Summary
Single-nucleus RNA sequencing (snRNA-seq), where nuclear transcriptomes are a proxy to cellular transcriptomes, has been used to profile human brain. snRNA-seq is sensitive to tissue processing, tissue quality, postmortem interval time, and cellular debris. This protocol outlines steps for the isolation of high-quality nuclei from surgically resected human brain tissue followed by a sucrose gradient yielding neuronal and non-neuronal nuclei enabling unbiased analysis of various cell types.
For complete details on the use and execution of this protocol, please refer to Ayhan et al. (2021).
Subject areas: Cell isolation, Single Cell, Cell separation/fractionation, Genomics, RNAseq, Microbiology, Neuroscience
Graphical abstract
Highlights
-
•
Protocol describing nuclei isolation from human brain for single-nuclei RNA-seq.
-
•
A sucrose gradient adapted for a low amount of tissue helps cleaning debris.
-
•
Various quality control measures ensuring RNA and nuclei integrity.
-
•
Details regarding biological safety and reproducible record keeping are considered.
Single-nucleus RNA sequencing (snRNA-seq), where nuclear transcriptomes are a proxy to cellular transcriptomes, has been used to profile human brain. snRNA-seq is sensitive to tissue processing, tissue quality, postmortem interval time, and cellular debris. This protocol outlines steps for the isolation of high-quality nuclei from surgically resected human brain tissue followed by a sucrose gradient yielding neuronal and non-neuronal nuclei enabling unbiased analysis of various cell types.
Before you begin
Timing: 30 min
-
1.
All experiments using human tissues must be performed with the informed consent of participants. For this study and our previous work (Ayhan et al., 2021), all hippocampal tissues were obtained from patients with temporal lobe epilepsy with the informed consent of patients and approval of the UTSW Institutional Review Board (IRB).
-
2.
All unfixed human tissue must be assumed to be infectious. Use appropriate PPE including lab coats, double gloves, and safety googles.
-
3.
As RNA polymerases and RNases show minimal activity at 4°C, it is critical to keep the dissected tissue at ice-cold temperatures after surgical resection. Prepare the solutions listed in materials and equipment and prechill all solutions to 4°C on ice. Cool all centrifuges, rotors, and tubes to 4°C.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Nuclei EZ lysis buffer | Sigma | Cat# NUC-101 |
| UltraPure™ BSA (50 mg/mL) | Thermo Fisher Scientific | Cat #AM2618 |
| SUPERase•In™ RNase Inhibitor (20 U/μL) | Thermo Fisher Scientific | Cat #AM2694 |
| Trypan Blue Solution, 0.4% | Thermo Fisher Scientific | Cat#15250061 |
| ProLong Diamond Antifade Reagent with DAPI | Life Technologies | Cat#P36971 |
| Neurobasal A Medium | Invitrogen | Cat#10888-022 |
| Ethidium Homodimer-1 | Invitrogen | Cat#E1169 |
| 1× PBS | Corning | Cat# 21040CM |
| Critical commercial assays | ||
| Nuclei PURE Prep Isolation Kit | Sigma | Cat# NUC201-1KT |
| RNA Screen Tape | Agilent Technologies | Cat# 5067-5576 |
| Chromium Single Cell 3′ kit | 10× Genomics | Cat#1000153 |
| Biological samples | ||
| Surgically resected human hippocampus tissue (mixed sex, 24–60 years old) | This study | N/A |
| Other | ||
| Eppendorf® Centrifuge 5810/5810R | Eppendorf | Cat# EP022628188 |
| 1.5 mL Eppendorf tubes | Eppendorf | Cat# 022431021 |
| 2 mL Eppendorf tubes | Eppendorf | Cat# EP022363344 |
| 15 mL conical tubes | Corning | Cat# CLS430791 |
| Dounce homogenizer | DWK Life Sciences | Cat# 8853000002 |
Materials and equipment
Neurobasal A medium
Aliquot ∼30 mL of Neurobasal A Medium in a 50 mL conical tube for each specimen to be transported from the operation room. Prechill on ice.
Nuclei EZ lysis buffer
Aliquot 8 mL for each sample to be processed and prechill on ice.
Nuclei suspension buffer (NSB)
| Reagent | Final concentration | Amount |
|---|---|---|
| 1× PBS | 1× | 4 mL |
| 5% BSA | 1% | 1 mL |
| RNase inhibitor | 0.2U/ul | 50 μL |
Prepare fresh and keep on ice.
Sucrose solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Nuclei PURE 2M Sucrose Cushion Solution | N/A | 2.7 mL |
| Nuclei PURE Sucrose Cushion Buffer | N/A | 0.3 mL |
Prepare fresh and keep on ice.
