Abstract
Background
Small cell lung cancer (SCLC) is a malignant disease with poor prognosis. At the time of diagnosis most patients are already in a metastatic stage. Current diagnosis is based on imaging, histopathology, and immunohistochemistry, but no blood-based biomarkers have yet proven to be clinically successful for diagnosis and screening. The precise mechanisms of SCLC are not fully understood, however, several genetic mutations, protein and metabolic aberrations have been described. We aim at identifying metabolite alterations related to SCLC and to expand our knowledge relating to this aggressive cancer.
Methods
A total of 30 serum samples of patients with SCLC, collected at the time of diagnosis, and 25 samples of healthy controls were included in this study. The samples were analyzed with nuclear magnetic resonance spectroscopy. Multivariate, univariate and pathways analyses were performed.
Results
Several metabolites were identified to be altered in the pre-treatment serum samples of small-cell lung cancer patients compared to healthy individuals. Metabolites involved in tricarboxylic acid cycle (succinate: fold change (FC) = 2.4, p = 0.068), lipid metabolism (LDL triglyceride: FC = 1.3, p = 0.001; LDL-1 triglyceride: FC = 1.3, p = 0.012; LDL-2 triglyceride: FC = 1.4, p = 0.009; LDL-6 triglyceride: FC = 1.5, p < 0.001; LDL-4 cholesterol: FC = 0.5, p = 0.007; HDL-3 free cholesterol: FC = 0.7, p = 0.002; HDL-4 cholesterol FC = 0.8, p < 0.001; HDL-4 apolipoprotein-A1: FC = 0.8, p = 0.005; HDL-4 apolipoprotein-A2: FC ≥ 0.7, p ≤ 0.001), amino acids (glutamic acid: FC = 1.7, p < 0.001; glutamine: FC = 0.9, p = 0.007, leucine: FC = 0.8, p < 0.001; isoleucine: FC = 0.8, p = 0.016; valine: FC = 0.9, p = 0.032; lysine: FC = 0.8, p = 0.004; methionine: FC = 0.8, p < 0.001; tyrosine: FC = 0.7, p = 0.002; creatine: FC = 0.9, p = 0.030), and ketone body metabolism (3-hydroxybutyric acid FC = 2.5, p < 0.001; acetone FC = 1.6, p < 0.001), among other, were found deranged in SCLC.
Conclusions
This study provides novel insight into the metabolic disturbances in pre-treatment SCLC patients, expanding our molecular understanding of this malignant disease.
Keywords: Small-cell lung cancer, Metabolomics, Serum metabolites, Pathways, Diagnostic signatures
Abbreviations:
- SCLC
Small cell lung cancer
- NSCLC
Non-small-cell lung cancer
- LS
Limited stage
- ES
Extensive stage
- TNM
Tumor, Nodes, Metastasis
- PCI
Prophylactic cranial irradiation
- CT
Computed tomography
- PET
Positron emission tomography
- TCA
Tricarboxylic acid
- NOESY
Nuclear Overhauser Effect Spectroscopy
- CPMG
Carr-Purcell-Meiboom-Gill
- HC
Healthy controls
- AUC
Area under the curve
- CV
Cross-validation
- VIP
Variable importance in projection
- FDR
False discovery rate
- PLS-DA
Partial least squared discriminant analysis
- BCAA
Branched-chain amino acid
- 3-HBA
3-hydroxybutyric acid
- VLDL
Very low-density lipoprotein
- IDL
Intermediate-density lipoprotein
- LDL
Low-density lipoprotein
- HDL
High-density lipoprotein
1. Introduction
Lung cancer is the second most common cancer type in both males and females and the leading cause of death among cancers globally, with an estimated 1.8 million in 2018 [1,2]. Histopathologically, lung cancer is divided into non-small cell lung carcinoma (NSCLC) and small-cell lung carcinoma (SCLC), with SCLC accounting for 15–20% of the incidents [3]. SCLC is highly malignant and is known for an aggressive tumor growth with poor prognosis [[4], [5], [6]]. Small cell lung cancer is traditionally categorized as limited stage (LS-SCLC) or extensive stage (ES-SCLC). However, in 2007 the International Association for the Study of Lung Cancer recommended the use of the Tumor, Lymph Node, Metastasis (TNM) staging system for SCLC [7]. Most SCLC patients are already in the metastatic ES-SCLC at diagnosis, and the reported two-year survival rate for ES-SCLC is only around 5% [5,6]. Patients are initially discovered when they consult their general practitioner with unspecific symptoms, which occur when the cancer is already advanced [8]. Diagnosis is confirmed by clinical imaging using computed tomography (CT) and positron emission tomography/CT in combination with cytological/histopathological biopsies of the tumor-suspected lesions of the lung [9].
Research fields like genomics and proteomics have already detected gene mutations and altered protein expressions [9] and these findings have helped pave the way for the testing of new treatments such as immune- and targeted therapies in SCLC [10]. However, the extensive research has not yet resulted in any successful screening modalities for SCLC [11]. It is expected that metabolomics can further explore the molecular mechanisms of SCLC [12]. Metabolomics is the scientific method investigating chemical processes involving metabolites, the small molecular substrates of cell metabolism, which are the end products of gene and protein regulations and are highly responsive to medication, disease progression or regression, and changes in environmental factors [13]. Since metabolites are the intermediates and end-products of cellular cancer processes, including cell differentiation, proliferation, regulation, and DNA repair, the “Hallmarks of Cancer” were changed in 2010 to include the reprogramming of energy metabolism as an emerging hallmark of cancer [14].
In the last two decades, the field of metabolomics have gradually accumulated important information towards the understanding of lung cancer [15]. However, the focus has primarily been on NSCLC, whereas SCLC has only recently gained attention [9,11]. The current metabolomic research on SCLC is limited, however, the metabolomic manifestation of SCLC appears to be partly comparable with the NSCLC metabolome, indicating both common and unique mechanisms [[16], [17], [18]]. Among most significant molecular aberrations found in lung cancers are those involved in cell growth such as energy metabolism, identified through changes in concentrations of metabolites related to tricarboxylic acid (TCA) cycle and high glucose metabolism including glucose, lactate, glutamic acid, and glutamine [[19], [20], [21], [22]], one-carbon metabolism, fatty acid metabolism, nucleotide biosynthesis, and amino acid metabolism [23,24].
This study aims to identify serum metabolite perturbations in newly diagnosed patients that may add new insight to the complex molecular mechanisms underpinning SCLC.
