Abstract
Phosphatidylserine (PS) is distributed asymmetrically in the plasma membrane of eukaryotic cells. Phosphatidylserine flippase (P4-ATPase) transports PS from the outer leaflet of the lipid bilayer to the inner leaflet of the membrane to maintain PS asymmetry. The β subunit TMEM30A is indispensable for transport and proper function of P4-ATPase. Previous studies have shown that the ATP11A and TMEM30A complex is the molecular switch for myotube formation. However, the role of Tmem30a in skeletal muscle regeneration remains elusive. In the current study, Tmem30a was highly expressed in the tibialis anterior (TA) muscles of dystrophin-null (mdx) mice and BaCl2-induced muscle injury model mice. We generated a satellite cell (SC)-specific Tmem30a conditional knockout (cKO) mouse model to investigate the role of Tmem30a in skeletal muscle regeneration. The regenerative ability of cKO mice was evaluated by analyzing the number and diameter of regenerated SCs after the TA muscles were injured by BaCl2-injection. Compared to the control mice, the cKO mice showed decreased Pax7+ and MYH3+ SCs, indicating diminished SC proliferation, and decreased expression of muscular regulatory factors (MYOD and MYOG), suggesting impaired myoblast proliferation in skeletal muscle regeneration. Taken together, these results demonstrate the essential role of Tmem30a in skeletal muscle regeneration.
Keywords: Tmem30a, Skeletal muscle regeneration, Knockout mouse model, Atp11a, Satellite cell
INTRODUCTION
Skeletal muscle is widely distributed in human tissue and plays a significant role in regulating basic life activities, such as respiration, metabolism, body temperature maintenance, and exercise. The strong regenerative capacity of skeletal muscle is important for maintaining skeletal muscle tissue homeostasis and ensuring normal physiological function under disease and trauma conditions. Regeneration of skeletal muscle occurs mainly through satellite cells (SCs), which are localized between the muscle and basement membranes during the resting state (Feige & Rudnicki, 2018; Sambasivan & Tajbakhsh, 2007). Resting SCs have a large nucleoplasmic ratio but very few mitochondria, and express marker genes such as Pax7, M-cadherin, syndecan 4, CD34, integrin-A7, CXCR4, and Myf5 (Alfaro et al., 2011; Kuang et al., 2006; Seale et al., 2000), but not MYOD (Cornelison & Wold, 1997; Zammit et al., 2004). In response to muscle injury or other pathological conditions, SCs are activated to proliferate and differentiate, and then repair damaged muscle tissue by fusing to existing myofibers or forming new ones. After activation, the SC nucleoplasmic ratio decreases, but pre-activation marker genes are still expressed. In particular, Myf5 expression is greatly increased in activated skeletal muscle stem cells, resulting in MYOD expression (Crist et al., 2012; Gayraud-Morel et al., 2012). SCs are regulated by a series of myogenic regulatory factors, such as MYOD, myogenin (MYOG), and Myf5, during differentiation. MYOD is a decisive master regulator of muscle lineage, and the expression of exogenous MYOD in non-muscle cells can lead to their transdifferentiation into muscle cells (Bichsel et al., 2013). MYOD and Myf5 can activate the expression of key downstream transcription factors, such as MYOG, and promote SC differentiation (Füchtbauer & Westphal, 1992; Londhe & Davie, 2011; Tapscott et al., 1988). These downstream transcription factors further activate the expression of muscle-specific genes, such as myosin, and promote the terminal differentiation of muscle cells. Among the terminally differentiated muscle cells, myoblasts fuse with each other to form myotube cells. These cells then form myofibers via orderly arrangement and finally muscle tissues, which play critical roles in many functions in vivo (Odelberg et al., 2000).
In mammalian plasma membranes, lipids are distributed asymmetrically, mainly in the form of phosphatidylethanolamine (PE) and phosphatidylserine (PS) in the cytoplasmic leaflet, and sphingomyelin (SM) and phosphatidylcholine (PC) in the exoplasmic leaflet (Bretscher, 1972). The asymmetric distribution of PS is largely dependent on P4-ATPase, which can transport PS from the outer leaflet of the lipid bilayer to the inner leaflet in the membrane (Holthuis & Levine, 2005). Mounting evidence suggests that P4-ATPase plays important roles in various disease conditions. For instance, mutations in ATP8B1 can cause progressive familial intrahepatic cholestasis type I and benign recurrent intrahepatic cholestasis (Bull et al., 1998). Mutations in ATP8A2 can lead to cerebellar ataxia, mental retardation, and disequilibrium (Tadini-Buoninsegni et al., 2019). Furthermore, ATP11A deficiency may affect myotube formation and cell surface PS maintenance (Tsuchiya et al., 2018), while ATP11C deficiency can cause congenital hemolytic anemia (Arashiki et al., 2016).
Similar to the sodium-potassium pump, P4-ATPase is also heterodimeric and requires the β-subunit for physiological function. The highly conserved transmembrane CDC50 protein family interacts with P4-ATPase and plays a crucial role in maintaining normal physiological function as well as proper cellular localization of P4-ATPase (Bryde et al., 2010; Saito et al., 2004; van der Velden et al., 2010). For proper cellular localization, P4-ATPase exportation from the endoplasmic reticulum requires the CDC50 subunit, which also plays an important role in the establishment of cell polarity. The CDC50 protein family contains three homologous proteins CDC50A, CDC50B, and CDC50C (also named TMEM30A, TMEM30B, and TMEM30C, respectively). TMEM30A is the main β-subunit that interacts with P4-ATPase (Sebastian et al., 2012). Indeed, increasing data support the key roles of TMEM30A in multiple tissues. For example, TMEM30A-deficient cells can cause severe damage in the formation of membrane ruffles, leading to cell migration inhibition (Kato et al., 2013; Paulusma et al., 2008). In addition, TMEM30A KO cells contain PS on their surface and are engulfed by macrophages (Segawa et al., 2014). Tmem30a deficiency can also lead to intrahepatic cholestasis due to the mislocalization of the PS transporter protein (Liu et al., 2017). Moreover, Tmem30a is essential for the function of retinal bipolar cells and photoreceptor cells in the retina (Yang et al., 2019; Zhang et al., 2017). Tmem30a deficiency inhibits vascular endothelial growth factor (VEGF)-induced signaling, leading to reduced endothelial cell proliferation and impaired retinal vascular development (Zhang et al., 2019). Recent research also reports that Tmem30a plays a role in insulin processing and secretion (Yang et al., 2021), and loss of Tmem30a in podocytes can lead to albuminuria and glomerulosclerosis in murine KO models (Liu et al., 2021). Neuronal-specific KO of Tmem30a results in exposure of PS on the neuronal outer membranes and loss of inhibitory post-synapses and seizures (Park et al., 2021).
Regeneration of skeletal muscle involves massive proliferation and migration of SCs and fusion of myoblasts, which requires the synthesis and fusion of many cell membranes. Abnormalities in cell membrane function are likely to affect skeletal muscle regeneration. Recent study found that the phospholipid flippase complex of ATP11A and TMEM30A regulates myoblast fusion and syncytium elongation by controlling the Ca2+ channel PIEZO1, a key regulator of myotube formation in C2C12 cells (Tsuchiya et al., 2018). As the ATP11A and TMEM30A complex is closely associated with myotube formation, we wondered whether this complex also plays a crucial role in the process of skeletal muscle regeneration. Previous study found that ATP11A-mediated PIEZO1 activation plays an important role in proper morphogenesis during myofiber regeneration (Tsuchiya et al., 2018). However, the in vivo functions of Tmem30a in skeletal muscle regeneration remain elusive.
