however, if endocytosis is involved, which would require ATP [80, 83–86]. To address this question, many biophysical studies have used simplified models of biological membranes composed of a small number of lipid types including phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidylglycerol (PG). For example, Mishra et al. reported that rhodamine-tagged Tat47–57 enters giant unilamellar vesicles (GUVs) composed of PS/PC (1:4 mole ratio) immeasurably slowly, but crosses a GUV composed of PS/PC/PE (1:2:1) lipids within 30 s [87]. In another experiment, fluorescently labeled Tat48–57 did not enter GUVs containing only PC with 20 mole % cholesterol, but translocated rapidly when PS or PE was included with PC [88]. Therefore, Tat-derived peptides can translocate across bilayers without ATP, but this depends on the lipid type. The mechanism by which bilayer translocation occurs remains unknown; our study attempted to delineate Tat’s effect upon and location in membrane mimics in order to elucidate the mechanism of Tat’s translocation.
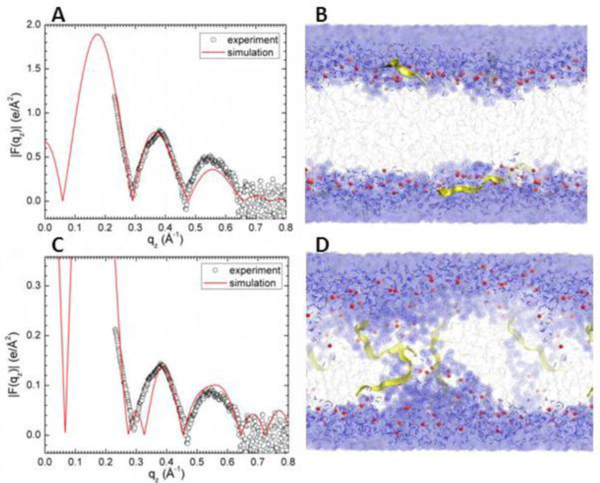
In both of our investigations we collaborated with Angel Garcia’s group, pairing MD simulation with our X-ray data. Grad student Kiyo Akabori led the experimental research effort in our lab. Using the Sim-to-Exp program [89], form factor data are calculated from the simulation’s trajectories; these are compared directly to the form factors that result from the X-ray data. Since peptides move slowly in atomistic simulations, grad student Kun Huang in the Garcia lab constrained the peptide positions using umbrella potentials at desired depths with a force constant of 3000 KJ/mol/nm2. Figure 15 shows one of these comparisons. In Fig. 15A, a mixture of Tat and the neutral lipid DOPC (33:1 lipid:peptide mole ratio) was simulated (red line) and compared to the X-ray form factor data (open black circles) of the same mixture. As shown, there is good visual agreement when Tat is constrained to 18 Å from the bilayer center (in the headgroup region). A snapshot of this simulation is shown in Fig. 15B. By contrast, when Tat was constrained to 5 Å from the bilayer center, i.e., near the center of the bilayer, the fit to the data was much worse and the chi-square was larger (see legend). Therefore, these results suggested that Tat does not translocate easily through DOPC. Fig. 16 shows the electron density profile corresponding to Fig. 15A. These comparisons informed construction of the cartoon shown in Fig. 17.
Figure 15.
MD simulated form factors (red solid lines in A and C of Tat/DOPC+Tat, x = 0.03), with Tat fixed at ZTat = 18 Å (A) and 5 Å (C) from the bilayer center compared to experimental form factors (open circles). Corresponding snapshots are shown in B and D in which the lipid chains are represented as gray sticks on a white background, Tats are yellow, phosphate groups are red and water is blue. Chi-square of the fit was 14.8 for A and 80.3 for C. Figure from Ref. [16].
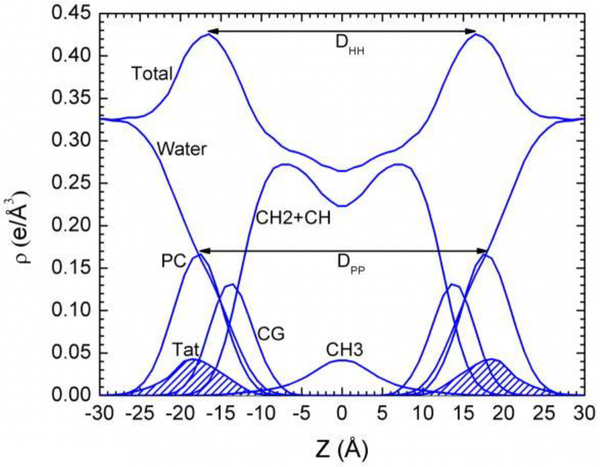
Figure 16.
Symmetrized total electron density profile (EDP) from the simulation with the form factor shown in Fig. 15A. Also shown are the component group contributions. Component groups are labeled: PC, phosphocholine: CG, carbonyl-glycerol; CH2 + CH, methylene and methane hydrocarbon region; CH3, terminal methyl; Tat peptide distribution is shaded. Figure from Ref. [16].
Figure 17.
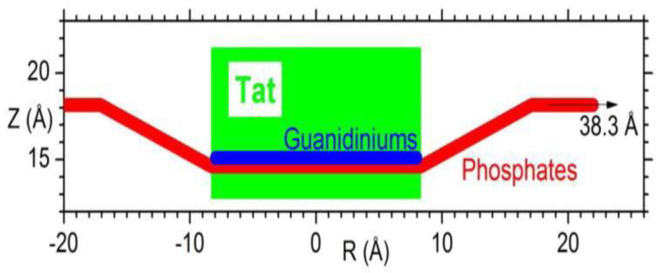
Location of Tat in DOPC bilayer. Tat is represented as a cylinder, Z is the distance from the bilayer center, and R is the in-plane distance from the center of Tat. The average Z of the lipid phosphates as a function of R and the arginine guanidiniums are shown in red and blue respectively. Figure from Ref. [16].
