Abstract
To determine clonal relationship among Chilean enterohemorrhagic Escherichia coli (EHEC) strains from different sources (clinical infections, animal reservoirs, and food), 54 EHEC isolates (44 of E. coli O157, 5 of E. coli O111, and 5 of E. coli O26) were characterized for virulence genes by colony blot hybridization and by pulsed-field gel electrophoresis (PFGE). By colony blotting, 12 different genotypes were identified among the 44 E. coli O157 isolates analyzed, of which the genetic profile stx1+ stx2+ hly+ eae+ was the most prevalent. All human O157 strains that were associated with sporadic cases of hemolytic-uremic syndrome (HUS) carried both the stx1 and stx2 toxin-encoding genes and were eaeA positive. Only 9 of 13 isolates from human controls were stx1+ stx2+, and 8 carried the eaeA gene. Comparison of profiles obtained by PFGE of XbaI-digested genomic DNA showed a great diversity among the E. coli O157 isolates, with 37 different profiles among 39 isolates analyzed. Cluster analysis of PFGE profiles showed a wide distribution of clinical isolates obtained from HUS cases and asymptomatic individuals and a clonal relationship among O157 isolates obtained from HUS cases and pigs. Analysis of virulence genes showed that a correlation exists among strains with the genotype stx1+ stx2+ eae+ and pathogenic potential. A larger difference in the PFGE restriction patterns was observed among the EHEC strains of serogroups O26 and O111. These results indicate that several different EHEC clones circulate in Chile and suggest that pigs are an important animal reservoir for human infections by EHEC. Guidelines have been proposed for better practices in the slaughter of animals in Chile.
Enterohemorrhagic Escherichia coli (EHEC), an important and emergent food-borne pathogen, has been associated with bloody and nonbloody diarrhea and hemolytic-uremic syndrome (HUS) (5, 13). In Chilean children, EHEC is the main cause of HUS, with incidence rates between 3 and 4.2 cases per 100,000 children below the age of 4 years (3, 20). Several virulence factors contribute to the pathogenicity of EHEC strains, including Shiga toxin 1 (Stx1) and/or Stx2, an eae locus that codes for the ability to produce an attaching-and-effacing lesion, and the EHEC hly operon that encodes an RTX (repeats in toxin) toxin designated EHEC hemolysin (Hly) and is required for expression of EHEC fimbrial antigen. EHEC strains belonging to serotype O157:H7, as well as other serogroups, are also associated with HUS disease (8, 13).
In the United States, most of the O157:H7 strains associated with human disease express Stx2, either alone or combined with Stx1 (11). In Chile, E. coli serogroup O157 bacteria expressing both Stx1 and Stx2 are the microorganisms most frequently isolated from children with HUS, although strains that express only Stx1 are also highly prevalent (15). A similar toxigenic pattern has been determined in EHEC strains obtained from asymptomatic children (15).
Epidemiologic analysis of outbreaks of EHEC strains has shown that human infections are usually linked to the consumption of improperly cooked or processed beef (5). Studies carried out in Chile have suggested that cows and pigs are important animal reservoirs of this human pathogen; e.g., 34% of the cows and 69% of the pigs slaughtered in Santiago were colonized with EHEC strains with a toxin profile similar to that of human isolates (2). O157 represented the most prevalent EHEC serogroup isolated from pigs, hamburger meat, ground beef, and sausage products purchased from different supermarkets in Santiago.
To determine clonal relatedness among bacterial isolates of EHEC, different typing methods have been used (9, 11, 15). Recent reports have shown that pulsed-field gel electrophoresis (PFGE) typing has a high degree of discriminatory power and reproducibility, superior to those of ribotyping and other molecular techniques (9). In this study, we used colony blot hybridization and PFGE typing to establish clonal relatedness among EHEC isolates obtained from sporadic cases of HUS, asymptomatic individuals, animal reservoirs, and food products, all within Santiago, Chile.
Fifty-four Chilean EHEC isolates were included in this study. Twenty-three human isolates from sporadic cases of HUS or asymptomatic controls obtained between March 1995 and March 1996. Twenty-three isolates from animals were obtained directly from the intestinal contents of pigs and cows slaughtered in Santiago from January to March 1994 (cows) and in March 1995 (pigs). EHEC strains isolated from eight food products obtained in October 1996 from hamburger meat, ground beef, or sausage products sold by different supermarkets in Santiago were also included. All samples were cultured on MacConkey agar, and 10 colonies per sample were tested for EHEC, both biochemically and by colony blot hybridization. We considered EHEC any strain containing genes that code for the production of at least one of the cytotoxins. Strains identified as EHEC were serogrouped by using commercial antisera (Probac, São Paulo, Brazil).
