Abstract
Islet function is critical for normal glucose homeostasis. Unlike adult β cells, fetal and neonatal islets are more proliferative and have decreased insulin secretion in response to stimuli. However, the underlying mechanisms governing functional maturity of islets have not been completely elucidated. Pancreatic islets comprise different cell types. The microenvironment of islets and interactions between these cell types are critical for β-cell development and maturation. Thus, the study of intact islets is optimal to identify novel molecular mechanisms controlling islet functional development. Transcriptomes and genome-wide histone landscapes of H3K4me3, H3K27me3, and H3K27Ac from intact islets isolated from 2- and 10-week-old Sprague-Dawley rats were integrated to elucidate genes and pathways modulating islet development, as well as the contribution of epigenetic regulation. A total of 4489 differentially expressed genes were identified; 2289 and 2200 of them were up- and down-regulated in 10-week islets, respectively. Ingenuity Pathway Analysis revealed critical pathways regulating functional maturation of islets, including nutrient sensing, neuronal function, immune function, cell replication, and extracellular matrix. Furthermore, we identified significant changes in enrichment of H3K4me3, H3K27me3, and H3K27Ac marks, which correlated with expression changes of genes critical for islet function. These histone marks were enriched at critical transcription factor-binding motifs, such as Hoxa9, C/EBP-β, Gata1, Foxo1, E2f1, E2f3, and Mafb. In addition, our chromatin immunoprecipitation sequencing data revealed multiple potential bivalent genes whose poised states changed with maturation. Collectively, our current study identified critical novel pathways for mature islet function and suggested a role for histone modifications in regulating islet development and maturation.
Keywords: pancreatic islets, transcriptome, histone modification, epigenetics, transcription factor binding motif, bivalent gene
The worldwide prevalence of diabetes, a major health care problem, is increasing rapidly and in 2019, approximately 463 million adults had diabetes (1). β-cell dysfunction and reduced insulin secretion play a key role in the pathogenesis of diabetes; however, the underlying mechanisms causing β-cell failure are complex and remain to be fully elucidated. Better understanding of the molecular mechanisms regulating β-cell development and function is critically important for diabetes prevention and treatment.
Unlike adult β cells, fetal and neonatal β cells have blunted glucose responsiveness and decreased insulin secretion in response to secretagogues (2-7). In rodents, glucose-stimulated insulin secretion changes to a more adult pattern after birth; however, at 2 weeks of age, insulin secretion has not achieved adult levels but is markedly higher than fetal levels (6). ß-cell replication is still robust at this age compared with adults. Blunted mitochondrial function and a low expression and activity of mitochondrial shuttle enzymes in β cells partly contribute to the immaturity of fetal and neonatal islets (7-12). High proliferation rates of fetal and neonatal β cells are also correlated with functional immaturity (13). The pancreatic islet functions as a miniorgan and is highly vascularized, innervated, and contains residential immune cells (14-17). The microenvironment of islets and the interactions among different cell types are critical for islet development and maturation, as well as maintaining glucose homeostasis. By integrating transcriptomes and proteomes from fetal and 2-week rat islets, we found the expression levels of more than 900 genes and proteins differed between the 2 ages (18). We also identified potential critical roles of AMP kinase (AMPK) signaling, aryl hydrocarbon receptor signaling, lipid homeostasis/signaling, neuronal function, and cell communication in regulating islet maturation, as well as many proteins that were differentially enriched in fetal and 2-week islets (18).
Epigenetic mechanisms play critical roles in pancreas development and islet cell differentiation (19-22). Histone methyltransferase Ezh2, which generates H3K27me3, can modulate the transition from endoderm to pancreas progenitors (19). However, deletion of Ezh2 at the pancreas progenitors increases the production of endocrine progenitors and β cells (21). Histone deacetylases expression and activity are highly regulated in the embryonic pancreas and play important roles at key points in exocrine and endocrine differentiation (20). Epigenetic regulation also maintains the identity and mature function of islet cells (22, 23). Increased DNA methylation of Aristaless-related homeobox (Arx) gene in β cells suppresses the expression of Arx and prevents β- to α-cell transdifferentiation (24). The interactions of different Swi/Snf chromatin remodeling complexes with Pdx1, a pancreatic and duodenal homeobox 1 transcription factor critical for β-cell function and development, can modulate Pdx1 activity in regulating the expression of β-cell-specific genes (25).
Histone modifications are a major class of epigenetic regulation. The amino termini of histones can be modified by methylation, acetylation, phosphorylation, ubiquitylation, sumoylation, and glycosylation (26, 27). The most common histone modifications are acetylation and methylation of lysine (K) residues of histone 3 (H3) and histone 4 (H4). Increased acetylation activates transcription, such as acetylation of H3K27, whereas decreased acetylation usually represses transcription. In contrast, methylation of histones can be associated with both transcription repression and activation. Trimethylation of H3K4 is implicated in activation of transcription, whereas trimethylation of H3K27 results in transcriptional silencing. Bivalent genes, identified by enrichment of both H3K4me3 and H3K27me3 marks in promoter regions, play critical roles in regulating pluripotency of embryonic stem cells, maintaining gene imprinting, and fine-tuning gene expression during development (28-31). In β cells, loss of H3K27me3 marks at poised/bivalent domains may contribute to β-cell dedifferentiation, dysfunction, and diabetes (32). Herein, we hypothesized that integrating transcriptomic data and genome-wide assessment of histone landscapes in islets isolated from rats at 2 weeks and 10 weeks of age would identify novel mechanisms mediating islet development and final maturation. In addition to identifying potential novel signaling pathways, we have demonstrated that alterations of histone modifications in key transcription factor binding motifs and genes regulate the development and functional maturation of pancreatic islet cells.
Materials and Methods
Islet Isolation
The animals and procedures used in this study were approved by the Animal Care Committee of The Children’s Hospital of Philadelphia and the Perelman School of Medicine at the University of Pennsylvania. Pancreata were excised from Sprague Dawley rats (Charles River, Raleigh, NC) at 2 and 10 weeks of age followed by islet isolation. Four pancreata per litter were pooled for each 2-week sample. Two pancreata per litter were pooled for each 10-week sample. Pancreatic islets were isolated as previously described (33). Briefly, pancreata were digested with Collagenase P (Millipore Sigma, St. Louis, MO) in Hanks’ balanced salt solution supplemented with 4 mM NaCO3 and 1% BSA. Digested tissues were then washed in cold supplemented Hanks’ balanced salt solution without collagenase. Islets were isolated by histopaque gradient centrifugation.
Total RNA Isolation and RNA-Sequencing Library Preparation
Total RNA was extracted from freshly isolated islets (n = 3, each 2-week islet sample was from 4 pooled pancreata and each 10-week islet sample was from 2 pooled pancreata) using TRIzol Reagent (Invitrogen), followed by Qiagen RNeasy Mini kit following the manufacturer’s instructions. Briefly, islet samples were lysed with TRIzol Reagent followed by chloroform extraction. Aqueous phase was mixed with 1 volume of 70% ethanol and transferred to RNeasy mini-column for RNA isolation. RNA samples with integrity numbers between 7 and 7.8 were used for RNA-Sequencing (RNA-Seq). Libraries were generated using Illumina TruSeq Stranded Total RNA LT Sample Prep Kit with Ribo-Zero Gold.
RNA-Seq and Gene Expression Analysis
RNA-Seq and gene expression were analyzed as previously described (34). Briefly, RNA-Seq libraries were single-end sequenced to 100 bp on an Illumina hiSeq2000. All RNA-Seq data were mapped using the Tophat package for the rat genome (rn5). Differential analysis was done using EdgeR. Differentially regulated genes were identified using a false discovery rate (FDR, q value) cutoff of 0.05. Functional analysis using Qiagen’s Ingenuity Pathway Analysis (IPA) was performed on genes with a fold change ≥1.5 and q value ≤ 0.05. The data were deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE104017.
Quantitative PCR Analysis
Quantitative PCR (qPCR) was performed to validate the changes in select genes in islets from 4 to 6 biological replicates. Reverse transcription was performed using iScript cDNA Synthesis Kit (BioRad, Hercules, CA). QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA) was used for qPCR. The following Taqman Gene Expression Assays (Applied Biosystems) were used: Gck (Rn00561265_m1), Mdh1 (Rn00583661_m1), Abat (Rn00578656_m1), Cdk2 (Rn01529540_m1), Col1a1 (Rn01463848_m1), Dnmt1 (Rn00709664_m1), Rgs9 (Rn00570117_m1), Slc3a2 (Rn01759899_g1), and Cltc (Rn00693501_m1). Cltc gene expression was used as the endogenous control. Relative fold changes were calculated using the ∆∆CT method.
Chromatin Preparation and Chromatin Immunoprecipitation
Chromatin was extracted from freshly isolated islets and chromatin immunoprecipitation (ChIP) with antibodies for H3K4me3 (07-473, Millipore Sigma), H3K27me3 (07-449, Millipore Sigma), and H3K27Ac (ab4729, Abcam, Cambridge, MA) were prepared as previously described (32, 33). Briefly, islets were cross-linked with 2.22% formaldehyde in PBS for 10 minutes at room temperature, and followed by incubation with 0.14M glycine to stop crosslinking. Samples were sonicated using a BioRuptor (Diagenode, Denville, NJ) to shear the chromatin with a high setting of cycling 30 seconds on, 30 seconds off for 5 minutes. The number of cycles to achieve appropriate shearing was monitored using a 2100 BioAnalyzer with a high-sensitivity DNA kit (Agilent, Santa Clara, CA). A total of 4 to 5 µg of chromatin were used for ChIP with antibodies for histone modifications. DNA from uncrosslinked chromatin was purified using Qiagen PCR purification kit (Germantown, MD).
ChIP-Seq and Data Analysis
Multiplexed ChIP-Seq libraries were created using the Illumina ChIP-seq DNA sample prep kit (Illumina, San Diego, CA) following manufacturer’s instructions and a previously published protocol (35, 36). Three biological replicates for each group were used to prepare libraries. Libraries for H3K4me3, H3K27me3, H3K27Ac, and input were sequenced to 50 bp on an Illumina hiSeq2000. ChIP-Seq data were analyzed as previously described (36). Briefly, the reads were mapped to rat genome assembly rn6. Histone-enriched sites were called by MACS2 using an FDR (q value) cutoff of 0.05, and histone marker differential enrichment analysis was performed with DiffBind (10.18129/B9.bioc.DiffBind). Functional analysis was performed using IPA. Enrichment analysis of transcription factor binding motifs was carried out using HOMER. Bivalent domains were identified by the enrichment of both H3K4me3 and H3K27me3 marks in which enrichment peaks have at least a 5-bp overlap. For genome browser tracks generation, bam files of replicates from the same condition are merged. Then, bamCompare from deepTools (doi:10.1093/nar/gkw257) with binSize 10–normalize using RPKM parameters was used to produce genome browser tracks that are normalized by sequence depth and input value. The Log2 ratio of sample to inputs is reported and visualized on IGV genome browser (http://software.broadinstitute.org/software/igv/). Sequence data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE147992.
