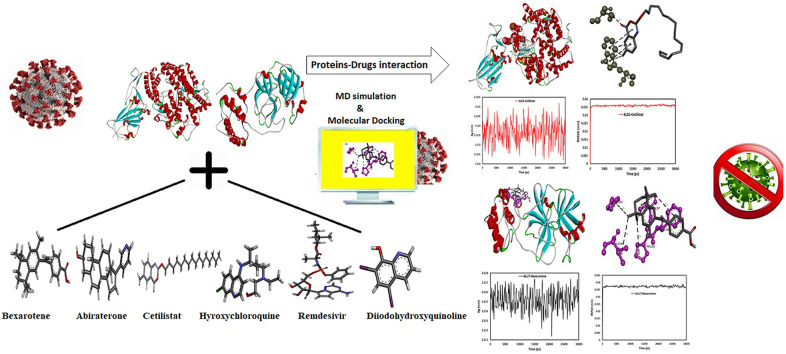
Abstract
By September 1, 2021, SARS-CoV-2, a respiratory virus that prompted Coronavirus Disease in 2019, had infected approximately 218,567,442 patients and claimed 4,534,151 lives. There are currently no specific treatments available for this lethal virus, although several drugs, including remdesivir and hydroxychloroquine, have been tested. The purpose of this study is to assess the activity of FDA-approved drugs cetilistat, abiraterone, diiodohydroxyquinoline, bexarotene, remdesivir, and hydroxychloroquine as potential SARS-CoV-2 main protease inhibitors. Additionally, this study aims to provide insight into the development of potential inhibitors that may inhibit ACE2, thereby preventing SARS-CoV-2 entry into the host cell and infection. To this end, remdesivir and hydroxychloroquine were used as comparator drugs. The calculations revealed that cetilistat, abiraterone, diiodohydroxyquinoline, and bexarotene inhibit main protease and ACE2 receptors more effectively than the well-known drug hydroxychloroquine when used against COVID-19. Meanwhile, bexarotene and cetilistat bind more tightly to the SARS-CoV-2 main protease and the ACE2 receptor, respectively, than remdesivir, a potential treatment for COVID-19 that is the first FDA-approved drug against this virus. As a result, the molecular dynamic simulations of these two drugs in the presence of proteins were investigated. The MD simulation results demonstrated that these drugs interact to stabilize the systems, allowing them to be used as effective inhibitors of these proteins. Meanwhile, bexarotene, abiraterone, cetilistat, and diiodohydroxyquinoline's systemic effects should be further investigated in suitable ex vivo human organ culture or organoids, animal models, or clinical trials.
Keywords: Molecular docking, Dynamic simulation, COVID-19, ACE2 receptor, Main protease
Graphical abstract
1. Introduction
SARS-CoV-2 (COVID-19) is a respiratory virus that originated in Wuhan, in 2019 [1,2]. This pandemic has spread aggressively throughout the world (Iran, Italy, India, Republic of Korea, United States of America, India, among others). Despite numerous significant efforts worldwide, there are currently no specific treatments for this lethal virus. Until September 1, 2021, this virus had infected 218,567,442 people worldwide and killed 4,534,151 people (https://www.worldometers.info/coronavirus/) due to lung failure caused by SARS-CoV-2 disease. COVID-19 attacks and destroys the protective proteins on T-lymphocytes in the human lung. This condition causes an accumulation of fluid in the lungs, resulting in breathing difficulties [3]. Numerous drug combinations have been used to treat this virus, including Ritonavir, Oseltamivir, and lopinavir [4], Darunavir, Remdesivir, Saquinavir [5], Sofosbuvir [6], Azithromycin and Hydroxychloroquine [7], and (Hydroxychloroquine and Chloroquine) [[8], [9], [10]]. The virus enters host cells via its surface spike glycoprotein and attacks the protein on the surface of human cells. It then begins its activity. Numerous drugs are directed against the main protease required for viral replication and the processing of polyproteins from the virus's RNA [11]. Thus, inhibiting this protease results in virus replication is prevented [12]. The kidney, lung cells, intestines, heart, lung cells, and arteries all have an outer membrane that contains an enzyme named ACE2. This critical enzyme plays a key role in cardiovascular/renal diseases and hypertension by regulating blood pressure and body fluid. The essential step in the infection of the host cell by SARS-CoV-2 is binding to the ACE2 receptor. Therefore, the ACE-2 receptor is a key target for researchers looking for therapeutic opportunities to combat this lethal virus. Anti-COVID-19 therapeutics that employ various strategies, including experimental and computational approaches, is critical for containing this pandemic disease [7,13]. Drug repurposing using a computer-aided drug design (CADD) approach, i.e., a computational drug repurposing study, is an appealing method for exploring potential therapeutic opportunities and new medications against COVID-19 that requires less time and expense. Multiple types of research have employed in silico approaches to investigate a promising candidate with a high binding affinity and interaction with the ACE-2 receptor and SARS-CoV-2 proteins. Bhardwaj et al. described the inhibitory activity of bioactive molecules derived from tea plants against SARS-main CoV-2's protease [14] and RNA-dependent RNA polymerase [15]. In another study, Bhardwaj et al. used a comparative in silico approach to evaluate the inhibitory activity of acridinedione analogs against the SARS-CoV-2 main protease [16]. Singh et al. [17] and Sharma et al. [18] recently reported on an in silico investigation of bioactive compounds from tea as potential inhibitors of SARS-CoV-2 nonstructural protein 15 and 16. In another research, Singh et al. [19] described a series of molecules based on α,β,γ-himachalenes scaffolds derived from a plant (Cedrus deodara) as potential SARS-CoV-2 nonstructural protein 1 inhibitors. Hussien et al. investigated the drug brincidofovir, which targets the ACE2 receptor and the main protease of SARS-CoV-2 [20]. Kalhor et al. used in silico strategies to determine the effect of the approved small molecule on the SARS-CoV-2 chimeric receptor-binding domain complexed with ACE2 [21]. Yuan et al. used a robust two-tier screening system in vitro to demonstrate the inhibitory activity of FDA-approved drugs cetilistat, abiraterone, diiodohydroxyquinoline, and bexarotene against COVID-19 infection [22]. A docking simulation approach was employed in this paper to investigate the interaction of four FDA-approved drugs, bexarotene (antineoplastic retinoid), abiraterone (synthetic androstane steroid), diiodohydroxyquinoline (anti-parasitic), and cetilistat (anti-pancreatic lipase), with the SARS-CoV-2 main protease and spike receptor domain complexed with ACE2 (ACE2-RBD). The results were compared to those obtained with hydroxychloroquine (an antimalarial medication) [7] and remdesivir (an antiviral drug). Additionally, the dynamic simulation elucidates the molecules' dynamic behavior. Several clinical trials indicated that antiviral, antimalarial, and anti-HIV medications were effective against the COVID-19 virus. Among them, remdesivir (Veklury®) is the first FDA-approved drug to treat COVID-19 disease in hospitalized adults and children aged 12 years and older who weigh at least 40 kg [23]. According to Nguyen et al., remdesivir may target RNA polymerase and the main protease, which may help explain why this drug is effective against COVID-19 [24]. Chloroquine and hydroxychloroquine were previously considered the most likely candidates for reducing the frequency of human SARS-CoV-2 infections [7]. Chloroquine was the first drug to be used on the front lines in China and abroad to treat severe SARS-CoV-2 infections.