In this study, we generated a novel SC-specific Tmem30a KO mouse model using Pax7CreER (Murphy et al., 2011) and investigated the roles of Temem30a in skeletal muscle regeneration. Our data showed that specific deletion of Temem30a in mouse SCs reduced the expression of key regenerative marker proteins, such as Pax7, MYH3, MYOD, and MYOG, which impaired the proliferation and differentiation of SCs. Taken together, our study showed that Tmem30a is indispensable for skeletal muscle regeneration.
MATERIALS AND METHODS
Animal model and genotyping
All animal experiment protocols (approval No.: LS2019-044) were approved by the Animal Care and Use Committee of Sichuan Provincial People’s Hospital, China. All experimental operations were performed in accordance with the approved study protocols and relevant regulations. All mice were housed in a specific pathogen-free (SPF) barrier facility with a standard diet and environment.
Floxed Tmem30a mice with a loxP site inserted to flank exon 3 of the Tmem30a gene have been described previously (Zhang et al., 2017). To generate a SC-specific KO mouse model, we mated Pax7CreER mice (Jackson Laboratory, https://www.jax.org/strain/017763) to Tmem30aloxP/loxP mice to yield heterozygous progeny (Tmem30aloxP/+; Pax7CreER). The Tmem30aloxP/+; Pax7CreER mice were mated to Tmem30aloxP/loxP mice to generate Tmem30aloxP/loxP; Pax7CreER mice (designated cKO mice). The Tmem30aloxP/loxP and Tmem30aloxP/+; Pax7CreER mice were designated as wild-type (WT) mice. To monitor the efficiency of Cre enzyme expression in SCs, the tdTomato reporter (strain name: B6.Cg-Gt (ROSA) 26Sortm14 (CAG-tdTomato) Hze/J, http://jaxmice.jax.Org/strain/007914.html) was used.
Mice were genotyped by polymerase chain reaction (PCR). Genomic DNA templates were obtained from mouse tails. Mice were screened using the primers: Tmem30a-loxP-F, 5'-ATTCCCCTTCAAGATAGCTAC-3', Tmem30a-loxP-R, 5'-AATGATCAACTGTAATTCCCC-3'; Cre-F, 5'- GAACGCACTGATTTCGACCA-3', Cre-R, 5'- GCTAACCAGCGTTTTCGTTC-3'; Rosa-tdt-wt-F, 5'-CACTTGCTCTCCCAAAGTCG-3', Rosa-tdt-wt-R, 5'-TAGTCTAACTCGCGACACTG-3'; and Rosa-tdt-R-KI, 5'-GTTATGTAACGCGGAACTCC-3'.
Tamoxifen injection and skeletal muscle injury
Tamoxifen salt (Sigma Aldrich, USA) was dissolved in anhydrous ethanol at a concentration of 10 mg/mL as a stock solution. On the day of injection, the stock solution was diluted in corn oil (Sigma Aldrich, USA) at a ratio of 1:9 as an injection solution. Mice received an intraperitoneal injection with a daily injection solution dosage of 15 µL/g body weight for five consecutive days.
We dissolved BaCl2 powder (Sigma Aldrich, USA, 10361-37-2) in normal saline to create a BaCl2 solution with a concentration of 1.5%. Skeletal muscle was injured by injection of 50 µl of 1.5% BaCl2 solution in the right tibialis anterior (TA) muscles.
Quantitative real-time PCR (qRT-PCR)
Extraction of RNA from the TA muscles was performed using TRIzol reagent (Life Technologies, USA, catalog # 15596-026). cDNA was synthesized using a reverse transcription kit (Ambion, USA, catalog # 18090010). In addition, qRT-PCR was performed with a SYBR Green kit (Bio-Rad Laboratories, USA; catalog # KK4607), and the values were normalized to the GAPDH transcript as an internal control. The following primers were used for qRT-PCR: Tmem30a-F, 5'-CAAACAGCAACGGCTACCC-3', Tmem30a-R, 5'-GTTGTTGGAGGTGACGAAGAT-3V; GAPDH-F, 5'-TGTGTCCGTCGTGGATCTGA-3', and GAPDH-R, 5'-TTGCTCTTGAAGTCGCAGGAG-3'.
Immunoblotting
The TA muscles were lysed in RIPA lysis buffer with protease inhibitor cocktail and ethylenediaminetetraacetic acid (EDTA) (TransGen, China). Protein concentration was determined using a BCA Protein Assay (TransGen, China). Equal amounts of protein were loaded onto a 10% polyacrylamide gel, then separated and transferred to nitrocellulose membranes (Merck Millipore, Germany). Blots were blocked with 8% non-fat dry milk in Tris-buffered saline and Tween 20 (TBST) for 2 h at room temperature and then incubated with primary antibodies in blocking solution overnight at 4 °C. The primary antibodies included TMEM30A (rabbit polyclonal, ab217330, Abcam, USA) and GAPDH (mouse monoclonal, 60004-1-Ig, Proteintech, USA). Primary antibodies were detected with either an anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibodies (Bio-Rad Laboratories, USA), and signals were developed using a SuperSignal West Pico Chemiluminescent Substrate (Pierce Protein Biology, USA). ImageJ (v1.8.0) was used to calculate the relative density of the protein.
Immunohistochemistry
For immunohistochemical analysis, mice were first anesthetized with ketamine (16 mg/kg body weight) and perfused transcardially with phosphate-buffered saline (PBS), followed by 4% paraformaldehyde (PFA) in PBS. The right hind limbs of the mice were removed from the skin and gastrocnemius muscle and immersed in 4% PFA overnight. The tibia and excess muscles were then removed, and the TA muscles were fixed for 3 h in 4% PFA, then dehydrated with 30% sucrose for 24 h. The TA muscles were then embedded in optimal cutting temperature solution (OCT), frozen, and sectioned at a thickness of 10 µm. The sections were blocked and permeabilized with 10% normal bovine serum and 0.2% 30%Triton X-100 in PBS for 2 h, then incubated overnight at 4 °C with primary antibodies. Subsequently, the sections were washed with PBS and stained with the Alexa-Fluor-488 or Alexa-Fluor-594-labeled goat anti-mouse or rabbit Ig secondary antibodies (diluted 1:500). For section immunofluorescence, the following antibodies were used: TMEM30A (rabbit polyclonal, ab217330, Abcam, USA), Pax7 (rabbit polyclonal, AF7584, Affinity, USA), Laminin (rabbit polyclonal, L9393, Sigma, USA), MYH3 (rabbit polyclonal, 22287-1-AP, Proteintech, USA), MYOD (rabbit polyclonal, 18943-1-AP, Proteintech, USA), and MYOG (rabbit polyclonal, bs-3550R, Bioss, USA).
Histological staining and cell quantification
For hematoxylin-eosin (H&E) and Sirius Red staining, mice were anesthetized with ketamine (16 mg/kg body weight) and perfused transcardially with PBS followed by 4% PFA. The isolated TA muscles were fixed in 4% PFA overnight at 4 °C, then embedded in paraffin, cut into 10 μm sections, and stained using H&E and Sirius Red staining protocols. The H&E and Sirius Red stained sections were viewed and imaged using a microscope and used to count and measure the number and diameter of regenerating SCs in injured skeletal muscle.
Statistical analysis
Statistical analysis was performed using Student’s t-test. Quantitative data are expressed as mean±standard error of the mean (SEM), as shown in the figure legends. Differences were considered statistically significant at P<0.05. Statistical analyses were performed using GraphPad Prism 6 software.