Results
Transcriptome Profiles Differ Between 2- and 10-week-old Islets
Next-generation sequencing of pancreatic islets isolated from 2- and 10-week rats detected 27 033 genes. In total, 4489 transcripts were differentially expressed in 10-week compared with 2-week islets (Supplemental Table S1) (37). Among them, 2289 and 2200 transcripts were significantly (q value ≤ 0.05, counts per million (CPM) > 2, fold-change ≥1.5) increased and decreased in 10-week islets, respectively. We demonstrated in our previous RNA-Seq study (34) that the changes for differentially expressed genes in the RNA-Seq dataset were consistent with the changes determined via qPCR. Therefore, in this study we only validated 8 differentially expressed genes, Gck, Mdh1, Abat, Cdk2, Col1a1, Dnmt1, Rgs9, and Slc3a2, by qPCR analysis. The changes in qPCR were consistent with the changes in our RNA-Seq dataset (Supplemental Figure S1) (38).
Pathways Critical for Islet Development and Maturation
To identify pathways regulating islet development and maturation, IPA was used for functional annotation of differentially expressed genes. More than 120 canonical pathways were altered in 10-week islets, including pathways regulating metabolism, neuronal function, immune function, cell cycle, and extracellular matrix (Table 1).
Table 1.
Canonical pathways altered in transcriptomes
| Categories | Ingenuity canonical pathways | P value | Activation z-score |
No. of genes |
|---|---|---|---|---|
| Nutrient sensing and metabolism | Insulin Secretion Signaling Pathway | 3.39E-05 | 3.18 | 50 |
| AMPK Signaling | 2.63E-03 | 0.87 | 39 | |
| Sphingosine-1-phosphate Signaling | 3.55E-03 | -0.85 | 24 | |
| Leptin Signaling | 3.98E-03 | 1.89 | 18 | |
| Mitochondrial L-carnitine Shuttle Pathway | 9.55E-03 | -0.82 | 6 | |
| Apelin Pancreas Signaling Pathway | 1.00E-02 | -1.51 | 11 | |
| Glutamate Degradation III (via 4-aminobutyrate) | 1.29E-02 | NA | 3 | |
| GPCR-Mediated Nutrient Sensing | 1.70E-02 | 0.22 | 21 | |
| Calcium Transport | 2.09E-02 | 2.00 | 4 | |
| Glycine Betaine Degradation | 2.09E-02 | -1.00 | 4 | |
| Ketolysis | 2.09E-02 | NA | 4 | |
| Zymosterol Biosynthesis | 2.34E-02 | NA | 3 | |
| Gluconeogenesis I | 2.45E-02 | 1.89 | 7 | |
| Ketogenesis | 3.02E-02 | NA | 4 | |
| cAMP-mediated Signaling | 3.24E-02 | -0.51 | 36 | |
| Methionine Salvage II | 3.72E-02 | NA | 2 | |
| HIF1α Signaling | 3.80E-02 | 0.19 | 36 | |
| Pyrimidine Ribonucleotides Interconversion | 4.79E-02 | -1.00 | 9 | |
| Neuronal function | Axonal Guidance Signaling | 5.01E-11 | NA | 106 |
| Synaptogenesis Signaling Pathway | 3.55E-07 | 0.62 | 67 | |
| Reelin Signaling in Neurons | 1.12E-02 | 2.71 | 24 | |
| Endocannabinoid Neuronal Synapse Pathway | 1.12E-02 | 0.85 | 24 | |
| G Protein Signaling Mediated by Tubby | 2.19E-02 | NA | 7 | |
| GABA Receptor Signaling | 2.34E-02 | NA | 18 | |
| β-adrenergic Signaling | 3.31E-02 | 0.47 | 24 | |
| Semaphorin Neuronal Repulsive Signaling Pathway | 4.27E-02 | -2.56 | 22 | |
| Immune function | Dendritic Cell Maturation | 9.33E-06 | 1.81 | 42 |
| IL-15 Production | 9.55E-05 | -1.30 | 29 | |
| CD28 Signaling in T Helper Cells | 2.04E-04 | 1.61 | 28 | |
| Natural Killer Cell Signaling | 5.37E-04 | 1.44 | 40 | |
| iCOS-iCOSL Signaling in T Helper Cells | 7.59E-04 | 2.52 | 25 | |
| Leukocyte Extravasation Signaling | 1.95E-03 | 1.30 | 37 | |
| Complement System | 2.34E-03 | 1.63 | 12 | |
| Neuroinflammation Signaling Pathway | 2.88E-03 | 2.65 | 51 | |
| Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes | 1.05E-02 | 2.06 | 20 | |
| PKCθ Signaling in T Lymphocytes | 1.10E-02 | 2.20 | 28 | |
| G-Protein Coupled Receptor Signaling | 3.09E-02 | NA | 42 | |
| Th1 Pathway | 3.72E-02 | 2.67 | 22 | |
| Calcium-induced T Lymphocyte Apoptosis | 3.72E-02 | 2.31 | 13 | |
| Role of NFAT in Regulation of the Immune Response | 4.27E-02 | 2.71 | 30 | |
| MIF Regulation of Innate Immunity | 4.79E-02 | 1.67 | 9 | |
| Cell cycle and replication | Kinetochore Metaphase Signaling Pathway | 3.98E-13 | -3.77 | 42 |
| Cell Cycle Control of Chromosomal Replication | 7.94E-11 | -4.71 | 26 | |
| Mitotic Roles of Polo-Like Kinase | 9.77E-06 | -1.94 | 21 | |
| Cell Cycle: G2/M DNA Damage Checkpoint Regulation | 1.91E-05 | 1.50 | 17 | |
| Role of BRCA1 in DNA Damage Response | 2.40E-05 | -1.21 | 23 | |
| Role of CHK Proteins in Cell Cycle Checkpoint Control | 5.37E-04 | 0.83 | 16 | |
| Protein Kinase A Signaling | 9.12E-04 | -1.31 | 58 | |
| Estrogen-mediated S-phase Entry | 1.82E-03 | -2.33 | 9 | |
| GADD45 Signaling | 3.89E-03 | NA | 7 | |
| Sonic Hedgehog Signaling | 4.17E-03 | -2.65 | 9 | |
| Cyclins and Cell Cycle Regulation | 4.57E-03 | -2.32 | 18 | |
| HIPPO Signaling | 1.66E-02 | -0.90 | 17 | |
| Cell Cycle Regulation by BTG Family Proteins | 2.24E-02 | -1.63 | 9 | |
| p53 Signaling | 3.09E-02 | -1.15 | 18 | |
| BEX2 Signaling Pathway | 3.55E-02 | -1.29 | 15 | |
| Microenvironment and extracellular matrix | GP6 Signaling Pathway | 2.57E-09 | -3.57 | 38 |
| Stellate Cell Activation | 1.00E-06 | NA | 45 | |
| Wnt/Ca + Pathway | 1.55E-04 | -2.83 | 18 | |
| Inhibition of Matrix Metalloproteases | 1.07E-03 | 0.90 | 12 | |
| HOTAIR Regulatory Pathway | 1.35E-03 | -3.18 | 32 | |
| Wnt/PCP Pathway | 2.82E-03 | -0.77 | 15 | |
| Regulation Of The Epithelial Mesenchymal Transition In Development Pathway | 2.95E-03 | -2.36 | 19 | |
| Wnt/β-catenin Signaling | 4.79E-03 | -1.13 | 32 | |
| Caveolar-mediated Endocytosis Signaling | 8.32E-03 | NA | 16 | |
| Osteoarthritis Pathway | 2.82E-02 | -1.35 | 34 | |
| Fibrosis Signaling Pathway | 2.82E-02 | -1.35 | 62 | |
| Miscellaneous | NER Pathway | 6.61E-03 | -1.50 | 21 |
| Gαq Signaling | 3.98E-02 | -2.40 | 26 |
Abbreviation: NA, not applicable.
Nutrient Sensing and Metabolism Changed During Islet Maturation
IPA analysis indicated that many nutrient sensing and metabolic pathways differed between 10- and 2-week islets. As predicted by activation z-score, the pathways activated included insulin secretion signaling, gluconeogenesis, leptin signaling, calcium transport, and multiple nutrient sensing and master metabolic regulator signaling, such as AMPK, HIF-1α, and GPCR-mediated nutrient sensing (Table 1). Several metabolic processes and signaling for lipids, amino acids, and nucleic acids were inhibited in 10-week islets, including apelin and sphingosie-1-phosphate signaling, mitochondrial L-carnitine shuttle pathway, zymosterol biosynthesis, ketogenesis/ketolysis, glycine, glutamate, and methionine metabolism, and pyrimidine ribonucleotides interconversion (Table 1). Given that 2-week islets are not functionally mature, it is not surprising that insulin secretion signaling is highly activated in 10-week islets with a z-score of 3.18. Fifty genes comprising this pathway were differentially expressed in 10-week islets, such as Gcgr, Ins1, Ins2, Kcna4, Prlr, and Slc2a2 (Supplemental Table S2) (37). Consistent with increased insulin secretion signaling, apelin signaling, which is an adipokine regulating glucose and lipid metabolism and blocks insulin secretion in pancreas (39, 40), was inhibited in 10-week islets (z-score = -1.51). Interestingly, in contrast to the inhibition of apelin signaling, leptin signaling, another adipokine critical in modulating glucose homeostasis, was activated in 10-week islets with a z-score of 1.89. Pancreatic β cells express leptin receptors and ß cells are an important target of leptin action (41). In addition to inhibiting insulin gene expression and secretion, leptin also regulates β-cell mass via modulating proliferation and apoptosis. Twenty-two genes comprising these 2 adipokine signaling pathways were differentially expressed, including Apln, Pik3c2g, Pik3cd, Adcy2, lepr, and Pomc (Supplemental Table S2) (37). Calcium homeostasis and signaling are critical for β-cell function and insulin release. Calcium also plays a role in mitochondrial function, endoplasmic reticulum stress, as well as regulating β-cell proliferation and survival (42). Calcium transport was activated in 10-week islets with a z-score of 2. Expression of Atp2a2, Atp2a3, Atp2b2, and Atp2b3 was increased in 10-week islets (Supplemental Table S2) (37). Similar to our previous finding comparing fetal and 2-week islets (18), AMPK signaling was activated in 10-week islets (z-score = 0.87). AMPK is a master metabolic regulator controlling glucose and lipid metabolism; it also plays a critical role in maintaining β-cell identity and inhibits expression of many β-cell-disallowed genes, such as Slc16a1, Ldha, Mgst1, and Pdgfra (43). Thirty-nine genes comprising this pathway were differentially expressed in 10-week islets, including Adipoq, Adra2a, Ccna2, Pfkfb2, and Ppm1e (Supplemental Table S2) (37).