While in vitro and preclinical evidence for these drugs was promising, clinical benefit has been limited, and the researchers concluded that hydroxychloroquine and chloroquine do not appear to be effective in reducing the mortality rate of COVID-19 patients hospitalized. Another solidarity trial, the French discovery trial, supported the findings (https://who.int/news-room/q-a-detail/coronavirus-disease-covid-19-hydroxychloroquine). Despite the study's limitations, researchers did not discourage using chloroquine or hydroxychloroquine, which were used in several countries to treat COVID-19. Hydroxychloroquine has a better clinical safety profile than chloroquine (over a long period), allows for a higher daily dose [25], and poses fewer concerns about drug-drug interactions [26]. Baildya et al. [27] and Beura et al. [28] employed an in silico approach to demonstrate the inhibitory activity of hydroxychloroquine on the COVID-19 main protease and the human ACE2 receptor. The drug-protein interaction was simulated in this study to better understand the mechanism of the proposed SARS-CoV-2 drugs and investigate the inhibitory effect of bexarotene, abiraterone diiodohydroxyquinoline, and cetilistat on the SARS-CoV-2 main protease. Additionally, this study aims to provide insight into the development of potential inhibitors that can inhibit ACE2 and thus prevent viral infection. The findings of this research can be used to introduce new candidate agents for ex vivo and in vivo surveys and provide necessary details for future research.
2. Methods
2.1. Docking of cetilistat, abiraterone, diiodohydroxyquinoline, bexarotene, remdesivir, and hydroxychloroquine
2.1.1. Receptors (proteins) preparation
AutoDock Vina [29] with MGL tools 1.5.4 was used to perform blind docking calculations. AutoDock Vina runs faster than AutoDock software and also makes more precise docking calculations [30]. The SARS-CoV-2 main protease and the ACE2-RBD were obtained from the Protein Data Bank (PDB ID: 6LU7, 6LZG) [31], and inhibitors were selected and removed. AutoDock Tools (v.1.5.4) was used to prepare the receptors. MGL Tools (v.1.5.4) combined non-polar hydrogen atoms and added Kollman charged atoms to protein crystal structures. SARS-CoV-2 main protease and ACE2-RBD coordinate files were then saved in PDBQT format. The 6LU7 was surrounded in a 55 × 68 × 65 box direction with a grid spacing of 1.00 Å and grid set centers of −26.02, 12.57, and 59.17, while the 6LZG was surrounded in a 64 × 74 × 110 box direction with a grid spacing of 1.00 Å and grid set centers of −25.41, 18.43, and −6.37.
2.1.2. Ligands (drugs) preparation
The three-dimensional SDF structures of cetilistat, abiraterone, diiodohydroxyquinoline, bexarotene, remdesivir, and hydroxychloroquine were obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov) and converted to PDB format. MGL Tools (v.1.5.4) was then used to save the drugs in PDBQT format, and docked results were visualized through the BIOVIA Discovery Studio software.
2.2. Molecular dynamics (MD) simulations
Molecular docking analysis revealed that bexarotene and cetilistat have the lowest energy in their interactions with the SARS-CoV-2 main protease and ACE2-RBD, respectively. As a result, the molecular dynamic simulations of these two drugs were investigated in the presence of proteins. Classical MD simulations [32,33] were conducted using the CHARMM27 force field [34,35]. SwissParam was used to generate the ligand topology [36]. TIP3P water [37] was used to solve the free protein and protein-ligand complexes in the cubic box with periodic boundary conditions in three directions. The solutes were positioned in the box's center, with a minimum distance of 1.0 nm between the solutes' surface and the box. Na+ ions were added to the system to neutralize the charge. The systems were balanced at 300 K and 1.0 bar after energy minimization through the steepest descent method. The Parrinello-Rahman barostat was used to maintain a pressure of 1.0 bar, and a temperature of 300 K was maintained using a modified Berendsen thermostat. The LINCS algorithm enabled the computation of bond lengths, while the particle-mesh Ewald scheme (PME) was used to compute long-range electrostatic forces (grid spacing 0.16 nm) [38]. The short-range nonbonded interactions were computed using cutoff ratios of 1.0 nm for van der Waals and Coulomb potentials. Finally, a 30 ns MD simulation with a time step of 2 fs was performed with random generation of velocities through a Maxwell distribution.
2.3. Interaction analysis by MM-PBSA binding energy
Using the Molecular Mechanics Poisson Boltzmann Surface Area (MM-PBSA) technique, the binding free energy of protein-ligand systems was calculated. This in silico method was a combined energy system described by the binding free energy, composed of electrostatic, SASA, van der Waals, and polar solvation energies. The MM-PBSA binding free energies were calculated using the GROMACS script g_mmpbsa [39]. Using the MM-PBSA method, the following equation was employed to determine the binding free energy of the interacting proteins:
| ΔGbinding = Gcomplex - (Greceptor + Gligand) |
ΔG binding denotes the total binding energy of the protein-ligand complexes; Greceptor and Gligand denote the binding energy of the free receptor and unbounded ligand, respectively.
