Abstract
Despite the critical role of Rab GTPases for intracellular transport, the vast majority of proteins within this family remain poorly characterized, including the Rab40 subfamily. Often recognized as atypical Rabs, the Rab40 family of proteins are unlike any other small GTPase because they contain a C-terminal suppressor of cytokine signaling (SOCS) box. It is well established that this SOCS domain in other proteins mediates an interaction with the scaffold protein Cullin5 in order to form a E3 ubiquitin ligase complex critical for protein ubiquitylation and turnover. Although the function of SOCS/Cullin5 complexes has been well defined in several of these other proteins, this is not yet the case for the Rab40 family of proteins. We have previously shown that the Rab40b family member plays an important role during three-dimensional (3D) breast cancer cell migration. To further this knowledge, we began to investigate the SOCS-dependent role of Rab40b during cell migration. Here, we describe an unbiased approach to identify potential Rab40b/Cullin5 substrates. We anticipate that this method will be useful for studying the function of other Rab40 family members as well as other SOCS box containing proteins.
Keywords: Rab GTPase, Rab40b, Cullin5, SOCS box, evolution, ubiquitylation, immunoprecipitation, mass spectrometry, proteomics
1. Introduction
With close to 70 members, Rab GTPases constitute the largest family of small monomeric GTPases within the human genome [1]. These proteins are members of the Ras superfamily, and the origins of Rab GTPases is believed to be tightly associated with the evolution of the eukaryotic cell. Rab GTPases are found in all eukaryote lineages and emerge phylogenetically with other defining eukaryote cell features such as the actin and microtubule cytoskeleton, internal membranes, and a nucleus [2] [3] [4]. Within eukaryotes, the Rab protein family has expanded dramatically, and vertebrate genomes in particular exhibit some of the largest numbers of Rab paralog family members in comparison to all other taxa [2] [5]. Functionally, Rab proteins are master regulators of intracellular membrane trafficking and act as molecular switches, cycling between a GTP-bound “active” state and a GDP-bound “inactive” state [6] [7]. In the active state, Rab proteins bind and recruit/activate effector proteins to specific intracellular membranes. Together, Rabs and their respective effectors act in a cooperative manner to regulate all stages of membrane and protein traffic [8] [9].
Since their discovery in the 1980s, characterization of about half of known Rab GTPases has helped uncover the impressive complexity and diversity of these proteins [7] [10]. Despite ongoing research and the critical role of Rabs in eukaryotic cells, the vast majority of Rab GTPases remain incompletely understood, both in terms of function and regulation. Given the implication of Rabs in disease pathogenesis [11], it is critical to uncover the function of these small GTPases. The Rab40 family of small GTPases is a prime example of a largely uncharacterized Rab subfamily, with a unique domain architecture that suggests these proteins play functional roles unlike any other Rab. Here we discuss the known functions of the Rab40 family, with special focus on Rab40b, and outline methods for studying the unique Rab40 SOCS box.
1.1. Origins and evolution of Rab40 subfamily
Rab40 paralogs have only been identified in the genomes of bilaterian metazoans (e.g. protostomes and deuterostomes), suggesting that the Rab40 gene family was present in the last bilaterian common ancestor. Prior studies further suggest that the Rab40 subfamily may have emerged as a duplication of Rab18 [5]. However, substantive sequence divergence differentiates Rab40 paralogs from all other Rab proteins [2] [5] [12] [13]. While most Rabs share only a conserved globular G-domain (housing the Switch I and Switch II regions), Rab40 GTPases have an extended C-terminal region that contains the conserved SOCS (suppressor of cytokine signaling) box (Figure 1A) [2]. This additional domain suggests that Rab40 may be a unique Rab GTPase with novel function, however there is little known about the cellular role or regulation of Rab40 proteins.
