Abstract
At present times, COVID-19 has become a global illness and infected people has increased exponentially and it is difficult to control due to the non-availability of large quantity of testing kits. Artificial intelligence (AI) techniques including machine learning (ML), deep learning (DL), and computer vision (CV) approaches find useful for the recognition, analysis, and prediction of COVID-19. Several ML and DL techniques are trained to resolve the supervised learning issue. At the same time, the potential measure of the unsupervised learning technique is quite high. Therefore, unsupervised learning techniques can be designed in the existing DL models for proficient COVID-19 prediction. In this view, this paper introduces a novel unsupervised DL based variational autoencoder (UDL-VAE) model for COVID-19 detection and classification. The UDL-VAE model involved adaptive Wiener filtering (AWF) based preprocessing technique to enhance the image quality. Besides, Inception v4 with Adagrad technique is employed as a feature extractor and unsupervised VAE model is applied for the classification process. In order to verify the superior diagnostic performance of the UDL-VAE model, a set of experimentation was carried out to highlight the effective outcome of the UDL-VAE model. The obtained experimental values showcased the effectual results of the UDL-VAE model with the higher accuracy of 0.987 and 0.992 on the binary and multiple classes respectively.
Keywords: COVID-19, Deep learning, Unsupervised learning, Variational autoencoder, Image classification
1. Introduction
Recently, 2019 novel coronavirus is referred as COVID-19 originated from Wuhan city, China in December 2019, and it has been progressed as a dreadful disease and considered a communal health problem around the globe. The COVID-19 is a global disorder and evolved from Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [1]. Coronaviruses (CoV) belong to the virus family and the disease is named COVID-19 which mimics the virus SARS-CoV as well as Middle East Respiratory Syndrome (MERS-CoV). COVID-19 has defined a novel viral infection developed in 2019 which has not been predicted in human beings. Also, it is considered to be a zoonotic disorder as the germs have affected both animals and users. Developers have found that SARS-CoV virus is formed out of decomposed cats and it has been spread among human beings while MERS-CoV virus is desecration from dromedaries and distributed from Arabian camel to humans. COVID-19 virus is actually dispersed from bats to human beings. This type of virus is transferred through respiratory organs from human to human and results in rapid virus transmission. It provokes slight symbols for most of the patients and some people have severe infection.
The physicians have applied X-ray images for pneumonia analysis, lung disorder, abscesses, and swollen lymph nodes. Every hospital has X-ray imaging devise and it is possible to take X-rays test for COVID-19 without any special testing tools. The demerits of X-ray testing are that it requires radiology expert, time-consuming, expensive. Therefore, developing an automatic analysis model is extremely significant to limit the overhead of medical professionals. In addition, positive findings are attained from X-ray as well as computed tomography (CT) images. However, it can be unfit for further diagnosis due to the prolonged diagnosis period of time. The accurate diagnosis rate of this method varies to greater extent. Hence, maximum count of repeated tests has to be conducted for gaining appropriate outcomes. But, it signifies a spherical allocation of images, it depicts the similar features with alternate viral lung disorder.
Previously, in case of fast progression of coronavirus, numerous works have been performed by many developers. In recent times, deep learning (DL) is considered to be the well-known model related to medical sector for diagnosing process [2]. DL is considered to be the combination of Machine Learning (ML) models which is extremely concentrated on computing automated feature extraction as well as image classification and capable of performing object prediction as well as medicinal image classification. The ML and DL techniques are assumed to be well-organized method used in mining, analyzing, and pattern identification from images. Regaining the betterment of clinical decision making as well as computer‐aided detection (CAD) is considered to be non-trivial due to the generation of new data. Moreover, DL is defined as the process in which Deep Convolutional Neural Networks (DCNN) was employed to perform automatic feature extraction, which exploits the convolution task and layers perform on non-linear data. A layer is operated by data transformation from high to abstract level. Basically, DL represents novel deep networks when compared with classical ML methods with the help of big data.
The contribution of the study can be defined as follows. This paper devises an efficient unsupervised DL based variational autoencoder (UDL-VAE) model for COVID-19 detection and classification. The UDL-VAE model performs adaptive Wiener filtering (AWF) based preprocessing, Inception v4 with Adagrad based feature extraction, and unsupervised VAE based classification. The application of Adagrad technique helps to adjust the hyperparameters of Inception v4 model, and thereby the classification performance can be improved. For facilitating the effective detection performance of the UDL-VAE method, a comprehensive experimental validation takes place to make sure the proficient performance of the UDL-VAE model.
2. Literature Review
Generally, ML methods are considered as a sub-section of Artificial Intelligence (AI) and used prominently for medical domains in feature extraction and image analysis. Sorensen et al. [3] processed the heterogeneity from collective Regions of Interest (ROI). Then, the features are classified by using a standard vector space-relied classifier. Zhang and Wang [4] offered a CT classification with 3 classical features called grayscale values, shape, as well as texture, and symmetric features. It can be achieved using Radial Basis Function Neural Network (RBFNN) and perform feature classification. Homem et al. [5] proposed a relative study utilizing JeffriesñMatusita (JñM) distance and KarhunenñLoËve transformation feature extraction processes. Albrecht et al. [6] developed a classifier with the average grayscale measure of images in several image classifiers. Yang et al. [7] suggested an automatic classifier for classifying the breast CT photographs utilizing morphological properties. Followed by, the performance is limited when same process is determined with diverse datasets. In addition, hand-engineered models are simulated to develop CNN as well as automated feature extraction methods. Thus, CNN structure is referred as DL structure used in extraction and classification of images.
Ozyurt et al. [8] employed a hybrid mechanism known as fused perceptual hash depends on CNN to reduce the classification time of liver CT photographs and maintain the performance. Xu et al. [9] applied a Transfer Learning (TL) procedure to overcome the clinical image imbalance problem. Followed by, the performance of GoogleNet, ResNet101, Xception, as well as MobileNetv2 are compared to gain better results. Lakshmanaprabu et al. [10] analyzed the CT scan of lung images utilizing optimal deep neural network (DNN) and Linear Discriminate Analysis (LDA). Gao et al. [11] changed original CT images to maximum and minimum attenuation pattern rescale. Consequently, images are subjected to re-sampling and classified by applying CNN.