3. Results
3.1. Molecular docking studies
While significant efforts are being made to develop drugs and vaccines against COVID-19, only a few available therapeutic agents are currently available. One of the most effective strategies for resolving this issue and combating this lethal virus is drug repurposing or using drugs already in clinical use for a different therapeutic indication. In drug discovery, molecular simulation approaches (docking and MD simulation) significantly reduce experimental time and cost [40]. Several research groups recently reported discovering inhibitors of the SARS-CoV-2 main protease and the ACE2 receptor [[41], [42], [43], [44]]. As is well known, the surface of the lung contains a large number of ACE2 receptors. Compounds that interact with the ACE2 receptor inhibit its function, decrease its surface area, and ultimately prevent virus entry into the host cell [45]. The main protease is a critical SARS-CoV-2 protein that plays a vital role in virus replication. Viral replication and infection may be inhibited by compounds that interact with this key protein [46]. Yuan et al. demonstrated that FDA-approved drugs cetilistat, abiraterone, diiodohydroxyquinoline, and bexarotene inhibited SARS-CoV-2 infection in vitro [22]. However, the question remains regarding the fundamental objective of this study. In this research, the binding affinity and interaction of cetilistat, abiraterone, diiodohydroxyquinoline, bexarotene, remdesivir, and hydroxychloroquine with the SARS-CoV-2 protease and ACE2 receptor were evaluated, which are responsible for virus replication and entry into the host cell, respectively. This computational study aids in clarifying the mechanism of the proposed SARS-CoV-2 drugs and investigates their inhibitory effect. As comparator drugs, remdesivir and hydroxychloroquine were used. Cetilistat is one of the pancreatic lipase inhibitors used to treat obesity. It inhibits fat digestion and absorption [47]. Diiodohydroxyquinoline is a quinolone derivative that is employed to treat amoebiasis as a luminal amebicide [48]. Diiodohydroxyquinoline is poorly absorbed into the circulatory system, and cetilistat is rapidly hydrolyzed into its metabolites when bile is present [47,49]. Approximately 15–20% of COVID-19 patients have been reported to exhibit gastrointestinal symptoms, with some also displaying infectious virus particles and detectable viral RNA [50]. The results of research on hamsters infected with SARS-CoV-2 revealed that their intestines expressed high levels of viral nucleocapsid protein and inflammation, as well as detectable viral RNA [51]. As with SARS, feces may be an essential source of SARS-CoV-2 infection [52,53], oral diiodohydroxyquinoline and cetilistat may be topical luminal antivirals in reducing viral shedding in the gastrointestinal tract. Bexarotene and abiraterone acetate are non-chemotherapeutic antineoplastic drugs with limited proven immunosuppressive effects [22]. Bexarotene is a third-generation retinoid that effectively treats breast cancer, non-small cell lung cancer, and cutaneous T cell lymphoma [[54], [55], [56], [57]]. Abiraterone acetate inhibited the androgen synthesizing enzyme CYP17A1 when combined with a corticosteroid. This procedure results in the treatment of refractory prostate cancer via androgen deprivation [58,59]. Yuan et al. reported that tamibarotene and AM580, both members of the same drug class as bexarotene, demonstrated remarkable antiviral activity against influenza viruses, Zika virus, coronaviruses (SARS-CoV and MERS-CoV), adenovirus, and enterovirus A71 [60]. The docked drugs with the SARS-CoV-2 main protease are depicted in Figs. S1–S6. The docking score of cetilistat, abiraterone, diiodohydroxyquinoline, bexarotene, remdesivir, and hydroxychloroquine with the SARS-CoV-2 main protease was calculated to determine their docking power on the crystal structure of the main protease. The binding free energy of bexarotene, remdesivir, abiraterone, cetilistat, diiodohydroxyquinoline, and hydroxychloroquine was −8.10,-7.80, −7.00, −6.60, −5.30, and −5.00 kcal/mol, respectively (Table 1 ).
Table 1.
Interaction energy between CoV-2 main protease and drug molecules.
| Drugs | ΔG (kcal/mol) |
|---|---|
| Abiraterone | −7.00 |
| Bexarotene | −8.10 |
| Cetilistat | −6.60 |
| Diiodohydroxyquinoline | −5.30 |
| Remdesivir | −7.80 |
| Hydroxychloroquine | −5.00 |
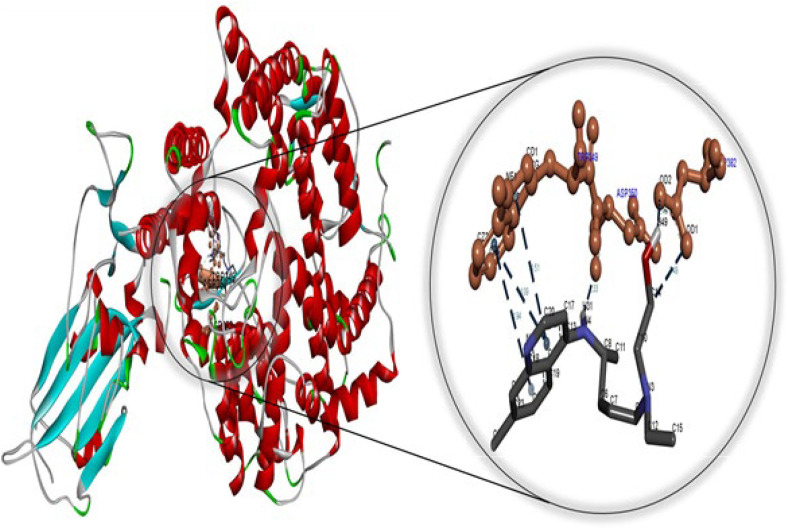
Cetilistat, abiraterone, diiodohydroxyquinoline, and bexarotene had lower binding free energy than hydroxychloroquine, which had in-vitro activity against COVID-19. The negative free energy values indicated a strong affinity for the binding pocket. These findings revealed that bexarotene formed a more stable complex with the SARS-CoV-2 main protease than remdesivir, the first FDA-approved drug against COVID-19, indicated by its lower free energy. In other words, the SARS-CoV-2 main protease favors bexarotene to the well-known drug remdesivir. The docking energies, atoms involved in bonding with ligands, the nature of interactions, and bond lengths are shown in Tables S1–S6. It is evident that hydrophobic interactions play a role in forming the complex between bexarotene and cetilistat and the SARS-CoV-2 main protease. Meanwhile, hydrophobic and hydrogen-bonding interactions play a significant role in forming the complex between abiraterone and hydroxychloroquine and SARS-CoV-2 protease. Additionally, electrostatic, hydrophobic, and hydrogen bonding interactions are dominant in the formation of the SARS-CoV-2 protease-diiodohydroxyquinoline and SARS-CoV-2 protease-remdesivir complexes. The amino acid residues surrounding drugs-SARS-CoV-2 protease systems are represented in Tables S1–S6. As illustrated in Fig. S2, the main protease system of bexarotene-SARS-CoV-2 is surrounded by the amino acids PHE294, ILE249, PRO252, PRO293, and VAL297. Overall, this research indicates that bexarotene with the highest binding free energy may inhibit viral replication and infection at levels comparable to those found in FDA-approved and clinically trialed drugs.
As illustrated in Tables S7–S12 and Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6 , docking results for cetilistat, bexarotene, abiraterone, diiodohydroxyquinoline, remdesivir, and hydroxychloroquine with ACE2-RBD were obtained.
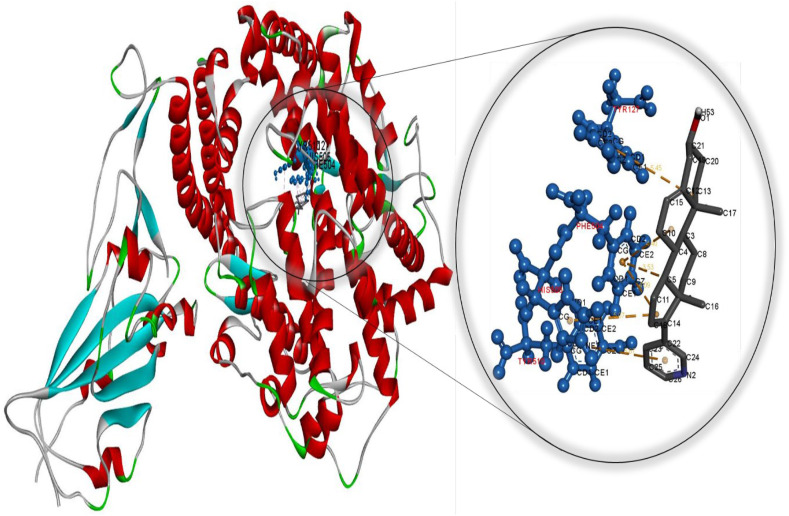
Fig. 1.
