Abstract
Artificial intelligence (AI) refers to the use of computational techniques to mimic human thought processes and learning capacity. The past decade has seen a rapid proliferation of AI developments for cardiovascular computed tomography (CT). These algorithms aim to increase efficiency, objectivity, and performance in clinical tasks such as image quality improvement, structure segmentation, quantitative measurements, and outcome prediction. By doing so, AI has the potential to streamline clinical workflow, increase interpretative speed and accuracy, and inform subsequent clinical pathways. This review covers state-of-the-art AI techniques in cardiovascular CT and the future role of AI as a clinical support tool.
INTRODUCTION
Computed tomography (CT) is widely used for the noninvasive diagnosis and risk stratification of cardiovascular disease. Through advancements in scanner technology, an increasing role in clinical pathways, and the generation of large 3D imaging datasets, cardiovascular CT is well-primed for artificial intelligence (AI) applications. The ultimate goal of these algorithms is to increase efficiency and objectivity in clinical tasks such as image quality improvement, segmentation of structures, quantitative measurements, and outcome prediction1. As these AI techniques permeate into daily clinical practice, cardiologists and radiologists alike would benefit from the fundamental tools and concepts with which to understand, evaluate, and facilitate the use of this technology. This review summarizes state-of-the-art AI techniques in cardiovascular CT and discusses the future role of AI as a clinical support tool.
ARTIFICIAL INTELLIGENCE BASICS
In 1950, Alan Turing designed the “Turing test” to determine whether a machine could exhibit intelligent behavior equivalent to, or indistinguishable from, that of a human2. Although nascent at the time, this concept of AI catalyzed the rapidly growing field of computer science which aims to mimic human thought processes, learning capacity, and knowledge storage. Machine learning (ML) is a subdiscipline of AI which enables computer algorithms to progressively learn and make predictions from data without being explicitly programmed for a given task. The two most popular categories of ML are: (i) supervised learning, in which algorithms are trained to predict a known outcome using labeled data, with examples being linear regression, support vector machines, and random forests; and (ii) unsupervised learning, whereby algorithms are trained to identify new patterns within an unlabeled dataset, as seen in clustering and principal component analysis3. Deep learning (DL) is a specific type of ML which uses multilayered artificial neural networks that can generate automated predictions from input data. The most commonly used DL networks in medical image analysis are convolutional neural networks (CNNs)3. These comprise multiple layers, including convolution layers which apply a filter to an input image to create a map summarizing the presence of detected features (measurable characteristics); and pooling layers which reduce the spatial volume of the feature map for computational efficiency. A fully convolutional network consists entirely of convolution layers, and is typically used for object detection and voxel-based classification.
Assessing the accuracy of AI algorithms in cardiovascular CT is a crucial step in their pathway to clinical implementation, and many image-based performance metrics exist. The Dice similarity coefficient (DSC), typically used to evaluate image segmentation, quantifies the similarity between the algorithm-predicted segmentation and the ground truth, and ranges from 0 (no similarity) to 1 (identical)4. The interclass correlation coefficient (ICC), commonly used to assess conformity among observers, can also rate the agreement between AI-based and human expert measurements. The area under the receiver operator characteristic curve (AUC) is widely used to evaluate and compare the performance of ML classification models, and represents the probability that a model ranks a random positive example more highly than a random negative example across all possible classification thresholds5.
CURRENT AI APPLICATIONS IN CARDIOVASCULAR CT
Image Quality Improvement
Given significant innovations in CT hardware over the past two decades, there is now a demand for image reconstruction algorithms to become more accurate and computationally efficient. This can be achieved through DL, whereby CNNs use lower quality images – rapidly obtained from raw data – as input to create a higher quality reconstructed image.
Denoising low-dose images
CNNs have been employed for the denoising of noncontrast cardiac CT scans acquired at a low radiation dose. Wolterink et al6. trained a CNN to transform low-dose CT images into routine-dose CT images, while a second discriminator CNN simultaneously distinguished the output of the generator from real routine-dose CTs and provided feedback during training. This resulted in substantial noise reduction when tested on clinical noncontrast cardiac CTs and enabled accurate coronary artery calcium (CAC) scoring from low-dose scans with high noise levels. Prior to this work, CNN-based noise reduction algorithms predicted the mean of voxel values from the reference routine-dose image and the denoised low-dose image7,8, resulting in smoothed images without the texture typical of routine-dose CT. In this study, feedback from the discriminator CNN prevented image smoothing and enabled more accurate quantification of low-density CAC. A potential limitation of this approach is the risk of introducing pathologies that are not actually present in the image, based on their presence in the training dataset. Hence, future work will need to focus on estimating the certainty of this method at each location in the image.
Low-dose CCTA permits coronary assessment at a significantly reduced radiation dose, however at the expense of increased data noise. Several CNN-based denoising approaches have been designed and evaluated in experimental studies. Green et al.9 trained a fully convolutional network for the per-voxel prediction of routine-dose Hounsfield unit (HU) values from simulated low-dose CCTA images. This resulted in a higher peak signal-to-noise ratio compared with state-of-the-art denoising algorithms, and more accurate reconstructions of the coronaries by clinical reporting software. Kang et al.10 used a CNN to learn the mapping between low- and high-dose cardiac phases in multiphase CCTA datasets, resulting in effective noise reduction in the low-dose CT images while preserving detailed texture and edge information. Following validation in further studies, the mentioned techniques for denoising non-contrast CT and CCTA may be generalizable to different CT acquisition protocols and image reconstruction software tools in clinical practice.
Motion artifact correction
Cardiac motion artifacts frequently reduce the interpretability of CCTA images and hinder CAD assessment. Lossau et al.11 trained CNNs to accurately estimate the direction and magnitude of coronary motion, before implementing them into an iterative motion compensation pipeline. This significantly reduced levels of motion artifact when applied to real clinical cases, especially in images with severe artifacts. Jung et al.12 used synthetic images without coronary motion as ground truth training data for their CNNs. During testing on clinical CCTA scans, a motion-corrected image patch output by the CNN was reinserted into the original 3D CT volume to compensate for motion artifacts of the entire coronary artery. This resulted in a significant reduction in the proportion of images with motion as graded by expert readers. The described algorithms have the potential for integration into commercial software used for routine CCTA interpretation, and may help maintain the high diagnostic value of this modality.
Segmentation of Structures
Accurate delineation of cardiac structures is fundamental to the diagnosis and monitoring of cardiovascular disease. Structural and functional parameters derived from noncontrast cardiac CT and CCTA correlate well with those from echocardiography and cardiac magnetic resonance imaging13,14. Although commercial software packages enable semi-automated segmentation and quantification of cardiac structures, these methods are operator-dependent and time-consuming. Hence, DL has been used to fully automate this task for various clinical purposes.
