Abstract
This study aims to investigate the influence of interobserver manual segmentation variability on the reproducibility of 2D and 3D unenhanced computed tomography (CT)- and magnetic resonance imaging (MRI)-based texture analysis. Thirty patients with cartilaginous bone tumors (10 enchondromas, 10 atypical cartilaginous tumors, 10 chondrosarcomas) were retrospectively included. Three radiologists independently performed manual contour-focused segmentation on unenhanced CT and T1-weighted and T2-weighted MRI by drawing both a 2D region of interest (ROI) on the slice showing the largest tumor area and a 3D ROI including the whole tumor volume. Additionally, a marginal erosion was applied to both 2D and 3D segmentations to evaluate the influence of segmentation margins. A total of 783 and 1132 features were extracted from original and filtered 2D and 3D images, respectively. Intraclass correlation coefficient ≥ 0.75 defined feature stability. In 2D vs. 3D contour-focused segmentation, the rates of stable features were 74.71% vs. 86.57% (p < 0.001), 77.14% vs. 80.04% (p = 0.142), and 95.66% vs. 94.97% (p = 0.554) for CT and T1-weighted and T2-weighted images, respectively. Margin shrinkage did not improve 2D (p = 0.343) and performed worse than 3D (p < 0.001) contour-focused segmentation in terms of feature stability. In 2D vs. 3D contour-focused segmentation, matching stable features derived from CT and MRI were 65.8% vs. 68.7% (p = 0.191), and those derived from T1-weighted and T2-weighted images were 76.0% vs. 78.2% (p = 0.285). 2D and 3D radiomic features of cartilaginous bone tumors extracted from unenhanced CT and MRI are reproducible, although some degree of interobserver segmentation variability highlights the need for reliability analysis in future studies.
Keywords: Artificial intelligence, Chondroma, Chondrosarcoma, Neoplasms, Radiomics, Texture analysis
Introduction
Cartilaginous tumors of the bone include a broad spectrum of lesions that range from benign to malignant entities [1, 2]. Reliable identification and grading are crucial, as clinical management varies widely. Specifically, asymptomatic benign enchondromas do not require any treatment, appendicular atypical cartilaginous tumors are managed with intralesional curettage or even watchful waiting, and appendicular higher grade lesions and axial skeleton chondrosarcomas are resected with free margins [3]. The diagnosis relies on a combination of clinical presentation, imaging, and biopsy [3, 4]. Imaging, and particularly magnetic resonance imaging (MRI), has good accuracy in discriminating atypical cartilaginous tumors from higher grade lesions [5] but is less reliable in differentiating the former from enchondromas [6]. Biopsy is considered the reference standard but has the disadvantages of sampling errors [7] and discrepancies even among specialized bone pathologists due to overlapping histological findings [8]. Additionally, the risk of biopsy-tract contamination remains a concern. Thus, the need for cutting-edge imaging-based tools, such as radiomics, is advocated to safely diagnose and grade cartilaginous bone tumors non-invasively [9].
Texture analysis is a post-processing method for quantification of tumor heterogeneity, which reflects adverse tumor biology but cannot be captured using conventional imaging modalities or sampling biopsies [10]. It belongs to the growing field of radiomics, which includes extraction, analysis, and interpretation of large amounts of quantitative parameters from medical images [11, 12]. To date, texture analysis has been used to discriminate tumor grades and types before treatment, monitor response to therapy, and predict outcome [13]. The resulting quantitative parameters, known as texture or radiomic features, may suffer however from interobserver variability, particularly with regard to tumor delineation while performing manual segmentation [14–16]. The influence of segmentation margins is also critical because of textural details of the peritumoral area, which may affect the reproducibility of texture features and therefore their diagnostic performance [17]. In literature, the intraclass correlation coefficient (ICC) is commonly employed to assess radiomic feature reproducibility [17–21].
The aim of this study is to investigate the influence of interobserver manual segmentation variability on the reproducibility of bidimensional (2D) and volumetric (3D) unenhanced computed tomography (CT)- and MRI-based texture analysis in cartilaginous bone tumors.
Materials and Methods
Design and Population
The local Institutional Review Board approved this retrospective study and waived the need for informed consent. According to the ICC guidelines by Koo et al. [22], we designed our study to meet the numerical requirements of a reliability analysis in terms of both patients and observers involved, namely 30 lesions and 3 different readers [22]. A search of the radiology information system was performed and 30 patients with cartilaginous bone tumors were recruited (median age 52 [range, 28–72] years), including 10 enchondromas, 10 atypical cartilaginous tumors, and 10 chondrosarcomas. Inclusion criteria were as follows: (i) enchondromas proven either by histology or minimum follow-up of 6 years without alteration in shape or size and typical imaging findings of lobulated morphology and T2-weighted hyperintensity on MRI; (ii) histology-proven atypical cartilaginous tumors; (iii) histology-proven primary conventional grades II–III or dedifferentiated chondrosarcomas; (iv) 1.5-T MRI including turbo spin echo T1-weighted and T2-weighted sequences and 64-slice CT performed within 1 month before biopsy, intralesional curettage, or surgical resection for tumors diagnosed by histology. Exclusion criteria were the presence of pathological fracture and ambiguous histology report.
Enchondromas were located in the femur (n = 5), fibula (n = 2), foot phalanx (n = 1), humerus (n = 1), and radius (n = 1); atypical cartilaginous tumors in the femur (n = 2), fibula (n = 2), and humerus (n = 6); chondrosarcomas in the calcaneus (n = 1), femur (n = 2), humerus (n = 1), pelvis (n = 2), spine (n = 3), and tibia (n = 1).
Image Segmentation
A musculoskeletal radiologist (S.G.) and two last-year radiology residents trained in musculoskeletal and oncologic imaging (I.E. and L.T.) independently performed manual image segmentation using the open-source software ITK-SNAP (v3.6) [23]. The readers knew the study would deal with cartilaginous bone tumors, but they were blinded to any other information regarding histological grade, disease course, and additional imaging studies. All tumors were segmented on axial CT scans and on axial MRI sequences as first choice and coronal or sagittal sequences as second choice. Manual contour-focused segmentation was performed on unenhanced bone-window CT and T1-weighted and T2-weighted MRI by drawing both a 2D region of interest (ROI) on the slice showing the largest tumor area and a 3D ROI including the whole tumor volume. The “polygon mode” ITK-SNAP tool was used for all segmentations. While segmenting the tumors on CT, the readers used the MRI sequences to aid contour identification of each tumor. Thereafter, margin shrinkage segmentation was computed by applying a marginal erosion to both 2D and 3D segmentations in order to evaluate the influence of segmentation margins on feature reproducibility (Fig. 1). In detail, ROI shrinkage was performed using the fslmaths erosion function of the FMRIB Software Library [24]. The default 2D and 3D kernels, which are 3 × 3 × 1 and 3 × 3 × 3 boxes centered at the target voxel, were employed as appropriate. During the erosion process, each voxel in the ROI is targeted sequentially, and its value is changed to 0 (i.e., removed from the ROI) if a zero-value voxel is found within the kernel. Therefore, the shrinkage was usually more extensive for 3D ROIs compared to 2D ones.