Molecular docking perspective of abiraterone – (ACE2-RBD).
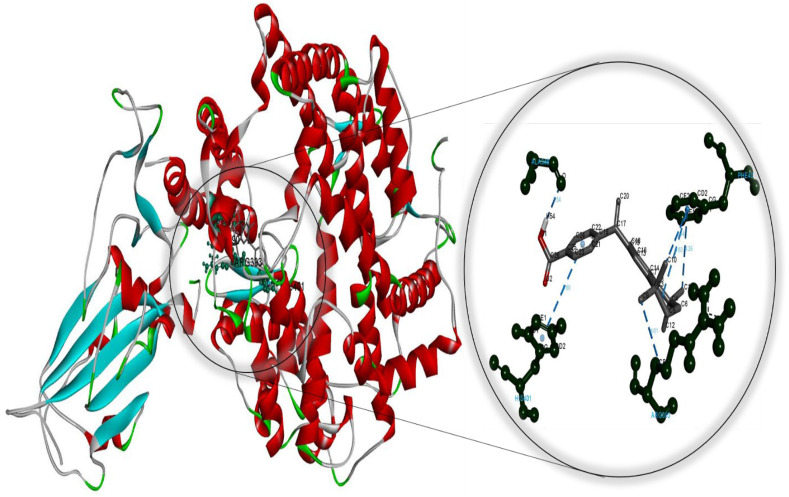
Fig. 2.
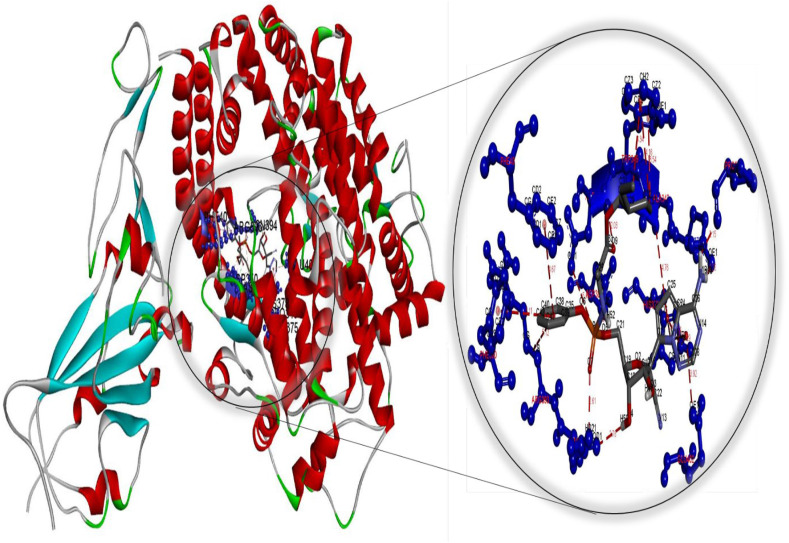
Molecular docking perspective of bexarotene – (ACE2-RBD).
Fig. 3.
Molecular docking perspective of cetilistat – (ACE2-RBD).
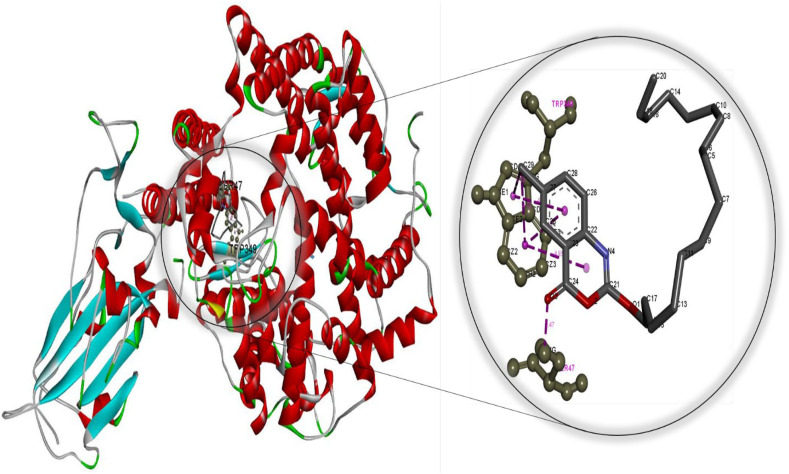
Fig. 4.
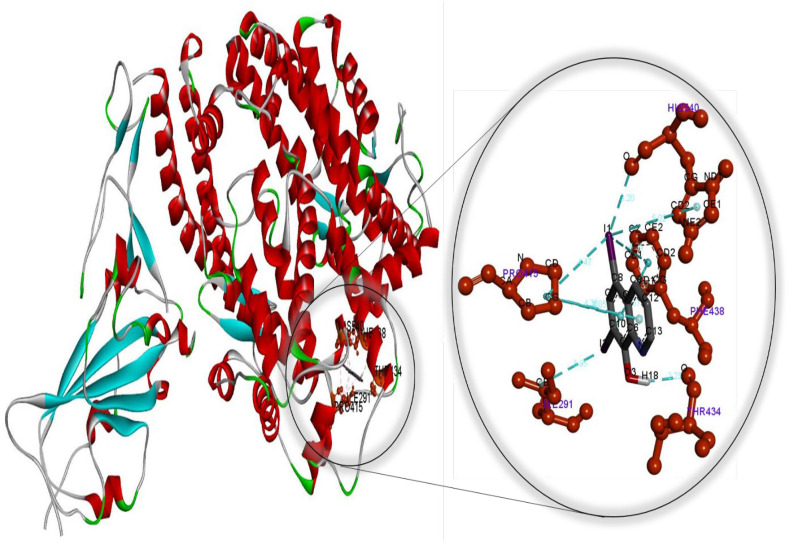
Molecular docking perspective of diiodohydroxyquinoline – (ACE2-RBD).
Fig. 5.
Molecular docking perspective of remdesivir – (ACE2-RBD).
Fig. 6.
Molecular docking perspective of hydroxychloroquine – (ACE2-RBD).