The sorbitol phenotypes of bacterial isolates were determined as previously described (10). Colony hybridizations were performed as previously described (4), by using biotinylated probes to detect the presence of the stx1, stx2, eae, and hly EHEC virulence genes and including positive and negative controls (4).
PFGE.
Genomic DNA for contour-clamped homogeneous electric field electrophoresis was prepared as previously described (19), with minor modifications. A 2-mm slice of an agarose plug in which EHEC genomic DNA was embedded was digested for 4 h with 20 U of XbaI (Gibco BRL) and 20 U of SfiI (New England BioLabs) for all strains and with 20 U of AvrII (New England BioLabs) for some strains. Restriction fragments of DNA were separated on a 1.5% agarose gel by PFGE with a CHEF-DR III apparatus (Bio-Rad Laboratories). A lambda ladder (New England BioLabs) was used as a molecular size marker. Southern blot hybridizations from SfiI-digested chromosomal DNAs of O157 isolates resolved by PFGE were performed as described by Sambrook et al. (16) by using a biotinylated CVD434 probe containing the central region of the eae gene.
Profiles derived from the PFGE analysis with SfiI and XbaI of O157 serogroup isolates were compared for the presence or absence of 48.5- to 436.5-kb bands. The analysis of PFGE patterns was performed by using Statistica version 4.5 from Statsoft Inc. The resulting matrix was used to construct a dendrographic tree employing the unweighted pair-group method with arithmetic means method included in this software. The discriminatory abilities of the typing systems used were estimated by Simpson’s index of diversity (D) (6).
The origin and characteristics of the 54 EHEC isolates studied are summarized in Table 1. Of the 44 O157 isolates, 31 (70.4%) were stx1+ stx2+, 10 (22.7%) were stx1+, and 3 (6.8%) were stx2+. The eae locus was detected in 33 of the 44 O157 isolates, and the Hly-encoding gene was detected in 36 of these strains. The genetic loci stx1, stx2, and eae were found in all five, O26 serogroup isolates analyzed, and the Hly-encoding region was detected in four of these isolates. For all five of the O111 isolates analyzed, the genotype was stx1+ stx2+ eae+, and three isolates carried the locus encoding EHEC Hly.
TABLE 1.
Chilean EHEC isolates characterized by DNA colony hybridization, serogrouping, and sorbitol phenotype
| Isolate origin (no. of isolates) | Genotype | No. of isolates | Serogroup | No. of sorbitol-negative strains |
|---|---|---|---|---|
| HUS (10) | stx1stx2eae hly | 4 | O157 | 4 |
| 1 | O26 | 1 | ||
| stx1stx2eae | 4 | O157 | 4 | |
| 1 | O26 | 1 | ||
| Asymptomatic subjects (13) | stx1stx2eae hly | 4 | O157 | 4 |
| 2 | O114 | 2 | ||
| 1 | O126 | 1 | ||
| stx1stx2eae | 2 | O157 | 2 | |
| stx1stx2hly | 1 | O157 | 1 | |
| stx1stx2 | 1 | O157 | 1 | |
| stx1hly | 1 | O157 | 1 | |
| stx1 | 1 | O157 | 1 | |
| Food (8) | stx1stx2eae hly | 5 | O157 | 4 |
| stx1stx2hly | 1 | O157 | 0 | |
| stx2eae hly | 1 | O157 | 0 | |
| stx2hly | 1 | O157 | 1 | |
| Porcine (19) | stx1stx2eae hly | 7 | O157 | 3 |
| 3 | O26 | 0 | ||
| 3 | O111 | 2 | ||
| stx1stx2eae | 2 | O111 | 0 | |
| stx1eae hly | 2 | O157 | 0 | |
| stx1eae | 1 | O157 | 1 | |
| stx1 | 1 | O157 | 1 | |
| Bovine (4) | stx1eae | 1 | O157 | 0 |
| stx1 | 3 | O157 | 2 |
All of the O157 strains isolated from symptomatic patients, including those from sporadic cases of HUS, carried both the stx1 and stx2 toxin genes and were eaeA positive, while 9 of 13 isolates from asymptomatic human controls were stx1+ stx2+ and 8 carried the eaeA gene. The locus encoding the production of hemolysin was detected in 5 of 8 and 7 of 13 O157 isolates from HUS cases and asymptomatic controls, respectively. Of the O157 porcine strains, 72.7% had a predominating genotype of stx1+ stx2+ and 94.7% had a predominating phenotype of eae+.