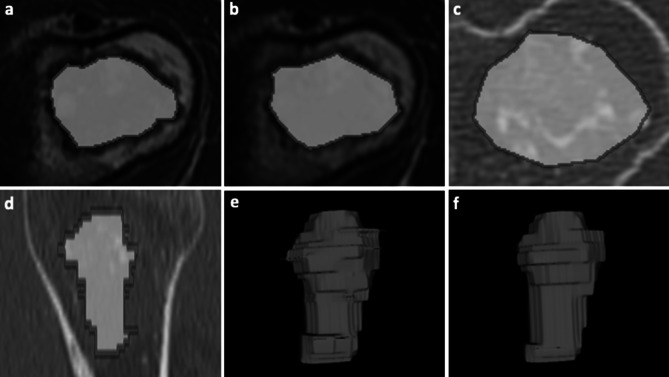
Fig. 1.
Contour-focused and margin shrinkage segmentation of an atypical cartilaginous tumor of the humerus in a 45-year-old woman. a–c 2D contour-focused segmentation was performed on axial T1-weighted MRI a, T2-weighted MRI b, and bone-window CT c on the slice showing the largest tumor extension. d 3D contour-focused segmentation was performed slice by slice in the axial plane to include the whole tumor volume, as shown in the sagittal CT image. Contour-focused segmentation provided the ROI including both green and red areas. Margin shrinkage segmentation provided the ROI including only the green area by computing a marginal erosion, which is shown in red. e–f Segmented tumor volumes obtained with 3D contour-focused e and margin shrinkage f segmentation are shown, where the latter has smoother margins as a result of marginal erosion
Texture Analysis
Image pre-processing consisted in resampling to a 2 × 2 isotropic pixel or 2 × 2 × 2 isotropic voxel, whole-image intensity normalization (mean value of 300 and standard deviation of 100), and discretization with a fixed bin width of 5. Original CT and MRI and 2D and 3D ROIs were used for feature extraction on PyRadiomics (v2.2.0) [25], an open-source Python software. The extracted features were grouped according to PyRadiomics official documentation (https://pyradiomics.readthedocs.io/en/latest/features.html), as follows:
18 first-order features, which describe the distribution of pixel or voxel gray-level values;
9 shape-based 2D and 14 shape-based 3D features, which respectively describe the 2D and 3D size and shape of the ROI;
22 Gy-level cooccurrence matrix (GLCM) features, which quantify how often pairs of pixels or voxels with certain values occur in a specified spatial range;
16 Gy-level size zone matrix (GLSZM) features, which quantify gray-level zones, i.e., the number of connected pixels or voxels sharing the same gray-level value;
16 Gy-level run length matrix (GLRLM) features, which quantify gray-level runs, i.e., the length in number of consecutive pixels or voxels having the same gray-level value;
14 Gy-level dependence matrix (GLDM) features, which quantify gray-level dependencies, i.e., the number of connected pixels or voxels within a set distance that are dependent on the center pixel and voxel.
In addition to the original CT and MRI, Laplacian of Gaussian (LoG)-filtered (sigma = 2, 3, 4, 5) and wavelet-transformed 2D and 3D images (all possible low- and high-pass filter combinations) were obtained for extraction of first-order and matrix features. Shape-based features are independent from gray-level value distribution and therefore were only computed on the original images. A total of 783 and 1132 features were extracted from original, LoG-filtered, and wavelet-transformed 2D and 3D images, respectively.
Statistical Analysis
Texture feature interobserver reliability was assessed using a two-way, random-effects, single-rater, absolute agreement ICC. Features were considered stable when achieving good (0.75 ≤ ICC < 0.9) to excellent (ICC ≥ 0.9) interobserver reliability [22]. Differences among variables were evaluated using Chi-square test. A 2-sided p-value < 0.05 indicated statistical significance [26]. Data analysis was performed using the pandas and numpy Python software and the “irr” R package [27, 28].
Machine Learning Analysis
To assess the potential value of CT and MRI texture features extracted from 2D and 3D annotations, an exploratory data analysis was performed with an Extra Trees (ET) ensemble model. The same pipeline was employed on all available datasets, consisting of feature selection through cross-validated recursive feature elimination (RFE) and random search hyperparameter tuning nested within a leave-one-out cross-validation on the entire dataset. RFE was conducted using tenfold cross-validation and an ET estimator with default hyperparameters. Then, in the training folds of the leave-one-out cross-validation, the synthetic oversampling technique was applied to balance the 3 classes (i.e., creating a synthetic instance to substitute the lesion in the test fold), followed by 100 iterations of ET hyperparameter random search. Given the presence of 3 classes with balanced cases, accuracy was used as the reference score for both RFE and ET tuning. The hyperparameter search space was as follows:
Number of trees = 100–1000
Criterion = entropy or Gini
Max depth = 1–10
Bootstrap = True or False
Max samples = 0–100%
Results
In 2D contour-focused vs. margin shrinkage segmentation, the stable feature rates were 74.71% (n = 585) vs. 71.65% (n = 561), 77.14% (n = 604) vs. 76.12% (n = 596), and 95.66% (n = 749) vs. 96.42% (n = 755) for CT and T1-weighted and T2-weighted images, respectively. The number of stable features derived from 2D contour-focused segmentation showed no difference in comparison with 2D margin shrinkage segmentation (p = 0.343). Table 1 details the number and percentage of stable features that were obtained with 2D contour-focused segmentation, grouped according to feature class and image type.
Table 1.
