Abstract
The hippocampus is a highly plastic brain region sensitive to environmental stress. It shows dynamic changes in epigenetic marks associated with stress related learning. Previous work has shown that acute stress induces substantial transient changes in histone H3 lysine 9 trimethylation (H3K9me3). Moreover, increased H3K9me3 is enriched in hippocampal gene deserts accumulating within endogenous retroviruses and transposable elements. We have found that in response to acute glucocorticoid treatment, a similar change in global H3K9me3 is observed. However, when localized we found that H3K9me3 is markedly decreased at B2 short interspersed nuclear elements but not within intracisternal-A particle endogenous retroviruses. Further, decreased H3K9me3 valence within B2 elements was associated with increased transcript abundance. These data demonstrate the capacity for acute glucocorticoids to mobilize transposable elements via epigenetic unmasking. Reconciled with previous findings following acute stress, this suggests the capacity for mobile elements to potentially function as novel regulators given their dynamic regulation by stress and glucocorticoids.
Keywords: Glucocorticoid, Epigenetic, Transposon, B2 SINE, lncRNA, Histone modification, Chromatin
1. Introduction
Stress is an ubiquitous environmental stimuli that is both perceived and adapted to by the brain. Stress has certainly been associated with pathogenesis of numerous neuropsychiatric disorders including anxiety, depression, and post-traumatic stress disorder (Bartlett et al., 2017; Guffanti et al., 2019). Physiologically, stress stimuli produce a signaling cascade through hypothalamic-pituitary-adrenal axis activation, resulting in the release of glucocorticoids which can diffuse and subsequently act on receptors within the brain including the hippocampus. Accumulating evidence shows that both stress and glucocorticoids affect the hippocampus via epigenetic mechanisms, i.e. histone modifications and non-coding RNA expression (Bartlett et al., 2019; Daskalakis et al., 2018a). Further, epigenetic mechanisms and stress have long been linked to the activity of transposable elements and more recent work has revealed an adaptive role for transposons in the nervous system and beyond (Hunter, 2020; Bartlett and Hunter, 2021; McClintock, 1984; Fedoroff, 2012). It has previously been shown that a single acute stress episode produces rapid, global increases in H3K9me3 in the CA1 and dentate gyrus (DG) of the hippocampal formation (Hunter et al., 2009). When changes in H3K9me3 following acute stress were localized, increases were observed in hippocampal gene deserts littered with transposable elements, notably B2 small interspersed nuclear elements (SINEs) and intracisternal-A particle endogenous retroviruses (IAP-ERV) (Hunter et al., 2012). Increased H3K9me3 at B2 SINE and IAP-ERV was associated with transcriptional repression as hippocampal mRNA levels in stressed animals were lower than that of naïve animals. This suggested that the hippocampal H3K9me3 response to acute stress was a transposable element silencing mechanism. Here we show that a single injection of acute corticosterone (CORT) in adrenalectomized animals is sufficient to increase global H3K9me3 levels in the CA1 and DG of the hippocampus. Further, we found that acute CORT treatment produced markedly different responses in H3K9me3 transposon-marking when localized than observed previously under acute stress conditions. For B2 SINE elements, H3K9me3 occupancy was reduced and permissive for increased expression following acute CORT treatment. For IAP-ERV elements, H3K9me3 occupancy was increased but transcript levels were comparable to that of control animals. Here, we show that acute glucocorticoid exposure is sufficient to induce large chromatin changes via H3K9me3 but that B2 SINE elements are epigenetically unmasked and transcriptionally active.
2. Materials and methods
2.1. Animals
All animal protocols were approved by the IACUC at the University of Massachusetts Boston. Adult male Sprague-Dawley rats were ordered adrenalectomized from Charles River Labs and maintained under social housing conditions with standard chow and 0.9% saline drinking water. One hour prior to sacrifice rats were i. p. injected with either ethanol or corticosterone dissolved in ethanol (3 mg/kg). For immunohistochemistry, rats were given a lethal dose of pentobarbitol (i.p.) and fully anesthetized prior to perfusion and fixation with 4% formaldehyde. Brains were extracted and allowed to reach isotonic state in two serial solutions of sucrose (30%). Brains were cut into 35 μm sections using a cryostat. For qRT-PCR and chromatin immunoprecipitation, rats were sacrificed via rapid decapitation. Brains were removed and hippocampi extracted, half the hippocampus was flash frozen for RNA extraction and the other fixed for chromatin immunoprecipitation (ChIP; counterbalanced within groups, i.e. left and right hippocampal tissues were used for both RNA and ChIP and mixed within-group across animals).
2.2. Cell culture
Rat C6 glioma cells were maintained at 37 °C, 5% CO2 in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% Fetal Bovine Serum (FBS) on poly-l-lysine coated dishes (Gibco; Sigma-Aldrich). Cells were grown to 70–80% confluence for experiments. For serum depletion, DMEM was not augmented with FBS. For histone and RNA extraction, after incubation, media was removed and cells were washed twice with phosphate buffered saline (PBS) containing protease inhibitors. Cells were then collected in PBS with protease inhibitors and pelleted. Pelleted cells were then stored at −80 °C until lysis. For ChIP, after 1 h in serum-depleted DMEM the protein-DNA interactions were fixed by crosslinking for 10 min on a shaker by adding fresh 16% formaldehyde to a final concentration of 1% to plate media. The reaction was quenched by adding glycine to a final concentration of 0.125 M for 10 min. Media was then discarded and cells were washed twice with PBS containing protease inhibitors (Pierce Protease Inhibitor tablets, Thermofisher). Cells were then collected in PBS with protease inhibitors and pelleted. Cell pellets were then stored at −80 °C until sonication.
2.3. Immunofluorescence
3-4 dorsal hippocampal sections per animal, selected from −3.1 to −3.8 mm caudal to bregma, were picked and processed together. Primary antibody for H3K9me3 diluted in PBS-0.5% Triton-X was incubated overnight at 4 °C (1:1000, Abcam: ab8898). Secondary antibody raised against rabbit IgG conjugated to Alexafluor-488 was incubated for 2 h at room temperature (1:200, Invitrogen: A-11008). Coverslips were applied with Vectashield hardmount with DAPI. Images were captured using Leica DFC and analyzed using ImageJ.