Shan et al. [12] developed DL based model for automatic segmentation of lung as well as defected regions using chest CT images. Xu et al. [13] concentrated on making basic screening models for differentiating COVID-19 pneumonia and Influenza-A viral pneumonia from healthy cases under the application of pulmonary CT images as well as DL module. Wang et al. [14] processed on the basis of COVID-19 radiographic modification from CT images and developed a DL method for extracting graphical features of COVID-19 to provide clinical analysis prior to reach pathologic condition and eliminate the fatal state of a patient. From Hamimi [15], MERSCoV has depicted that features in chest X-ray as well as CT are depictions of pneumonia. Xie et al. [16] utilized Data Mining (DM) models for classifying SARS and pneumonia use of X-ray images.
In [17], a new AI-powered pipeline using DL model is developed to automatically detect COVID-19 and classification process takes place using CT scans. In addition, the segmentation network is integrated to the classifier model for COVID-19 identification and lesion categorization. In [18], a novel Multiple Kernels-ELM-based Deep Neural Network (MKs-ELM-DNN) model is projected to detect COVID-19 using CT images with CNN model. In [19], a set of 9 classifier models are employed for COVID-19 diagnosis such as VGG16, DenseNet121, DenseNet169, DenseNet201 and MobileNet, artificial neural network (ANN), decision tree (DT), and random forest (RF) using CT images. Some other COVID-19 diagnosis models are existed in the literature [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37]. Even though there are numerous methods developed in diagnosing COVID-19, still there is a requirement of identifying COVID-19 from Chest X-ray images. Followed by, X-ray devices are used in scanning defected regions of a body such as fractures, bone misplacement, lung disorder, and lesions. The CT scanning is defined as the expanded version of X-ray used for examining soft tissues in the internal organs. The time delay in detecting COVID-19 pneumonia results in mortality. Hence, an efficient COVID-19 diagnosing method is applied for prediction and classification of disease.
3. Unsupervised Learning based COVID-19 Diagnosis Model
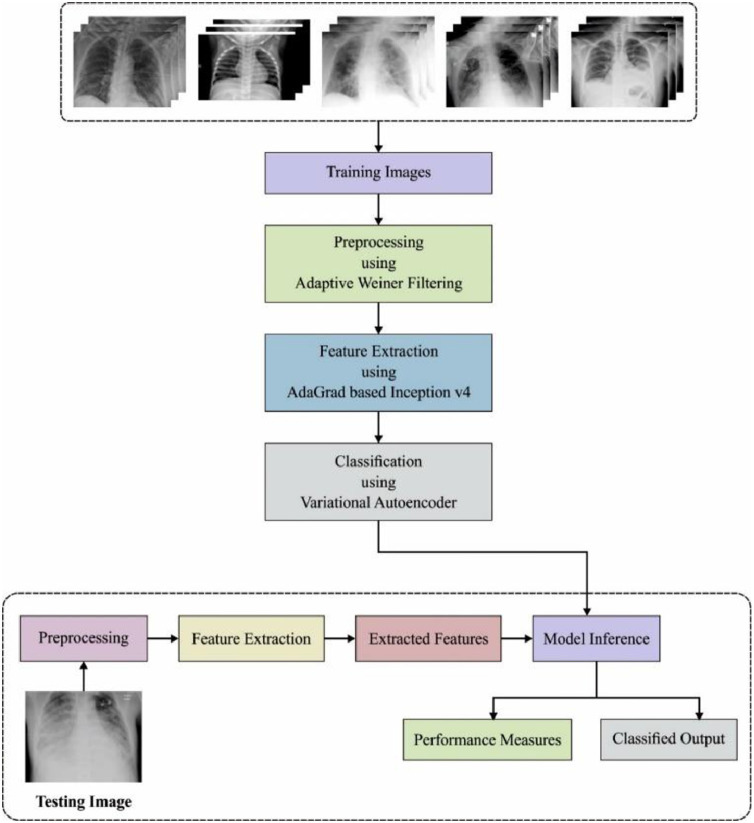
The proposed unsupervised learning based COVID-19 detection and classification model is demonstrated in Fig. 1 . The figure indicated that the medical image quality can be raised by the use of AWF technique. Then, Inception v4 with Adagrad model extracts out the suitable set of feature vectors from the preprocessed image. At last, unsupervised VAE method is implemented to define the appropriate class labels of the input medical images.
Fig. 1.
The working process of proposed UDL-VAE model.
3.1. Adaptive Wiener Filtering based Preprocessing
The remarkable development of Wiener Filter (WF) has performed the image processing due to the low pass features and intends to develop inevitable blurring of lines as well as edges. When the signal is defined as a non-Gaussian process like natural images, WF has surpassed the nonlinear estimators. The count of iterations for resolving the problem has applied a nonstationary scheme in which the features of signal as well as noise are enabled to modify. The WF expressed is describes a shift-invariant filter, and a similar filter is applied for the entire image. The filter can be made as spatially variant by applying local spatially differing method of noise attribute and represented as,
| (1) |
The filter development is costlier due to the filter is changed from pixel to pixel in an image.
Lee suggested an effective execution of noise-adaptive WF by changing the signal as stationary procedure and leads in the filter formation,
| (2) |
Where denotes the local mean of signal , and implies local signal variance. Actually, a local mean operator is adopted by normalized local production function (LPF) in which support region of filter describes the position of mean operator. It can be pointed that the filter development is depicted as same as unsharp masking, even though the latter is not adaptive locally.
The WF result from a minimization relied on mean square error (MSE) criteria is related to 2nd order statistics of input data. The application of WF is to filter signals of interest and offers suboptimal results. The stationary white noise Wiener solution is developing visible function, , based on the magnitude of image gradient vector, in which 0 for “huge” gradients and 1 for absence of gradient. As a result, the generalized Backus-Gilbert condition has attained better result:
| (3) |
The Backus-Gilbert model is defined as a regularization model that varies from alternate models and prefers to enhance the stability and smoothness of the solution. Even though the Backus-Gilbert statement is varied from standard linear regularization models, the variations among the models are minimum.