2D contour-focused segmentation. Number and percentage of stable features with good (0.75 ≤ ICC < 0.9) and excellent (ICC ≥ 0.9) interobserver reliability grouped according to feature class and image type. GLCM, gray-level cooccurrence matrix; GLDM, gray-level dependence matrix; GLRLM, gray-level run length matrix; GLSZM, gray-level size zone matrix; ICC, intraclass correlation coefficient; LoG, Laplacian of Gaussian
| 2D | Feature class | Image type | Total features (n) | ICC ≥ 0.75 (n) | ICC ≥ 0.75 (%) | ICC ≥ 0.90 (n) | ICC ≥ 0.90 (%) |
|---|---|---|---|---|---|---|---|
| CT | First order | LoG | 72 | 63 | 87.50 | 29 | 40.28 |
| Original | 18 | 16 | 88.89 | 8 | 44.44 | ||
| Wavelet | 72 | 53 | 73.61 | 20 | 27.78 | ||
| GLCM | LoG | 88 | 78 | 88.64 | 31 | 35.23 | |
| Original | 22 | 13 | 59.09 | 5 | 22.73 | ||
| Wavelet | 88 | 60 | 68.18 | 27 | 30.68 | ||
| GLDM | LoG | 56 | 49 | 87.50 | 18 | 32.14 | |
| Original | 14 | 10 | 71.43 | 2 | 14.29 | ||
| Wavelet | 56 | 34 | 60.71 | 10 | 17.86 | ||
| GLRLM | LoG | 64 | 58 | 90.63 | 27 | 42.19 | |
| Original | 16 | 13 | 81.25 | 2 | 12.50 | ||
| Wavelet | 64 | 43 | 67.19 | 12 | 18.75 | ||
| GLSZM | LoG | 64 | 46 | 71.88 | 20 | 31.25 | |
| Original | 16 | 9 | 56.25 | 3 | 18.75 | ||
| Wavelet | 64 | 32 | 50.00 | 14 | 21.88 | ||
| Shape | Original | 9 | 8 | 88.89 | 7 | 77.78 | |
| Overall | 783 | 585 | 74.71 | 235 | 30.01 | ||
| T1w | First order | LoG | 72 | 65 | 90.28 | 42 | 58.33 |
| Original | 18 | 15 | 83.33 | 8 | 44.44 | ||
| Wavelet | 72 | 52 | 72.22 | 27 | 37.50 | ||
| GLCM | LoG | 88 | 81 | 92.05 | 48 | 54.55 | |
| Original | 22 | 17 | 77.27 | 10 | 45.45 | ||
| Wavelet | 88 | 67 | 76.14 | 50 | 56.82 | ||
| GLDM | LoG | 56 | 43 | 76.79 | 29 | 51.79 | |
| Original | 14 | 10 | 71.43 | 7 | 50.00 | ||
| Wavelet | 56 | 38 | 67.86 | 30 | 53.57 | ||
| GLRLM | LoG | 64 | 51 | 79.69 | 34 | 53.13 | |
| Original | 16 | 12 | 75.00 | 9 | 56.25 | ||
| Wavelet | 64 | 46 | 71.88 | 35 | 54.69 | ||
| GLSZM | LoG | 64 | 50 | 78.13 | 26 | 40.63 | |
| Original | 16 | 8 | 50.00 | 6 | 37.50 | ||
| Wavelet | 64 | 40 | 62.50 | 19 | 29.69 | ||
| Shape | Original | 9 | 9 | 100.00 | 8 | 88.89 | |
| Overall | 783 | 604 | 77.14 | 388 | 49.55 | ||
| T2w | First order | LoG | 72 | 68 | 94.44 | 61 | 84.72 |
| Original | 18 | 16 | 88.89 | 15 | 83.33 | ||
| Wavelet | 72 | 60 | 83.33 | 48 | 66.67 | ||
| GLCM | LoG | 88 | 86 | 97.73 | 79 | 89.77 | |
| Original | 22 | 22 | 100.00 | 18 | 81.82 | ||
| Wavelet | 88 | 84 | 95.45 | 71 | 80.68 | ||
| GLDM | LoG | 56 | 56 | 100.00 | 48 | 85.71 | |
| Original | 14 | 12 | 85.71 | 10 | 71.43 | ||
| Wavelet | 56 | 53 | 94.64 | 31 | 55.36 | ||
| GLRLM | LoG | 64 | 64 | 100.00 | 60 | 93.75 | |
| Original | 16 | 15 | 93.75 | 13 | 81.25 | ||
| Wavelet | 64 | 63 | 98.44 | 45 | 70.31 | ||
| GLSZM | LoG | 64 | 64 | 100.00 | 47 | 73.44 | |
| Original | 16 | 15 | 93.75 | 12 | 75.00 | ||
| Wavelet | 64 | 62 | 96.88 | 41 | 64.06 | ||
| Shape | Original | 9 | 9 | 100.00 | 8 | 88.89 | |
| Overall | 783 | 749 | 95.66 | 607 | 77.52 | ||
In 3D contour-focused vs. margin shrinkage segmentation, the stable feature rates were 86.57% (n = 980) vs. 83.66% (n = 947), 80.04% (n = 906) vs. 71.47% (n = 809), and 94.97% (n = 1075) vs. 65.72% (n = 744) for CT and T1-weighted and T2-weighted images, respectively. The number of stable features derived from 3D contour-focused segmentation was higher compared to 3D margin shrinkage segmentation (p < 0.001). Table 2 details the number and percentage of stable features that were obtained with 3D contour-focused segmentation, grouped according to feature class and image type.
Table 2.