Pathways Regulating Neuronal Function Changed During Islet Maturation
Sympathetic, parasympathetic, and sensory nerves innervate pancreatic islets and are important for regulating islet hormone secretion and glucose metabolism (15). Multiple pathways regulating neuronal function and synaptogenesis differed between 10- and 2-week islets (Table 1). More than 100 genes comprising these pathways were differentially expressed, such as Gria4, Syt4, Thbs4, Dcx, and Mapk10 (Supplemental Table S3) (37). Semaphorins are growth cone guidance molecules regulating neuron development and regeneration. They also regulate islet morphogenesis in fetal pancreas (44); thus, it is not surprising that semaphorin signaling was inhibited in 10-week islets (z-score = -2.56). Twenty-two genes comprising this pathway were differentially expressed, including Crmp1, Dpysl5, Mapt, and Sema6c (Supplemental Table S3) (37). GABA receptor signaling also differed between 10- and 2-week islets. Eighteen genes mediating GABA signaling were differentially expressed, such as Gabrd, Kcnq2, Slc6a11, and Ubd (Supplemental Table S3) (34). GABA can be produced by β cells and co-released with insulin. It inhibits glucagon secretion by α cells and increases insulin release by β cells (45). GABA signaling also stimulates β-cell replication and protects against apoptosis (46).
Pathways Regulating Immune Function Were Altered During Islet Maturation
Both resident macrophages and T cells are localized in the pancreas and are critical for islet development and normal function (16, 17, 47). Many pathways important for immune function were all activated in 10-week islets, including Th1 pathway, iCOS-iCOSL signaling, PKCθ signaling, and dendritic cell maturation (Table 1). These pathways regulate the function of T cells, macrophages, dendritic cells, and natural killer cells, and are involved in both innate and adaptive immune systems. T helper (Th) cells are separated into 2 subtypes: Th1 and Th2 cells based on their cytokine profiles and activities. Imbalance between Th1 and Th2 activities is implicated for many diseases, including type 1 and type 2 diabetes (48-50). We previously demonstrated that transient recruitment of Th2 lymphocytes and macrophages in fetal islets mediates intrauterine growth restriction-induced β-cell failure in adult rats (33). Surprisingly, the Th1 pathway was activated in 10-week islets with a z-score of 2.67. This was accompanied by the activation of dendritic cell maturation and natural killer cell signaling, suggesting the importance of Th1 signaling in the maturation and function of islets. A total of 71 genes comprising these pathways were differentially expressed in 10-week islets, such as RT1-CE4, Il18, Pik3c2g, Pik3r3, Tbx21, and Stat4 (Supplemental Table S4) (37). Interestingly, IL-15 production was the only immune-related pathway inhibited in 10-week islets with a z-score = -1.30. IL-15 is associated with autoimmune and inflammatory diseases. Increased expression of IL-15 in β cells is associated with type 1 diabetes (51, 52). Twenty-nine genes comprising this pathway were differentially expressed, including Alk, Epha3, Epha5, Itk, and Zap70 (Supplemental Table S4) (37).
Pathways Regulating Cell Cycle and Replication
It is not surprising that many pathways modulating cell cycle and replication were inhibited in 10-week islets, including cell-cycle control of chromosomal replication, kinetochore metaphase signaling pathway, Sonic hedgehog signaling, and cyclins and cell-cycle regulation (Table 1). These changes are consistent with a significantly higher rate of islet cell replication in late gestation and the neonatal period (53).
Pathways Regulating Microenvironment and Extracellular Matrix Changed During Islet Maturation
The islet extracellular matrix and microenvironment play a critical role in cell development and function (54, 55). Several pathways regulating extracellular matrix and cytoskeleton organization differed between 2- and 10-week islets (Table 1). Wnt signals, which are transduced by Wnt/β-catenin pathway, Wnt/calcium pathway, and Wnt/PCP pathway, regulate development and function of islets, β-cell proliferation, as well as insulin production and secretion (56, 57). Interestingly, while activation of Wnt signaling promotes β-cell proliferation and expansion (57), inhibition of Wnt signaling drives β cells toward final maturation (58). All 3 pathways were inhibited in 10-week islets (z-score = -1.13, -2.83, and -0.77, respectively) (Table 1). Forty-seven genes mediating Wnt signaling were differentially expressed in 10-week islets, including Fzd1, Fzd9, Smo, Wnt5a, and Wnt5b (Supplemental Table S5) (37). Epithelial-mesenchymal transition (EMT) signaling, a process by which islet cells can give rise to highly proliferative islet precursor cells (59), was inhibited in 10-week islets (z-score = -2.36) (Table 1). The process of EMT involves changes in adherence junction complexes, as well as actin cytoskeleton reorganization. Nineteen genes comprising this pathway were differentially expressed, such as Brca1, Gli1, and Wnt3 (Supplemental Table S5) (37). Consistently, the HOX transcript antisense RNA (HOTAIR) regulatory pathway, which functions through Wnt/β-catenin and EMT (60), was inhibited in 10-week islets with a z-score = -3.18 (Table 1). HOTAIR is a long noncoding RNA. It also regulates gene expression via interacting with polycomb repressive complex 2, a H3K27 histone methyltransferase, and LSD1/CoREST/REST complex, an H3K4 histone demethylase (61). Although rat HOTAIR is yet to be identified, our finding suggests a potential role of HOTAIR in regulating rat islet development and maturation. Thirty-two genes comprising this pathway were differentially expressed in 10-week islets, including Col1a1, Dnmt3b, Ezh2, Mmp2, and Wnt3 (Supplemental Table S5) (37). Glycoprotein VI (GP6) is a major receptor for collagen and plays a critical role in platelet activation and thrombus formation. However, its effects on islet maturation and function remain to be determined. GP6 signaling was inhibited in 10-week islets with a z-score of -3.57 (Table 1). Thirty-eight genes comprising this pathway were differentially expressed in 10-week islets, including Col1a1, Lama2, Lcp2, Pik3cd, and Syk (Supplemental Table S5) (37).
The Landscape of Histone Modifications Differs Between 2- and 10-week-old Islets
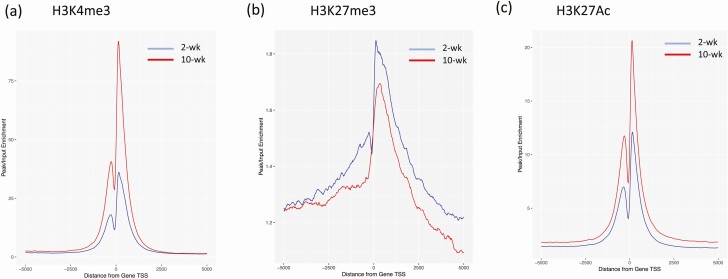
We performed ChIP-Seq analysis for H3K4me3, H3K27me3, and H3K27Ac histone modifications in 2- and 10-week islets, and annotated the loci enriched with histone marks by proximity to RefSeq and known genes. As we previously reported (36), the majority of the histone marks were located in intergenic or intronic regions, and less than 10% of modifications were located in promoters and transcriptional start sites (TSSs), suggesting many potentially novel TSS, TSS for noncoding RNAs, and regulatory regions. To assess the distribution and enrichment of each histone mark in 2- and 10-week islets, we created TSS plots (Fig. 1). The TSS plots showed higher enrichment of H3K4me3 and H3K27Ac in 10-week islets, whereas enrichment of H3K27me3 was higher in 2-week islets, except near TSSs. These results were consistent with the finding of inhibition of the HOTAIR regulatory pathway in 10-week islets (Table 1) because HOTAIR acts as a scaffold for polycomb repressive complex 2 and LSD1/CoREST/REST complex (61).
Figure 1.
Transcription start site plots (TSS plots) of histone modifications in islets. TSS plots show the distribution and enrichment of each histone mark. (A) represents H3K4me3; (B) represents H3K27me3; (C) represents H3K27Ac. Blue lines indicate 2-week islets, and red lines indicate 10-week islets.
Histone Modification Changes in Islets Correlated With Transcription Changes
To avoid the complexity of gene overlapping, we focused on the alterations of histone marks within 5 kb of the transcriptional start site. Thousands of genes showed histone mark enrichment that differed between 2- and 10-week islets. Integrating our ChIP-Seq and RNA-Seq datasets, we found histone mark enrichment that differed at many genes in 10-week islets that correlated with their RNA expression (Table 2, Supplemental Tables S6, S7, and S8) (37), suggesting these genes were potentially regulated by histone modifications. Interestingly, we found 198 genes potentially regulated by all 3 histone modifications, whereby enrichment of 3 histone marks correlated with expected changes in gene expression in 10-week islets (Table 2, Supplemental Table S9, Supplemental Figure S2) (37, 38). These included 63 up-regulated genes, such as Trpa1, Kcnma1, and Nkx6-1, and 135 down-regulated genes, such as Dlk1, Igf2bp2, and Col1a1. Trpa1 is a nonselective cation channel. Its expression increased 24-fold in 10-week islets, which was associated with enrichment of H3K4me3 and H3K27Ac marks and decreased enrichment of H3K27me3 marks. Activation of Trpa1 results in calcium influx and increases insulin release in β-cells (62, 63). Kcnma1 is a component of the BK channel, large conductance calcium-activated potassium channels, which plays an important role in islet function. The BK channel can protect islet cells from oxidative stress-induced apoptosis (64). BK channel knockout mice also display glucose intolerance and reduced insulin secretion (64). Homeobox protein Nkx6.1 (Nkx6-1), which was up-regulated in 10-week islets, is a transcription factor critical for β-cell development and mature islet function. It regulates insulin biosynthesis and secretion, as well as β-cell proliferation (65). Decreased Nkx6-1 expression is associated with the development of type 2 diabetes (66). Dlk1 is important for islet development. The expression of Dlk1 in islets is high in the fetus and newborn but significantly decreased in adults (67). Dlk1 can promote β-cell proliferation and induce pancreatic duct cells into β-like cells (68, 69). The expression of Dlk1 significantly decreased in 10-week islets compared with 2-week islets, accompanied by decreased H3K4me3 and H3K27Ac modifications and an enrichment of H3K27me3. Igf2bp2, which was decreased in 10-week islets, binds to the IGF2 mRNA and promotes its translation (70). Although IGF2 can regulate adult β-cell mass and preserve glucose-stimulated insulin secretion in aging and in response to metabolic stress (71), overexpression of local IGF2 in islets may result in β-cell dedifferentiation, islet dysfunction, and predisposition of diabetes (72). Col1a1 is a major component of type I collagen and decreased in 10-week islets. Type I collagen is present in the extracellular matrix surrounding islets and near islet capillaries (73). Although it plays a role in maintaining glucose-stimulated insulin secretion, long-term exposure to type I collagen may cause the formation of cystic structures and transdifferentiation of islets to duct epithelial-like structures (74).