2. Methods
2.1. Study participants
This cross-sectional study included medical data and pre-treatment blood samples of 31 patients diagnosed with SCLC between March 2015 until September 2017 at the Department of Oncology, Aalborg University Hospital, Denmark. The study was primarily dealing with thrombogenic alterations in SCLC patients, and the inclusion and exclusion criteria were previously described by Pedersen et al. [25]. Briefly, for inclusion criteria patients had to be eligible to receive chemotherapy consisting of platinum and a topoisomerase inhibitor according to the standard treatment guidelines in Denmark. In all cases, a histopathologically and/or cytologically confirmed SCLC was required. The exclusion criteria were: prior systemic chemotherapy for lung cancer, concomitant anticoagulation treatment (platelet inhibitors, acetylsalicylic acid, and clopidogrel were allowed), active or high risk of overt bleeding of clinical importance, severe coagulopathy such as hemophilia, severe liver dysfunction with impaired coagulation, acute peptic ulcer, intracranial hemorrhage or surgery in the central nervous system within the last three months, treatment with any other investigational agents, and participation in other clinical trials. Clinical data, administration of medications, treatment details, and radiological evaluation, and routine laboratory testing were collected at the time of diagnosis (at baseline). Staging of the SCLC was based on the 7th edition of the tumor, lymph node, metastasis (TNM) classification of lung cancer [7]. The present study was approved by the North Denmark Region Committee on Health Research Ethics (N-20140055), reported to the Danish Data Protection Authority (2021-000206), and performed in accordance with the Declaration of Helsinki. All included participants provided written informed consent before enrolment in the study. In addition, blood obtained from 25 age- and gender-matched healthy individuals from the blood bank at Aalborg University Hospital were used for comparison. In Denmark, blood donors are healthy volunteers without any biochemical anomalies or evidence of illness and only rarely using medicine (most medicine is not allowed). Donors were also required to complete a questionnaire regarding their physical health including smoking status and current medication.
2.2. Sample collection and preparation
Patient blood samples were collected at the time of diagnosis. Blood samples were collected in 10 mL clot activator tubes (BD Vacutainer®, UK) and centrifuged at 2500×g for 15 min at room temperature. The subsequent serum was snap-frozen in liquid nitrogen and stored at −80 °C until analysis. For this study, only samples obtained at the time of diagnosis were analyzed.
2.3. Nuclear magnetic resonance spectroscopy
Prior to NMR analysis, serum samples were thawed for 1 h, gently mixed with an equal volume (350 μL) of sodium phosphate buffer (0,075 M, pH 7.4, 20% D2O in H2O, 6 mM NaN3, 4.6 mM 3-(trimethylsilyl)-2,2,3,3-tetradeuteropropanoic acid (TSP-d4)) and transferred to 5 mm NMR tubes. NMR spectra were recorded on a Bruker Avance III 600 MHz spectrometer fitted with a BBI probe (Bruker Biospin Corporation, Billerica, MA). Sample handling and data acquisition was performed in automation using IconNMR on Topspin 3.6.2 and Samplejet autosampler (Bruker Biospin). One-dimensional nuclear Overhauser effect (NOESY) spectra (pulse program “noesygppr1d”) and Carr-Purcell-Meiboom-Gill (CPMG) (“cpmgpr1d) were recorded at 310 K using acquisition parameters from Dona et al. (2014) [26]. The NOESY spectra were recorded with 96k data points, 30 ppm spectral width, whereas CPMG spectra were recorded with 72k data points and 20 ppm spectral width. Both experiments were recorded with 32 scans and with water suppression (25 Hz) during relaxation delay (4 s) and mixing time (NOESY, 10 ms). The free induction decays were Fourier transformed after zero fillings to 128k data points and 0.3 Hz line broadening. In accordance with B.I.Methods (Bruker Biospin), reference samples for temperature calibration, water suppression determination and external quantitative referencing were routinely recorded and processed in automation.
Quantification of 155 metabolites was automatically performed using B.I.Quant-PS™ 2.0 and B.I.LISA™ (Bruker Biospin). A comprehensive list of included metabolites and abbreviations can be found in the supplementary table (Table S1). Metabolic changes were compared to the Edinburgh Human Metabolic Network [27] and Kyoto Encyclopedia of Genes and Genomes [28] databases to enrich the findings with references and metabolic pathways from existing research.
Metabolite annotation was performed using 2D 1H–1H total correlation spectroscopy and 1H–13C heteronuclear single-quantum correlation spectra, the Human Metabolome Database (HMDB) [29], and results from literature [[30], [31], [32], [33], [34]].
2.4. Statistics
A supervised partial least squares discriminant analysis (PLS-DA) was conducted to identify metabolite differences between healthy controls and SCLC patients. Variable Importance in Projection (VIP)-score algorithm was used to select most important metabolites in sample grouping. A VIP score rank of ≥1 was considered important. Multivariate analyses were performed in the open-source software program RStudio [35] using the R-package “mixOmics” [36].
Prior to multivariate analysis, the data was normalized, Pareto scaled, and mean centered. A 10-Fold Cross-Validation (CV) repeated 1000 times was applied for validation of the PLS-DA modelling. The classification error rate was averaged across all folds and the optimal number of components was determined by the balanced error rate of the maximum distance and the Mahalanobis distance error rate [36]. Data was checked for normality by Shapiro test and nonparametric Mann-Whitney U Test and post-hoc false discovery rate (FDR) correction were performed to identify metabolites presenting statistically significant differences among groups. Fold change analysis was performed based on median ratio between sick and healthy individuals. Receiver operating characteristics (ROC) analysis was conducted in GraphPad Prism version 9.1.1 (GraphPad Software, La Jolla, CA, USA) to test the discriminatory efficacy of the identified diagnostics metabolites. Finally, visualization of altered metabolite pathways was performed in MetScape 3.1.3 [37].
3. Results
3.1. Subject characteristics
A total of 30 SCLC patients fulfilled the inclusion criteria and were enrolled in this study. Demographics and clinical characteristics of the study participants are described in Table 1. At the time of inclusion, 24 SCLC-patients were diagnosed with an extensive disease where 23 of these had lymph node metastases. Based on TNM staging, 19 patients were classified as stage IV.
Table 1.
