Abstract
African swine fever (ASF) is a contagious viral transboundary animal disease affecting domestic pigs caused by ASF virus (ASFV). This study was conducted in order to determine the genetic characteristics, risk factors and socioeconomic impact of an ASF outbreak in 2019 in Ngara, western Tanzania. Tissue samples from dead domestic pigs with clinical picture suggestive of ASF infection were collected for ASF confirmation and genetic characterization of ASFV. Data on the risk factors and socioeconomic impact associated with the ASF outbreak were collected from consenting farmers using a semi-structured questionnaire. Disease confirmation was done by detection of genomic ASFV DNA using polymerase chain reaction. Partial amplification of the ASFV genome, dideoxynucleotide sequencing of the PCR products followed by bioinformatics analyses was conducted to determine the ASFV genotypes. Phylogenetic reconstruction of the variable 3′-end of the B646L gene clustered the ASFV isolate into genotype X. Analysis of the intergenic fragment sequences between 173R and 1329L genes showed that the viral strains TAN/19/Ngara and Kenya 1950 similarly lacked a 36 bp fragment that is present in strain Ken05/Tk1. Feeding pigs of uncooked swill was shown to be significantly associated with ASF spread (OR = 3.08, C.I.95% = 1.06–8.99, P = 0.0009). Occurrence of ASF outbreak resulted in loss of income and investment as most farmers kept pigs for the purpose of income generation. Food security was disturbed due to high pig mortality following occurrence of ASF outbreak. A total of 93,630,000 Tanzanian shillings (approximately 41,065 USD) was estimated to be lost as a result of pigs’ mortality in 219 households. The findings of the present study associate ASFV genotype X with the 2019 ASF outbreak in Ngara and feeding pigs with uncooked swill with spread of the disease.
Keywords: African swine fever virus, Genotypes, Outbreak determinants, Pigs, Small-scale farming system
Introduction
Domestic pigs in Tanzania are mainly kept for subsistence by smallholder farmers (Kimbi et al., 2015). Domestic pig farming is mainly conducted in rural areas particularly in the southern highland regions including Mbeya, Iringa, Rukwa and Ruvuma (Karimuribo et al., 2011). Both urban and rural farmers benefit from pig production by earning their livelihoods and enhancing their food security (URT, 2012). Diseases including African swine fever (ASF), foot and mouth disease (FMD) and porcine cysticercosis have been substantial limiting factors to domestic pig farming in Tanzania (URT, 2012). African swine fever, a highly lethal and contagious disease of domestic pigs has been the most often reported disease of domestic pigs to the World organisation for Animal Health (OIE) by the Government of Tanzania (Wilson & Swai, 2013).
The causative agent of ASF is an ASF virus (ASFV), a sole member of the family Asfarviridae, genus Asfivirus (Costard et al., 2013). The virus is large, enveloped with a linear double stranded DNA genome and is the only known DNA arbovirus (Alonso et al., 2018). The virus circulates in four cycles; the domestic cycle, the pig-tick cycle, the sylvatic circle between common warthogs and soft ticks of the genus Ornithodoros and the wild-boar habitat cycle (Chenais et al., 2019). Pigs contact the disease through direct contact with infected pigs or fomites in domestic pig cycle and get infected through infected tick bites in the tick-domestic pig cycle (Boinas et al., 2011). The sylvatic cycle involves asymptomatically infected wild pigs including bush pigs, warthogs and ticks which act as both reservoir and transmission vectors (Jori et al., 2013). In the wild boar-habitat cycle the Eurasian wild boars get infected directly through contact with other infected boars. Boars also indirectly get infected through contact with carcasses of infected pigs or contact with habitat contaminated by the carcasses (Chenais et al., 2019). In eastern and southern Africa, presence of warthogs (Phacochoerus africanus) and Ornithodoros moubata ticks maintain the presence of ASFV in nature (Costard et al., 2009;Costard et al., 2013).