According to molecular docking studies, cetilistat, bexarotene, abiraterone, diiodohydroxyquinoline, remdesivir, and hydroxychloroquine do not interact with SARS-CoV-2 spike protein but do bind to the ACE2 receptor, preventing SARS CoV-2 spike protein from binding to ACE2. Cetilistat, bexarotene, abiraterone, remdesivir, diiodohydroxyquinoline, and hydroxychloroquine had binding free energies of −8.70, −8.40, −8.10, −8.00, −6.70, and −5.30 kcal/mol, respectively (Table 2 ).
Table 2.
Interaction energy between (ACE2-RBD) and drug molecules.
| Drugs | ΔG (KJ/mol) |
|---|---|
| Abiraterone | −8.10 |
| Bexarotene | −8.40 |
| Cetilistat | −8.70 |
| Diiodohydroxyquinoline | −6.70 |
| Remdesivir | −8.00 |
| Hydroxychloroquine | −5.30 |
Cetilistat, bexarotene, and abiraterone had a lower binding free energy than remdesivir and hydroxychloroquine. Additionally, diiodohydroxyquinoline had a lower binding free energy than hydroxychloroquine. These findings indicated that cetilistat, bexarotene, and abiraterone form a more stable complex with ACE2 than remdesivir and hydroxychloroquine, indicated by their lower binding free energy values.
Meanwhile, the diiodohydroxyquinoline-ACE2 complex formed is more stable than the hydroxychloroquine-ACE2 complex. These findings strongly suggest that cetilistat, which had the lowest binding free energy, interacts efficiently with ACE2 receptors and disrupts SARS-CoV-2 attachment. Tables S7–S12 summarize the results of docking simulations, including the nature of interactions, the atoms involved in bonding with drugs, docking energies, and bond lengths. As can be seen, hydrophobic interactions play a role in the formation of the cetilistat-ACE2 system. Furthermore, one hydrogen bond was formed between the cetilistat-ACE2 system. SER47 and TRP349 amino acid residues surrounded the cetilistat-ACE2 system. Both hydrophobic and hydrogen bonding interactions were dominant in the formation of a complex between bexarotene and ACE2. The bexarotene-ACE2 system was surrounded by amino acids ALA348, HIS401, ARG393, PHE40, and PHE390. PHE504, TYR510, TYR127, and HIS505 were involved in the formation of hydrophobic interactions between abiraterone and ACE2 of SARS-CoV-2. The diiodohydroxyquinoline - ACE2 system resulted in the formation of one hydrogen and halogen bond. Moreover, the formation of the diiodohydroxyquinoline - ACE2 complex involved hydrophobic interactions. Five amino acids influenced the hydrophobic interactions between diiodohydroxyquinoline and ACE2: THR434, HIS540, PHE438 ILE291, and PRO415. By blocking the angiotensin-converting enzyme-2 receptor, the proposed FDA-approved drugs cetilistat, bexarotene, abiraterone, and diiodohydroxyquinoline may inhibit the attachment of SARS-CoV-2 to this receptor and thus neutralize viral entry into the host cell. As a result, these drugs may be promising candidates for inhibiting covid-19 infection at concentrations comparable to those found in FDA-approved and clinically tested drugs such as remdesivir and hydroxychloroquine. On the whole, these drugs interacted with both the main protease and the ACE2 receptor, indicating that they could be used as multitarget drugs against COVID-19, and their systemic effects should be further investigated in suitable ex vivo human organ culture or organoids, animal models, or clinical trials. The researchers hope that this study will aid in the development of potential SARS-CoV-2 vaccines and therapeutics.
3.2. Molecular dynamics simulations
3.2.1. Radius of gyration (Rg)
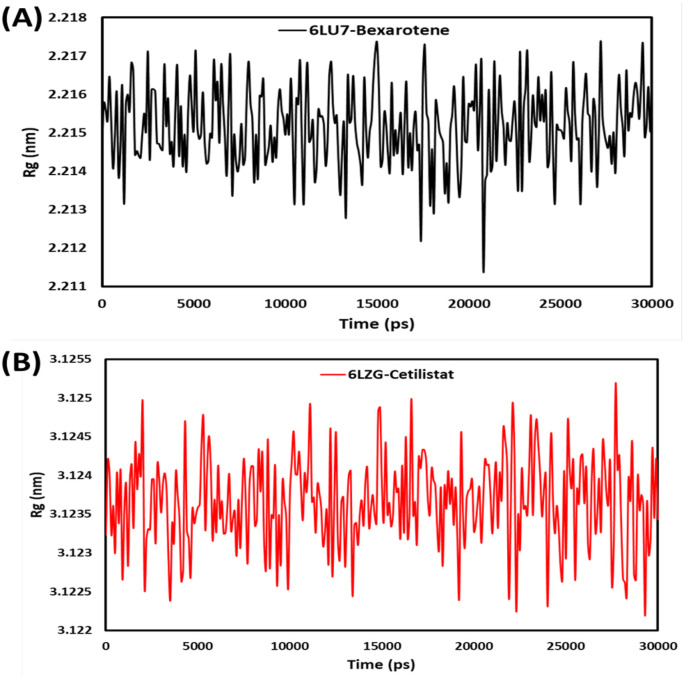
The radius of gyration (Rg) of both ACE2-cetilistat and protease-bexarotene complexes quantifies the molecule's overall extension during a 30ns MD run (Fig. 7 ). A low Rg value demonstrates better structural entirety and folding treatment [61]. Throughout the 30ns MD simulation, two complexes maintain a stable mean Rg of 2.2 nm for 6LU7-Bex and 3.2 nm for 6LZG-Ceti. During simulation, a slight enhancement in the Rg value of the 6LZG-Ceti complex was observed, indicating its structural integrity. The MD simulation results entirely support that bexarotene and cetilistat form stable complexes with 6LU7 and 6LZG, indicating their inhibitory properties for the main protease and ACE2 receptor, respectively.
Fig. 7.
Radius of gyration (Rg) for (A) bexarotene – CoV-2 main protease and (B) cetilistat – (ACE2-RBD) during 30 ns MD simulation.