By using this typing method, we distinguished 12 different genotypes among all of the O157 isolates analyzed. The genetic profile stx1+ stx2+ hly+ eae+ was the most prevalent (21 of 44 isolates), followed by the genetic profile stx1+ stx2+ hly eae+ (7 of 44 isolates). A negative sorbitol fermentation phenotype was associated with 81% of stx1+ stx2+ hly+ eae+ O157 isolates and with 100% of human isolates.
Comparison of E. coli strains by PFGE.
The profiles of chromosomal DNA fragments generated by using XbaI were compared in 48 of the 54 isolates (39 strains were serogroup O157) in accordance with the criteria of Tenover et al. (18). Among the 39 O157 isolates, we found 37 different XbaI PFGE patterns. Thirty-six O157 strains were considered genetically unrelated because their PFGE patterns showed more than six nonmatching bands, consistent with the occurrence of more than two independent genetic events. Only two clinical O157 isolates, obtained from epidemiologically unrelated HUS cases, showed the same restriction pattern by Xba I-PFGE analysis. This finding was confirmed by using the restriction enzymes SfiI and AvrII. By PFGE, we could discriminate between two O157 strains although they had the same virulence genotype and sorbitol phenotype. Among the O157 isolates of animal origin, two porcine strains (isolated from different animals from different farms) were genetically indistinguishable by PFGE and by eae RFLP of SfiI-digested chromosomal DNA. These two porcine strains differed greatly from the clinical isolates described above.
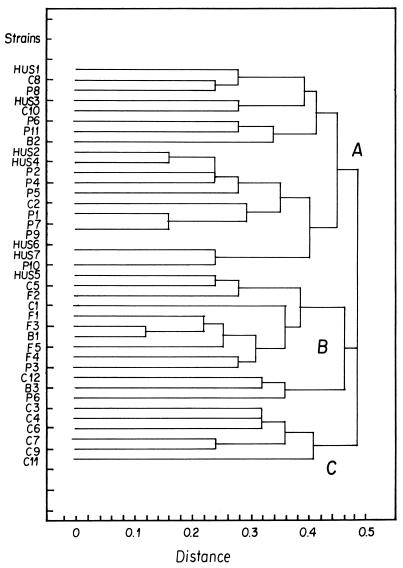
From the dendrogram analysis of XbaI-digested strains, we observed that O157 isolates were distributed among three different branches of the phylogenetic tree (clusters A, B, and C in Fig. 1). All but one isolate of clinical origin were in cluster A; the exception was an isolate from an HUS case, whereas the majority of isolates from asymptomatic subjects were in cluster C (P = 0.027, Fisher two-tailed test).
FIG. 1.
Dendrogram analysis of Chilean O157 EHEC strains isolated from food products, animal reservoirs, children with HUS, and healthy controls. The tree was constructed applying the unweighted pair-group method with arithmetic means to a matrix resulting from comparison of XbaI PFGE patterns. A, B, and C are genetically related clusters of strains. HUS1 to HUS7, EHEC isolates from HUS patients; C1 to C12, EHEC isolates from asymptomatic patients; P1 to P11, EHEC isolates from pigs; B1 to B3, EHEC isolates from bovines; F1 to F6, EHEC isolates from food products.
Among O157 isolates of animal and food origin, porcine isolates were located predominantly in cluster A, and isolates of food and bovine origin were predominantly in cluster B (P = 0.0009, Fisher two-tailed test).
Analysis of virulence genes revealed a correlation of the eae+ strains with clusters A and B but not with cluster C (Table 2). In strains of clusters A and B, the predominant genotype was stx1+ stx2+ hly+ eae+, whereas none of the cluster C isolates carried the four virulence genes combined together.
TABLE 2.
Summary of virulence genes associated with different clusters of Chilean E. coli O157 isolates
| Cluster (no. of isolates) | % (no.) of isolates with:
|
||
|---|---|---|---|
| stx1+stx2+ | hly+ | eae+ | |
| A (20) | 75 (15) | 60 (12) | 95 (19) |
| B (13) | 69 (9) | 77 (10) | 69 (9) |
| C (6) | 50 (3) | 33 (2) | 17 (1) |
Major differences in PFGE restriction patterns were also observed among EHEC strains belonging to serogroups O26 and O111, but because of the low number of isolates, the dendrographic analysis was not performed.