Figure 1. Evolution and structure of Rab40 gene family.
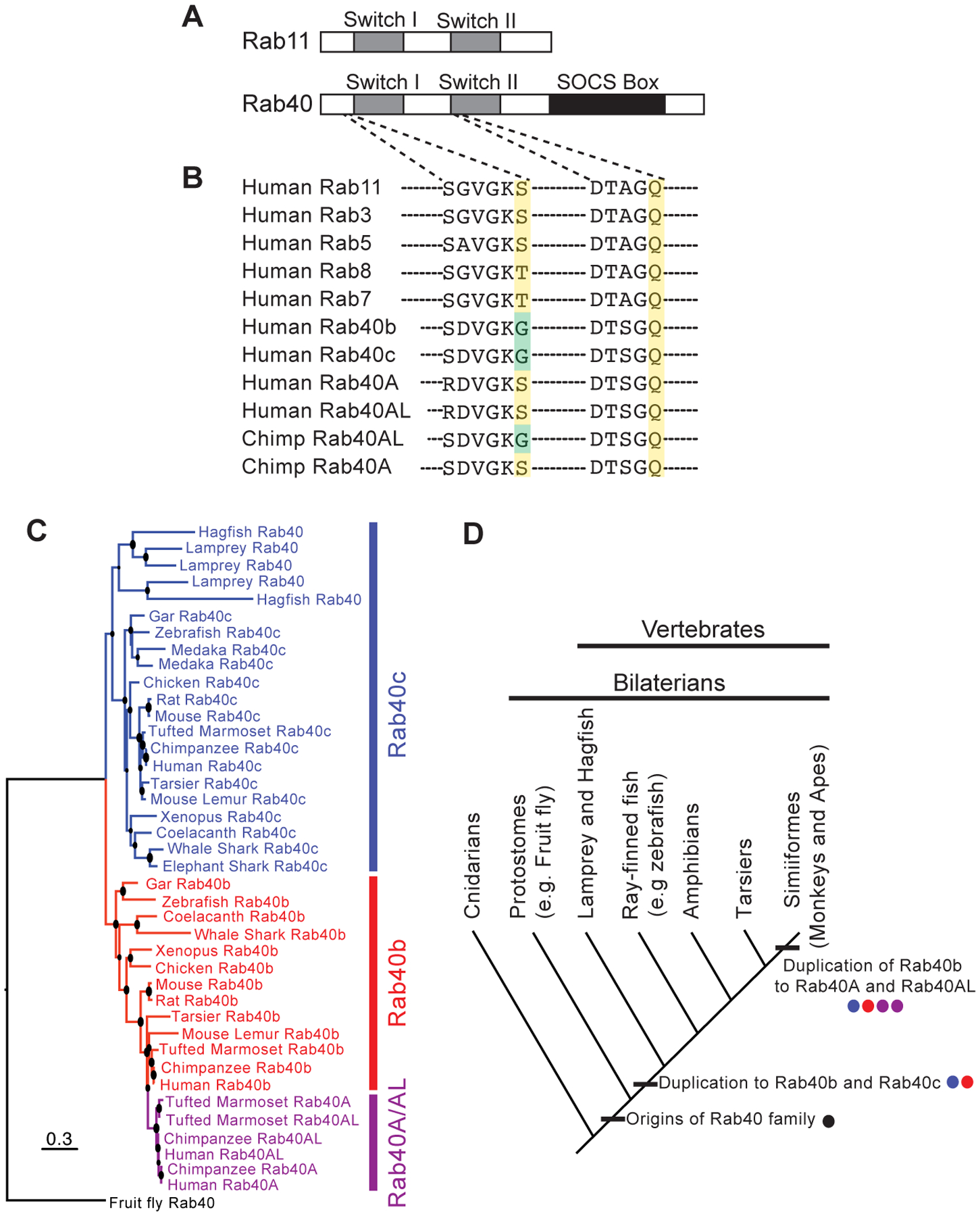
A Domain architecture of Rab40 compared to Rab11 shows extended C-terminal SOCS box domain. B Sequence alignment of Rab40 subfamily and a subset of well-studied Rabs highlighting GEF binding region (left) and Switch II region (right). Rab40 proteins exhibit a non-canonical Glycine (green) within the GEF binding motif (conserved Serine/Threonine in yellow) and maintain the conserved Glutamine critical for GTPase activity (yellow). C Maximum likelihood gene tree (PhyML) of Rab40 coding sequences shows duplications of Rab40 paralogs in vertebrate taxa. Note that Cyclostome Rab40 sequences form a clade with Gnathostome Rab40c sequences, and that Rab40a/al result from a duplication of Rab40b. Larger node sizes correspond to greater bootstrap support. D Hypothesis of Rab40 gene family evolution in vertebrates. Rab40 originates in the lineage leading to bilaterians, and duplicates in the lineage leading to vertebrates resulting in Rab40b and Rab40c paralogs. A subsequent duplication in the lineage leading to monkeys and apes results in the addition of Rab40a and Rab40al paralogs.
In addition to an extended C-terminal domain, Rab40 proteins also exhibit non-canonical amino acids at functionally critical sites in the domains responsible for GTPase activity and guanine nucleotide exchange factor (GEF) binding. For instance, most Rab GTPases exhibit a conserved Serine or Threonine residue at a critical site in the GTPase domain upstream of Switch I (Figure 1B) [14]. This site is crucial for GEF binding and is commonly mutagenized by researchers to lock Rabs in a GDP bound (dominant negative) state in order to study Rab function. Surprisingly, the Rab40 gene family has evolved novel amino acids at this conserved residue (Figure 1B). Drosophila Rab40 exhibits a Histidine at this site, in mosquito this same site is occupied by a Glutamine (not shown), while in humans the Rab40b and Rab40c paralogs have a Glycine at this site (Figure 1B). Even more remarkable is that primate Rab40a and human Rab40al paralogs have re-evolved a Serine residue at this critical site. The fact that this residue is highly conserved across millions of years of Rab protein evolution, and variable in Rab40 genes suggests exciting possibilities that Rab40 proteins may have unique GTPase activity that is entirely unlike any other Rab protein. Such possibilities, however, will have to remain speculative until further insight into the function of these Rab40 proteins is better understood. On the other hand, note that the Rab40 subfamily do contain the conserved catalytic Glutamine residue in the Switch II region, strongly suggesting that they still function as GTPases (Figure 1B).
The evolution of the Rab40 gene family is characterized by expansion and lineage specific losses in vertebrates (Figure 1C). That Rab40 proteins are functional in the cell seems highly likely given the conservation of Rab40 genes in the genomes of most bilaterian organisms. While a single Rab40 gene is found in non-vertebrate bilaterians, a duplication event at the base of the vertebrate lineage resulted in Rab40b and Rab40c paralogs that are conserved across most vertebrate taxa (Figure 1D). Preliminary analyses date this duplication to before the last vertebrate common ancestor, suggesting that Rab40b and Rab40c may have emerged from genome duplications at the root of the vertebrate phylogeny [15]. This dating is based on the preliminary finding that Cyclostomes (lamprey and hagfish) retain a Rab40c paralog. However, the node supporting this claim has low support in our gene trees, and further work is needed to rule out the possibility that Rab40b and Rab40c evolved in the lineage leading to Gnathostomes (sharks, bony fish, and tetrapods) after it split from Cyclostomes.