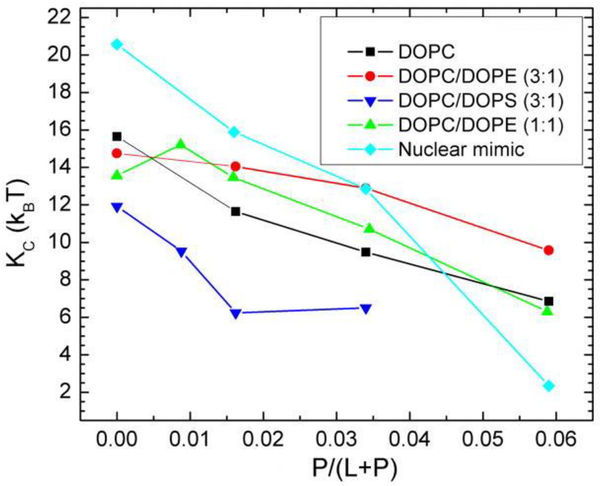
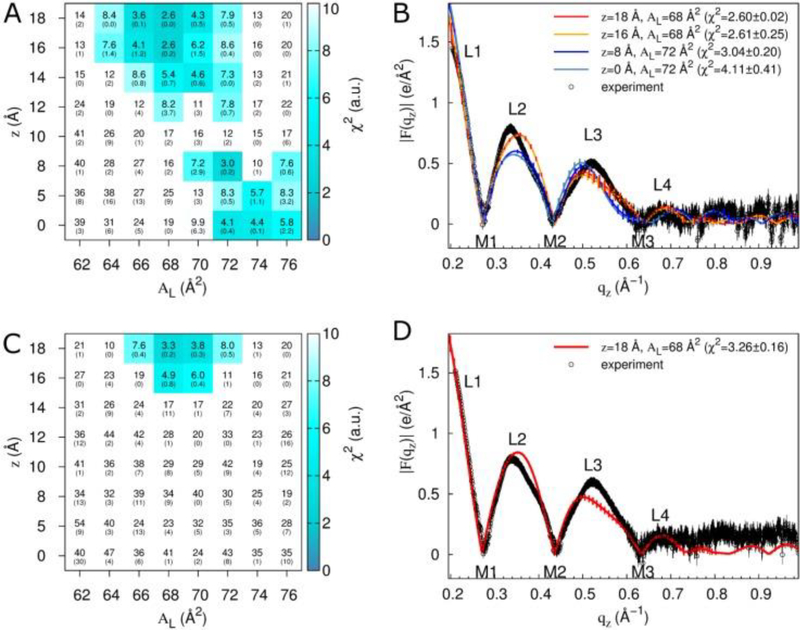
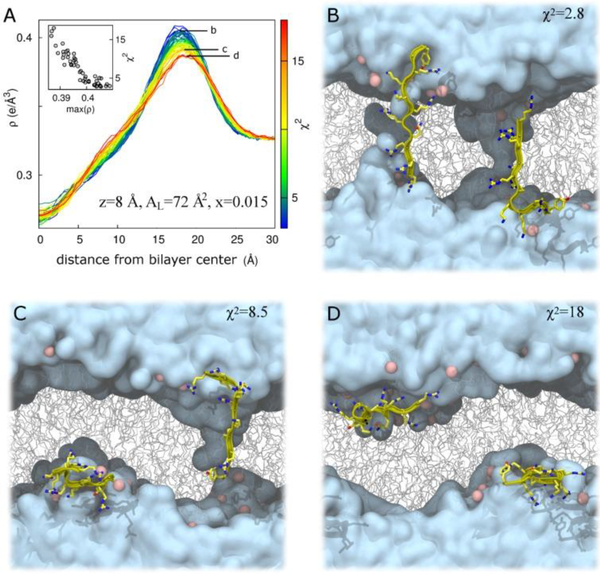
In the same investigation, four other membrane mimics were also studied [16] (see Fig. 18). In all of these mixtures, Tat peptide decreased the bending modulus KC as shown in Fig. 18. Thus, like FP23, Tat lowers the energy required to bend a membrane which could be crucial for its translocation through membranes. While Ref. [16] showed a clear preference for the headgroup location for Tat in DOPC, two locations, in the headgroup, and near the bilayer center, were found for the DOPC:DOPE 1:1 mixture as detailed in the Supplementary Material to Ref. [16]. To pursue this further, the second investigation [17] focused on the DOPC:DOPE 1:1 lipid mixture. Due to the efforts of Garcia’s postdoc, Chris Neale, a matrix of Tat positions was investigated, also varying the area/lipid AL and the Tat concentration. In total Chris carried out over 500 100-nsec atomistic GROMACS simulations. As shown in Fig. 19A, at the lower concentration of Tat in the lipid mixture, two clusters of blue squares, indicating a low chi-square, appear. One cluster is near zTat = 16–18 Å and a second cluster is near zTat = 0–8 Å. The smallest chi-squares are shown on the form factor graphs in 19B and D, where the solid lines represent the simulated form factors and the open black circles the experimental form factors. This indicates that it is equally probable to find Tat near the center of the bilayer as in the headgroup region for a lipid:peptide molar ratio of 67:1. At the higher concentration, 33:1, Tat remains in the headgroup region, possibly due to crowding. When Tat is constrained at 8 Å from the bilayer center, if bilayer undulations are large, then Tat could be located in the headgroup region. This is shown in Fig. 20C and D. However, in those cases, the chi-square of the fit increases dramatically, thus ruling out these possibilities. What is remarkable is that in the case of the favored position in Fig 20B, Tat spans the bilayer, interacting with the headgroups of both bilayer leaflets and drawing water and ions into the bilayer’s hydrophobic core. When Tat is restrained at 8 Å from the DOPC:DOPE (1:1) bilayer center, the hydrocarbon thickness 2DC decreases by 2.5 Å, which leads to a 6 Å2 increase in AL when volume is also measured.
Figure 18.
Bilayer bending modulus, KC, vs. P/(L+P) mole fraction. D-spacings for DOPC/Tat mixtures varied from 64 to 68 Å, for DOPC/DOPE/Tat mixtures from 64 to 69 Å, for DOPC/DOPS/Tat (3:1) mixtures from 57 Å to >100 Å (pure DOPS was unbound), and for nuclear mimic/Tat mixtures from unbound (control) to 64 Å. Estimated uncertainty in all values is ± 2. Nuclear mimic is composed of POPC/POPE/POPS/SoyPI/Cholesterol 69:15:2:4:11 based on Ref. [90]. Figure from Ref. [16].
Figure 19.
