Abstract
Detonation of an improvised nuclear device highlights the need to understand the risk of mixed radiation exposure as prompt radiation exposure could produce significant neutron and gamma exposures. Although the neutron component may be a relatively small percentage of the total absorbed dose, their large relative biological effectiveness (RBE) can induce larger biological DNA damage and cell killing. The objective of this study was to use the hematopoietically humanized mouse model to measure chromosome DNA damage in human lymphocytes in vivo exposed to neutrons (0.3 Gy) and X rays (1 Gy), 24 hours after exposure. The human dicentric (DIC) and cytokinesis-block micronucleus (CBMN) assays were performed to measure chromosomal aberrations in human lymphocytes in vivo from the blood and spleen, respectively. The mBAND assay based on fluorescent in-situ hybridization labeling was used to detect neutron-induced chromosome 1 inversions in the blood lymphocytes of the neutron-irradiated mice. Cytogenetics endpoints, MN and DIC show that there was no significant difference in yields between the two irradiation types at the doses tested, indicating that neutron-induced chromosomal DNA damage in vivo was more biologically effective (RBE ~ 3.3) compared to X rays. The mBAND assay, which is considered a specific biomarker of high-LET neutron exposure, confirmed the presence of clustered DNA damage in the neutron-irradiated mice but not in the X-irradiated mice, 24 hours after exposure.
Keywords: X rays, Neutrons, humanized mice, chromosomal aberrations
Introduction
Prompt radiation exposure from an improvised nuclear device (IND) detonation will involve a mixture of low linear energy transfer (LET) photons and high-LET neutrons in the Me-V range. Neutrons are highly energetic uncharged particles that generate low secondary protons which induce more severe DNA damage than photons whereby equivalent doses of neutrons and photons (γ or X rays) do not produce equal biologic effects, due to the differences in the patterns of energy deposited (Cary et al. 2012). The relative biological effect (RBE) value of photons and neutrons can be very different: the RBE of neutrons has been reported to be as low as 1 to greater than 10 depending on the tissue type, cell radiosensitivity, neutron energy and the biological endpoint measured (Ryan et al. 2006; Seth et al. 2014). Monte-Carlo simulations of neutron / photon transport after a 10kT urban groundburst in Washington DC, specifically in terms of organ doses at 700 m from the explosion suggest that neutrons will contribute an estimated 14% of the total organ dose in the colon and lung, and 27% in bone marrow (Kramer et al. 2016). Of course, neutron contributions numbers will vary depending on the specifics of the explosion and the shielding. Therefore, in a nuclear accident or detonation scenario, there is an important need to detect and estimate the size of the neutron component in order to assess radiation risk and injury of potentially exposed individuals to aid treatment decisions.
It is widely accepted that unrepaired or misrepaired DNA double-strand breaks (DSBs) play an important role in the formation of chromosome aberrations that can lead to genomic instability, mutations and cell death (lliakis et al. 2004; Vijg and Suh 2013). Neutrons are highly energetic uncharged particles that induce, via secondary protons, more localized, structurally complex clusters of DSBs than gamma rays. These clustered damages have a higher probability of becoming lethal because they are more likely to be misrepaired during the slow stage of rejoining, as they are more difficult and slower to repair, leading to increased interactions for chromosome exchange and the formation of complex chromosome aberrations (Hada and Georgakilas 2008; Kysela et al. 1993; Sachs and Brenner 1993). A series of mechanistic, in-vitro and epidemiological studies (Bauchinger and Schmid 1998; Brenner and Sachs 1994; Hada et al. 2011; Hande et al. 2003; Mitchell et al. 2004) have suggested that chromosomal inversions represent a high-specificity biomarker for the presence of densely ionizing radiation (IR) such as neutrons. Multicolor banding (mBAND) fluorescence in situ hybridization technique (Chudoba et al. 2004; Chudoba et al. 1999) has been used to label chromosome 1 and 2 inversions in peripheral blood lymphocytes after past exposure to high- LET radiation (Mitchell et al. 2004).
Cytogenetic analyses of dicentrics (DIC) and micronuclei (MN) in irradiated human T-lymphocytes are established biomarkers in biological dosimetry for retrospective dose reconstruction after exposure to IR (Kang et al. 2016; Testa et al. 2019; Zeegers et al. 2017). The advantage of these two cytogenetic biodosimetry assays is that they can be used to measure persistent DNA damage in vivo in mature peripheral blood lymphocytes weeks after radiation exposure (Ghandhi et al. 2018; Preston et al. 1974). Neutrons have been shown to be 2-6 times more effective than photons in inducing cytogenetic damage (Huber et al. 1994; Wuttke et al. 1998; Xu et al. 2015a) such that roughly half of the biological effect observed will be due to neutrons with the other half due to photons (Garty et al. 2017). Previously at the Columbia IND-Neutron Facility (CINF)(Xu et al. 2015a), we generated dose-response curves for MN frequency using the cytokinesis-block micronucleus assay (Fenech 1993, 2007), and showed that neutron-induced MN yields in human blood T-lymphocytes ex vivo up to 1.5 Gy, were enhanced 3- to 5-fold over equivalent x-ray doses. A similar ratio was also seen with dicentrics in ex-vivo irradiated blood (our unpublished data).
The objective of the present work was to quantify chromosomal DNA damage in human T-lymphocytes in vivo in hematopoietically humanized (Hu-NSG) mice (Lee et al. 2018; Pujol-Canadell et al. 2019) exposed to total body neutron (0.3 Gy) and X ray (1 Gy) irradiations. Cytogenetic assays, micronucleus (MN) and dicentric (DIC) were used to analyze chromosome aberrations in human T-lymphocytes in vivo, 24 hours after exposure. The results show here that the frequency of MN and DIC were similar after neutron (0.3 Gy) and x-ray (1 Gy) exposure, indicating that for these cytogenetic endpoints, neutrons were about three times more biologically effective compared to X rays. In addition, mBAND labeling of chromosome 1 was used to demonstrate the presence of inversions on chromosome 1 following exposure to neutrons.
Materials and Methods
Human peripheral blood samples were collected by venipuncture in 6 mL lithium-heparinized Vacutainer® tubes (BD Vacutainer™, Franklin Lakes, NJ) from apparently healthy adult donors (2 males/females) aged between 30 and 45 with informed consent and approval by the Columbia University Irving Medical Center Institutional Review Board (IRB protocol AAAE-2671). All donors were non-smokers and had no X rays within the last 12 months. All animal and experimental procedures were conducted in accordance with applicable federal and state guidelines and approved by the Columbia University Institutional Animal Care and Use Committee (IACUC; protocol AAAQ2410). As described in our earlier work (Lee et al. 2018; Pujol-Canadell et al. 2019), female immunodeficient NOD. Cg-Prkdcscid//2rgtm1WJl/SzJ (NSG) mice aged 6 to 8 weeks (The Jackson Laboratory; Bar Harbor, ME), were injected with commercially available human cord blood CD34+ cells (Cincinnati Children’s hospital Medical Center; Cincinnati, OH), and followed by long-term engraftment. At 16 weeks, the level of human cell engraftment in the mouse peripheral blood was assessed by flow cytometry for human cell surface markers CD45 (leukocyte common antigen), CD3 (T-cells) and CD19 (B-cells) and mouse-specific CD45 (all purchased from BioLegend® Inc., San Diego, CA). Only mice validated with > 25% human CD45+ cells were used for the radiation studies.
