Background
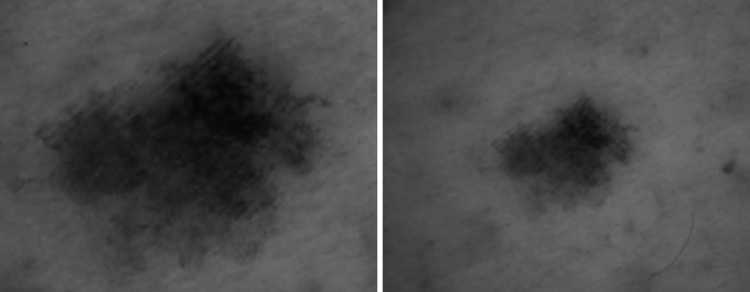
The use of digital imaging in dermatology is increasing due to several factors including teledermatology, longitudinal imaging to observe changes in skin lesions, advanced imaging (e.g., dermoscopy), and the emergence of artificial intelligence (AI) image classifiers for dermatology. Currently, digital imaging in dermatology predominantly uses consumer file formats (e.g., JPEG and TIFF) which lack patient and clinical metadata. Further, consumer file formats suffer from a lack of color consistency. Figure 1 shows photographs of the same lesion image at a different zoom level and color space. The intent is to illustrate that different diagnoses may result from images of the same lesion. The availability of metadata related to acquisition parameters (e.g., zoom, color space, compression) could potentially improve diagnostic accuracy and confidence. There is evolving realization that adopting the Digital Imaging and Communication in Medicine (DICOM) standard for dermatological imaging may improve availability of important patient, clinical, and acquisition metadata [1].
Fig. 1.
Identical lesion under two different magnification (digital) zoom conditions and color representation; assuming same/similar magnification (µm/pixel or total height/width in mm), the two lesions may have different meaning
In addition to improving human interpretation of images, metadata may improve AI prediction models. Many recent AI challenges have attempted to classify images based on pixel data alone. However, the addition of metadata to images of skin lesions has shown to improve diagnostic accuracy compared to images alone. These metadata included patient age, sex, and location of the lesion on the body [2]. We believe that richer metadata may further improve AI prediction accuracy and having those data readily available and securely attached to the pixel data is then clearly advantageous.
In the present paper, we describe our experiences of a proof-of-concept study packaging metadata into a DICOM file format for the 2020 International Skin Imaging Collaborative/Society for Imaging Informatics in Medicine (ISIC/SIIM) sponsored and Kaggle-hosted melanoma detection challenge [3]. The ISIC/SIIM challenge participants were given the option of downloading the datasets as DICOM files with embedded metadata, or in a consumer image Joint Photographic Experts Group (JPEG) or machine learning (TFRecord) format with a comma separated value (CSV) metadata file [4]. The aims of this study are two-fold. Firstly, we aim to identify the benefits and describe the challenges in using DICOM for dermoscopic image standardization. Secondly, we aim to test how many challenge participants are interested and willing to use DICOM for dermatology-related AI challenges.
Material and Methods
Setting
For more than a quarter century, the International Society for Digital Imaging of the Skin (ISDIS), has spearheaded efforts to bring digital information processing technologies into greater awareness of dermatologists [5]. Over the past five years, the Society has increased their efforts in machine learning. As a part of these efforts, the International Skin Imaging Collaborative (ISIC), together with the machine learning community, has hosted five grand challenges in melanoma detection [6–10]. These annual competitions have contributed to improvements in computer-aided diagnosis of malignancies of the skin (e.g., melanoma) [10].
The 2020 grand challenge was co-organized by ISIC and SIIM [3]. The dataset for this challenge consists of 44,108 skin lesion images which were contributed by six global dermatology sites. The dataset has been fully described elsewhere. [4] One critical aspect of the 2020 grand challenge dataset was to associate each image with a patient, thereby providing contextual lesion images. Contextual lesion images may contribute to melanoma diagnosis using the “ugly duckling” sign [11, 12]. The intention of the challenge was to ascertain if the machine learning algorithms could use the contextual lesion images to improve predictive accuracy. The decision was made to encode the dataset in DICOM PS3.10 file format given that it supports patient identification metadata [13].