Fits of simulation to experiment for a systematic evaluation of zTat and AL in DOPC:DOPE (1:1). Data are from simulations with two (A,B) or four (C,D) molecules of Tat47–57 (simulation mole fractions of 0.015 and 0.030, respectively; comparisons are to experimental data at mole fractions of 0.016 and 0.034, respectively). (A,C) Color and inset numbers represent χ2 values (standard deviations of averages from repeat simulations in parentheses). (B,D) Selected form factors (χ2 values in parentheses). Out-of-plane scattering lobes (L) and minima (M) are numbered based on experimental data, which are averaged over two (B) and nine (D) experiments. Black vertical lines depict experimental error estimates (see Methods). Vertical lines on the simulation data depict standard deviations between two starting configurations. Experimental data in (B) and (D) were scaled to the simulations at z = 18 Å, AL = 68 Å2. Figure from Ref. [17].
Figure 20.
Bilayer undulations increase χ2 in DOPC:DOPE (1:1) at z = 8 Å, AL = 72 Å2 for a Tat47–57 mole fraction of 0.015. (A) Total system electron density, ρ, as a function of absolute distance from the global bilayer center along its global normal for N = 52 simulations, colored by χ2 value representing a range from best (blue) to worst (red) fit to experiment. Inset, χ2 as a function of the maximum value of ρ. Labels b, c, and d in part A identify electron density curves for systems whose snapshots are shown in parts B, C, and D, respectively. (B–D) Snapshots after 100 ns of simulation in repeats with χ2 values of 2.8 (B); 8.5 (C); and 18 (D). Water is cyan, lipids are gray, lipid headgroup phosphorus atoms are pink, and protein is yellow. Protein is rendered in front of all other molecules for clarity. Figure from Ref. [17].
Our main results that Tat47–57 in DOPC remains in the headgroup region, and that it is located either in the headgroup region or near the center of the bilayer in DOPC:DOPE (1:1) mixtures, supports a previous suggestion that non-bilayer forming lipids are required for Tat translocation across membranes (Ref. [91] and references within). To compare to literature, Bradshaw’s lab has published two neutron studies of Tat in membranes. Ref. [92] studied mixtures of Tat with DOPC:DOPS (80:20 molar ratio) and found Tat to be located in the headgroup near the glycerol/carbonyl region, similar to our result for Tat in pure DOPC [16]. However, in pure DOPC, Bradshaw’s lab found Tat to be located deep in the hydrocarbon region, ~6 Å from the bilayer center [93]. This is in direct contradiction to our result, where we found that DOPE is necessary for Tat internalization in the hydrocarbon core. The reason for these different results could stem from the degree of hydration. In their experiment, the highest hydration level was 98% relative humidity (RH). We have previously shown that even at 99% RH, oriented bilayers are still 10 Å (~10 water molecules) below their full hydration value [23]. When there is not enough water between bilayers in a stack, then a peptide may be forced into the hydrocarbon region. In our experiment, there is more than sufficient water for the peptide to partition naturally. In another study, Shin’s lab used neutron reflectivity (NR) of Tat in a DPPC bilayer to show that Tat entered into the hydrocarbon region [94]. However, high temperature and high concentration manipulation were required for this result, since their NR measurements occurred at 25 °C, where DPPC is in the gel phase, so their result may not be physiologically relevant.
8. HIV-1 matrix-31 membrane binding peptide
Retroviral replication occurs as the virus acquires its lipid envelope at the infected cell’s plasma membrane (PM) [95], as shown in Fig. 1. Many complex steps occur during viral budding, which is mediated by the viral structural protein Gag [96]. Human immunodeficiency virus (HIV-1) Gag is synthesized as a precursor polyprotein, Pr55Gag, which is cleaved into four major domains upon release: N-terminal matrix (MA), capsid (CA), nucleocapsid (NC) and P6 [97]. The membrane binding domain is MA, which was the subject of our study [18]. This binding depends on a conserved highly basic region (HBR) in the 131 amino acid MA protein [98, 99], as well as on the myristoyl moiety at MA’s N-terminus. The binding is enhanced when a conformational switch removes the myristate moiety from its sequestered domain [100], as the HBR interacts with acidic phospholipids on the inner leaflet of the PM [98, 100]. Gag is targeted to the PM due to the interaction between the MA HBR and phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2 or PIP2), where it is concentrated. Specific binding of HIV-1 Gag to PIP2 has been demonstrated by several experimental approaches [72, 101, 102]. In our work, interactions between the HIV-1 Gag MA membrane binding interface with lipid bilayers were studied using X-ray diffuse scattering to obtain the membrane bendng rigidity, chain order, area/lipid and bilayer thickness. In a collaboration with the Lösche/Heinrich lab, we used NR to determine the peptide position in bilayers. The model peptide, called MA31myr, includes the main membrane interaction sites, myristoyl and the HBR, of the full-length MA protein. For contrast we also studied MA31 without the myristoyl group. We compared the interaction of MA31 and MA31myr with PIP2 or PS-containing membranes in an effort to understand the requirement of PIP2 for viral budding.
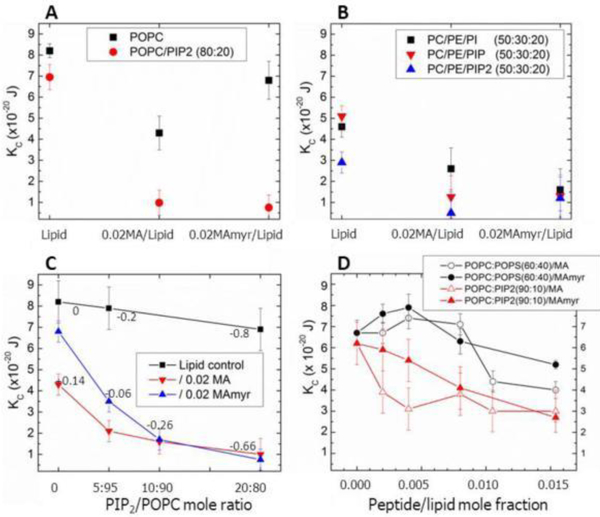
Similarly to the above studies, we first determined KC, the bending modulus of different lipid/peptide interactions. As shown in Fig. 21A, there was a greater decrease in KC when MA31 was added to the neutral lipid POPC than when MA31myr was added (black squares). This was at first surprising, but it indicates that the myristoyl group has a stabilizing effect on a neutral membrane. However, when 20 mole % of PIP2 was added to POPC, the difference caused by the myristoyl group disappeared, indicating that the electrostatic interaction dominated. The negative charge on PIP2 caused a decrease in KC in the presence of either peptide. Without the peptides, adding PIP2 to POPC caused only a small decrease in KC (21C, black squares). Fig. 21B again shows that differences between MA31myr and MA31 were largely suppressed by the addition of any of the negatively charged lipids, PI, PIP or PIP2 to POPC/POPE mixtures. In Fig. 21C, all differences between MA31myr and MA31 are gone when the overall net negative charge is at least −0.26. Fig. 21D shows a dramatic difference between the behavior of PS-containing membranes and PIP2-containing membranes: those with PS first stiffen the membrane as either peptide is added, while those with PIP2 are generally softened (lowered KC), with either peptide. These lipid mixtures were designed to achieve equal overal net negative charge.