Eq. (3) indicates the explicit trade-off among resolution as well as stability. provides a filter which is identity mapping while offers a smoother Wiener solution. Under the selection of spatially variant solution, a function of image location is . It is comprised of a method with undesired properties where the filter is changed from point to point and it results in computation overhead. The “signal equivalent” model with a filter in linear integration of stationary WF as well as identity map:
| (4) |
It can be pointed that is similar to Wiener solution for and in case of a filter is considered as identity map. It is evident that Eq. (4) is represented as
| (5) |
Eq. (5) implies that method is considered as a linear integration of stationary LPF elements as well as nonstationary high pass units [38]. The addition of in stationary Wiener solution is depicted in Eq. (5) as
| (6) |
3.2. Inception v4 with Adagrad based Feature Extraction
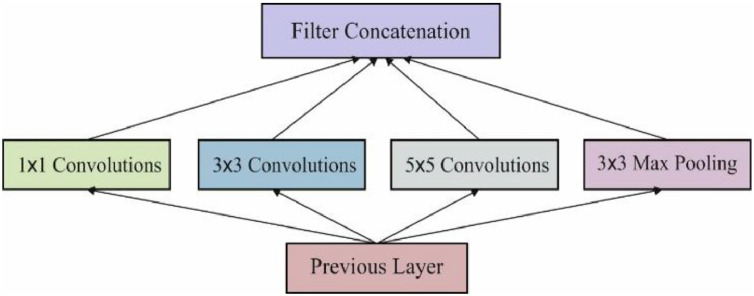
Here, preprocessed medical image is subjected to feature extraction in which the required features are extracted from given image using Inception v4 method Szegedy et al. [39]. Basically, CNN is comprised of numerous layers namely, convolutional layers, down-sampling layers, as well as activation layers. The inputs of CNN are determined by means of 1D, 2D as well as 3D. CNNs having 1D inputs classify the images directly in spectral domain whereas, in 2D, the inputs extract features from adjacent pixels and using neighbors of pixel to be divided as input; and finally, CNNs with 3D inputs filter complicated features from spectral as well as spatial domains. Then, CNNs with spatial data is capable of accomplishing optimal performance by means of classification accuracy. Fig. 2 shows the structure of Inception module. Here, 2 CNNs were developed on the basis of Directed Acyclic Graphs (DAG) structure in which fundamental layers are defined in the following.
Fig. 2.
The inception module.
Assume as the vector of pixels in input image , and single neuron computes the process and generates the simulation outcome . Hence, neuron function is described in the following:
| (7) |
Where f denotes the weight filter, b implies the bias, and indicates the activation function called nonlinear function. A neuron is commonly related to particular spatial position as well as a dimension d. It refers that convolutional block is executed n entire position of spectral dimensionality. For DAG, the input feature is defined as the position with multi‐dimensional filter as well as bias b, the output is depicted as given below:
| (8) |
It is pointed that activation function is performed with square images under different image-processing issues, however, it is operated in random inputs and filters. When the top-bottom and left-right paddings as well as down-sampling strides are depicted in Eq. (8):
| (9) |
The size of output for convolutional layer in DAG structure is represented as
For every layer, a single activation function has been employed. Also, a sigmoid function as well as Rectified Linear Unit (ReLU) are the prominently applied activation functions. And ReLU is expressed in Eq. (10).
| (10) |
Additionally, the convolutional layers, down-sampling process are included in the layers for enhancing the receptive field of neurons. Initially, down sampling is achieved by using pooling layers and stride for skipping few convolutions [40]. The major responsibility of CNNs in image classification is to detect the class labels of sample by reducing the loss function . A typically employed log-loss function is used and depicted as:
| (11) |
Where represents the positive label values. Here, a softmax function has been employed for top layer and generate the result with probability distribution which means that . After applying , weights and biases are computed by loss reduction. Finally, optimization is estimated by using a Gradient Descent (GD) method. The primary derivative of loss function is attained by means of upgrading weight with a learning rate in all iteration ad shown in the following:
| (12) |
The primary derivative is attained by using Back Propagation (BP) chain rule.
The former models of Inception schemes are applied for training diverse blocks where the repeated blocks are divided as massive subnetworks and place the entire storage region. Therefore, Inception method is simply tuned and depicted that massive credible modifications are performed on the basis of filter count in different layers which do not affect the supremacy of fully equipped system. The optimization of training rate is performed by proper layer sizes tuned in reaching optimal trade-off in processing under diverse subnetworks. In contrast, by Tensor Flow, present Inception models are described without the repeated partitions. It is due to the application of present storage area to perform backpropagation (BP), accomplished by activating the required tensors for gradient processing and demonstrate estimation for reducing the tensors.
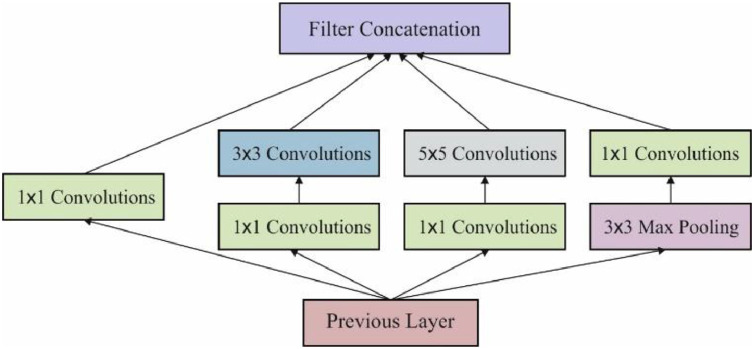
Then, Inception-v4 is projected to drop out the unwanted estimation and develop estimation for Inception blocks of different grid sizes. Fig. 3 demonstrates the process of Inception module with dimensionality reduction. In the residual inception blocks of residual versions from Inception method, minimum Inception blocks are projected across common Inception. Between the distinct versions of Inception techniques, the step time of Inception-v4 is found to be low. Batch normalization is found to be the actual variance among the residual as well as non-residual Inception blocks.
Fig. 3.
The Inception Module with Dimensionality Reduction.
Under residual scaling, the residual version begins to provide unstability and the network becomes inactive in the presence of filter count >1000, representing that the last layer earlier to the average pooling generates only 0’s under various number of iterations. It can be discarded by diminishing the rate of learning or adjusting the extra batch normalization layer. On the other hand, hyperparameters undergo tuning for controlling the effective performance of the Inception method. The procedure of selecting the hyperparameters is the main feature of the DL approaches. In this case, the hyperparameters of the Inception-v4 model are tuned by the use of Adagrad optimization model.