RESULTS
Tmem30a was highly expressed in regenerating skeletal muscle
Previous studies have suggested that both Duchenne muscular dystrophy (DMD) patients and dystrophin-null (mdx) mice exhibit sustained activation of SCs and muscle regeneration (Bulfield et al., 1984; Sacco et al., 2010). Therefore, we examined the expression of Tmem30a in the skeletal muscles of mdx mice. Results showed that Tmem30a was highly expressed (Figure 1A, C, D), thus suggesting the potential role of Tmem30a in skeletal muscle regeneration. To verify whether Tmem30a is highly expressed during skeletal muscle regeneration, the TA muscles of WT mice were injured with an injection of BaCl2. At 5 days post-injury (dpi), the TA muscles were harvested and subjected to qRT-PCR, immunoblotting, and immunohistochemical analysis. The mRNA expression of Tmem30a showed a 6-fold increase in the injured muscle (Figure 1E). The TMEM30A protein levels were also higher in 5 dpi mice (Figure 1B, F). The immunohistochemical results verified that the TMEM30A protein was up-regulated in 5 dpi mice (Figure 1G). Therefore, Tmem30a may play a role in skeletal muscle regeneration.
Figure 1.
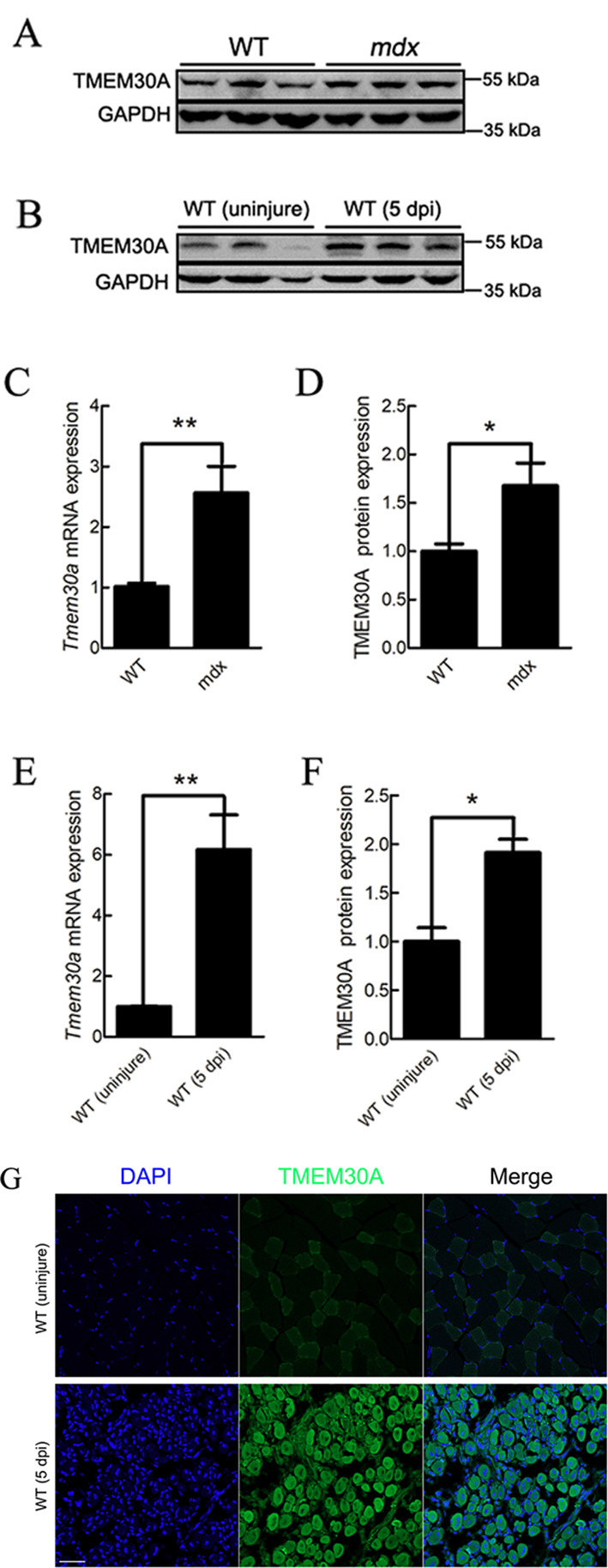
Tmem30a was up-regulated during skeletal muscle regeneration
A, D: Immunoblot analysis of TMEM30A protein expression in 3-month-old wild-type (WT) and mdx mice. Relative protein density was calculated by ImageJ. B, F: Immunoblot analysis of TMEM30A protein expression in uninjured WT mice and in WT mice at 5 days post-injury (dpi). Relative protein density was calculated by ImageJ. C: mRNA expression analysis of Tmem30a in 3-month-old WT and mdx mice using qRT-PCR. E: mRNA expression analysis of Tmem30a in uninjured WT mice and in WT mice at 3 dpi using qRT-PCR. Data are mean±SEM. n=3 in each group. Significance was calculated using two-tailed Student’s t-test. *: P<0.05;**: P<0.01. G: Immunofluorescence analysis of TMEM30A expression in TA muscles of uninjured WT mice and of WT mice at 5 dpi. TMEM30A immunostaining is shown in green, and nuclei are shown in blue (counterstained with DAPI). Scale bar: 50 μm.
Generation of inducible SC-specific Tmem30a-deleted mice
Previous studies have demonstrated that Tmem30a global KO results in embryonic lethality (Zhang et al., 2017). To investigate the role of Tmem30a in skeletal muscle regeneration, we crossed Pax7CreER mice expressing Cre recombinase from the Pax7 locus (Murphy et al., 2011) with mice bearing floxed Tmem30a alleles to generate Tmem30aloxP/loxP; Pax7CreER (cKO) mice (Figure 2A). PCR was used to identify the genotypes of the offspring (Figure 2B).
Figure 2.