Figure 21.
Bending modulus, KC, as a function of lipid and peptide concentration. (A) 0.02 MA31 or MA31myr in POPC, in POPC/PIP2 (80:20), (B) 0.02 MA31 and MA31myr in POPC/POPE/PI (50:30:20), in POPC/POPE/PIP (50:30:20), in POPC/POPE/PIP2 (50:30:20), (C) 0.02 MA31 or MA31myr in POPC with increasing PIP2, (D) Increasing MA31 or MA31myr in POPC/POPS (60:40) or POPC/PIP2 (90:10). Numbers in Fig. 21C indicate the net membrane charge/lipid plus protein. Lipid control indicates samples with no peptides. Error bars represent standard deviations of the average KC values from 2 to 5 different samples. Figure from Ref. [18]. To compare KC units in Joules to kT units, divide by 0.43 × 10−20 J.
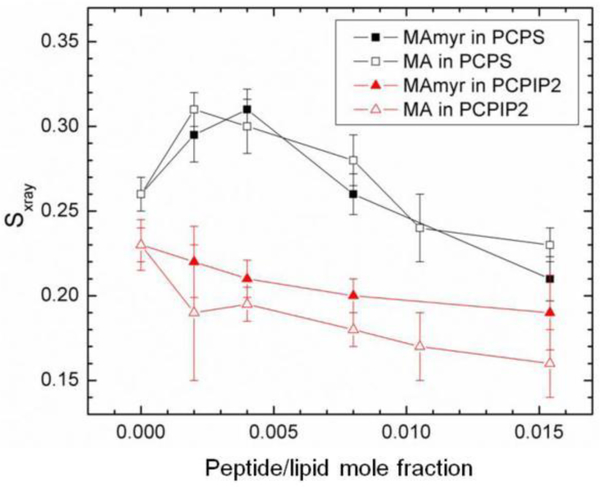
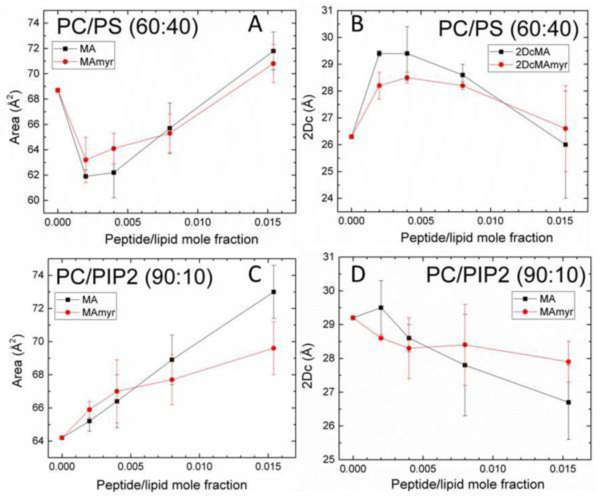
Fig. 22 shows the chain order Sxray results as a function of peptide concentration for the series of samples in Fig. 21D. Chain order initially increased, then decreased with increasing concentration of both peptides in POPC:POPS (60:40) membranes, similar to the trend observed in the KC results. The structural results in Fig. 23 followed a similar trend; first a membrane thickening followed by a membrane thinning at higher peptide concentration. The thickening is due to an increase in chain order which causes the lipid chains to extend, but at higher concentrations this reverses, causing the membrane to thin back to the control value. In contrast to POPC/POPS, a very gradual decrease in membrane order occurred in the POPC/PIP2 (90:10) samples (Fig. 22), which is consistent with the 2.5 Å membrane thinning that occurred (Fig. 23D). Concomitant with membrane thinning, there was an increase in AL with added peptide(s) in the POPC/PIP2 samples.
Figure 22.
Sxray order parameter for MA31myr or MA in POPC/POPS (60:40), or POPC/PIP2 (90:10). Sxray values were corrected for misalignment of layers. Error bars indicate standard deviations from the average Sxray from 2 or more images. Figure from Ref. [18].
Figure 23.
Structural results vs. increasing peptide mole ratio: (A) area/lipid AL in POPC/POPS (60:40), (B) hydrocarbon thickness 2DC in POPC/POPS (60:40), (C) AL in POPC/PIP2 (90:10), (D) 2DC in POPC/PIP2 (90:10). Error bars indicate the standard deviations of the average of 2–5 fits. Figure from Ref. [18].