Important for humans is that a subsequent duplication of Rab40b in the lineage leading to Simiiform primates (e.g. monkeys and apes, ~60K ybp) resulted in two additional Rab40 paralogs: Rab40a and Rab40al (Figure 1C, D). Thus, the human genome contains four Rab40 paralogs: Rab40a, Rab40al, Rab40b, and Rab40c. Rab40al lacks introns, a pattern that suggests it may have been duplicated by reverse transcription transposition [2]. Intriguingly, both Rab40a and Rab40al are found on the X chromosome. As noted above, Rab40a and Rab40al have also re-evolved a conserved Serine at a critical functional site. While all primate Rab40a genes exhibit a Serine at this site, only human Rab40al proteins exhibit a Serine residue suggesting a complex evolutionary history for these proteins that may involve gene conversion in the human lineage.
Additional lineage specific duplications and deletions in the Rab40 gene subfamily include independent duplications of three Rab40 paralogs, and the potential loss of Rab40b, in Cyclostomes. Teleost fish experienced a whole genome duplication and could retain up to four Rab40 paralogs (e.g. two each of Rab40b and Rab40c). Teleost taxa surveyed, however, appear to have lost paralogs to return to a diploid state. Zebrafish retains a single Rab40b and Rab40c paralog. In contrast, the teleost fish Medaka lost both Rab40b paralogs, but retains two Rab40c paralogs (Figure 1C).
1.2. Conservation and function of the Rab40 SOCS box
Evolution of the Rab40 subfamily is defined by conservation of the C-terminal SOCS box through speciation and duplication events. Although originally identified in the SH2-containing SOCS family, the number of SOCS box containing proteins has expanded, and includes families such as WD40 proteins, SPRY domain proteins, and ankyrin repeat proteins [16] [17] [18]. The SOCS box is conserved across these protein families as shown by an alignment in Figure 2A. This ~40 amino acid motif functions as a substrate recognition module, as part of the larger E3 ubiquitin ligase containing the scaffold protein Cullin5 (Cul5), RING protein Rbx2, and adaptor proteins Elongin B and Elongin C (Figure 2B) [19] [20]. Together, this Cullin-RING ligase (CRL) complex regulates protein turnover via ubiquitylation and subsequent proteasomal degradation [18] [21]. It should also be noted that when the Rab40 subfamily was first grouped with other SOCS box proteins, old nomenclature was utilized to name these proteins which is no longer readily used (RAR2A/Rab40a, RAR2/Rab40al, RAR/Rab40b and RAR3/Rab40c).
Figure 2. Conservation and function of the Rab40 SOCS box.

A. Alignment of Rab40 SOCS box and other human SOCS proteins shows sequence conservation. Green highlights the Cul5 box, where the LPLP motif is conserved across all Rab40 sequences and multiple human SOCS box containing proteins. Conserved residues across all human SOCS box containing proteins are summarized at the bottom. B. Example of a Cullin-RING ligase (CRL) that includes the scaffold protein Cul5, a SOCS-containing adaptor protein, RING protein Rbx2, and adaptor proteins Elongin B and Elongin C (left). This CRL complex facilitates ubiquitylation of target substrates. Strong evidence from our lab and others shows that Rab40b (and other members of the Rab40 subfamily) is a legitimate SOCS-containing adaptor protein for Cul5 (right).
Within the SOCS box, there are two defined motifs including the BC box (critical for binding the adaptor proteins Elongin B and C) and the Cul5 box (Figure 2A) [22] [23]. The Cul5 box has the consensus sequence φxxLPφPxxφxx (where φ is a hydrophobic residue and x is any amino acid). The LPφ P motif is thought to be the primary determinant of Cul5 binding specificity [16] [24] [25]. The Rab40 Cul5 box contains residues ‘LPLP’, which is 100% conserved across all bilaterian Rab40 sequences and conserved in a number of non-Rab40 SOCS containing proteins (Figure 2A). This conservation through bilaterian evolution suggests that the SOCS box is functional in Rab40 proteins, however what role it may be playing is still unclear.
1.3. Known functions of Rab40 paralogs
Despite a novel domain architecture and the retention of duplicate paralogs in vertebrate lineages, there is limited knowledge on the function of the Rab40 subfamily. Based on the current research that is available, it seems likely that the four family members are not fully redundant. While Rab40a, Rab40b, and Rab40c paralogs have been shown to interact with Cul5, it still remains to be determined whether these Rab40/Cul5 complexes have unique roles within the cell. Below is a summary of the Rab40 literature as it stands.
Rab40a and Rab40al
Rab40a and Rab40al (97.8% sequence identity), which appear to be unique to Simiiformes, are arguably the least studied amongst the four paralogs. The Rab40a/Cul5 complex has been suggested to target the small GTPase RhoU for ubiquitylation and degradation, which can be protected by the Cdc42 effector protein PAK4 [26]. This study only tested RhoU binding to Rab40a, so it is unclear whether RhoU may be a target of other Rab40/Cul5 complexes. In 2012, a study identified a mutation in Rab40al that is associated with the rare X-linked disorder, Martin-Probst Syndrome (MPS) [27]. This missense mutation (variant p. D59G) is located within the highly conserved GTPase domain between β2 and β3 strands. However, several reports since have argued against the pathogenicity of p.D59G, so the linkage between Rab40al and MPS remains controversial [28] [29].
Rab40b
Work from our lab identified Rab40b as a small GTPase required for targeted matrix metalloproteinase (MMP, specifically MMP2 and MMP9) secretion at invadopodia structures during 3D breast cancer cell migration [30] [31]. While depletion of Rab40a did not have any effect on MMP2 or MMP9 secretion, knockdown of Rab40c did reduce MMP2 secretion, suggesting that there may be some functional overlap between Rab40c and Rab40b in this particular context.