Demographics and patient characteristics of the study population. All values are presented as mean ± standard deviation. SD = standard deviation, TNM staging = Tumor, Lymph Node, and Metastasis.
| Characteristics of SCLC patients and healthy controls | |||
|---|---|---|---|
| SCLC patients |
Healthy controls |
||
| n = 30 | n = 25 | ||
| Demographics | |||
| Sex (Male/females, n) | 16/14 | 12/13 | |
| Mean age (±SD) | 65 ± 9 | 63 ± 3 | |
| Smokers/Non-smokers | 29/1 | 0/25 | |
| Disease characteristics | |||
| Disease stage, n (%) | |||
| Limited stage | 6 (20%) | ||
| Extended stage | 24 (80%) | ||
| TNM stage, n (%) | |||
| IIB | 1 (3%) | ||
| IIIA | 7 (23%) | ||
| IIIB | 3 (10%) | ||
| IV | 19 (63%) | ||
3.2. Altered metabolic signatures in small cell lung cancer patients
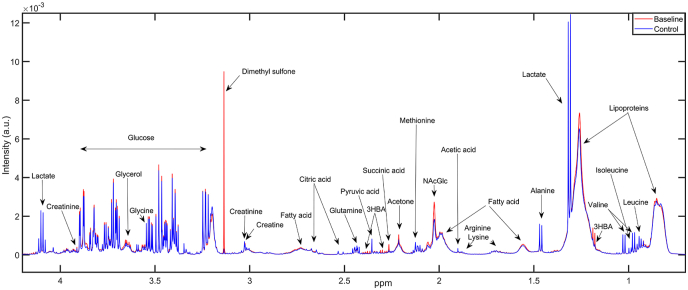
Several intensities were found different between healthy control and SCLC individuals, indicating altered metabolite fingerprints among cancer patients (Fig. 1). Visual inspection of the averaged CPMG spectra of newly diagnosed patients (Baseline, red) and healthy controls (Control, blue) revealed distinctive patterns between the groups within several metabolic intensities corresponding to fatty acids, lipoproteins, creatinine, glycerol, glycine, acetone, and succinic acid, among others.
Fig. 1.
Averaged metabolic fingerprints of healthy controls (Control, blue) and newly diagnosed small cell lung cancer patients (Baseline, red) with annotated peak intensities in the ∼4.5–0.5 ppm range on the Carr-Purcell Meiboom-Gill (CPMG) spectra belonging to amino acids, glucose, lactate, lipoproteins, fatty acids, and ketone bodies, among others. The intense dimethyl sulfone signal was due to one patient presenting abnormally high concentration. Abbreviations: ppm = parts per million, 3-HBA = 3-hydroxybutyric acid, NAcGlc = N-Acetyl-Glycoprotein fragment. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
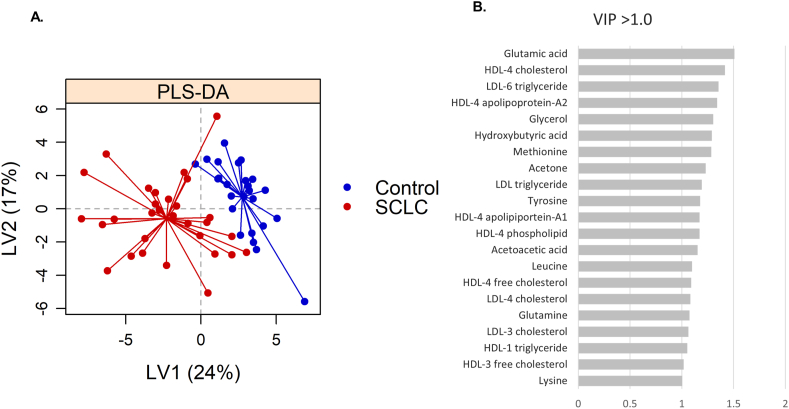
To illustrate the maximum variance between groups, a supervised PLS-DA modelling was performed on the 155 quantified metabolites. A clear separation was found between healthy controls and newly diagnosed SCLC patients (Fig. 2A), with an error of cross validation of ≤0.20. Most significant metabolites based on VIP-scores≥1.0 are shown in Fig. 2B and include amino acids (glutamic acid/glutamate, glutamine, isoleucine, leucine, lysine, methionine, ornithine, tyrosine, and succinic acid), ketone bodies (3-hydroxybutyric acid, acetone, and acetoacetic acid), glucose, glycerol, and lipoprotein subfractions of LDL and HDL (Table 2).
Fig. 2.
Distinct patterns between healthy controls and Small-Cell Lung Cancer (SCLC) patients. A. Partial least squares discriminant analysis (PLS-DA) scores plot of healthy control samples (blue) versus newly diagnosed SCLC patients (red) on latent variable 1 (LV) and 2. B. Most significant metabolites based on VIP-scores ranking (VIP >1.0). VIP = Variable importance in projection, LV = latent variables. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 2.
Metabolic signatures significantly altered in SCLC patients at the time of diagnosis compared to healthy controls. Testing was based on non-parametric Mann-Whitney U Test with FDR correction. The medians of significant metabolites were compared between groups to determine fold changes (FC). All metabolite concentrations are in mmol/L except for the lipoprotein subfractions which are displayed in mg/dL. Abbreviation: FDR = false discovery rate, LDL = low-density lipoprotein, HDL = high-density lipoprotein, based on population and square root. FC = median fold change, n.a. = not available, ROC = receiver operating characteristics curve, AUC = area under the curve a. p-values of Mann Whitney U Test. b. FDR corrected p-values. c. p-values of ROC-based AUCs.