Early reports of ASF were made in 1920s in eastern Africa from which was followed by its spread to disease-free regions within Africa and to continents other than Africa (Costard et al., 2013). Human activities contribute to the transboundary spread of the disease. These include improper disposal of wastes from ships and aircrafts, illegal transportation of infected pigs and pig products by traders for commercial purposes and by tourists for personal use. Some non-African countries including Portugal, Caribbean and South America have successfully eradicated the disease through drastic measures (Galindo & Alonso, 2017). Currently, ASF is endemic in a number of sub-Saharan African countries and in the island of Sardinia, Italy. Recent ASF outbreaks outside Africa have been reported in Europe, Caucasus region and Asia (Galindo & Alonso, 2017; Garigliany et al., 2019; Taylor et al., 2020). The 2018 outbreaks in Asia started in China and later on spread to a number of Asian countries (Ge et al., 2018; Li & Tian, 2018; Lu, Pan & Zhang, 2020).
Molecular epidemiology of ASF is aided by genotyping of ASFV. African swine fever virus genomic DNA varies in length from 170 to 194 kbp (Alonso et al., 2018). Factors accounting for the length variations include differences in number of tandem repeats in the central variable region (CVR) of the genome and insertions and deletions at the left and right terminal regions of the genome (Tulman et al., 2009). Twenty four p72 ASFV genotypes (I–XXIV) have been described based on partial B646L (coding for the p72 capsid protein) gene sequencing (Boshoff et al., 2007; Gallardo et al., 2009; Quembo et al., 2018). Sequence analysis of full-length gene E183L (coding for p54 envelope protein) and central variable region (CVR) within the B602L gene is used in clustering isolates into groups and subtypes (Gallardo et al., 2009; Lubisi et al., 2007). Thirty one subgroups of ASFV have been identified by sequence analysis of the especially discriminative genetic marker, the B602L gene based on variations in the encoded tetrameric amino acid repeats (Nix et al., 2006). Prior to the description of genotype XXIII and XXIV in Ethiopia and Mozambique respectively, 22 genotypes were known to circulate in southern and eastern Africa (Achenbach 2017, Quembo et al., 2018). Genotype I was known to circulate in eastern and southern Africa, the Caribbean, South America, West Africa and Europe (Costard et al., 2013). Later on transboundary transmissions of the virus led to introduction of genotype IX to Democratic Republic of the Congo, western Africa and genotype II to the Republic of Georgia in 2007 that subsequently spread into Europe, Russia and Asia including China (Costard et al., 2013; Gallardo et al., 2011; Ge et al., 2018). The highly virulent p72 genotype II ASFV, identical to the Georgia 2007/1 isolate has been described to circulate persistently in Tanzania. In addition to genotype II, genotypes IX, X, XV and XVI have been described in ASF outbreaks in Tanzania (Lubisi et al., 2005; Misinzo et al., 2010; Misinzo et al., 2014; Yona et al., 2020).
In this study, we report diagnosis and molecular characterization of the ASFV strains from the 2019 ASF outbreak in Ngara district, western Tanzania and its associated risk factors and socio economic impact.
Materials and methods
Study area
This study was conducted in eight villages of Ngara district namely Rusumo, Rwakaremera, Kyenda, Kasulo, Kagali, Nyakibiza, Nyakariba and Bulengo (Fig. 1). Ngara is one of the eight districts of Kagera region, divided into four divisions that are further divided into 72 villages. The district has a total area of 3744 square kilometers. Pig farming in the region is conducted by small-scale farmers who mainly depend on family labor to feed the domestic pigs, clean shelters, sell the live animals and their products. Several national parks are located in Kagera region including Burigi and Rumanyika in Karagwe district and Ibanda in Kyerwa district. The national parks provide the potential natural habitat for wild pigs and ticks which are reservoir hosts of ASFV.
Fig. 1.
Map showing Rusumo village (shaded in grey) in Ngara district, western Tanzania where outbreak of African swine fever occurred in 2019. The red circles denote some of the pig keeping households where risk factors and socioeconomic impact data of the ASF outbreak were collected.
Sampling of domestic pigs
An outbreak of ASF was suspected in Ngara district, Kagera region in 2019 following reports of pig mortalities from a severe hemorrhagic disease. Postmortem examination was conducted on dead domestic pigs followed by collection of spleen, lymph nodes, lungs and kidney tissue samples were transported to the laboratory under a cold chain. Upon reaching the laboratory, tissue samples were prepared by chopping 1 g of each sample in a separate petri dish in presence of 5 mL, sterile phosphate buffer saline (PBS). The homogenized tissues were centrifuged at 6000 g for five minutes at room temperature. Afterwards, the tissue supernatant was transferred into cryovials and stored at −80 °C until DNA extraction.