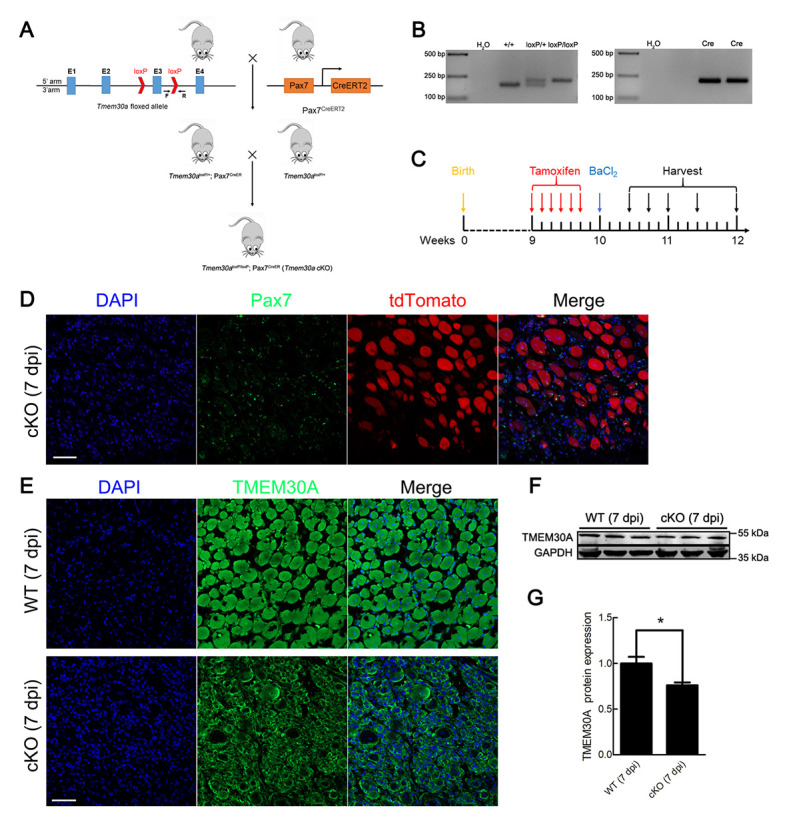
Generation of Tmem30a conditional knockout (cKO) mice
A: Breeding strategy of cKO mice by crossing Tmem30aloxP/loxP with Pax7CreER mice. B: Genotyping of mice by PCR and agarose gel electrophoresis. C: Axis of tamoxifen intraperitoneal injection, BaCl2 injection, and TA muscle harvesting in mice. D: In 7 dpi cKO mice, expression of Pax7CreER was monitored by ROSA-tdTomato reporter (red). SCs were labeled with Pax7 antibody (green). Nuclei were counterstained with DAPI (blue). Scale bar: 50 μm. E: TMEM30A protein from 7 dpi WT and cKO mice was labeled with TMEM30A antibody (green) and DAPI (blue). Scale bar: 50 μm. F, G: Immunoblot analysis of TMEM30A protein expression in 7 dpi WT and cKO mice. Relative protein band density was calculated by ImageJ. Data are mean±SEM. n=3 in each group. Significance was calculated by two-tailed Student’s t-test. *: P<0.05.
Cre reporter line ROSA26-tdTomato was used to verify the specific expression of Pax7CreER in SCs. We crossed ROSA26-tdTomato reporter mice with cKO mice to generate Tmem30aloxP/loxP; Pax7CreER; Rosa-tdTomato mice (Supplementary Figure S1). Cre enzyme expression was a prerequisite for the expression of tdTomato. A loxP-flanked STOP sequence before the tdTomato expression cassette was deleted in the Cre-expressing cells, and tdTomato was subsequently expressed. Next, 7 dpi TA muscle sections expressing tdTomato were stained with a specific antibody against Pax7. Results showed that tdTomato was expressed distinctly in Pax7-positive SCs, suggesting that Pax7CreER drives deletion of Tmem30a in SCs specifically (Figure 2D). Immunofluorescence staining of TMEM30A in 7 dpi mice revealed that TMEM30A was barely expressed in the SCs of the cKO mice (Figure 2E). Immunoblot analysis of proteins extracted from the TA muscles at the injury site further confirmed that TMEM30A expression in the cKO mice was reduced to 76% that of the controls (Figure 2F, G). This KO efficiency is meaningful, as Tmem30a in the cKO mouse model was only deleted in the SCs and Tmem30a is expressed in other non-SCs, which account for a large proportion of the TA muscles.
Loss of Tmem30a led to impaired skeletal muscle regeneration
We first investigated whether deletion of Tmem30a in SCs affects muscle development. Deletion of Tmem30a was induced with an intraperitoneal injection of tamoxifen on postnatal days 1 to 3 (P1–P3). No visible gross abnormalities were observed and no visible differences in body weight were detected between the WT and cKO mice from P30 to P60 (Supplementary Figure S2).
To investigate the role of Tmem30a in skeletal muscle regeneration, cKO and WT mice were induced with an intraperitoneal injection of tamoxifen at 9 weeks of age, followed by BaCl2-induced damage (Figure 2C). The TA muscles were subjected to histological analysis at 3, 5, 7, 10, and 14 dpi. Based on H&E staining, the histological appearance of skeletal muscle was compared between the WT and cKO mice. Uninjured skeletal muscle cells exhibited a well-organized pattern with dark-blue stained nuclei mainly located at the cell periphery (Figure 3A). At 3 dpi, inflammatory cells and necrotic myofibers were prominently observed, as well as a small number of proliferating SCs in WT mice (Figure 3A). At 5 dpi, myofibers with centralized nuclei proliferated from SCs replaced most of the necrotic myofibers in WT mice. In contrast, few proliferating myofibers were observed in the Tmem30a cKO mice (Figure 3A, B). At 7–10 dpi, newly formed myofibers with larger diameters were observed in WT mice, whereas these myofibers were barely visible in the cKO mice (Figure 3A, C). At 14 dpi, the WT mice displayed tightly packed, well-formed myofibers, whereas the cKO mice exhibited numerous small myofibers with single nuclei (Figure 3A).
Figure 3.
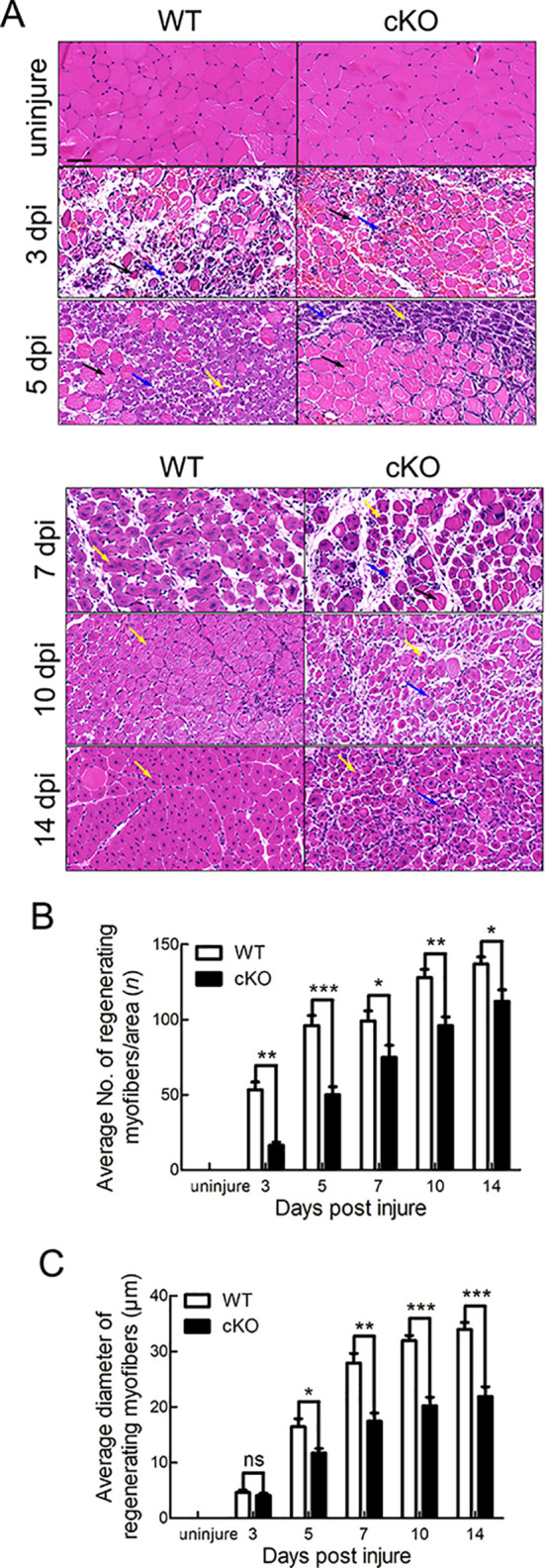
Myoblast regeneration was impaired in Tmem30a cKO mice after injury
A: H&E staining of TA muscles from uninjured WT and cKO mice and WT and cKO mice at 3, 5, 7, 10, and 14 dpi. Representative H&E-stained images of TA muscles from WT and cKO mice are shown. Blue arrows represent inflammatory cells; black arrows represent necrotic myofibers; yellow arrows represent proliferating SCs. Scale bar: 50 μm. B: Average number of regenerating myofibers per field in uninjured mice and mice at 3, 5, 7, 10, and 14 dpi. Reduced number of regenerative myofibers was observed in cKO mice. C: Average diameter of regenerating myofibers in uninjured mice and mice at 3, 5, 7, 10, and 14 dpi. Reduced diameter of regenerative myofibers was observed in cKO mice. Data are mean±SEM. n=3 in each group. Significance was calculated by two-tailed Student’s t-test. *: P<0.05;**: P<0.01;***: P<0.001; ns: Not significant.