Cardiac chambers
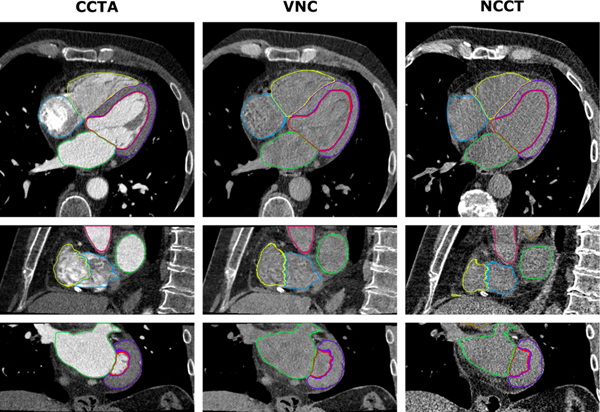
Bruns et al15 recently trained a CNN to automatically segment all 4 cardiac chambers and the left ventricular (LV) myocardium on noncontrast CT images reconstructed from a dual-layer detector CT scanner in 18 patients (Figure 1). This DL model showed good agreement with manual reference segmentations derived from CCTA: DSCs for volumes of the LV cavity, right ventricle, left atrium, right atrium, and LV myocardium were 0.90, 0.92, 0.92, 0.91, and 0.84, respectively. When applied to an independent multicenter dataset of single-energy noncontrast CT images, DL segmentation was overall graded as accurate by expert visual assessment. However, there was a significant underestimation of the volumes of all cardiac structures by noncontrast CT compared to CCTA as reference. The most likely explanation is that the absence of contrast agent influences the CNN’s interpretation of the position of the heart borders due to partial-volume effects.
Figure 1. Automatic segmentation of cardiac structures using deep learning.
Cardiac CT acquisitions from a dual-layer detector CT scanner were reconstructed into coronary CT angiography (CCTA), corresponding virtual noncontrast (VNC), and true noncontrast CT (NCCT) images. Manual reference segmentations of 7 cardiac structures on CCTA images (left) were used to train CNNs for automatic segmentation in VNC (middle) and NCCT (right) images. Shown is a case example of DL-based segmentations in axial (top), sagittal (middle), and coronal (bottom) views. Reproduced with permission from Bruns et al.15
CCTA provides greater demarcation of cardiac chambers, and Zreik et al.16 were among the first to apply DL to this modality for automatic LV segmentation. Trained using 60 CCTA datasets, this CNN based on LV localization and voxel classification yielded high performance (DSC 0.85) compared to expert manual annotations. Recently, Guo et al.17 developed a fully convolutional network which accurately contoured the LV myocardium in 100 patients (DSC 0.92), with physician-approved clinical contours used as ground truth training data. Baskaran et al.18 applied a fully convolutional network in 166 patients to automatically segment and quantify all 4 cardiac chambers and the LV myocardium, achieving DSCs of 0.94, 0.93, 0.93, 0.92, and 0.92 for LV volume, right ventricular volume, left atrial volume, right atrial volume, and LV mass, respectively, compared to manual annotations.
The ability of DL to accurately segment cardiac chambers on CT would enable computer-aided detection of abnormal morphology and function, thereby enhancing clinical risk stratification. Increased LV volume and reduced LV ejection fraction on CCTA have been shown to predict all-cause mortality, while a greater LV mass is associated with risk of myocardial infarction and cardiac death19. The prognostic value of AI-enabled fully automated chamber quantification should be explored in future studies.
Heart valves
Segmentation of the heart valves is of great interest due to the widespread adoption of transcatheter valve replacement procedures, which require precise modeling of the native valve and sizing of the implant. Grbic and colleagues20 combined ML algorithms with marginal space learning – a 3D learning-based object detection method – to automatically generate patient-specific models of all 4 heart valves from 4D CT cine images. Expert manual annotations were used as the ground truth for training and testing. For the aortic and mitral valves, the differences between estimated and ground truth parameters of valve position and orientation were slightly above the inter-user variability. This discrepancy was largely attributable to datasets with low contrast agent, image noise, and poor image quality, which may limit the clinical performance of this ML approach in such scans. Liang et al.21 developed a ML algorithm which automatically reconstructed patient-specific 3D geometries of the aortic valve using leaflet meshes. The mean discrepancy between automatic and manually generated geometries as measured by a landmark-to-mesh distance was 0.69 mm, compared to the inter-observer error of 0.65 mm.
Al et al.22 leveraged a regression-based ML algorithm to accurately localize 8 aortic valve landmarks within 12 milliseconds on pre-procedural CTA of patients undergoing transcatheter aortic valve implantation (TAVI). Zheng et al.23 used marginal space learning to automatically segment the aorta from rotational C-arm CT images and extract a real-time 3D aorta model which was super-imposed on 2D fluoroscopic images during live TAVI cases. Ultimately, the described patient-specific computational valve models could be used to assess the biomechanical interaction between a bioprosthesis and the surrounding tissue in order to determine the proper device and positioning for an individual patient to maximize the likelihood of success.
Coronary arteries
Coronary artery segmentation from CCTA enables luminal stenosis grading, plaque assessment, and blood flow simulation. The key first step in this process is automated coronary centerline extraction, with commercial software packages employing mathematical models such as minimum cost paths between proximal and distal artery points or centerline tracking by iterative prediction and correction24. Recent studies have shown that this step can be augmented by AI, using random forest25 or CNN26 classifiers which verify centerline extraction results, or a CNN which guides the centerline tracker27; resulting in improved speed and accuracy. Following centerline extraction, the coronary artery lumen is typically segmented by software using cross-sectional CCTA images, transforming the entire length of the vessel into a single volume. Wolterink et al.28 achieved this using a CNN which automatically generated patient-specific luminal meshes of the entire coronary tree, with a DSC of 0.75 compared to expert annotations and an average speed of 45 seconds including centerline extraction. Overall, AI techniques for coronary artery segmentation have demonstrated high performance and computational efficiency compared to conventional methods, and may reduce the degree of manual input required by clinicians during routine CCTA interpretation.
Quantitative Measurements
Coronary artery calcium scoring
The presence of CAC on noncontrast cardiac CT is direct marker of coronary atherosclerosis and strong predictor of major adverse cardiovascular events (MACE)29. Given the sheer volume of these scans performed in clinical practice, rapid and fully automated computer-aided CAC scoring would reduce the workload of physicians and technicians.