Step-by-step method details
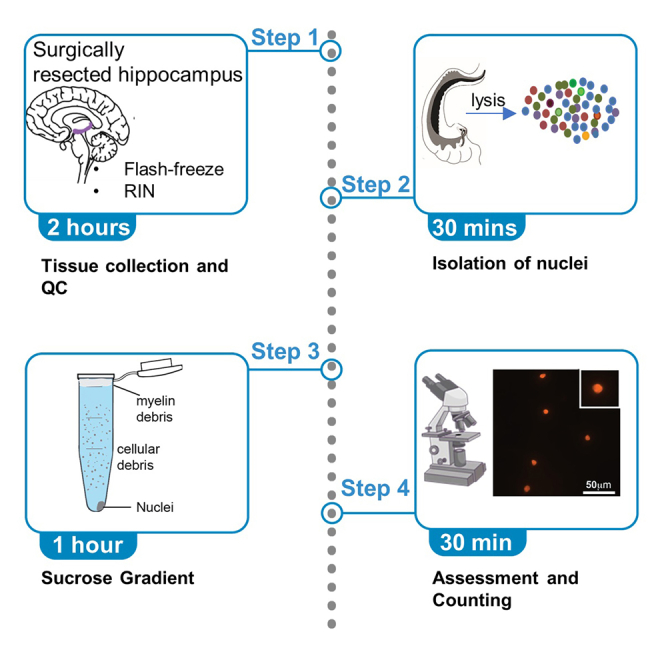
Tissue collection, transport, and banking
Timing: 2 h
Here we describe the best practices to transport and store the resected hippocampal tissue after surgical removal.
-
1.The neurosurgeon collects the anterior and posterior hippocampal tissue by initially resecting the lateral temporal cortex to expose the hippocampus after opening the temporal horn of the ventricle.
-
a.Following division of the amygdalo-hippocampal sulcus, the hippocampus is reflected inferiorly on its vascular pedicle until the fibria and adjacent choroidal fissure are visualized. After opening the alveus just lateral to the fimbria, an anterior-posterior line of dissection is carried along the hippocampal axis until the arteries feeding the hippocampus are identified and sequentially divided, freeing the hippocampus from its pedicle in an en bloc fashion. The tail of the hippocampus is severed posterior to the lateral geniculate nucleus.Note: Only specimens greater than 3.5 cm in length in the anterior-posterior dimension are included.
-
b.An approximately 0.5 cm segment is removed from the anterior and posterior poles of this specimen for processing. Anterior specimens are taken from a location anterior to the uncal notch.
-
a.
-
2.
Send the middle portion of the hippocampus for neuropathological analysis.
CRITICAL: This pathological analysis is a key step of the tissue exclusion criteria. Specimens with mesial temporal sclerosis (MTS) are excluded unless the scope of the study includes studying the pathogenesis related to MTS. Other neuropathological findings such as astrogliosis are noted and shared along with the study results.
-
3.
Carefully drop anterior and posterior hippocampal specimens into separate 50 mL conical tubes containing ice-cold Neurobasal A Medium after surgical resection. Place the tubes on ice and transport the tissue back to the laboratory within 20 min for processing.
-
4.
Wash tissue in ice cold 1× PBS. Using a scalpel, dissect out ∼25% of each specimen for RNA integrity analysis.
CRITICAL: Since it is not possible to get RNA integrity (RIN) values once tissue is processed as single nuclei, it is important to isolate RNA from a small piece of adjacent tissue. RIN values for each specimen will provide insights into RNA quality for a given sample and can be used as a covariate for computational analysis.
-
5.
Transfer specimens into prechilled and prelabelled Eppendorf tubes. Flash-freeze the tissue in liquid nitrogen.
-
6.
RNA isolation and RIN: Isolate total RNA from the dissected piece (from Step 4) using a standard RNA isolation. Determine RIN values for each sample using a RNA Screen Tape Analysis system (Agilent Technologies) according to the manufacturer’s instructions
CRITICAL: Label the tubes with both case identification number and date of the tissue collection. Update the tissue inventory spreadsheet accordingly. Using the date and time of the surgery can be a more reliable way to track the samples and helps avoid any confusion between the lab and surgical department.
Nuclei isolation
Timing: 30 min
This section includes steps for the complete digestion of the plasma membrane yielding single-nuclei dissociation of human hippocampus. Similar approaches have been used in previous studies (Habib et al., 2017). We adopted this dissociation protocol and the coupled it with a modified sucrose gradient.
-
7.
Using a scalpel, dissect the surgically resected hippocampal tissue into smaller than ∼0.3 cm3 on a petri dish placed on ice. This step should be performed quickly without allowing the tissue to thaw.
-
8.Lyse the tissue pieces
-
a.Transfer tissue pieces to a Dounce homogenizer containing 2 mL of ice-cold Nuclei EZ lysis buffer.
-
b.Use pestle A for 20 strokes, followed by 20 strokes with pestle B while on ice.