Connective tissue fibrosis can reflect the status of skeletal muscle regeneration (Murphy et al., 2011). We histologically analyzed Sirius Red-stained TA muscle sections. We obtained similar results as H&E staining regarding the number and diameter of regenerating myofibers (Supplementary Figure S3A). The number of regenerating myofibers in the cKO mice was significantly less than that in the WT mice at 5–14 dpi (Supplementary Figure S3A, B). Similarly, in the cKO mice, more small-diameter regenerating myofibers were observed at 7–14 dpi (Supplementary Figure S3A, C). In summary, pathological section-staining revealed that loss ofTmem30a in SCs impaired skeletal muscle regeneration.
Tmem30a deficiency impaired SC proliferation
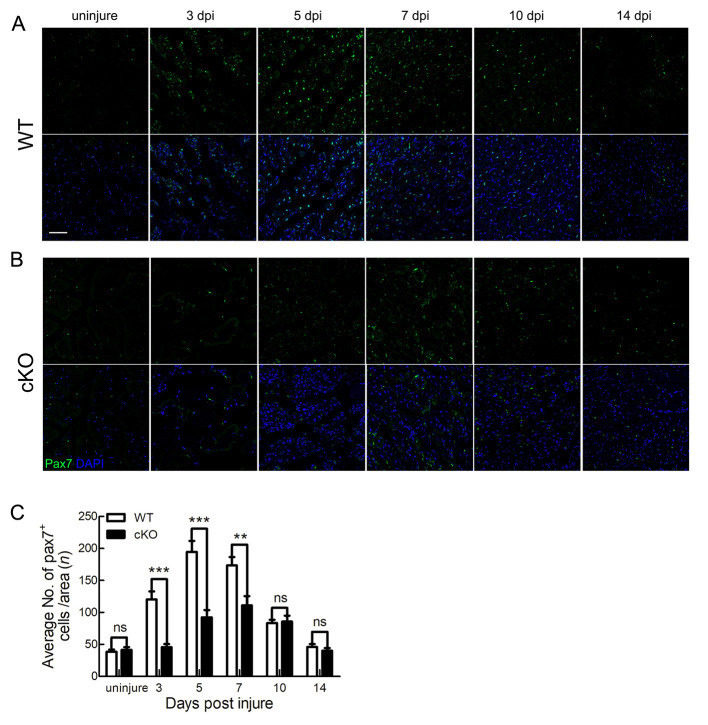
To further investigate the mechanism by which deletion of Tmem30a delays skeletal muscle regeneration, we examined Pax7, a transcription factor involved in the regulation of SC proliferation (Diao et al., 2012). During skeletal muscle regeneration, SCs first proliferate rapidly and Pax7 expression increases. Most SCs begin to differentiate into myoblasts and myofibers (which down-regulate Pax7), while the remaining Pax7+ SCs located under the basement membrane of the myofibers return to quiescence (Seale et al., 2000). Here, we performed immunohistochemical analysis of TA muscles at 3, 5, 7, 10, and 14 dpi. No differences in Pax7+ SCs were noted between the WT and cKO mice when SCs were at the resting state (Figure 4A–C). In the WT mice, Pax7+ SCs were mainly concentrated in the regenerating myofiber region and proliferated rapidly during the early stage of injury, with a peak level at 5 dpi (Figure 4A). Subsequently, Pax7 expression gradually decreased. By 14 dpi, the number of quiescent Pax7+ SCs returned to uninjured levels (Figure 4A). In comparison to the WT mice, the cKO mice showed a significant decrease in the number of Pax7+ SCs at 3, 5, 7, 10, and 14 dpi (Figure 4B). Quantification of Pax7+ SCs revealed a significant reduction in Pax7 expression in cKO mice at 3–5 dpi, a critical period of SC proliferation (Figure 4C).
Figure 4.
Pax7 expression was decreased in Tmem30a cKO mice
A, B: Immunohistochemical results of TA muscles in uninjured WT and cKO mice and in WT and cKO mice at 3, 5, 7, 10, and 14 dpi. Pax7 immunostaining is shown in green, and nuclei are shown in blue (counterstained with DAPI). Scale bar: 50 μm. C: Average number of Pax7+ cells per field in uninjured mice and in mice at 3, 5, 7, 10, and 14 dpi. Data are mean±SEM. n=3 in each group. Significance was calculated by two-tailed Student’s t-test. **: P<0.01,***: P<0.001; ns: Not significant.
We next explored the expression of MYH3, an embryonic myosin heavy chain expressed in embryonic and regenerating myofibers only (Schiaffino et al., 2015). In the WT mice, regenerating myofibers began to express MYH3 at 3 dpi, followed by a gradual increase in expression until 14 dpi, after which the mature regenerated myofibers no longer expressed MYH3 (Figure 5A). In contrast, the cKO mice exhibited delayed and lower expression of MYH3 (Figure 5B, D). The number of regenerating myofibers was also investigated. In the cKO mice, few large-diameter myofibers with centralized nuclei were observed and those myofibers were morphologically disorganized (Figure 5C). Furthermore, fewer myofibers with centralized nuclei were observed in the cKO mice than in the WT mice at the different periods after injury (Figure 5E). The diameters of myofibers with centralized nuclei were smaller in the cKO mice than in the WT mice (Figure 5F). These data suggest that deletion of Tmem30a inhibits Pax7 and MYH3 expression in activated SCs, thus affecting the SC proliferation process.
Figure 5.