Wolterink and colleagues30 applied a random forest ML algorithm to CAC scoring CT scans to distinguish between true and false calcifications based on size, shape, and intensity features; with the final automatic Agatston score correlating strongly with manual measurements (r=0.94). Building on this framework, the investigators31 added a multi-class classifier to label CAC in each artery, along with an ambiguity detection system which flagged difficult cases for expert review. The resulting ICCs between fully manual and automatic CAC volume scores were 0.98, 0.69, and 0.95 for the LAD, LCX, and RCA, respectively; improved by expert review of automatically flagged cases to 1.00, 0.95, and 0.99, respectively. The combination of automatic scoring and expert review enabled more rapid computation compared to fully manual scoring (45 vs. 128 seconds per scan). These findings suggest that in clinical practice, results computed by ML algorithms may be improved upon by the knowledge and experience of an expert reader. The optimal learning strategy may indeed involve combining the complementary strengths of human and AI.
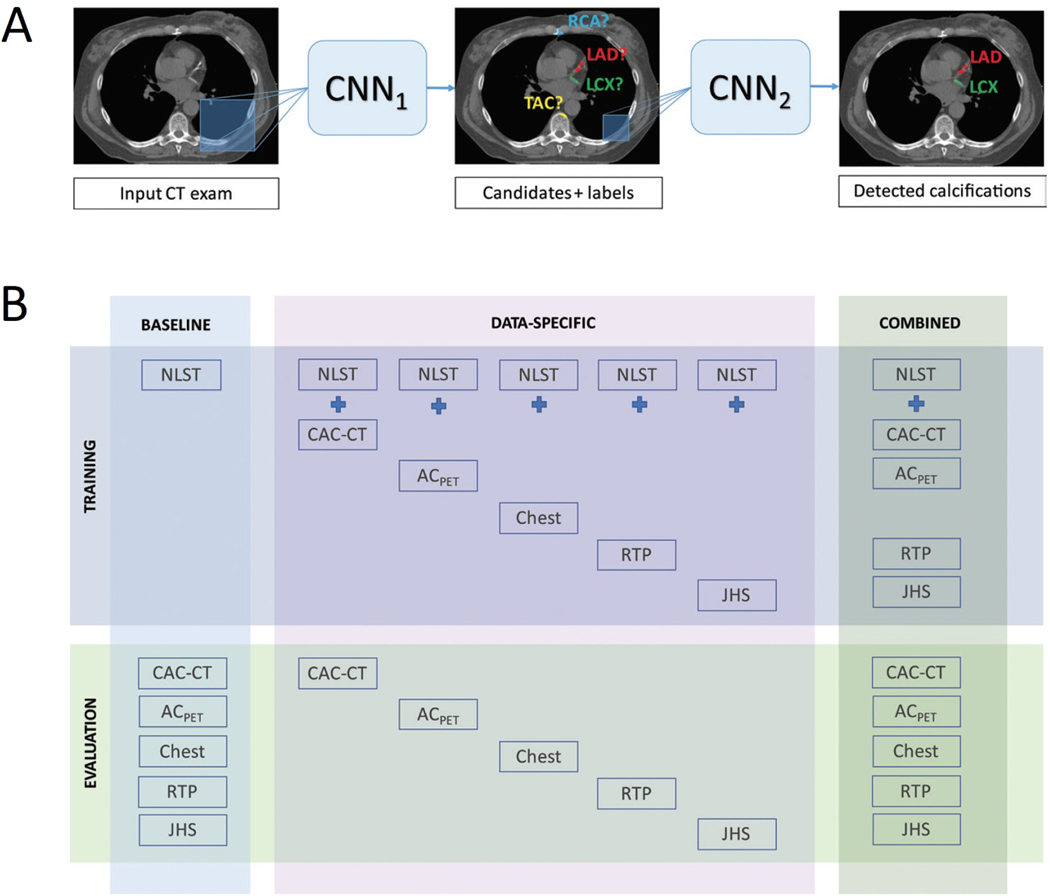
Recently, van Velzen et al.32 trained and tested a DL method for automated CAC scoring in 7,240 patients scanned with various noncontrast cardiac and chest CT protocols. Two consecutive CNNs were used to quantify the Agatston score: the first CNN detected candidate calcifications in each image and assigned them to a coronary artery; while the second CNN classified these calcifications as true or false-positives (Figure 2). The DL algorithm provided reliable automatic CAC scores compared to the manual reference, with ICCs of 0.79–0.97 across the range of scan types. In cardiovascular risk categorization by the Agatston score, DL had excellent agreement with manual CAC scoring (Cohen’s kappa = 0.90). These finding demonstrate the feasibility of DL-based CAC scoring in a real-world setting across a diverse set of noncontrast chest CT images. This may translate into a much wider clinical applicability of AI-enabled automatic CAC scoring.
Figure 2. Automatic coronary artery calcium scoring using deep learning.
(A) The deep learning algorithm consists of two convolutional neural networks (CNNs): the first CNN detects candidate calcifications by voxels with attenuation of greater than 130 Hounsfield units and assigns them to a coronary artery; while the second CNN detects true calcified voxels among candidates detected by the first CNN. (B) The baseline algorithm was trained with National Lung Screening Trial (NLST) scans, and its performance was evaluated in each CT protocol type. Five data-specific algorithms were trained, according to CT protocol type, and evaluated in the respective CT type. The combined algorithm was trained and evaluated using all available CT protocol types. CT types used for training were NLST CT scans, coronary artery calcium scoring CT (CAC-CT), PET attenuation correction CT (ACPET), diagnostic chest CT, radiation therapy treatment planning (RTP) CT, and Jackson Heart Study (JHS) CT scans. Reproduced with permission from van Velzen et al.32. LAD = left anterior descending artery; LCX = left circumflex artery; RCA = right coronary artery; TAC = thoracic aorta calcification.
Epicardial adipose tissue quantification
Located between the myocardium and visceral pericardium, epicardial adipose tissue (EAT) is a metabolically active fat depot which modulates coronary arterial function33. EAT volume and attenuation quantified semi-automatically from routine CAC scoring CT using research software have a well-established association with CAD and future MACE34–36. Recently, a fully automated DL algorithm for EAT quantification was evaluated in large multicenter study of 850 patients who underwent CAC scoring CT37. DL measurements correlated strongly with expert manual annotations (R = 0.97 for volume and R= 0.96 for attenuation; both p<0.001) and were computed at a fraction of the speed of experts (6 seconds vs 15 minutes per case).