-
c.Transfer the homogenate to a 15 mL conical tube, add 2 mL of ice-cold Nuclei EZ lysis buffer and mix by pipetting with a glass pipette. Incubate on ice for 5 min.
-
d.Collect the nuclei by centrifugation at 500 × g for 5 min at 4°C.
-
e.Discard supernatant and carefully resuspend nuclei in 4 mL of ice-cold Nuclei EZ lysis buffer. Incubate on ice for 5 min.
-
f.Repeat the centrifugation at 500 × g for 5 min at 4°C and discard the supernatant.
-
a.
-
9.
Resuspended the nuclei in 500 μL of nuclei suspension buffer (NSB) consisting of 1×PBS, 1%BSA and 0.2 U/ul RNAse inhibitor.
-
10.
Filter nuclei suspension using a Flowmi Cell Strainer (Bel-Art, H13680-0040). Based on the amount of debris in the suspension, it might be hard to filter. Try filtering smaller volumes and using a new filter for every ∼100 μL volume.
Note: At this point of the protocol tissue lysis is complete and nuclei are dissociated in the suspension. However, using adult human brain as starting tissue results in a large amount of debris. Filtering is effective in removing debris larger than nuclei however smaller size cellular and/or myelin debris often remains after filtering. We implemented the following sucrose gradient to obtain a cleaner nuclei suspension.
Sucrose gradient
Timing: 1 h
Preparing a nuclei suspension with minimal debris is key to minimize technical noise in snRNA-seq datasets. Typically, ultracentrifugation at higher speeds has been used to separate subcellular compartments such as nuclei (https://www.protocols.io/view/nuclei-isolation-from-human-brain-using-sucrose-gr-scneave). Working with relatively smaller tissue amounts from human hippocampus, we noticed that there is a high degree of nuclei loss during ultracentrifugation potentially due to the larger volume of tubes used in the ultracentrifuge. Adopting the modified sucrose gradient using a tabletop centrifuge as is detailed below minimized the nuclei loss.
-
11.Remove the debris using a density gradient centrifugation
-
a.Add 500 μL of the sucrose solution (see materials and equipment) to a 2 mL Eppendorf tube.
-
b.Add 900 μL of the sucrose solution to 500 μL of isolated nuclei in NSB.
-
c.Layer 1400 μL nuclei suspension (from Step 11b) to the top of the sucrose solution (from Step 11a).
-
d.Centrifuge this gradient at 13,000 × g for 45 min at 4°C.
-
a.
-
12.Resuspend and wash nuclei pellet
-
a.Carefully remove the myelin layer on the top and the supernatant containing the cellular debris (Figure 1A).
-
b.Resuspend the nuclei pellet in 500 μL NSB and transfer the solution to a 1.5 mL Eppendorf tube.
-
c.Centrifuge at 500 × g for 5 min at 4°C
-
d.Remove the supernatant and resuspend the pellet in 100–300 μL NSB
-
e.Filter the suspension through a 40-μm Flowmi Cell Strainer (#H13680-0040, Bel-Art).
-
a.
Figure 1.
Sucrose gradient for isolation of high-quality nuclei
(A) Images showing phases separated by sucrose gradient.
(B) Nuclei isolated using this protocol stained by Ethidium Homodimer-1. Insets show closeup displaying individual nuclei without DNA blebbing. Scale bar = 50 microns.
Quality control and nuclei count
Timing: 30 min
Here we describe how we evaluate the quality of the nuclei suspension and calculate the nuclei concentration. In terms of quality control, we consider this protocol successful if 1) nuclei appear as a single-nuclei solution and are not clumped together, and 2) nuclei look intact. For these quality control steps the following staining approaches were used.
-
13.Assess nuclei integrity with Ethidium Homodimer-1 (EthD-1) staining.
-
a.Stain 10 μL of nuclei suspension (from 11e) with 4 μM EthD-1 (Figure 1B). EthD-1 emits red fluorescence upon binding to DNA and will show DNA blebbing if the sample quality is low. Visualize stained nuclei under an epifluorescence microscope in the red channel.
-
a.
-
14.Count nuclei with Trypan Blue staining
-
a.Stain 10 μL of nuclei suspension (from Step 11e) with 10 μL of premade 0.4% Trypan Blue.
-
b.Load the stained sample on a hemocytometer and visualize the nuclei under a bright field microscope. Count blue/purple stained nuclei.
-
a.
-
15.
Adjust the final concentration of nuclei to 1000 nuclei/μL with NSB.
-
16.