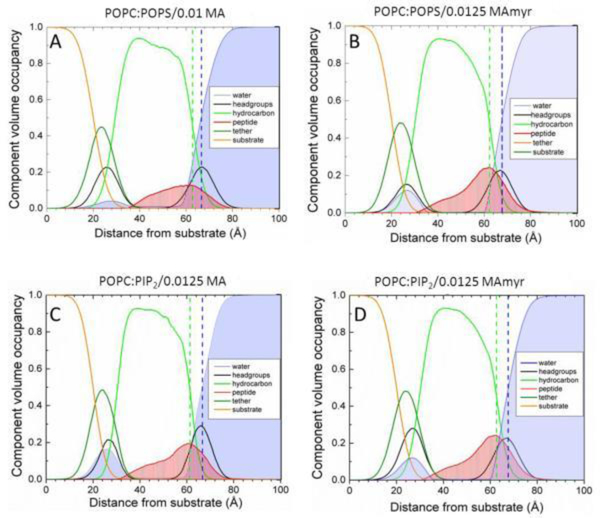
In this work, we used both X-ray diffuse scattering and neutron reflectivity to determine the location of both peptides in membrane mimics. Neutron reflectivity is superior for obtaining peptide position in a bilayer due to a greater contrast between the protein, lipid headgroup and lipid hydrocarbon chains [103], while X-ray scattering is well suited to reporting changes in membrane thickness and elastic properties. NR results (Fig. 24) show MA31myr located both in the headgroup (HG) region as well as 69 ± 17% in the hydrocarbon (HC) region in both membrane mimics. There were no significant differences between MA31 and MA31myr, within the 68% confidence limits. The striking result is that the MA peptides reside quite deeply in the interior of charged membrane mimics, deeper than predicted from hydrophobicity scales [104]. This can be understood in terms of the cartoon in Fig. 25, which shows a proposed distribution of the peptides along the membrane normal. If the charged amino acid side chains are self-neutralized, this can allow greater hydrophobic penetration. As arginines and lysines are neutralized, their side chains become hydrophobic moieties.
Figure 24.
Scattering density profiles obtained from neutron reflectivity. (A) POPC/POPS (60:40 mol ratio)/0.01 MA31,(B) POPC/POPS (60:40)/0.0125 MA31myr, (C) POPC/PIP2 (90:10)/0.0125 MA31, (D) POPC/PIP2 (90:10)/0.0125 MA31myr. Bilayer components are shown in the legends; Gibbs dividing surfaces HC (green dashed line) and water (blue dashed line) are shown. Figure from Ref. [18].
Figure 25.
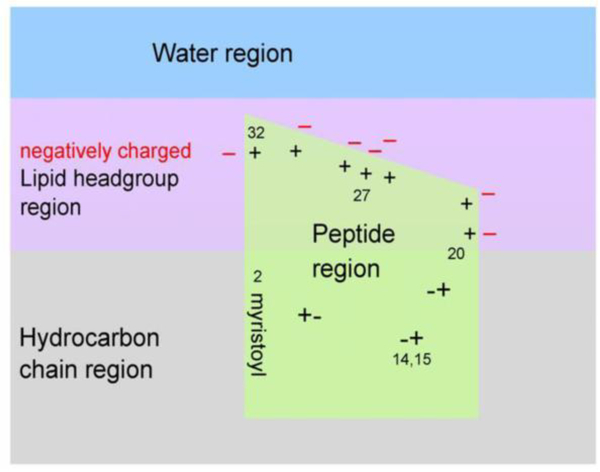
Cartoon representation of MA31myr insertion into a negatively charged membrane, based upon neutron reflectivity results. The location of the 7 positively charged amino acid residues in the HBR from AA20–32 are visualized by + signs with nearby negatively charged lipid headgroups (−signs). The charged amino acids in the AA2–19 stretch are visualized as +−pairs. The last AA32 is visualized near bulk water to allow the remainder of the MA131protein to reside in water. Figure from Ref. [18].
Our main conclusions are that substitution of PIP2 by PS lipids of equivalent charge changed the effect of MA31’s binding on many membrane properties. In addition to the recognized function of PIP2 of providing a stereo-specific anchor for the MA protein to the membrane [101], reducing the bending modulus reduces the free energy for restructuring the membrane for viral budding. Our other results show that only when a strong charge neutralization does not dominate the interaction does the myristoyl group aid MA31 binding. Overall, these results emphasize the importance of the lipid composition in MA31 peptide binding.
9. Concluding Remarks
This review article has summarized the HIV-1 work published in the Tristram-Nagle/Nagle lab during over a decade of funding from the National Institutes of Health. The results show particularly that elastic properties of membranes are sensitive to the effects of the membrane-binding portions of proteins from the HIV-1 virus. How the bending modulus of the control membrane mimics responds to the peptides depends strongly on the lipid composition, as well as on charge, length and hydrophobicity of the peptides. In addition, chain order is a second sensitive measure of interactions. Thus, physics has provided a unique window using probe-free scattering techniques into detailed information concerning peptide/lipid interactions. Structural information is also obtained from the X-ray and neutron scattering, regarding bilayer thickness and depth of insertion of parts of proteins into the relevant membranes. When X-ray data are combined with MD simulation, molecular details of the interactions are observed and the simulation is validated. Information provided by these studies may be used to guide the design of new drugs to thwart the HIV virus.
Supplementary Material
10. Acknowledgements
I would like to thank all of the students, postdocs and faculty members who were co-authors on the summarized papers for their contributions. I would like to thank personally Dr. Jean Chin, former program officer at the Biochemistry and Biophysics of Membranes study section at the NIH, without whose encouragement and support this work could not have been completed. This work was funded primarily by the National Institute of General Medical Sciences of the National Institutes of Health under award R01GM44976 (S.T.-N., John F. Nagle, co-PIs) as well as many other grants listed in the individual papers. X-ray scattering data were taken at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation and the National Institutes of Health/National Institute of General Medical Sciences under National Science Foundation Award DMR-0225180. Parts of this research were performed at the NIST Center for Nanoscale Science and Technology and were supported by the NIST IMS program Precision Measurements for Integral Membrane Proteins.