It refers the parameter‐based learning rates and corresponding learning rates of variables that are improvised sequentially as small and large parameters which are upgraded irregularly. Hence, update rule for Adagrad is provided as,
| (13) |
| (14) |
Where 0 defines Hadamard (element‐wise) product as well as implies the element‐wise square of applied gradient. Also, division and square root are evaluated from element‐wise model. Then, ith component of secondary vector refers the learning rate applied for upgrading the parameter in iteration . A major inefficiency of Adagrad optimizer is static development of accumulator in complete training process and every iteration of ith coordinate the respective squared partial derivative of cost function has been included and concludes in infinite learning rates like and terminates the training operations.
3.3. Unsupervised VAE based Classification
At the last stage, VAE model is applied to allocate the proper class label of the applied medical images. Basically, VAE is relied on the traditional Auto-Encoder (AE). In prior to developing VAE, it is crucial to learn the AE process. In general, AE is a type of unsupervised learning method which desires to extract secondary features concealed in the actual data. Hinton had applied unsupervised automated encoder to pre‐train and resolve the issues of gradient diminishing.
VAE is a type of deep Bayesian system which is the combination of NN in conjunction with statistics. When compared with former AE, it enforces the latent codes and follows certain distribution like Gaussian distribution. Actually, encoder portion of entire NN is developed to imply the conditional probability , in which denotes the actual data, refers the weights of encoder, and shows the latent codes. Inversely, VAE promotes the distribution of latent codes to the standard normal distribution. Finally, latent codes are comprised of stable statistical features which result in better convenience for decoder and efficacy of this model [41]. Next, decoder manages to reform the actual data by applying latent codes Consequently, NN having the weight 0 indicates a conditional probability . In order to retain the uncertainty within the system, inputs of decoder undergo sampling from scattering of latent codes . Therefore, encoder is modeled in 2 resultant parameters namely, mean as well as variance vector of latent codes . Followed by, 2 parameters describe the normal distribution for sample.
3.4. Kullback-Leibler (KL) divergence
To make distribution of latent code model for specific distribution, a major problem is to estimate the space among 2 distribution and divergence is established for estimating the variations among 2 probability distributions. Hence, the closer distributions are smaller the measure of . When there are 2 unseen distribution and , then divergence is attained by applied expression:
| (15) |
This is referred as Kullback Leibler or divergence from and .
3.5. Establishment of loss function
In this model, there are 2 objectives in VAE namely, reform the actual data and create latent codes in certain distribution. Hence, the loss function has been classified into 2 portions. The initial part is to estimate the distance from produced and actual data. Afterward, MSE function has been applied and referred as desired value of squared difference among 2 parameters. MSE is applicable to measure the difference between actual and reformed data. The latter portion is the loss to relate the distance among distribution of latent code as well as remarkable Gaussian distribution. The divergence is used and resulted in final loss process of VAE has been devised as,
| (16) |
Where indicates the actual data, defines the reformed data, represents a parameter, defines the distribution of latent codes produced from actual data by encoding device, and depicts the distribution of latent codes. Therefore, demonstrates the numerical expression.
4. Experimental Validation
The performance validation of the presented UDLVAE model has been tested against COVID Chest X-ray dataset [42]. It contains the images under distinct classes namely Normal, COVID-19, SARS, ARDS, and Streptococcus. The parameter setting of the proposed model is given as follows. Batch size: 500, max. epochs:15, learning rate: 0.05, dropout rate: 0.2, and momentum: 0.9. A few sample test images are illustrated in Fig. 4 . Besides, the dataset is split is five different folds.
Fig. 4.
The Sample Images.
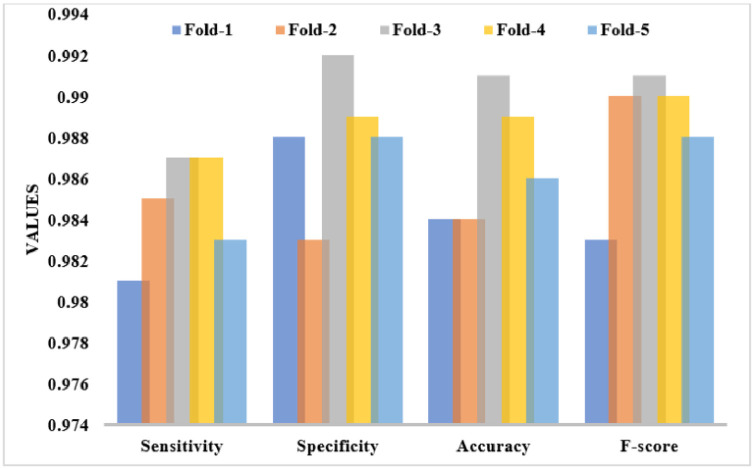
Table 1 and Fig. 5 tabulates the binary classification results of the UDLVAE model interms of distinct performance measures with varying folds. The obtained values denoted that the UDLVAE model has reached to effective classification outcome under diverse folds. On the applied fold_1, the UDLVAE model has accomplished a sens. of 0.981, spec. of 0.988, acc. of 0.984, and F-measure of 0.983. Besides, on the applied fold_3, the UDLVAE method has demonstrated a sens. of 0.987, spec. of 0.992, acc. of 0.991, and F-measure of 0.991. Also, on the applied fold_5, the UDLVAE technique has exhibited a sens. of 0.983, spec. of 0.988, acc. of 0.986, and F-measure of 0.988.
Table 1.
The Performance analysis of UDLVAE Model for Binary Class under different folds
| Cross Folds | Sens. | Spec. | Acc. | F-measure |
|---|---|---|---|---|
| Fold_1 | 0.981 | 0.988 | 0.984 | 0.983 |
| Fold_2 | 0.985 | 0.983 | 0.984 | 0.990 |
| Fold_3 | 0.987 | 0.992 | 0.991 | 0.991 |
| Fold_4 | 0.987 | 0.989 | 0.989 | 0.990 |
| Fold_5 | 0.983 | 0.988 | 0.986 | 0.988 |
| Average | 0.985 | 0.988 | 0.987 | 0.988 |
Fig. 5.
The Binary-class results analysis of the UDLVAE model.
