Abstract
Artificial or augmented intelligence, machine learning, and deep learning will be an increasingly important part of clinical practice for the next generation of radiologists. It is therefore critical that radiology residents develop a practical understanding of deep learning in medical imaging. Certain aspects of deep learning are not intuitive and may be better understood through hands-on experience; however, the technical requirements for setting up a programming and computing environment for deep learning can pose a high barrier to entry for individuals with limited experience in computer programming and limited access to GPU-accelerated computing. To address these concerns, we implemented an introductory module for deep learning in medical imaging within a self-contained, web-hosted development environment. Our initial experience established the feasibility of guiding radiology trainees through the module within a 45-min period typical of educational conferences.
Keywords: Machine learning, Deep learning, Medical education, Medical imaging
Background
Practical applications for machine learning (ML) in medical imaging are under intense development, both in academia and industry. Deep learning (DL), a subfield of ML that utilizes so-called deep convolutional neural networks, has become increasingly popular in recent years due to its excellent performance in image analysis tasks. Although clinically useful applications of DL remain scarce, the massive investment into research and development in this space suggest that DL applications will soon arrive to clinical practice. With the emergence of any new tool comes the responsibility to develop a practical understanding of the technology underlying that tool to facilitate effective clinical use. The need for a DL curriculum in radiology training — and medical education, more broadly — is clear.
No standards currently exist for the development of a DL curriculum in radiology education, nor is there a standard curriculum for the broader fields of clinical and/or imaging informatics in medical education; however, a number of recent publications have emphasized the needs and explored various approaches to integrating these important concepts into clinical education [1–10]. Data curation and annotation — on the front end — along with analysis and clinical application of model results — on the back end — are generally intuitive topics for radiologists to understand and can be presented with a traditional educational approach. However, the technical details of model development and deployment — including training, validation, and inference — may be foreign concepts to radiologists, benefiting from a more hands-on experience to promote insight. This practical knowledge will help radiologists better interpret model predictions as well as identify and analyze model failures.
The technical aspects of a DL model draw on concepts from linear algebra, statistics, and computer science. An ideal introductory module for DL in medical imaging should engage a learner at their knowledge level and foster excitement that may motivation deeper study. The “Hello World Deep Learning in Medical Imaging” module — hereafter referred to as the “Hello World” module — developed by Lakhani et al. provides a straight-forward conceptual introduction to training, validation, and testing of a DL model [11]. This module is designed to run in a Python language programming environment.
Given the growing interest in the science of DL at our institution and its associated training programs, as well as within the broader community, we sought to adapt the “Hello World” module for a more general audience, including non-coders and beginning programmers with limited programming experience. The goal of the work described herein was to facilitate the engagement of this audience with the “Hello World” module by providing a web-hosted environment with minimal setup required and by extending the module to further engage the user with simple experiments and code modifications. To bypass the necessity of installing Python, setting up an environment with the necessary libraries, downloading the dataset, and obtaining GPU access, we selected the web-hosted, cloud-based Kaggle Notebooks platform (https://www.kaggle.com/notebooks) for implementation of the “Hello World” module. The Kaggle platform is easy to use, free, and recognized within the radiology community after it hosted the 2017–2019 Radiological Society of North America (RSNA) and 2019 Society for Imaging Informatics in Medicine-American College of Radiology (SIIM-ACR) machine learning challenges. This decision allowed us to focus more time and effort on running the code and explaining the concepts highlighted in the “Hello World” module when leading the live, interactive demonstrations.
Methods
Module Design and Dataset
The “Hello World” module takes users through the process of training a deep learning model to distinguish chest radiographs from abdominal radiographs. It is implemented in the Python programming language, utilizing the Keras deep learning framework, the TensorFlow “back end” deep learning library, and the Jupyter Notebook environment. The dataset and code were obtained from Paras Lakhani’s GitHub repository [12]. The dataset was modified to match the directory structure specified in the original article and uploaded to Kaggle Datasets (https://www.kaggle.com/wfwiggins203/hello-world-deep-learning-siim). A Kaggle Notebook was created from the original Jupyter Notebook and linked to the Kaggle Dataset.