Table 2.
Genes potentially regulated by histone modifications
| Histone mark | Total gene no. | Up-regulated gene no. | Down-regulated gene no. |
|---|---|---|---|
| H3K4me3 | 1298 | 547 | 751 |
| H3K27me3 | 615 | 284 | 331 |
| H3K27Ac | 1126 | 494 | 632 |
| All 3 marks | 198 | 63 | 135 |
To identify molecular pathways potentially regulated by histone modifications that may play critical roles in islet maturation, IPA analysis was used to map histone mark alterations at identified gene loci into functional networks. IPA analysis of genes potentially regulated by H3K4me3 modification revealed enrichment for multiple pathways regulating metabolism, immune system, cell cycle, and extracellular matrix, such as insulin secretion, glycolysis, crosstalk between dendritic cells and natural killer cells, and GP6 signaling (Table 3). Several pathways regulating neuronal function were enriched in the genes potentially regulated by H3K27me3 modification, including endocannabinoid neuronal synapse pathway, reelin signaling, semaphorin neuronal repulsive signaling, and THOP1 signaling (Table 4). The enrichment of pathways regulating metabolism, cell cycle, and extracellular matrix were observed for the genes potentially regulated by H3K27Ac modifications, such as insulin secretion, leptin signaling, AMPK signaling, and Wnt signaling (Table 5). Natural killer cell signaling, insulin secretion, GP6 signaling, and fibrosis were top pathways identified for the genes potentially regulated by all 3 histone marks (Table 6). Of note, these findings overlapped with key pathways identified by our RNA-Seq data (Table 1).
Table 3.
Top canonical pathways potentially regulated by H3K4me3 histone modification
| Ingenuity canonical pathways | P value | Activation z-score |
|---|---|---|
| Crosstalk between Dendritic Cells and Natural Killer Cells | 4.90E-02 | 2.24 |
| Glycolysis I | 1.12E-02 | 2.24 |
| Interferon Signaling | 4.17E-02 | 2.24 |
| Insulin Secretion Signaling Pathway | 6.03E-03 | 2.13 |
| PKCθ Signaling in T Lymphocytes | 3.98E-02 | 2.11 |
| Gluconeogenesis I | 4.79E-02 | 2.00 |
| iCOS-iCOSL Signaling in T Helper Cells | 1.58E-02 | 1.90 |
| Neuroinflammation Signaling Pathway | 3.24E-02 | 1.70 |
| Inhibition of Matrix Metalloproteases | 1.70E-02 | 1.63 |
| IL-15 Production | 7.59E-04 | -2.00 |
| HOTAIR Regulatory Pathway | 4.79E-06 | -2.04 |
| Cyclins and Cell Cycle Regulation | 3.89E-03 | -2.11 |
| PEDF Signaling | 3.16E-02 | -2.12 |
| Sonic Hedgehog Signaling | 3.89E-03 | -2.24 |
| Regulation Of The Epithelial Mesenchymal Transition | 4.68E-02 | -2.31 |
| Fibrosis Signaling Pathway | 8.91E-03 | -2.41 |
| Semaphorin Neuronal Repulsive Signaling Pathway | 2.24E-02 | -2.50 |
| Kinetochore Metaphase Signaling Pathway | 1.55E-06 | -2.67 |
| GP6 Signaling Pathway | 6.31E-04 | -3.00 |
| Cell Cycle Control of Chromosomal Replication | 3.39E-05 | -3.46 |
Table 4.
Top canonical pathways potentially regulated by H3K27me3 histone modification
| Ingenuity canonical pathways | P value | Activation z-score |
|---|---|---|
| Endocannabinoid Neuronal Synapse Pathway | 4.57E-04 | 2.11 |
| G Beta Gamma Signaling | 3.72E-02 | 1.89 |
| Reelin Signaling in Neurons | 4.79E-02 | 1.89 |
| Insulin Secretion Signaling Pathway | 5.89E-05 | 1.70 |
| Calcium Signaling | 1.74E-02 | 1.51 |
| HOTAIR Regulatory Pathway | 2.51E-04 | -1.39 |
| Semaphorin Neuronal Repulsive Signaling Pathway | 1.86E-02 | -1.41 |
| Corticotropin Releasing Hormone Signaling | 1.29E-03 | -1.90 |
| Intrinsic Prothrombin Activation Pathway | 4.17E-03 | -2.24 |
| Neuroprotective Role of THOP1 | 2.95E-02 | -2.24 |
| GP6 Signaling Pathway | 5.62E-05 | -2.31 |
Table 5.
Top canonical pathways potentially regulated by H3K27Ac histone modification
| Ingenuity canonical pathways | P value | Activation z-score |
|---|---|---|
| Insulin Secretion Signaling Pathway | 8.13E-05 | 2.60 |
| Leptin Signaling | 7.59E-03 | 2.00 |
| Gluconeogenesis I | 3.16E-02 | 2.00 |
| 3-phosphoinositide Biosynthesis | 2.45E-02 | 1.39 |
| AMPK Signaling | 1.23E-02 | 1.29 |
| Wnt/PCP Pathway | 6.76E-03 | -2.12 |
| Wnt/Ca + Pathway | 8.13E-03 | -2.12 |
| GP6 Signaling Pathway | 1.00E-02 | -2.31 |
| Semaphorin Neuronal Repulsive Signaling Pathway | 1.91E-02 | -2.31 |
| Cyclins and Cell Cycle Regulation | 1.35E-02 | -2.33 |
| Mitotic Roles of Polo-Like Kinase | 1.78E-06 | -2.45 |
| Estrogen-mediated S-phase Entry | 1.07E-03 | -2.45 |
| HOTAIR Regulatory Pathway | 1.41E-03 | -2.67 |
| Regulation Of The Epithelial Mesenchymal Transition In Development Pathway | 1.86E-03 | -2.71 |
| IL-15 Production | 1.15E-02 | -2.89 |
| Cell Cycle Control of Chromosomal Replication | 3.16E-13 | -4.02 |
Table 6.
Top canonical pathways potentially regulated by all 3 histone modifications
| Ingenuity canonical pathways | P value | Activation z-score |
|---|---|---|
| Natural Killer Cell Signaling | 2.34E-02 | 1.34 |
| Insulin Secretion Signaling Pathway | 1.55E-02 | 0.82 |
| cAMP-mediated Signaling | 4.07E-02 | -0.45 |
| IL-15 Production | 3.31E-03 | -1.34 |
| HOTAIR Regulatory Pathway | 1.05E-02 | -1.34 |
| Fibrosis Signaling Pathway | 3.31E-02 | -1.89 |
| Intrinsic Prothrombin Activation Pathway | 3.89E-04 | -2.00 |
| GP6 Signaling Pathway | 4.57E-04 | -2.45 |
Histone Modifications at Critical Transcription Factor Binding Motifs Differ During Maturation
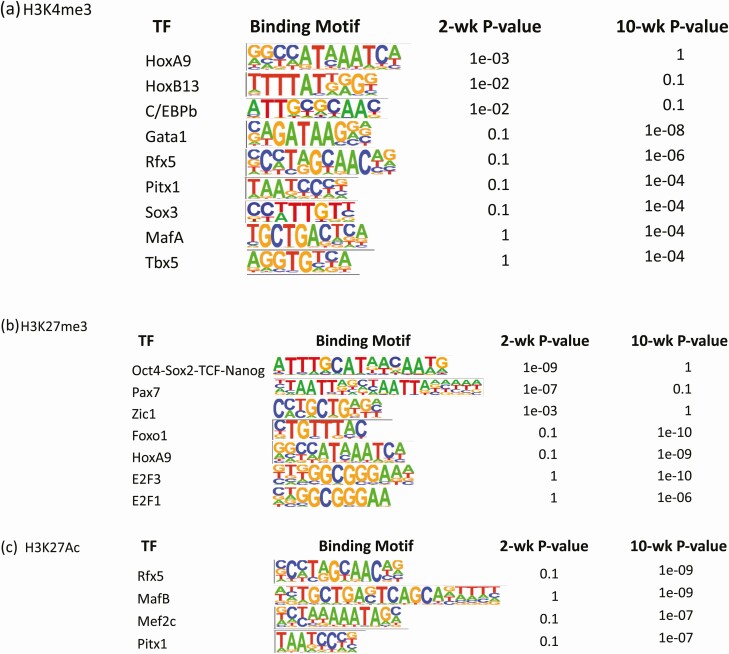
To further investigate whether changes in histone modifications in 10-week islets were located near transcription factor-binding sites that may facilitate or block transcription factor binding and result in differential gene expression, we performed HOMER binding motif analysis for all 3 histone marks. The binding motifs for HoxA9, HoxB13, and CCAAT enhancer binding protein-β (C/EBP-β) were enriched with H3K4me3 marks in 2-week islets but the H3K4me3 marks were lost in 10-week islets (Fig. 2A), suggesting their target genes were down-regulated, such as Mafb, Cdc6, Adipoq, Antxr1, Bhmt, Ccna2, Cdt1, Col1a1, Col1a2, and Col5a2. In contrast, Gata1, Rfx5, Pitx1, Sox3, MafA, and Tbx5 were associated with H3K4me3 mark enrichment in 10-week islets (Fig. 2A), suggesting up-regulation of their target genes, including Ins1, Pdx1, Slc2a2, Cybb, Fyb, Itgax, Ms4a1, Wnt3, Spock2, and Plek. HoxA9 and HoxB13 are 2 transcription factors important for embryonic development and determine of region specificity in the anteroposterior body axis. Their expression levels also differ in different types of islet cells. β cells have lower expression levels of HoxA9 and HoxB13 compared with α cells (75). C/EBPs are critical for normal β-cell function. However, overexpression of C/EBP-β is associated with endoplasmic reticulum stress and β-cell failure (76). C/EBP-β also inhibits the expression of the insulin gene in β cells (77). Gata1 is critical for the development of hematopoietic system and T lymphocytes (78). Activation of Gata1 signaling suggests the importance of hematopoietic and immune systems in the development and normal function of islets and β cells. RFX5 is a member of regulatory factor X gene family and plays a critical role in major histocompatibility complex II expression (79). Although the importance of RFX5 in islets remains to be investigated, another 2 transcription factors in the same family, RFX3 and RFX6, have been shown to regulate β-cell differentiation and maintaining adult β-cell identity, as well as regulation of insulin secretion and glucokinase expression (80, 81). MafA is essential for normal islet function. This key transcription factor regulates insulin expression and secretion and maintains mature β-cell phenotype (82, 83).