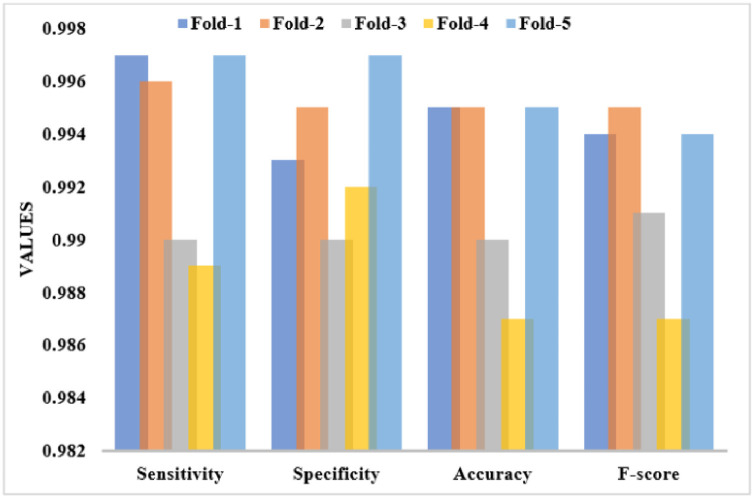
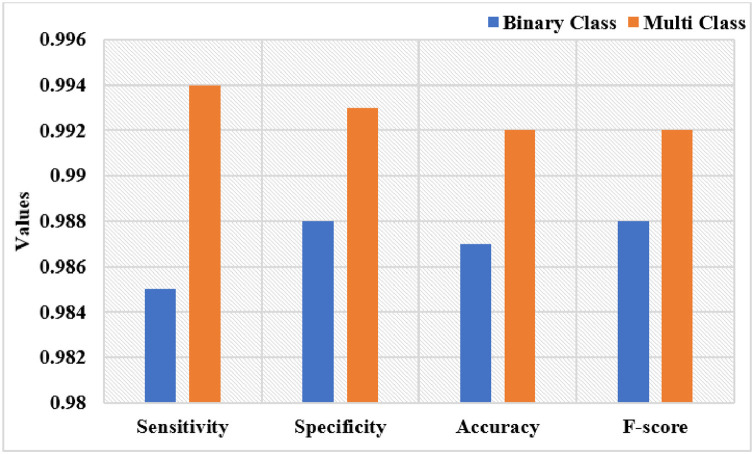
Table 2 and Fig. 6 examines the Multi-class classification results of the UDLVAE model with respect to distinct performance measures with varying folds. The attained values referred that the UDLVAE model has achieved effective classification results under diverse folds. On the applied fold_1, the UDLVAE method has showcased a sens. of 0.997, spec. of 0.993, acc. of 0.995, and F-measure of 0.994. Additionally, on the applied fold_3, the UDLVAE technique has accomplished a sens. of 0.990, spec. of 0.990, acc. of 0.990, and F-measure of 0.991. Eventually, on the applied fold_5, the UDLVAE approach has outperformed a sens. of 0.997, spec. of 0.997, acc. of 0.995, and F-measure of 0.994. An average classification results analysis of the UDLVAE model with other existing methods takes place, as given in Fig. 7 . The resultant values depicted that the UDLVAE model has accomplished a higher average sens. of 0.985, spec. of 0.988, acc. of 0.987, and F-measure of 0.988 on the classification of binary classes. On the other hand, the UDLVAE model has resulted in a maximum sens. of 0.994, spec. of 0.993, acc. of 0.992, and F-measure of 0.992.
Table 2.
The Performance analysis of UDLVAE Model for Multi-Class under different folds.
| Cross Folds | Sens. | Spec. | Acc. | F-measure |
|---|---|---|---|---|
| Fold_1 | 0.997 | 0.993 | 0.995 | 0.994 |
| Fold_2 | 0.996 | 0.995 | 0.995 | 0.995 |
| Fold_3 | 0.990 | 0.990 | 0.990 | 0.991 |
| Fold_4 | 0.989 | 0.992 | 0.987 | 0.987 |
| Fold_5 | 0.997 | 0.997 | 0.995 | 0.994 |
| Average | 0.994 | 0.993 | 0.992 | 0.992 |
Fig. 6.
The Multi-class results analysis of the UDLVAE model.
Fig. 7.
The Average Analysis of Proposed UDLVAE Model.
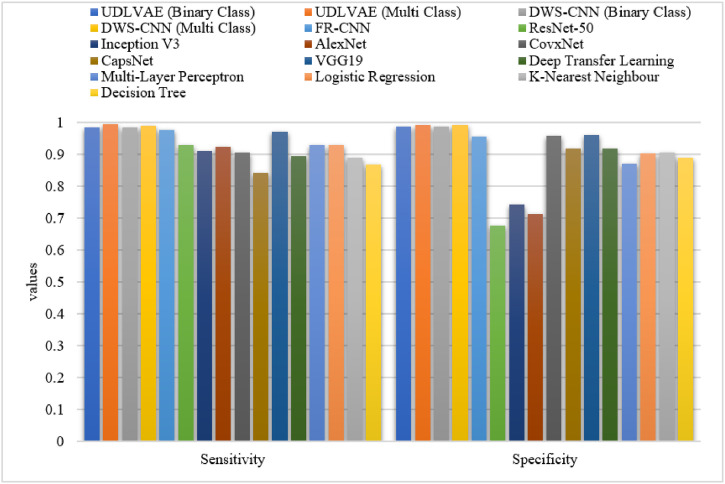
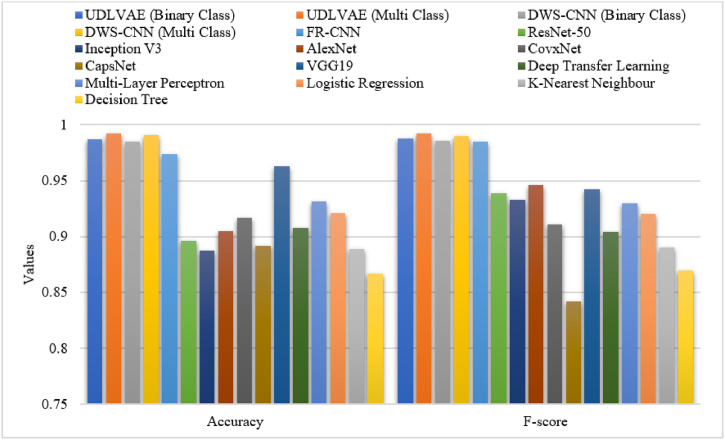
Table 3 and Fig. 8 Fig. 9 provide an elaborate comparative results analysis of the UDLVAE model with other existing techniques. On examining the predictive outcome interms of sensitivity, the CapsNet and DT models have achieved minimal sensitivity values of 0.842 and 0.87 respectively. Afterward, the KNN, DTL, and CovxNet models have reached slightly increased and nearer sensitivity values of 0.89, 0.896, and 0.905 respectively. Afterward, the Inception v3 and AlexNet methods have reached a moderately closer sensitivity value of 0.91 and 0.925 respectively. At the same time, the ResNet-50, MLP, and LR models have accomplished an identical sensitivity value of 0.93. Along with that, the VGG19 and FR-CNN models have resulted in considerably improved sensitivity values of 0.971 and 0.977 respectively. Eventually, the DWS-CNN model has exhibited the higher result sensitivity values of 0.984 and 0.991 on the prediction of binary and multiple class labels respectively. Finally, the UDLVAE model has showcased superior outcomes by offering a maximum sensitivity of 0.985 and 0.994 on the prediction of binary and multiple class labels respectively.