3D contour-focused segmentation. Number and percentage of stable features with good (0.75 ≤ ICC < 0.9) and excellent (ICC ≥ 0.9) interobserver reliability grouped according to feature class and image type. GLCM, gray-level cooccurrence matrix; GLDM, gray-level dependence matrix; GLRLM, gray-level run length matrix; GLSZM, gray-level size zone matrix; ICC, intraclass correlation coefficient; LoG, Laplacian of Gaussian
| 3D | Feature class | Image type | Total features (n) | ICC ≥ 0.75 (n) | ICC ≥ 0.75 (%) | ICC ≥ 0.90 (n) | ICC ≥ 0.90 (%) |
|---|---|---|---|---|---|---|---|
| CT | First order | LoG | 72 | 64 | 88.89 | 44 | 61.11 |
| Original | 18 | 14 | 77.78 | 9 | 50.00 | ||
| Wavelet | 144 | 114 | 79.17 | 93 | 64.58 | ||
| GLCM | LoG | 88 | 86 | 97.73 | 65 | 73.86 | |
| Original | 22 | 22 | 100.00 | 19 | 86.36 | ||
| Wavelet | 176 | 169 | 96.02 | 153 | 86.93 | ||
| GLDM | LoG | 56 | 50 | 89.29 | 24 | 42.86 | |
| Original | 14 | 13 | 92.86 | 8 | 57.14 | ||
| Wavelet | 112 | 98 | 87.50 | 71 | 63.39 | ||
| GLRLM | LoG | 64 | 62 | 96.88 | 30 | 46.88 | |
| Original | 16 | 14 | 87.50 | 9 | 56.25 | ||
| Wavelet | 128 | 112 | 87.50 | 86 | 67.19 | ||
| GLSZM | LoG | 64 | 46 | 71.88 | 19 | 29.69 | |
| Original | 16 | 11 | 68.75 | 2 | 12.50 | ||
| Wavelet | 128 | 93 | 72.66 | 67 | 52.34 | ||
| Shape | Original | 14 | 12 | 85.71 | 7 | 50.00 | |
| Overall | 1132 | 980 | 86.57 | 706 | 62.37 | ||
| T1w | First order | LoG | 72 | 67 | 93.06 | 43 | 59.72 |
| Original | 18 | 12 | 66.67 | 7 | 38.89 | ||
| Wavelet | 144 | 121 | 84.03 | 89 | 61.81 | ||
| GLCM | LoG | 88 | 77 | 87.50 | 47 | 53.41 | |
| Original | 22 | 16 | 72.73 | 10 | 45.45 | ||
| Wavelet | 176 | 151 | 85.80 | 125 | 71.02 | ||
| GLDM | LoG | 56 | 42 | 75.00 | 24 | 42.86 | |
| Original | 14 | 9 | 64.29 | 7 | 50.00 | ||
| Wavelet | 112 | 85 | 75.89 | 60 | 53.57 | ||
| GLRLM | LoG | 64 | 50 | 78.13 | 31 | 48.44 | |
| Original | 16 | 9 | 56.25 | 6 | 37.50 | ||
| Wavelet | 128 | 99 | 77.34 | 77 | 60.16 | ||
| GLSZM | LoG | 64 | 47 | 73.44 | 21 | 32.81 | |
| Original | 16 | 10 | 62.50 | 5 | 31.25 | ||
| Wavelet | 128 | 97 | 75.78 | 55 | 42.97 | ||
| Shape | Original | 14 | 14 | 100.00 | 11 | 78.57 | |
| Overall | 1132 | 906 | 80.04 | 618 | 54.59 | ||
| T2w | First order | LoG | 72 | 70 | 97.22 | 53 | 73.61 |
| Original | 18 | 17 | 94.44 | 11 | 61.11 | ||
| Wavelet | 144 | 126 | 87.50 | 94 | 65.28 | ||
| GLCM | LoG | 88 | 81 | 92.05 | 69 | 78.41 | |
| Original | 22 | 21 | 95.45 | 15 | 68.18 | ||
| Wavelet | 176 | 169 | 96.02 | 145 | 82.39 | ||
| GLDM | LoG | 56 | 55 | 98.21 | 41 | 73.21 | |
| Original | 14 | 14 | 100.00 | 9 | 64.29 | ||
| Wavelet | 112 | 106 | 94.64 | 73 | 65.18 | ||
| GLRLM | LoG | 64 | 64 | 100.00 | 53 | 82.81 | |
| Original | 16 | 16 | 100.00 | 11 | 68.75 | ||
| Wavelet | 128 | 122 | 95.31 | 92 | 71.88 | ||
| GLSZM | LoG | 64 | 62 | 96.88 | 46 | 71.88 | |
| Original | 16 | 16 | 100.00 | 11 | 68.75 | ||
| Wavelet | 128 | 122 | 95.31 | 70 | 54.69 | ||
| Shape | Original | 14 | 14 | 100.00 | 9 | 64.29 | |
| Overall | 1132 | 1075 | 94.97 | 802 | 70.85 | ||
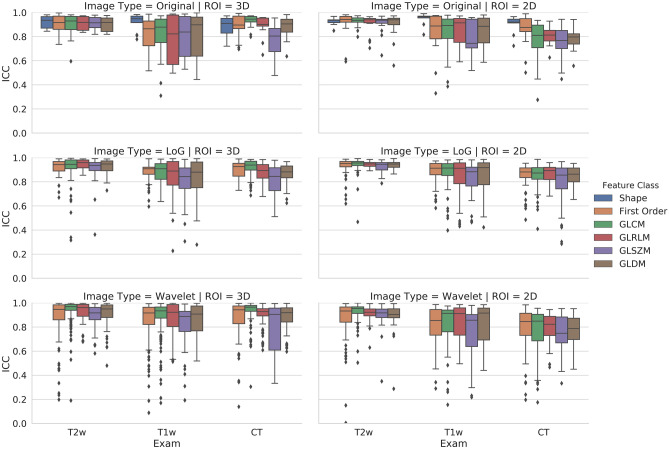
The rate of stable features derived from CT was higher for 3D compared to 2D contour-focused segmentation (p < 0.001), while no difference was found for features derived from T1-weighted and T2-weighted MRI between 3D and 2D contour-focused segmentation (p = 0.142 and 0.554, respectively). In Fig. 2, box and whisker plots show the interobserver reliability of feature classes derived from 3D and 2D contour-focused segmentation, grouped according to image type.
Fig. 2.
3D and 2D contour-focused segmentation. Box and whisker plots show the interobserver reliability of feature classes grouped according to image type
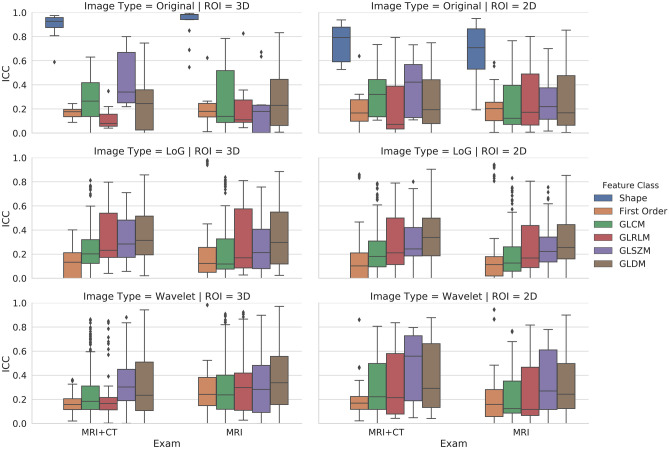
In 2D vs. 3D contour-focused segmentation, matching stable features derived from CT and MRI were 65.77% (n = 515) vs. 68.73% (n = 778), and those derived from T1-weighted and T2-weighted images were 75.99% (n = 595) vs. 78.18% (n = 885), respectively (p = 0.191 and 0.285). Tables 3 and 4 respectively detail the number and percentage of matching stable features obtained with 2D and 3D contour-focused segmentation, as well as overall interobserver reliability across different imaging modalities and MRI sequences, grouped according to feature class and image type. In Fig. 3, box and whisker plots show the overall interobserver reliability of matching feature classes derived 3D and 2D contour-focused segmentation of CT and MRI, as well as MRI including T1-weighted and T2-weighted sequences, grouped according to image type. Most shape-based 2D and 3D features were stable even across different imaging modalities and MRI sequences.