Figure 2.
Enrichment of transcription factor binding motifs altered in the regions enriched with histone modifications. (A) Enriched binding motifs for H3K4me3; (B) Enriched binding motifs for H3K27me3; (C) Enriched binding motifs for H3K27Ac. TF, transcription factor.
The binding motifs for Oct4-Sox2-TCF-Nanog, Pax7, and Zic1 were enriched with H3K27me3 marks in 2-week islets but not in 10-week islets (Fig. 2B), suggesting up-regulation of their target genes in 10-week islets, including Nqo1, Ass1, F2rl1, Galnt13, and Kcnk2. Foxo1, HoxA9, E2f1, and E2f3 were associated with the enrichment of H3K27me3 in 10-week islets (Fig. 2B), indicating down-regulation of their target genes, such as Angpt2, Cdkn1c, Depdc1, Foxm1, and Gpd1. Oct4, Sox2, and homeobox protein Nanog are master regulators for cell pluripotency (84). It is surprising that the binding motif of Oct4-Sox2-TCF-Nanog lost the enrichment of the silencing mark H3K27me3 in 10-week islets, suggesting the pluripotency and cell-renewal potential of mature islet cells. Pax7 is important in neural crest development and gastrulation and regulates the expression of neural crest markers including Sox9 and Sox10 (85). Although the function of Pax7 in islets is still unclear, it can regulate skeletal muscle myogenesis via chromatin landscape remodeling (86). Foxo1 is a transcription factor critical for islet development and function. It inhibits β-cell proliferation, protects β cells against oxidative damage, and regulates β-cell compensation in insulin resistance (87-89). However, activation of Foxo1 is also associated with fatty-acid and endoplasmic reticulum stress-induced apoptosis, glucose intolerance, and islet dysfunction (90, 91). E2f1 and E2f3, 2 transcription factors regulating cell cycles, can promote the proliferation of β cells (92, 93). Enrichment of H3K27me3 at their binding motifs in 10-week islets is consistent with the phenotype of the decreased proliferation rate in mature islets.
Similar to H3K4me3 enrichment, the binding motifs for Rfx5 and Pitx1 were also enriched with H3K27Ac in 10-week islets (Fig. 2C). Furthermore, there was an enrichment of H3K27Ac at MafB and Mef2c binding motifs as well (Fig. 2C). MafB plays a critical role in islet α- and β-cell maturation (94). It also regulates the expression of several genes involving glucose sensing and insulin secretion (95).
Consistent with the HOMER-binding motif analysis, many transcription factors discussed here were also identified as critical upstream regulators modulating gene expression changes observed in 10-week islets in our transcriptome datasets (Table 7).
Table 7.
Critical upstream regulators identified in the transcriptome and binding motif analysis
| Upstream regulator | Activation z-score |
P value | Target gene no. |
|---|---|---|---|
| HOXA9 | -1.31 | 1.86E-02 | 29 |
| CEBPB | -1.06 | 2.15E-06 | 77 |
| HOXB13 | -1.13 | 3.73E-02 | 7 |
| GATA1 | 1.82 | 8.55E-03 | 38 |
| RFX5 | 1.98 | 3.99E-03 | 6 |
| SOX3 | 0.78 | 1.17E-02 | 16 |
| MAFA | 1.94 | 2.71E-02 | 4 |
| TBX5 | 1.07 | 2.96E-02 | 13 |
| PAX7 | 0.62 | 1.25E-02 | 15 |
| FOXO1 | -3.11 | 4.46E-04 | 60 |
| E2F3 | -4.18 | 9.84E-12 | 52 |
| E2F1 | -4.25 | 3.04E-12 | 108 |
| MAFB | 1.61 | 3.58E-03 | 11 |
| MEF2C | 1.28 | 3.91E-02 | 19 |
Poised States of Potential Bivalent Genes Were Altered
Bivalent genes play critical roles during development as well as in regulating β-cell function (28, 32). Our ChIP-Seq data revealed more than 1000 potential bivalent genes. Lack of expression or very low expression of these genes was consistent with a poised state of transcription. The poised state of multiple potential bivalent genes changed during islet maturation (Table 8, Supplemental Figure S3) (38). Some genes lost their bivalency in 10-week islets, such as Reg3g, Ccl5, and Fgf21, and some gained de novo bivalency, including Dmgdh, Cdh16, and Fabp7. Reg3g lost its bivalency in 10-week islets and expression was increased. Reg3g can promote β-cell regeneration, balance TH1 and TH2 cytokine levels, and induce differentiation of regulatory T-cells and immature dendritic cells to prevent autoimmunity of β cells (96). Ccl5 also lost its bivalency in 10-week islets. Through interacting with GPR75, Ccl5 can increase intracellular calcium level in β cells, stimulate insulin secretion, and improve glucose tolerance (97). Fgf21, a secreted endocrine factor, functions as a major metabolic regulator (98). It protects against type 2 diabetes in mice by increasing insulin expression and secretion (99). Loss of Fgf21 induces insulin resistance and islet dysfunction in adult mice (100). Dmgdh catalyzes the degradation of dimethylglycine to glycine. Dimethylglycine deficiency is associated with development of diabetes (101). In 10-week islets, Dmgdh gained de novo bivalency and its expression was significantly decreased. Cdh16 is an atypical cadherin. It functions as the principal mediator for homotypic cell–cell adhesion and plays a role in the morphogenesis during tissue development (102). Fabp7 regulates fatty acid uptake, transport, and metabolism. The expression changes of these potential bivalent genes during islet maturation may be critical in maintaining normal islet function, but also suggest that these genes maybe particularly susceptible to environmental insults.
Table 8.
Potential bivalent genes critical for islet maturation
| Gene | Gene name | Bivalency | logFC | FDR | |
|---|---|---|---|---|---|
| 2-week | 10-week | ||||
| Reg3g | Regenerating islet-derived protein 3 gamma | Y | N | 5.88 | 1.19E-07 |
| Ccl5 | C-C motif chemokine ligand 5 | Y | N | 3.33 | 4.04E-07 |
| Chst5 | Carbohydrate sulfotransferase 5 | Y | N | 2.75 | 4.19E-07 |
| Fgf21 | Fibroblast growth factor 21 | Y | N | 2.98 | 1.65E-05 |
| Tp63 | Tumor protein p63 | Y | N | 4.56 | 1.39E-04 |
| Slco4c1 | Solute carrier organic anion transporter family, member 4C1 | Y | N | 2.64 | 2.52E-04 |
| Zbp1 | Z-DNA binding protein 1 | Y | N | 3.49 | 3.75E-04 |
| Sh2d4b | SH2 domain containing 4B | Y | N | 2.15 | 2.15E-03 |
| Gzma | Granzyme A | Y | N | 2.56 | 1.08E-02 |
| Cxcr2 | C-X-C motif chemokine receptor 2 | Y | N | 2.22 | 1.77E-02 |
| Bpifa2f | BPI fold containing family A, member 2F | Y | N | 2.16 | 2.09E-02 |
| Kmo | Kynurenine 3-monooxygenase | Y | N | 2.48 | 2.50E-02 |
| Ankrd34a | Ankyrin repeat domain 34A | Y | N | 1.15 | 2.64E-02 |
| Tnn | Tenascin N | Y | N | 3.00 | 3.95E-02 |
| Serpina11 | Serpin family A member 11 | Y | N | 1.54 | 4.10E-02 |
| Mxd3 | Max dimerization protein 3 | N | Y | -2.16 | 2.40E-07 |
| Gdpd2 | Glycerophosphodiester phosphodiesterase domain containing 2 | N | Y | -3.11 | 2.01E-05 |
| Camp | Cathelicidin antimicrobial peptide | N | Y | -5.50 | 2.71E-05 |
| Rtkn2 | Rhotekin 2 | N | Y | -1.83 | 5.26E-05 |
| Fam71f1 | Family with sequence similarity 71, member F1 | N | Y | -2.78 | 6.44E-05 |
| Slc26a3 | Solute carrier family 26 member 3 | N | Y | -2.78 | 7.31E-04 |
| Ebf4 | EBF family member 4 | N | Y | -1.68 | 8.02E-04 |
| LOC303566 | Homologous recombination factor with OB-fold | N | Y | -1.54 | 1.43E-03 |
| Dmgdh | Dimethylglycine dehydrogenase | N | Y | -3.37 | 1.61E-03 |
| Tssk4 | Testis-specific serine kinase 4 | N | Y | -1.81 | 2.43E-03 |
| Fabp7 | Fatty acid binding protein 7 | N | Y | -2.90 | 2.71E-03 |
| Bglap | Bone gamma-carboxyglutamate protein | N | Y | -2.77 | 4.62E-03 |
| Vnn3 | Vanin 3 | N | Y | -1.96 | 6.28E-03 |
| Chad | Chondroadherin | N | Y | -1.59 | 6.29E-03 |
| Sfta2 | Surfactant associated 2 | N | Y | -2.45 | 8.01E-03 |
| Folh1 | Folate hydrolase 1 | N | Y | -1.87 | 9.84E-03 |
| Hoxd10 | Homeo box D10 | N | Y | -2.22 | 1.41E-02 |
| Cdh16 | Cadherin 16 | N | Y | -1.45 | 2.38E-02 |
| Tsga10ip | Testis-specific 10 interacting protein | N | Y | -1.33 | 4.57E-02 |
Abbreviations: FDR, false discovery rate; N, no; Y, yes.
Discussion
Epigenetic mechanisms are pivotal in modulating gene expression, which are implicated in the development and maturation of pancreatic islets and β cells, as well as in the pathogenesis of β-cell failure and type 2 diabetes (22, 23, 103-110). In the present study, we sought to further elucidate molecular mechanisms modulating islet and β-cell development to reach their fully mature function. The rodent islet functions as a mini-organ and contains ~80% β cells. We proposed that integrating the transcriptome and histone landscapes of H3K4me3, H3K27me3, and H3K27Ac marks in intact islets would identify key pathways and genes that regulate β-cell and/or other islet cell development. To that end, we found that expression of 4489 genes differed between 2- and 10-week islets, and many of these were associated with altered histone modifications. As predicted, canonical pathways modulating cell-cycle and islet cell proliferation, which were identified in our previous transcriptome and proteome study comparing fetal and 2-week islets (18), were further inhibited in 10-week islets. Similarly, we found AMPK signaling and pathways modulating neuronal function continue to regulate islet development during final maturation. Moreover, in the current study we identified a number of novel pathways contributing to islet/β-cell development and maturation, such as adipokine signaling, the Th1 pathway, Wnt signaling, and the HOTAIR regulatory pathway.