Dataset Contribution and Curation
The contributing sites all used different (sometimes proprietary) image management software. Furthermore, the provisioning of the metadata that was included in the challenge dataset took substantial effort to collect and curate [4, 14]. Images, in JPEG format, were first ingested into the ISIC Archive using the web interface [15]. As part of the ingestion, the original contributor was asked to also upload a CSV file of metadata containing: patient identifier, age, sex, anatomic site, and diagnosis.
To avoid disclosing unwanted metadata, all exchangeable image file format (EXIF) fields were removed from the JPEG images using Exiftool [16]. Subsequently, the compressed images were encapsulated as DICOM instances using the PixelMed Java toolkit developed by one of the authors [17]. For this proof-of-concept, the metadata described in Table 1 was added to the DICOM header.
Table 1.
Description of metadata used for encoding grand challenge dataset in DICOM Part 10 file format
| Metadata | Description |
|---|---|
| Naming of images | A unique name of the format ISIC_[7-digits] is assigned automatically during data ingestion into the ISIC Archive. To allow cross-referencing to the ISIC Archive, the image name assigned by the ISIC archive was maintained and stored in the Study ID (0020,0010) attribute |
| Patient identifiers | To avoid releasing any internal data about the source of an image and to homogenize format, all patient IDs provided with the original metadata were mapped to a common format, IP_[8-digits], which was stored in the Patient ID (0010,0020) attribute |
| Patient’s sex | Patient’s sex (0010,0040) was coded as a binary variable. For images, where the patient’s sex was not known, this attribute was set to an empty string according to rules for encoding of Type 2 attributes |
| Patient’s age | DICOM encodes age as a four-character string comprised of a three-digit integer followed by a letter to indicate the unit (e.g., 018M for 18 months or 047Y for 47 years). The ISIC Archive only provides an approximate age (binned into five-year brackets) to decrease the likelihood of patient identification. While it is not fully compliant, the (approximate) age value was encoded according to the four-character Age String (AS) notation in the Patient’s Age (0010,1010) attribute |
| Anatomical site of skin lesion | The Systematized Nomenclature of Medicine Clinical Terms (SCT) coding scheme [30] was used as the coding scheme for the anatomical site of lesion. This coding scheme was challenging as the ISIC Archive has its own coding scheme, tailored to the needs of the field and availability of anatomic site mapping of historical data. From a dermatologist’s perspective, the palms of the hands as well as the soles of the feet show similar lesion characteristics, and these two locations are currently coded using a single token [31, 32]. For similar reasons, oral and genital regions are folded into a single token as well. Since these two ISIC Archive-based anatomic site terms do not have corresponding SCT codes, and given limited time available to query the medical record where each image was acquired to achieve further granularity, we decided (for this proof-of-concept study) to code these lesions with the generic parent term, “Skin.” The values stored in the attribute as a text string in the Body Part Examined (0018,0015) and a code in the Anatomical Region Sequence (0008,2218) attributes as shown in Table 2 |
| Image (binary) data | DICOM supports JPEG streams directly (using lossy image compression Transfer Syntaxes), and the JPEG binary data was stored as a byte stream in the Pixel Data (7FE0,0010) attribute |
To produce DICOM files whose format could be shared by both the training and test data sets, the diagnosis classification of benign or malignant was not added to the DICOM header, and instead only available in the CSV metadata file. The code used for the encapsulation was written in Python Software Package [18] which is publicly available [19] and has been tested on Microsoft Windows 10 and Apple Mac OS X 10.15 Catalina systems.
We performed a practical test of the usability of the DICOM files by displaying them using two publicly available DICOM image viewers, IrfanView [https://www.irfanview.com] and RadiAnt [https://www.radiantviewer.com]. To test extracting the images, a Python script using the pydicom package was used [20]. For each DICOM file, the contents of the Pixel Data (7FE0,0010) attribute (i.e., the encapsulated compressed byte stream) was written into a binary file and the resultant (JPEG) file displayed with common image viewing software applications.