DNA extraction and ASF detection
Frozen aliquots (100 μL) of preserved tissue supernatant were allowed to thaw and genomic DNA extraction was performed using Qiamp nucleic acid extraction kit (Qiagen, Hilden, Germany), according to protocol supplied by the manufacturer. Detection of ASFV DNA was done using primers PPA1 and PPA2 to amplify the C-terminal region of P72 major capsid protein encoded by the B646L gene. The expected PCR product size was 257 bp (Aguero et al., 2003).
Molecular characterization of ASFV
Genotyping was done by nucleotide sequence analysis of the variable 3′ end of the B646L gene, the CVR region and intergenic fragment between 173R and 1329L genes. Amplification of the C-terminal region of the B646L gene was done using p72U and p72D primers to amplify a 478 bp DNA fragment (Bastos et al., 2003). Amplification of the tandem repeats sequence (TRS) located between the I73R and I329L genes was done using primers ECO1A and ECO1B that amplify a 356 bp DNA fragment (Gallardo et al., 2014). Amplification of the CVR region was done using ORF9L-F/ORF9L-R primers (Nix et al., 2006). Sequencing was done using dideoxynucleotide cycle sequencing using a Big Dye Terminator kit v3.0 (Applied Biosystem Foster City, CA). Sequencing was performed using p72U, p72D, ECO1A, ECO1B, ORF9L-F and ORF9L-R primers. The similarity search against other ASFV sequences at GenBank database and their alignment with other Tanzanian ASFV nucleotide sequences was done using BLASTn. Clustal omega was used to perform multiple sequence alignment of nucleotide sequences of the tandem repeat sequences (TRS) between I73R and I329L genes. Phylogenetic analysis was performed using the Maximum Likelihood method with 1000 bootstrap replications, and evolutionary distances were calculated by the Kimura 2-parameter method as implemented in MEGA X (Kumar et al., 2018).
Risk factors and socioeconomic impact data collection
A semi-structured questionnaire was designed and used for collection of data on risk factors and socioeconomic impact of the disease outbreak from affected farmers. Questionnaires addressing different factors including demographic characteristics of respondents, general domestic pig husbandry, ASF-related risk factors and ASF outbreak resultant financial loss were administered to a total of 219 farmers. The questions were asked in the local language (Kiswahili) and involved both closed and open-ended answers. Demographic data including gender, age, occupation, educational and marital status of respondents were collected. Farmers were also asked about practices that could potentially result in spread of ASF. These included sharing of farm equipment, sharing of boars and occurrence of disease in neighboring farmers. Furthermore, general information on pig husbandry such as types of pig breeds kept, whether pigs were kept in confinement or not, keeping of health records and handling of wastes was collected. Epidemiological data was entered and validated in excel spreadsheets. Data analysis was done using epi info software version 3.5.1. Frequencies, percentages, means and counts were used to determine distributions and magnitudes of the variables among the respondents. Association between ASF occurrence and risk factors was determined using unconditional logistic regression analysis. The statistical significance was established at 95% confidence interval and critical p-value of 0.05. Economic loss associated with the outbreak was estimated from total financial value of all pigs died of ASF in Tanzanian shillings.
Results
Clinical signs and postmortem findings
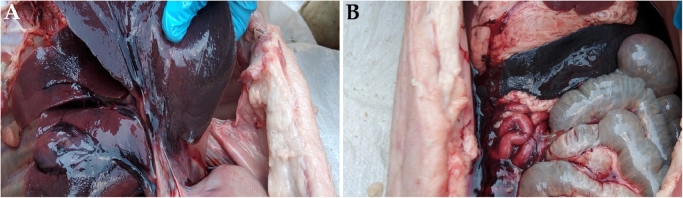
Sick pigs in visited farms showed ASF clinical signs including high fever (41–42 °C), staggering gait, anorexia and erythemic skin on the ears, flanks, belly and genitalia. Pathological lesions suggestive of ASF were observed during postmortem including gastroenteritis, enlargement of spleen (splenomegaly), petechiation of the kidney cortex and hemorrhages in lymph nodes (Fig. 2).