Table 3.
2D matching features. Number and percentage of matching stable features obtained with 2D contour-focused segmentation, as well as number and percentage of matching stable features with good (ICC ≥ 0.75) overall interobserver reliability across different imaging modalities and MRI sequences, grouped according to feature class and image type. GLCM, gray-level cooccurrence matrix; GLDM, gray-level dependence matrix; GLRLM, gray-level run length matrix; GLSZM, gray-level size zone matrix; ICC, intraclass correlation coefficient; LoG, Laplacian of Gaussian
| 2D | Feature class | Image type | Total features (n) | Matching features (n) | Matching features (%) | ICC ≥ 0.75 (n) | ICC ≥ 0.75 (%) |
|---|---|---|---|---|---|---|---|
| CT + MRI (T1w + T2w) | First order | LoG | 72 | 61 | 84.72 | 4 | 6.56 |
| Original | 18 | 15 | 83.33 | 0 | 0 | ||
| Wavelet | 72 | 45 | 62.50 | 3 | 6.67 | ||
| GLCM | LoG | 88 | 74 | 84.09 | 2 | 2.70 | |
| Original | 22 | 11 | 50.00 | 0 | 0 | ||
| Wavelet | 88 | 55 | 62.50 | 2 | 3.64 | ||
| GLDM | LoG | 56 | 41 | 73.21 | 4 | 9.76 | |
| Original | 14 | 7 | 50.00 | 0 | 0 | ||
| Wavelet | 56 | 29 | 51.79 | 6 | 20.69 | ||
| GLRLM | LoG | 64 | 48 | 75.00 | 1 | 2.08 | |
| Original | 16 | 10 | 62.50 | 1 | 10.00 | ||
| Wavelet | 64 | 36 | 56.25 | 7 | 19.44 | ||
| GLSZM | LoG | 64 | 40 | 62.50 | 1 | 2.50 | |
| Original | 16 | 7 | 43.75 | 0 | 0 | ||
| Wavelet | 64 | 28 | 43.75 | 3 | 10.71 | ||
| Shape | Original | 9 | 8 | 88.89 | 4 | 50.00 | |
| Overall | 783 | 515 | 65.77 | 38 | 7.38 | ||
| MRI (T1w + T2w) | First order | LoG | 72 | 63 | 87.50 | 8 | 12.70 |
| Original | 18 | 15 | 83.33 | 0 | 0 | ||
| Wavelet | 72 | 50 | 69.44 | 6 | 12.00 | ||
| GLCM | LoG | 88 | 80 | 90.91 | 2 | 2.50 | |
| Original | 22 | 17 | 77.27 | 1 | 5.88 | ||
| Wavelet | 88 | 65 | 73.86 | 2 | 3.08 | ||
| GLDM | LoG | 56 | 43 | 76.79 | 2 | 4.65 | |
| Original | 14 | 9 | 64.29 | 1 | 11.11 | ||
| Wavelet | 56 | 37 | 66.07 | 6 | 16.22 | ||
| GLRLM | LoG | 64 | 51 | 79.69 | 1 | 1.96 | |
| Original | 16 | 12 | 75.00 | 2 | 16.67 | ||
| Wavelet | 64 | 46 | 71.88 | 2 | 4.35 | ||
| GLSZM | LoG | 64 | 50 | 78.13 | 1 | 2.00 | |
| Original | 16 | 8 | 50.00 | 0 | 0 | ||
| Wavelet | 64 | 40 | 62.50 | 2 | 5.00 | ||
| Shape | Original | 9 | 9 | 100.00 | 4 | 44.44 | |
| Overall | 783 | 595 | 75.99 | 40 | 6.72 | ||
Table 4.
3D matching features. Number and percentage of matching stable features obtained with 3D contour-focused segmentation, as well as number and percentage of matching stable features with good (ICC ≥ 0.75) overall interobserver reliability across different imaging modalities and MRI sequences, grouped according to feature class and image type. GLCM, gray-level cooccurrence matrix; GLDM, gray-level dependence matrix; GLRLM, gray-level run length matrix; GLSZM, gray-level size zone matrix; ICC, intraclass correlation coefficient; LoG, Laplacian of Gaussian
| 3D | Feature class | Image type | Total features (n) | Matching features (n) | Matching features (%) | ICC ≥ 0.75 (n) | ICC ≥ 0.75 (%) |
|---|---|---|---|---|---|---|---|
| CT + MRI (T1w + T2w) | First order | LoG | 72 | 57 | 79.17 | 0 | 0 |
| Original | 18 | 10 | 55.56 | 0 | 0 | ||
| Wavelet | 144 | 97 | 67.36 | 0 | 0 | ||
| GLCM | LoG | 88 | 75 | 85.23 | 4 | 5.33 | |
| Original | 22 | 16 | 72.73 | 0 | 0 | ||
| Wavelet | 176 | 147 | 83.52 | 6 | 4.08 | ||
| GLDM | LoG | 56 | 37 | 66.07 | 5 | 13.51 | |
| Original | 14 | 8 | 57.14 | 0 | 0 | ||
| Wavelet | 112 | 72 | 64.29 | 6 | 8.33 | ||
| GLRLM | LoG | 64 | 48 | 75.00 | 1 | 2.08 | |
| Original | 16 | 7 | 43.75 | 0 | 0 | ||
| Wavelet | 128 | 81 | 63.28 | 3 | 3.70 | ||
| GLSZM | LoG | 64 | 34 | 53.13 | 0 | 0 | |
| Original | 16 | 5 | 31.25 | 1 | 20.00 | ||
| Wavelet | 128 | 72 | 56.25 | 6 | 8.33 | ||
| Shape | Original | 14 | 12 | 85.71 | 11 | 91.67 | |
| Overall | 1132 | 778 | 68.73 | 43 | 5.53 | ||
| MRI (T1w + T2w) | First order | LoG | 72 | 65 | 90.28 | 8 | 12.31 |
| Original | 18 | 12 | 66.67 | 0 | 0 | ||
| Wavelet | 144 | 116 | 80.56 | 14 | 12.07 | ||
| GLCM | LoG | 88 | 75 | 85.23 | 10 | 13.33 | |
| Original | 22 | 16 | 72.73 | 2 | 12.50 | ||
| Wavelet | 176 | 149 | 84.66 | 16 | 10.74 | ||
| GLDM | LoG | 56 | 42 | 75.00 | 6 | 14.29 | |
| Original | 14 | 9 | 64.29 | 1 | 11.11 | ||
| Wavelet | 112 | 83 | 74.11 | 10 | 12.05 | ||
| GLRLM | LoG | 64 | 50 | 78.13 | 3 | 6.00 | |
| Original | 16 | 9 | 56.25 | 1 | 11.11 | ||
| Wavelet | 128 | 96 | 75.00 | 10 | 10.42 | ||
| GLSZM | LoG | 64 | 47 | 73.44 | 2 | 4.26 | |
| Original | 16 | 10 | 62.50 | 0 | 0 | ||
| Wavelet | 128 | 92 | 71.88 | 6 | 6.52 | ||
| Shape | Original | 14 | 14 | 100.00 | 12 | 85.71 | |
| Overall | 1132 | 885 | 78.18 | 101 | 11.41 | ||
Fig. 3.