Rab40c
Rab40c is the most studied member of Rab40 subfamily. This includes the very first study citing a functional role for a Rab40/Cul5 complex [32]. The authors demonstrated that Rab40c interacts with Cul5 to form a CRL complex that poly-ubiquitylates Rap2 in order to regulate non-canonical Wnt signaling during Xenopus gastrulation and embryogenesis. Although the notation of XRab40 throughout the paper lends itself to some confusion, it is clear from the methods and supplementary material that the authors of the Xenopus study were indeed working with Rab40c. However, it should be noted that both Rab40b (Gene ID: 779634; XB-GENE-490554) and Rab40c (Gene ID: 100492927; XB-GENE-6461049) are annotated within the Xenopus genome. A more recent study identified the protein Varp as a potential Rab40c/Cul5 substrate in melanocytes [33]. Other functions of Rab40c (assumed to be non-SOCS related) have been reported as well, including Rab40c function during oligodendrocyte vesicle transport [34] and lipid droplet biogenesis [35]. Taken together, it is clear that we lack a complete understanding of Rab40c function, and like the other members of the Rab40 subfamily, whether there are clear SOCS-dependent and -independent roles.
2. Rab40b/Cul5 complex
To further define Rab40b-dependent mechanisms regulating MMP2/9 secretion, it seems critical to a establish whether there is a role for Rab40b’s SOCS box during cell migration. Based on previous literature described above, there is precedence for Rab40b acting as a SOCS adaptor protein for the CRL containing Cul5 (Figure 2B) [18] [32], and we recently demonstrated an interaction between Rab40b and Cul5 in MDA-MB-231 cells (data not shown). Interestingly, the interaction between Rab40b and Cul5 appears to be GTP independent, as locking Rab40b in either a GTP or GDP state had no effect on its ability to interact with Cul5 (data not shown).
We next asked whether we could disrupt Rab40b/Cul5 binding in order to study the function of this complex during cell migration. Based on previous biochemical data in other SOCS box proteins (including Rab40c), we designed a Rab40b construct with all four LPLP residues mutated to alanine (AAAA), which we have designated as Rab40b SOCSAAAA (Figure 3A) [23] [24] [25]. While not a complete loss of complex formation, we found a significant decrease in Rab40b SOCSAAAA binding to Cul5 compared to Rab40b WT (Figure 3B).
Figure 3. Rab40b SOCSAAAA mutant disrupts Cul5 complex interaction.

A. Mutation of LPLP➔AAAA within Rab40b Cul5 box to generate Rab40b SOCSAAAA. B. To test the effect of Rab40b SOCSAAAA binding to Cul5, a pull-down assay was performed. Briefly, MDA-MB-231 lysates overexpressing human FLAG-Rab40b (WT or SOCSAAAA) were incubated with purified human GST-Cul5 or GST alone (control) followed by a standard GST pull-down experiment. Eluates were immunoblotted with α-FLAG mouse antibody. Graph below shows results from three independent experiments, normalized to WT binding. * indicates significant p-value=0.0176. C. Mass spectrometry results from two independent experiments shows disruption of CRL complex with Rab40b SOCSAAAA. These are raw spectral counts (defined as the total number of spectra identified for a protein).
2.1. Using the Rab40b SOCSAAAA mutant to uncover function
With generation of the Rab40b SOCSAAAA mutant, we became poised to ask a number of critical questions regarding the role of Rab40b/Cul5 during cell migration. Although we know that these two proteins interact, this complex has not previously been shown to directly regulate MMP secretion, actin dynamics, or cell migration. One way that Rab40b/Cul5 could regulate these processes is through ubiquitylation of downstream substrates. However, putative ubiquitylation targets of the Rab40b/Cul5 complex are not known. To address this gap, we sought to identify potential substrates, with the ultimate goal of studying their function and regulation during cell migration.
Pinpointing bona fide ubiquitylation substrates is challenging because most substrates are quickly degraded by the proteasome or processed by deubiquitylases. To overcome this challenge, we utilized the Rab40b SOCSAAAA mutant as a way to capture protein substrates. We hypothesized that Rab40b SOCSAAAA would still bind substrates, but that without Cul5 interaction, ubiquitylation and subsequent turnover/release of the substrate would be inhibited. In effect, we expected that these proteins bound to Rab40b SOCSAAAA would be “stuck” and would accumulate in the cell compared to a WT context. Indeed, using mass spectrometry and proteomic analysis we identified a subset of Rab40b binding proteins that were enriched in Rab40b SOCSAAAA vs Rab40b WT. Detailed here are the materials and methods. In the Conclusion section, we share our results and our future goals for using this method to test for functional differences in the Rab40 family.