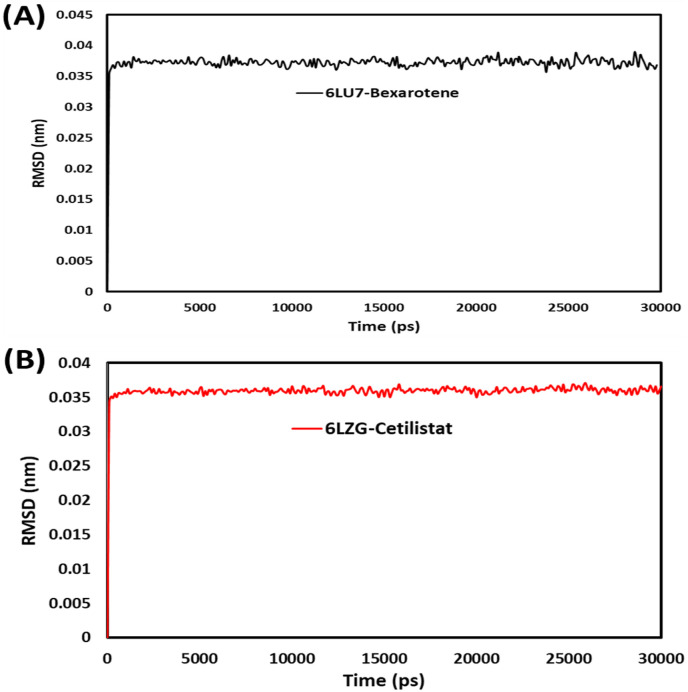
3.2.2. RMSD
RMSD analysis revealed insights and structural changes in the protein that confirm the protein's stability and equilibrium during simulation. The RMSD plot of the backbone atoms in the 6LU7 complex with bexarotene and the 6LZG complex with cetilistat is shown in Fig. 8 . RMSD was calculated for the 6LU7-Bex and 6LZG-Ceti structures that converged during the 30ns MD simulation. The results indicated that both structures stabilized after 1000ps. 6LU7-Bex averaged 0.37 A0, while 6LZG-Ceti averaged 0.36 A0 throughout the 30ns simulation. The low RMSD values indicated that bexarotene and cetilistat were stable in MD simulations with proteins.
Fig. 8.
RMSD plots for (A) bexarotene – CoV-2 main protease and (B) cetilistat – (ACE2-RBD) during 30 ns MD simulation.
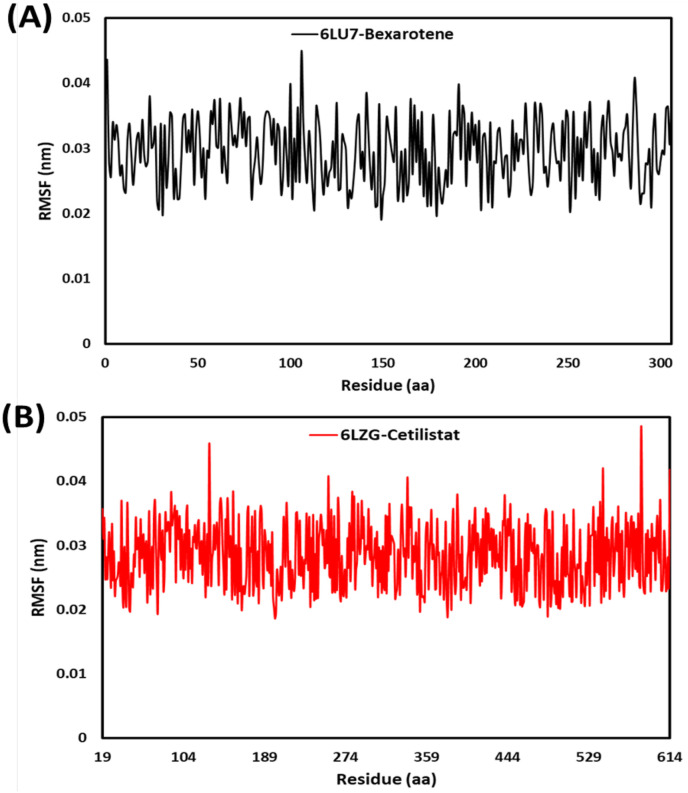
3.2.3. RMSF
RMSF analysis was used to determine the flexibility of the total protein concerning its average structure. Low RMSF values demonstrated narrowed movements, whereas high RMSF values demonstrated increased flexibility [62]. Ligand binding poses energy, and interaction has a direct correlation with residual fluctuation (RMSF) values. The RMSF plots for each residue in the 6LU7 complex with bexarotene and 6LZG complex with cetilistat are shown in Fig. 9 . The RMSF values for the 6LU7-Bex and 6LZG-Ceti complexes were extremely low; as a result, they exhibited minimal movement, indicating that both complexes were stable. RMSF values were 0.29 and 0.28 A0 on average for 6LU7-Bex and 6LZG-Ceti during a 30 ns simulation. During the simulation, it was observed that residues in the loop region of 6LU7 were more fluctuated than those in the alpha-helix and beta-sheet regions. This indicated that the protein remained stable throughout the 30ns simulation period [31]. Except for the loop region, the residues 584 in alpha-helix and 131 in beta-sheet region of 6LZG exhibited significant variation up to 0.48 A0 and 0.45 A0, respectively. Overall, the RMSF values for both proteins indicated that the complexes 6LU7-Bex and 6LZG-Ceti were stable [63].
Fig. 9.
RMSF plots for (A) bexarotene – CoV-2 main protease and (B) cetilistat – (ACE2-RBD) during 30 ns MD simulation.
3.3. MM-PBSA binding free energy
MmPbSaStat.py was used to calculate the average free binding energy of the 6LU7-Bex and 6LZG-Ceti (Table 3 ). The average free binding energy and the standard deviation/error of the files were calculated using the information obtained from g_mmpbsa. The binding free energy can evaluate the durability of the ligand-receptor interaction, which is a critical parameter in drug discovery. The lower the binding energy, the more effectively the ligand and protein bind [14]. Except for the polar solvation energy, the van der Waals, SASA, and electrostatic energies were used to bind 6LU7 and 6LZG to bexarotene and cetilistat. van der Waals energy augmentation to the overall binding free energy was greater than that of electrostatic contribution energy. The binding free energies of 6LU7-Bex and 6LZG-Ceti were determined to be −63.40 ± 11.70 and −95.92 ± 12.04 kJ/mol, respectively, indicating that the studied drugs interacted uniquely with the 6LU7 and 6LZG proteins. The binding energy against time graphs for 6LU7-Bex and 6LZG-Ceti are depicted in Fig. S7. The findings above indicate that bexarotene and cetilistat may be candidates for inhibiting the main protease of SARS-CoV-2 and ACE2 receptors.
Table 3.
Binding free energy (MM-PBSA) calculations for 6LU7-Bexarotene and 6LZG-Cetilistat.
| system | ΔEvan der Waal (kJ/mol) | ΔEElectrostatic (kJ/mol) | ΔEPolar solvation (kJ/mol) | ΔESASA (kJ/mol) | ΔEBinding (kJ/mol) |
|---|---|---|---|---|---|
| 6LU7-Bex | −73.14 ± 11.10 | −11.88 ± 17.51 | 32.02 ± 21.65 | −10.40 ± 1.45 | −63.40 ± 11.70 |
| 6LZG-Ceti | −181.08 ± 16.80 | −4.24 ± 12.22 | 110.38 ± 18.32 | −20.98 ± 2.05 | −95.92 ± 12.04 |
4. Conclusion
The docking studies revealed that the binding free energies of FDA-approved drugs cetilistat, abiraterone, diiodohydroxyquinoline, and bexarotene are more potent than those of hydroxychloroquine, which has been used against COVID-19. In other words, these drugs formed more stable complexes with the main protease of SARS-CoV-2 and the ACE2 receptor. Additionally, cetilistat and bexarotene have a higher affinity for and interactions with the ACE2 receptor and the main SARS-CoV-2 proteases than remdesivir, the first FDA-approved anti-COVID-19 drug. In this study, Gromacs software was used to simulate the interaction of bexarotene and cetilistat with the SARS-CoV-2 main protease and ACE2-RBD. Furthermore, the radius of gyration of ACE2-cetilistat and protease-bexarotene have values that indicate the systems' stability. Moreover, low RMSF and RMSD values indicate that bexarotene and cetilistat are stable in the presence of proteins during MD simulation. Finally, these drugs may be tested in vivo as a potent and promising anti-COVID-19 candidate.