Irradiation and dosimetry
Healthy human volunteers were recruited at the Center for Radiological Research, Columbia University Irving Medical Center. Blood sample aliquots (1 mL) were prepared in 15 mL conical bottom tubes (Santa Cruz Biotechnology, Dallas, TX) and exposed up to 3 Gy X rays using a highly filtered X-RAD 320 biological irradiator (Precision X-Ray Inc., North Branford, CT), operating at 320 kVp, 12.5 mA with a dose rate of 0.88 Gy/min and Half Value layer of 4 mm Cu (Pujol-Canadell et al. 2019). The humanized mice were randomly assigned to dose groups, 0, 1, 2 and 3 Gy and X-irradiated using a custom irradiation pie (Precision X-Ray). The control mice were sham irradiated. The delivered dose was measured using a Radcal® ion chamber (Monrovia, CA) calibrated annually by RadCal and placed in the pie along with the mice. Following irradiation, the blood samples were placed at 37°C in a humidified atmosphere with 5% CO2 for 24 hours and the mice were housed in micro-isolator cages.
For the neutron/photon studies, mouse irradiations were performed at the Columbia University Radiological Research Accelerator Facility (RARAF) using the accelerator-based IND-spectrum neutron irradiator (Xu et al. 2015a; Xu et al. 2015b) and a filtered (HVL: 3.1 mm Cu) Westinghouse Coronado orthovoltage X-ray machine operating at 250 kVp, 15mA and the use of a 0.5mm Cu + 1mm Al filter. The dosimetry for the neutron irradiations was performed using a custom A-150 muscle tissue-equivalent gas ionization chamber (Xu et al. 2012; Xu et al. 2015a; Xu et al. 2015b). A compensated Geiger-Muller dosimeter was used to measure the gamma-ray dose associated with the neutron exposure (typically 18% of the total dose). With a total beam current of 18 μA on the beryllium target, the resulting dose rate was 1.55 Gy/h of neutrons and 0.34 Gy/h of γ rays. A detailed description of the mouse irradiation protocol for neutron exposures using custom holders in a “Ferris wheel” system setup has been described previously (Broustas et al. 2017). For the photon exposures, up to 3 mice at a time were placed, in the same custom holders, at the center of the irradiation field. The mice were exposed at a dose rate of 1.23 Gy/min, determined using a Victoreen model 570 condenser R meter with a 250r chamber. Hu-NSG mice were randomly assigned to three treatment groups: shamirradiated, 0.3 Gy neutron and 1 Gy of X-ray.
Sample collection and blood counts
All mice were euthanized 24 h after irradiation by CO2 asphyxiation prior to blood collection. Peripheral whole blood samples (0.4-0.6 ml) were collected by cardiac puncture using a heparinized 1 ml syringe (BD Precisionglide™; Becton-Dickson, Franklin Lakes, NJ) and transferred into 1.5 ml Eppendorf tube. Each spleen was isolated, free of large connecting ligaments and placed into a 1.5 ml Eppendorf tube containing 1 ml of sample buffer (SB) with 2% FBS and 0.01% lithium heparin in DPBS (ThermoFisher Scientific™, Waltham, MA). Mononuclear cells from the mouse blood and spleen were isolated as described previously (Turner et al. 2019). Briefly, each spleen was placed onto a 40-micron mesh cell strainer (Corning; #352340), and homogenized gently using the rubber end of a 1 ml syringe plunger and SB-heparin. The resulting cell suspension was filtered again using a second 40-micron mesh cell strainer and isolated using lymphocyte separation media (Histopaque-1083; Invitrogen) and RBC lysis. Human leukocyte T-/B-cell counts in blood and spleen were measured by flow cytometry from 20 μl of heparinized blood or splenocyte suspension using a CytoFLEX flow cytometer (Beckman Coulter Inc., Pasedena, CA) surface staining protocol (Lee et al. 2018; Pujol-Canadell et al. 2019). Antibodies for CD3 clone UCHT1 (marker for T-cells) and CD19 clone 2H7 (marker for B-cells) were purchased from Biolegend; San Diego, CA).
Cytokinesis-Block Micronucleus Assay
Isolated mouse blood and spleen mononuclear cells and aliquots (100 μL) human peripheral blood samples were added to 1.4 mL 2D Matrix microtubes containing pre-warmed RPMI-1640 medium supplemented with 15% heat-inactivated fetal calf serum, 2% Pen/Strep and 2% phytohaemagglutinin (PHA) (all purchased from Thermofisher). Human blood cells were cultured at 37°C in a humidified atmosphere with 5% CO2 for 44 hours, after which time the media was exchanged for fresh media containing cytochalasin B (Cyt-B; Sigma-Aldrich, St. Louis, MO) at a final concentration of 6 μg/ml. After 28 h of incubation, the samples were swollen with 0.075 M KCI solution and fixed in 4:1 methanol: glacial acetic acid. Cells were dropped onto clean glass slides and allowed to air-dry before counter-staining with DAPI Vectashield® mounting medium (#H-1200; Vector Laboratories, Inc., Burlingame, CA).
Dicentric Assay
Human lymphocytes were isolated from aliquots of whole blood by Histopaque 1083 separation and cultured for a period of 54 h in RPMI medium supplemented with 4% PHA, 10% heat inactivated fetal bovine serum and antibiotics followed by addition of Colcemid (0.1 μg/ml) for 24 h and the cells were harvested at 72 h (from the start of stimulation with PHA). Cells were treated with a hypotonic solution (0.56% KCI) for 15 min at 37°C and fixed in three changes of ice-cold acetic acid: methanol (1:3) solution. An aliquot of fixed cells (25-30 μl) was dropped onto glass sides and air-dried. For chromosomal analysis, slides were rehydrated in PBS, fixed in 4% formaldehyde, washed with PBS, digested with HCI-pepsin solution, washed with PBS and dehydrated in a 70%–85%–100% ethanol series. Chromosomes were denatured for 3 min by heat (80°C) in the presence of 20 μl of centromere and telomere probes (PNA Bio Inc, CA) in 90% formamide, 2 × saline sodium citrate buffer and incubated in the dark for 3 h. After hybridization, excess unbound probes were removed by washing twice in 70% formamide and TBS, with 0.05% Tween™-20 solutions at room temperature and counterstained with DAPI.