Table 3.
The Comparison study of UDLVAE with other existing methods.
| Methods | Sensitivity | Specificity | Accuracy | F-measure |
|---|---|---|---|---|
| UDLVAE (Binary Class) | 0.985 | 0.988 | 0.987 | 0.988 |
| UDLVAE (Multi Class) | 0.994 | 0.993 | 0.992 | 0.992 |
| DWS-CNN (Binary Class) | 0.984 | 0.986 | 0.985 | 0.986 |
| DWS-CNN (Multi Class) | 0.991 | 0.992 | 0.991 | 0.990 |
| FR-CNN | 0.977 | 0.955 | 0.974 | 0.985 |
| ResNet-50 | 0.930 | 0.677 | 0.896 | 0.939 |
| Inception V3 | 0.910 | 0.742 | 0.887 | 0.933 |
| AlexNet | 0.925 | 0.714 | 0.905 | 0.946 |
| CovxNet | 0.905 | 0.958 | 0.917 | 0.911 |
| CapsNet | 0.842 | 0.918 | 0.892 | 0.842 |
| VGG19 | 0.971 | 0.960 | 0.963 | 0.942 |
| Deep Transfer Learning | 0.896 | 0.920 | 0.908 | 0.904 |
| Multi-Layer Perceptron | 0.930 | 0.872 | 0.931 | 0.930 |
| Logistic Regression | 0.930 | 0.903 | 0.921 | 0.920 |
| K-Nearest Neighbour | 0.890 | 0.907 | 0.889 | 0.890 |
| Decision Tree | 0.870 | 0.889 | 0.867 | 0.870 |
Fig. 8.
The Sensitivity and Specificity analysis of UDLVAE with other models.
Fig. 9.
The Accuracy and F-measure analysis of UDLVAE with other models.
On investigative the predictive outcome with respect to specificity, the ResNet-50 and AlexNet methods have achieved the worst specificity values of 0.677 and 0.714 correspondingly. Similarly, the Inception, MLP, and DT models have obtained slightly enhanced and nearer specificity values of 0.742, 0.872, and 0.889 correspondingly. Next, the LR and KNN techniques have reached a moderately closer specificity value of 0.903 and 0.907 correspondingly. Simultaneously, the CapsNet, DTL, and CovxNet models have accomplished even superior specificity values of 0.918, 0.920, and 0.958 respectively. Likewise, the FR-CNN and VGG19 models have resulted in considerably enhanced specificity values of 0.955 and 0.960 respectively. Also, the DWS-CNN technique has exhibited higher result specificity values of 0.986 and 0.992 on the prediction of binary and multiple class labels respectively. At last, the UDLVAE model has showcased superior outcomes by offering a maximum specificity of 0.988 and 0.993 on the prediction of binary and multiple class labels respectively.
On determining the predictive outcome interms of accuracy, the DT and Inception V3 models have reached lesser accuracy values of 0.867 and 0.887 respectively. Afterward, the KNN, CapsNet, and ResNet-50 techniques have reached somewhat increased and nearer accuracy values of 0.889, 0.892, and 0.896 correspondingly. Besides, the AlexNet and DTL approaches have reached a moderately closer accuracy value of 0.905 and 0.908 respectively. At the same time, the CovxNet, LR, and MLP models have accomplished even higher accuracy values of 0.917, 0.921, and 0.931 correspondingly. Similarly, the VGG19 and FR-CNN manners have resulted in considerably improved accuracy values of 0.963 and 0.974 respectively. But, the DWS-CNN approach has demonstrated the superior result accuracy values of 0.985 and 0.991 on the prediction of binary and multiple class labels respectively. Finally, the UDLVAE model has showcased superior outcomes by offering a maximum accuracy of 0.987 and 0.992 on the prediction of binary and multiple class labels correspondingly.
On examining the predictive result with respect to F-measure, the CapsNet and DT techniques have obtained minimum F-measure values of 0.842 and 0.87. Similarly, the KNN, DTL, and CovxNet techniques have attained slightly increased and nearer F-measure values of 0.89, 0.904, and 0.911 respectively. Afterward, the LR and MLP models have achieved a moderately closer F-measure value of 0.92 and 0.93 correspondingly. Simultaneously, the Inception V3, ResNet-50, and VGG19 methods have accomplished even higher F-measure values of 0.933, 0.939, and 0.942 correspondingly. At the same time, the AlexNet and FR-CNN techniques have resulted in considerably improved F-measure values of 0.946 and 0.985 respectively. Also, the DWS-CNN algorithm has portrayed the higher outcome F-measure values of 0.986 and 0.990 on the prediction of binary and multiple class labels correspondingly. Finally, the UDLVAE method has showcased superior outcomes by offering a superior F-measure of 0.988 and 0.992 on the prediction of binary and multiple class labels correspondingly.
5. Conclusion
This paper has developed a novel UDL-VAE method for COVID-19 detection and classification. The UDL-VAE model performs AWF based preprocessing, Inception v4 with Adagrad based feature extraction, and unsupervised VAE based classification. Primarily, the medical image quality can be raised by the use of AWF technique. Secondly, Inception v4 with Adagrad model extracts out the useful set of feature vectors from the preprocessed image. The application of Adagrad technique helps to adjust the hyperparameters of Inception v4 model, and thereby the classification performance can be improved. Lastly, unsupervised VAE model is applied to define the appropriate class labels of the input medical images. For facilitating the effective detection performance of the UDL-VAE method, a comprehensive experimental validation takes place to make sure the proficient performance of the UDL-VAE method. The obtained experimental values showcased the effectual results of the UDL-VAE model with the higher accuracy of 0.987 and 0.992 on the binary and multiple classes respectively. In future, metaheuristic optimization based learning rate schedulers can be designed for hyperparameter settings. In addition, the presented model can be employed to diagnose COVID-19 using other imaging modalities like computed tomography (CT). As a part of future extension, it can be incorporated to the internet of things (IoT) and cloud based environment to enable e-healthcare applications.