11. References
- 1.Freed EO 1998HIV-1 Gag proteins: Diverse functions in the virus life cycle Virology 2511–15 [DOI] [PubMed] [Google Scholar]
- 2.Tristram-Nagle S and Nagle JF 2007. HIV-1 fusion peptide decreases bending energy and promotes curved fusion intermediates Biophysical Journal 93 2048–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyatskaya Y, Liu YF, Tristram-Nagle S, Katsaras J and Nagle JF 2001. Method for obtaining structure and interactions from oriented lipid bilayers Physical Review E 63 0119071–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y and Nagle JF 2004. Diffuse scattering provides material parameters and electron density profiles of biomembranes Phys Rev E Stat Nonlin Soft Matter Phys 69 040901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenwood AI et al. 2008CRAC motif peptide of the HIV-1 gp41 protein thins SOPC membranes and interacts with cholesterol Biochim Biophys Acta 17781120–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tristram-Nagle S et al. 2010HIV fusion peptide penetrates, disorders, and softens T-cell membrane mimics Journal of Molecular Biology 402139–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caillé A 1972X-ray scattering by smectic-a crystals Comptes Rendus Hebdomadaires Des Seances De L Academie Des Sciences Serie B 274891–893 [Google Scholar]
- 8.Holyst R 1991Landau-peierls instability, X-ray-diffraction patterns, and surface freezing in thin smectic films Physical Review A 443692–3709 [Google Scholar]
- 9.Podgornik R and Parsegian VA 1992. Thermal mechanical fluctuations of fluid membranes in confined geometries - the case of soft confinement Langmuir 8 557–562 [Google Scholar]
- 10.Lei N, Safinya CR and Bruinsma RF 1995. Discrete harmonic model for stacked membranes - Theory and experiment Journal De Physique Ii 5 1155–1163 [Google Scholar]
- 11.Petrache HI et al. 1998Interbilayer interactions from high-resolution x-ray scattering Physical Review E 577014–7024 [Google Scholar]
- 12.Gouliaev N and Nagle JF 1998. Simulations of interacting membranes in the soft confinement regime Physical Review Letters 81 2610–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shchelokovskyy P, Tristram-Nagle S and Dimova R 2011. Effect of the HIV-1 fusion peptide on the mechanical properties and leaflet coupling of lipid bilayers New Journal of Physics 13 025004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pabst G, Rappolt M, Amenitsch H and Laggner P 2000. Structural information from multilamellar liposomes at full hydration: Full q-range fitting with high quality x-ray data Physical Review E 62 4000–9 [DOI] [PubMed] [Google Scholar]
- 15.Boscia AL et al. 2013Membrane structure correlates to function of LLP2 on the cytoplasmic tail of HIV-1 gp41 protein Biophysical Journal 105657–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akabori K et al. 2014HIV-1 Tat membrane interactions probed using X-ray and neutron scattering, CD spectroscopy and MD simulations Biochimica Et Biophysica Acta-Biomembranes 18383078–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neale C, Huang K, Garcia AE and Tristram-Nagle S 2015. Penetration of HIV-1 Tat47–57 into PC/PE bilayers assessed by MD simulation and X-ray scattering Membranes (Basel) 5 473–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Neil L et al. 2016HIV-1 matrix-31 membrane binding peptide interacts differently with membranes containing PS vs. PI(4,5)P-2 Biochimica Et Biophysica Acta-Biomembranes 18583071–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salditt T, Li C, Spaar A and Mennicke U 2002. X-ray reflectivity of solid-supported, multilamellar membranes European Physical Journal E 7 105–16 [Google Scholar]
- 20.Li DP, Hu SX and Li M 2006. Full q-space analysis of X-ray scattering of multilamellar membranes at liquid-solid interfaces Physical Review E 73 031916(1–8) [DOI] [PubMed] [Google Scholar]
- 21.Boscia AL et al. 2014X-ray structure, thermodynamics, elastic properties and MD simulations of cardiolipin/dimyristoylphosphatidylcholine mixed membranes Chemistry and Physics of Lipids 1781–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braun AR et al. 2012alpha-Synuclein Induces Both Positive Mean Curvature and Negative Gaussian Curvature in Membranes Journal of the American Chemical Society 1342613–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu N, Kučerka N, Liu YF, Tristram-Nagle S and Nagle JF 2005. Anomalous swelling of lipid bilayer stacks is caused by softening of the bending modulus Physical Review E 71 041904(1–8) [DOI] [PubMed] [Google Scholar]
- 24.Kučerka N et al. 2005Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles Biophys J 882626–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrache HI, Tristram-Nagle S and Nagle JF 1998. Fluid phase structure of EPC and DMPC bilayers Chemistry and Physics of Lipids 95 83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pogozheva ID, Tristram-Nagle S, Mosberg HI and Lomize AL 2013. Structural adaptations of proteins to different biological membranes Biochimica Et Biophysica Acta-Biomembranes 1828 2592–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raghunathan M et al. 2012Structure and Elasticity of Lipid Membranes with Genistein and Daidzein Bioflavinoids Using X-ray Scattering and MD Simulations Journal of Physical Chemistry B 1163918–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones DR, Suzuki K and Piller SC 2002. A 100-amino acid truncation in the cytoplasmic tail of glycoprotein 41 in the reference HIV type 1 strain RF AIDS Res Hum Retroviruses 18 513–517 [DOI] [PubMed] [Google Scholar]
- 29.Su Y, Chong HHU, Qiu ZL, Xiong SW and He YX 2015. Mechanism of HIV-1 resistance to short-peptide fusion inhibitors targeting the Gp41 pocket Journal of Virology 89 5801–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steckbeck JD, Sun CQ, Sturgeon TJ and Montelaro RC 2013. Detailed topology mapping reveals substantial exposure of the “cytoplasmic” C-terminal tail (CTT) sequences in HIV-1 Env proteins at the cell surface Plos One 8 e65220(1–9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blumenthal R, Clague MJ, Durell SR and Epand RM 2003. Membrane fusion Chem Rev 103 53–69 [DOI] [PubMed] [Google Scholar]
- 32.Veronese FD et al. 1985Characterization of gp41 as the transmembrane protein coded by the HTLV-III/LAV envelope gene Science 2291402–1405 [DOI] [PubMed] [Google Scholar]
- 33.Lasky LA et al. 1987Delineation of a region of the human immunodeficiency virus type 1 gp120 glycoprotein critical for interaction with the CD4 receptor Cell 50975–985 [DOI] [PubMed] [Google Scholar]
- 34.Choe H et al. 1996The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates Cell 851135–1148 [DOI] [PubMed] [Google Scholar]
- 35.Bosch ML et al. 1989Identification of the fusion peptide of primate immunodeficiency viruses Science 244694–697 [DOI] [PubMed] [Google Scholar]