Modifications to the Original Code
The original “Hello World” module takes users through the process of establishing training, validation and test datasets for a binary classification task — distinguishing chest radiographs from abdominal radiographs. Using the Keras framework with the TensorFlow backend, the authors employ the InceptionV3 model architecture [13] and a technique called “transfer learning” to train a model that performs this task with high accuracy. In transfer learning, a model is initialized with weights (or parameters) obtained by pretraining a model on a different task [14]. Custom layers are then added to the “top” of the network to achieve the target task.
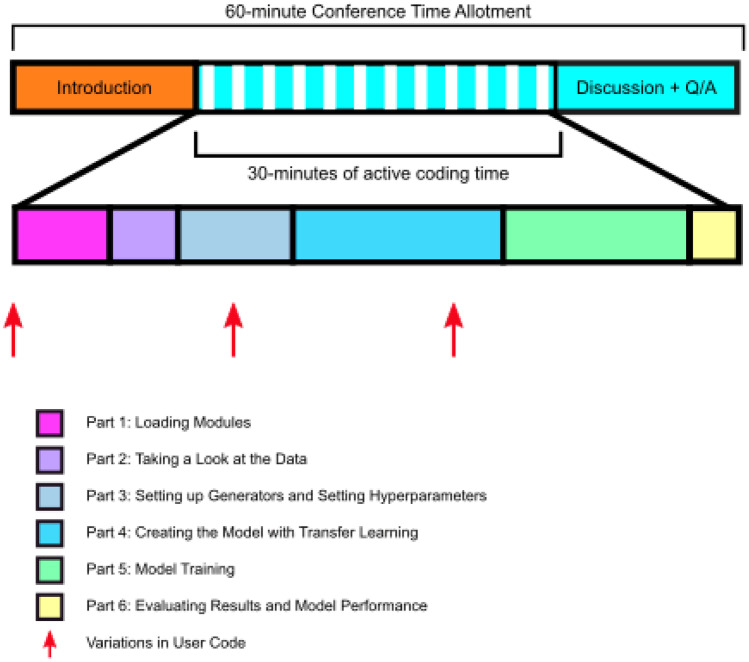
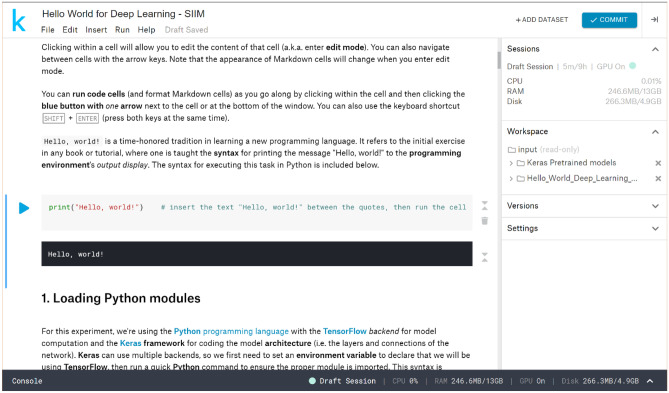
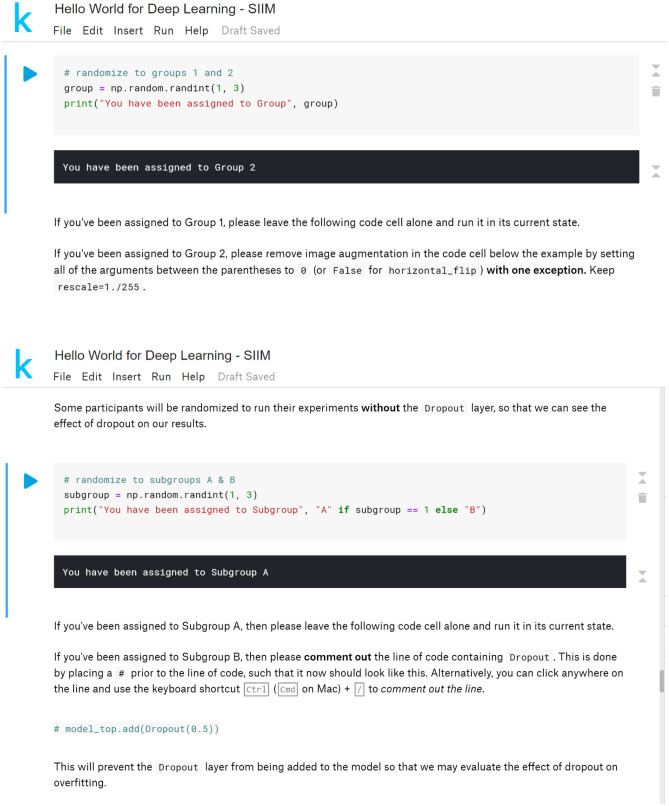
A graphical representation of the time allotted for each step in the module walkthrough is presented in Fig. 1. We modified the code for our implementation of the module, adding steps to increase participant engagement, highlight important concepts, and facilitate independent use and deeper study of the module. In addition, the code was split into smaller blocks of code placed within individual code cells to further break down each of the steps in the module. Steps requiring user input were added to increase engagement with the module, rather than have users simply run the code. An initial step to print “Hello, World!” to the output was added in homage to the name of the module as well as the time-honored tradition of performing this task when first learning a new programming language (Fig. 2). Two randomization steps were added at later points in the module, where users were prompted to make changes to the code based on the group to which they were randomized (Fig. 3). In addition to fostering participant interaction with the module, these steps highlighted important concepts in deep learning: data augmentation and “dropout.”
Fig. 1.
Overview of module walk-through timing
Fig. 2.
Code editor interface and “Hello World” task
Fig. 3.
Randomization steps with instructions for user code editing
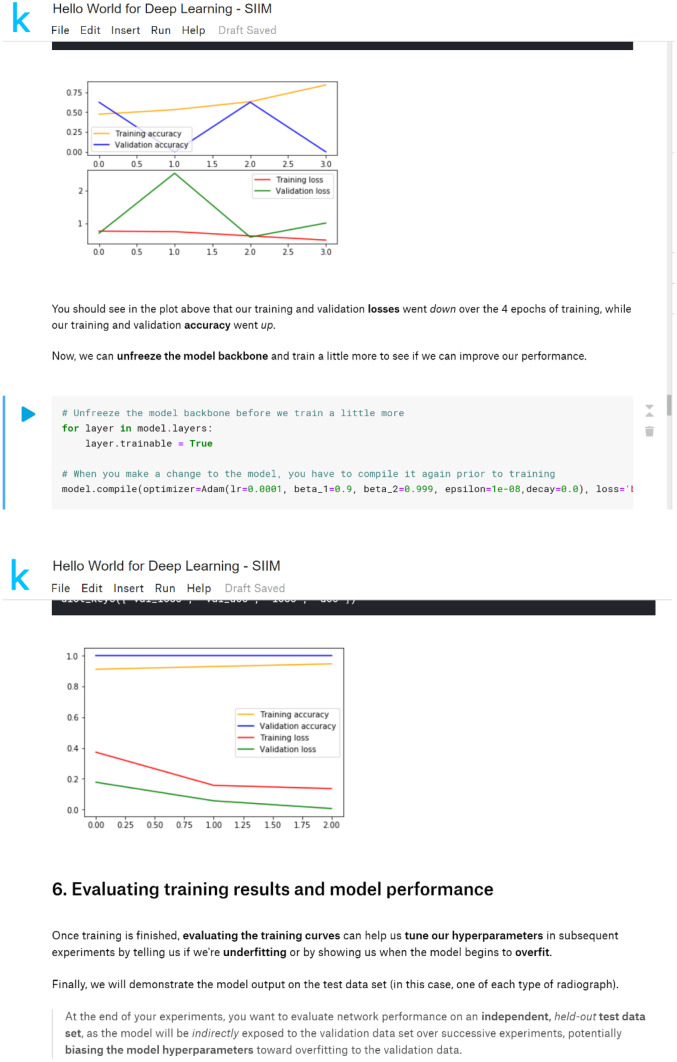
Finally, the code was modified to introduce the concept of “fine tuning” a model when applying the transfer learning technique. In the “Hello World” module, the initial weights were obtained from pretraining the model on the ImageNet general image classification task, then custom layers were added to the “top” of the network to generate the binary classification of chest radiograph or abdominal radiograph. We added a step to “freeze” (i.e., make untrainable) the pretrained portion of the network for the first few epochs of model training. Another step was added to then “unfreeze” the network (i.e., make trainable) for “fine tuning” of the pretrained weights to the target task (Fig. 4).