The high discriminatory power of this DNA fingerprinting method was confirmed by the diversity index, Simpson’s D value, obtained for XbaI PFGE types of EHEC serogroup O157 (D value of 0.973) in comparison with the discrimination based on the presence of virulence genes (D value of 0.777). By using PFGE typing, this study shows that during the study period, there was great bacterial diversity among the EHEC strains of human, animal, or food origin.
A relationship between the genetic makeup of EHEC isolates and their pathogenic potential for humans has been proposed (11). Some investigators have suggested that individuals infected with Stx2-producing EHEC serogroup O157 are at a higher risk of progress to HUS (7, 11). This hypothesis is not supported by the toxigenic profiles of EHEC isolates from Chilean patients with HUS or acute diarrhea. In these patients, the cytotoxin profiles were similar, with a predominant genotype of stx1+ stx2+ or stx1+ alone (3, 14). The EHEC serogroup O157 strains included in the present study showed a predominant genotype of stx1+ stx2+, genetic characteristics also detected among porcine strains and isolates obtained from contaminated food products. The genotype stx2+ alone was found in only a few O157 isolates obtained from different origins. Thus, the role of each Shiga-like toxin in the pathogenesis of infection caused by EHEC strains and, more specifically, in the development of HUS remains controversial.
Previous results have shown a clear association between the enterohemolytic phenotype of EHEC strains and their pathogenic potential for humans, especially among O157 isolates (17). In this study, we detected the hly locus in 50 to 60% of the serogroup O157 isolates obtained from humans, regardless of the severity of the clinical manifestations, suggesting that the relationship of the expression of the Hly phenotype with the risk of developing HUS needs further investigation.
The most significant correlation of a specific genotype with HUS disease was the presence of the eae locus among O157 isolates; the eae locus was detected in 100% of the strains from HUS cases versus 61.5% of the strains isolated from the asymptomatic group and 17% of the E. coli O157 isolates associated with cluster C. Interestingly, in animal reservoirs, this genotype was also found in 100% of colonized pigs while it was absent among bovine isolates.
These results support previous findings indicating that in Chile, pigs appear to be an important animal reservoir for EHEC strains with a high pathogenic potential for humans (2). Our results of XbaI PFGE typing analysis of the 39 Chilean O157 isolates of human, food, and animal origin support this hypothesis. Most of the isolates obtained from pigs clustered with HUS-associated O157 strains within the same branch of the phylogenetic tree. In contrast, O157 isolates from human control cases and of bovine or contaminated-food origin were located in different clusters. Despite the frequent isolation of EHEC strains from pigs, there are only a few reports that associate porcine EHEC with HUS (1) or that identify porcine products like sausages as a source of infection (12). Moreover, a previous report indicating that a minority of EHEC isolates from pigs possessed the eae gene (1) contrasts with our findings, in which all porcine isolates were eae positive.
Surprisingly, the PFGE analysis of Chilean EHEC isolates demonstrated greater clonal diversity than expected. Only two clinical isolates obtained from epidemiologically unrelated HUS cases and two isolates of porcine origin (but from different farms) showed the same restriction pattern. By using XbaI PFGE typing, we could differentiate EHEC isolates that share virulence markers identified by DNA probes, confirming the high discriminatory power of this technique.
In conclusion, these results demonstrate that EHEC strains were widespread in Chile and infect animals and humans, as well as contaminate food products. There seems to be simultaneous circulation of several different EHEC clones in Chile, a country where pigs appear to be an important animal reservoir of EHEC strains with high pathogenic potential for humans. However, confirmation of this hypothesis needs more-detailed molecular microbiological studies of clonal relatedness among EHEC strains isolated from human and pig reservoirs.
Based on our findings, guidelines have been proposed to the Ministry of Health for the slaughtering of animals in Chile in order to reduce the contamination of meat products with EHEC and prevent human infection.
Acknowledgments
This research was supported by Proyecto Fondecyt 1950736-95.
We are grateful to Center for Vaccine Development, Molecular Pathogenesis Structure/Unit Section group members Ying-Kang Deng, Adel Taalat, and Tim Conn for their valuable help, technical assistance, and encouragement. We gratefully acknowledge Miguel O’Ryan for critically reading the manuscript.