3D and 2D contour-focused segmentation. Box and whisker plots show the overall interobserver reliability of matching feature classes derived from CT and MRI, as well as T1-weighted and T2-weighted MRI sequences, grouped according to image type
Regarding the machine learning pipeline, the number of selected features ranged from 1 (from 2D annotations on T2-weighted images) to 236 (2D annotations on CT images). The accuracy of the ET models was fair to good, ranging between 77% (2D annotations on CT images) and 90% (3D annotations on T2-weighted images). Table 5 reports the results of each annotation and image type combination.
Table 5.
Feature selection process and exploratory machine learning pipeline in the reproducible feature datasets. The results of each annotation and image type combination are reported
| Annotation type | Imaging modality | Selected features (n) | Accuracy (%) |
|---|---|---|---|
| 2D | T1w | 5 | 83 |
| T2w | 1 | 83 | |
| CT | 236 | 77 | |
| 3D | T1w | 67 | 87 |
| T2w | 14 | 90 | |
| CT | 108 | 80 |
Discussion
The main finding of our study is that the rates of stable radiomic features extracted from unenhanced CT and MRI were 75% or higher for 2D and 80% or higher for 3D contour-focused segmentation. 3D CT-based texture analysis provided more stable features than 2D approach, while no difference in feature stability rates was found between 2 and 3D MRI-based texture analyses. Overall, a certain degree of segmentation variability highlighted the need to include a reliability analysis in future studies.
Despite its great potential as a non-invasive biomarker to quantify several tumor characteristics, radiomics still faces challenges to clinical implementation, both standalone and paired to machine learning [13, 29]. A great variability in radiomic features has emerged as a major issue across studies, and segmentation is the most critical step [12]. Image segmentation represents the basis of radiomic image analysis pipelines and can be time-consuming if performed manually. Therefore, methodological analyses are advisable prior to conducting radiomic studies in order to assess the robustness of different segmentation approaches and avoid biases due to non-reproducible, noisy features. These analyses have been previously performed in kidney [30, 31], lung, and head and neck [15] lesions. With regard to cartilaginous bone tumors, radiomic studies to date have focused on discriminating among benign, atypical, and malignant lesions [32–35], differentiating chondrosarcoma from other entities such as skull chordoma [36], or predicting recurrence of chondrosarcoma [37]. To our knowledge, our work is the first comprehensively addressing the influence of interobserver manual segmentation variability on the reproducibility of 2D and 3D CT- and MRI-based texture analysis in cartilaginous bone tumors. Nonetheless, Fritz et al. [33] and Gitto et al. [34] performed an interobserver reliability assessment as a feature-reduction method in their radiomic analysis, which provided a model for prediction of tumor grade. In particular, Fritz et al. found that most 2D features derived from unenhanced (15 out of 19) and contrast-enhanced (18 out of 19) T1-weighted MRI had at least good agreement between two observers, using an ICC cutoff of 0.6 [33]. In this study, however, the number of extracted features was only 19 per sequence, the impact of different feature classes was not analyzed, and filtered and transformed images were not used. Despite these issues, a common conclusion that can be drawn from this and our studies is that most MRI radiomic features of cartilaginous bone tumors have good reproducibility, even though a certain degree of segmentation variability exists. In a more recent study by Gitto et al., stability was assessed as a feature-reduction method and CT radiomic features were considered stable if ICC 95% confidence interval lower bound was 0.75 or higher. This resulted in a lower feature stability rate (30%) [34] compared to our current study.
In our study, all imaging modalities demonstrated good reproducibility both employing 2D and 3D annotations, with a robust feature percentage ranging from 75 to 96% for the former and 80 to 95% for the latter. Stable features also proved quite informative for predictive modeling at our preliminary analysis, with accuracies of 77–90%. Given the limited sample size and presence of 3 class labels, this result is promising and supports the use of radiomic data in this research domain. These findings are encouraging for future radiomic analyses, even though they confirm the need for a preliminary assessment of feature stability, and in line with recent literature emphasizing the importance of reproducibility in artificial intelligence and radiology [38]. The higher spatial resolution of CT did not seem to influence feature reproducibility and was probably offset by the better contrast resolution of T1-weighted and T2-weighted images. Furthermore, margin shrinkage did not lead to improvements in terms of feature reproducibility, contrary to a previous investigation on renal cell carcinoma CT images [17]. It should be noted that in this investigation, however, the authors reported that margin shrinkage produced less informative features even with improved reproducibility [17].
We found higher rates of stable features derived from CT for 3D compared to 2D segmentation, but no difference in the rates of 2D and 3D MRI-derived stable features. This finding is in favor of a 2D approach in future radiomic studies dealing with MRI-based texture analysis of cartilaginous bone tumors, as this is less time-consuming and easier to be employed in clinical practice, particularly in large atypical cartilaginous tumors and chondrosarcomas. Furthermore, most 2D (66–76%) and 3D (69–78%) stable features matched between CT and MRI, as well as T1-weghted and T2-weighted images. Finally, shape-based features were stable even across different imaging modalities and MRI sequences, and were thus reproducible and independent descriptors of tumor size and shape. On the other hand, overall interobserver reliability of other feature classes was unsurprisingly low across different imaging modalities and MRI sequences, indicating that their quantitative values depend on the specific image used.