| Metabolic signatures of small cell lung cancer | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Metabolites [mmol/L or mg/dL] | Controls |
SCLC |
FC | ap-value | bq-value | Sensitivity (%) | Specificity (%) | AUC (95% CL) | cp-value |
| Median (min-max) | Median (min-max) | ||||||||
| 3-Hydroxybutyric acid | 0.037 (0.00–0.27) | 0.091 (0.03–0.86) | 2.5 | <0.001 | 0.001 | 83 | 72 | 0.86 (0.76–0.95) | <0.001 |
| Acetoacetic acid | 0.000 (0.00–0.05) | 0.010 (0.00–0.26) | n.a. | 0.001 | 0.011 | 57 | 88 | 0.81 (0.60–0.86) | 0.003 |
| Acetone | 0.019 (0.00–0.04) | 0.030 (0.01–0.15) | 1.6 | <0.001 | 0.005 | 80 | 68 | 0.73 (0.68–0.91) | <0.001 |
| Glutamic acid | 0.069 (0.00–0.14) | 0.115 (0.06–0.27) | 1.7 | <0.001 | 0.004 | 80 | 72 | 0.81 (0.70–0.92) | <0.001 |
| Glutamine | 0.660 (0.51–0.81) | 0.587 (0.34–0.79) | 0.9 | 0.007 | 0.047 | 73 | 64 | 0.71 (0.58–0.85) | 0.007 |
| Glycerol | 0.000 (0.00–0.42) | 0.292 (0.00–0.88) | n.a. | 0.001 | 0.096 | 73 | 72 | 0.76 (0.63–0.89) | 0.001 |
| HDL-3 free cholesterol | 2.261 (1.24–3.56) | 1.685 (0.16–4.02) | 0.7 | 0.002 | 0.023 | 70 | 68 | 0.74 (0.61–0.87) | 0.002 |
| HDL-4 apolipoprotein-A1 | 68.871 (48.96–84.58) | 56.793 (29.06–81.18) | 0.8 | 0.005 | 0.038 | 60 | 84 | 0.72 (0.59–0.86) | 0.005 |
| HDL-4 apolipoprotein-A2 | 18.006 (10.45–22.06) | 13.358 (6.33–21.86) | 0.7 | 0.001 | 0.006 | 67 | 76 | 0.77 (0.65–0.90) | <0.001 |
| HDL-4 cholesterol | 18.812 (13.67–23.48) | 14.295 (5.85–23.12) | 0.8 | <0.001 | 0.006 | 70 | 76 | 0.78 (0.66–0.90) | <0.001 |
| HDL-4 free cholesterol | 3.522 (2.25–4.77) | 2.720 (0.74–4.74) | 0.8 | 0.008 | 0.047 | 73 | 64 | 0.71 (0.58–0.85) | 0.007 |
| HDL-4 phospholipid | 26.706 (19.20–33.12) | 22.264 (12.67–32.48) | 0.8 | 0.005 | 0.027 | 67 | 76 | 0.73 (0.60–0.87) | 0.003 |
| LDL triglyceride | 15.950 (9.55–32.90) | 21.471 (11.00–55.92) | 1.3 | 0.001 | 0.006 | 80 | 76 | 0.78 (0.65–0.91) | <0.001 |
| LDL-1 triglyceride | 4.664 (2.73–11.31) | 6.264 (3.42–19.49) | 1.3 | 0.012 | 0.038 | 70 | 76 | 0.70 (0.55–0.84) | 0.012 |
| LDL-2 triglyceride | 1.945 (1.06–5.15) | 2.730 (0.95–3.95) | 1.4 | 0.009 | 0.054 | 73 | 72 | 0.70 (0.56–0.85) | 0.010 |
| LDL-4 cholesterol | 13.585 (0.00–28.42) | 6.995 (0.00–22.19) | 0.5 | 0.007 | 0.047 | 70 | 60 | 0.71 (0.58–0.85) | 0.007 |
| LDL-6 triglyceride | 3.934 (2.20–6.11) | 5.854 (3.04–15.33) | 1.5 | <0.001 | 0.003 | 73 | 72 | 0.81 (0.70–0.92) | <0.001 |
| Leucine | 0.100 (0.07–0.20) | 0.078 (0.05–0.21) | 0.8 | <0.001 | 0.004 | 77 | 80 | 0.81 (0.69–0.93) | <0.001 |
| Lysine | 0.208 (0.00–0.33) | 0.167 (0.00–0.29) | 0.8 | 0.004 | 0.035 | 60 | 72 | 0.73 (0.59–0.86) | 0.004 |
| Methionine | 0.085 (0.06–0.13) | 0.065 (0.00–0.13) | 0.8 | <0.001 | 0.005 | 80 | 72 | 0.80 (0.68–0.92) | <0.001 |
| Threonine | 0.116 (0.00–0.37) | 0.000 (0.00–0.30) | n.a. | 0.005 | 0.044 | 73 | 60 | 0.71 (0.57–0.85) | 0.009 |
| Tyrosine | 0.068 (0.04–0.11) | 0.049 (0.00–0.11) | 0.7 | 0.002 | 0.021 | 73 | 68 | 0.75 (0.62–0.88) | 0.002 |
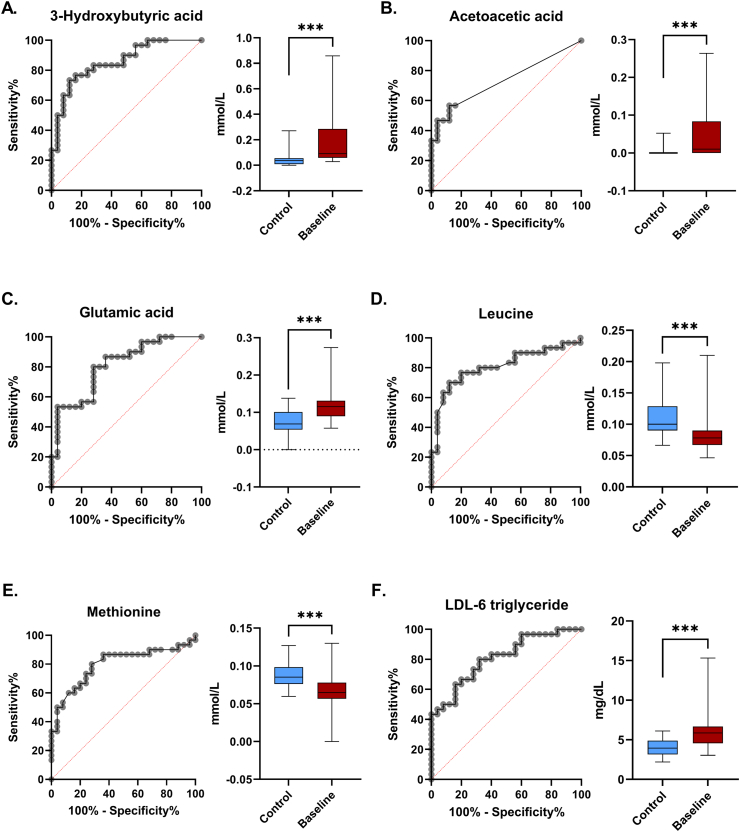
Fig. 3 represents the most significant metabolite differences found between healthy controls and SCLC patients: 3-HBA (AUC = 0.86, 95% CI. 0.76–0.95, p < 0.001), acetoacetic acid (AUC = 0.81, 95% CI. 0.6–0.86, p = 0.003), glutamic acid (AUC = 0.81, 95% CI. 0.7–0.92, p < 0.001), leucine (AUC = 0.81, 95% CI. 0.69–0.93, p < 0.001), methionine (AUC = 0.80, 95% CI. 0.68–0.92, p < 0.001), and LDL-6 triglyceride (AUC = 0.81, 95% CI. 0.700.92, p < 0.001).
Fig. 3.