Our findings suggest that adipokine signaling is associated with islet maturation. Adipokines are cytokines secreted by adipose tissue, and the β cell has been identified as a direct target of many of them (39-41). Adipokines regulate β-cell proliferation, viability, and insulin secretion, and are associated with the pathogenesis of type 2 diabetes (111, 112). Intriguingly, different adipokines may have distinct roles in the development and maturation of islet cells. Although both leptin and apelin may decrease insulin secretion (39, 41), we found leptin signaling was activated in 10-week islets whereas apelin signaling was inhibited. Thus, a fine balance among different adipokines may be pivotal for the functional maturation of islet cells.
One of the more interesting observations was the differences in the signaling regulating immune function between 10- and 2-week islets. Both macrophages and T cells reside in the pancreas and play critical roles in islet development and normal function (16, 17, 47). Multiple pathways regulating both innate and adaptive immunity were activated in 10-week islets, including signaling pathways modulating differentiation and function of T cells, macrophages, dendritic cells, natural killer cells, and the complement system. To our surprise, the Th1 pathway was activated in 10-week islets. Interferon-γ, TNF-β, and IL-2 are major cytokines mediating Th1 responses, and the Th2 response is mainly mediated by IL-4, IL-5, IL-10, and IL-13 (113). Although the Th1 response is generally pro-inflammatory and the Th2 response is anti-inflammatory, we previously demonstrated that IL-4-mediated Th2 response in fetal islets contributes to intrauterine growth restriction-induced β-cell failure in adult rats (33). Furthermore, both Th1 and Th2 responses may be important for maintaining normal islet function because an imbalance between Th1 and Th2 activities are implicated for both type 1 and type 2 diabetes (48-50, 114). Thus, activation of Th1 pathway in 10-week islets suggests a potential novel role of Th1 signaling in the maturation and function of islets. Future investigation into the mechanisms by which Th1 cytokines regulate the development and maturation of islet cells will be key.
Another intriguing observation were the differences in Wnt signaling and HOTAIR regulatory pathways in 2- and 10-week islets. Wnt signaling modulates pancreatic development and β-cell function (56, 57), as well as regulates glucose metabolism and energy homeostasis (115). Interestingly, fine-tuning of Wnt signaling may play a critical role in islet development and maturation. Activation of Wnt signaling promotes β-cell proliferation and expansion (57), whereas inhibition of Wnt signaling drives β cells toward final maturation (58). Our finding that Wnt/β-catenin signaling, Wnt/calcium signaling, Wnt/PCP signaling, and HOTAIR regulatory pathways were all inhibited in 10-week islets indicates the complexity and importance of Wnt signaling in islet development and maturation. Furthermore, the change in the HOTAIR regulatory pathway with maturation, which can modulate histone methyltransferase and demethylase activities, also suggests that histone modifications play a central role in regulating islet development.
Histone modifications are an important epigenetic mechanism regulating gene expression. Different modifications, as well as which amino acid residue is modified, have different influences on gene transcriptional activity. In our present study, the ChIP-Seq analysis identified key genes and signaling pathways underlying islet and β-cell development and maturation, which were consistent with the results of the transcriptome analysis. These results revealed that the expression of many genes was potentially regulated by histone modifications, including 198 genes that may be modulated by all three histone modifications. Interestingly, the alterations of histone modifications were enriched at many transcription factor-binding motifs, such as Hoxa9, C/EBP-β, Gata1, Foxo1, E2f1, E2f3, and Mafb. These transcription factors were also identified as critical upstream regulators in our transcriptome study and are critical to the development and maintenance of normal islet function.
Bivalency of developmental genes is an essential process in embryonic stem cells and during development; however, their role in the maturational process as well as in fully differentiated cells remains to be fully elucidated (28, 29). Our ChIP-Seq data revealed many potential bivalent genes in islets whose poised states changed during islet maturation. These included genes important for β-cell function, such as Reg3g, Ccl5, and Fgf21. Further investigation is needed to determine whether these genes are truly bivalent as well as their roles in regulating islet development and final maturation.
Although animal studies cannot completely translate to the human, β-cell function and development in rats is very similar to what has been observed in the human, which makes it a useful animal model to study the molecular mechanisms underlying islet development and the pathogenesis of diabetes. In addition to β cells, the rodent islet is also highly vascularized, innervated by neurons, and contains other hormone producing cells, such as α cells (glucagon), δ cells (somatostatin), ε cells (ghrelin), and PP cells (pancreatic polypeptides), as well as residential immune cells. Because our study was conducted in intact islets, our findings may not be specific to β cells. For example, MafB, an upstream regulator identified in our study, plays a critical role in both α- and β-cell differentiation and maturation and regulates the expression of several genes involving glucose sensing and secretion of insulin and glucagon (94, 116). However, the rodent islet functions as a miniorgan. The microenvironment of islets and the interactions among different cell types are critical for β-cell development and maturation, as well as maintaining glucose homeostasis. Thus, a study in intact islets may identify novel molecular mechanisms controlling islet functional development.
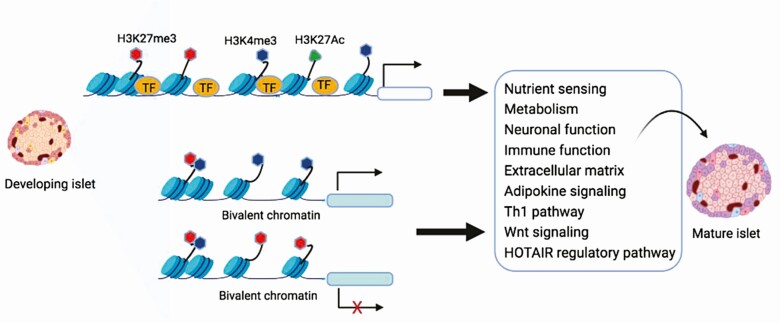
In conclusion, our current study integrated transcriptome and histone landscapes to elucidate cellular mechanisms controlling islet development and final maturation in intact islets. We identified novel pathways that were associated with maturation of islets such as adipokine signaling, Th1 pathway, Wnt signaling, and HOTAIR regulatory pathways (Fig. 3). We also demonstrated alterations in histone modifications at key transcription factor-binding motifs and genes as a potential epigenetic mechanism modulating gene expression during islet development (Fig. 3). Our study will serve as a valuable resource of chromatin landscapes in rat islets and for further elucidating critical signaling pathways for β-cell and islet maturation, which provides the basis for research on diabetes prevention and treatment.
Figure 3.
Islet development and maturation summary diagram. Alterations of histone modifications at key transcription factor-binding motifs and genes may serve as a potential epigenetic mechanism modulating gene expression during islet development and maturation. Adipokine signaling, Th1 pathway, Wnt signaling, and HOTAIR regulatory pathways were identified as novel pathways associated with maturation of islets.
Acknowledgments
The authors thank Dr. Lane J. Jaeckle Santos for assisting with harvesting of the pancreas and islet isolation. Fig. 3 was created in BioRender.com.
Funding : This research was supported by NIDDK: R01 DK055704 (R.A.S.), R01 DK114054 (R.A.S.), and The Novo Nordisk Foundation grant NNF17CC0027852 (K.J.W.).
Author Contributions: Conceptualization, Y.-C.L. and R.A.S.; methodology, Y.-C.L.; validation, Y.-C.L.; formal analysis of RNA-Seq data, K.J.W.; formal analysis of ChIP-Seq data, X.M.L. and P.Z.W.; qPCR analysis, Y.-C.L. and W.O.B.; data curation, Y.-C.L., K.J.W., X.M.L., P.Z.W., R.A.S.; writing/original draft preparation, Y.-C.L.; writing/review and editing, R.A.S.; supervision, R.A.S.; project administration, Y.-C.L. and R.A.S.; funding acquisition, R.A.S. All authors have read and agreed to the published version of the manuscript.
Glossary
Abbreviations
- AMPK
AMP kinase
- BK
channels
- C/EBP-β
CCAAT enhancer binding protein-β
- ChIP-Seq
chromatin immunoprecipitation sequencing
- EMT
epithelial-mesenchymal transition
- FDR
false discovery rate
- H3
histone 3
- H4
histone 4
- HOTAIR
HOX transcript antisense RNA
- IPA
ingenuity pathway analysis
- K
lysine
- qPCR
quantitative PCR
- RNA-Seq
RNA sequencing
- Th
T helper
Additional Information
Disclosures: The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Data Availability
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in References.