REFERENCES
- 1.Beutin L, Geier D, Zimmermann S, Karch H. Virulence markers of Shiga-like toxin-producing Escherichia coli strains originating from healthy domestic animals of different species. J Clin Microbiol. 1995;33:631–635. doi: 10.1128/jcm.33.3.631-635.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borie C, Monreal Z, Guerrero P, Sánchez M L, Martínez J, Arellano C, Prado V. Prevalence and characterization of enterohaemorrhagic Escherichia coli isolated from healthy cattle and pigs slaughtered in Santiago, Chile. Arch Med Vet. 1997;29:205–212. [Google Scholar]
- 3.Cordovèz A, Prado V, Maggi L, Cordero J, Martínez J, Misraji A, Ríos R, Soza G, Ojeda A, Levine M M. Enterohemorrhagic Escherichia coli associated with hemolytic-uremic syndrome in Chilean children. J Clin Microbiol. 1992;30:2153–2157. doi: 10.1128/jcm.30.8.2153-2157.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gicquelais K G, Baldini M M, Martínez J, Maggi L, Martin W C, Prado V, Kaper J B, Levine M M. Practical and economical method for using biotinylated DNA probes with bacterial colony blots to identify diarrhea-causing Escherichia coli. J Clin Microbiol. 1990;28:2585–2590. doi: 10.1128/jcm.28.11.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffin P M, Tauxe R V. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 6.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;6:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karpman D, Connell H, Svensson M, Scheutz F, Alm P, Svanborg C. The role of lipopolysaccharide and shiga-like toxin in a mouse model of Escherichia coli O157:H7 infection. J Infect Dis. 1997;175:611–620. doi: 10.1093/infdis/175.3.611. [DOI] [PubMed] [Google Scholar]
- 8.Levine M M, Xu J-G, Kaper J B, Lior H, Prado V, Tall B, Nataro J, Karch H, Wachsmuth K. A DNA probe to identify enterohemorrhagic Escherichia coli of O157:H7 and other serotypes that cause hemorrhagic colitis and hemolytic uremic syndrome. J Infect Dis. 1987;156:175–182. doi: 10.1093/infdis/156.1.175. [DOI] [PubMed] [Google Scholar]
- 9.Martin I E, Tyler S D, Tyler K D, Khakhria R, Johnson W M. Evaluation of ribotyping as epidemiologic tool for typing Escherichia coli serogroup O157 isolates. J Clin Microbiol. 1996;34:720–723. doi: 10.1128/jcm.34.3.720-723.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ojeda A, Prado V, Martínez J, Arellano C, Borczyk A, Johnson W, Lior H, Levine M M. Sorbitol-negative phenotype among enterohemorrhagic Escherichia coli strains of different serotypes and from different sources. J Clin Microbiol. 1995;33:2199–2201. doi: 10.1128/jcm.33.8.2199-2201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ostroff S M, Tarr P I, Neill M A, Lewis J H, Hargett-Bean N, Kobayashi J M. Toxin genotypes and plasmid profiles as determinants of systemic sequelae in Escherichia coli O157:H7 infections. J Infect Dis. 1989;160:994–998. doi: 10.1093/infdis/160.6.994. [DOI] [PubMed] [Google Scholar]
- 12.Paton A W, Ratcliff R M, Doyle R M, Seymour-Murray J, Davos D, Lanser J A, Paton J C. Molecular microbiological investigation of an outbreak of hemolytic-uremic syndrome caused by dry fermented sausage contaminated with Shiga-like toxin-producing Escherichia coli. J Clin Microbiol. 1996;34:1622–1627. doi: 10.1128/jcm.34.7.1622-1627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prado V, O’Ryan M. Acute gastroenteritis in Latin America. Infect Dis Clin N Am. 1994;8:77–106. [PubMed] [Google Scholar]
- 14.Prado V, Cordero J, Garreud C, Olguín H, Arellano C, Nachar C L, Misraji A, Martínez J, Tous M, Rivas M, Levine M M. Enterohemorrhagic E. coli (EHEC), the most important cause of hemolytic uremic syndrome in children. Rev Med Chile. 1995;123:13–22. [PubMed] [Google Scholar]
- 15.Prado V, Martínez J, Arellano C, Levine M M. Temporal variation of genotypes and serotypes of enterohemorrhagic E. coli isolated from chilean children with asymptomatic infections or hemolytic uremic syndrome. Rev Med Chile. 1997;125:291–297. [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Schmidt H, Beutin L, Karch H. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun. 1995;63:1055–1061. doi: 10.1128/iai.63.3.1055-1061.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trucksis M, Wolfson J S, Hooper D C. A novel locus conferring fluoroquinolone resistance in Staphylococcus aureus. J Bacteriol. 1991;173:5854–5860. doi: 10.1128/jb.173.18.5854-5860.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vizcaya M C, Sandoval C, Salin M P, Prado V. Hemolytic-uremic syndrome impact in different metropolitan health care areas. Rev Chil Infect. 1996;13:223–230. [Google Scholar]