- 36.Gallaher WR 1987Detection of a fusion peptide sequence in the transmembrane protein of human immunodeficiency virus Cell 50327–328 [DOI] [PubMed] [Google Scholar]
- 37.Tristram-Nagle SA 2007Preparation of oriented, fully hydrated lipid samples for structure determination using X-ray scattering Methods Mol Biol 40063–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rawicz W, Olbrich KC, McIntosh T, Needham D and Evans E 2000. Effect of chain length and unsaturation on elasticity of lipid bilayers Biophysical Journal 79 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagle JF 2013Introductory lecture: basic quantities in model biomembranes Faraday Discuss 16111–29; 113–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagle JF, Jablin MS, Tristram-Nagle S and Akabori K 2015. What are the true values of the bending modulus of simple lipid bilayers? Chem Phys Lipids 185 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jablin MS, Akabori K and Nagle JF 2014. Experimental support for tilt-dependent theory of biomembrane mechanics Phys Rev Lett 113 248102. [DOI] [PubMed] [Google Scholar]
- 42.Nagle JF 2017Experimentally determined tilt and bending moduli of single-component lipid bilayers Chem Phys Lipids 20518–24 [DOI] [PubMed] [Google Scholar]
- 43.Nagle JF 2017X-ray scattering reveals molecular tilt is an order parameter for the main phase transition in a model biomembrane Phys. Rev. E 96030401(1–4) [DOI] [PubMed] [Google Scholar]
- 44.Israelachvili J Intermolecular and Surface Forces. 1992Academic Press: London. [Google Scholar]
- 45.Leibler S 1986Curvature instability in membranes J. Physique 47507–516 [Google Scholar]
- 46.Fournier JB 1996Nontopological saddle-splay and curvature instabilities from anisotropic membrane inclusions Phys. Rev. Lett 764436–4439 [DOI] [PubMed] [Google Scholar]
- 47.Bivas I, Meleard P 2003Bending elasticity and bending fluctuations of lipid bilayer containing an additive Phys. Rev. E 67012901. [DOI] [PubMed] [Google Scholar]
- 48.Rand RP and Fuller NL 1994. Structural dimensions and their changes in a reentrant hexagonal-lamellar transition of phospholipids Biophys J 66 2127–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rand RP, Fuller NL, Gruner SM and Parsegian VA 1990. Membrane curvature, lipid segregation, and structural transitions for phospholipids under dual-solvent stress Biochemistry 29 76–87 [DOI] [PubMed] [Google Scholar]
- 50.Leikin S, Kozlov MM, Fuller NL and Rand RP 1996. Measured effects of diacylglycerol on structural and elastic properties of phospholipid membranes Biophys J 71 2623–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salzwedel K, West JT and Hunter E 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity Journal of Virology 73 2469–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Epand RM, Sayer BG and Epand RF 2003. Peptide-induced formation of cholesterol-rich domains Biochemistry 42 14677–14689 [DOI] [PubMed] [Google Scholar]
- 53.Epand RF et al. 2006Juxtamembrane protein segments that contribute to recruitment of cholesterol into domains Biochemistry 456105–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mills TT et al. 2008Order parameters and areas in fluid-phase oriented lipid membranes using wide angle X-ray scattering Biophys J 95669–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mills TT et al. 2008Liquid-liquid domains in bilayers detected by wide angle X-ray scattering Biophys J 95682–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan J, Tristram-Nagle S and Nagle JF 2009. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation Phys Rev E Stat Nonlin Soft Matter Phys 80 021931(1–12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klauda JB, Kučerka N, Brooks BR, Pastor RW and Nagle JF 2006. Simulation-based methods for interpreting X-ray data from lipid bilayers Biophysical Journal 90 2796–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brugger B et al. 2006The HIV lipidome: a raft with an unusual composition Proc Natl Acad Sci U S A 1032641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koufos E, Chang EH, Rasti ES, Krueger E and Brown AC 2016. Use of a cholesterol recognition amino acid consensus peptide to inhibit binding of a bacterial toxin to cholesterol Biochemistry 55 4787–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller CM, Brown AC and Mittal J 2014. Disorder in cholesterol-binding functionality of CRAC peptides: A molecular dynamics study Journal of Physical Chemistry B 118 13169–13174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vishwanathan SA et al. 2008Hydrophobic substitutions in the first residue of the CRAC segment of the gp41 protein of HIV Biochemistry 47124–130 [DOI] [PubMed] [Google Scholar]
- 62.Mukai M, Glover KJ and Regen SL 2016. Evidence for surface recognition by a cholesterol-recognition peptide Biophysical Journal 110 2577–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen SSL, Lee SF and Wang CT 2001. Cellular membrane-binding ability of the C-terminal cytoplasmic domain of human immunodeficiency virus type 1 envelope transmembrane protein gp41 Journal of Virology 75 9925–9938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Costin JM, Rausch JM, Garry RF and Wimley WC 2007. Viroporin potential of the lentivirus lytic peptide (LLP) domains of the HIV-1 gp41 protein Virology Journal 4 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kliger Y and Shai Y 1997. A leucine zipper-like sequence from the cytoplasmic tail of the HIV-1 envelope glycoprotein binds and perturbs lipid bilayers Biochemistry 36 5157–5169 [DOI] [PubMed] [Google Scholar]
- 66.Koenig BW, Ferretti JA and Gawrisch K 1999. Site-specific deuterium order parameters and membrane-bound behavior of a peptide fragment from the intracellular domain of HIV-1 gp41 Biochemistry 38 6327–6334 [DOI] [PubMed] [Google Scholar]
- 67.Moreno MR, Gludici M and Villalain J 2006. The membranotropic regions of the endo and ecto domains of HIV gp41 envelope glycoprotein Biochimica Et Biophysica Acta-Biomembranes 1758 111–123 [DOI] [PubMed] [Google Scholar]
- 68.Murakami T and Freed EO 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions Proceedings of the National Academy of Sciences of the United States of America 97 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilk T, Pfeiffer T and Bosch V 1992. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product Virology 189 167–177 [DOI] [PubMed] [Google Scholar]