2.4. Histone extraction and western blotting
Total histone extractions were conducted according to the manufacturer's protocol (Epigentek). Total histone extracts were aliquoted and stored at −80 °C until blotting. Samples were diluted in laemmli buffer with 5% beta-mercaptoethanol heated to 100 °C for 5 min and then cooled to RT. 3 μg of histones were loaded into a precast 4–20% stacked polyacrylamide gel (Bio-rad). The gel was run in 1× running buffer (Invitrogen) for 45 min at 60 V and then for 1 h at 120 V. The gel was then washed with diH2O for 5 min and then allowed to equilibrate for 10 min in transfer buffer, 1X Tris-Glycine with 15% methanol (Boston Bioproducts). PVDF membrane was activated in methanol and allowed to equilibrate for 10 min in transfer buffer. The transfer ran for 1 h at 100 V at 4 °C. The membrane was then blocked for 30 min with 5% (w.v.) non-fat milk 0.1% (w.v.) BSA in 0.1% (v.v.) Tween-20 Tris Buffered Saline (TBS-T). Following blocking, the membrane incubated overnight at 4 °C with the primary antibody for H3K9me3 diluted in blocking solution (1:1000, Abcam: ab8898). The membrane was then washed 3× with TBS-T for 5 min and then incubated with the secondary antibody diluted in blocking solution for 1 h at RT (1:10 K, ThermoFisher: 31460). The membrane was then washed 3× with TBS-T for 5 min and allowed to incubate with ECL substrate for 5 min. Membranes were visualized using biorad chemidoc. The membrane was then stripped by briefly rinsing with 0.1 M NaOH and washing 2× with TBS-T for 5 min. The membrane was then blocked again before reprobing with pan-H3 primary antibody diluted in blocking solution for 1 h at RT (1:25 K, Abcam: ab1791). The membrane was then washed 3× with TBS-T for 5 min before incubation with secondary antibody diluted in blocking solution for 1 h at RT (1:10 K, ThermoFisher: 31460). The membrane was then washed 3× with TBS-T for 5 min before incubation with ECL substrate for 3 min followed by visualization. Blot images were analyzed using NIH ImageJ to calculate relative H3K9me3 to pan-H3 ratio for each sample.
2.5. qRT-PCR
RNA extractions were carried out according to manufacturer's protocol and cDNA generated likewise (Qiagen RNAeasy; Qiagen Quantitect reverse transcription kit). RNA and cDNA concentrations were determined using the Nanodrop 2000 (Thermoscientific). For qRT-PCRs, Sybr Green Master mix was used with the following primers and allowed to run for 40 cycles: B2 FWD 5′AGATGGCTCAGCGGTTAAGA-3’; B2 REV 5′-GACACACCAGAAGAGGGTATCA-3’; IAP FWD 5′-AATTTCTTGAACGGCTCAGCCAGG-3’; IAP REV 5′-TGTTAGCAAACGGTCAGTCGTCCT-3’; 7SK FWD 5′- TCGGTCAAGGGTATACGAGTAG-3’; 7SK REV 5′- TTTGGATGTGTCTGGAGTCTTG-3’. Per1, Myo GRE primers were adopted from Polman et al., 2012). Data was then analyzed using the ΔCt method.
2.6. Chromatin immunoprecipitation
Chromatin immunoprecipitation was carried out on hippocampal tissue as previously described (Bartlett and Hunter, 2019). In short, following decapitation and dissection, tissue was fixed immediately with 1% formaldehyde for 15 min under continuous rotation. Cross-linking reactions were quenched with 0.125 M glycine for 5 min. Tissue was then washed 5 times with ice-cold PBS containing protease inhibitors and pellets flash frozen and stored at −80 °C until processing. Chromatin was prepared using the Covaris ultra-shearing kit according to manufacturer's instructions using the Covaris M220 ultrasonicator to achieve a chromatin smear between 200 and 600bp. Sheared chromatin was then used for H3K9me3 immunoprecipitation with magnetic protein A beads (10 μg/reaction; Abcam: ab8898; 20 μL/reaction; Millipore). For GR-ChIP, 10 μg of primary antibody and 20 μL of protein A magnetic beads were used for immunoprecipitation (Santa Cruz: G-5 sc-393232; Millipore). Cleared, precipitated chromatin was analyzed using the percent input method following qRT-PCR.
2.7. Statistics
We used R for preparation of graphs and statistical analysis. Data are presented as mean ± standard error (SEM). Data was analyzed using a t-test to compare control and CORT treated groups.
3. Results
3.1. Corticosterone induces increase in global H3K9me3 in the hippocampus
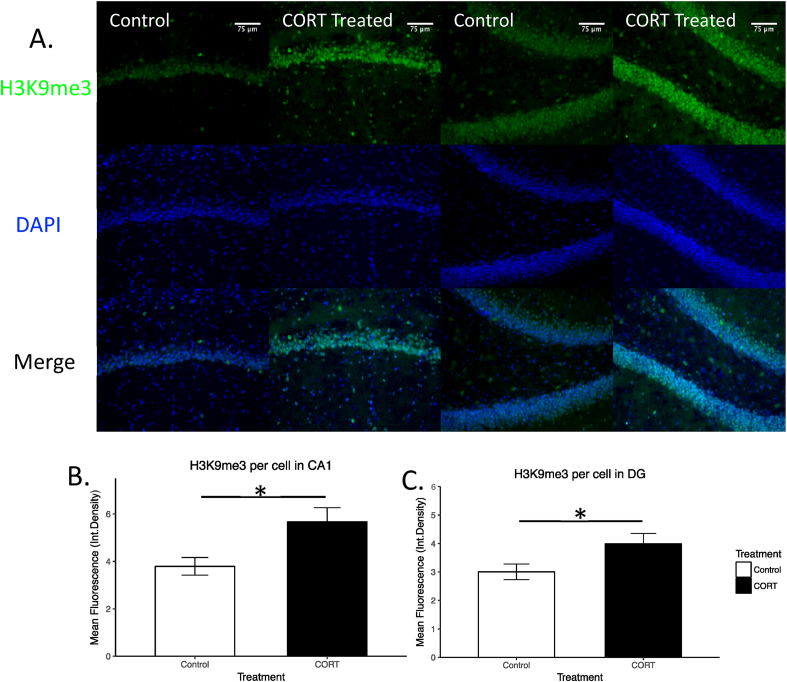
We used immunohistochemistry to test whether CORT increased H3K9me3 levels in the hippocampal formation in adrenalectomized rats. Following acute CORT injection, H3K9me3 levels were increased in the granular regions of the CA1 (Fig. 1A and B) and DG (Fig. 1A and C) subfields compared to vehicle treated animals (p < 0.05, n = 8 per group).
Fig. 1.