Multi-color band fluorescence in situ hybridization (mBAND)
Multicolor band (mBAND) analysis was conducted using chromosome 1 given that it is the largest chromosome in the human genome, representing ~8.2% of the genome (Morton 1991). The mBAND technique was performed essentially according to the manufacturer’s protocol (MetaSystems, MA) using DNA probes specific for human chromosomes 1. Briefly, slides were treated for 1 min with 0.001% acidic pepsin solution (in 0.01 N HCI) at 37°C for 1-2 min followed by two washes of 5 min each in phosphate buffered saline. The slides were post-fixed for 10 min in a solution of formaldehyde/MgCl2 (1% formaldehyde/50mM MgCl2 in PBS). The slides after denaturation (2X SSC at 70°C for 20 min) and after cooling to ambient temperature (1 min in 0.07N NaOH) were dehydrated in graded series of ethanol (30%, 70%, 90% and 100%) and air dried. The mBAND probe was denatured separately by incubation at 75°C for 5 min followed by incubation at 37°C for 30 min to allow the annealing of repetitive DNA sequences. An aliquot of 10 μl probe was placed on the slide and covered with a coverslip. The slides were kept in a humidified hybridization chamber at 37°C for at least 72 h. The unbound probe was removed by washing the slides in pre-warmed (75°C) 1X SSC (pH 7.0-7.5) for 5 min followed by incubation in 4XSSCT (4X SSC with 0.1% Tween 20) for 5 min. Indirectly labeled probe (Cy5), if needed, was amplified by incubation with antibodies biotinylated anti-streptavidin and Cy5 conjugated streptavidin (Invitrogen, Carlsbad, CA) sequentially for 30 min followed by two washes of 3 min each in 4XSSCT and in PBS. The nuclei were counterstained with DAPI.
Scoring and Analysis
Images were captured using Zeiss fluorescent microscope (Axioplan 2; Carl Zeiss Microimaging Inc., Thornwood, NY), CoolCube 1 Digital High Resolution CCD Camera and Metafer 4 Master Station (MetaSystems, MA). Micronuclei per binucleate cell yields were quantified using 10x air objective lens and using Metafer MSearch platform. Metaphase cell images were acquired using a Zeiss Plan-Apochromat 63x/1.40 oil immersion objective and AutoCapt version 3.9.1. Human chromosome spreads were easily differentiated from mouse ones, given the marked different in morphology of chromosomes from the two species, i.e. human spreads contain more diverse chromosomes, both in size and position of the centromere, compared to mouse spreads that contain similar sized acrocentric chromosomes. For the mBand assay, normal and aberrant chromosomes were identified by unique chromosome specific processed color generated by ISIS software based on the combinatorial labeling of five fluorochromes (FITC, Spectrum Orange, Texas Red, DEAC and Cy5) and their pixel intensities. Representative images that were used to score micronuclei, dicentrics and inversions are presented in Figure 1.
Fig. 1:
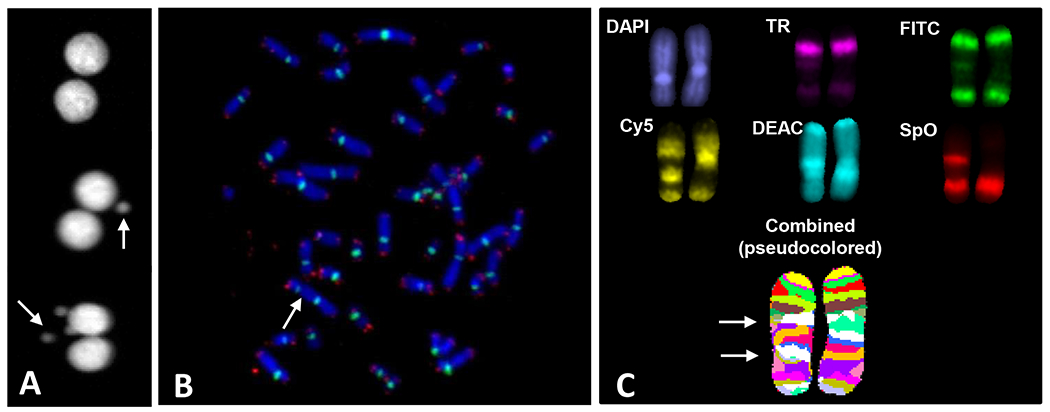
Representative images of micronuclei (panel A), PNA-FISH of centromeres (green) and telomeres (red) (panel B), and mBAND of chromosome 1 (panel C). Arrows point to micronuclei (panel A), a dicentric (panel B) and an inversion (panel C).
Statistics
The statistical analyses were performed using GraphPad Prism 7 (GraphPad software Inc., La Jolla, CA). The Kruskal-Wallis test was first used to compare the data among the study groups. Comparison between the control and irradiated groups was performed by the Mann-Whitney U test. Two-tailed p values less than 0.05 were considered statistically significant.
Results
Micronuclei frequency and radiation sensitivity in human lymphocytes
Previous studies by our group and that of others have identified differences in the sensitivity of human and mouse T-cells by specific mitogens and cytochalasin (Cyt-B) block (Erexson and Kligerman 1987; Kim et al. 1997; Turner et al. 2015). The optimal assay time for the mouse CBMN assay is ~ 50 h (0.5% PHA; 3 μg/ml Cyt-B) whereas the human CBMN assay is ~ 70-72 h (2% PHA; 6 μg/ml Cyt-B) (Fenech 1993, 2007). These humanized mouse studies, therefore used the human cytokinesis-block micronucleus assay (CBMN) to compare the intrinsic radiosensitivity of human lymphocytes in blood and spleen collected from humanized mice compared to peripheral blood samples collected from healthy human volunteers. Figure 2 shows the dose-response curves for micronuclei frequency (micronuclei per binucleated cell) in X-irradiated human lymphocytes in vivo from blood and spleen in humanized mice and human peripheral blood exposed ex vivo, at 24 h after radiation exposure. The data indicate that MN formation in engrafted human lymphocyte cells in vivo from blood and spleen is dose dependent and indicate a similar radiation sensitivity to cells exposed using the human blood ex vivo model over the dose range tested here (up to 3 Gy). At the highest TBI dose 3 Gy, quantification of MN yields in the peripheral blood showed comparatively reduced MN levels.
Fig. 2.