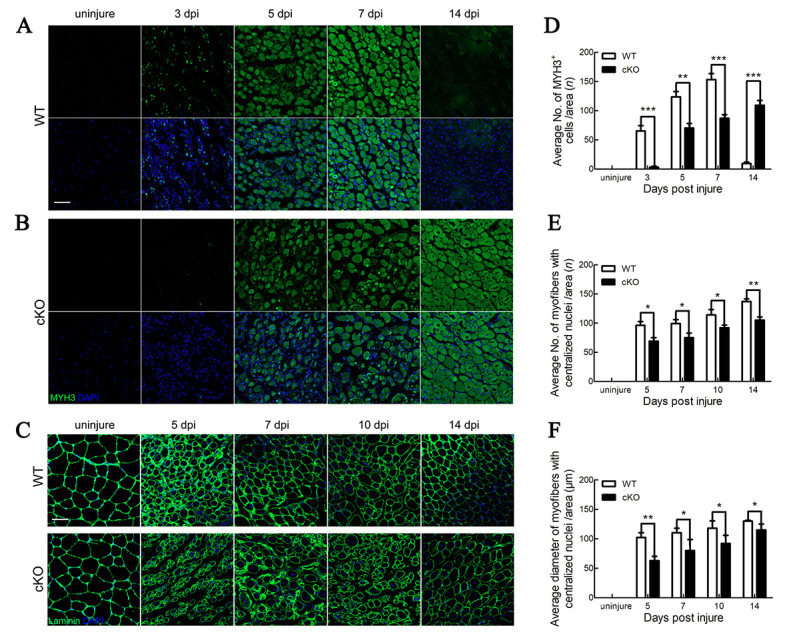
Deletion of Tmem30a impaired SC proliferation
A, B: Expression of MYH3 in uninjured WT and cKO mice and in WT and cKO mice at 3, 5, 7, and 14 dpi by immunohistochemical staining. MYH3 was labeled with MYH3 antibody (green). Nuclei were counterstained with DAPI (blue). Scale bar: 50 μm. C: Immunohistochemical staining analysis of size and number of regenerating myofibers in TA muscles of uninjured WT and cKO mice and of WT and cKO mice at 5, 7, 10, and 14 dpi. Laminin immunostaining is shown in green, and nuclei are shown in blue (counterstained with DAPI). Scale bar: 50 μm. D: Average number of MYH3+ cells per field in uninjured mice and in mice at 3, 5, 7, and 14 dpi. E: Average number of regenerating myofibers per field in uninjured mice and in mice at 5, 7, 10, and 14 dpi. F: Average diameter of regenerating myofibers in uninjured mice and in mice at 5, 7, 10, and 14 dpi. Data are mean±SEM. n=3 in each group. Significance was calculated by two-tailed Student’s t-test. *: P<0.05;**: P<0.01;***: P<0.001.
Tmem30a was essential for myoblast differentiation
As previously noted, ATP11A plays an essential role during myofiber fusion (Tsuchiya et al., 2018). Here, we further explored the function of Tmem30a in the differentiation of myoblasts. The expression of MYOD, a master regulator of muscular regulatory factors (MRFs), was evaluated using immunofluorescent staining. Results showed that fewer MYOD+ cells were observed in the cKO mice (Figure 6A), with a ~70% reduction in number (Figure 6C). We also examined the expression of MYOG, which can promote differentiation of SCs. Fewer MYOG+ cells were detected in the cKO mice than in the WT mice at 7 dpi (Figure 6B, D). These results indicate that deletion of Tmem30a in SCs impairs MRF expression. To investigate the effects of reduced MRF expression on myoblast differentiation, we histologically analyzed the TA muscles at 14 dpi. Both H&E and Sirius Red staining revealed abnormally fused myofibers in the cKO mice (Figure 6E). These results suggest that Tmem30a deficiency decreases MRF expression, which ultimately impairs myoblast differentiation.
Figure 6.
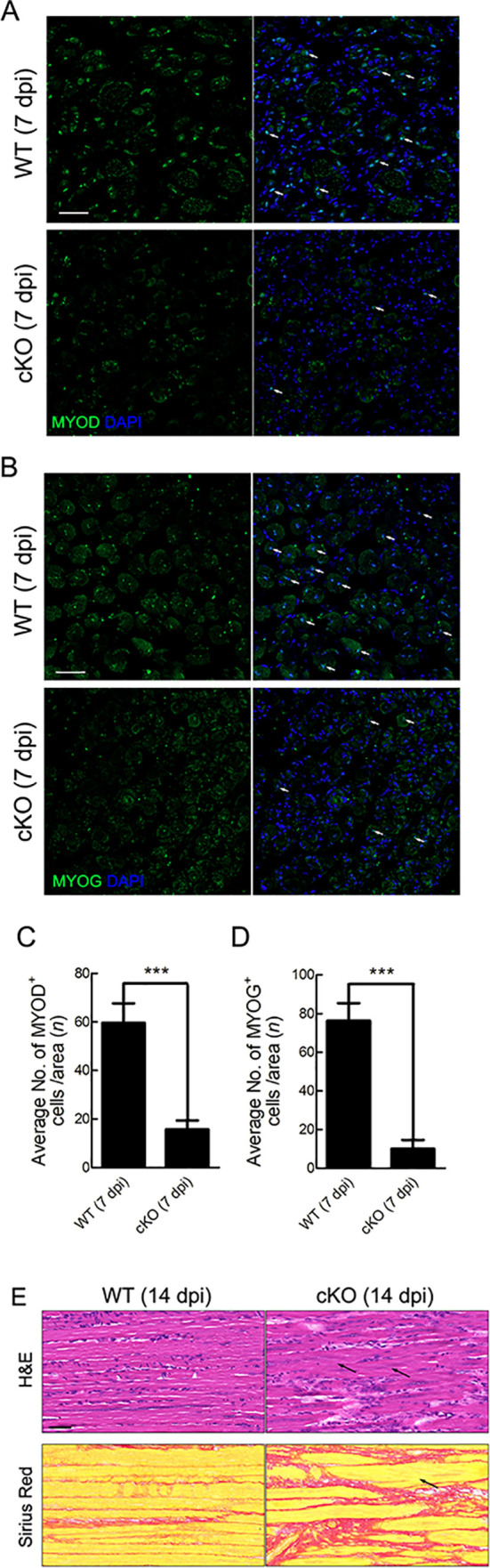
Specific deletion of Tmem30a in SCs impaired differentiation of myoblasts
A: Immunofluorescence analysis of MYOD expression in TA muscles of WT and cKO mice at 7 dpi. MYOD immunostaining is shown in green, and nuclei are shown in blue (counterstained with DAPI). Arrows depict cells positive for MYOD. Scale bar: 50 μm. B: Average number of MYOD+ cells per field at 7 dpi. C: Immunofluorescence analysis of MYOG expression in TA muscles of WT and cKO mice at 7 dpi. MYOG immunostaining is shown in green, and nuclei are shown in blue (counterstained with DAPI). Arrows depict cells positive for MYOG. Scale bar: 50 μm. D: Average number of MYOG+ cells per field at 7 dpi. Data are mean±SEM. n=3 in each group. Significance was calculated by two-tailed Student’s t-test. ***: P<0.001. E: H&E and Sirius Red staining of TA muscles from WT and cKO mice at 14 dpi. Arrows depict abnormal myofiber fusion.n=3 in each group. Scale bar: 50 μm.
DISCUSSION
In this study, we investigated the role of Tmem30a in skeletal muscle regeneration by generating SC-specific Tmem30a KO mice. The important function of Tmem30a in skeletal muscle regeneration was suggested by the up-regulation of Tmem30a expression in the mdx and 5 dpi mice (Figure 1). Furthermore, loss of Tmem30a impacted skeletal muscle regeneration by reducing activated SCs and inhibiting proper myoblast fusion.
Studies have indicated that SCs proliferate mainly by asymmetric division after skeletal muscle injury (Gurevich et al., 2016; Kuang et al., 2007; Shinin et al., 2006; Webster et al., 2016). Asymmetric division of SCs is primarily maintained by intracellular polarity (Dumont et al., 2015). Asymmetric distribution of PS on the plasma membrane mediated by flippase is essential for cell polarity (Chen et al., 2006; Das et al., 2012). Thus, in Tmem30a cKO mice, the absence of a functional flippase complex due to Tmem30a deficiency may result in the disruption of the asymmetric distribution of PS on the plasma membrane, which may disturb cell polarity. Ultimately, the asymmetric division of SCs may be affected, leading to inhibition of proliferation. Thus, the relationship between PS distribution and SC regeneration warrants further investigation. As shown in Figures 3B, 5E, the number of regenerating myofibers was reduced in the cKO mice and SC proliferation was impaired.