Fig. 2.
Postmortem findings in one of the domestic pigs with African swine fever. (A) Hemorrhagic lungs and (B) enlargement of spleen (splenomegaly).
Genetic characterization of ASFV from the 2019 ASF outbreaks in Ngara district
The nucleotide sequences generated in this study for the three regions; B646L (3′ variable end of the major capsid protein p72), I739R-I329L (intergenic region) and B602L (central variable region) were submitted to Genbank nucleotide database and given accession numbers; MW659863, MW 659,865 and MW659867 respectively. A phylogenetic tree (Fig. 3), representing evolutionary relationships between the 2019 ASFV strain and previously identified Tanzanian ASFV strains (Table. 1) was constructed. The ASFV strain from the 2019 outbreak in Ngara, TAN/19/Ngara clustered into ASFV p72 genotype X. The TAN/19/Ngara TRS region between I73R and I329L genes had 99.76% and 90.76% nucleotides identity to Kenya 1950 and Ken05/Tk1 isolates, respectively. Both TAN/19/Ngara and Kenya 1950 isolates lacked a 51 bp fragment (Fig. 4) compared to Ken05/Tk1 isolate (Bishop et al., 2015). Amplicons of 180 bp were obtained by PCR using primers ORF9L-F/ORF9L-R for TAN/19/Ngara. The numbers of amino acid repeats were 10 for TAN/19/Ngara with signature BNDBNDBNAA.
Fig. 3.
Evolutionary relationships of representative strains of African swine fever virus based on the Maximum Likelihood phylogeny of the partial p72 gene sequences. The phylogenetic analysis was performed using MEGA X (http://www.megasoftware.net) and the Kimura 2-parameter substitution model, determined by a model selection analysis. Phylogeny was inferred following 1,000 bootstrap replications, and the node values show percentage bootstrap support (only the values above 50% are shown). The square black spots indicate the African swine fever virus sequence from this study and the scale bar indicates nucleotide substitution per site.
Fig 4.
Partial nucleotide sequence alignment of the intergenic region between 173R and 1329L genes in African swine fever virus (ASFV) isolates belonging to B646L p72 genotype X from Kenya, DRC, Burundi and Tanzania. Both TAN/19/Ngara and Kenya1950 lack a 36 base pair fragment compared to Ken05/Tk1.
Risk factors and socioeconomic impact
In this study a total of 219 domestic pig farmers from pig keeping households were interviewed, owning a total of 832 pigs. Most of them were male (70.32%) of the prime working age (25–54 years). Majority were married (95.43%) and had primary education (73.06%). The primary and secondary occupations for all 219 respondents were practicing crop farming and livestock keeping, respectively. Pigs were found to be the leading livestock kept (80.82%). Majority of farmers kept cross pig breed (49.8%) and the leading pig keeping system was confinement where pigs were permanently kept in their pig pens and were stall-fed (65.3%) followed by semi-confinement where pigs were partially kept in pigpens or were just tethered to restrict their movement (34.7%). The building materials mostly used for constructing pig pens were logs and wood. All respondents claimed to have never seen ticks in their pig pens and warthogs around their premises.
Majority of respondents had heard about ASF (65.3%) before and the main source of information was through other farmers (67.1%). Out of 219 interviewed farmers 62 had encountered ASF between May and September 2019. Most of the affected farmers (69.4%) mentioned anorexia and fever as observed ASF clinical signs. Most farmers did not take any action following occurrence of ASF and some of the affected farmers (2.3%) applied treatment to their pigs before ASF was confirmed. Among the eight villages of Ngara district included in this study five of them had respondents who had encountered ASF in year 2019. The five villages included Rwakaremera, Rusumo, Kyenda, Kasulo and Kagali. Affected farmers had a total of 728 pigs before the outbreak, 654 of which got sick and 649 pigs ultimately died resulting in 89.8% morbidity and 89.1% mortality. A total of 93,630,000 Tanzanian shillings was estimated to be lost as a result of pigs’ death during the outbreak. The amount approximately equals to 41,065 USD (Exchange rate: 1 USD = 2280) in October 2019.