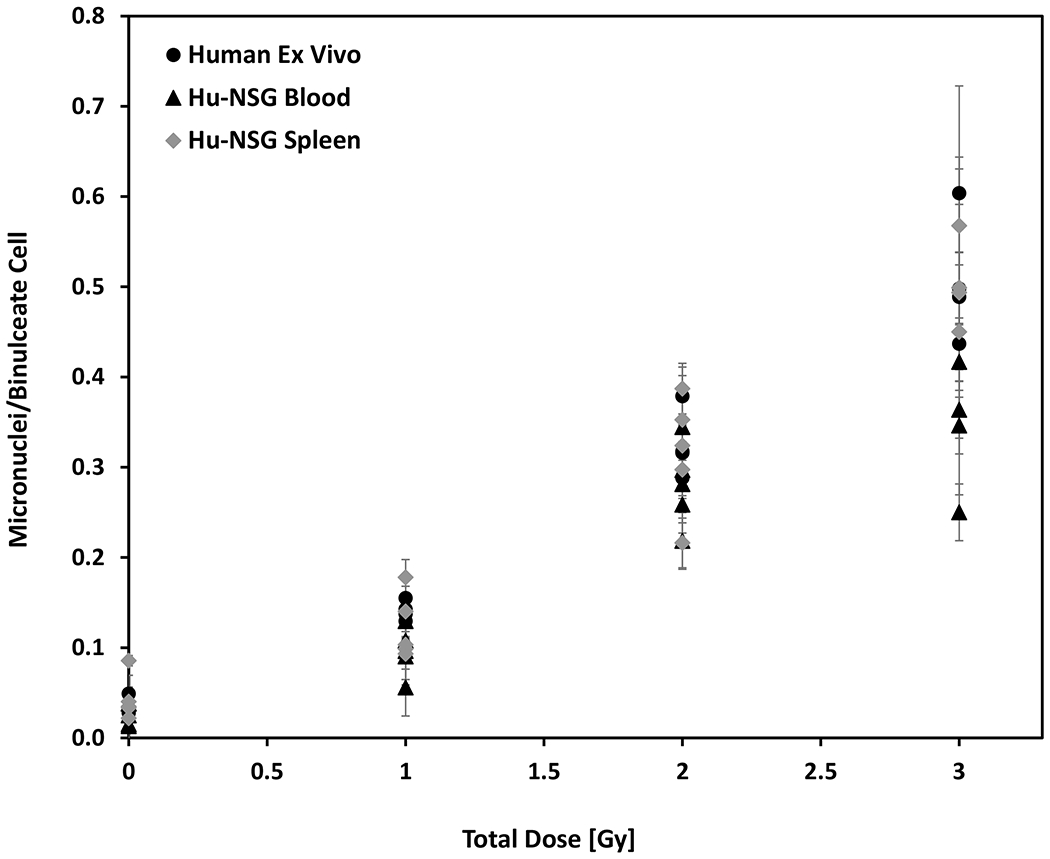
Micronuclei frequency in human lymphocytes in vivo from Hu-NSG mouse blood and spleen compared with human peripheral blood exposed ex vivo to X-rays, in the dose range 0 to 3 Gy. Error bars show mean ± SEM
X ray and neutron exposure in humanized mice
i). Hematology
Figure 3 compares the radiation-induced loss of human CD3+ (T-cells) and CD19+ (B-cells) in mouse blood and spleen samples, 24 hours after exposure. The results show that T- and B-cell levels in the peripheral blood were significantly lower (p<0.05; Mann-Whitney U-test) in the X-irradiated mice compared to the non-irradiated mice, although there was no significant difference between the two irradiated groups. In the spleen, overall there was no significant change in human lymphocytes in the irradiated mice compared to the control mice.
Fig. 3.
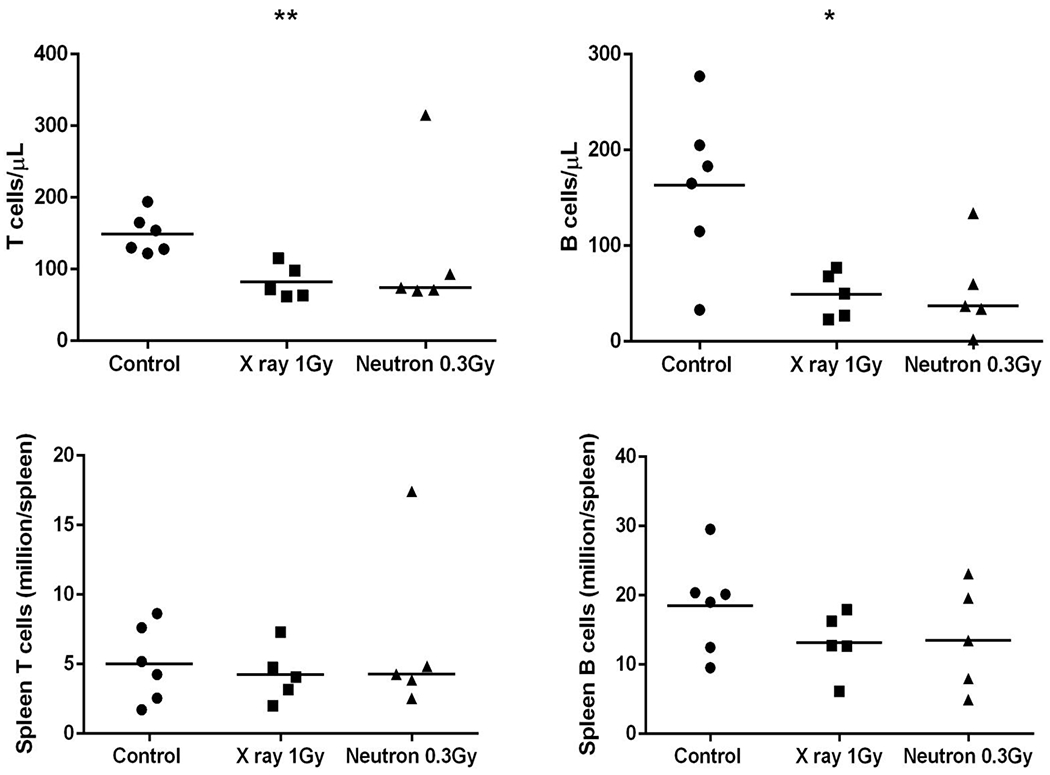
Human hematopoietic cell counts in the peripheral blood and spleen after exposure of humanized mice to X ray (1 Gy) and neutron (0.3 Gy) irradiations. The horizontal line indicates the median values. Comparison between the control and irradiated groups was performed by the Mann-Whitney U test (*p<0.05, ** p<0.01).
ii). Induction of Micronuclei
Figure 4 shows the micronucleus frequency per binucleate cell (MN/BNC) in human lymphocytes isolated from the mouse spleen using the CBMN assay. The frequency of induced MN/BNC was similar in human lymphocytes exposed to 1 Gy X ray and 0.3 Gy neutron irradiation. The Kruskal-Wallis test was first performed to compare the data among the three study groups (p<0.05). Comparison between the control and irradiated groups was performed by the Mann-Whitney U test (** p<0.01). The data show that there was no significant difference in MN/BN after 1 Gy X rays and 0.3 Gy of neutrons (shown in Fig. 4a). The results suggest an RBE of ~ 3.3 for the MN endpoint, and dose response with X rays and neutrons (shown in Fig. 4b).