Most significant metabolites found perturbed in SCLC patients (AUC≥0.80). Box plots and receiver operating characteristic curves (ROC) are presented. A. ROC-curve and boxplot of 3-hydroxybutyric acid: increased in SCLC patients compared to healthy controls. B. ROC-curve and boxplot of acetoacetic acid: increased in SCLC patients. C. ROC-curve and boxplot of glutamic acid: increased in SCLC patients. D. ROC-curve and boxplot of leucine: decreased in SCLC patients. E. ROC-curve and boxplot of methionine: decreased in SCLC patients. F. ROC-curve and boxplot of LDL-6 triglyceride: increased in SCLC patients. LDL = low-density lipoprotein. Significance is indicated by; *** <0.001, ** <0.01.
3.3. Reprogrammed metabolic pathways in small-cell lung cancer
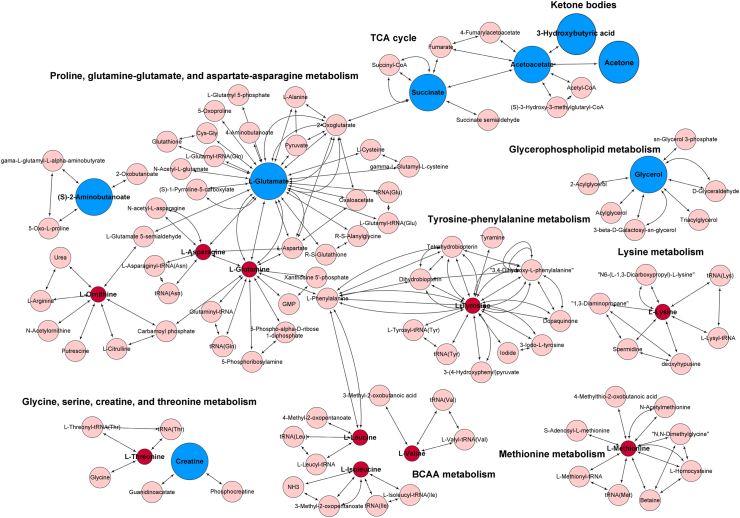
To extrapolate important metabolic changes possibly related to SCLC, pathway analysis was performed on altered small metabolites with non-corrected p-values <0.1, excluding lipoproteins due to missing KEGG IDs. We found eight additional metabolites of interest including glucose (FC = 1.1, p = 0.067), valine (FC = 0.9, p = 0.085), isoleucine (FC = 0.8, p = 0.023), creatine (FC = 1.9, p = 0.015), ornithine (p = 0.017), asparagine (p = 0.011), succinic acid (FC = 2.4, p = 0.014), and 2-aminobutyric acid (p = 0.017) (Table S2). Fig. 4 shows several metabolic pathways to be altered in SCLC, including ketone body metabolism, TCA cycle, amino acids, and glycerophospholipid metabolisms.
Fig. 4.
Altered metabolic pathways in small cell lung cancer (SCLC) patients. Mapped metabolites are based on their KEGG IDs to classify their involvement in different pathways. Red and blue nodes indicate altered metabolites in the study and pink nodes represent metabolites involved in the pathway that were not investigated in the study. Red color codes represent decreased metabolite, while blue increased metabolite concentrations in SCLC compared to healthy controls. BCCA: Branched-chain amino acid, TCA: tricarboxylic acid. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
This study provides a detailed NMR-based metabolomics analysis of serum from healthy controls and newly diagnosed patients with the purpose of identifying distinguished metabolites that help underpinning the impaired complex molecular mechanisms of SCLC. We identified changes in amino acid, energy, and lipid metabolisms of newly diagnosed SCLC patients compared to healthy individuals.
The altered amino acids included glutamic acid, glutamine, leucine, isoleucine, valine, lysine, methionine, threonine, and tyrosine (Fig. 4, Table 1). Increased glutamic acid showed great potential in discriminating between SCLC patients and healthy controls (FC = 1.7, p < 0.001, and AUC = 0.81 with 95% CI. For AUC: 0.70–0.92, p < 0.001). Similar perturbations have previously been described in serum and plasma of NSCLC patients [[38], [39], [40]], and described in a small SCLC study, giving confidence to our findings. The role of glutamic acid in cancer is well-described and has become an important target in cancer treatment research [41,42]. Among others, glutamic acid plays key roles in biosynthetic pathways of cells, including purine and pyrimidine synthesis, as nitrogen and carbon source in TCA cycle anaplerosis, protein and amino acid synthesis, and lipid metabolism [43]. Furthermore, we identified glutamine to be slightly decreased in SCLC patients (FC = 0.9, p = 0.007). Glutamine has previously been found to be decreased in serum of NSCLC [38,44], however, to our knowledge, this is the first time we provide evidence of similar alterations in SCLC. Glutamine is known to be important for the cellular uptake of essential amino acids and activation of the oncogenic mTOR pathway, involved in cell proliferation and inhibition of apoptosis, which is known to be associated with SCLC pathogenesis [9,45], hence our results may reflect alterations in these mechanisms.
Interestingly, we found the branched-chain amino acids leucine, isoleucine, and valine to be decreased in SCLC patients (Fig. 4), which might be a distinctive signature for SCLC since previous studies have reported increased plasma and serum isoleucine and leucine in NSCLC [19,22,38,44]. BCAAs are involved in various metabolic pathways, including glutamic acid and glutamine metabolism, as well as downstream catabolism to acetyl-CoA, which fuels the TCA cycle, fatty acid, and cholesterol synthesis, and interferes with tumor growth metabolism through the activation of the mTOR pathway [46,47].
The three main ketone bodies 3-HBA, acetoacetic acid, and acetone were found significantly increased in SCLC patients, with FCs of 2.5 and 1.6 for 3-HBA and acetone, respectively (p ≤ 0.001). Enhanced ketone bodies are a common finding in various cancer types including lung cancer [16]. Ketone bodies are primarily responsible for delivering energy to cells; however, 3-HBA also participates in regulation of various cellular processes including protein acetylation, cell signaling, and augmented cell resistance to reactive oxygen species formation [48,49]. It is worth considering that the increased serum ketone body levels found could be explained by a loss of appetite, which is a well-known symptom observed in cancer, leading to a lower food intake or patients fasting before blood sample collection [50]. Blood samples in this study were collected at different times of the day and patients were not asked to be fasting beforehand; however, this does not exclude the possibility that patients could have been fasting prior to sample collection. The normal range of ketone bodies in the blood is around 200–400 μM, which correlates with the serum levels of healthy individuals found in this study. These values can increase up to five-fold during fasting [49], which is similar to the mean concentrations measured in SCLC patients in this study. It is still interesting that some cancer cells, due to their abnormal mitochondria, are unable to use ketones as fuel and some studies have even suggested a potential positive effect of a ketogenic diet against cancer progression [50,51]. However, it is possible that the increased ketogenesis in cancer cells was due to altered lipogenesis, gluconeogenesis, and cholesterogenesis, which are all mechanisms tightly linked to ketone regulation, as well as due to the high competition of primary energy compounds between cancer and healthy cells [51,52].