Immunofluorescent imaging of H3K9me3 in control and CORT treated rats. Representative images of the CA1 and DG subregions of the hippocampus are shown (A.). H3K9me3 mean fluorescence was higher in CA1 (B.) and DG (C.) following CORT treatment (*p < 0.05, mean ± SEM).
3.2. Corticosterone induces selective H3K9me3 unmasking of B2 SINEs in the hippocampus
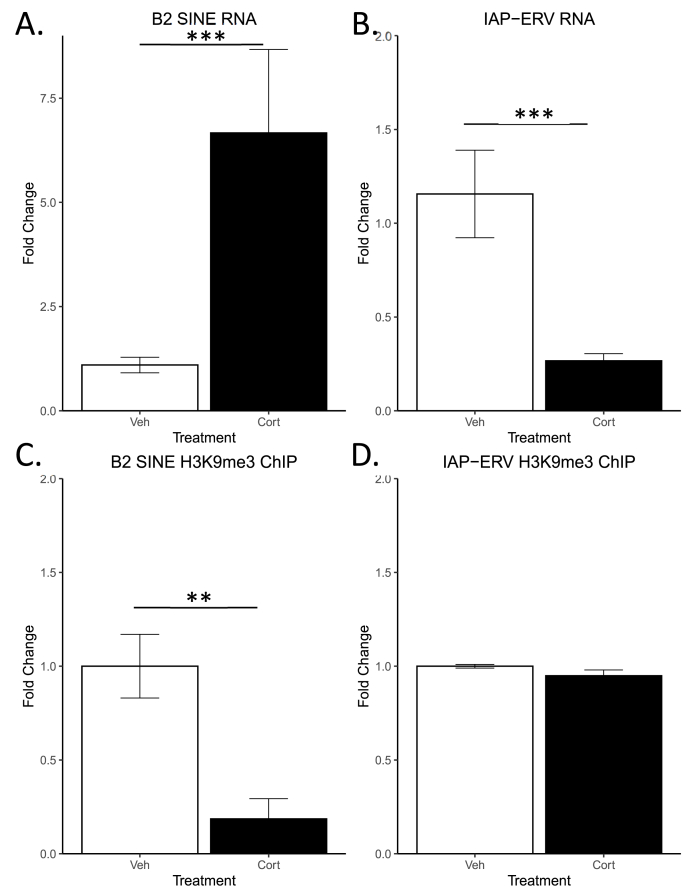
We used qRT-PCR to observe differences in retrotransposable element expression levels following acute CORT treatment. We found that acute CORT increased B2 SINE expression (Fig. 2A, p < 0.001, n = 7 per group) but decreased IAP-ERV expression (Fig. 2B, p < 0.001, n = 7 per group). Concurrently, ChIP-qPCR was employed to assess differences in H3K9me3 occupancy at these transposons. We also observed that H3K9me3 was markedly reduced at B2 SINE elements following acute CORT treatment (Fig. 2C, p < 0.01, n = 7 per group) but was unchanged at IAP-ERV elements (Fig. 2D).
Fig. 2.
B2 SINE and IAP-ERV transcript abundance and H3K9me3 ChIP in the hippocampi of control and CORT treated rats. B2 SINE RNA was markedly increased in CORT treated animals (A.). IAP-ERV RNA was markedly decreased in CORT treated animals (B.). B2 SINE H3K9me3 association was reduced in CORT treated animals (C.) No difference in H3K9me3-IAP-ERV association was observed across groups (D.) (***p < 0.001,**p < 0.01, mean ± SEM).
3.3. Acute corticosterone increases global H3K9me3 in C6 cells
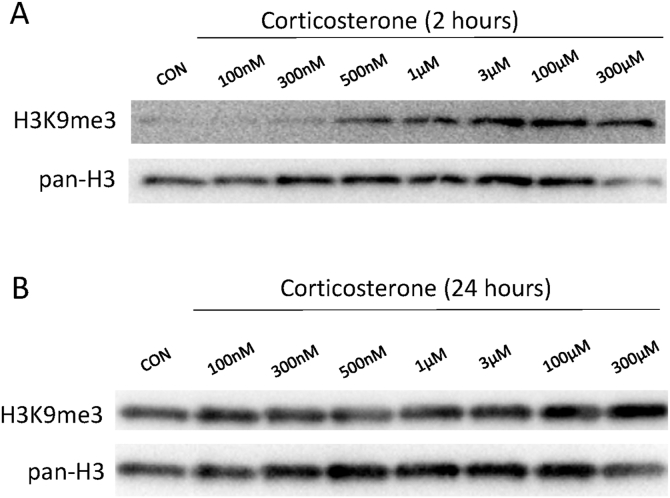
To determine if corticosterone treatment resulted in large changes in heterochromatin, cells were treated with a range of corticosterone concentrations and collected for western blotting of histone extractions. Acute corticosterone treatment, 2 h, produced a logistic curve like dose response for H3K9me3 relative to H3 (Fig. 3A). Conversely, long term corticosterone treatment, 24 h, failed to produce changes in H3K9me3 levels independent of dose (Fig. 3B).
Fig. 3.
Corticosterone increases global H3K9me3 in C6 cells following 2-h incubation.
After 24hr incubation, western blotting for H3K9me3 of histone extracts reveals does not reveal a dose response to CORT (Fig. 3A). However, after 2-h incubation, western blotting for H3K9me3 of histone extracts does appear to produce a logistic dose response curve (Fig. 3B).
3.4. Acute corticosterone induces retrotransposon RNA expression via the glucocorticoid receptor
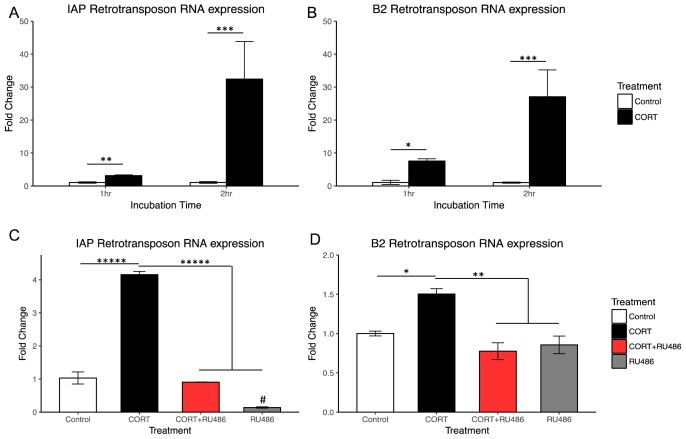
To determine if corticosterone activated transcription of repetitive transposable elements, cells were treated with corticosterone or vehicle for one or 2 h. After both 1 h and 2 h, corticosterone induced IAP-ERV transposon expression (Fig. 4A). Similarly, corticosterone also induced B2 SINE transposon expression (Fig. 4B). Additionally, 1 h co-treatment with GR specific antagonist, RU486, blocks corticosterone induction of IAP (Fig. 4C) and B2 SINE expression (Fig. 4D).