Following the challenge, the encoded files were more formally tested for conformance to the DICOM standard using dciodvfy [http://www.dclunie.com/dicom3tools/dciodvfy.html]. Based on the errors reported, further tests of readability were performed using other publicly available viewers, Horos [http://horosproject.org/], ClearCanvas [http://clearcanvas.github.io/], and PixelMed DicomImageViewer [17]. Test of transmission using the DICOM network protocol (to simulate a camera application sending to a Picture Archive and Communication Systems [PACS]) was performed using dcmtk storescu (http://dicom.offis.de/dcmtk.php.en) sending to DicomImageViewer and Horos.
Results
Encapsulation
The encapsulation process on currently available hardware (dual-core Intel CPU with > 2 GHz clock speed and at least 4 GB memory) took approximately 68 min with an average of 0.1 s per image (range 0.89–1.17 s), for the 44,108 images.
The resultant DICOM file size was approximately 1.5 kB per image greater than the original JPEG file due to the addition of the metadata. Given an average image file size of 1.8 MB, the storage overhead of the metadata was considered negligible.
Fidelity
The validation of the DICOM files using the dciodvfy utility identified problematic data values containing invalid characters in the SOP Class UID (0008,0016), Modality (0008,0060), Series Number (0020,0011) and Instance Number (0020,0013) attributes. This was caused by embedded quotes propagated from the original script that encapsulated the files, and then using Python as a wrapper. Visual inspection of a dump of the DICOM attributes revealed that the same embedded quote problem was present in other attributes that allow such quotes, such as Patient’s Name (0010,0010) and Patient’s ID (0010,0020). Both IrfanView and RadiAnt applications were able to read the DICOM objects, and both programs displayed the images contained in the DICOM files correctly (aspect ratio, colors, etc.). Other viewers failed to import or display the incorrectly encoded images. The storescu DICOM network send also failed, reporting an unknown SOP Class.
Programmatic extraction (de-encapsulation) of the compressed JPEG data to a file using the pydicom module worked for all DICOM files, which could then be read in Python (e.g., using the Python Imaging library) or opened with any common JPEG viewing software. A summary of the fidelity test are shown in Table 2.
Table 2.
Results of fidelity tests
| Fidelity test | Application | Success | Cause of error | Remedy |
|---|---|---|---|---|
| Validate conformance | dciodvfy | No | Embedded quotes | Remove quotes |
| Display image | IrfanView | Yes | ||
| Display image | RadiAnt | Yes | ||
| Display image | DicomImageViewer | No | Embedded quotes | Remove quotes |
| Display image | Horos | No | Embedded quotes | Remove quotes |
| Display image | ClearCanvas | No | Embedded quotes | Remove quotes |
| Store image | Java DICOM Toolkit storescu | No | Embedded quotes | Remove quotes |
| De-encapsulation | Pydicom module | Yes | None |
Since the pixel data in the JPEG images are not decompressed and recompressed, but rather the original JPEG byte stream is simply embedded into the Pixel Data (7FE0,0010) attribute, there was no further loss in image quality beyond the original lossy compression.
A limitation due to missing granularity of anatomic site tokens affected 150 images for “oral/genital” and 483 images for “palms/soles” which equated to a total of 1.4% of images. In addition, 878 images selected for this year’s challenge did not have their anatomic site provided in the metadata; hence, a total of 1511 images (i.e., 3.3%) had their Anatomic Region Sequence (0008,2218) attribute set to a generic code for Skin (SCT 39,937,001) (see Table 3).
Table 3.
Comparison of anatomic site coding between the ISIC Archive and DICOM metadata attributes
| ISIC Archive term (token) | SNOMED CT code | SNOMED description |
|---|---|---|
| Head/neck | 70,762,009 | Skin of head |
| Lower extremity | 281,739,007 | Skin of part of lower limb |
| Oral/genital | 39,937,001 | Skin |
| Palms/soles | 39,937,001 | Skin |
| Torso | 86,381,001 | Skin of trunk |
| Upper extremity | 281,733,008 | Skin of part of upper limb |
| Not available or unknown | 39,937,001 | Skin |
Utilization in Grand Challenge
For the duration of the challenge (May 28 through August 17, 2020) a total of 6290 users downloaded the dataset from the Kaggle website [http://kaggle.com/c/siim-isic-melanoma-classification/data].