Eight risk factors were tested for their association with the disease outbreaks. Feeding pigs with uncooked swill was found to be significantly associated with ASF (p < 0.05). Furthermore, swill feeding and occurrence of ASF in neighboring farms are the most likely confounding factors in occurrence and spread of the disease. This is because all ASF victims reported to practice swill feeding and had ASF occurred in their neighboring farms. Details on association between the risk factors for spread of the disease are shown in Table 2.
Discussion
In this study, ASFV clustering to B646L p(72) genotype X was confirmed as a causative agent of hemorrhagic fever outbreaks in domestic pigs in Ngara district of Tanzania in 2019. Genotype X has been described as a sylvatic cycle-associated genotype as it has been identified in domestic pigs, ticks and warthogs in Burundi, Kenya and Tanzania (Lubisi et al., 2005). Early identification of genotype X in Tanzania was done in northeastern Tanzania in Serengeti national park where it was recovered from ticks and warthogs (Lubisi et al., 2005; Wambura et al., 2006; Misinzo et al., 2012a). Furthermore, it was recovered in regions with close proximity to national parks such as Tarakea, a region bordering Kenya and very close to Kilimanjaro National Park (Misinzo et al., 2014). The genotype was also described in other parts of northern and northwestern Tanzania including Mwanza, Kigoma and Ngara (Misinzo et al., 2014; Yona et al., 2020). Identification of genotype X in Ngara, 2019 ASF outbreak in this study indicates restriction of the genotype in northern Tanzania. Phylogenetic analysis of the TAN/19/Ngara showed it has high nucleotide similarity with ASFV isolates from Kenya, Ken05/Tk1 and Kenya 1950. In 2018 the isolate from Burundi, BUR/18/Rutana showed high similarity with both of the mentioned Kenyan isolates (Hakizimana et al., 2020). The shown close relationship between isolates from East African countries put emphasis on the potential contribution of the transboundary nature of the virus in spreading ASF.
Feeding pigs of uncooked swill was found to be significantly associated with the outbreak. Swill feeding and occurrence of ASF in neighboring farms were the most likely confounding factors as they were reported in all ASF victims. The later could have been caused by movement of people between farms and sharing of equipment as these factors have been associated in ASF outbreaks in other studies (Fasina et al., 2010; Yona, 2017). All ASF affected farmers were reported to not take appropriate action after the outbreak including slaughtering, burying the carcasses and decontaminating the premises. As a result, the outbreak has caused a significant financial loss of about 41,065 USD as a result of pig mortality. More financial losses resulted from some of the farmers’ attempts to treat the disease before ASF was confirmed and losses from selling of pigs at lower prices. This posed a considerable threat to growth in pig keeping industry considering it was shown to be leading livestock kept (80.82%) followed by goat keeping (11.42%). The disease outbreak affected the farmers’ livelihood stability and growth considering subsistence farming is mainly aimed at providing for household expenses. The expenses include provision for medical expenses and school fees. Educating the community on the significant danger ASF poses to pig industry and biosecurity measures required to limit the spread of ASFV can help in prevention of the disease spread. This education could preferably be incorporated in the primary education curriculum as the findings of this study showed that the majority of interviewed domestic pig farmers (73.06%) had a primary level education. The community needs to understand the importance of adhering to biosecurity measures taken so as to prevent ASF transmission. The government can help in ASF prevention and control by routinely surveying the disease occurrence and strict enforcement of regulations put forward in prevention of ASF transmission between regions. Compensation following compulsory slaughter of ASF infected pigs is recommended whenever outbreaks occur as it enables them to adhere to biosecurity measures against ASF spread (Fasina et al., 2010; Fasina et al.,2020).