Altered lipid- and lipoprotein metabolism of SCLC was observed in this study through elevated levels of glycerol and LDL triglycerides, and lowered serum concentration of HDL-4 subfractions in SCLC patients. Our results are in line with previous findings, where serum and plasma glycerol levels were found altered in lung cancer patients [16,38,40]. Glycerol is involved in glycolysis and biosynthesis of lipids, including triglycerides and phospholipids [53]. Cancer cells have a high need for energy and lipids for cell membrane synthesis in order to proliferate. Therefore, it is not surprising that glycerol was found to be disturbed in SCLC patients. Same mechanisms might also explain the changes in lipoprotein concentrations we observed in LDL and HDL subfractions. While the triglyceride subfractions of LDL subtypes were mainly found increased (FC ≥ 1.3, p ≤ 0.012), LDL-4 cholesterol was found consistently decreased in SCLC patients. While similar LDL profiles with high serum LDL triglycerides and low LDL-cholesterols have been identified in a large case-control study with several cancer types [54], to our knowledge, this is the first SCLC study to show this pattern. Interestingly, Zhou, T. et al. 2017 [55] found that elevated serum-levels of LDL at diagnosis correlated with a worse overall outcome for SCLC patients. The precise involvement of LDL in cancer is not fully established and it is currently an emerging area of research. However, a possible involvement of LDL in cancer is considered to be by providing lipids to cancer cells [56], which is supported by findings of increased LDL uptake and LDL-receptor expression in cancer cells, including lung cancer [57]. Moreover, LDL is known to play a role in regulating pathways commonly altered in cancer including STAT3-and the mTOR-pathway. We further identified a consistent decrease in HDL-4 subfractions and in the cholesterol of HDL-3 in SCLC patients (FC ≤ 0.8, p ≤ 0.008). Previous studies have found HDL to be lower in plasma of lung cancer patients, and LDL and VLDL to be increased in the plasma of lung cancer patients [56]. Moreover, a large cohort study indicated that low plasma levels of HDL cholesterol were associated with a greater risk of lung cancer [58].
A relatively small cohort of individuals (25 healthy and 30 SCLC individuals) is one of the limitations of this study and may introduce a risk of bias. Furthermore, the subjects were mostly ES-SCLC (80%) in TNM Stage IV (63%). Previous studies have concluded that an effective screening modality must be capable of identifying tumors earlier than when the primary tumor is visible through CT-imaging [59]. To identify metabolite signatures of early SCLC, additional comparisons between disease states (LS vs ED) or TNM stages (early vs locally advanced vs metastatic disease), are warranted. Therefore, a study with a larger cohort of LS-SCLC patients is recommended. The age distribution of controls was narrower than the patients because there is an upper limit of age being a blood donor. However, the mean age was almost the same, and this difference is probably of minor importance. Furthermore, the patients were smokers and the controls non-smokers; we cannot exclude that this may result in (some) metabolic differences in itself.
Therapeutic approaches could potentially be guided from the metabolic changes, but the mechanisms are probably complex. Ideally, combining these findings with genetic and/or protein alterations in SCLC cells would provide for more detailed information on the molecular mechanisms of SCLC linked to these metabolic changes.
In this study, we demonstrate significant disturbances in serum metabolic profile of SCLC patients, thus providing novel scientific basis for future research in clinical routine settings. Understanding the metabolic landscape of SCLC could provide essential knowledge about disease mechanisms and characteristics, and ultimately help improve the survival of patients through better treatment options, and perhaps pave the way for the discovery of metabolite biomarkers for early detection of SCLC.
Data availability
Data is available upon request.
CRediT authorship contribution statement
Shona Pedersen: Conceptualization, Data curation, Investigation, Resources, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Joachim Bavnhøj Hansen: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Software, Writing – original draft, Writing – review & editing. Raluca Georgiana Maltesen: Investigation, Visualization, Validation, Writing – review & editing. Weronika Maria Szejniuk: Validation, Supervision. Trygve Andreassen: Methodology, Investigation, Software. Ursula Falkmer: Resources, Validation. Søren Risom Kristensen: Conceptualization, Validation, Writing – review & editing.
Acknowledgements
The authors are greatly appreciative to Lona Rosborg Nielsen, Helle Dalsgaard Holst, Helle Hylander, and Mette Jespersgaard for their assistance in enrolment and blood sample collection from patients and controls.