Fig. 4.
RNA expression of IAP-ERV and B2 SINE retrotransposons is increased by CORT treatment in C6 cells via the glucocorticoid receptor.
Fold change compared to control at both 1hr and 2hr is shown for 100 μM CORT treated cells (mean ± SEM). Expression experiments were independently replicated three times to verify the effect, the data shown and analyzed is from the first experiment. After 1hr incubation with 100 μM CORT, IAP-ERV expression is significantly increased by an average of 3.11 fold relative to control (Fig. 4A., Student's t-test, n = 3 per group, p < 0.01). After 2hr incubation with 100 μM CORT, IAP-ERV expression is significantly increased by an average of 32.44 fold relative to control (Fig. 4A., Student's t-test, n = 3 per group, p < 0.001). Likewise, after 1hr incubation with 100 μM CORT, B2 SINE expression is significantly increased by an average of 7.53 fold relative to control (Fig. 4B., Student's t-test, n = 3 per group, p < 0.05). After 2hr incubation with 100 μM CORT, B2 SINE expression is significantly increased by an average of 27.08 fold relative to control (Fig. 4B., Student's t-test, n = 3 per group, p < 0.001). After 1hr incubation with 100 μM CORT, IAP-ERV expression is significantly increased relative to control (Fig. 4C, Tukey's post-hoc, n = 3 per group, p < 0.0001). Co-treatment with GR-specific antagonist, RU486, blocks CORT induction (Fig. 4C, Tukey's post-hoc, n = 3 per group, p < 0.0001). Interestingly, RU486 alone appears to reduce IAP expression compared to control (Fig. 4C, Tukey's post-hoc, n = 3 per group, p < 0.01). Likewise, after 1hr incubation with 100 μM CORT, B2 SINE expression is significantly increased relative to control (Fig. 4D, Tukey's post-hoc, n = 3 per group, p < 0.05). Co-treatment with GR-specific antagonist, RU486, blocks CORT induction (Fig. 4D, Tukey's post-hoc, n = 3 per group, p < 0.01).
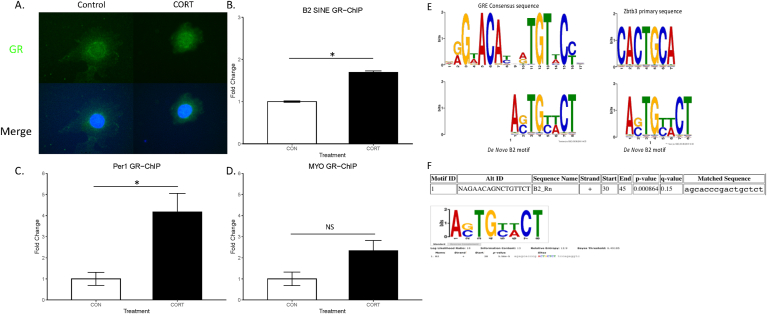
3.5. Acute corticosterone increases glucocorticoid receptor binding to B2 SINE elements
Given the relatively short length of B2 SINE elements, we investigated the idea that the GR may bind to these loci. First, we confirmed previous reports showing that GR activation induces nuclear GR accumulation (Koufali et al., 2003) (Fig. 5A). An exploratory in silico analysis using the MEME suite revealed a potential putative binding site within the B2 SINE consensus sequence (Fig. 5E&F) (Bailey et al., 2009) (Bailey et al., 2009). Therefore, we predicted that acute corticosterone treatment (1 h) may increase GR binding at B2 SINE loci. We observed an increase in GR association to the Per1 GRE following corticosterone treatment (p < 0.05, n = 3; Fig. 5B). We observed no difference in GR association to the Myo between groups (Fig. 5C). These observations are in alignment with previous findings of robust GR association to the Per1 GRE following GR activation but no such association to the Myo gene (Polman et al., 2012, 2013). We observed large increases in GR association to B2 SINE genomic loci following 1 h corticosterone treatment (p < 0.001, n = 3; Fig. 5D). A de novo motif discovery was conducted using MEME with default settings allowing for up to 10 motifs on the B2_Rn RepBase consensus sequence (Bailey et al., 2009; Jurka et al., 2005). De novo motifs were then compared against known eukaryotic vertebrate DNA motifs using TomTom (Gupta et al., 2007). This revealed significant alignment between one de novo motif and the GRE consensus sequence and Zbtb3 primary motif sequence (p < 0.05; Fig. 5E). Previous reports have shown that as much as 58% of GRE containing GR binding sites contain Zbtb3 motifs (Polman et al., 2012, 2013). A complimentary in silico approach using FIMO revealed significant alignment with the Homer GR consensus motif within the B2 SINE consensus sequence (p < 0.001; Fig. 5F) (Grant et al., 2011). The FIMO alignment overlapped with the location of the de novo B2 motif within the B2 consensus sequence between bp 30–45 on the + strand, together suggesting a strong likelihood of GR binding to B2 SINE elements (Fig. 5F).
Fig. 5.
Acute corticosterone incubation increases glucocorticoid receptor binding at B2 SINE elements.
C6 cells were incubated with either 100 μM CORT or vehicle for 1 h prior to cross-linking. As shown above, GR localization is observed in the nucleus following corticosterone treatment (Fig. 5A). Interestingly, GR association with B2 SINE elements is higher in corticosterone treated cells compared to vehicle treated cells (Fig. 5B; mean ± SEM, student's t-test, n = 3 per group, p < 0.001) suggesting that GR activation leads to transient binding at B2 SINE loci. GR association with a known Per1 GRE is higher in corticosterone treated cells compared to vehicle treated cells (Fig. 5C, student's t-test, n = 3 per group, p < 0.05). Conversely, GR association with Myo gene did not vary between conditions (Fig. 5C). A de novo motif discovery from B2 SINE showed significant overlap with glucocorticoid response element (GRE) consensus sequence as well as GR cofactor Zbtb3 primary sequence (Fig. 5E). Similarly, motif search for the consensus GRE identified a GR-binding site within B2 SINEs (Fig. 5F).