3. Materials
3.1. Crosslinking of FLAG antibody to Protein G Sepharose
α-FLAG mouse antibody (Sigma Aldrich, Clone M2)
Mouse IgG (Jackson Labs, 015-000-003)
Protein G Sepharose (PGS, GE Healthcare, 17-0618-01)
Phosphate Buffered Saline (PBS) pH 7.4
Tris buffer: 20mM Tris pH 8.0, 100mM NaCl
Borate buffer: 200mM Na-Borate pH 9.0 (Notes 1)
Dimethyl pimelimidate dihydrochloride (DMP) crosslinker (Notes 2)
Ethanolamine buffer: 200mM Ethanolamine pH 8.0 (Notes 3)
Reaction buffer: 20mM HEPES pH 7.4, 150mM NaCl
Equipment: rotator, microcentrifuge at 4C
3.2. FLAG-Rab40b immunoprecipitation
Ice-cold Reaction buffer for cell lysis: 20mM HEPES pH 7.4, 150mM NaCl, 1% Triton X-100, 1mM Phenylmethylsulfonyl Fluoride (PMSF), 1mM Phosphatase Inhibitors (Calbiochem 524625), 10mM Iodoacetamide (DUBi)
Reagents for locking Rab40b WT and Rab40b SOCSAAAA in a GTP/active state: 5mM Ethylenediaminetetraacetic acid (EDTA), 5mM Guanosine 5′-[β,γ-imido]triphosphate trisodium salt hydrate (GMP-PNP), 15mM MgCl2 (Notes 4)
α-FLAG beads and mice IgG beads prepared in 3.1
Ice-cold Reaction buffer for washing: 20mM HEPES pH 7.4, 150mM NaCl, 0.1% Triton X-100, 1mM MgCl2
Gel loading tips (or any ultra-thin tip)
Elution buffer: 10mM Tris pH 7.4, 1% Sodium Dodecyl Sulfate (SDS), 100uM Dithiothreitol (DTT)
Equipment: rotator, microcentrifuge at 4C, benchtop/large capacity centrifuge at 4C, heat bath at 55C
4. Methods
4.1. Crosslinking of FLAG antibody to Protein G Sepharose
This protocol is written to make 1mL of α-FLAG beads and 500μL of mouse IgG beads. 200μL of beads are used for each reaction (ie. 50μg of antibody). This can be scaled up or down.
Measure α-FLAG mouse antibody and mouse IgG concentration.
Prepare a 50% PGS bead solution in PBS. Keep cold.
Combine 300ug of α-FLAG antibody and 250μg of mouse IgG with 1mL and 500μL of 50% PGS beads, respectively. For these first steps, 1.5mL tubes are sufficient. Notice switch to 15mL conical in step 8.
Incubate for 2 hours at room temperature (RT) while rotating.
Spin down antibody-bead solution. 2000rpm for 5 minutes at 4C. Unless otherwise noted, all spins should be performed at these settings.
Wash beads 1X with 1mL of Tris buffer. Spin and discard supernatant (sup).
Wash beads 5X with 1mL of Borate buffer. Spin and discard sup between each wash.
Resuspend beads in 1mL of Borate buffer. Transfer solution to 15mL conical.
Make 1M stock of DMP crosslinker in Borate buffer. Add 50μL of 1M DMP stock to each set of beads. Cover 15mL conical in foil, DMP is light sensitive.
Incubate at RT for 30 minutes while rotating.
Spin and discard sup.
Wash beads 1X with 5mL of Ethanolamine buffer. Spin and discard sup.
Quench crosslinking reaction for 3 hours with 5mL of Ethanolamine buffer. RT while rotating.
Spin and discard sup.
Wash 5X with 1mL of Reaction buffer. Spin and discard sup between each wash.
On the last discard, use gel loading tips (or something similar) to get as much buffer off as possible. Resuspend α-FLAG beads in 500μL of Reaction buffer. Resuspend mouse IgG beads in 250μL of Reaction buffer.
4.2. FLAG-Rab40b immunoprecipitation
Before performing this immunoprecipitation large-scale, we recommend testing the beads in a small-scale reaction first to make sure α-FLAG and mouse IgG are covalently bonded and to troubleshoot any issues with elution. See Notes 5 for more details.
Grow 4×10cm dishes of MDA-MB-231 FLAG-Rab40b WT cells and FLAG-Rab40b SOCSAAAA cells.
Once confluent, wash plates with PBS. Then, lyse cells (250μL per plate) with ice-cold Reaction buffer for cell lysis (can either scrape or lyse from trypsinized pellet).
Incubate at 4C for 1 hour while rotating.
Pellet lysed cells by centrifugation. 12000rpm for 10 minutes at 4C.
Take supernatant. Save a small sample for checking FLAG expression later (if needed). Measure lysate concentration.
Set 3 tubes. These can either be 1.5mL tubes or 15mL conicals, depending on your lysate concentration and volume. Calculate the volume needed for 1.5mg of lysate. Tube 1 gets 1.5mg of FLAG-Rab40b WT lysate. Tube 2 gets 1.5mg of FLAG-Rab40b 4A lysate. Tube 3 gets 0.75mg of FLAG-Rab40b WT lysate and 0.75mg of FLAG-Rab40b SOCSAAAA lysate. For each tube, make up the difference in volume with Reaction buffer to ensure equal concentration and equal volume. The goal volume should be about 1mL.
Pre-clear lysates by adding 200μL of 50% PGS solution. Incubate at RT for 30 minutes while rotating.
Spin and move sups to new tubes.
For lysates in tubes 1 and 2, follow the steps 10–12 for GMP-PNP locking. It is not necessary to lock tube 3 (mouse IgG control).
Add EDTA to 5mM. Incubate at RT for 10 minutes while rotating. This will chelate Magnesium ions and will remove any GTP/GDP nucleotide bound to Rab40b.
Add GMP-PNP to 5mM. Incubate at RT for 10 minutes while rotating. (Notes 6)
Add MgCl2 to 15mM (3X fold EDTA concentration). Incubate at 37C for 10 min while rotating.
Add 200μL of α-FLAG beads to tubes 1 and 2. Add 100μL of mouse IgG beads to tube 3.
Incubate at RT for 2 hours while rotating.
Spin and remove sup. Take a sample of flow through for later analysis (if needed).
Wash 5X with 10mL of ice-cold Reaction buffer for washing. Spin and discard sup between each wash.
Resuspend beads in 1mL of ice-cold Reaction buffer for washing. Transfer beads to 1.5mL tubes. Spin and discard sup. On this last discard, use gel loading tips (or something similar) to get as much buffer off as possible.