Cell migration is inhibited by severe defects in the formation of membrane ruffles in TMEM30A-deficient cells (Kato et al., 2013; Paulusma et al., 2008). After muscle injury, most activated SCs migrate to damaged myofibers, where they further differentiate and fuse together (Tidball, 2011). Reduced migration of SCs impairs skeletal muscle regeneration (Li et al., 2021). Here, loss of Tmem30a decreased the number of regenerating myofibers that could fuse with damaged myofibers, possibly by reducing SC migration, which resulted in smaller diameter regenerating myofibers in the cKO mice (Figures 3, 5F).
Calcium signaling is a critical element in the development, maintenance, and regeneration of skeletal muscle (Tu et al., 2016) and perturbing Ca2+ release from internal stores impairs the regenerative process (Tu et al., 2016). Tmem30a-mutant cells exhibit abnormally high intracellular calcium levels (Cao et al., 2019), indicating disturbance of calcium efflux. Therefore, Tmem30a regulation of skeletal muscle regeneration may be related to intracellular Ca2+ concentrations. Recent research suggests that Atp11a and Tmem30a regulate myotube fusion through control of the calcium channel (Tsuchiya et al., 2018). Moreover, abolishing Ca2+ transients in regenerating muscle cells decreases the number of activated SCs in regenerating skeletal muscle (Tu & Borodinsky, 2014). Calcium and calcineurin can also regulate skeletal muscle differentiation by activating the MYOD transcription factor (Friday et al., 2003). Therefore, in the current study, the intracellular Ca2+ concentration in the cKO mice may be abnormal, resulting in the observed decrease in MYOD expression (Figure 6A, C). MYOG can be activated by MYOD and serves as a master regulator of MRFs to promote myoblast differentiation. This explains why MYOG expression was markedly lower than that of MYOD in the cKO mice (Figure 6B, D).
In the current study, we discovered a novel role of Tmem30a in SC regeneration. Notably, Tmem30a deficiency in SCs inhibited their activation and proliferation and myoblast differentiation was also impaired by reduced MYOD and MYOG expression. Collectively, our results indicate that Tmem30a is an important factor in skeletal muscle regeneration.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHOR CONTRIBUTIONS
X.J.Z conceived the research. K.X.S., X.L., Y.J.S, X.Y.J., J.L.W., L.Z., and Y.M.Y. conducted the experiments. X.J.Z. and K.X.S. wrote the manuscript. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the members of the Animal Facility of the Institute of Laboratory Animal Research of the Sichuan Academy of Medical Sciences and Sichuan Provincial People’s Hospital for excellent animal welfare and husbandry. The authors would like to thank Chengdu LiLai Biotechnology Co., Ltd. for technical assistance with histological analysis.
Funding Statement
This study was supported by the National Natural Science Foundation of China (81770950, 81970841), Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (2019-12M-5-032), and Department of Science and Technology of Sichuan Province (21ZDYF4279, 2020JDZH0026, 2021JDZH0022)
References
- 1.Alfaro LAS, Dick SA, Siegel AL, Anonuevo AS, McNagny KM, Megeney LA, et al CD34 promotes satellite cell motility and entry into proliferation to facilitate efficient skeletal muscle regeneration. Stem Cells. 2011;29(12):2030–2041. doi: 10.1002/stem.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arashiki N, Takakuwa Y, Mohandas N, Hale J, Yoshida K, Ogura H, et al ATP11C is a major flippase in human erythrocytes and its defect causes congenital hemolytic anemia. Haematologica. 2016;101(5):559–565. doi: 10.3324/haematol.2016.142273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bichsel C, Neeld D, Hamazaki T, Chang LJ, Yang LJ, Terada N, et al Direct reprogramming of fibroblasts to myocytes via bacterial injection of MyoD protein. Cellular Reprogramming. 2013;15(2):117–125. doi: 10.1089/cell.2012.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bretscher MS Asymmetrical lipid bilayer structure for biological membranes. Nature: New Biology. 1972;236(61):11–12. doi: 10.1038/newbio236011a0. [DOI] [PubMed] [Google Scholar]
- 5.Bryde S, Hennrich H, Verhulst PM, Devaux PF, Lenoir G, Holthuis JCM CDC50 proteins are critical components of the human class-1 P4-ATPase transport machinery . Journal of Biological Chemistry. 2010;285(52):40562–40572. doi: 10.1074/jbc.M110.139543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulfield G, Siller WG, Wight PA, Moore KJ X chromosome-linked muscular dystrophy (mdx) in the mouse. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(4):1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bull LN, van Eijk MJT, Pawlikowska L, DeYoung JA, Juijn JA, Liao M, et al A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nature Genetics. 1998;18(3):219–224. doi: 10.1038/ng0398-219. [DOI] [PubMed] [Google Scholar]
- 8.Cao CJ, Wang Y, Husain S, Soteropoulos P, Xue CY A mechanosensitive channel governs lipid flippase-mediated echinocandin resistance in Cryptococcus neoformans . mBio. 2019;10(6):e01952–19. doi: 10.1128/mBio.01952-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen S, Wang JY, Muthusamy BP, Liu K, Zare S, Andersen RJ, et al Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic. 2006;7(11):1503–1517. doi: 10.1111/j.1600-0854.2006.00485.x. [DOI] [PubMed] [Google Scholar]
- 10.Cornelison DDW, Wold BJ Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Developmental Biology. 1997;191(2):270–283. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- 11.Crist CG, Montarras D, Buckingham M Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules . Cell Stem Cell. 2012;11(1):118–126. doi: 10.1016/j.stem.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Das A, Slaughter BD, Unruh JR, Bradford WD, Alexander R, Rubinstein B, et al Flippase-mediated phospholipid asymmetry promotes fast Cdc42 recycling in dynamic maintenance of cell polarity. Nature Cell Biology. 2012;14(3):304–310. doi: 10.1038/ncb2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diao YR, Guo X, Li YF, Sun K, Lu LN, Jiang L, et al Pax3/7BP is a Pax7- and Pax3-binding protein that regulates the proliferation of muscle precursor cells by an epigenetic mechanism. Cell Stem Cell. 2012;11(2):231–241. doi: 10.1016/j.stem.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 14.Dumont NA, Wang YX, von Maltzahn J, Pasut A, Bentzinger CF, Brun CE, et al Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nature Medicine. 2015;21(12):1455–1463. doi: 10.1038/nm.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feige P, Rudnicki MA Muscle stem cells. Current Biology. 2018;28(10):R589–R590. doi: 10.1016/j.cub.2018.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friday BB, Mitchell PO, Kegley KM, Pavlath GK Calcineurin initiates skeletal muscle differentiation by activating MEF2 and MyoD. Differentiation. 2003;71(3):217–227. doi: 10.1046/j.1432-0436.2003.710303.x. [DOI] [PubMed] [Google Scholar]
- 17.Füchtbauer EM, Westphal H MyoD and myogenin are coexpressed in regenerating skeletal muscle of the mouse . Developmental Dynamics. 1992;193(1):34–39. doi: 10.1002/aja.1001930106. [DOI] [PubMed] [Google Scholar]
- 18.Gayraud-Morel B, Chrétien F, Jory A, Sambasivan R, Negroni E, Flamant P, et al Myf5 haploinsufficiency reveals distinct cell fate potentials for adult skeletal muscle stem cells. Journal of Cell Science. 2012;125(7):1738–1749. doi: 10.1242/jcs.097006. [DOI] [PubMed] [Google Scholar]