Anatomical assessment of coronary artery stenosis
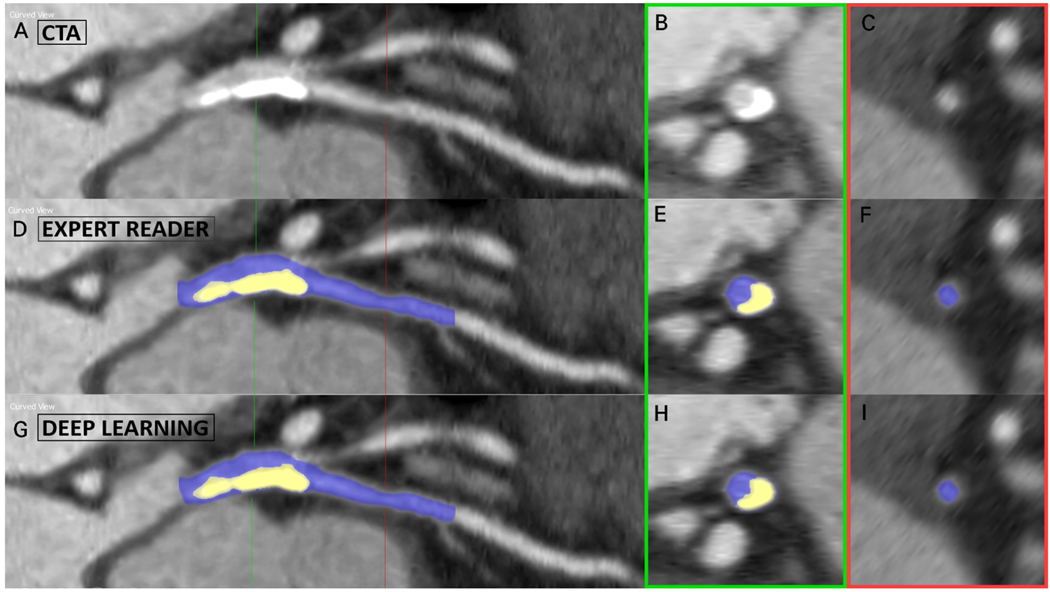
Current clinical reporting of CCTA is based on visual grading of coronary artery stenosis severity, which is subject to significant inter-reader variability38. Several AI approaches have been developed to provide automated and objective quantification. Kang et al.39 used a support vector machine-based ML algorithm for the detection of diameter stenosis ≥25% based on geometric and shape features of coronary lesions. This method performed with high sensitivity (93%), specificity (95%), and accuracy (94%) for the detection of stenosis ≥25%, with an AUC of 0.94 when compared to expert visual assessment as reference. Kelm et al.25 leveraged a random forest ML algorithm to calculate the lumen cross-sectional area based on local image features along the vessel centerline. Stenotic segments were then identified and graded as significant (≥50%) or non-significant (<50%) based on the estimated lumen radius. This technique achieved a per-vessel sensitivity of 98% for the detection of significant noncalcified stenosis, at an average speed of 1.8 seconds per case. Automated evaluation of coronary stenosis can also be achieved using DL, with Hong and colleagues40 training an end-to-end CNN to automatically quantify minimal luminal area and percentage diameter stenosis in 156 CCTA datasets with 716 diseased coronary segments (Figure 3). The resultant DL measurements correlated strongly with manual annotations (r=0.984 for minimal luminal area and r=0.957 for stenosis; both p<0.001), at an average computation speed of 31 seconds per segment.
Figure 3. Deep learning-based coronary stenosis quantification.
Case example of lumen and calcified plaque segmentation on coronary CT angiography (CTA) in a lesion extending from the left main into the left anterior descending artery. Top row: curved planar reformation (A) and cross-sections of the lesion (B) and healthy lumen (C). Middle row: (D-F) manual segmentations performed by the expert reader. Bottom row: (G-I) automated segmentations performed by the deep learning algorithm. Using these segmentations, the minimum luminal area and percentage diameter stenosis were automatically computed.
AI-powered detection and quantification of coronary stenosis may benefit clinical workflow in CCTA reporting. By functioning as a “second reader” and providing rapid, objective results, ML or DL algorithms may reduce variability and interpretative error. Further, these algorithms could pre-screen CCTA scans for the clinician, flagging those with obstructive disease to be prioritized for reporting. A new frontier for AI in cardiovascular CT is the fully automated quantification of coronary plaque volumes and composition, which would further enhance risk prediction and enable assessment of treatment response; with research efforts are currently underway.
Functional assessment of coronary artery stenosis
The hemodynamic significance of a given coronary stenosis on CCTA can also be estimated using AI techniques. Non-invasive fractional flow reserve (FFR) derived from CCTA (CT-FFR) is currently performed in clinical practice with the assistance of DL methods which identify lumen boundaries and thus enable assessment of computational fluid dynamics (CFD; Figure 4). In a real-world study, this method (AUC 0.94) outperformed conventional CCTA stenosis assessment (AUC 0.83) and positron emission tomography (AUC 0.87; both p<0.01) for the detection of per-vessel ischemia when compared to invasive FFR as the reference standard41.
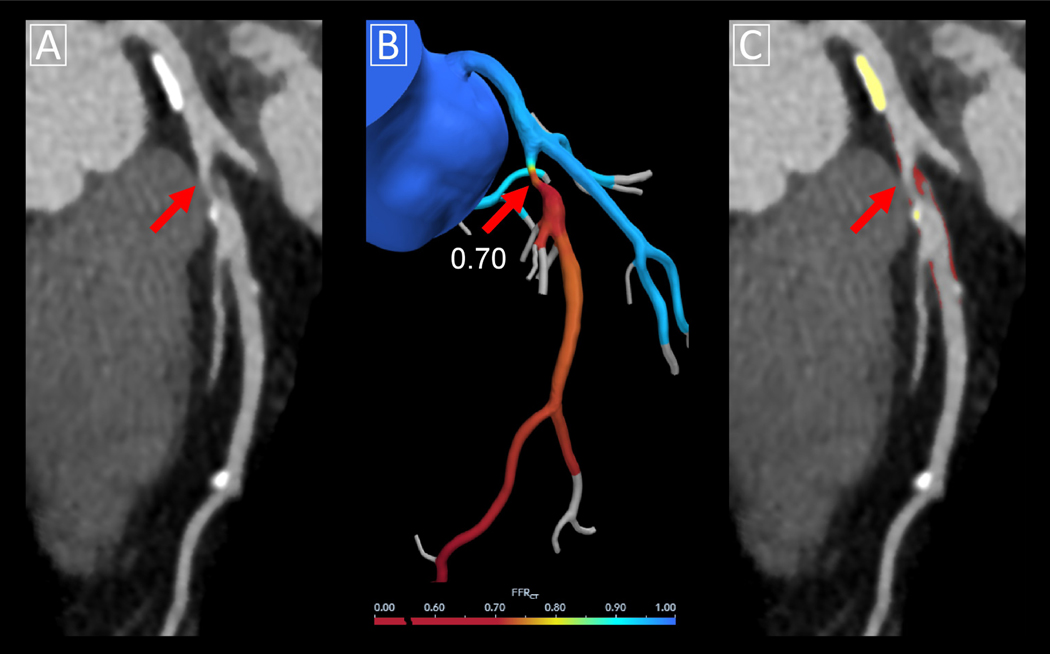
Figure 4. CT-FFR using computational fluid dynamics aided by deep learning.