Some limitations of our study should be acknowledged. First, it has a retrospective design as a prospective analysis is not strictly necessary for radiomic studies [13]. The retrospective design accounts for the exclusion of contrast-enhanced images, as they were not performed for all enchondromas. Contrast-enhanced and dynamic contrast-enhanced MRI improve the accuracy of cartilaginous bone tumor assessment [39–41] and future radiomic studies focusing on these sequences are warranted. Finally, due to its scope, this was a single-institution study and generalizability of our findings needs to be confirmed on more varied datasets.
Conclusions
In conclusion, radiomic features of cartilaginous bone tumors extracted from 2D and 3D segmentations on CT and MRI examinations are reproducible, although some degree of segmentation variability highlights the need to perform a preliminary reliability analysis in radiomic studies. 3D and 2D MRI-based texture analyses provide similar rates of stable features. Thus, a 2D approach can be favored in future studies, as this is easier to implement in clinical practice.
Abbreviations
- 2D
Bidimensional
- 3D
Volumetric
- CT
Computed tomography
- ET
Extra Trees
- GLCM
Gray-level cooccurrence matrix
- GLDM
Gray-level dependence matrix
- GLRLM
Gray-level run length matrix
- GLSZM
Gray-level size zone matrix
- ICC
Intraclass correlation coefficient
- LoG
Laplacian of Gaussian
- MRI
Magnetic resonance imaging
- RFE
Recursive feature elimination
- ROI
Region of interest
Funding
Open access funding provided by Università degli Studi di Milano within the CRUI-CARE Agreement.
Declarations
Compliance with ethical standards
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The local Institutional Review Board approved this retrospective study and waived the need for informed consent.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Salvatore Gitto and Renato Cuocolo contributed equally to this work.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Murphey MD, Walker EA, Wilson AJ, Kransdorf MJ, Temple HT, Gannon FH. From the archives of the AFIP: imaging of primary chondrosarcoma: radiologic-pathologic correlation. Radiographics. 2003;23:1245–1278. doi: 10.1148/rg.235035134. [DOI] [PubMed] [Google Scholar]
- 2.Albano D, Messina C, Gitto S, Papakonstantinou O, Sconfienza L. Differential Diagnosis of Spine Tumors: My Favorite Mistake. Semin Musculoskelet Radiol. 2019;23:26–35. doi: 10.1055/s-0038-1675551. [DOI] [PubMed] [Google Scholar]
- 3.Casali PG, Bielack S, Abecassis N, Aro HT, Bauer S, Biagini R, Bonvalot S, Boukovinas I, Bovee JVMG, Brennan B, Brodowicz T, Broto JM, Brugières L, Buonadonna A, De Álava E, Dei Tos AP, Del Muro XG, Dileo P, Dhooge C, Eriksson M, Fagioli F, Fedenko A, Ferraresi V, Ferrari A, Ferrari S, Frezza AM, Gaspar N, Gasperoni S, Gelderblom H, Gil T, Grignani G, Gronchi A, Haas RL, Hassan B, Hecker-Nolting S, Hohenberger P, Issels R, Joensuu H, Jones RL, Judson I, Jutte P, Kaal S, Kager L, Kasper B, Kopeckova K, Krákorová DA, Ladenstein R, Le Cesne A, Lugowska I, Merimsky O, Montemurro M, Morland B, Pantaleo MA, Piana R, Picci P, Piperno-Neumann S, Pousa AL, Reichardt P, Robinson MH, Rutkowski P, Safwat AA, Schöffski P, Sleijfer S, Stacchiotti S, Strauss SJ, Sundby Hall K, Unk M, Van Coevorden F, van der Graaf WTA, Whelan J, Wardelmann E, Zaikova O, Blay JY: Bone sarcomas: ESMO–PaedCan–EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 29:iv79–iv95, 2018 [DOI] [PubMed]
- 4.Cannavò L, Albano D, Messina C, Corazza A, Rapisarda S, Pozzi G, Di Bernardo A, Parafioriti A, Scotto G, Perrucchini G, Luzzati A, Sconfienza LM. Accuracy of CT and MRI to assess resection margins in primary malignant bone tumours having histology as the reference standard. Clin Radiol. 2019;74:736.e13–736.e21. doi: 10.1016/j.crad.2019.05.022. [DOI] [PubMed] [Google Scholar]
- 5.Douis H, Singh L, Saifuddin A. MRI differentiation of low-grade from high-grade appendicular chondrosarcoma. Eur Radiol. 2014;24:232–240. doi: 10.1007/s00330-013-3003-y. [DOI] [PubMed] [Google Scholar]
- 6.Crim J, Schmidt R, Layfield L, Hanrahan C, Manaster BJ. Can imaging criteria distinguish enchondroma from grade 1 chondrosarcoma? Eur J Radiol. 2015;84:2222–2230. doi: 10.1016/j.ejrad.2015.06.033. [DOI] [PubMed] [Google Scholar]
- 7.Hodel S, Laux C, Farei-Campagna J, Götschi T, Bode-Lesniewska B, Müller DA. The impact of biopsy sampling errors and the quality of surgical margins on local recurrence and survival in chondrosarcoma. Cancer Manag Res. 2018;10:3765–3771. doi: 10.2147/CMAR.S178768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eefting D, Schrage YM, Geirnaerdt MJA, Le Cessie S, Taminiau AHM, Bovée JVMG, Hogendoorn PCW. Assessment of Interobserver Variability and Histologic Parameters to Improve Reliability in Classification and Grading of Central Cartilaginous Tumors. Am J Surg Pathol. 2009;33:50–57. doi: 10.1097/PAS.0b013e31817eec2b. [DOI] [PubMed] [Google Scholar]
- 9.van de Sande MAJ, van der Wal RJP, Navas Cañete A, van Rijswijk CSP, Kroon HM, Dijkstra PDS, Bloem JL. Radiologic differentiation of enchondromas, atypical cartilaginous tumors, and high-grade chondrosarcomas—Improving tumor-specific treatment: A paradigm in transit? Cancer. 2019;125:3288–3291. doi: 10.1002/cncr.32404. [DOI] [PubMed] [Google Scholar]