Declaration of Competing Interest
All the authors of the manuscript declared that there are no potential conflicts of interest.
Acknowledgments
This work is financially supported by the Ministry of Science and Higher Education of the Russian Federation (Government Order FENU-2020-0022).
Edited by: Maria De Marsico
References
- 1.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60(6):84–90. May. [Google Scholar]
- 2.Pustokhin D.A., Pustokhina I.V., Dinh P.N., Phan S.V., Nguyen G.N., Joshi G.P. An effective deep residual network based class attention layer with bidirectional LSTM for diagnosis and classification of COVID-19. Journal of Applied Statistics. 2020:1–18. doi: 10.1080/02664763.2020.1849057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le D.N., Parvathy V.S., Gupta D., Khanna A., Rodrigues J.J., Shankar K. IoT enabled depthwise separable convolution neural network with deep support vector machine for COVID-19 diagnosis and classification. International Journal of Machine Learning and Cybernetics. 2021:1–14. doi: 10.1007/s13042-020-01248-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorensen L., Loog M., Lo P., Ashraf H., Dirksen A., Duin R.P., De Bruijne M. Image dissimilarity-based quantification of lung disease from CT. International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer, Berlin, Heidelberg; 2010. pp. 37–44. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W.L., Wang X.Z. Feature extraction and classification for human brain CT images. In 2007 International Conference on Machine Learning and Cybernetics; IEEE; 2007. pp. 1155–1159. [Google Scholar]
- 6.Homem M.R.P., Mascarenhas N.D.A., Cruvinel P.E. The linear attenuation coefficients as features of multiple energy CT image classification. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment, 2000;452(1-2):351–360. [Google Scholar]
- 7.Albrecht A., Hein E., Steinhˆfel K., Taupitz M., Wong C.K. Bounded-depth threshold circuits for computer-assisted CT image classification. Artificial Intelligence in Medicine. 2002;24(2):179–192. doi: 10.1016/s0933-3657(01)00101-4. [DOI] [PubMed] [Google Scholar]
- 8.Yang X., Sechopoulos I., Fei B. Automatic tissue classification for high-resolution breast CT images based on bilateral filtering. In Medical Imaging 2011: Image Processing. 2011;7962:79623H. doi: 10.1117/12.877881. International Society for Optics and Photonics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozyurt F., Tuncer T., Avci E., Koc M., Serhatlioglu I. A novel liver image classification method using perceptual hash-based convolutional neural network. Arabian Journal for Science and Engineering. 2019;44(4):3173–3182. [Google Scholar]
- 10.Xu G., Cao H., Udupa J.K., Yue C., Dong Y., Cao L., Torigian D.A. A novel exponential loss function for pathological lymph node image classification. In MIPPR 2019: Parallel Processing of Images and Optimization Techniques; and Medical Imaging. 2020;11431 International Society for Optics and Photonics. [Google Scholar]
- 11.Lakshmanaprabu S.K., Mohanty S.N., Shankar K., Arunkumar N., Ramirez G. Optimal deep learning model for classification of lung cancer on CT images. Future Generation Computer Systems. 2019;92:374–382. [Google Scholar]
- 12.Gao M., Bagci U., Lu L., Wu A., Buty M., Shin H.C., Xu Z. Holistic classification of CT attenuation patterns for interstitial lung diseases via deep convolutional neural networks. Computer Methods in Biomechanics and Biomedical Engineering: Imaging & Visualization. 2018;6(1):1–6. doi: 10.1080/21681163.2015.1124249. ... &. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan, F., Gao, Y., Wang, J., Shi, W., Shi, N., Han, M., Xue, Z., and Shi, Y. Lung Infection Quantification of COVID-19 in CT Images with Deep Learning. arXiv preprint arXiv:2003.04655, 1-19, 2020.
- 14.Xu, X., Jiang, X., Ma, C., Du, P., Li, X., Lv, S., Yu, L., Chen, Y., Su, J., Lang, G., Li, Y., Zhao, H., Xu, K., Ruan, L., and Wu, W. Deep Learning System to Screen Coronavirus Disease 2019 Pneumonia. arXiv preprint arXiv:2002.09334, 1-29, 2020. [DOI] [PMC free article] [PubMed]
- 15.Wang, S., Kang, B., Ma, J., Zeng, X., Xiao, M., Guo, J., Cai, M., Yang, J., Li, Y., Meng, X., and Xu, B. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). medRxiv preprint doi: https://doi.org/ 10.1101/2020.02.14.20023028, 1-26, 2020. [DOI] [PMC free article] [PubMed]
- 16.Hamimi A.MERS-CoV. Middle East respiratory syndrome corona virus: Can radiology be of help? Initial single center experience. The Egyptian Journal of Radiology and Nuclear Medicine. 2016;47(1):95–106. [Google Scholar]
- 17.Xie X., Li X., Wan S., Gong Y. In: Data Mining: Theory, Methodology, Techniques, and Applications. Williams Graham J., Simoff Simeon J., editors. Springer-Verlag; Berlin, Heidelberg: 2006. Mining X-ray images of SARS patients; pp. 282–294. ISBN: 3540325476. [Google Scholar]
- 18.Pennisi, M., Kavasidis, I., Spampinato, C., Schininà, V., Palazzo, S., Rundo, F., Cristofaro, M., Campioni, P., Pianura, E., Di Stefano, F. and Petrone, A., 2021. An Explainable AI System for Automated COVID-19 Assessment and Lesion Categorization from CT-scans. arXiv preprint arXiv:2101.11943. [DOI] [PMC free article] [PubMed]
- 19.Turkoglu, M., 2021. COVID-19 Detection System Using Chest CT Images and Multiple Kernels-Extreme Learning Machine Based on Deep Neural Network. IRBM. [DOI] [PMC free article] [PubMed]
- 20.Agarwal M., Saba L., Gupta S.K., Carriero A., Falaschi Z., Paschè A., Danna P., El-Baz A., Naidu S., Suri J.S. A Novel Block Imaging Technique Using Nine Artificial Intelligence Models for COVID-19 Disease Classification, Characterization and Severity Measurement in Lung Computed Tomography Scans on an Italian Cohort. Journal of Medical Systems. 2021;45(3):1–30. doi: 10.1007/s10916-021-01707-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamshidi M., Lalbakhsh A., Talla J., Peroutka Z., Hadjilooei F., Lalbakhsh P., Jamshidi M., La Spada L., Mirmozafari M., Dehghani M., Sabet A. Artificial intelligence and COVID-19: deep learning approaches for diagnosis and treatment. IEEE Access. 2020;8:109581–109595. doi: 10.1109/ACCESS.2020.3001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hemdan, E.E.D., Shouman, M.A. and Karar, M.E., 2020. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv preprint arXiv:2003.11055.