- 19.Gurevich DB, Nguyen PD, Siegel AL, Ehrlich OV, Sonntag C, Phan JMN, et al Asymmetric division of clonal muscle stem cells coordinates muscle regeneration in vivo. Science. 2016;353(6295):aad9969. doi: 10.1126/science.aad9969. [DOI] [PubMed] [Google Scholar]
- 20.Holthuis JCM, Levine TP Lipid traffic: floppy drives and a superhighway. Nature Reviews Molecular Cell Biology. 2005;6(3):209–220. doi: 10.1038/nrm1591. [DOI] [PubMed] [Google Scholar]
- 21.Kato U, Inadome H, Yamamoto M, Emoto K, Kobayashi T, Umeda M Role for phospholipid flippase complex of ATP8A1 and CDC50A proteins in cell migration. Journal of Biological Chemistry. 2013;288(7):4922–4934. doi: 10.1074/jbc.M112.402701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuang S, Chargé SB, Seale P, Huh M, Rudnicki MA Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. The Journal of Cell Biology. 2006;172(1):103–113. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuang SH, Kuroda K, Le Grand F, Rudnicki MA Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129(5):999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li CC, Vargas-Franco D, Saha M, Davis RM, Manko KA, Draper I, et al Megf10 deficiency impairs skeletal muscle stem cell migration and muscle regeneration. FEBS Open Bio. 2021;11(1):114–123. doi: 10.1002/2211-5463.13031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu LM, Zhang LL, Zhang L, Yang F, Zhu XD, Lu ZJ, et al Hepatic Tmem30a Deficiency causes intrahepatic cholestasis by impairing expression and localization of bile salt transporters . The American Journal of Pathology. 2017;187(12):2775–2787. doi: 10.1016/j.ajpath.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Liu WJ, Peng L, Tian WL, Li Y, Zhang P, Sun KX, et al Loss of phosphatidylserine flippase β-subunit Tmem30a in podocytes leads to albuminuria and glomerulosclerosis . Disease Models & Mechanisms. 2021;14(6):dmm048777. doi: 10.1242/dmm.048777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Londhe P, Davie JK Sequential association of myogenic regulatory factors and E proteins at muscle-specific genes. Skeletal Muscle. 2011;1(1):14. doi: 10.1186/2044-5040-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138(17):3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Odelberg SJ, Kollhoff A, Keating MT Dedifferentiation of mammalian myotubes induced by msx1. Cell. 2000;103(7):1099–1109. doi: 10.1016/S0092-8674(00)00212-9. [DOI] [PubMed] [Google Scholar]
- 30.Park J, Choi Y, Jung E, Lee SH, Sohn JW, Chung WS Microglial MERTK eliminates phosphatidylserine-displaying inhibitory post-synapses. The EMBO Journal. 2021;40(15):e107121. doi: 10.15252/embj.2020107121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulusma CC, Folmer DE, Ho-Mok KS, de Waart DR, Hilarius PM, Verhoeven AJ, et al ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology. 2008;47(1):268–278. doi: 10.1002/hep.21950. [DOI] [PubMed] [Google Scholar]
- 32.Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, et al Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143(7):1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito K, Fujimura-Kamada K, Furuta N, Kato U, Umeda M, Tanaka K Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae . Molecular Biology of the Cell. 2004;15(7):3418–3432. doi: 10.1091/mbc.e03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambasivan R, Tajbakhsh S Skeletal muscle stem cell birth and properties. Seminars in Cell & Developmental Biology. 2007;18(6):870–882. doi: 10.1016/j.semcdb.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 35.Schiaffino S, Rossi AC, Smerdu V, Leinwand LA, Reggiani C Developmental myosins: expression patterns and functional significance. Skeletal Muscle. 2015;5(1):22. doi: 10.1186/s13395-015-0046-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, Rudnicki MA Pax7 is required for the specification of myogenic satellite cells. Cell. 2000;102(6):777–786. doi: 10.1016/S0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 37.Sebastian TT, Baldridge RD, Xu P, Graham TR Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2012;1821(8):1068–1077. doi: 10.1016/j.bbalip.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segawa K, Kurata S, Yanagihashi Y, Brummelkamp TR, Matsuda F, Nagata S Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science. 2014;344(6188):1164–1168. doi: 10.1126/science.1252809. [DOI] [PubMed] [Google Scholar]
- 39.Shinin V, Gayraud-Morel B, Gomès D, Tajbakhsh S Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nature Cell Biology. 2006;8(7):677–687. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 40.Tadini-Buoninsegni F, Mikkelsen SA, Mogensen LS, Molday RS, Andersen JP Phosphatidylserine flipping by the P4-ATPase ATP8A2 is electrogenic. Proceedings of the National Academy of Sciences of the United States of America. 2019;116(33):16332–16337. doi: 10.1073/pnas.1910211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242(4877):405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- 42.Tidball JG Mechanisms of muscle injury, repair, and regeneration. Comprehensive Physiology. 2011;1(4):2029–2062. doi: 10.1002/cphy.c100092. [DOI] [PubMed] [Google Scholar]
- 43.Tsuchiya M, Hara Y, Okuda M, Itoh K, Nishioka R, Shiomi A, et al Cell surface flip-flop of phosphatidylserine is critical for PIEZO1-mediated myotube formation. Nature Communications. 2018;9(1):2049. doi: 10.1038/s41467-018-04436-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tu MK, Borodinsky LN Spontaneous calcium transients manifest in the regenerating muscle and are necessary for skeletal muscle replenishment. Cell Calcium. 2014;56(1):34–41. doi: 10.1016/j.ceca.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tu MK, Levin JB, Hamilton AM, Borodinsky LN Calcium signaling in skeletal muscle development, maintenance and regeneration. Cell Calcium. 2016;59(2-3):91–97. doi: 10.1016/j.ceca.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Velden LM, Wichers CGK, van Breevoort AED, Coleman JA, Molday RS, Berger R, et al Heteromeric interactions required for abundance and subcellular localization of human CDC50 proteins and class 1 P4-ATPases. Journal of Biological Chemistry. 2010;285(51):40088–40096. doi: 10.1074/jbc.M110.139006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webster MT, Harvey T, Fan CM Quantitative 3D time lapse imaging of muscle progenitors in skeletal muscle of live mice. Bio-Protocol. 2016;6(24):e2066. doi: 10.21769/BioProtoc.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang YM, Liu WJ, Sun KX, Jiang L, Zhu XJ Tmem30a deficiency leads to retinal rod bipolar cell degeneration . Journal of Neurochemistry. 2019;148(3):400–412. doi: 10.1111/jnc.14643. [DOI] [PubMed] [Google Scholar]
- 49.Yang YM, Sun KX, Liu WJ, Li X, Tian WL, Shuai P, et al The phosphatidylserine flippase β-subunit Tmem30a is essential for normal insulin maturation and secretion . Molecular Therapy. 2021;29(9):2854–2872. doi: 10.1016/j.ymthe.2021.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? Journal of Cell Biology. 2004;166(3):347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L, Yang YM, Li SJ, Zhang SS, Zhu X, Tai ZF, et al Loss of Tmem30a leads to photoreceptor degeneration . Scientific Reports. 2017;7(1):9296. doi: 10.1038/s41598-017-09506-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang SS, Liu WJ, Yang YM, Sun KX, Li SJ, Xu HJ, et al TMEM30A deficiency in endothelial cells impairs cell proliferation and angiogenesis. Journal of Cell Science. 2019;132(7):jcs225052. doi: 10.1242/jcs.225052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.