Case example of CT-FFR using computational fluid dynamics. (A) Multiplanar reformat of coronary CT angiography demonstrating obstructive stenosis in the proximal left anterior descending (red arrow) due to mixed plaque with high-risk features. (B) Corresponding CT-FFR value was 0.70. Lumen boundaries were identified using a DL algorithm embedded in commercial software. (C) Quantitative plaque analysis of the same lesion was performed using semi-automated research software, with calcified plaque shown in yellow overlay and noncalcified plaque in red overlay. CT-FFR = computed tomography-fractional flow reserve.
However, current CFD models are complex and computationally demanding, requiring clinical CT-FFR analysis to be performed off-site by supercomputers in core laboratories42. To address these limitations, Itu et al.43 developed a fully automated DL-based approach for the calculation of CT-FFR on-site. The DL algorithm was trained on a large dataset of synthetically generated coronary models, with CFD-based results used a ground truth training data. DL-based CT-FFR yielded a sensitivity of 82%, specificity of 85%, and accuracy of 83% when compared to invasive FFR; with much faster execution times than conventional computational fluid dynamics (2.4 vs 196.3 seconds per case). CT-FFR by this DL method was further validated in a multicenter study of 351 patients and 525 vessels44, where it improved the diagnostic accuracy of CCTA and was comparable to computational fluid dynamics-based CT-FFR. Although currently a research prototype, the DL application has the potential to deliver CT-FFR assessment at a standard workstation in a point-of-care manner. This would allow for interactive interpretation by physicians and the ability to instantaneously observe the effect of changes in vessel segmentation on FFR results. Of note, low CCTA image quality and extensive coronary calcium have been shown to reduce the diagnostic performance of DL-based CT-FFR44,45, potentially limiting its clinical applicability in these settings.
Outcome Prediction
The ability to correctly identify high-risk patients who may benefit from intensification of preventative measures remains a major challenge in cardiovascular medicine. Conventional risk assessment involves the classification of patients into predefined risk categories for population-based comparisons, often leading to misclassification46. In contrast, ML enables precise, patient-specific, continuous risk prediction. ML algorithms can integrate many CT parameters with clinical data to provide comprehensive and individualized prognostication. This will become increasingly important as the amount of extractable data from cardiovascular CT images increases and electronic health records evolve at a rapid pace.
Long-term cardiovascular event risk
In 66,636 asymptomatic individuals without known CAD from the CAC Consortium, Nakanishi et al.47 used a supervised ML model (LogitBoost) integrating 31 noncontrast CT metrics (including calcium scores of the coronaries, aortic valve, mitral valve, and thoracic aorta) with 46 clinical variables for the prediction of CAD-related death at 10 years. ML (AUC 0.86) provided superior risk prediction compared to the conventional atherosclerotic cardiovascular disease (ASCVD) risk score (AUC 0.83) or Agatston score (AUC 0.82; both p<0.001). This comprehensive ML model also outperformed ML with either clinical data alone or CT data alone (Figure 3).
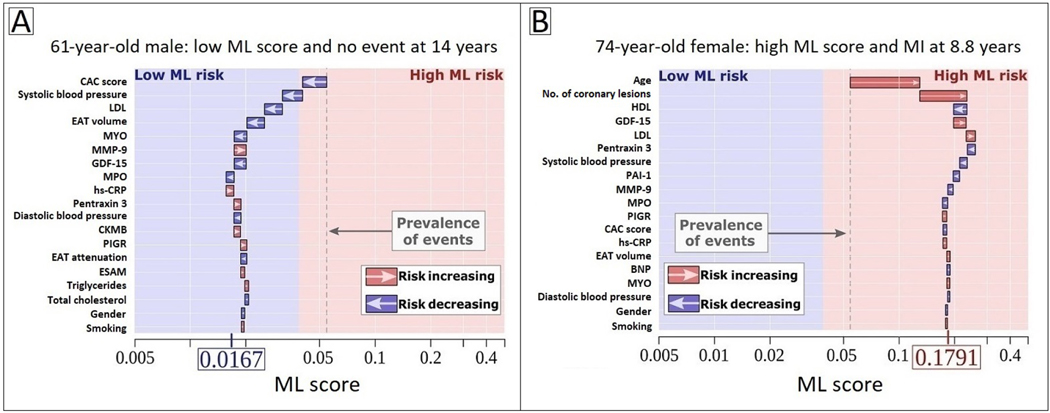
EAT quantified by the previously described DL algorithm37 has also shown prognostic value. In 2,068 asymptomatic subjects from the EISNER (Early Identification of Subclinical Atherosclerosis by Noninvasive Imaging Research) trial, DL-based EAT volume was associated with MACE on 14-year follow-up (hazard ratio [HR] 1.35 per doubling, p=0.009), independently of the ASCVD risk score and CAC score. EAT attenuation was inversely associated with MACE risk (HR 0.83 per 5 Hounsfield unit increase, p=0.01)48. In a subsequent EISNER substudy, DL-based EAT measures were combined with serum biomarkers and clinical data using a supervised ML algorithm (XGBoost). The comprehensive ML model (AUC 0.81) outperformed the CAC score (AUC 0.75) and ASCVD risk score (AUC 0.74; both p=0.02) for the long-term prediction of hard cardiac events49. Further, the ML algorithm had the ability to clarify its decision-making process by specifically describing the contribution of each variable to individualized prediction – an example of ‘explainable AI’ (Figure 5). Together, these studies suggest that rapid, automated measurements of EAT have the potential for integration into routine reporting of CAC scoring CT, providing real-time, complementary information on cardiovascular risk.
Figure 5. Explainable AI through individualized machine learning risk prediction.
(A) 62-year-old male with no event at 14 years and (B) 74-year-old female with an MI at 8.8 years. The X-axis denotes the ML risk score which is the predicted probability of events. The arrows represent the influence of each variable on the overall prediction; blue and red arrows indicate whether the respective parameters decrease (blue) or increase (red) the risk of future events. The combination of all variables’ influence provides the final ML risk score. The subject in (A) has a low ML risk score (0.0167), with an ASCVD risk score of 7.25% and CAC score of 0. The subject in (B) has a high ML risk score (0.1791), with an ASCVD risk score of 30.4% and CAC score of 324. The blue and red background colors indicate low versus high ML risk according to study-specific threshold, and the gray dashed line corresponds to the base risk obtained from the prevalence of events in the population (4.7%). Reproduced with permission from Tamarappoo et al.49
Abbreviations: ASCVD, atherosclerotic cardiovascular disease; BNP, brain natriuretic peptide; CAC, coronary artery calcium; CKMB, creatine kinase MB; CRP, C-reactive protein; EAT, epicardial adipose tissue; ESAM, endothelial cell-selective adhesion molecule; GDF-15, growth differentiation factor 15; HDL, high-density lipoprotein; LDL, low-density lipoprotein; MCP-1, monocyte chemoattractant protein 1; ML, machine learning; MMP-9, matrix metalloprotease 9; MPO: myeloperoxidase; PAI-1, plasminogen activator inhibitor 1; PIGR, polymeric immunoglobulin receptor.