- 70.Postler TS and Desrosiers RC 2013. The Tale of the long tail: the cytoplasmic domain of HIV-1 gp41 Journal of Virology 87 2–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kalia V, Sarkar S, Gupta P and Montelaro RC 2003. Rational site-directed mutations of the LLP-1 and LLP-2 lentivirus lytic peptide domains in the intracytoplasmic tail of human immunodeficiency virus type 1 gp4l indicate common functions in cell-cell fusion but distinct roles in virion envelope incorporation Journal of Virology 77 3634–3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chan R et al. 2008Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides Journal of Virology 8211228–11238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kučerka N et al. 2008Lipid bilayer structure determined by the simultaneous analysis of neutron and x-ray scattering data Biophysical Journal 952356–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cantor RS 1999Lipid composition and the lateral pressure profile in bilayers Biophysical Journal 762625–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kondo N, Miyauchi K, Meng FX, Iwamoto A and Matsuda Z 2010. Conformational changes of the HIV-1 envelope protein during membrane fusion are inhibited by the replacement of its membrane-spanning domain Journal of Biological Chemistry 285 14681–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shang L, Yue L and Hunter E 2008. Role of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein in cell-cell fusion and virus infection Journal of Virology 82 5417–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.West JT, Johnston PB, Dubay SR and Hunter E 2001. Mutations within the putative membrane-spanning domain of the simian immunodeficiency virus transmembrane glycoprotein define the minimal requirements for fusion, incorporation, and infectivity Journal of Virology 75 9601–9612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frankel AD and Pabo CO 1988. Cellular uptake of the Tat protein from human immunodeficiency virus Cell 55 1189–1193 [DOI] [PubMed] [Google Scholar]
- 79.Green M and Loewenstein PM 1988. Autonomous functional domains of chemically synthesized human immunodeficiency virus Tat trans-activator protein Cell 55 1179–1188 [DOI] [PubMed] [Google Scholar]
- 80.Vives E, Brodin P and Lebleu B 1997. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus Journal of Biological Chemistry 272 16010–16017 [DOI] [PubMed] [Google Scholar]
- 81.Zhao M and Weissleder R 2004. Intracellular cargo delivery using tat peptide and derivatives Medicinal Research Reviews 24 1–12 [DOI] [PubMed] [Google Scholar]
- 82.Zorko M and Langel U 2005. Cell-penetrating peptides: mechanism and kinetics of cargo delivery Advanced Drug Delivery Reviews 57 529–545 [DOI] [PubMed] [Google Scholar]
- 83.Ter-Avetisyan G et al. 2009Cell entry of arginine-rich peptides is independent of endocytosis Journal of Biological Chemistry 2843370–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Richard JP et al. 2005Cellular uptake of unconjugated TAT peptide involves clathrin-dependent endocytosis and heparan sulfate receptors Journal of Biological Chemistry 28015300–15306 [DOI] [PubMed] [Google Scholar]
- 85.Herce HD and Garcia A E 2007. Molecular dynamics simulations suggest a mechanism for translocation of the HIV-1 TAT peptide across lipid membranes Proceedings of the National Academy of Sciences of the United States of America 104 20805–20810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Herce HD, Garcia AE and Cardoso MC 2014. Fundamental molecular mechanism for the cellular uptake of guanidinium-rich molecules Journal of the American Chemical Society 136 17459–17467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mishra A, Gordon VD, Yang LH, Coridan R and Wong GCL 2008. HIV TAT forms pores in membranes by inducing saddle-splay curvature: Potential role of bidentate hydrogen bonding Angewandte Chemie-International Edition 47 2986–2989 [DOI] [PubMed] [Google Scholar]
- 88.Ciobanasu C, Siebrasse JP and Kubitscheck U 2010. Cell-penetrating HIV-11 Tat peptides can generate pores in model membranes Biophysical Journal 99 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kučerka N, Katsaras J and Nagle JF 2010. Comparing membrane simulations to scattering experiments: Introducing the SIMtoEXP software Journal of Membrane Biology 235 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jarasch ED, Reilly CE, Comes P, Kartenbeck J and Franke WW 1973. Isolation and characterization of nuclear membranes from calf and rat thymus Hoppe Seylers Z Physiol Chem 354 974–986 [DOI] [PubMed] [Google Scholar]
- 91.Schmidt N, Mishra A, Lai GH and Wong GCL 2010. Arginine-rich cell-penetrating peptides Febs Letters 584 1806–1813 [DOI] [PubMed] [Google Scholar]
- 92.Chen XC, Sa’adedin F, Deme B, Rao PF and Bradshaw J 2013. Insertion of TAT peptide and perturbation of negatively charged model phospholipid bilayer revealed by neutron diffraction Biochimica Et Biophysica Acta-Biomembranes 1828 1982–1988 [DOI] [PubMed] [Google Scholar]
- 93.Chen XC et al. 2017Efficient internalization of TAT peptide in zwitterionic DOPC phospholipid membrane revealed by neutron diffraction Biochimica Et Biophysica Acta-Biomembranes 1859910–916 [DOI] [PubMed] [Google Scholar]
- 94.Choi D et al. 2012Insertion mechanism of cell-penetrating peptides into supported phospholipid membranes revealed by X-ray and neutron reflection Soft Matter 88294–8297 [Google Scholar]
- 95.Gelderblom HR, Hausmann EHS, Ozel M, Pauli G and Koch MA 1987. Fine-structure of human-immunodeficiency-virus (Hiv) and immunolocalization of structural proteins Virology 156 171–176 [DOI] [PubMed] [Google Scholar]
- 96.Chukkapalli V and Ono A 2011. Molecular determinants that regulate plasma membrane association of HIV-1 Gag Journal of Molecular Biology 410 512–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ono A 2009HIV-1 assembly at the plasma membrane: Gag trafficking and localization Future Virology 4241–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou WJ, Parent LJ, Wills JW and Resh MD 1994. Identification of a membrane-binding domain within the amino-terminal region of human-immunodeficiency-virus type-1 Gag protein which interacts with acidic phospholipids Journal of Virology 68 2556–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ono A, Orenstein JM and Freed EO 2000. Role of the gag matrix domain in targeting human immunodeficiency virus type 1 assembly Journal of Virology 74 2855–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tang C et al. 2004Entropic switch regulates myristate exposure in the HIV-1 matrix protein Proceedings of the National Academy of Sciences of the United States of America 101517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Saad JS et al. 2006Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly Proceedings of the National Academy of Sciences of the United States of America 10311364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shkriabai N et al. 2006Interactions of HIV-1 Gag with assembly cofactors Biochemistry 454077–4083 [DOI] [PubMed] [Google Scholar]
- 103.Heinrich F and Lösche M 2014. Zooming in on disordered systems: Neutron reflection studies of proteins associated with fluid membranes Biochimica Et Biophysica Acta-Biomembranes 1838 2341–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wimley WC and White SH 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces Nature Structural Biology 3 842–848 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.