Fig. 4.
Transfer learning and fine-tuning steps
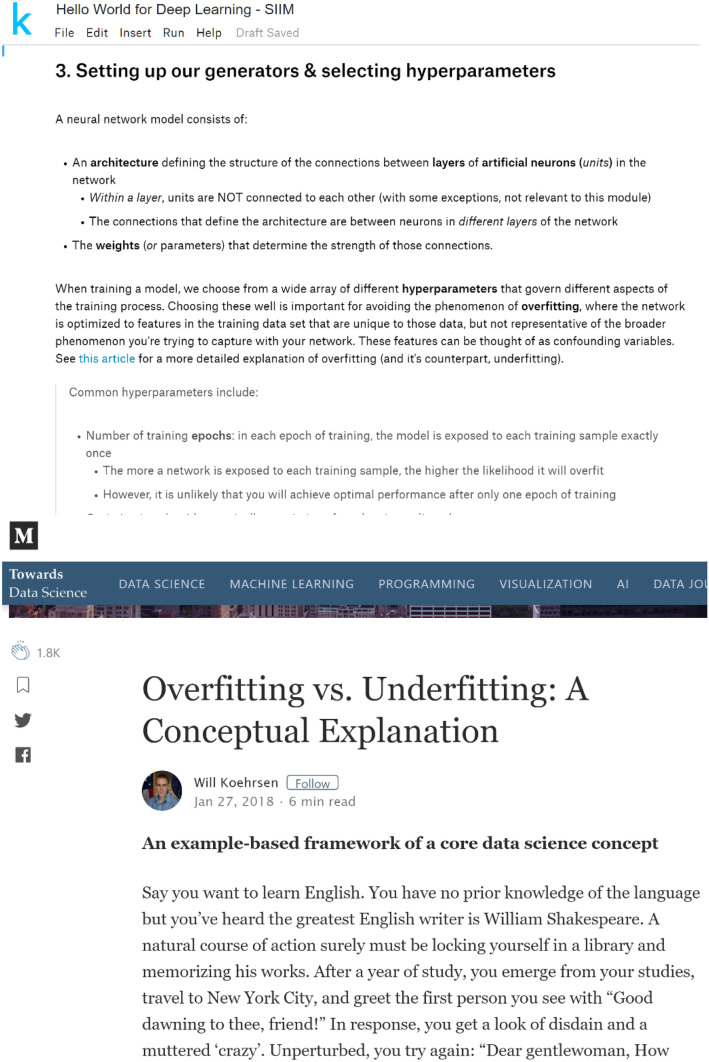
Furthermore, detailed explanations of each step in the module were generated, along with links to more in-depth external resources (Fig. 5) to facilitate independent review of the code and deeper study of the materials upon returning to the module after the live demonstration.
Fig. 5.
Link to external resource for further learning (with screen capture from resource)
Feedback
An informal survey was given to participants following each of the in-person demonstrations to obtain feedback on the module and platform.
Kaggle Notebook and Source Code Repository
The Kaggle Notebook is available at https://www.kaggle.com/wfwiggins203/hello-world-for-deep-learning-siim. If you would like to experiment with the Notebook, you must first create a Kaggle account and then “copy and edit” the original Notebook.
The source code for the Kaggle Notebook is available in Dr. Wiggins’ GitHub repository (https://github.com/wfwiggins/Hello_World_Deep_Learning).
Results and Discussion
Live demonstration of the Kaggle “Hello World” module was performed in-person during 45-min teaching conferences for two radiology training programs within our institution and one program at a neighboring institution. Presentations occurred during regularly scheduled conferences for which attendance was required at both institutions; however, active participation in the hands-on component was optional. A total of 23 residents and 6 fellows participated at our institution; 20 residents participated at the neighboring institution. A fourth demonstration was performed via an hour-long webinar for a radiology trainee-oriented journal club, for which there were 72 live participants. A recording of this webinar is available at the referenced link [15]. At the time of this writing, our version of the Kaggle notebook was accessed by 281 unique users. One user made a copy of the notebook publically available under their own profile with an additional 16 unique users accessing that version for a total of 297 unique users.