The NMR experiments were performed at the MR Core Facility, Norwegian University of Science and Technology (NTNU). MR Core facility is funded by the Faculty of Medicine and Health Sciences at NTNU and Central Norway Regional Health Authority.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.metop.2021.100127.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
None.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization . 2018. WHO: cancer - key facts.https://www.who.int/news-room/fact-sheets/detail/cancer Available at: Accessed. [Google Scholar]
- 3.Rudin C.M., Poirier J.T. Small-cell lung cancer in 2016: shining light on novel targets and therapies. Nat Rev Clin Oncol. 2017;14(2):75–76. doi: 10.1038/nrclinonc.2016.203. [DOI] [PubMed] [Google Scholar]
- 4.Dowell J.E. Small cell lung cancer: are we making progress? Am J Med Sci. 2010;339(1):68–76. doi: 10.1097/MAJ.0b013e3181bccef5. [DOI] [PubMed] [Google Scholar]
- 5.Lu T., Yang X., Huang Y., Zhao M., Li M., Ma K. Trends in the incidence, treatment, and survival of patients with lung cancer in the last four decades. Canc Manag Res. 2019;11:943–953. doi: 10.2147/CMAR.S187317. Jan 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunn P.A., Minna J.D., Augustyn A., Gazdar A.F., Ouadah Y., Krasnow M.A. Small cell lung cancer: can recent advances in biology and molecular biology Be translated into improved outcomes? J Thorac Oncol. 2016;11(4):453–474. doi: 10.1016/j.jtho.2016.01.012. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobin L.H., Gospodarwicz M.K., Wittekind C. Wiley-Blackwell; Chichester, West Sussex, UK: 2009. TNM classification of malignant tumours. [Google Scholar]
- 8.Tsoukalas N., Aravantinou-Fatorou E., Baxevanos P., Tolia M., Tsapakidis K., Galanopoulos M. Advanced small cell lung cancer (SCLC): new challenges and new expectations. Ann Transl Med. 2018;6(8):145. doi: 10.21037/atm.2018.03.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudin C.M., Brambilla E., Faivre-Finn C., Sage J. Small-cell lung cancer. Nat Rev Dis Primers. 2021;7(1) doi: 10.1038/s41572-020-00235-0. Jan 14. 3-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang S., Zhang Z., Wang Q. Emerging therapies for small cell lung cancer. J Hematol Oncol. 2019;12(1):47. doi: 10.1186/s13045-019-0736-3. 05 02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gazdar A.F., Bunn P.A., Minna J.D. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Canc. 2017;17(12):725–737. doi: 10.1038/nrc.2017.87. Nov 12. [DOI] [PubMed] [Google Scholar]
- 12.Vignoli A., Ghini V., Meoni G., Licari C., Takis P.G., Tenori L. High-Throughput metabolomics by 1D NMR. Angew Chem. 2019;58(4):968–994. doi: 10.1002/anie.201804736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derveaux E. In: Lung cancer Rijeka: IntechOpen. Louis Evelyne., editor. 2018. Diagnosis of lung cancer: what metabolomics can contribute. Ch. 4. [Google Scholar]
- 14.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. Mar 4. [DOI] [PubMed] [Google Scholar]
- 15.Tang Y., Li Z., Lazar L., Fang Z., Tang C., Zhao J. Metabolomics workflow for lung cancer: discovery of biomarkers. Clin Chim Acta. 2019;495:436–445. doi: 10.1016/j.cca.2019.05.012. Aug. [DOI] [PubMed] [Google Scholar]
- 16.Bamji-Stocke S., van Berkel V., Miller D.M., Frieboes H.B. A review of metabolism-associated biomarkers in lung cancer diagnosis and treatment. Metabolomics. 2018;14(6):81. doi: 10.1007/s11306-018-1376-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hori S., Nishiumi S., Kobayashi K., Shinohara M., Hatakeyama Y., Kotani Y. A metabolomic approach to lung cancer. Lung Canc. 2011;74(2):284–292. doi: 10.1016/j.lungcan.2011.02.008. Nov. [DOI] [PubMed] [Google Scholar]
- 18.Lim S.L., Jia Z., Lu Y., Zhang H., Ng C.T., Bay B.H. Metabolic signatures of four major histological types of lung cancer cells. Metabolomics. 2018;14(9):118. doi: 10.1007/s11306-018-1417-x. 08 31. [DOI] [PubMed] [Google Scholar]
- 19.Deja S., Porebska I., Kowal A., Zabek A., Barg W., Pawelczyk K. Metabolomics provide new insights on lung cancer staging and discrimination from chronic obstructive pulmonary disease. J Pharmaceut Biomed Anal. 2014;100:369–380. doi: 10.1016/j.jpba.2014.08.020. Nov. [DOI] [PubMed] [Google Scholar]
- 20.Rocha C.M., Barros A.S., Goodfellow B.J., Carreira I.M., Gomes A., Sousa V. NMR metabolomics of human lung tumours reveals distinct metabolic signatures for adenocarcinoma and squamous cell carcinoma. Carcinogenesis. 2015;36(1):68–75. doi: 10.1093/carcin/bgu226. Jan. [DOI] [PubMed] [Google Scholar]
- 21.Fan T.W.M., Lane A.N., Higashi R.M., Farag M.A., Gao H., Bousamra M. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM) Mol Canc. 2009;8:41. doi: 10.1186/1476-4598-8-41. Jun 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Louis E., Adriaensens P., Guedens W., Bigirumurame T., Baeten K., Vanhove K. Detection of lung cancer through metabolic changes measured in blood plasma. J Thorac Oncol. 2016;11(4):516–523. doi: 10.1016/j.jtho.2016.01.011. Apr. [DOI] [PubMed] [Google Scholar]
- 23.Yu L., Li K., Zhang X. Next-generation metabolomics in lung cancer diagnosis, treatment and precision medicine: mini review. Oncotarget. 2017;8(70):115774–115786. doi: 10.18632/oncotarget.22404. Dec 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang L., Fang S., Gu W. The molecular mechanism of metabolic remodeling in lung cancer. J Canc. 2020;11(6):1403–1411. doi: 10.7150/jca.31406. 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedersen S., Kristensen A.F., Falkmer U., Christiansen G., Kristensen S.R. Increased activity of procoagulant factors in patients with small cell lung cancer. PloS One. 2021;16(7) doi: 10.1371/journal.pone.0253613. Jul 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dona A.C., Jiménez B., Schäfer H., Humpfer E., Spraul M., Lewis M.R. Precision high-throughput proton NMR spectroscopy of human urine, serum, and plasma for large-scale metabolic phenotyping. Anal Chem. 2014;86(19):9887–9894. doi: 10.1021/ac5025039. 10-07. [DOI] [PubMed] [Google Scholar]
- 27.Ma H., Sorokin A., Mazein A., Selkov A., Selkov E., Demin O. The Edinburgh human metabolic network reconstruction and its functional analysis. Mol Syst Biol. 2007;3:135. doi: 10.1038/msb4100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wishart D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vázquez-Fresno R. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–D617. doi: 10.1093/nar/gkx1089. Jan 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanifa M.A., Skott M., Maltesen R.G., Rasmussen B.S., Nielsen S., Frøkiær J. Tissue, urine and blood metabolite signatures of chronic kidney disease in the 5/6 nephrectomy rat model. Metabolomics. 2019;15(8):112. doi: 10.1007/s11306-019-1569-3. [DOI] [PubMed] [Google Scholar]