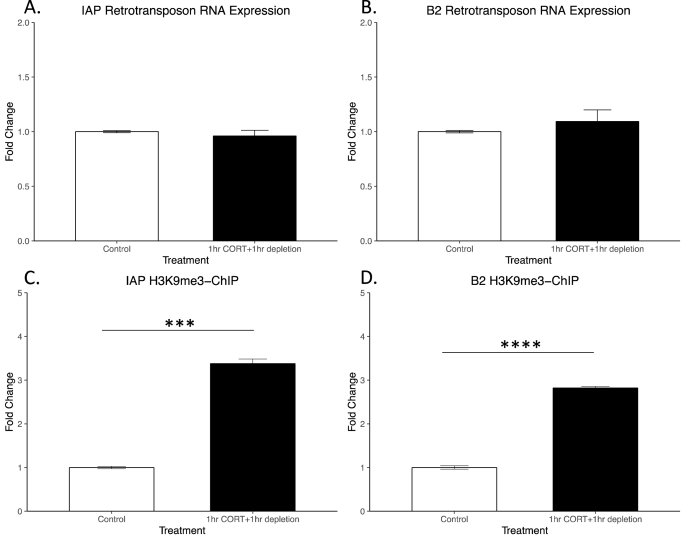
3.6. Acute corticosterone followed by serum depletion attenuates retrotransposon RNA expression
We previously observed that corticosterone was sufficient to induce IAP and B2 SINE RNA expression in a GR dependent manner (Fig. 4). Therefore, we aimed to determine if changes in heterochromatin, i.e. H3K9me3, acted in a compensatory fashion in response to corticosterone induced retrotransposon activation. The rationale for this experiment was that in vivo circulating glucocorticoids rise in response to acute stress but under normal circumstances quickly return to baseline. Therefore, we attempted to model this transient increase in glucocorticoids in culture. We observed that following 1 h of corticosterone treatment and then 1 h of serum depletion effectively attenuated retrotransposon RNA expression (Fig. 6A and B). This attenuation in expression coincided with increased H3K9me3 at retrotransposon loci as shown via H3K9me3 ChIP-PCR (p < 0.001, n = 3; Fig. 6C and D). Increased H3K9me3 occupancy at retrotransposon loci following serum depletion suggests that H3K9me3 acts to suppress corticosterone induced retrotransposon activation.
Fig. 6.
Serum depletion following corticosterone incubation attenuates retrotransposon expression and increases H3K9me3 occupancy at retrotransposon loci.
C6 cells were incubated with either 100 μM CORT or vehicle serum augmented media for 1 h then media for both conditions was changed with serum-depleted DMEM. After 1 h in serum-depleted media, cells were harvested for RNA extraction. As shown above, IAP-ERV (Fig. 6A) and B2 SINE (Fig. 6B) expression is reduced to control levels following 1hr depletion. Increased H3K9me3 accumulation at RTs is observed after 1hr 100 μM CORT incubation followed by 1hr serum depletion. Percent input is shown for H3K9me3-ChIP followed by qPCR (mean ± SEM). H3K9me3 binding of IAP-ERV elements is significantly higher after 1hr 100 μM CORT incubation followed by 1hr serum depletion compared to control (Fig. 6C, student's t-test, n = 3 per group, p < 0.001). H3K9me3 binding of B2 elements is significantly higher after 1hr 100 μM CORT incubation followed by 1hr serum depletion compared to control (Fig. 6D, student's t-test, n = 3 per group, p < 0.0001). Increased H3K9me3 at RT after 1hr 100 μM CORT incubation followed by 1hr serum depletion is correlated with reduced RT expression, returning to control levels, suggesting that H3K9me3 silences RT expression following CORT depletion.
4. Discussion
The hippocampus is a brain region that dynamically regulates gene accessibility via chromatin modifications in response to acute stress and glucocorticoids. Previous reports have demonstrated large increases in H3K9me3 in the CA1 and DG regions of the hippocampal formation following acute stress (Hunter et al., 2009, 2012, 2013). Our data suggests that these changes may be driven at least in part by hypothalamic-pituitary-adrenal axis activation via CORT. Further, CORT-induced changes in H3K9me3 appear specific and are not observed across stress and glucocorticoid sensitive brain regions (Fig. S1). Yet, the increase in hippocampal H3K9me3 following acute stress localizes to transposable elements and appears permissive for transcriptional repression (Hunter et al., 2012). We have observed that in fact H3K9me3 occupancy at B2 SINEs is reduced by CORT treatment and remains unchanged at IAP-ERVs. The H3K9me3 unmasking of H3K9me3 at B2 SINEs was also associated with increased RNA expression. One possible explanation for reconciling these findings is that the time course for these studies vary significantly. The acute stress work on H3K9me3 valence at retrotransposons implemented an acute stress period, 45 min, followed by 45 min home cage recovery prior to sacrifice. Therefore, at time of sacrifice CORT levels have largely returned to baseline. This study utilized a high dose of CORT followed by sacrifice an hour later. Perhaps, Hunter et al. observed a compensatory homeostatic response of retrotransposon re-silencing by H3K9me3 after stress- and/or glucocorticoid-induced activation (Hunter et al., 2012). This is in line with numerous studies demonstrating glucocorticoid action with the recovery phase of the stress response. Interestingly, observed differential effects of CORT on B2 SINEs and IAP-ERVs activation and silencing, possibly suggesting different time courses across transposon families. Evidence of family specific regulation by glucocorticoids was observed in the rat hippocampus as we did not observe CORT dependent increases in the expression of either LINE1 ORF1 or LINE1 ORF2 or expression of LINE1s in C6 cells (Supplementary Fig. 4). Further studies are needed to establish the time course of both CORT and stress activation of transposons as well as the time course of transposon re-silencing.
In line with our finding that CORT facilitates epigenetic activation of B2 SINEs, activation of transposable elements by environmental stress have been observed in several contexts. For instance, in response to repeated cocaine exposure H3K9me3 valence was reduced at transposable element loci and associated with increased transposon expression in the nucleus accumbens (Maze et al., 2011). Similarly, Linker et al. have consistently observed transient transcriptional activation of B2 SINEs in the mouse hippocampus following novel environment exposure. First, they showed that B2 SINE RNA was enriched in fos + DG cells following novel environment exposure (Lacar et al., 2016). Subsequently, this group confirmed that these B2 SINE RNAs were in fact bona fide transcripts featuring the necessary machinery for independent transcriptional activation - not simply an artifact of “read-through” transcriptional events (Linker et al., 2020). These findings suggest the capacity for hippocampal B2 SINE transposons to be transcriptionally activated on the same time scale as immediate early genes by acute stress, supporting a potential role for rapid activation B2 SINEs by corticosteroids. Likewise, cellular stress, i.e. heat shock, has also been shown to rapidly activate B2 SINEs though whether this activation is due to epigenetic unmasking remains unanswered (Allen et al., 2004; Kugel and Goodrich, 2006). Within this context, B2 SINE was shown to regulate “housekeeping gene” expression of at least one known sexually dimorphic gene, beta-actin (Espinoza et al., 2004; Yakovchuk et al., 2009). While these experiments have only included males, preliminary findings have suggested that hippocampal B2 SINE transcript abundance may be sexually dimorphic (Lambert et al., 2019). Future studies leveraging next-generation sequencing approaches are needed to determine both the presence of internal promoters within CORT activated transposon transcripts, so called bona fide transcripts, as well as the precise genomic loci of transposon-derived RNA (Linker et al., 2020; Guffanti et al., 2018).