Fig. 4.
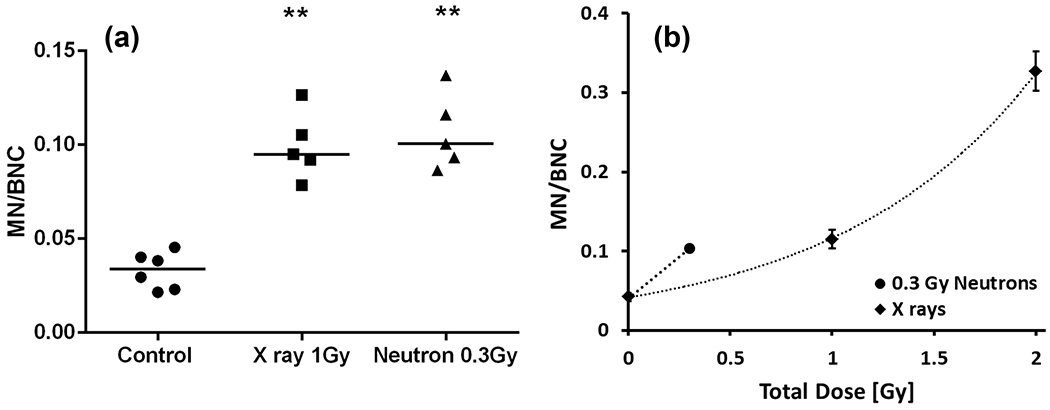
Micronuclei frequency in human lymphocytes isolated from the spleen, 24 h after exposure to total expected body X ray (1 Gy) and neutron (0.3 Gy) irradiations. (a) Dot blot analysis shows the micronucleus frequency per binucleate cell. The horizontal line indicates the median values. Comparison between the control and irradiated groups was performed by the Mann-Whitney U test (** p<0.01). (b) MN frequency fitted with an exponential curve after exposure to X rays. Error bars show mean ± SEM. Neutron-induced MN yields indicate an RBE of ~ 3.3.
iii). Induction of chromosomal aberrations
Peripheral blood samples from X- and neutron irradiated humanized mice were assayed for the induction of dicentrics (shown in Fig. 5). Exposure to 1 Gy X rays resulted in 0.13 dicentrics per metaphase cell; these frequencies were similar to yields in samples exposed to 0.3 Gy of neutrons. Samples from neutron-irradiated animals were also examined for chromosomal inversions that are thought to be a hallmark of neutron exposure. Inversions are produced by the symmetric rejoining of breaks along the length of a single chromosome and are not detected by routine cytogenetic assays. Using mBAND probes for human chromosome 1, inversions were observed at frequencies of 0.05 inversions per metaphase (shown in Fig. 5).
Fig. 5.
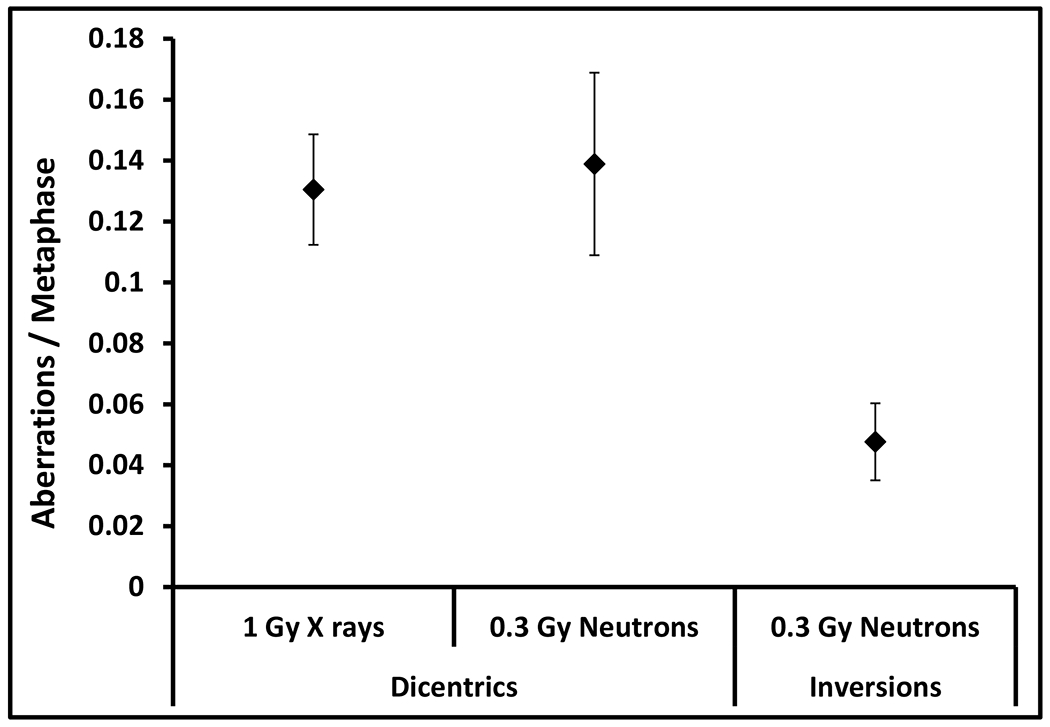
Frequencies of dicentrics (interchromosomal) and inversions (intrachromosomal) observed in peripheral blood of humanized mice following exposure to X rays (1 Gy) and neutrons (0.3 Gy). Error bars show mean ± SD
Discussion/Conclusion
The primary focus of the present work was to evaluate cytogenetic damage in human lymphocytes in vivo in hematopoietically humanized (Hu-NSG) mice, after separate exposure to neutrons and X rays. The Hu-NSG mouse model combines the many advantages of ex-vivo irradiated human blood and in-vivo mouse models to represent a potentially valuable resource for radiation biodosimetry studies (Lee et al. 2018; Pujol-Canadell et al. 2019; Wang et al. 2020). Here, we specifically utilized the presence of a large number of mature human T-cells present in the circulating blood and the spleen (Shultz et al. 2012), to evaluate DIC frequency and chromosome 1 inversions in the peripheral blood and MN yields in the spleen after neutron and x-ray exposure. The 24-hour time point was chosen to represent the earliest time point when blood samples are likely collected after a radiological incident such as an explosive improvised nuclear device.