Among these, 2350 users submitted their solution for scoring. And of these, 255 users (10.8%) downloaded the data exclusively in DICOM format, 1356 users (57.7%) downloaded DICOM intentionally, and including those who downloaded the entire bundle a total of 2039 users (86.7%) downloaded DICOM.
Discussion
This proof-of-concept has demonstrated the ease of encoding of dermoscopic images in a DICOM PS3.10 file format, including the feasibility of encoding metadata within the image file. Though consumer file formats (e.g., JPEG) are currently widely used for dermatology imaging, they lack the ability to support the additional and rich metadata that would allow dermatology as a field to more fully utilize the potential value provided by skin lesion images across the diverse range of applications.
Despite failing validation of conformance with the DICOM standard, the encapsulated DICOM PS3.10 files could be displayed by two viewing applications, which ignored the incorrect metadata. The binary JPEG compressed pixel data was extractable from the PS3.10 file using a common DICOM reading library (pydicom). Other tools that depended on the metadata (particularly the SOP Class UID that specifies the type of stored image) failed to import or display the images. The SOP Class UID allows the recipient to distinguish between objects that are images versus those that are not (e.g., reports or presentation states or radiotherapy plans) and to determine what type of image is encoded (e.g., a photographic image as opposed to a CT or ultrasound). Some viewers ignore this and will display any file that appears to contain pixel data, whereas others are more selective. Image management systems, such as PACS, are dependent on SOP Class UID and generally will reject objects of unrecognized type or if the patient identifying attributes do not match a registered patient. The DICOM network protocol for transferring images depends on a valid SOP Class UID, so sending applications will also reject these images. Other invalid values, such as quotes present in numeric attributes like Instance or Series Number may also cause failure of ingestion if the recipient uses the number (e.g., to sort images or series in numerical order).
These failures demonstrate that it is important to use a validation tool to verify conformance as opposed to simply reading and displaying files in a small number of programs. DICOM validation tools are publicly available and commonly used at interoperability testing events (such as IHE Connectathons).
Further, since the point of using DICOM is to correctly identify and describe the images using embedded metadata, the incorrect encoding (e.g., inclusion of embedded quotes) of the patient demographic attributes was also concerning. The inclusion of embedded quotes would cause a failure to match other images of the same patient, or information from the medical record obtained via HL7. Standalone validation tools will not generally detect extraneous but legal characters, such as quotes in names and identifiers. Though the melanoma detection challenge for which the DICOM images were created was completed with the invalid DICOM files, it is planned to distribute a corrected set of images, to avoid propagating these errors further into the field.
Adopting DICOM for dermatology imaging and as the standard for metadata encapsulation may provide a number of benefits. Firstly, clinicians who need to base their assessment on digital images can be expected to commit fewer errors if the available data contains information about color and size of the lesion. By extending the overall high level of comfort and ease by which data from imaging at different time points can be linked together (e.g., for ensuring that images can always be viewed in context), the risk of making mistakes after transmitting dermoscopic images between the image acquiring clinic and a secondary service provider is also greatly reduced. Next, dermoscopic image research has been shown to benefit from metadata, and if images regularly contained this information embedded in standardized attributes across image providers, the current cost for efforts in dataset curation, particularly if data is collected from several clinics, could be greatly minimized. Finally, DICOM has grown into a reliable, secure, and trusted technology, and extant software can help improve teaching of and communication about skin lesion diagnoses [21].
DICOM is well established within the medical and computer vision communities. However, dermatology (and within it dermoscopy) are relatively immature in terms of using imaging standards. DICOM offers a rich set of available metadata attributes [22]. Through efforts of the DICOM Dermatology Working Group 19, some additional dermatology (and dermoscopy-specific) attributes will be established as part of the DICOM standard [23]. We believe switching from consumer file formats to DICOM should be recommended for many participants involved in dermoscopy, including vendors, clinics, and research centers. In addition to advantages for processing DICOM files as input, outputs of machine learning algorithms (segmentations, labels) can also be stored in DICOM format, allowing clinicians to display auxiliary data (salience maps, etc.) on top of lesions, being a requirement for bringing AI advancements to the clinic. [24].