Numerous stress-related disorders, for example PTSD and MDD, show evidence of epigenetic influences related to acute stress or trauma (Daskalakis et al., 2018b). Additionally, the hallmark of these disorders is the improper regulation of peripheral endocrine HPA axis response (Yehuda and Seckl, 2011; Adam et al., 2010). Many large genome-wide association studies have failed to identify significant gene(s) associated with neuropsychiatric disease pathology (consortium and Spar, 2015; Duncan et al., 2018). Therefore, understanding the basic mechanisms by which glucocorticoids interact with the deep non-coding genome is paramount to understanding the complex molecular physiology associated with HPA axis dysregulation and may be pertinent to understanding neuropsychiatric disease states (Bartlett and Hunter, 2018). Here, we have demonstrated that glucocorticoids can dynamically induce retrotransposon expression and that epigenetic machinery appears to control the glucocorticoid dependent window of expression (Fig. 5). The current study suggests that perhaps the findings of Maze et al. and Hunter et al. demonstrating the ability of the environment to induce changes in H3K9me3 at repetitive transposon loci in both the nucleus accumbens following chronic cocaine exposure and in the hippocampus following acute stress, respectively, represent compensatory mechanisms to regulate environmentally induced repetitive element expression (10, 20). This idea is further supported by the findings of Lacar & Linker et al. showing by single cell sequencing that in the dentate gyrus following novel environment stress exposure recently active neurons, c-fos positive, show robust B2 SINE expression compared to other local neurons (21). In light of our own findings, it is tempting to posit that perhaps stress induced B2 SINE expression on the same temporal scale as immediate early gene expression may in part be driven by circulating glucocorticoids.
It is worth noting that for many neuropsychiatric diseases, large genome wide association studies have failed to identify significant, potentially casual, links between protein coding gene mutations and subsequent pathology (consortium and Spar, 2015; Duncan et al., 2018). This suggests that epigenetic mechanisms and or the non-coding genome may likely prove fruitful in furthering our understanding of the molecular underpinnings of neuropsychiatric disease. For instance, many transposable elements feature cis-regulatory elements that can function as promoters and drive neighboring gene expression (Mager and Stoye, 2015; Chuong et al., 2017). Therefore, changes in the chromatin landscape at these non-coding elements can lead to altered proximal gene expression (Lunyak et al., 2007). Transposable elements have appeared to co-evolved with host genomes and adapted to recognize host transcription factors (Rebollo et al., 2012; Sundaram et al., 2014). For instance, many Alu SINE elements, the primate specific B2 SINE-like analog, feature both direct and inverted steroid response motifs while others appear necessary for steroid receptor binding to motif “half-sites” (Babich et al., 1999; Bolotin et al., 2011; Polak and Domany, 2006; Jacobsen et al., 2009). Recent work has suggested an emerging role for transposable element RNA in regulating local inflammatory responses and housekeeping gene expression (Allen et al., 2004; Espinoza et al., 2004; Yakovchuk et al., 2009; Kaneko et al., 2011; Tarallo et al., 2012). In the former context, accumulation of Alu retrotransposon RNA following DICER1 loss of function facilitated NLRP3 inflammasome activation and subsequent macular degeneration (Tarallo et al., 2012; Bam et al., 2017). Interestingly, DICER loss of function has been associated with increased inflammation and PTSD diagnosis and also has been shown to be predictive of PTSD status comorbid with depression (Bam et al., 2017; Wingo et al., 2015). In the latter, both Alu and B2 SINE elements were found upregulated in response to heat shock (Liu et al., 1995; Fornace and Mitchell, 1986). When elucidated, these RNA were found to directly interact with RNA Pol II to pause transcription at a number of housekeeping genes (Allen et al., 2004; Espinoza et al., 2004; Yakovchuk et al., 2009). Further, the processing of SINE RNA appears to be necessary to relieve housekeeping gene repression and most recently aberrant SINE processing has been associated with Alzheimer's disease (Zovoilis et al., 2016; Cheng et al., 2021). It stands to reason that potentially in other contexts, i.e. acute stress or corticosterone challenge, these RNAs are acting as master regulators of gene expression changes and may represent a novel basis for understanding stress-related brain disorders.
5. Conclusions
While large scale hippocampal chromatin reorganization is observed in response to acute CORT treatment these changes are similar but distinct from those seen in response to acute restraint stress. Acute CORT depletes H3K9me3 occupancy at B2 SINEs and increases B2 SINE transcript abundance while remaining unchanged at IAP-ERVs. IAP-ERVs transcripts are downregulated by CORT treatment but do not show differences in H3K9me3 occupancy. This may suggest that silencing of IAP-ERVs is facilitated via other epigenetic mechanisms or that any change in H3K9me3 occupancy is localized to a region outside our amplicon. The precise timings of these events as well as their potential relevance for affecting cellular processes remain open questions. Data from C6 cells show that CORT treatment rapidly induces global changes H3K9me3 and B2 SINE and IAP-ERV RNA expression. GR binding to the genomic B2 SINE was observed and may represent one potential mechanism by which CORT drives retrotransposon RNA expression. After CORT depletion, retrotransposon RNA expression returns to baseline and is accompanied by H3K9me3 occupancy at these loci suggesting a compensatory epigenetic response to CORT induced-expression. Together these data highlight the capacity for stress and glucocorticoids to interact with the deep genome via the glucocorticoid receptor.
CRediT authorship contribution statement
A.A. Bartlett: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft, Writing – review & editing. H. DeRosa: Investigation, Writing – review & editing. M. Clark: Investigation, Writing – review & editing. H.E. Lapp: Investigation, Writing – review & editing. G. Guffanti: Conceptualization, Writing – review & editing. R.G. Hunter: Conceptualization, Supervision, Resources, Funding acquisition, Writing – review & editing.