Add 100μL of Elution buffer to tubes 1, 2, and 3.
Incubate beads with Elution buffer for 15 minutes at 55C.
Spin and collect eluates.
Repeat steps 18–20 for a second elution step.
Before freezing or proceeding to 4.3, take 15μL of each elution and run a gel to test the efficiency of the immunoprecipitation. Probe with α-FLAG antibody to check for FLAG-Rab40b in eluates.
4.3. Proteomics
The following protocol was performed by the University of Colorado School of Medicine Biological Mass Spectrometry Facility. In brief, samples from 4.2 were digested with trypsin in-solution and were analyzed on a Q Exactive HF mass spectrometer.
Trypsin digestion
The samples were digested according to the FASP protocol using a 10 kDa molecular weight cutoff filter. In brief, the samples were mixed in the filter unit within 8 M urea in 100mM ammonium bicarbonate (ABC), pH 8.5 and centrifuged at 14 000g for 15 min. The proteins were reduced by addition of 100 μL of 10 mM DTT in 8 M urea in 100 mM ABC, pH 8.5, incubation for 30 min at RT and the device was centrifuged. Subsequently, 100 μl of 25 mM iodoacetamide in 8 M urea in 100 mM ABC, pH 8.5 were added to the samples, incubation for 15 min at RT in dark followed by centrifugation. Afterward, three washing steps with 100 μL of 8 M urea in 100 mM ABC, pH 8.5 solution were performed, followed by three washing steps with 100 μL of 50 mM ABC buffer. Protein digestion was carried out with the presence 0.02 % of ProteaseMax (Promega, Madison, WI) detergent at 37°C overnight. Peptides were recovered from the filter using 25mM ABC. Samples were dried in Speed-Vac and Thermo Scientific Pierce® Detergent Removal Resin was used to remove residue of detergent from the sample.
Mass Spectrometry
Samples were analyzed on a Q Exactive HF quadrupole orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) coupled to an Easy nLC 1000 UHPLC (Thermo Fisher Scientific) through a nanoelectrospray ion source. Peptides were separated on a self-made C18 analytical column (100 μm internal diameter, × 20 cm length) packed with 2.7 μm Phenomenex Cortecs particles. After equilibration with 3 μL 5% acetonitrile 0.1% formic acid, the peptides were separated by a 120 min a linear gradient from 4% to 30% acetonitrile with 0.1% formic acid at 400nL/min. LC mobile phase solvents and sample dilutions used 0.1% formic acid in water (Buffer A) and 0.1% formic acid in acetonitrile (Buffer B) (Optima™ LC/MS, Fisher Scientific, Pittsburgh, PA). Data acquisition was performed using the instrument supplied Xcalibur™ (version 4.0) software. The mass spectrometer was operated in the positive ion mode, in the data–dependent acquisition mode. The full MS scans were obtained with a range of m/z 300 to 1800, a mass resolution of 120,000 at m/z 200, and a target value of 1.00E+06 with the maximum injection time of 50 ms. HCD collision was performed on the 15 most significant peaks, and tandem mass spectra were acquired at a mass resolution of 30,000 at m/z 200 and a target value of 1.00E+05 the maximum injection time of 100 ms. Isolation of precursors was performed with a window of 1.2 Th. The dynamic exclusion time was 20s. The normalized collision energy was 32. We excluded precursor ions with single, unassigned, or eight and higher charge states from fragmentation selection.
MS/MS spectra were extracted from raw data files and converted into mgf files using Proteome Discoverer 2.2. These mgf files were then independently searched against the mouse uniprotKB database (release date 2018.02) using an in-house Mascot™ server (Version 2.6, Matrix Science). Mass tolerances were +/− 10 ppm for MS peaks, and +/− 25 ppm for MS/MS fragment ions. The parameters included; trypsin as digestion enzyme with up to two missed cleavages permitted, carbamidomethyl (C) as a fixed modification and Pyro-glu (N-term), Oxidation (M).
Scaffold (version 4.8.0, Proteome Software, Portland, OR, USA) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least two identified unique peptides. To determine Rab40b-interacting proteins (both WT and SOCSAAAA), we established the following criteria. First, only proteins >3-fold IgG control (spectral counts) were analyzed. Second, any hits identified as non-specific based on the CRAPome database were dismissed, as well as any additional DNA, RNA, and mitochondrial proteins [36]. Finally, a 1.5-fold enrichment cutoff (spectral counts) was used to identify proteins preferentially bound to Rab40b SOCSAAAA vs Rab40b WT.
5. Notes
Make 200mM Boric Acid solution in water, and pH to 9.0 with NaOH. It is best to make this buffer the day of, as borate has a tendency to precipitate over time.
DMP is moisture sensitive and light sensitive. Powder stock should be stored with a desiccant. Once DMP is solubilized, keep in the dark for crosslinking reaction.
Ethanolamine hydrochloride was used for this experiment. Do not use any buffers that contain primary amines, as these will compete with the crosslinking reaction.
GMP-PNP was used for this experiment as a non-hydrolyzable form of GTP. It is possible to use other alternatives such as GTPγS, which we have successfully used for other GTP locking experiments.
It is important to do a small-scale immunoprecipitation test with the beads before moving on to the large-scale experiment. This is to make sure that both FLAG and IgG are covalently attached to the beads, and to troubleshoot any potential issues with elution off the beads. We recommend using 10μg of α-FLAG beads and 10μg of mouse IgG beads for the small-scale test. The amount of lysate needed is dependent on expression of your protein of interest, but for reference, 250μg of lysate was used in our small-scale experiment. For this smaller test, steps from 4.2 can be significantly shortened or skipped: GMP-PNP loading is not needed, lysate and beads can be incubated for just 1 hour, washing steps can be reduced to 1mL, etc. We do recommend keeping the pre-clearing step.