In this study, we have encountered substantial challenges with anatomic site labeling, color fidelity, and lesion size estimation in particular, which would be worthwhile to examine carefully while developing DICOM standards for dermatology applications.
Codes for anatomical structures defined in DICOM do already support dermatology [25]. Unfortunately, these are not often used and may be insufficiently granular or comprehensive enough. Recently developed hierarchical anatomical terminology sets may provide a solution [26, 27]. However, codification of these anatomical terms is required before they can be included in DICOM. The merging of disjoint anatomic sites (palms and soles, or orogenital regions) also proved problematic, and the addition of such combined sites to standard coding schemes may be necessary for clinical purposes.
Faithful reproduction of color in digital images, especially for machine learning applications, has been recognized as a crucial issue [28]. The core technological challenge lies in detection and numeric representation of relative energy density of a priori selected visible light wavelengths in order to simulate color similar to human perception. Any attempt to replicate this process using technology requires making certain assumptions, each of which determines the amount of fidelity and interpretability of the measured signal. The most common schema storing color information in digital applications is the red–green–blue (RGB) encoding. Each of the three principle RGB components is roughly mapped to the preferred wavelengths of the three types of retinal cones. RGB encoding represents each of the three wavelengths by a number, typically an integer between 0 and 255, meaning that each component has an eight-bit dynamic range or resolution. Little is known about whether machine learning algorithms might benefit from a different encoding schema, a greater dynamic range, or calibrated energy step functions. The widely accepted mechanism for achieving consistency of color rendering, regardless of how accurately it reflects the original scene, is the use of the International Color Consortium (ICC) profiles, and this is the mechanism adopted by DICOM [29].
In addition to the coding, the conditions under which an image is taken greatly impact the output of the sensor of a digital device, that is how, e.g., the charge-coupled (CCD) or complementary metal–oxide–semiconductor (CMOS) chip together with the integrated circuits regulate the interpretation of the internal sensor data. Metadata attributes such as which chip was used to convert the light entering the lens into digital information, whether the dynamic range was compressed, or whether gain was applied can help in making sense of this data in any subsequent application, although these are not typically available.
Encoding the physical distance between adjacent pixels in both the horizontal and vertical plane in the Pixel Spacing (0028,0030) attribute allows distance measurements of a subject lesion using software measurement tools. Pixel spacing was missing from this particular image set. Distance measurements are however critical to clinical decision making, which likely makes it relevant for AI classifiers as well.
Conclusion
Despite these challenges, we believe that this initial demonstration of using DICOM suggests it as a more suitable format for clinical and research applications, compared to consumer file formats. At no point were apparent barriers either cost-prohibitive from a user perspective or came with unexpected additional effort. Most clinical service providers are likely to already be in possession of DICOM technology such as clinical viewing workstations that are able to review radiology images, which also reduces the barriers to potential near-term adoption of these imaging standards for dermatology.
Taken together, we want to urge the community to consider adopting DICOM as a standard for future studies, especially those involving machine learning applications.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Liam Caffery and Jochen Weber contributed equally to this work.