Coronary stenosis measures on CCTA have also been incorporated into ML models for prognostication. In 10,030 patients from the CONFIRM (COronary CT Angiography EvaluatioN For Clinical Outcomes: An InteRnational Multicenter) registry, Motwani et al.50 trained a LogitBoost algorithm combining 44 visually-determined parameters of stenosis location and severity with 25 clinical variables for the prediction of 5-year all-cause mortality. The ML model (AUC 0.79) outperformed conventional CCTA-derived segments scores (AUC 0.64 for both segment involvement score and segment stenosis score) and the Framingham risk score (AUC 0.61; p<0.001 for all) (Figure 5).
Comprehensive ischemia risk scoring
In a substudy of the NXT (Analysis of Coronary Blood Flow. Using CT Angiography: Next Steps) trial, Dey et al.51 integrated CCTA-derived quantitative plaque and vessel-based metrics with clinical data using ML to generate a per-vessel “Ischemia Risk Score” for the prediction of a positive invasive FFR. This score yielded a significantly higher AUC (0.84) compared with stenosis severity (0.76, p=0.005) or clinical pre-test probability of CAD (AUC 0.63, p<0.001). In a subsequent multicenter study of patients undergoing CCTA for suspected CAD52, the Ischemia Risk Score predicted early revascularization at both the per-vessel and per-patient level, independently of risk factors, symptoms, and presence of obstructive CAD; and provided incremental value when added to a traditional risk model with clinical data and stenosis severity.
BARRIERS TO AI IMPLEMENTATION
In order for the described AI techniques to become a reality in clinical practice, several challenges need to be overcome. First, large-scale, labeled datasets with high quality CT image data are required for training and testing new algorithms. In healthcare, there are often legal barriers around data sharing, with only a limited number of datasets being made publicly available. Encouragingly, international collaborative cardiac imaging databases such as the Cardiac Atlas Project53 have managed to clear many of the ethical, legal, and organizational hurdles to data sharing and distribution. Beyond data availability, standardization of CT acquisition protocols and quantitative parameters is required when aggregating data from different centers as input for an AI model. Recent initiatives such as the Radiological Society of North America’s Quantitative Imaging Biomarkers Alliance have been established to improve the standardization and performance of quantitative imaging metrics.
Second, bias can arise in AI algorithms over time, through learning from disparities in patient demographics or healthcare systems. Hence, algorithms need to be monitored by institutions and regulatory authorities for biases in predictive performance and also for biases in the way their predictions are used in clinical care. Further, ML models trained and tested at a single institution may not be generalizable to different cohorts; underscoring the importance of developing and validating AI models in multi-center, multi-vendor studies with diverse patient demographics. Formal external validation of these models in independent datasets not used for training should also be performed.
Third, AI algorithms can often be viewed as “black boxes” which autonomously learn and make decisions. An understanding of why decisions are made by an algorithm, the rigor of evaluation, and when and why errors occur should all be considered before any algorithm is adopted in clinical practice. Recently, explainable AI models have emerged which have the ability to provide some explanation or justification for their decision-making. Such algorithms may highlight a recognizable variable or combination of variables that contributed to its decision (for example an increase in age or number of calcified coronary lesions; Figure 5B).
Finally, there is the legal consideration of clinical clearance for AI-powered software applications. Current diagnostic software tools are categorized by the United States Food and Drug Administration as Class II (medium-risk) medical devices; to be concordant with regulatory requirements, the final results often need to be validated by the user. If fully automated systems were to be ultimately used for end-to-end diagnosis or risk stratification, they would most likely be categorized as Class III (high-risk) devices, which are held to much higher performance and validation standards.
FUTURE IMPLICATIONS
The described AI applications for cardiovascular CT have the potential to improve the efficiency of clinical workflow, provide objective and reproducible quantitative results, increase interpretative speed, and inform subsequent clinical pathways. With respect to image quality, DL algorithms which reduce data noise and cardiac motion artifacts can speed up image reconstruction while maintaining the high diagnostic value of cardiovascular CT. For patients undergoing CAC scoring scans, rapid and fully automated measurements of CAC score, EAT, and cardiac chambers could potentially be integrated into routine clinical reporting, reducing the workload of physicians and technicians while providing real-time, complementary information on cardiovascular risk. For patients undergoing CCTA, AI-enabled quantification of anatomical and functional stenosis severity has been shown to be feasible, fast, and accurate. Finally, ML models combining CT-derived biomarkers with all available clinical data have consistently provided superior outcome prediction compared to conventional methods.
In order for AI to reach its full potential in cardiovascular CT, cardiologists and radiologists need to be actively involved in the development and implementation of this technology. Physicians can participate in annotation of image datasets as the expert reader, beta-testing of AI-powered software in the clinical setting, or validating ML model predictions in real-world studies. They could advise their institutions on investment in AI tools and advocate for responsible data sharing. Eventually, clinicians will need to determine whether they can trust and work alongside AI platforms, and explain their reliance on or rejection of AI-based results to patients.
In the near future, one can foresee an increasing number of AI algorithms embedded in standard cardiovascular CT image analysis software and routine clinical reporting, automatically performing measurements which are then seamlessly integrated with electronic health record data. Personalized AI-based risk scores combining all available parameters will be computed in real-time, providing long-term prognostication or flagging patients at risk of impending cardiac events. Ultimately, specific AI models may be able to guide patient therapy, with algorithms utilizing clinical and CT data to objectively quantify the potential benefit to an individual.
Acknowledgments
Funding: This work was supported in part by grants from the National Heart, Lung, and Blood Institute [1R01HL133616 and 1R01HL148787-01A1].
Footnotes
Disclosure statement: Outside of the current work, Drs Dey and Slomka receive software royalties from Cedars-Sinai Medical Center. The remaining authors have no relevant conflicts of interest to disclose.