The findings of this study have confirmed presence of ASFV in Ngara located in northwestern part of Tanzania in 2019 outbreaks. The virus was genotyped and was found to be of p72 genotype X with the CVR signature BNDBNDBNAA. Presence of genotype X in the 2019 ASF outbreak indicates persistence of this genotype circulation in Tanzania, which indicates the endemicity of the disease in Tanzania. Feeding pigs with uncooked swill was shown to be significantly associated with the outbreak. Furthermore, most farmers did not take action to contain and prevent spread of the disease following the outbreak. Given the unavailability of both vaccine and treatment against ASF and its transboundary nature, the recommended disease control method is adherence to sanitary measures. These include, prevention of movement, slaughtering, proper disposal of infected animals and decontamination of affected premises. The Government should impose surveillance of risks of spreading the diseases associated with illegal transfer of infected pig and pig products for personal and commercial uses. Molecular characterization studies should be conducted in all outbreaks occurring in Tanzania so as to broaden the epidemiological knowledge of the virus within the country.
Funding
This study was funded by the Government of the United Republic of Tanzania through the World Bank [WB-ACE II Grant PAD1436, IDA [Credit5799-TZ] to the SACIDS Africa Centre of Excellence for Infectious Diseases at Sokoine University of Agriculture.
Ethical statement
Ethical approval for animals sampling was sought from the Ethical Committee of Sokoine University of Agriculture. Written consent to participate was obtained from farmers and veterinarians before sampling of tissues from slaughtered pigs.
Declaration of Competing Interest
The authors declare that they have no personal or financial relationships that may have inappropriately influenced them in writing this article.
Acknowledgements
The authors are especially grateful to Ngara District Veterinary Officer Dr. Richard Ngowi for his skillful field assistance. Furthermore, appreciation is extended to Miriam R. Makange, Mhoja Ndalahwa and Anna Rogath for their laboratory technical assistance.
References
- Achenbach J.E., Gallardo C., Nieto-Pelegrín E., Rivera-Arroyo B., Degefa-Negi T., Arias M. Identification of a New Genotype of African Swine Fever Virus in Domestic Pigs from Ethiopia. Transboundary Emerging Diseases. 2017;64(5):1393–1404. doi: 10.1111/tbed.12511. [DOI] [PubMed] [Google Scholar]
- Aguero M., Fernandez J., Romero L., SanchezMascaraque C., Arias M., Sanchez-Vizcaino J. Highly sensitive PCR assay for routine diagnosis of African swine fever virus in clinical samples. Journal of Clinical Microbiology. 2003;41(9):4431–4434. doi: 10.1128/JCM.41.9.4431-4434.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso C., Borca M., Dixon L., Revilla Y., Rodriguez F., Escribano J.M. ICTV. ICTV Virus Taxonomy Profile : Asfarviridae. 2018;99(5):613–614. doi: 10.1099/jgv.0.001049. [DOI] [PubMed] [Google Scholar]
- Bastos A.D.S., Penrith M.L., Crucière C., Edrich J.L., Hutchings G., Roger F. Genotyping field strains of African swine fever virus by partial p72 gene characterisation. Archives of Virology. 2003;148(4):693–706. doi: 10.1007/s00705-002-0946-8. [DOI] [PubMed] [Google Scholar]
- Bishop R.P., Fleischauer C., de Villiers E.P., Okoth E.A., Arias M., Gallardo C. Comparative analysis of the complete genome sequences of Kenyan African swine fever virus isolates within p72 genotypes IX and X. Virus genes. 2015;50(2):303–309. doi: 10.1007/s11262-014-1156-7. [DOI] [PubMed] [Google Scholar]