Continue with preparation of droplet-based single-nuclei RNA-seq libraries using the Chromium Single Cell 3′ kit (10× Genomics) according to the manufacturer’s protocol
Expected outcomes
This protocol results in obtaining a total of ∼50,000–100,000 nuclei per 0.3 cm3 tissue sample. The final number of nuclei varies based on how much white matter is present in the starting tissue piece. In general, anterior hippocampal samples contained more white matter tracks. Single-nuclei RNA-sequencing libraries prepared following these preparations consistently enabled sampling of excitatory neurons, inhibitory neurons, astrocytes, oligodendrocytes, microglia and oligodendrocyte progenitor cells.
Limitations
A sucrose gradient, washes and filtering steps are implemented to obtain intact nuclei without cellular and myelin debris. However, some degree of ambient RNA will still exist in the nuclei suspension and will result in barcoded droplets containing low RNA count during snRNA-seq. These artifacts will then be removed from the dataset by filtering based on the number of transcripts and genes found associated with each cell barcode. Please see (Ayhan et al., 2021) for an examples of filtering criteria.
Here, we used a commercially available sucrose buffer for the sucrose gradient. Theoretically, homemade sucrose solution could be used (https://www.protocols.io/view/nuclei-isolation-from-human-brain-using-sucrose-gr-scneave). However, centrifuge times, speed and the molar concentration for the gradient will need to be optimized for the tabletop centrifuge. Similarly, we used a commercially available lysis buffer. It is possible to use a homemade lysis buffer (e.g., 10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, and 0.1% Nonidet™ P40 Substitute) but lysis times will need to be tested.
Troubleshooting
Problem 1
Nuclei form clumps rather than a single-nuclei solution (step 14b).
Potential solution
Nuclei clumping indicates that nuclei are over lysed. This could be due to lysis time that was too long for the tissue of interest. Repeat the experiment using shorter times for lysis and/or fewer number of strokes with the Dounce homogenizer. In fact, the number of strokes and lysis will have to be optimized for different tissue types and amount of tissue used. Alternatively, this could also occur if the reagents used were warmer than ice-cold. If the tissue sample is too valuable to be discarded, repeating a filtering step might help to remove clumps if there is enough non clumping single-nuclei in the solution.
Problem 2
No visible pellet after sucrose gradient (step 12d).
Potential solution
If the starting tissue amount is low, the pellet containing nuclei after the sucrose gradient will be too small to visualize. In that case, resume the protocol as directed without touching the potential invisible pellet. Adjust the final resuspension volume accordingly as the number of nuclei will be low.
Problem 3
Cell bodies are found in the nuclei suspension (step 14b).
Potential solution
The presence of cell bodies in the trypan blue stained nuclei suspension indicates incomplete lysis. Increasing lysis time and/or cutting the tissue into smaller pieces before starting lysis is recommended.
Problem 4
Number of genes detected is lower than expected after single-nuclei sequencing (downstream result).
Potential solution
Low quality tissue preparations can lead to low quality data after sequencing. The steps for tissue processing should be considered carefully. Addition of RNase inhibitors to the lysis buffer can also help with protecting the RNA during lysis.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Genevieve Konopka (Genevieve.konopka@UTSouthwestern.edu).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
The authors thank the patients for consenting to participate in this research and Haley Moore for providing the images shown in Figure 1A. G.K. is a Jon Heighten Scholar in Autism Research at UT Southwestern. This work was supported by the NIH (NS106447), a UT BRAIN Initiative Seed Grant (366582), the Chilton Foundation, and the National Center for Advancing Translational Sciences of the NIH under the Center for Translational Medicine’s award number UL1TR001105 to G.K. and B.C.L., the NIH (T32DA007290 and T32HL139438) to F.A., the NIH (NS107357) to B.C.L., and the Chan Zuckerberg Initiative, an advised fund of Silicon Valley Community Foundation (HCA-A-1704-01747) and the James S. McDonnell Foundation 21st Century Science Initiative in Understanding Human Cognition – Scholar Award (220020467) to G.K. Some images in the Graphical Abstract are reprinted with permission from Ayhan et al. (2021).
Author contributions
B.C.L. resected the hippocampal tissue. F.A. designed and optimized the protocol. C.D. assisted with tissue collection and banking. F.A. and G.K. wrote the manuscript. G.K. supervised the study.
Declaration of interests
The authors declare no competing interests.
Data and code availability
This study did not generate any unique data sets or code. Please see (Ayhan et al., 2021) for data sets generated using this protocol.
References
- Ayhan F., Kulkarni A., Berto S., Sivaprakasam K., Douglas C., Lega B.C., Konopka G. Resolving cellular and molecular diversity along the hippocampal anterior-to-posterior axis in humans. Neuron. 2021;109:2091–2105.e6. doi: 10.1016/j.neuron.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N., Avraham-Davidi I., Basu A., Burks T., Shekhar K., Hofree M., Choudhury S.R., Aguet F., Gelfand E., Ardlie K. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any unique data sets or code. Please see (Ayhan et al., 2021) for data sets generated using this protocol.