Declaration of interests
To: Journal of Cardiovascular Computed Tomography
Re: “Artificial Intelligence in Cardiovascular CT: Current Status and Future Implications”
Authors: Andrew Lin, Márton Kolossváry, Manish Motwani, Ivana Išgum, Pál Maurovich-Horvat, Piotr J Slomka, Damini Dey
☐ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Outside of the current work, Damini Dey and Piotr Slomka receive software royalties from Cedars-Sinai Medical Center. The remaining authors have no relevant conflicts of interest to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lin A, Kolossváry M, Išgum I, Maurovich-Horvat P, Slomka PJ, Dey D. Artificial intelligence: improving the efficiency of cardiovascular imaging. Expert Review of Medical Devices. 2020:null-null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turing AM. Computing Machiner and Intelligence. Mind. 1950;LIX(236):433–460. [Google Scholar]
- 3.Dey D, Slomka PJ, Leeson P, et al. Artificial Intelligence in Cardiovascular Imaging: JACC State-of-the-Art Review. J Am Coll Cardiol. 2019;73(11):1317–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudre CH, Li W, Vercauteren T, Ourselin S, Jorge Cardoso M. Generalised Dice Overlap as a Deep Learning Loss Function for Highly Unbalanced Segmentations. Paper presented at: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; 2017//, 2017; Cham. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognition. 1997;30(7):1145–1159. [Google Scholar]
- 6.Wolterink JM, Leiner T, Viergever MA, Isgum I. Generative Adversarial Networks for Noise Reduction in Low-Dose CT. IEEE Trans Med Imaging. 2017;36(12):2536–2545. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Zhang Y, Zhang W, et al. Low-dose CT via convolutional neural network. Biomed Opt Express. 2017;8(2):679–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang E, Min J, Ye JC. A deep convolutional neural network using directional wavelets for low-dose X-ray CT reconstruction. Med Phys. 2017;44(10):e360–e375. [DOI] [PubMed] [Google Scholar]
- 9.Green M, Marom EM, Konen E, Kiryati N, Mayer A. 3-D Neural denoising for low-dose Coronary CT Angiography (CCTA). Computerized Medical Imaging and Graphics. 2018;70:185–191. [DOI] [PubMed] [Google Scholar]
- 10.Kang E, Koo HJ, Yang DH, Seo JB, Ye JC. Cycle-consistent adversarial denoising network for multiphase coronary CT angiography. Med Phys. 2019;46(2):550–562. [DOI] [PubMed] [Google Scholar]
- 11.Lossau T, Nickisch H, Wissel T, et al. Motion estimation and correction in cardiac CT angiography images using convolutional neural networks. Computerized Medical Imaging and Graphics. 2019;76:101640. [DOI] [PubMed] [Google Scholar]
- 12.Jung S, Lee S, Jeon B, Jang Y, Chang H. Deep Learning Cross-Phase Style Transfer for Motion Artifact Correction in Coronary Computed Tomography Angiography. IEEE Access. 2020;8:81849–81863. [Google Scholar]
- 13.Murphy DJ, Lavelle LP, Gibney B, O’Donohoe RL, Rémy-Jardin M, Dodd JD. Diagnostic accuracy of standard axial 64-slice chest CT compared to cardiac MRI for the detection of cardiomyopathies. Br J Radiol. 2016;89(1059):20150810–20150810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greupner J, Zimmermann E, Grohmann A, et al. Head-to-Head Comparison of Left Ventricular Function Assessment with 64-Row Computed Tomography, Biplane Left Cineventriculography, and Both 2- and 3-Dimensional Transthoracic Echocardiography. Journal of the American College of Cardiology. 2012;59(21):1897. [DOI] [PubMed] [Google Scholar]
- 15.Bruns S, Wolterink JM, Takx RAP, et al. Deep learning from dual-energy information for whole-heart segmentation in dual-energy and single-energy non-contrast-enhanced cardiac CT. Med Phys. 2020. [DOI] [PubMed] [Google Scholar]
- 16.Zreik M, Leiner T, De Vos B, van Hamersvelt R, Viergever M, Isgum I. Automatic segmentation of the left ventricle in cardiac CT angiography using convolutional neural networks. IEEE International Symposium on Biomedical Imaging. 2016:40–43. [Google Scholar]
- 17.Jun Guo B, He X, Lei Y, et al. Automated left ventricular myocardium segmentation using 3D deeply supervised attention U-net for coronary computed tomography angiography; CT myocardium segmentation. Medical Physics. 2020;47(4):1775–1785. [DOI] [PubMed] [Google Scholar]
- 18.Baskaran L, Maliakal G, Al’Aref SJ, et al. Identification and Quantification of Cardiovascular Structures From CCTA: An End-to-End, Rapid, Pixel-Wise, Deep-Learning Method. JACC: Cardiovascular Imaging. 2020;13(5):1163–1171. [DOI] [PubMed] [Google Scholar]
- 19.Klein R, Ametepe ES, Yam Y, Dwivedi G, Chow BJ. Cardiac CT assessment of left ventricular mass in mid-diastasis and its prognostic value. Eur Heart J Cardiovasc Imaging. 2017;18(1):95–102. [DOI] [PubMed] [Google Scholar]
- 20.Grbic S, Ionasec R, Vitanovski D, et al. Complete valvular heart apparatus model from 4D cardiac CT. Medical Image Analysis. 2012;16(5):1003–1014. [DOI] [PubMed] [Google Scholar]
- 21.Liang L, Kong F, Martin C, et al. Machine learning-based 3-D geometry reconstruction and modeling of aortic valve deformation using 3-D computed tomography images. Int J Numer Method Biomed Eng. 2017;33(5): 10.1002/cnm.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al WA, Jung HY, Yun ID, Jang Y, Park H-B, Chang H-J. Automatic aortic valve landmark localization in coronary CT angiography using colonial walk. PloS one. 2018;13(7):e0200317-e0200317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng Y, Barbu A, Georgescu B, Scheuering M, Comaniciu D. Four-Chamber Heart Modeling and Automatic Segmentation for 3-D Cardiac CT Volumes Using Marginal Space Learning and Steerable Features. IEEE Transactions on Medical Imaging. 2008;27(11):1668–1681. [DOI] [PubMed] [Google Scholar]
- 24.Lesage D, Angelini ED, Bloch I, Funka-Lea G. A review of 3D vessel lumen segmentation techniques: Models, features and extraction schemes. Medical Image Analysis. 2009;13(6):819–845. [DOI] [PubMed] [Google Scholar]
- 25.Kelm BM, Mittal S, Zheng Y, et al. Detection, grading and classification of coronary stenoses in computed tomography angiography. Med Image Comput Comput Assist Interv. 2011;14(Pt 3):25–32. [DOI] [PubMed] [Google Scholar]
- 26.Gülsün M, Funka-Lea G, Sharma P, Rapaka S, Zheng Y. Coronary Centerline Extraction via Optimal Flow Paths and CNN Path Pruning. 2016. [Google Scholar]
- 27.Wolterink JM, van Hamersvelt RW, Viergever MA, Leiner T, Isgum I. Coronary artery centerline extraction in cardiac CT angiography using a CNN-based orientation classifier. Med Image Anal. 2019;51:46–60. [DOI] [PubMed] [Google Scholar]