To evaluate the intrinsic radiosensitivity of human lymphocytes in blood and spleen from humanized mice (LD50/30 of approximately 3-4 Gy)(Wang et al. 2013) compared to the human blood ex vivo model, the human CBMN assay was used to measure the dose response for MN frequency (up to 3 Gy), 24 hours after exposure. Figure 2 shows a good correspondence between the numbers of MN formed in human lymphocytes in vivo in the humanized mice and ex vivo from human donors over the dose range tested, suggesting similar radiation sensitivity. In the mouse peripheral blood samples, there was a notable reduction in MN frequency/low numbers of BN cells in the isolated lymphocytes at the 3 Gy dose, suggesting that a large number of the highly damaged cells may have died or redistributed to the spleen (Pecaut et al. 2001). It is also possible that the highly damaged cells did not survive the 3-day assay culture time. Based on these observations and our earlier work which demonstrated a neutron RBE of ~ 4 for MN frequency at 1 Gy using the human blood ex vivo model (Xu et al. 2015a), total body exposures of 1 Gy photons and 0.3 Gy neutrons were chosen for this work.
Evidence for increased sensitivity of the CD45+ lymphocytes in the mouse blood compared to the spleen is shown in the hematology data presented in Figure 3. There is a significant (p<0.05) reduction in T-/B-cells in the peripheral blood after neutron and x-ray exposures compared to the control (n = 5 mice, engrafted with 3 different stem cell donors) compared to the spleen where the blood counts were not significantly different across the three groups. Based on our own experience, one possible explanation here is that there is variability between the mice, particularly the size of the spleen after radiation exposure. Although, there was no significant change in T-/B-cell numbers in the mouse spleens, there was a significant (p< 0.01) induction of MN yields in the neutron and x-ray irradiated mice compared to the control mice (shown in Fig. 4a). The data show that the neutron RBE for the MN yields in human lymphocytes is about 3.3 compared to the levels induced by X rays (shown in Fig. 4b), indicating that for the MN endpoint, neutrons produce increased residual DNA damage that is unrepaired or misrepaired, 24 hours after exposure. Novel insights observed here using the humanized mouse model, could be used in the development of future studies to investigate mixed exposures with different percentages of neutrons and photons at longer times after exposure. More recently, we have used a high-throughput automated CBMN assay in human lymphocytes and developed a machine learning approach to reconstruct neutron and photon doses in complex exposure scenarios (Shuryak et al. 2020; Shuryak et al. 2021).
The results of the dicentric analysis were in keeping with that observed for MN, in that, yields were similar in samples from mice exposed to either 1 Gy X rays or 0.3 Gy neutrons. Based on track structure calculations, it has been predicted that there would be a higher incidence of double strand breaks (DSBs) in close proximity to each other following exposure to high LET radiation (e.g. neutrons and heavy ions) relative to sparsely IR (e.g. X rays) (Brenner and Sachs 1994; Burkart et al. 1999; Sachs et al. 1997a; Sachs et al. 1997b). It follows that the close proximity of breaks would result in increased frequencies of intra chromosomal exchanges particularly intra-arm exchanges. In fact, it has been proposed that intrachromosomal exchanges, particularly inversions, may constitute a cytogenetic fingerprint for densely IR. Interestingly, the persistence of these intrachromosomal exchanges (inter- and intra-arm) has been reported in the lymphocytes of workers several years after exposure to high doses of Plutonium (Hande et al. 2005; Hande et al. 2003; Mitchell et al. 2004). The detection of these inversions following exposure to neutron irradiation agrees with previous reports and further supports the suitability of the Hu-NSG model for biodosimetric studies. It should be stressed that while mBAND analysis is capable of detecting inversions and other gross chromosomal aberrations involving the painted chromosome, including large insertions and deletions, translocations, and dicentrics, smaller insertions and deletions and other aberrations in the metaphase not involving the painted chromosome may remain undetected.
Peripheral blood lymphocytes are sensitive to the lethal effects of IR that are largely mediated through the induction of DSBs and apoptotic cell death (Pugh et al. 2014). We have shown previously that radiation-induced apoptosis/cell death can contribute to the depletion of mouse peripheral blood leukocytes in vivo (Turner et al. 2015). DSB repair kinetics has been shown to be associated with different repair pathways that allow a biphasic fast (initial few hours) and slow component (hours to days) of repair after exposure to IR (Beels et al. 2010; Iliakis et al. 2004; Lee et al. 2019). Defects in DNA repair machinery can increase cell vulnerability to DNA-damaging agents and accumulation of mutations in the genome, leading to the development of various disorders including cancers. It has been demonstrated that DSBs induced by neutrons are repaired with different repair kinetics from DSBs induced by X-rays such that neutron-induced DNA clusters are repaired more slowly by the fast repair component leaving more breaks to be misrepaired in the slow component (Kysela et al. 1993; Vandersickel et al. 2014). Theoretical analysis and experimental evidence suggest an increased complexity and severity of complex DNA damage with increasing LET that has high mutagenic or carcinogenic potential (Hada and Georgakilas 2008). Preliminary measurements in the present study (not shown) identified increased levels of phospho-p53 protein in CD45+ human lymphocytes in the spleen after exposure to X rays compared to neutrons, normalized to the non-irradiated control. Further studies are required to evaluate the effect of these two different radiation types on specific biomarkers involved in the kinetics of DNA repair and cell killing.
In conclusion, we have extended our earlier work to show that neutrons are more biologically effective for the cytogenetic endpoints DIC and MN in human lymphocytes in vivo, compared to x-ray irradiation, highlighting the fact that complex chromosome aberrations are repaired with different repair kinetics compared to more simple DNA double strand breaks. We show here that the humanized mouse model supports the quantification of cytogenetic endpoints at an early time point after radiation exposure. In the future, it will be important to extend these measurements out to 7-14 days to confirm that the kinetics of repair are expected for human in vivo exposures. These studies highlight the importance of different types of interactions between different radiation qualities not only from the biodosimetry viewpoint, but also at the radiobiological and cellular level.
Acknowledgments
Acknowledgement
We wish to thank Ms. Maria Taveras for the recruitment and collection of blood samples from the healthy blood donor volunteers and Ms. Mashkura Chowdhury for help with the preparation of the humanized mice.
Funding Sources
This work was supported by the Center for High-Throughput Minimally-Invasive Radiation Biodosimetry, National Institute of Allergy and Infectious Diseases grant number U19AI067773. The content of the manuscript is solely the responsibility of the authors. The funding body was not involved in the design of the study, sample collection, analysis and interpretation of the data or the writing of this manuscript.