Declaration of competing interest
No authors have any conflict of interest to disclose, and there has been no financial support that might influence this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2021.100397.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Adam E.K., Doane L.D., Zinbarg R.E., Mineka S., Craske M.G., Griffith J.W. Prospective prediction of major depressive disorder from cortisol awakening responses in adolescence. Psychoneuroendocrinology. 2010;35:921–931. doi: 10.1016/j.psyneuen.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T.A., Von Kaenel S., Goodrich J.A., Kugel J.F. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- Babich V., Aksenov N., Alexeenko V., Oei S.L., Buchlow G., Tomilin N. Association of some potential hormone response elements in human genes with the Alu family repeats. Gene. 1999;239:341–349. doi: 10.1016/s0378-1119(99)00391-1. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. Meme suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bam M., Yang X., Zumbrun E.E., Ginsberg J.P., Leyden Q., Zhang J., Nagarkatti P.S., Nagarkatti M. Decreased AGO2 and DCR1 in PBMCs from War Veterans with PTSD leads to diminished miRNA resulting in elevated inflammation. Transl. Psychiatry. 2017;7:e1222. doi: 10.1038/tp.2017.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett A.A., Hunter R.G. Transposons, stress and the functions of the deep genome. Front Neuroendocrinol. 2018;49:170–174. doi: 10.1016/j.yfrne.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Bartlett A.A., Hunter R.G. Chromatin immunoprecipitation techniques in neuropsychiatric research. Methods Mol. Biol. 2019;2011:633–645. doi: 10.1007/978-1-4939-9554-7_36. [DOI] [PubMed] [Google Scholar]
- Bartlett A.A., Singh R., Hunter R.G. Anxiety and epigenetics. Adv. Exp. Med. Biol. 2017;978:145–166. doi: 10.1007/978-3-319-53889-1_8. [DOI] [PubMed] [Google Scholar]
- Bartlett A.A., Hunter R.G. In: Stress: Genetics, Epigenetics and Genomics. Fink G., editor. Vol. 4. Academic Press; 2021. pp. 119–124.http://www.sciencedirect.com/science/article/pii/B9780128131565000108 [Google Scholar]
- Bartlett A.A., Lapp H.E., Hunter R.G. Epigenetic mechanisms of the glucocorticoid receptor. Trends Endocrinol. Metabol.: TEM (Trends Endocrinol. Metab.) 2019;30(11):807–818. doi: 10.1016/j.tem.2019.07.003. [DOI] [PubMed] [Google Scholar]
- Bolotin E., Chellappa K., Hwang-Verslues W., Schnabl J.M., Yang C., Sladek F.M. Nuclear receptor HNF4α binding sequences are widespread in Alu repeats. BMC Genom. 2011;12:560. doi: 10.1186/1471-2164-12-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Saville L., Gollen B., Veronesi A.A., Mohajerani M., Joseph J.T., Zovoilis A. Increased Alu RNA processing in Alzheimer brains is linked to gene expression changes. EMBO Rep. 2021;22 doi: 10.15252/embr.202052255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong E.B., Elde N.C., Feschotte C. Nat. Rev. Genet. 2017;18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONVERGE consortium, Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalakis N.P., Provost A.C., Hunter R.G., Guffanti G. Noncoding RNAs: stress, glucocorticoids, and posttraumatic stress disorder. Biol. Psychiatr. 2018;83:849–865. doi: 10.1016/j.biopsych.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Daskalakis N.P., Provost A.C., Hunter R.G., Guffanti G. Noncoding RNAs: stress, glucocorticoids, and posttraumatic stress disorder. Biol. Psychiatr. 2018;83:849–865. doi: 10.1016/j.biopsych.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Duncan L.E., Ratanatharathorn A., Aiello A.E., Almli L.M., Amstadter A.B., Ashley-Koch A.E., Baker D.G., Beckham J.C., Bierut L.J., Bisson J., Bradley B., Chen C.-Y., Dalvie S., Farrer L.A., Galea S., Garrett M.E., Gelernter J.E., Guffanti G., Hauser M.A., Johnson E.O., Kessler R.C., Kimbrel N.A., King A., Koen N., Kranzler H.R., Logue M.W., Maihofer A.X., Martin A.R., Miller M.W., Morey R.A., Nugent N.R., Rice J.P., Ripke S., Roberts A.L., Saccone N.L., Smoller J.W., Stein D.J., Stein M.B., Sumner J.A., Uddin M., Ursano R.J., Wildman D.E., Yehuda R., Zhao H., Daly M.J., Liberzon I., Ressler K.J., Nievergelt C.M., Koenen K.C. Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Mol. Psychiatr. 2018;23:666–673. doi: 10.1038/mp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza C.A., Allen T.A., Hieb A.R., Kugel J.F., Goodrich J.A. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- Fedoroff N.V. McClintock's challenge in the 21st century. Proc. Natl. Acad. Sci. U. S. A. 2012;109:20200–20203. doi: 10.1073/pnas.1215482109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace A.J., Mitchell J.B. Induction of B2 RNA polymerase III transcription by heat shock: enrichment for heat shock induced sequences in rodent cells by hybridization subtraction. Nucleic Acids Res. 1986;14:5793–5811. doi: 10.1093/nar/14.14.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C.E., Bailey T.L., Noble W.S. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti G., Bartlett A., Klengel T., Klengel C., Hunter R., Glinsky G., Macciardi F. Novel bioinformatics approach identifies transcriptional profiles of lineage-specific transposable elements at distinct loci in the human dorsolateral prefrontal cortex. Mol. Biol. Evol. 2018;35:2435–2453. doi: 10.1093/molbev/msy143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti G., Bartlett A., DeCrescenzo P., Macciardi F., Hunter R. Transposable elements. Curr. Top Behav. Neurosci. 2019;42:221–246. doi: 10.1007/7854_2019_112. [DOI] [PubMed] [Google Scholar]
- Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter R.G. Stress, adaptation, and the deep genome: why transposons matter. Integr. Comp. Biol. 2020;60:1495–1505. doi: 10.1093/icb/icaa050. [DOI] [PubMed] [Google Scholar]