Data availability
Because the majority of work is performed using licensed software, these are not transferable. Additional scripts written by members of the group may be made available upon request.
Code availability
Most of the work is done with licensed software therefore those are not transferable. Other scripts written in the group could be made available on request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors thank the Razi University Research Council for support of this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2021.100745.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A. Severe acute respiratory syndrome-related coronavirus: the species and its viruses – a statement of the Coronavirus Study Group. bioRxiv. 2020 doi: 10.1101/2020.02.07.937862. In press. [DOI] [Google Scholar]
- 2.Gautret P., Lagier J.-C., Parola P., Hoang V.T., Meddeb L., Mailhe M. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.-Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muralidharan N., Sakthivel R., Velmurugan D., Gromiha M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 Protease against COVID-19. J Biomol Struct Dyn. 2020;39:2673–2678. doi: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- 5.Khan S.A., Zia K., Ashraf S., Uddin R., Ul-Haq Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J Biomol Struct Dyn. 2020;39:2607–2616. doi: 10.1080/07391102.2020.1751298. [DOI] [PubMed] [Google Scholar]
- 6.Elfiky A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gautret P., Lagier J.-C., Parola P., Meddeb L., Mailhe M., Doudier B. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. Epub 2020 Feb 19. [DOI] [PubMed] [Google Scholar]
- 10.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71:732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tam N.M., Pham M.Q., Ha N.X., Nam P.C., Phung H.T.T. Computational estimation of potential inhibitors from known drugs against the main protease of SARS-CoV-2. RSC Adv. 2021;11:17478–17486. doi: 10.1039/d1ra02529e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.-Y. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yalçın S., Yalçınkaya S., Ercan F. In silico detection of inhibitor potential of Passiflora compounds against SARS-Cov-2 (Covid-19) main protease by using molecular docking and dynamic analyses. J Mol Struct. 2021;1240 doi: 10.1016/j.molstruc.2021.130556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. 2021;39:3449–3458. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Bioactive molecules of Tea as potential inhibitors for RNA-dependent RNA polymerase of SARS-CoV-2. Front Med. 2021:8. doi: 10.3389/fmed.2021.684020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhardwaj V.K., Singh R., Das P., Purohit R. Evaluation of acridinedione analogs as potential SARS-CoV-2 main protease inhibitors and their comparison with repurposed anti-viral drugs. Comput Biol Med. 2021;128 doi: 10.1016/j.compbiomed.2020.104117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh R., Bhardwaj V.K., Sharma J., Purohit R., Kumar S. In-silico evaluation of bioactive compounds from tea as potential SARS-CoV-2 nonstructural protein 16 inhibitors. J. Tradit. Compl. Med. 2021 doi: 10.1016/j.jtcme.2021.05.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma J., Bhardwaj V.K., Singh R., Rajendran V., Purohit R., Kumar S. An in-silico evaluation of different bioactive molecules of tea for their inhibition potency against non structural protein-15 of SARS-CoV-2. Food Chem. 2021;346 doi: 10.1016/j.foodchem.2020.128933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh R., Bhardwaj V.K., Das P., Purohit R. A computational approach for rational discovery of inhibitors for non-structural protein 1 of SARS-CoV-2. Comput Biol Med. 2021 doi: 10.1016/j.compbiomed.2021.104555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussien M.A., Abdelaziz A.E. Molecular docking suggests repurposing of brincidofovir as a potential drug targeting SARS-CoV-2 ACE2 receptor and main protease. Netw. 2020;9:1–8. doi: 10.1007/s13721-020-00263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalhor H., Sadeghi S., Abolhasani H., Kalhor R., Rahimi H. Repurposing of the approved small molecule drugs in order to inhibit SARS-CoV-2 S protein and human ACE2 interaction through virtual screening approaches. J Biomol Struct Dyn. 2020:1–6. doi: 10.1080/07391102.2020.1824816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan S., Chan J.F., Chik K.K., Chan C.C., Tsang J.O., Liang R. Discovery of the FDA-approved drugs bexarotene, cetilistat, diiodohydroxyquinoline, and abiraterone as potential COVID-19 treatments with a robust two-tier screening system. Pharmacol Res. 2020;159 doi: 10.1016/j.phrs.2020.104960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Food U., Administration D. 2020. FDA approves first treatment for COVID-19. [Google Scholar]
- 24.Nguyen H.L., Thai N.Q., Truong D.T., Li M.S. Remdesivir strongly binds to both RNA-dependent RNA polymerase and main protease of SARS-CoV-2: evidence from molecular simulations. J Phys Chem B. 2020;124:11337–11348. doi: 10.1021/acs.jpcb.0c07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marmor M.F., Kellner U., Lai T.Y., Melles R.B., Mieler W.F. Recommendations on screening for chloroquine and hydroxychloroquine retinopathy (2016 revision) Ophthalmology. 2016;123:1386–1394. doi: 10.1016/j.ophtha.2016.01.058. [DOI] [PubMed] [Google Scholar]
- 26.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin Infect Dis. 2020;71:732–739. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baildya N., Ghosh N.N., Chattopadhyay A.P. Inhibitory activity of hydroxychloroquine on COVID-19 main protease: an insight from MD-simulation studies. J Mol Struct. 2020;1219 doi: 10.1016/j.molstruc.2020.128595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beura S., Chetti P. In-silico strategies for probing chloroquine based inhibitors against SARS-CoV-2. J Biomol Struct Dyn. 2020;11:28876–28891. doi: 10.1080/07391102.2020.1772111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh S., Srivastava H.K., Kishor G., Singh H., Agrawal P., Raghava G.P. Evaluation of protein-ligand docking methods on peptide-ligand complexes for docking small ligands to peptides. BioRxiv. 2017 [Google Scholar]
- 31.Prasanth D., Murahari M., Chandramohan V., Panda S.P., Atmakuri L.R., Guntupalli C. In silico identification of potential inhibitors from Cinnamon against main protease and spike glycoprotein of SARS CoV-2. J Biomol Struct Dyn. 2020:1–15. doi: 10.1080/07391102.2020.1779129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berendsen H.J., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 1995;91:43–56. [Google Scholar]