For each demonstration, the participants were asked to create an account on the Kaggle website prior to the session. At the beginning of the session, the participants created their own copy of the Kaggle Notebook to follow along with the demonstration. Once within the Notebook environment, the participants were oriented to basic details and functions of the environment itself before running the code cell-by-cell.
In our initial two experiences, the participants were required to establish Internet connectivity within the Kaggle Notebook environment to later download the pretrained weights for the DL model. The initial Internet connection requires two-factor authentication, which limited participation for one trainee with poor cellular service reception. Additionally, several users experienced errors during the download step, which required a reboot of the Notebook and re-running the preceding code cells to resolve.
Following these initial experiences, two modifications to the module were made to obviate the need for Internet connectivity and to increase efficiency of training the model during the live demonstrations. First, the pretrained weights for the Keras implementation of the InceptionV3 model were added as a separate dataset to circumvent the errors incurred when downloading the pretrained weights to the Kaggle Notebook environment. Second, Kaggle’s experimental cloud GPU support was added to increase the speed of model training and, thus, more comfortably complete the live demonstrations within the target time limit of 45 min — typical for a radiology teaching conference.
The third in-person demonstration was completed with ample time to discuss concepts introduced in the module in further detail. One participant encountered an error related to the GPU support, which was alleviated by restarting the Kaggle Notebook and re-running the preceding code cells.
The webinar-based demonstration posed new challenges, as presenters were unable to circulate among participants to monitor progress or assist with troubleshooting. This was addressed by establishing an online forum in which the participants could submit screenshots to demonstrate progress or errors encountered. To our knowledge, none of the webinar participants encountered any major errors and all were able to complete the module within the scheduled hour, which included approximately 15 min of didactic material and other discussions. A recording of this session is available on the ACR Resident & Fellow Section’s AI Journal Club YouTube Channel [15].
In each of the in-person demonstrations, informal feedback was mostly positive. Constructive criticism tended to highlight time constraints and errors, both of which were addressed prior to later demonstrations. The participants in the webinar gave informal feedback via the discussion forum with several requesting tips on how to modify the code for other datasets and tasks, suggesting a desire to continue learning with the module. To our knowledge, 7 of the residents who participated at our institution went on to participate in research projects involving deep learning for medical imaging, though we did not determine whether this was a direct effect of their participation in the module.
Through our experience with the “Hello World” module, we learned that we were able to engage multiple groups of radiology trainees in an educational, interactive experience that introduced important concept in deep learning for medical imaging within a single, 45-min session. However, there are some limitations to our approach that should be considered for future iterations on this work. While we built features into the module designed to facilitate deeper, independent engagement with the material presented, we did not implement any mechanism for assessing further engagement with the material or deeper investigation, nor did we perform longitudinal assessment of participants’ retention of the concepts introduced in this module. Furthermore, future iterations of this work should be integrated within the context of a broader ML and/or informatics curriculum.
Conclusion
We successfully developed a web-hosted, self-contained implementation of an introductory module for deep learning in medical imaging, which is freely available for use and presentable within a 45-min period typical of radiology educational conferences. This module aims to foster deeper study and experimentation with deep learning, which may facilitate deeper understanding of these tools when clinical applications of DL start to emerge in clinical radiology practice.
Acknowledgements
The authors would like to thank Drs. James Condon and Andreas Rauschecker for their suggestions regarding the design of the Kaggle Notebook implementation. We would also like to thank Dr. Judy Wawira Gichoya and the ACR Resident and Fellow Section for the invitation to present the module at their AI Journal Club.