References
- 1. Saeedi P, Petersohn I, Salpea P, et al. ; IDF Diabetes Atlas Committee . Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res Clin Pract. 2019;157:107843. [DOI] [PubMed] [Google Scholar]
- 2. Grasso S, Messina A, Saporito N, Reitano G. Serum-insulin response to glucose and aminoacids in the premature infant. Lancet. 1968;2(7571):755-756. [DOI] [PubMed] [Google Scholar]
- 3. Asplund K, Westman S, Hellerström C. Glucose stimulation of insulin secretion from the isolated pancreas of foetal and newborn rats. Diabetologia. 1969;5(4):260-262. [DOI] [PubMed] [Google Scholar]
- 4. Lavine RL, Chick WL, Like AA, Makdisi TW. Glucose tolerance and insulin secretion in neonatal and adult mice. Diabetes. 1971;20(3):134-139. [DOI] [PubMed] [Google Scholar]
- 5. Hole RL, Pian-Smith MC, Sharp GW. Development of the biphasic response to glucose in fetal and neonatal rat pancreas. Am J Physiol. 1988;254(2 Pt 1):E167-E174. [DOI] [PubMed] [Google Scholar]
- 6. Hellerström C, Swenne I. Functional maturation and proliferation of fetal pancreatic beta-cells. Diabetes. 1991;40Suppl 2:89-93. [DOI] [PubMed] [Google Scholar]
- 7. Freinkel N, Lewis NJ, Johnson R, Swenne I, Bone A, Hellerström C. Differential effects of age versus glycemic stimulation on the maturation of insulin stimulus-secretion coupling during culture of fetal rat islets. Diabetes. 1984;33(11):1028-1038. [DOI] [PubMed] [Google Scholar]
- 8. Rorsman P, Arkhammar P, Bokvist K, et al. Failure of glucose to elicit a normal secretory response in fetal pancreatic beta cells results from glucose insensitivity of the ATP-regulated K+ channels. Proc Natl Acad Sci U S A. 1989;86(12):4505-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tan C, Tuch BE, Tu J, Brown SA. Role of NADH shuttles in glucose-induced insulin secretion from fetal beta-cells. Diabetes. 2002;51(10):2989-2996. [DOI] [PubMed] [Google Scholar]
- 10. Welsh N, Svensson C, Welsh M. Content of adenine nucleotide translocator mRNA in insulin-producing cells of different functional states. Diabetes. 1989;38(11):1377-1380. [DOI] [PubMed] [Google Scholar]
- 11. Taniguchi S, Tanigawa K, Miwa I. Immaturity of glucose-induced insulin secretion in fetal rat islets is due to low glucokinase activity. Horm Metab Res. 2000;32(3):97-102. [DOI] [PubMed] [Google Scholar]
- 12. Jermendy A, Toschi E, Aye T, et al. Rat neonatal beta cells lack the specialised metabolic phenotype of mature beta cells. Diabetologia. 2011;54(3):594-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Puri S, Roy N, Russ HA, et al. Replication confers β cell immaturity. Nat Commun. 2018;9(1):485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cleaver O, Dor Y. Vascular instruction of pancreas development. Development. 2012;139(16):2833-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woods SC, Porte D Jr. Neural control of the endocrine pancreas. Physiol Rev. 1974;54(3):596-619. [DOI] [PubMed] [Google Scholar]
- 16. Calderon B, Carrero JA, Ferris ST, et al. The pancreas anatomy conditions the origin and properties of resident macrophages. J Exp Med. 2015;212(10):1497-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Radenkovic M, Uvebrant K, Skog O, et al. Characterization of resident lymphocytes in human pancreatic islets. Clin Exp Immunol. 2017;187(3):418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lien YC, Won KJ, Simmons RA. Transcriptomic and quantitative proteomic profiling reveals signaling pathways critical for pancreatic islet maturation. Endocrinology. 2020;161(12):bqaa187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu CR, Cole PA, Meyers DJ, Kormish J, Dent S, Zaret KS. Chromatin “prepattern” and histone modifiers in a fate choice for liver and pancreas. Science. 2011;332(6032):963-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haumaitre C, Lenoir O, Scharfmann R. Histone deacetylase inhibitors modify pancreatic cell fate determination and amplify endocrine progenitors. Mol Cell Biol. 2008;28(20):6373-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu CR, Li LC, Donahue G, et al. Dynamics of genomic H3K27me3 domains and role of EZH2 during pancreatic endocrine specification. Embo J. 2014;33(19):2157-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Golson ML, Kaestner KH. Epigenetics in formation, function, and failure of the endocrine pancreas. Mol Metab. 2017;6(9):1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Arda HE, Li L, Tsai J, et al. Age-dependent pancreatic gene regulation reveals mechanisms governing human β cell function. Cell Metab. 2016;23(5):909-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dhawan S, Georgia S, Tschen SI, Fan G, Bhushan A. Pancreatic β cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell. 2011;20(4):419-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKenna B, Guo M, Reynolds A, Hara M, Stein R. Dynamic recruitment of functionally distinct Swi/Snf chromatin remodeling complexes modulates Pdx1 activity in islet β cells. Cell Rep. 2015;10(12):2032-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707-719. [DOI] [PubMed] [Google Scholar]
- 27. Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693-705. [DOI] [PubMed] [Google Scholar]
- 28. Vastenhouw NL, Schier AF. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol. 2012;24(3):374-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315-326. [DOI] [PubMed] [Google Scholar]
- 30. Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27(12):1318-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maupetit-Méhouas S, Montibus B, Nury D, et al. Imprinting control regions (ICRs) are marked by mono-allelic bivalent chromatin when transcriptionally inactive. Nucleic Acids Res. 2016;44(2):621-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lu TT, Heyne S, Dror E, et al. The polycomb-dependent epigenome controls β cell dysfunction, dedifferentiation, and diabetes. Cell Metab. 2018;27(6):1294-1308.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jaeckle Santos LJ, Li C, Doulias PT, Ischiropoulos H, Worthen GS, Simmons RA. Neutralizing Th2 inflammation in neonatal islets prevents β-cell failure in adult IUGR rats. Diabetes. 2014;63(5):1672-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rashid CS, Lien YC, Bansal A, et al. Transcriptomic analysis reveals novel mechanisms mediating islet dysfunction in the intrauterine growth-restricted rat. Endocrinology. 2018;159(2):1035-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bramswig NC, Everett LJ, Schug J, et al. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J Clin Invest. 2013;123(3):1275-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lien YC, Wang PZ, Lu XM, Simmons RA. Altered transcription factor binding and gene bivalency in islets of intrauterine growth retarded rats. Cells. 2020;9(6):1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lien Y-C, Lu XM, Won K-J, Wang PZ, Osei-Bonsu W, Simmons RA. Data from: Transcriptome and histone landscape reveal critical signaling and altered transcription factor binding and gene bivalency during pancreatic islet maturation. Figshare 2021. Deposited 6 May 2021. 10.6084/m9.figshare.14535768.v1 [DOI] [Google Scholar]
- 38. Lien Y-C, Lu XM, Won K-J, Wang PZ, Osei-Bonsu W, Simmons RA. Figures from: Transcriptome and histone landscape reveal critical signaling and altered transcription factor binding and gene bivalency during pancreatic islet maturation. Figshare 2021. Deposited 6 May 2021. 10.6084/m9.figshare.14547585.v1 [DOI] [Google Scholar]
- 39. Guo L, Li Q, Wang W, et al. Apelin inhibits insulin secretion in pancreatic beta-cells by activation of PI3-kinase-phosphodiesterase 3B. Endocr Res. 2009;34(4):142-154. [DOI] [PubMed] [Google Scholar]
- 40. Sörhede Winzell M, Magnusson C, Ahrén B. The apj receptor is expressed in pancreatic islets and its ligand, apelin, inhibits insulin secretion in mice. Regul Pept. 2005;131(1-3):12-17. [DOI] [PubMed] [Google Scholar]
- 41. Marroquí L, Gonzalez A, Ñeco P, et al. Role of leptin in the pancreatic β-cell: effects and signaling pathways. J Mol Endocrinol. 2012;49(1):R9-17. [DOI] [PubMed] [Google Scholar]
- 42. Klec C, Ziomek G, Pichler M, Malli R, Graier WF. Calcium signaling in ß-cell physiology and pathology: a revisit. Int J Mol Sci. 2019;20(24):6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kone M, Pullen TJ, Sun G, et al. LKB1 and AMPK differentially regulate pancreatic β-cell identity. Faseb J. 2014;28(11):4972-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pauerstein PT, Tellez K, Willmarth KB, et al. A radial axis defined by semaphorin-to-neuropilin signaling controls pancreatic islet morphogenesis. Development. 2017;144(20):3744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bansal P, Wang S, Liu S, Xiang YY, Lu WY, Wang Q. GABA coordinates with insulin in regulating secretory function in pancreatic INS-1 β-cells. Plos One. 2011;6(10):e26225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Q, Ren L, Wan Y, Prud’homme GJ. GABAergic regulation of pancreatic islet cells: Physiology and antidiabetic effects. J Cell Physiol. 2019;234(9):14432-14444. [DOI] [PubMed] [Google Scholar]
- 47. Geutskens SB, Otonkoski T, Pulkkinen MA, Drexhage HA, Leenen PJ. Macrophages in the murine pancreas and their involvement in fetal endocrine development in vitro. J Leukoc Biol. 2005;78(4):845-852. [DOI] [PubMed] [Google Scholar]
- 48. Azar ST, Tamim H, Beyhum HN, Habbal MZ, Almawi WY. Type I (insulin-dependent) diabetes is a Th1- and Th2-mediated autoimmune disease. Clin Diagn Lab Immunol. 1999;6(3):306-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xia C, Rao X, Zhong J. Role of T lymphocytes in type 2 diabetes and diabetes-associated inflammation. J Diabetes Res. 2017;2017:6494795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sia C. Imbalance in Th cell polarization and its relevance in type 1 diabetes mellitus. Rev Diabet Stud. 2005;2(4):182-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen J, Feigenbaum L, Awasthi P, et al. Insulin-dependent diabetes induced by pancreatic beta cell expression of IL-15 and IL-15Rα. Proc Natl Acad Sci U S A. 2013;110(33):13534-13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bobbala D, Chen XL, Leblanc C, et al. Interleukin-15 plays an essential role in the pathogenesis of autoimmune diabetes in the NOD mouse. Diabetologia. 2012;55(11):3010-3020. [DOI] [PubMed] [Google Scholar]
- 53. Kaung HL. Growth dynamics of pancreatic islet cell populations during fetal and neonatal development of the rat. Dev Dyn. 1994;200(2):163-175. [DOI] [PubMed] [Google Scholar]
- 54. Lucas-Clerc C, Massart C, Campion JP, Launois B, Nicol M. Long-term culture of human pancreatic islets in an extracellular matrix: morphological and metabolic effects. Mol Cell Endocrinol. 1993;94(1):9-20. [DOI] [PubMed] [Google Scholar]
- 55. Navarro-Alvarez N, Rivas-Carrillo JD, Soto-Gutierrez A, et al. Reestablishment of microenvironment is necessary to maintain in vitro and in vivo human islet function. Cell Transplant. 2008;17(1-2):111-119. [DOI] [PubMed] [Google Scholar]
- 56. Liu Z, Habener JF. Wnt signaling in pancreatic islets. Adv Exp Med Biol. 2010;654:391-419. [DOI] [PubMed] [Google Scholar]
- 57. Rulifson IC, Karnik SK, Heiser PW, et al. Wnt signaling regulates pancreatic beta cell proliferation. Proc Natl Acad Sci U S A. 2007;104(15):6247-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vethe H, Ghila L, Berle M, et al. The effect of wnt pathway modulators on human iPSC-derived pancreatic beta cell maturation. Front Endocrinol. 2019;10:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gershengorn MC, Hardikar AA, Wei C, Geras-Raaka E, Marcus-Samuels B, Raaka BM. Epithelial-to-mesenchymal transition generates proliferative human islet precursor cells. Science. 2004;306(5705):2261-2264. [DOI] [PubMed] [Google Scholar]