- 33.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 34.Bjelkmar P., Larsson P., Cuendet M.A., Hess B., Lindahl E. Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J Chem Theor Comput. 2010;6:459–466. doi: 10.1021/ct900549r. [DOI] [PubMed] [Google Scholar]
- 35.Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 37.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. Int. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 38.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅ log (N) method for Ewald sums in large systems. Int. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 39.Kumari R., Kumar R. Consortium OSDD, Lynn A. g_mmpbsa– A GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 40.Vardhan S., Sahoo S.K. In silico ADMET and molecular docking study on searching potential inhibitors from limonoids and triterpenoids for COVID-19. Comput Biol Med. 2020;124 doi: 10.1016/j.compbiomed.2020.103936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.García-Fernández R., Ziegelmüller P., González L., Mansur M., Machado Y., Redecke L. Two variants of the major serine protease inhibitor from the sea anemone Stichodactyla helianthus, expressed in Pichia pastoris. Protein Expr. Purif. 2016;123:42–50. doi: 10.1016/j.pep.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 42.Verma D., Kapoor S., Das S., Thakur K. 2020. Potential inhibitors of SARS-CoV-2 main protease (M pro) identified from the library of FDA approved drugs using molecular docking studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohammadi S., Heidarizadeh M., Entesari M., Esmailpour A., Esmailpour M., Moradi R. In silico investigation on the inhibiting role of nicotine/caffeine by blocking the S protein of SARS-CoV-2 versus ACE2 receptor. Microorganisms. 2020;8:1600. doi: 10.3390/microorganisms8101600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wanga N, Hana S, Liua R, Mengb L, Hea H, Zhanga Y, et al. Chloroquine and hydroxychloroquine as ACE2 blockers to inhibit viropexis of COVID-19 Spike pseudotype virus. [DOI] [PMC free article] [PubMed]
- 45.Imai Y., Kuba K., Penninger J.M. The discovery of angiotensin-converting enzyme 2 and its role in acute lung injury in mice. Exp Physiol. 2008;93:543–548. doi: 10.1113/expphysiol.2007.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hasan M., Parvez M.S.A., Azim K.F., Imran M.A.S., Raihan T., Gulshan A. Main protease inhibitors and drug surface hotspots for the treatment of COVID-19: a drug repurposing and molecular docking approach. Biomed Pharmacother. 2021;140 doi: 10.1016/j.biopha.2021.111742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bryson A., De La Motte S., Dunk C. Reduction of dietary fat absorption by the novel gastrointestinal lipase inhibitor cetilistat in healthy volunteers. Br J Clin Pharmacol. 2009;67:309–315. doi: 10.1111/j.1365-2125.2008.03311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rickards A. The treatment of amoebiasis with" Diodoquin". Am. J. Trop. 1949;52:33–38. [PubMed] [Google Scholar]
- 49.Padwal R. Cetilistat, a new lipase inhibitor for the treatment of obesity. Curr Opin Invest Drugs. 2008;9:414–421. [PubMed] [Google Scholar]
- 50.Cheung K.S., Hung I.F., Chan P.P., Lung K., Tso E., Liu R. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology. 2020;159:81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan J.F.-W., Zhang A.J., Yuan S., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID-19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20:660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hung I., Cheng V., Wu A., Tang B., Chan K., Chu C. Viral loads in clinical specimens and SARS manifestations. Emerg Infect Dis. 2004;10:1550. doi: 10.3201/eid1009.040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boehm M.F., Zhang L., Zhi L., McClurg M.R., Berger E., Wagoner M. Design and synthesis of potent retinoid X receptor selective ligands that induce apoptosis in leukemia cells. J Med Chem. 1995;38:3146–3155. doi: 10.1021/jm00016a018. [DOI] [PubMed] [Google Scholar]
- 55.Gniadecki R., Assaf C., Bagot M., Dummer R., Duvic M., Knobler R. The optimal use of bexarotene in cutaneous T-cell lymphoma. Br J Dermatol. 2007;157:433–440. doi: 10.1111/j.1365-2133.2007.07975.x. [DOI] [PubMed] [Google Scholar]
- 56.Dragnev K.H., Petty W.J., Shah S.J., Lewis L.D., Black C.C., Memoli V. A proof-of-principle clinical trial of bexarotene in patients with non–small cell lung cancer. Clin Canc Res. 2007;13:1794–1800. doi: 10.1158/1078-0432.CCR-06-1836. [DOI] [PubMed] [Google Scholar]
- 57.Esteva F.J., Glaspy J., Baidas S., Laufman L., Hutchins L., Dickler M. Multicenter phase II study of oral bexarotene for patients with metastatic breast cancer. J Clin Oncol. 2003;21:999–1006. doi: 10.1200/JCO.2003.05.068. [DOI] [PubMed] [Google Scholar]
- 58.Duc I., Bonnet P., Duranti V., Cardinali S., Rivière A., De Giovanni A. In vitro and in vivo models for the evaluation of potent inhibitors of male rat 17α-hydroxylase/C17, 20-lyase. J Steroid Biochem Mol Biol. 2003;84:537–542. doi: 10.1016/s0960-0760(03)00078-5. [DOI] [PubMed] [Google Scholar]
- 59.Arasaratnam M., Crumbaker M., Bhatnagar A., McKay M.J., Molloy M.P., Gurney H. Inter-and intra-patient variability in pharmacokinetics of abiraterone acetate in metastatic prostate cancer. Canc. Chemother. Pharmacol. 2019;84:139–146. doi: 10.1007/s00280-019-03862-x. [DOI] [PubMed] [Google Scholar]
- 60.Yuan S., Chu H., Chan J.F.-W., Ye Z.-W., Wen L., Yan B. SREBP-dependent lipidomic reprogramming as a broad-spectrum antiviral target. Nat Commun. 2019;10:1–5. doi: 10.1038/s41467-018-08015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Erva R.R., Rajulapati S.B., Durthi C.P., Bhatia M., Pola M. Molecular dynamic simulations of Escherichia coli L-asparaginase to illuminate its role in deamination of asparagine and glutamine residues. 3 Biotech. 2016;6:2. doi: 10.1007/s13205-015-0339-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Priya R., Sumitha R., Doss C.G.P., Rajasekaran C., Babu S., Seenivasan R. Molecular docking and molecular dynamics to identify a novel human immunodeficiency virus inhibitor from alkaloids of Toddalia asiatica. 2015;11:S414. doi: 10.4103/0973-1296.168947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Basit A., Ali T., Rehman S.U. Truncated human angiotensin converting enzyme 2; a potential inhibitor of SARS-CoV-2 spike glycoprotein and potent COVID-19 therapeutic agent. J Biomol Struct Dyn. 2021;39(10):3605–3614. doi: 10.1080/07391102.2020.1768150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Because the majority of work is performed using licensed software, these are not transferable. Additional scripts written by members of the group may be made available upon request.