Footnotes
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Statement of Ethics
The Columbia University Irving Medical Center Institutional Review Board (IRB; protocol number AAAE-2671) approved the recruitment and consent of healthy human volunteers and procedures for the collection of peripheral blood samples. All animal studies and experimental procedures were approved by the Columbia University Institutional Animal Care and Use Committee (IACUC; protocol number AAAQ2410) and were conducted in facilities accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
References
- Bauchinger M, Schmid E: LET dependence of yield ratios of radiation-induced intra- and interchromosomal aberrations in human lymphocytes. International journal of radiation biology 74:17–25 (1998). [DOI] [PubMed] [Google Scholar]
- Beels L, Werbrouck J, Thierens H: Dose response and repair kinetics of gamma-H2AX foci induced by in vitro irradiation of whole blood and T-lymphocytes with X- and gamma-radiation. International journal of radiation biology 86:760–768 (2010). [DOI] [PubMed] [Google Scholar]
- Brenner DJ, Sachs RK: Chromosomal “fingerprints” of prior exposure to densely ionizing radiation. Radiation research 140:134–142 (1994). [PubMed] [Google Scholar]
- Broustas CG, Xu Y, Harken AD, Garty G, Amundson SA: Comparison of gene expression response to neutron and x-ray irradiation using mouse blood. BMC genomics 18:2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart W, Jung T, Frasch G: Damage pattern as a function of radiation quality and other factors. Cr Acad Sci Iii-Vie 322:89–101 (1999). [DOI] [PubMed] [Google Scholar]
- Cary LH, Ngudiankama BF, Salber RE, Ledney GD, Whitnall MH: Efficacy of radiation countermeasures depends on radiation quality. Radiation research 177:663–675 (2012). [DOI] [PubMed] [Google Scholar]
- Chudoba I, Hickmann G, Friedrich T, Jauch A, Kozlowski P, Senger G: mBAND: a high resolution multicolor banding technique for the detection of complex intrachromosomal aberrations. Cytogenetic and genome research 104:390–393 (2004). [DOI] [PubMed] [Google Scholar]
- Chudoba I, Plesch A, Lorch T, Lemke J, Claussen U, Senger G: High resolution multicolor-banding: a new technique for refined FISH analysis of human chromosomes. Cytogenetics and cell genetics 84:156–160 (1999). [DOI] [PubMed] [Google Scholar]
- Erexson GL, Kligerman AD: A modified mouse peripheral blood lymphocyte culture system for cytogenetic analysis. Environmental and molecular mutagenesis 10:377–386 (1987). [DOI] [PubMed] [Google Scholar]
- Fenech M: The cytokinesis-block micronucleus technique: a detailed description of the method and its application to genotoxicity studies in human populations. Mutation research 285:35–44 (1993). [DOI] [PubMed] [Google Scholar]
- Fenech M: Cytokinesis-block micronucleus cytome assay. Nature protocols 2:1084–1104 (2007). [DOI] [PubMed] [Google Scholar]
- Garty G, Xu Y, Elliston C, Marino SA, Randers-Pehrson G, Brenner DJ: Mice and the A-Bomb: Irradiation Systems for Realistic Exposure Scenarios. Radiation research 187:465–475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandhi SA, Turner HC, Shuryak I, Dugan GO, Bourland JD, Olson JD, Tooze JA, Morton SR, Batinic-Haberle I, Cline JM, Amundson SA: Whole thorax irradiation of non-human primates induces persistent nuclear damage and gene expression changes in peripheral blood cells. PloS one 13:e0191402 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hada M, Georgakilas AG: Formation of clustered DNA damage after high-LET irradiation: a review. Journal of radiation research 49:203–210 (2008). [DOI] [PubMed] [Google Scholar]
- Hada M, Zhang Y, Feiveson A, Cucinotta FA, Wu H: Association of inter- and intrachromosomal exchanges with the distribution of low-and high-LET radiation-induced breaks in chromosomes. Radiation research 176:25–37 (2011). [DOI] [PubMed] [Google Scholar]
- Hande MP, Azizova TV, Burak LE, Khokhryakov VF, Geard CR, Brenner DJ: Complex chromosome aberrations persist in individuals many years after occupational exposure to densely ionizing radiation: An mFISH study. Gene Chromosome Canc 44:1–9 (2005). [DOI] [PubMed] [Google Scholar]
- Hande MP, Azizova TV, Geard CR, Burak LE, Mitchell CR, Khokhryakov VF, Vasilenko EK, Brenner DJ: Past exposure to densely ionizing radiation leaves a unique permanent signature in the genome. American journal of human genetics 72:1162–1170 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R, Schraube H, Nahrstedt U, Braselmann H, Bauchinger M: Dose-response relationships of micronuclei in human lymphocytes induced by fission neutrons and by low LET radiations. Mutation research 306:135–141 (1994). [DOI] [PubMed] [Google Scholar]
- Iliakis G, Wang H, Perrault AR, Boecker W, Rosidi B, Windhofer F, Wu W, Guan J, Terzoudi G, Pantelias G: Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenetic and genome research 104:14–20 (2004). [DOI] [PubMed] [Google Scholar]
- Kang CM, Yun HJ, Kim H, Kim CS: Strong Correlation among Three Biodosimetry Techniques Following Exposures to Ionizing Radiation. Genome integrity 7:11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Han DU, Lim JT, Jo SK, Kim TH: Induction of micronuclei in human, goat, rabbit peripheral blood lymphocytes and mouse splenic lymphocytes irradiated in vitro with gamma radiation. Mutation research 393:207–214 (1997). [DOI] [PubMed] [Google Scholar]
- Kramer K, Li A, Madrigal J, Sanchez B, Millage K: Monte Carlo Modeling of the Prompt Radiation Emitted by a Nuclear Device in the National Capital Region, Revision 1. Technical Report (2016). [Google Scholar]
- Kysela BP, Arrand JE, Michael BD: Relative contributions of levels of initial damage and repair of double-strand breaks to the ionizing radiation-sensitive phenotype of the Chinese hamster cell mutant, XR- V15B. Part II. Neutrons. International journal of radiation biology 64:531–538 (1993). [DOI] [PubMed] [Google Scholar]
- Lee Y, Pujol Canadell M, Shuryak I, Perrier JR, Taveras M, Patel P, Koller A, Smilenov LB, Brenner DJ, Chen EI, Turner HC: Candidate protein markers for radiation biodosimetry in the hematopoietically humanized mouse model. Scientific reports 8:13557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Wang Q, Shuryak I, Brenner DJ, Turner HC: Development of a high-throughput gamma-H2AX assay based on imaging flow cytometry. Radiation oncology 14:150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell CR, Azizova TV, Hande MP, Burak LE, Tsakok JM, Khokhryakov VF, Geard CR, Brenner DJ: Stable intrachromosomal biomarkers of past exposure to densely ionizing radiation in several chromosomes of exposed individuals. Radiation research 162:257–263 (2004). [DOI] [PubMed] [Google Scholar]