- Hunter R.G., McCarthy K.J., Milne T.A., Pfaff D.W., McEwen B.S. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter R.G., Murakami G., Dewell S., Seligsohn M., Baker M.E.R., Datson N.A., McEwen B.S., Pfaff D.W. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc. Natl. Acad. Sci. U.S.A. 2012;109:17657–17662. doi: 10.1073/pnas.1215810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter R.G., McEwen B.S., Pfaff D.W. Environmental stress and transposon transcription in the mammalian brain. Mobile Genet. Elem. 2013;3 doi: 10.4161/mge.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen B.M., Jambal P., Schittone S.A., Horwitz K.B. ALU repeats in promoters are position-dependent Co-response elements (coRE) that enhance or repress transcription by dimeric and monomeric progesterone receptors. Mol. Endocrinol. 2009;23:989–1000. doi: 10.1210/me.2009-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kaneko H., Dridi S., Tarallo V., Gelfand B.D., Fowler B.J., Cho W.G., Kleinman M.E., Ponicsan S.L., Hauswirth W.W., Chiodo V.A., Karikó K., Yoo J.W., Lee D., Hadziahmetovic M., Song Y., Misra S., Chaudhuri G., Buaas F.W., Braun R.E., Hinton D.R., Zhang Q., Grossniklaus H.E., Provis J.M., Madigan M.C., Milam A.H., Justice N.L., Albuquerque R.J.C., Blandford A.D., Bogdanovich S., Hirano Y., Witta J., Fuchs E., Littman D.R., Ambati B.K., Rudin C.M., Chong M.M.W., Provost P., Kugel J.F., Goodrich J.A., Dunaief J.L., Baffi J.Z., Ambati J. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koufali M.-M., Moutsatsou P., Sekeris C.E., Breen K.C. The dynamic localization of the glucocorticoid receptor in rat C6 glioma cell mitochondria. Mol. Cell. Endocrinol. 2003;209:51–60. doi: 10.1016/j.mce.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Kugel J.F., Goodrich J.A. Beating the heat: a translation factor and an RNA mobilize the heat shock transcription factor HSF1. Mol. Cell. 2006;22:153–154. doi: 10.1016/j.molcel.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Lacar B., Linker S.B., Jaeger B.N., Krishnaswami S., Barron J., Kelder M., Parylak S., Paquola A., Venepally P., Novotny M., O'Connor C., Fitzpatrick C., Erwin J., Hsu J.Y., Husband D., McConnell M.J., Lasken R., Gage F.H. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun. 2016;7:11022. doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert K., Hunter R.G., Bartlett A.A., Lapp H.E., Kent M. In search of optimal resilience ratios: differential influences of neurobehavioral factors contributing to stress-resilience spectra. Front. Neuroendocrinol. 2019:100802. doi: 10.1016/j.yfrne.2019.100802. [DOI] [PubMed] [Google Scholar]
- Linker S.B., Randolph-Moore L., Kottilil K., Qiu F., Jaeger B.N., Barron J., Gage F.H. Identification of bona fide B2 SINE retrotransposon transcription through single-nucleus RNA-seq of the mouse hippocampus. Genome Res. 2020;30:1643–1654. doi: 10.1101/gr.262196.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.M., Chu W.M., Choudary P.V., Schmid C.W. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak V.V., Prefontaine G.G., Núñez E., Cramer T., Ju B.-G., Ohgi K.A., Hutt K., Roy R., García-Díaz A., Zhu X., Yung Y., Montoliu L., Glass C.K., Rosenfeld M.G. Developmentally regulated activation of a SINE B2 repeat as a domain boundary in organogenesis. Science. 2007;317:248–251. doi: 10.1126/science.1140871. [DOI] [PubMed] [Google Scholar]
- Mager D.L., Stoye J.P. Mammalian endogenous retroviruses. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0009-2014. [DOI] [PubMed] [Google Scholar]
- Maze I., Feng J., Wilkinson M.B., Sun H., Shen L., Nestler E.J. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3035–3040. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Polak P., Domany E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC Genom. 2006;7:133. doi: 10.1186/1471-2164-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polman J.A.E., Welten J.E., Bosch D.S., de Jonge R.T., Balog J., van der Maarel S.M., de Kloet E.R., Datson N.A. A genome-wide signature of glucocorticoid receptor binding in neuronal PC12 cells. BMC Neurosci. 2012;13:118. doi: 10.1186/1471-2202-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polman J.A.E., Kloet D., Ronald E., Datson N.A. Two populations of glucocorticoid receptor-binding sites in the male rat hippocampal genome. Endocrinology. 2013;154:1832–1844. doi: 10.1210/en.2012-2187. [DOI] [PubMed] [Google Scholar]
- Rebollo R., Farivar S., Mager D.L. C-GATE - catalogue of genes affected by transposable elements. Mobile DNA. 2012;3:9. doi: 10.1186/1759-8753-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V., Cheng Y., Ma Z., Li D., Xing X., Edge P., Snyder M.P., Wang T. Genome Res. 2014;24:1963–1976. doi: 10.1101/gr.168872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarallo V., Hirano Y., Gelfand B.D., Dridi S., Kerur N., Kim Y., Cho W.G., Kaneko H., Fowler B.J., Bogdanovich S., Albuquerque R.J.C., Hauswirth W.W., Chiodo V.A., Kugel J.F., Goodrich J.A., Ponicsan S.L., Chaudhuri G., Murphy M.P., Dunaief J.L., Ambati B.K., Ogura Y., Yoo J.W., Lee D., Provost P., Hinton D.R., Núñez G., Baffi J.Z., Kleinman M.E., Ambati J. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingo A.P., Almli L.M., Stevens J.S., Stevens J.J., Klengel T., Uddin M., Li Y., Bustamante A.C., Lori A., Koen N., Stein D.J., Smith A.K., Aiello A.E., Koenen K.C., Wildman D.E., Galea S., Bradley B., Binder E.B., Jin P., Gibson G., Ressler K.J. DICER1 and microRNA regulation in post-traumatic stress disorder with comorbid depression. Nat. Commun. 2015;6:10106. doi: 10.1038/ncomms10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovchuk P., Goodrich J.A., Kugel J.F. B2 RNA and Alu RNA repress transcription by disrupting contacts between RNA polymerase II and promoter DNA within assembled complexes. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5569–5574. doi: 10.1073/pnas.0810738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R., Seckl J. Minireview: stress-related psychiatric disorders with low cortisol levels: a metabolic hypothesis. Endocrinology. 2011;152:4496–4503. doi: 10.1210/en.2011-1218. [DOI] [PubMed] [Google Scholar]
- Zovoilis A., Cifuentes-Rojas C., Chu H., Hernandez A., Lee J. Destabilization of B2 RNA by EZH2 activates the stress response. Cell. 2016;167:1788–1802. doi: 10.1016/j.cell.2016.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.