Declarations
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lindqwister AL, Hassanpour S, Lewis PJ, Sin JM. AI-RADS: an artificial intelligence curriculum for residents. Acad Radiol. 2020. Epub 2020/10/20. 10.1016/j.acra.2020.09.017. PubMed PMID: 33071185; PubMed Central PMCID: PMCPMC7563580. [DOI] [PMC free article] [PubMed]
- 2.Park SH, Do KH, Kim S, Park JH, Lim YS. What should medical students know about artificial intelligence in medicine? J Educ Eval Health Prof. 2019;16:18. Epub 2019/07/19. 10.3352/jeehp.2019.16.18. PubMed PMID: 31319450; PubMed Central PMCID: PMCPMC6639123. [DOI] [PMC free article] [PubMed]
- 3.Siddiqui KM, Weiss DL, Dunne AP, Branstetter BF. Integrating imaging informatics into the radiology residency curriculum: rationale and example curriculum. J Am Coll Radiol. 2006;3(1):52–7. Epub 2007/04/07. 10.1016/j.jacr.2005.08.016. PubMed PMID: 17412006. [DOI] [PubMed]
- 4.Simpson SA, Cook TS. Artificial intelligence and the trainee experience in radiology. J Am Coll Radiol. 2020. Epub 2020/10/04. 10.1016/j.jacr.2020.09.028. PubMed PMID: 33010211. [DOI] [PubMed]
- 5.Slanetz PJ, Daye D, Chen PH, Salkowski LR. Artificial intelligence and machine learning in radiology education is ready for prime time. J Am Coll Radiol. 2020;17(12):1705–7. Epub 2020/05/20. 10.1016/j.jacr.2020.04.022. PubMed PMID: 32428437. [DOI] [PubMed]
- 6.Tajmir SH, Alkasab TK. Toward augmented radiologists: changes in radiology education in the era of machine learning and artificial intelligence. Acad Radiol. 2018;25(6):747–50. Epub 2018/03/31. 10.1016/j.acra.2018.03.007. PubMed PMID: 29599010. [DOI] [PubMed]
- 7.Tejani AS. Identifying and addressing barriers to an artificial intelligence curriculum. Journal of the American College of Radiology. 10.1016/j.jacr.2020.10.001. [DOI] [PubMed]
- 8.Vey BL, Cook TS, Nagy P, Bruce RJ, Filice RW, Wang KC, et al. A survey of imaging informatics fellowships and their curricula: current state assessment. J Digit Imaging. 2019;32(1):91–6. Epub 2018/10/31. 10.1007/s10278-018-0147-y. PubMed PMID: 30374655; PubMed Central PMCID: PMCPMC6382631. [DOI] [PMC free article] [PubMed]
- 9.Wiggins WF, Caton MT, Magudia K, Glomski S-hA, George E, Rosenthal MH, et al. Preparing radiologists to lead in the era of artificial intelligence: designing and implementing a focused data science pathway for senior radiology residents. Radiology: Artificial Intelligence. 2020;2(6):e200057. 10.1148/ryai.2020200057. [DOI] [PMC free article] [PubMed]
- 10.Wood MJ, Tenenholtz NA, Geis JR, Michalski MH, Andriole KP. The need for a machine learning curriculum for radiologists. J Am Coll Radiol. 2019;16(5):740–2. Epub 2018/12/12. 10.1016/j.jacr.2018.10.008. PubMed PMID: 30528932. [DOI] [PubMed]
- 11.Lakhani P, Gray DL, Pett CR, Nagy P, Shih G. Hello world deep learning in medical imaging. J Digit Imaging. 2018;31(3):283–9. Epub 2018/05/05. 10.1007/s10278-018-0079-6. PubMed PMID: 29725961; PubMed Central PMCID: PMCPMC5959832. [DOI] [PMC free article] [PubMed]
- 12.Lakhani P. Hello World Introduction to Deep Learning for Medical Image Classification https://github.com/paras42/Hello_World_Deep_Learning [updated 4/16/20183/19/2019].
- 13.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z, editors. Rethinking the inception architecture for computer vision. Proceedings of the IEEE conference on computer vision and pattern recognition; 2016.
- 14.Rawat W, Wang Z. Deep convolutional neural networks for image classification: a comprehensive review. Neural computation. 2017;29(9):2352–2449. doi: 10.1162/neco_a_00990. [DOI] [PubMed] [Google Scholar]
- 15.RFS AI Journal Club: Hands-on session for non technical beginner with model building on Kaggle https://youtu.be/BsypYX8rhBI [updated 3/1/20196/28/2021].