- 28.Wolterink J, Leiner T, Išgum I. Graph Convolutional Networks for Coronary Artery Segmentation in Cardiac CT Angiography. In: International Workshop on Graph Learning in Medical Imaging. Springer; 2019:62–69. [Google Scholar]
- 29.Budoff MJ, Young R, Burke G, et al. Ten-year association of coronary artery calcium with atherosclerotic cardiovascular disease (ASCVD) events: the multi-ethnic study of atherosclerosis (MESA). European Heart Journal. 2018;39(25):2401–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolterink J, Leiner T, Takx R, Viergever M, Isgum I. An automatic machine learning system for coronary calcium scoring in clinical non-contrast enhanced, ECG-triggered cardiac CT. Vol 9035. Progress in Biomedical Optics and Imaging - Proceedings of SPIE. 90352014. [Google Scholar]
- 31.Wolterink JM, Leiner T, Takx RA, Viergever MA, Isgum I. Automatic Coronary Calcium Scoring in Non-Contrast-Enhanced ECG-Triggered Cardiac CT With Ambiguity Detection. IEEE Trans Med Imaging. 2015;34(9):1867–1878. [DOI] [PubMed] [Google Scholar]
- 32.van Velzen SGM, Lessmann N, Velthuis BK, et al. Deep Learning for Automatic Calcium Scoring in CT: Validation Using Multiple Cardiac CT and Chest CT Protocols. Radiology. 2020;295(1):66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin A, Dey D, Wong DTL, Nerlekar N. Perivascular Adipose Tissue and Coronary Atherosclerosis: from Biology to Imaging Phenotyping. Current Atherosclerosis Reports. 2019;21(12):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahabadi AA, Berg MH, Lehmann N, et al. Association of epicardial fat with cardiovascular risk factors and incident myocardial infarction in the general population: the Heinz Nixdorf Recall Study. J Am Coll Cardiol. 2013;61(13):1388–1395. [DOI] [PubMed] [Google Scholar]
- 35.Dey D, Wong ND, Tamarappoo B, et al. Computer-aided non-contrast CT-based quantification of pericardial and thoracic fat and their associations with coronary calcium and Metabolic Syndrome. Atherosclerosis. 2010;209(1):136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goeller M, Achenbach S, Marwan M, et al. Epicardial adipose tissue density and volume are related to subclinical atherosclerosis, inflammation and major adverse cardiac events in asymptomatic subjects. J Cardiovasc Comput Tomogr. 2018;12(1):67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Commandeur F, Goeller M, Razipour A, et al. Fully Automated CT Quantification of Epicardial Adipose Tissue by Deep Learning: A Multicenter Study. Radiology: Artificial Intelligence. 2019;1(6):e190045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arbab-Zadeh A, Hoe J. Quantification of coronary arterial stenoses by multidetector CT angiography in comparison with conventional angiography methods, caveats, and implications. JACC Cardiovasc Imaging. 2011;4(2):191–202. [DOI] [PubMed] [Google Scholar]
- 39.Kang D, Dey D, Slomka PJ, et al. Structured learning algorithm for detection of nonobstructive and obstructive coronary plaque lesions from computed tomography angiography. J Med Imaging (Bellingham). 2015;2(1):014003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong Y, Commandeur F, Cadet S, et al. Deep learning-based stenosis quantification from coronary CT Angiography. Proc SPIE Int Soc Opt Eng. 2019;10949:109492I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Driessen Roel S, Danad I, Stuijfzand Wijnand J, et al. Comparison of Coronary Computed Tomography Angiography, Fractional Flow Reserve, and Perfusion Imaging for Ischemia Diagnosis. Journal of the American College of Cardiology. 2019;73(2):161–173. [DOI] [PubMed] [Google Scholar]
- 42.Tesche C, De Cecco CN, Albrecht MH, et al. Coronary CT Angiography–derived Fractional Flow Reserve. Radiology. 2017;285(1):17–33. [DOI] [PubMed] [Google Scholar]
- 43.Itu L, Rapaka S, Passerini T, et al. A machine-learning approach for computation of fractional flow reserve from coronary computed tomography. J Appl Physiol (1985). 2016;121(1):42–52. [DOI] [PubMed] [Google Scholar]
- 44.Coenen A, Kim YH, Kruk M, et al. Diagnostic Accuracy of a Machine-Learning Approach to Coronary Computed Tomographic Angiography-Based Fractional Flow Reserve: Result From the MACHINE Consortium. Circ Cardiovasc Imaging. 2018;11(6):e007217. [DOI] [PubMed] [Google Scholar]
- 45.Tesche C, Otani K, De Cecco CN, et al. Influence of Coronary Calcium on Diagnostic Performance of Machine Learning CT-FFR: Results From MACHINE Registry. JACC Cardiovasc Imaging. 2020;13(3):760–770. [DOI] [PubMed] [Google Scholar]
- 46.Wynants L, van Smeden M, McLernon DJ, et al. Three myths about risk thresholds for prediction models. BMC Medicine. 2019;17(1):192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakanishi R, Slomka P, Rios R, et al. Mahine learning adds to clincial and CAC assessment in predicting 1-year CHD and CVD deaths. JACC Cardiovasc Imaging. 2020. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eisenberg E, McElhinney Priscilla A, Commandeur F, et al. Deep Learning–Based Quantification of Epicardial Adipose Tissue Volume and Attenuation Predicts Major Adverse Cardiovascular Events in Asymptomatic Subjects. Circulation: Cardiovascular Imaging. 2020;13(2):e009829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamarappoo BK, Lin A, Commandeur F, et al. Machine learning integration of circulating and imaging biomarkers for explainable patient-specific prediction of cardiac events: A prospective study. Atherosclerosis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Motwani M, Dey D, Berman DS, et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur Heart J. 2017;38(7):500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dey D, Gaur S, Ovrehus KA, et al. Integrated prediction of lesion-specific ischaemia from quantitative coronary CT angiography using machine learning: a multicentre study. Eur Radiol. 2018;28(6):2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwan A, McElhinney P, Tamarappoo B, et al. Prediction of revascularization by coronary CT angiography using a machine learning ischemia risk score. European Radiology. 2020. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fonseca CG, Backhaus M, Bluemke DA, et al. The Cardiac Atlas Project--an imaging database for computational modeling and statistical atlases of the heart. Bioinformatics (Oxford, England). 2011;27(16):2288–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]