- Boinas F.S., Wilson A.J., Hutchings G.H., Martins C., Dixon L.J. The persistence of African swine fever virus in field- infected ornithodoros erraticus during the ASF endemic period in Portugal. PloS one. 2011;6(5):20383. doi: 10.1371/journal.pone.0020383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshoff C.I., Bastos A.D.S., Gerber L.J., Vosloo W. Genetic characterisation of African swine fever viruses from outbreaks in southern Africa (1973 –1999) Veterinary Microbiology. 2007;121:45–55. doi: 10.1016/j.vetmic.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Chenais E., Depner K., Guberti V., Dietze K., Viltrop A., Ståhl K. Epidemiological considerations on African swine fever in Europe 2014 –2018. Porcine Health Management. 2019;5(6):1–10. doi: 10.1186/s40813-018-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costard S., Mur L., Lubroth J., Sánchez-Vizcaíno J.M., Pfeiffer D.U. Epidemiology of African swine fever virus. Virus Research. 2013;173:191–197. doi: 10.1016/j.virusres.2012.10.030. [DOI] [PubMed] [Google Scholar]
- Costard S., Wieland B., Glanville W., Jori F., Rowlands F., Rowlands R. African swine fever : How can global spread be prevented ? African swine fever : How can global spread be prevented ? Philosophical Transactions of the Royal Society B: Biological Sciences. 2009;364:2683–2696. doi: 10.1098/rstb.2009.0098. (August) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasina F.O., Kissinga H., Mlowe F., Mshang S., Matogo B. Drivers, risk factors and dynamics of African swine fever outbreaks, southern highlands, Tanzania. Pathogens (Basel, Switzerland) 2020;9(156):1–20. doi: 10.3390/pathogens9030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasina F.O., Shamaki D., Makinde A.A., Lombin L.H., Lazarus D.D., Rufai S.A. Surveillance for African swine fever in Nigeria. 2006 –2009. 2010;57:244–253. doi: 10.1111/j.1865-1682.2010.01142.x. [DOI] [PubMed] [Google Scholar]
- Galindo I., Alonso C. African swine fever virus : A review. Viruses. 2017;9(103):1–10. doi: 10.3390/v9050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo C., Fernández-Pinero J., Pelayo V., Gazaev I., Markowska-Daniel I., Pridotkas G. Genetic variation among African swine fever genotype II viruses, eastern and central Europe. Emerging Infectious Diseases. 2014;20(9):1544–1547. doi: 10.3201/eid2009.140554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo C., Anchuelo R., Pelayo V., Poudevigne F., Leon T., Nzoussi J. African swine fever virus p72 genotype IX in domestic pigs, Congo, 2009. Emerging Infectious Diseases. 2011;17:1556–1558. doi: 10.3201/eid1708.101877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo C., Mwaengo D.M., Macharia J.M., Arias M., Taracha E.A., Soler A. Enhanced discrimination of African swine fever virus isolates through nucleotide sequencing of the p54, p72. and pB602L (CVR) genes. 2009;38(1):85–95. doi: 10.1007/s11262-008-0293-2. [DOI] [PubMed] [Google Scholar]
- Garigliany M., Desmecht D., Tignon M., Cassart D., Lesenfant C., Paternostre J. Phylogeographic Analysis of African swine fever Virus, Western Europe, 2018. Emerging Infectious Diseases. 2019;25(1):2018–2020. doi: 10.3201/eid2501.181535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S., Li J., Fan X., Liu F., Li L., Wang Q. Molecular characterization of African swine fever virus, China, 2018. Emerging Infectious Diseases. 2018;24(11):2131–2133. doi: 10.3201/eid2411.181274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakizimana, J.N., .Nyabongo, L., Ntirandekura, J.B., .Yona, C., Ntakirutimana, D., Kamana, O. et al., (2020). Genetic analysis of African Swine Fever Virus From the 2018 Outbreak in South-Eastern Burundi. Frontiers in Veterinary Science, 7, 578474.. [DOI] [PMC free article] [PubMed]
- Jori F., Vial L., Penrith M., Pérez-Sánchez R., Etter E., Albina E. Review of the sylvatic cycle of African swine fever in sub-Saharan Africa and the Indian ocean. Virus Research. 2013;173:212–227. doi: 10.1016/j.virusres.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Karimuribo E.D., Chenyambuga S.W., Makene V.W., Mathias S. Characteristics and production constraints of rural-based small-scale pig farming in Iringa region, Tanzania. Livestock Research for Rural Development. 2011;23:173. [Google Scholar]