- Morton NE: Parameters of the Human Genome. Proceedings of the National Academy of Sciences of the United States of America 88:7474–7476 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecaut MJ, Nelson GA, Gridley DS: Dose and dose rate effects of whole-body gamma-irradiation: I. Lymphocytes and lymphoid organs. In vivo 15:195–208 (2001). [PubMed] [Google Scholar]
- Preston RJ, Brewen JG, Gengozian N: Persistence of radiation-induced chromosome aberrations in marmoset and man. Radiation research 60:516–524 (1974). [PubMed] [Google Scholar]
- Pugh JL, Sukhina AS, Seed TM, Manley NR, Sempowski GD, van den Brink MR, Smithey MJ, Nikolich-Zugich J: Histone deacetylation critically determines T cell subset radiosensitivity. Journal of immunology 193:1451–1458 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol-Canadell M, Young E, Smilenov L: Use of a Humanized Mouse Model System in the Validation of Human Radiation Biodosimetry Standards. Radiation research 191:439–446 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan LA, Wilkins RC, McFarlane NM, Sung MM, McNamee JP, Boreham DR: Relative biological effectiveness of 280 keV neutrons for apoptosis in human lymphocytes. Health physics 91:68–75 (2006). [DOI] [PubMed] [Google Scholar]
- Sachs RK, Brenner DJ: Effect of LET on chromosomal aberration yields. I. Do long-lived, exchange-prone double strand breaks play a role? International journal of radiation biology 64:677–688 (1993). [DOI] [PubMed] [Google Scholar]
- Sachs RK, Brenner DJ, Chen AM, Hahnfeldt P, Hlatky LR: Intra-arm and interarm chromosome intrachanges: tools for probing the geometry and dynamics of chromatin. Radiation research 148:330–340 (1997a). [PubMed] [Google Scholar]
- Sachs RK, Chen AM, Brenner DJ: Review: proximity effects in the production of chromosome aberrations by ionizing radiation. International journal of radiation biology 71:1–19 (1997b). [DOI] [PubMed] [Google Scholar]
- Seth I, Schwartz JL, Stewart RD, Emery R, Joiner MC, Tucker JD: Neutron exposures in human cells: bystander effect and relative biological effectiveness. PloS one 9:e98947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Brehm MA, Garcia-Martinez JV, Greiner DL: Humanized mice for immune system investigation: progress, promise and challenges. Nature reviews Immunology 12:786–798 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuryak I, Turner HC, Perrier JR, Cunha L, Canadell MP, Durrani MH, Harken A, Bertucci A, Taveras M, Garty G, Brenner DJ: A High Throughput Approach to Reconstruct Partial-Body and Neutron Radiation Exposures on an Individual Basis. Scientific reports 10:2899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuryak I, Turner HC, Pujol-Canadell M, Perrier JR, Garty G, Brenner DJ: Machine learning methodology for high throughput personalized neutron dose reconstruction in mixed neutron + photon exposures. Scientific reports 11:4022 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa A, Palma V, Patrono C: Dicentric Chromosome Assay (DCA) and Cytokinesis-Block Micronucleus (CBMN) Assay in the Field of Biological Dosimetry. Methods in molecular biology 2031:105–119 (2019). [DOI] [PubMed] [Google Scholar]
- Turner HC, Lee Y, Weber W, Melo D, Kowell A, Ghandhi SA, Amundson SA, Brenner DJ, Shuryak I: Effect of dose and dose rate on temporal gamma-H2AX kinetics in mouse blood and spleen mononuclear cells in vivo following Cesium-137 administration. BMC molecular and cell biology 20:13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner HC, Shuryak I, Taveras M, Bertucci A, Perrier JR, Chen C, Elliston CD, Johnson GW, Smilenov LB, Amundson SA, Brenner DJ: Effect of Dose Rate on Residual gamma-H2AX Levels and Frequency of Micronuclei in X-Irradiated Mouse Lymphocytes. Radiation research 183:315–324 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandersickel V, Beukes P, Van Bockstaele B, Depuydt J, Vral A, Slabbert J: Induction and disappearance of gammaH2AX foci and formation of micronuclei after exposure of human lymphocytes to (6)(0)Co gamma-rays and p(66)+ Be(40) neutrons. International journal of radiation biology 90:149–158 (2014). [DOI] [PubMed] [Google Scholar]
- Vijg J, Suh Y: Genome instability and aging. Annual review of physiology 75:645–668 (2013). [DOI] [PubMed] [Google Scholar]
- Wang C, Nakamura S, Oshima M, Mochizuki-Kashio M, Nakajima-Takagi Y, Osawa M, Kusunoki Y, Kyoizumi S, Imai K, Nakachi K, Iwama A: Compromised hematopoiesis and increased DNA damage following non-lethal ionizing radiation of a human hematopoietic system reconstituted in immunodeficient mice. International journal of radiation biology 89:132–137 (2013). [DOI] [PubMed] [Google Scholar]
- Wang Q, Lee Y, Shuryak I, Pujol Canadell M, Taveras M, Perrier JR, Bacon BA, Rodrigues MA, Kowalski R, Capaccio C, Brenner DJ, Turner HC: Development of the FAST-DOSE assay system for high-throughput biodosimetry and radiation triage. Scientific reports 10:12716 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuttke K, Muller WU, Streffer C: The sensitivity of the in vitro cytokinesis-blocked micronucleus assay in lymphocytes for different and combined radiation qualities. Strahlentherapie und Onkologie : Organ der Deutschen Rontgengesellschaft [et al. ] 174:262–268 (1998). [DOI] [PubMed] [Google Scholar]
- Xu Y, Garty G, Marino SA, Massey TN, Randers-Pehrson G, Johnson GW, Brenner DJ: Novel neutron sources at the Radiological Research Accelerator Facility. Journal of Instrumentation 7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Randers-Pehrson G, Turner HC, Marino SA, Geard CR, Brenner DJ, Garty G: Accelerator-Based Biological Irradiation Facility Simulating Neutron Exposure from an Improvised Nuclear Device. Radiation research 184:404–410 (2015a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YP, Randers-Pehrson G, Marino SA, Garty G, Harken A, Brenner DJ: Broad energy range neutron spectroscopy using a liquid scintillator and a proportional counter: Application to a neutron spectrum similar to that from an improvised nuclear device. Nucl Instrum Meth A 794:234–239 (2015b). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeegers D, Venkatesan S, Koh SW, Low GK, Srivastava P, Sundaram N, Sethu S, Banerjee B, Jayapal M, Belyakov O, Baskar R, Balajee AS, Hande MP: Biomarkers of Ionizing Radiation Exposure: A Multiparametric Approach. Genome integrity 8:6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