Although we know that Rab40b binding to Cul5 is GTP independent (data not shown), it is possible that potential ubiquitylation substrates only bind to Rab40b in a GTP dependent manner. To increase the success rate of capturing GTP-dependent substrates, we locked Rab40b in an active GTP bound state, via GMP-PNP loading (see Notes 4). Briefly, we stripped any nucleotide bound (during lysis) with EDTA, then re-loaded the GTP binding pocket with GMP-PNP. Finally, MgCl2 was added in excess to quench the reaction and stabilize nucleotide binding.
6. Results and Conclusion
After analyzing protein hits from two independent experiments (including cut off criteria described in 4.3), we identified a set of Rab40b binding proteins that were enriched in Rab40b SOCSAAAA compared to Rab40b WT. From the first run, 43.1% (81/188) of proteins were enriched in SOCSAAAA vs WT. In the second run, 52.5% (32/61) of proteins were enriched in SOCSAAAA vs WT. In Importantly, all of the core components of the Cul5 CRL were detected in the WT background and noticeably disrupted with the SOCSAAAA mutant in both experiments (Figure 3C). This disruption of the complex gives us confidence in the overall success of the experiment and the ability to identify true ubiquitylation targets. We are now validating these SOCSAAAA binding proteins through a number of different experiments. In general, we first begin by measuring overall protein levels of substrates in Rab40b SOCSAAAA and Rab40b knockout cells. This is to directly test the prediction that substrates levels should increase if we disrupt the complex responsible for its degradation and turnover. We follow this up with in vitro binding and ubiquitylation assays to confirm that Rab40b/Cul5 directly regulates a given substrate. Ultimately, the goal is to understand how ubiquitylation, or lack thereof, of a particular substrate impacts 3D cell migration. Given our previous evidence for Rab40b being a positive regulator of cell migration (decreased MMP2/9 secretion in Rab40b depleted cells), we hypothesize that Rab40b/Cul5 degrades negative regulators of cell migration processes.
Here we have presented an unbiased approach for identifying potential ubiquitylation targets of the Rab40b/Cul5 complex. The results and knowledge gained from this study will greatly surpass our original goal of elucidating novel Rab40b SOCS-dependent function during breast cancer cell migration, by allowing us to consider bigger picture questions. For example, this method can be broadly applied to other members of the Rab40 family and will help uncover the functional differences between the Rab40/Cul5 complexes. We also anticipate that this workflow will be useful for identifying ubiquitylation substrates of other SOCS box proteins.
7. Acknowledgements
We are grateful to Cayla Jewett for critical reading of this manuscript. We thank the University of Colorado School of Medicine Biological Mass Spectrometry Facility, specifically Monika Dzieciatkowska for performing the mass spectrometry and proteomic analysis. This work was supported by NIHT32GM008730 to EDD, NIHT32CA174648 to EL, and NIH1R01GM122768 to RP.
8. References
- 1.Stenmark H, Olkkonen VM (2001) The Rab GTPase family. Genome Biol 2:REVIEWS3007–7. doi: 10.1186/gb-2001-2-5-reviews3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coppola U, Ristoratore F, Albalat R, D’Aniello S (2019) The evolutionary landscape of the Rab family in chordates. Cellular and Molecular Life Sciences 76:4117–4130. doi: 10.1007/s00018-019-03103-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jékely G (2003) Small GTPases and the evolution of the eukaryotic cell. Bioessays 25:1129–1138. doi: 10.1002/bies.10353 [DOI] [PubMed] [Google Scholar]
- 4.Surkont J, Pereira-Leal JB (2016) Are There Rab GTPases in Archaea? Mol Biol Evol 33:1833–1842. doi: 10.1093/molbev/msw061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klöpper TH, Kienle N, Fasshauer D, Munro S (2012) Untangling the evolution of Rab G proteins: implications of a comprehensive genomic analysis. BMC Biol 10:71–17. doi: 10.1186/1741-7007-10-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zerial M, McBride H (2001) Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2:107–117. doi: 10.1038/35052055 [DOI] [PubMed] [Google Scholar]
- 7.Stenmark H (2009) Rab GTPases as coordinators of vesicle traffic. Nature Publishing Group 10:513–525. doi: 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- 8.Pfeffer SR (2001) Rab GTPases: specifying and deciphering organelle identity and function. Trends in Cell Biology 11:487–491. doi: 10.1016/s0962-8924(01)02147-x [DOI] [PubMed] [Google Scholar]
- 9.Grosshans BL, Ortiz D, Novick P (2006) Rabs and their effectors: achieving specificity in membrane traffic. Proceedings of the National Academy of Sciences 103:11821–11827. doi: 10.1073/pnas.0601617103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz SL, Cao C, Pylypenko O, et al. (2008) Rab GTPases at a glance. J Cell Sci 121:246–246. doi: 10.1242/jcs.03495 [DOI] [PubMed] [Google Scholar]
- 11.Hutagalung AH, Novick PJ (2011) Role of Rab GTPases in Membrane Traffic and Cell Physiology. Physiol Rev 91:119–149. doi: 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pereira-Leal JB, Seabra MC (2001) Evolution of the rab family of small GTP-binding proteins. Journal of Molecular Biology 313:889–901. doi: 10.1006/jmbi.2001.5072 [DOI] [PubMed] [Google Scholar]