References
- 1.Caffery LJ, Clunie D, Curiel-Lewandrowski C, Malvehy J, Soyer HP, Halpern AC. Transforming dermatologic imaging for the digital era: metadata and standards. Vol. 31, Journal of Digital Imaging. 2018. p. 568–77. [DOI] [PMC free article] [PubMed]
- 2.Liu Z, Sun J, Smith M, Smith L, Warr R. Incorporating clinical metadata with digital image features for automated identification of cutaneous melanoma. Vol. 169, British Journal of Dermatology. 2013. p. 1034–40. [DOI] [PubMed]
- 3.SIIM-ISIC Melanoma Classification | Kaggle [Internet]. [cited 2020 Jul 16]. Available from: https://www.kaggle.com/c/siim-isic-melanoma-classification
- 4.Rotemberg V, Kurtansky N, Betz-Stablein B, Caffery L, Chousakos E, Codella N, Combalia M, Dusza S, Guitera P, Gutman D, Halpern A, Helba B, Kittler H, Kose K, Langer S, Lioprys K, Malvehy J, Musthaq S, Nanda J, Reiter O, Shih G, Stratigos A, Tschandl P, Weber J, Soyer HP. Publisher correction: Author correction: a patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci Data. 2021;8:88. doi: 10.1038/s41597-021-00879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.ISDIS, Metadata Working Group. ISIC Melanoma Project. 2017; Available from: http://isdis.net/isic-project/isic-melanoma-project-metadata-working-group/
- 6.Codella N, Gutman D, Celebi ME, Helba B, Marchetti M, W. Dusza S, Skin lesion analysis toward melanoma detection: a challenge at the, , et al. International Symposium on Biomedical Imaging (ISBI), Hosted by the International Skin Imaging Collaboration (ISIC) Proceedings of ISBI. 2017;2018:2017. [Google Scholar]
- 7.Codella N, Rotemberg V, Tschandl P, Celebi ME, Dusza S, Gutman D, et al. Skin lesion analysis toward melanoma detection 2018: a challenge hosted by the International Skin Imaging Collaboration (ISIC). ArXiv190203368 Cs [Internet]. 2019 Mar 29 [cited 2020 Jul 7]; Available from: http://arxiv.org/abs/1902.03368
- 8.Marchetti MA, Codella NCF, Dusza SW, Gutman DA, Helba B, Kalloo A, et al. Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. Vol. 78, Journal of the American Academy of Dermatology. 2018. p. 270–277.e1. [DOI] [PMC free article] [PubMed]
- 9.Rotemberg V, Halpern A, Dusza S, Codella N. The role of public challenges and data sets towards algorithm development, trust, and use in clinical practice. Semin Cutan Med Surg. 2019;1(38):E38–42. doi: 10.12788/j.sder.2019.013. [DOI] [PubMed] [Google Scholar]
- 10.Tschandl P, Codella N, Akay BN, Argenziano G, Braun RP, Cabo H, et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: an open, web-based, international, diagnostic study. Vol. 20, The Lancet Oncology. 2019. p. 938–47. [DOI] [PMC free article] [PubMed]
- 11.Gaudy-Marqueste C, Wazaefi Y, Bruneu Y, Triller R, Thomas L, Pellacani G, et al. Ugly duckling sign as a major factor of efficiency in melanoma detection. Vol. 153, JAMA Dermatology. 2017. p. 279–84. [DOI] [PubMed]
- 12.Scope A, Dusza SW, Halpern AC, Rabinovitz H, Braun RP, Zalaudek I, et al. The “Ugly Duckling” sign: agreement between observers. Vol. 144, Archives of Dermatology. 2008. p. 58–64. [DOI] [PubMed]
- 13.National Electrical Manufacturers Association. Digital Imaging and Communications in Medicine (DICOM) Standard PS3.10 2018d—media storage and file format for media interchange [Internet]. 2018 [cited 2020 Jul 17]. Available from: http://dicom.nema.org/medical/dicom/2018d/output/html/part10.html
- 14.Combalia M, Codella NCF, Rotemberg V, Helba B, Vilaplana V, Reiter O, et al. BCN20000: dermoscopic lesions in the wild. ArXiv190802288 Cs Eess [Internet]. 2019 Aug 30 [cited 2020 Jul 7]; Available from: http://arxiv.org/abs/1908.02288
- 15.ISIC Archive [Internet]. [cited 2020 Jul 7]. Available from: https://www.isic-archive.com/