- 60. Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12(1):1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cao DS, Zhong L, Hsieh TH, et al. Expression of transient receptor potential ankyrin 1 (TRPA1) and its role in insulin release from rat pancreatic beta cells. Plos One. 2012;7(5):e38005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Islam MS. Molecular regulations and functions of the transient receptor potential channels of the islets of langerhans and insulinoma cells. Cells. 2020;9(3):685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Düfer M, Neye Y, Hörth K, et al. BK channels affect glucose homeostasis and cell viability of murine pancreatic beta cells. Diabetologia. 2011;54(2):423-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Taylor BL, Liu FF, Sander M. Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep. 2013;4(6):1262-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Guo S, Dai C, Guo M, et al. Inactivation of specific β cell transcription factors in type 2 diabetes. J Clin Invest. 2013;123(8):3305-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Carlsson C, Tornehave D, Lindberg K, et al. Growth hormone and prolactin stimulate the expression of rat preadipocyte factor-1/delta-like protein in pancreatic islets: molecular cloning and expression pattern during development and growth of the endocrine pancreas. Endocrinology. 1997;138(9):3940-3948. [DOI] [PubMed] [Google Scholar]
- 68. Wang Y, Lee K, Moon YS, et al. Overexpression of Pref-1 in pancreatic islet β-cells in mice causes hyperinsulinemia with increased islet mass and insulin secretion. Biochem Biophys Res Commun. 2015;461(4):630-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rhee M, Lee SH, Kim JW, et al. Preadipocyte factor 1 induces pancreatic ductal cell differentiation into insulin-producing cells. Sci Rep. 2016;6:23960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cao J, Mu Q, Huang H. The roles of insulin-like growth factor 2 mRNA-binding protein 2 in cancer and cancer stem cells. Stem Cells Int. 2018;2018:4217259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Modi H, Jacovetti C, Tarussio D, et al. Autocrine action of IGF2 regulates adult β-cell mass and function. Diabetes. 2015;64(12):4148-4157. [DOI] [PubMed] [Google Scholar]
- 72. Casellas A, Mallol C, Salavert A, et al. Insulin-like growth factor 2 overexpression induces β-cell dysfunction and increases beta-cell susceptibility to damage. J Biol Chem. 2015;290(27):16772-16785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Riopel M, Wang R. Collagen matrix support of pancreatic islet survival and function. Front Biosci. 2014;19:77-90. [DOI] [PubMed] [Google Scholar]
- 74. Wang R, Li J, Rosenberg L. Factors mediating the transdifferentiation of islets of Langerhans to duct epithelial-like structures. J Endocrinol. 2001;171(2):309-318. [DOI] [PubMed] [Google Scholar]
- 75. Mizusawa N, Hasegawa T, Ohigashi I, et al. Differentiation phenotypes of pancreatic islet beta- and alpha-cells are closely related with homeotic genes and a group of differentially expressed genes. Gene. 2004;331:53-63. [DOI] [PubMed] [Google Scholar]
- 76. Matsuda T, Takahashi H, Mieda Y, et al. Regulation of pancreatic β cell mass by cross-interaction between CCAAT enhancer binding protein β induced by endoplasmic reticulum stress and amp-activated protein kinase activity. Plos One. 2015;10(6):e0130757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lu M, Seufert J, Habener JF. Pancreatic beta-cell-specific repression of insulin gene transcription by CCAAT/enhancer-binding protein beta. Inhibitory interactions with basic helix-loop-helix transcription factor E47. J Biol Chem. 1997;272(45):28349-28359. [DOI] [PubMed] [Google Scholar]
- 78. Weiss MJ, Orkin SH. GATA transcription factors: key regulators of hematopoiesis. Exp Hematol. 1995;23(2):99-107. [PubMed] [Google Scholar]
- 79. Villard J, Reith W, Barras E, et al. Analysis of mutations and chromosomal localisation of the gene encoding RFX5, a novel transcription factor affected in major histocompatibility complex class II deficiency. Hum Mutat. 1997;10(6):430-435. [DOI] [PubMed] [Google Scholar]
- 80. Ait-Lounis A, Bonal C, Seguín-Estévez Q, et al. The transcription factor Rfx3 regulates beta-cell differentiation, function, and glucokinase expression. Diabetes. 2010;59(7):1674-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Piccand J, Strasser P, Hodson DJ, et al. Rfx6 maintains the functional identity of adult pancreatic β cells. Cell Rep. 2014;9(6):2219-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang H, Brun T, Kataoka K, Sharma AJ, Wollheim CB. MAFA controls genes implicated in insulin biosynthesis and secretion. Diabetologia. 2007;50(2):348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nishimura W, Takahashi S, Yasuda K. MafA is critical for maintenance of the mature beta cell phenotype in mice. Diabetologia. 2015;58(3):566-574. [DOI] [PubMed] [Google Scholar]
- 84. Morey L, Santanach A, Di Croce L. Pluripotency and epigenetic factors in mouse embryonic stem cell fate regulation. Mol Cell Biol. 2015;35(16):2716-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Basch ML, Bronner-Fraser M, García-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441(7090):218-222. [DOI] [PubMed] [Google Scholar]
- 86. Lilja KC, Zhang N, Magli A, et al. Pax7 remodels the chromatin landscape in skeletal muscle stem cells. Plos One. 2017;12(4):e0176190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kitamura T, Nakae J, Kitamura Y, et al. The forkhead transcription factor Foxo1 links insulin signaling to Pdx1 regulation of pancreatic beta cell growth. J Clin Invest. 2002;110(12):1839-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kitamura YI, Kitamura T, Kruse JP, et al. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005;2(3):153-163. [DOI] [PubMed] [Google Scholar]
- 89. Okamoto H, Hribal ML, Lin HV, Bennett WR, Ward A, Accili D. Role of the forkhead protein FoxO1 in beta cell compensation to insulin resistance. J Clin Invest. 2006;116(3):775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Martinez SC, Tanabe K, Cras-Méneur C, Abumrad NA, Bernal-Mizrachi E, Permutt MA. Inhibition of Foxo1 protects pancreatic islet beta-cells against fatty acid and endoplasmic reticulum stress-induced apoptosis. Diabetes. 2008;57(4):846-859. [DOI] [PubMed] [Google Scholar]
- 91. Kikuchi O, Kobayashi M, Amano K, et al. FoxO1 gain of function in the pancreas causes glucose intolerance, polycystic pancreas, and islet hypervascularization. Plos One. 2012;7(2):e32249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Fajas L, Annicotte JS, Miard S, Sarruf D, Watanabe M, Auwerx J. Impaired pancreatic growth, beta cell mass, and beta cell function in E2F1 (-/-)mice. J Clin Invest. 2004;113(9):1288-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rady B, Chen Y, Vaca P, et al. Overexpression of E2F3 promotes proliferation of functional human β cells without induction of apoptosis. Cell Cycle. 2013;12(16):2691-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Artner I, Blanchi B, Raum JC, et al. MafB is required for islet beta cell maturation. Proc Natl Acad Sci U S A. 2007;104(10):3853-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Artner I, Hang Y, Mazur M, et al. MafA and MafB regulate genes critical to beta-cells in a unique temporal manner. Diabetes. 2010;59(10):2530-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xia F, Cao H, Du J, Liu X, Liu Y, Xiang M. Reg3g overexpression promotes β cell regeneration and induces immune tolerance in nonobese-diabetic mouse model. J Leukoc Biol. 2016;99(6):1131-1140. [DOI] [PubMed] [Google Scholar]
- 97. Liu B, Hassan Z, Amisten S, et al. The novel chemokine receptor, G-protein-coupled receptor 75, is expressed by islets and is coupled to stimulation of insulin secretion and improved glucose homeostasis. Diabetologia. 2013;56(11):2467-2476. [DOI] [PubMed] [Google Scholar]
- 98. Kharitonenkov A, Shiyanova TL, Koester A, et al. FGF-21 as a novel metabolic regulator. J Clin Invest. 2005;115(6):1627-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pan Y, Wang B, Zheng J, et al. Pancreatic fibroblast growth factor 21 protects against type 2 diabetes in mice by promoting insulin expression and secretion in a PI3K/Akt signaling-dependent manner. J Cell Mol Med. 2019;23(2):1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. So WY, Cheng Q, Xu A, Lam KS, Leung PS. Loss of fibroblast growth factor 21 action induces insulin resistance, pancreatic islet hyperplasia and dysfunction in mice. Cell Death Dis. 2015;6:e1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Magnusson M, Wang TJ, Clish C, et al. Dimethylglycine deficiency and the development of diabetes. Diabetes. 2015;64(8):3010-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wendeler MW, Praus M, Jung R, Hecking M, Metzig C, Gessner R. Ksp-cadherin is a functional cell-cell adhesion molecule related to LI-cadherin. Exp Cell Res. 2004;294(2):345-355. [DOI] [PubMed] [Google Scholar]
- 103. Simmons RA. Developmental origins of beta-cell failure in type 2 diabetes: the role of epigenetic mechanisms. Pediatr Res. 2007;61(5 Pt 2):64R-67R. [DOI] [PubMed] [Google Scholar]
- 104. Pinney SE, Simmons RA. Epigenetic mechanisms in the development of type 2 diabetes. Trends Endocrinol Metab. 2010;21(4):223-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Akbari M, Hassan-Zadeh V. The inflammatory effect of epigenetic factors and modifications in type 2 diabetes. Inflammopharmacology. 2020;28(2):345-362. [DOI] [PubMed] [Google Scholar]
- 106. Eliasson L, Regazzi R. Micro(RNA) management and mismanagement of the islet. J Mol Biol. 2020;432(5):1419-1428. [DOI] [PubMed] [Google Scholar]
- 107. De Jesus DF, Kulkarni RN. “Omics” and “epi-omics” underlying the β-cell adaptation to insulin resistance. Mol Metab. 2019;27S:S42-S48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ling C, Rönn T. Epigenetics in human obesity and type 2 diabetes. Cell Metab. 2019;29(5):1028-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Rosen ED, Kaestner KH, Natarajan R, et al. Epigenetics and epigenomics: implications for diabetes and obesity. Diabetes. 2018;67(10):1923-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hardikar AA, Satoor SN, Karandikar MS, et al. Multigenerational undernutrition increases susceptibility to obesity and diabetes that is not reversed after dietary recuperation. Cell Metab. 2015;22(2):312-319. [DOI] [PubMed] [Google Scholar]
- 111. Gómez-Banoy N, Lo JC. Adipokines as key players in β cell function and failure. Clin Sci. 2019;133(22):2317-2327. [DOI] [PubMed] [Google Scholar]
- 112. Dunmore SJ, Brown JE. The role of adipokines in β-cell failure of type 2 diabetes. J Endocrinol. 2013;216(1):T37-T45. [DOI] [PubMed] [Google Scholar]
- 113. Kaiko GE, Horvat JC, Beagley KW, Hansbro PM. Immunological decision-making: how does the immune system decide to mount a helper T-cell response? Immunology. 2008;123(3):326-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Mahlangu T, Dludla PV, Nyambuya TM, et al. A systematic review on the functional role of Th1/Th2 cytokines in type 2 diabetes and related metabolic complications. Cytokine. 2020;126:154892. [DOI] [PubMed] [Google Scholar]
- 115. Elghazi L, Gould AP, Weiss AJ, et al. Importance of β-catenin in glucose and energy homeostasis. Sci Rep. 2012;2:693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Artner I, Le Lay J, Hang Y, et al. MafB: an activator of the glucagon gene expressed in developing islet alpha- and beta-cells. Diabetes. 2006;55(2):297-304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in References.