- 23.Fong S.J., Li G., Dey N., Crespo R.G., Herrera-Viedma E. Composite Monte Carlo decision making under high uncertainty of novel coronavirus epidemic using hybridized deep learning and fuzzy rule induction. Applied soft computing. 2020;93 doi: 10.1016/j.asoc.2020.106282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louati H., Bechikh S., Louati A., Hung C.C., Said L.B. Neurocomputing; 2021. Deep Convolutional Neural Network Architecture Design as a Bi-level Optimization Problem. [Google Scholar]
- 25.Ganesan V., Rajarajeswari P., Govindaraj V., Prakash K.B., Naren J. Machine Intelligence and Soft Computing. Springer; Singapore: 2021. Post-COVID-19 Emerging Challenges and Predictions on People, Process, and Product by Metaheuristic Deep Learning Algorithm; pp. 275–287. [Google Scholar]
- 26.Jaiswal A., Gianchandani N., Singh D., Kumar V., Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. Journal of Biomolecular Structure and Dynamics. 2020:1–8. doi: 10.1080/07391102.2020.1788642. [DOI] [PubMed] [Google Scholar]
- 27.Alazab M., Awajan A., Mesleh A., Abraham A., Jatana V., Alhyari S. COVID-19 prediction and detection using deep learning. International Journal of Computer Information Systems and Industrial Management Applications. 2020;12:168–181. [Google Scholar]
- 28.Ezzat D., Hassanien A.E., Ella H.A. An optimized deep learning architecture for the diagnosis of COVID-19 disease based on gravitational search optimization. Applied Soft Computing. 2020 doi: 10.1016/j.asoc.2020.106742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasinathan P., Montoya O.D., Gil-González W., Arul R., Moovendan M., Dhivya S., Kanimozhi R., Angalaeswari S. APPLICATION OF SOFT COMPUTING TECHNIQUES IN THE ANALYSIS OF COVID–19: A REVIEW. European Journal of Molecular & Clinical Medicine. 2020;7(6):2480–2503. [Google Scholar]
- 30.Altan A., Karasu S. Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm and deep learning technique. Chaos, Solitons & Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar R., Arora R., Bansal V., Sahayasheela V.J., Buckchash H., Imran J., Narayanan N., Pandian G.N., Raman B. Accurate prediction of COVID-19 using chest x-ray images through deep feature learning model with smote and machine learning classifiers. MedRxiv. 2020 [Google Scholar]
- 32.Roy S., Menapace W., Oei S., Luijten B., Fini E., Saltori C., Huijben I., Chennakeshava N., Mento F., Sentelli A., Peschiera E. Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE Transactions on Medical Imaging. 2020;39(8):2676–2687. doi: 10.1109/TMI.2020.2994459. [DOI] [PubMed] [Google Scholar]
- 33.Zhou T., Lu H., Yang Z., Qiu S., Huo B., Dong Y. The ensemble deep learning model for novel COVID-19 on CT images. Applied Soft Computing. 2021;98 doi: 10.1016/j.asoc.2020.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nour M., Cömert Z., Polat K. A novel medical diagnosis model for COVID-19 infection detection based on deep features and Bayesian optimization. Applied Soft Computing. 2020;97 doi: 10.1016/j.asoc.2020.106580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ezzat, D. and Ella, H.A., 2020. GSA-DenseNet121-COVID-19: a hybrid deep learning architecture for the diagnosis of COVID-19 disease based on gravitational search optimization algorithm. arXiv preprint arXiv:2004.05084. [DOI] [PMC free article] [PubMed]
- 36.Islam M.Z., Islam M.M., Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Informatics in medicine unlocked. 2020;20 doi: 10.1016/j.imu.2020.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahaman MM, Li C, Yao Y, Kulwa F, Rahman MA, Wang Q, Qi S, Kong F, Zhu X, Zhao X. Identification of COVID-19 samples from chest X-Ray images using deep learning: A comparison of transfer learning approaches. Journal of X-ray Science and Technology. 2020 Jan 1;(Preprint):1–9. doi: 10.3233/XST-200715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nath M.K., Kanhe A., Mishra M. In 2020 IEEE 5th International Conference on Computing Communication and Automation (ICCCA) 2020. A Novel Deep Learning Approach for Classification of COVID-19 Images; pp. 752–757. IEEE. [Google Scholar]
- 39.Westin C.F., Knutsson H., Kikinis R. Handbook of Medical Imaging Processing and Analysis. Academic press; 2000. Adaptive image filtering; pp. 3208–3212. [Google Scholar]
- 40.Szegedy C, Iofe S, Vanhoucke V, Alemi AA. In Thirty-First AAAI Conference on Artifcial Intelligence, Association for the Advancement of Artifcial Intelligence,USA. 2017. Inception-v4, inception-resnet and the impact of residual connections on learning; pp. 1–3. [Google Scholar]
- 41.Sikkandar M.Y., Alrasheadi B.A., Prakash N.B., Hemalakshmi G.R., Mohanarathinam A., Shankar K. Deep learning based an automated skin lesion segmentation and intelligent classification model. Journal of ambient intelligence and humanized computing. 2020:1–11. [Google Scholar]
- 42.COVID-19 Image Data Collection: Prospective Predictions Are the Future Joseph Paul Cohen and Paul Morrison and Lan Dao and Karsten Roth and Tim Q Duong and Marzyeh Ghassemi arXiv:2006.11988, https://github.com/ieee8023/covid-chestxray-dataset, 2020