- 13.Pereira-Leal JB, Seabra MC (2000) The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily 1 1Edited by M. Yaniv. Journal of Molecular Biology 301:1077–1087. doi: 10.1006/jmbi.2000.4010 [DOI] [PubMed] [Google Scholar]
- 14.Pylypenko O, Hammich H, Yu I-M, Houdusse A (2018) Rab GTPases and their interacting protein partners: Structural insights into Rab functional diversity. 1–27. doi: 10.1080/21541248.2017.1336191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasahara M, Naruse K, Sasaki S, et al. (2007) The medaka draft genome and insights into vertebrate genome evolution. Nature 447:714–719. doi: 10.1038/nature05846 [DOI] [PubMed] [Google Scholar]
- 16.Hilton DJ, Richardson RT, Alexander WS, et al. (1998) Twenty proteins containing a C-terminal SOCS box form five structural classes. Proceedings of the National Academy of Sciences 95:114–119. doi: 10.1073/pnas.95.1.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hilton DJ (1999) Negative regulators of cytokine signal transduction. Cell Mol Life Sci 55:1568–1577. doi: 10.1007/s000180050396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okumura F, Joo-Okumura A, Nakatsukasa K, Kamura T (2016) The role of cullin 5-containing ubiquitin ligases. Cell Division 1–16. doi: 10.1186/s13008-016-0016-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petroski MD, Deshaies RJ (2005) Function and regulation of cullin–RING ubiquitin ligases. Nat Rev Mol Cell Biol 6:9–20. doi: 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- 20.Linossi EM, Nicholson SE (2012) The SOCS box-Adapting proteins for ubiquitination and proteasomal degradation. IUBMB Life 64:316–323. doi: 10.1002/iub.1011 [DOI] [PubMed] [Google Scholar]
- 21.Kile BT, Schulman BA, Alexander WS, et al. (2002) The SOCS box: a tale of destruction and degradation. Trends in Biochemical Sciences 27:235–241. [DOI] [PubMed] [Google Scholar]
- 22.Mahrour N, Redwine WB, Florens L, et al. (2008) Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. Journal of Biological Chemistry 283:8005–8013. doi: 10.1074/jbc.M706987200 [DOI] [PubMed] [Google Scholar]
- 23.Kamura T, Sato S, Haque D, et al. (1998) The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes & Development 12:3872–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamura T, Maenaka K, Kotoshiba S, et al. (2004) VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes & Development 18:3055–3065. doi: 10.1101/gad.1252404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim YK, Kwak MJ, Ku B, et al. (2013) Structural basis of intersubunit recognition in elongin BC-cullin 5-SOCS box ubiquitin-protein ligase complexes. Acta Cryst (2013) D69, 1587–1597 [doi:101107/S0907444913011220] 1–11. doi: 10.1107/S0907444913011220 [DOI] [PubMed] [Google Scholar]
- 26.Dart AE, Box GM, Court W, et al. (2015) PAK4 promotes kinase-independent stabilization of RhoU to modulate cell adhesion. J Cell Biol 211:863–879. doi: 10.1083/jcb.201501072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bedoyan JK, Schaibley VM, Peng W, et al. (2012) Disruption of RAB40AL function leads to Martin–Probst syndrome, a rare X-linked multisystem neurodevelopmental human disorder. J Med Genet 49:332–340. doi: 10.1136/jmedgenet-2011-100575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ołdak M, Ruszkowska E, Pollak A, et al. (2014) A note of caution on the diagnosis of Martin-Probst syndrome by the detection of the p.D59G mutation in the RAB40AL gene. Eur J Pediatr 174:693–696. doi: 10.1007/s00431-014-2452-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ołdak M, Ścieżyńska A, Młynarski W, et al. (2014) Evidence Against RAB40ALBeing the Locus for Martin-Probst X-Linked Deafness-Intellectual Disability Syndrome. Human Mutation 35:1171–1174. doi: 10.1002/humu.22620 [DOI] [PubMed] [Google Scholar]
- 30.Jacob A, Jing J, Lee J, et al. (2013) Rab40b regulates trafficking of MMP2 and MMP9 during invadopodia formation and invasion of breast cancer cells. J Cell Sci 126:4647–4658. doi: 10.1242/jcs.126573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacob A, Linklater E, Bayless BA, et al. (2016) The role and regulation of Rab40b-Tks5 complex during invadopodia formation and cancer cell invasion. J Cell Sci 129:4341–4353. doi: 10.1242/jcs.193904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee RHK, Iioka H, Ohashi M, et al. (2007) XRab40 and XCullin5 form a ubiquitin ligase complex essential for the noncanonical Wnt pathway. EMBO J 26:3592–3606. doi: 10.1038/sj.emboj.7601781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yatsu A, Shimada H, Ohbayashi N, Fukuda M (2015) Rab40C is a novel Varp-binding protein that promotes proteasomal degradation of Varp in melanocytes. Biology Open 4:267–275. doi: 10.1242/bio.201411114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Gabin AG, Almazan G, Larocca JN (2004) Vesicle transport in oligodendrocytes: Probable role of Rab40c protein. J Neurosci Res 76:758–770. doi: 10.1002/jnr.20121 [DOI] [PubMed] [Google Scholar]
- 35.Tan R, Wang W, Wang S, et al. (2013) Small GTPase Rab40c Associates with Lipid Droplets and Modulates the Biogenesis of Lipid Droplets. PLoS ONE 8:e63213–11. doi: 10.1371/journal.pone.0063213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mellacheruvu D, Wright Z, Couzens AL, et al. (2013) The CRAPome: a contaminant repository for affinity purification–mass spectrometry data. Nat Methods 10:730–736. doi: 10.1038/nmeth.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]