- Kimbi E., Lekule F., Mlangwa J., Mejer H., Thamsborg S. Smallholder pigs production systems in Tanzania. Journal of Agricultural Science and Technology. 2015;A5:47–60. [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Tian K. African swine fever in China. Veterinary Record. 2018;183:300–301. doi: 10.1136/vr.k3774. [DOI] [PubMed] [Google Scholar]
- Lu G., Pan J., Zhang G. African swine fever virus in Asia: Its rapid spread and potential threat to unaffected countries. Journal of Infection. 2020;80:350–371. doi: 10.1016/j.jinf.2019.11.011. [DOI] [PubMed] [Google Scholar]
- Lubisi B., Bastos A.D.S., Dwarka R.M., Vosloo W. Molecular epidemiology of African swine fever in East Africa by. Archives of Virology. 2005;150:2439–2452. doi: 10.1007/s00705-005-0602-1. (June) [DOI] [PubMed] [Google Scholar]
- Lubisi B.A., Bastos A.D.S., Dwarka R.M., Vosloo W. Intra-genotypic resolution of African swine fever viruses from an East African domestic pig cycle : A combined p72 -CVR approach. Virus genes. 2007;35:729–735. doi: 10.1007/s11262-007-0148-2. [DOI] [PubMed] [Google Scholar]
- Misinzo G., Kasanga C.J., Ngeleja C.M., Masambu J., Annette K., Van Doorsselaere J. African swine fever virus. Emerging Infectious Diseases. 2012;18(12):2081–2083. doi: 10.3201/eid1812.121083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misinzo G., Kwavi D.E., Sikombe C.D., Makange M., Peter E., Muhairwa A.P. Molecular characterization of African swine fever virus from domestic pigs in northern Tanzania during an outbreak in 2013. Tropical animal health and production. 2014;46:1199–1207. doi: 10.1007/s11250-014-0628-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misinzo G., Magambo J., Masambu J., Yongolo M.G., Doorsselaere J.Van, Nauwynck H.J. Genetic characterization of African swine fever viruses from a 2008 outbreak in Tanzania. Transboundary and Emerging Diseases. 2010;58:86–92. doi: 10.1111/j.1865-1682.2010.01177.x. [DOI] [PubMed] [Google Scholar]
- Nix R.J., Gallardo C., Hutchings G., Blanco E., Dixon L.K. Molecular epidemiology of African swine fever virus. Archives of Virology. 2006;151:2475–2494. doi: 10.1007/s00705-006-0794-z. [DOI] [PubMed] [Google Scholar]
- Quembo C.J., Jori F., Vosloo W., Heath L. Genetic characterization of African swine fever virus isolates from soft ticks at the wildlife/domestic interface in Mozambique and identification of a novel genotype. Transboundary and Emerging Diseases. 2018;65(2):420–431. doi: 10.1111/tbed.12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.A., Condoleo R., Simons R.R.L., Gale P., Kelly L.A., Snary E.L. The risk of infection by African swine fever virus in European swine through boar movement and legal trade of pigs and pig meat. Frontiers in Veterinary Sciences. 2020;6:1–19. doi: 10.3389/fvets.2019.00486. (January) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulman E.R., Delhon G.A., Ku B.K., Rock D.L. African swine fever virus. Current Topics in Microbiology and Immunology. 2009;328:43–87. doi: 10.1007/978-3-540-68618-7_2. [DOI] [PubMed] [Google Scholar]
- URT No titlenational sample census of agriculture 2007/2008 small holder agriculture volume III: Livestock sector - national report. Dar es Salaam. 2012 [Google Scholar]
- Wambura P.N., Masambu J., Msami H. Molecular diagnosis and epidemiology of African swine fever outbreaks in Tanzania. Veterinary Research Communications. 2006;30(6):667–672. doi: 10.1007/s11259-006-3280-x. [DOI] [PubMed] [Google Scholar]
- Wilson R.T., Swai E. A review of pig pathology in Tanzania. Tropical and Animal Health Production. 2013;45(6):1393–1404. doi: 10.1007/s11250-013-0426-z. [DOI] [PubMed] [Google Scholar]
- Yona, C. (2017). Studies on epidemiology and socio-economic impact associuated wth African swine fever 2015-2017 outbreaks in Tanzania. Sokoine University of Agriculture.
- Yona C.M., Vanhee M., Simulundu E., Makange M., Nauwynck H.J., Misinzo G. Persistent domestic circulation of African swine fever virus in Tanzania, 2015–2017. BMC Veterinary Research. 2020;16(1) doi: 10.1186/s12917-020-02588-w. 369. [DOI] [PMC free article] [PubMed] [Google Scholar]