- 31.Maltesen R.G., Wimmer R., Rasmussen B.S. A longitudinal serum NMR-based metabolomics dataset of ischemia-reperfusion injury in adult cardiac surgery. Scientific Data. 2020;7(1):198. doi: 10.1038/s41597-020-0545-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maltesen R.G., Hanifa M.A., Kucheryavskiy S., Pedersen S., Kristensen S.R., Rasmussen B.S. Predictive biomarkers and metabolic hallmark of postoperative hypoxaemia. Metabolomics. 2016;12(5):87. [Google Scholar]
- 33.Maltesen R. Aalborg Universitetsforlag. Ph.d.-serien for Det Teknisk-Naturvidenskabelige Fakultet, Aalborg Universitet; 2016. Postoperative Lung Injury- the path from Initiation to Clinical Diagnosis: a molecular view on a complex pathophysiological process. [Google Scholar]
- 34.Maltesen R.G., Rasmussen B.S., Pedersen S., Hanifa M.A., Kucheryavskiy S., Kristensen S.R. Metabotyping patients' journeys reveals early predisposition to lung injury after cardiac surgery. Sci Rep. 2017;7(1):40275. doi: 10.1038/srep40275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.RStudio Team . Vol. 1.4. Integrated Development Environment for R; 2021. p. 1106. (RStudio). [Google Scholar]
- 36.Rohart F., Gautier B., Singh A., Lê Cao K.A. mixOmics: an R package for 'omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13(11) doi: 10.1371/journal.pcbi.1005752. Nov 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karnovsky A., Weymouth T., Hull T., Tarcea V.G., Scardoni G., Laudanna C. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;28(3):373–380. doi: 10.1093/bioinformatics/btr661. Feb 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puchades-Carrasco L., Jantus-Lewintre E., Pérez-Rambla C., García-García F., Lucas R., Calabuig S. Serum metabolomic profiling facilitates the non-invasive identification of metabolic biomarkers associated with the onset and progression of non-small cell lung cancer. Oncotarget. 2016;7(11):12904–12916. doi: 10.18632/oncotarget.7354. Mar 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L., Zheng J., Ahmed R., Huang G., Reid J., Mandal R. A high-performing plasma metabolite panel for early-stage lung cancer detection. Cancers. 2020;12(3) doi: 10.3390/cancers12030622. Mar 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyamoto S., Taylor S.L., Barupal D.K., Taguchi A., Wohlgemuth G., Wikoff W.R. Systemic metabolomic changes in blood samples of lung cancer patients identified by gas chromatography time-of-flight mass spectrometry. Metabolites. 2015;5(2):192–210. doi: 10.3390/metabo5020192. Apr 09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohamed A., Deng X., Khuri F.R., Owonikoko T.K. Altered glutamine metabolism and therapeutic opportunities for lung cancer. Clin Lung Canc. 2014;15(1):7–15. doi: 10.1016/j.cllc.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dutta S., Ray S., Nagarajan K. Glutamic acid as anticancer agent: an overview. Saudi Pharmaceut J. 2013;21(4):337–343. doi: 10.1016/j.jsps.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kodama M., Oshikawa K., Shimizu H., Yoshioka S., Takahashi M., Izumi Y. A shift in glutamine nitrogen metabolism contributes to the malignant progression of cancer. Nat Commun. 2020;11(1):1320. doi: 10.1038/s41467-020-15136-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Louis E., Adriaensens P., Guedens W., Vanhove K., Vandeurzen K., Darquennes K. Metabolic phenotyping of human blood plasma: a powerful tool to discriminate between cancer types? Ann Oncol. 2016;27(1):178–184. doi: 10.1093/annonc/mdv499. Jan. [DOI] [PubMed] [Google Scholar]
- 45.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lieu E.L., Nguyen T., Rhyne S., Kim J. Amino acids in cancer. Exp Mol Med. 2020;52(1):15–30. doi: 10.1038/s12276-020-0375-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green C.R., Wallace M., Divakaruni A.S., Phillips S.A., Murphy A.N., Ciaraldi T.P. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nat Chem Biol. 2016;12(1):15–21. doi: 10.1038/nchembio.1961. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newman J.C., Verdin E. β-hydroxybutyrate: much more than a metabolite. Diabetes Res Clin Pract. 2014;106(2):173–181. doi: 10.1016/j.diabres.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng S., Wang H., Liu J., Aa J., Zhou F., Wang G. Multi-dimensional roles of ketone bodies in cancer biology: opportunities for cancer therapy. Pharmacol Res. 2019;150:104500. doi: 10.1016/j.phrs.2019.104500. Dec. [DOI] [PubMed] [Google Scholar]
- 50.Weber D.D., Aminzadeh-Gohari S., Tulipan J., Catalano L., Feichtinger R.G., Kofler B. Ketogenic diet in the treatment of cancer – where do we stand? Mol Metabol Canc Metabol. 2020;33:102–121. doi: 10.1016/j.molmet.2019.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poff A.M., Ari C., Arnold P., Seyfried T.N., D'Agostino D.P. Ketone supplementation decreases tumor cell viability and prolongs survival of mice with metastatic cancer. Int J Canc. 2014;135(7):1711–1720. doi: 10.1002/ijc.28809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng S., Wang H., Liu J., Aa J., Zhou F., Wang G. Multi-dimensional roles of ketone bodies in cancer biology: opportunities for cancer therapy. Pharmacol Res. 2019;150:104500. doi: 10.1016/j.phrs.2019.104500. [DOI] [PubMed] [Google Scholar]
- 53.Xue L., Chen H., Jiang J. Implications of glycerol metabolism for lipid production. Prog Lipid Res. 2017;68:12–25. doi: 10.1016/j.plipres.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 54.Muntoni S., Atzori L., Mereu R., Satta G., Macis M.D., Congia M. Serum lipoproteins and cancer. Nutr Metabol Cardiovasc Dis. 2009;19(3):218–225. doi: 10.1016/j.numecd.2008.06.002. Mar. [DOI] [PubMed] [Google Scholar]
- 55.Zhou T., Zhan J., Fang W., Zhao Y., Yang Y., Hou X. Serum low-density lipoprotein and low-density lipoprotein expression level at diagnosis are favorable prognostic factors in patients with small-cell lung cancer (SCLC) BMC Canc. 2017;17(1):269. doi: 10.1186/s12885-017-3239-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rocha C.M., Carrola J., Barros A.S., Gil A.M., Goodfellow B.J., Carreira I.M. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of blood plasma. J Proteome Res. 2011;10(9):4314–4324. doi: 10.1021/pr200550p. Sep. 2. [DOI] [PubMed] [Google Scholar]
- 57.Vitols S., Peterson C., Larsson O., Holm P., Aberg B. Elevated uptake of low density lipoproteins by human lung cancer tissue in vivo. Canc Res. 1992;52(22):6244–6247. Nov 15. [PubMed] [Google Scholar]
- 58.Merino Salvador M., Gómez de Cedrón M., Moreno Rubio J., Falagán Martínez S., Sánchez Martínez R., Casado E. Lipid metabolism and lung cancer. Crit Rev Oncol Hematol. 2017;112:31–40. doi: 10.1016/j.critrevonc.2017.02.001. Apr. [DOI] [PubMed] [Google Scholar]
- 59.Silva M., Galeone C., Sverzellati N., Marchianò A., Calareso G., Sestini S. Screening with low-dose computed tomography does not improve survival of small cell lung cancer. J Thorac Oncol. 2016;11(2):187–193. doi: 10.1016/j.jtho.2015.10.014. Feb. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available upon request.