- 16.ExifTool by Phil Harvey [Internet]. [cited 2020 Jul 16]. Available from: https://exiftool.org/
- 17.PixelMed PublishingTM Java DICOM Toolkit [Internet]. [cited 2020 Jul 16]. Available from: http://www.pixelmed.com/dicomtoolkit.html
- 18.G Van Rossum, FL Drake Jr. Python Reference Manual [Internet]. Centrum voor Wiskunde en Informatica, Amsterdam; 1995 [cited 2020 Jul 7]. Available from: http://citebay.com/how-to-cite/python/
- 19.ISIC-Research/dicomwrapper [Internet]. ISIC-Research; 2020 [cited 2020 Jul 16]. Available from: https://github.com/ISIC-Research/dicomwrapper
- 20.Pydicom | [Internet]. [cited 2020 Jul 16]. Available from: https://pydicom.github.io/
- 21.Kahn CE, Carrino JA, Flynn MJ, Peck DJ, Horii SC. DICOM and radiology: past, present, and future. J Am Coll Radiol. 2007;4(9):652–657. doi: 10.1016/j.jacr.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 22.National Electrical Manufacturers Association. Registry of DICOM Data Elements [Internet]. [cited 2020 Jul 16]. Available from: http://dicom.nema.org/dicom/2013/output/chtml/part06/chapter_6.html
- 23.National Electrical Manufacturers Association. Digital Imaging and Communications in Medicine (DICOM) Supplement 221: dermoscopy [Internet]. [cited 2020 Sep 29]. Available from: https://www.dicomstandard.org/News/current/docs/sups/sup221.pdf
- 24.Caffery LJ, Rotemberg V, Weber J, Soyer HP, Malvehy J and Clunie D. The role of DICOM in artificial intelligence for skin disease. Front Med (Lausanne). 2020; 7: 619787. [DOI] [PMC free article] [PubMed]
- 25.National Electrical Manufacturers Association. Digital Imaging And Communications in Medicine (DICOM) CID 4029 Dermatology Anatomic Sites [Internet]. [cited 2020 Sep 29]. Available from: http://dicom.nema.org/medical/dicom/current/output/chtml/part16/sect_CID_4029.html
- 26.Navarrete‐Dechent C, Liopyris K, Molenda MA, Braun R, Curiel‐Lewandrowski C, Dusza SW, et al. Human surface anatomy terminology for dermatology: a Delphi consensus from the International Skin Imaging Collaboration. J Eur Acad Dermatol Venereol [Internet]. [cited 2020 Sep 14];n/a(n/a). Available from: https://onlinelibrary.wiley.com 10.1111/jdv.16855 [DOI] [PMC free article] [PubMed]
- 27.Kenneweg KA, Halpern AC, Chalmers RJG, Soyer HP, Weichenthal M, Molenda MA. Developing an international standard for the classification of surface anatomic location for use in clinical practice and epidemiologic research. J Am Acad Dermatol. 2019;80(6):1564–1584. doi: 10.1016/j.jaad.2018.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Can Karaimer H, Brown MS. Improving color reproduction accuracy on cameras. In 2018 [cited 2020 Jul 16]. p. 6440–9. Available from: https://openaccess.thecvf.com/content_cvpr_2018/html/Karaimer_Improving_Color_Reproduction_CVPR_2018_paper.html
- 29.Phil Green MK. Color management: understanding and using ICC profiles | Wiley [Internet]. [cited 2020 Sep 29]. Available from: https://www.wiley.com/en-us/Color+Management+%3A+Understanding+and+Using+ICC+Profiles-p-9780470058251
- 30.Donnelly K. SNOMED-CT: The advanced terminology and coding system for eHealth. Stud Health Technol Inform. 2006;121:279–290. [PubMed] [Google Scholar]
- 31.Altamura D, Altobelli E, Micantonio T, Piccolo D, Fargnoli MC, Peris K. Dermoscopic patterns of acral melanocytic nevi and melanomas in a white population in central Italy. Arch Dermatol. 2006;142(9):1123–1128. doi: 10.1001/archderm.142.9.1123. [DOI] [PubMed] [Google Scholar]
- 32.Braun RP, Thomas L, Dusza SW, Gaide O, Menzies S, Dalle S, et al. Dermoscopy of acral melanoma: a multicenter study on behalf of the international dermoscopy society. Vol. 227, Dermatology. 2013. p. 373–80. [DOI] [PubMed]