- 10.Davnall F, Yip CSP, Ljungqvist G, Selmi M, Ng F, Sanghera B, Ganeshan B, Miles KA, Cook GJ, Goh V. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging. 2012;3:573–589. doi: 10.1007/s13244-012-0196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Codari M, Melazzini L, Morozov SP, van Kuijk CC, Sconfienza LM, Sardanelli F. Impact of artificial intelligence on radiology: a EuroAIM survey among members of the European Society of Radiology. Insights Imaging. 2019;10:105. doi: 10.1186/s13244-019-0798-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures. They Are Data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ. CT Texture Analysis: Definitions, Applications, Biologic Correlates, and Challenges. Radiographics. 2017;37:1483–1503. doi: 10.1148/rg.2017170056. [DOI] [PubMed] [Google Scholar]
- 14.Berenguer R, Pastor-Juan M, Canales-Vázquez J, Castro-García M, Villas MV, Mansilla Legorburo F, Sabater S. Radiomics of CT Features May Be Nonreproducible and Redundant: Influence of CT Acquisition Parameters. Radiology. 2018;288:407–415. doi: 10.1148/radiol.2018172361. [DOI] [PubMed] [Google Scholar]
- 15.Pavic M, Bogowicz M, Würms X, Glatz S, Finazzi T, Riesterer O, Roesch J, Rudofsky L, Friess M, Veit-Haibach P, Huellner M, Opitz I, Weder W, Frauenfelder T, Guckenberger M, Tanadini-Lang S. Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol. 2018;57:1070–1074. doi: 10.1080/0284186X.2018.1445283. [DOI] [PubMed] [Google Scholar]
- 16.Bologna M, Corino VDA, Montin E, Messina A, Calareso G, Greco FG, Sdao S, Mainardi LT. Assessment of Stability and Discrimination Capacity of Radiomic Features on Apparent Diffusion Coefficient Images. J Digit Imaging. 2018;31:879–894. doi: 10.1007/s10278-018-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kocak B, Ates E, Durmaz ES, Ulusan MB, Kilickesmez O. Influence of segmentation margin on machine learning–based high-dimensional quantitative CT texture analysis: a reproducibility study on renal clear cell carcinomas. Eur Radiol. 2019;29:4765–4775. doi: 10.1007/s00330-019-6003-8. [DOI] [PubMed] [Google Scholar]
- 18.Gitto S, Cuocolo R, Albano D, Morelli F, Pescatori LC, Messina C, Imbriaco M, Sconfienza LM: CT and MRI radiomics of bone and soft-tissue sarcomas: a systematic review of reproducibility and validation strategies. Insights Imaging 12:68, 2021 [DOI] [PMC free article] [PubMed]
- 19.Schwier M, van Griethuysen J, Vangel MG, Pieper S, Peled S, Tempany C, Aerts HJWL, Kikinis R, Fennessy FM, Fedorov A. Repeatability of Multiparametric Prostate MRI Radiomics Features. Sci Rep. 2019;9:9441. doi: 10.1038/s41598-019-45766-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ugga L, Cuocolo R, Solari D, Guadagno E, D’Amico A, Somma T, Cappabianca P. del Basso de Caro ML, Cavallo LM, Brunetti A: Prediction of high proliferative index in pituitary macroadenomas using MRI-based radiomics and machine learning. Neuroradiology. 2019;61:1365–1373. doi: 10.1007/s00234-019-02266-1. [DOI] [PubMed] [Google Scholar]
- 21.Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, Ashrafinia S, Bakas S, Beukinga RJ, Boellaard R, Bogowicz M, Boldrini L, Buvat I, Cook GJR, Davatzikos C, Depeursinge A, Desseroit M, Dinapoli N, Dinh CV, Echegaray S, El Naqa I, Fedorov AY, Gatta R, Gillies RJ, Goh V, Götz M, Guckenberger M, Ha SM, Hatt M, Isensee F, Lambin P, Leger S, Leijenaar RTH, Lenkowicz J, Lippert F, Losnegård A, Maier-Hein KH, Morin O, Müller H, Napel S, Nioche C, Orlhac F, Pati S, Pfaehler EAG, Rahmim A, Rao AUK, Scherer J, Siddique MM, Sijtsema NM, Socarras Fernandez J, Spezi E, Steenbakkers RJHM, Tanadini-Lang S, Thorwarth D, Troost EGC, Upadhaya T, Valentini V, van Dijk LV, van Griethuysen J, van Velden FHP, Whybra P, Richter C, Löck S. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koo TK, Li MY. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J Chiropr Med. 2016;15:155–163. doi: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 25.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin J, Pieper S, Aerts HJWL. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Leo G, Sardanelli F: Statistical significance: p value, 0.05 threshold, and applications to radiomics—reasons for a conservative approach. Eur Radiol Exp 4:18, 2020 [DOI] [PMC free article] [PubMed]
- 27.R Core Team: R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, 2020
- 28.van der Walt S, Colbert SC, Varoquaux G. The NumPy Array: A Structure for Efficient Numerical Computation. Comput Sci Eng. 2011;13:22–30. doi: 10.1109/MCSE.2011.37. [DOI] [Google Scholar]
- 29.Cuocolo R, Caruso M, Perillo T, Ugga L, Petretta M. Machine Learning in oncology: A clinical appraisal. Cancer Lett. 2020;481:55–62. doi: 10.1016/j.canlet.2020.03.032. [DOI] [PubMed] [Google Scholar]
- 30.Kocak B, Durmaz ES, Kaya OK, Ates E, Kilickesmez O. Reliability of Single-Slice–Based 2D CT Texture Analysis of Renal Masses: Influence of Intra- and Interobserver Manual Segmentation Variability on Radiomic Feature Reproducibility. AJR Am J Roentgenol. 2019;213:377–383. doi: 10.2214/AJR.19.21212. [DOI] [PubMed] [Google Scholar]
- 31.Kocak B, Durmaz ES, Erdim C, Ates E, Kaya OK, Kilickesmez O. Radiomics of Renal Masses: Systematic Review of Reproducibility and Validation Strategies. AJR Am J Roentgenol. 2020;214:129–136. doi: 10.2214/AJR.19.21709. [DOI] [PubMed] [Google Scholar]
- 32.Gitto S, Cuocolo R, Albano D, Chianca V, Messina C, Gambino A, Ugga L, Cortese MC, Lazzara A, Ricci D, Spairani R, Zanchetta E, Luzzati A, Brunetti A, Parafioriti A, Sconfienza LM: MRI radiomics-based machine-learning classification of bone chondrosarcoma. Eur J Radiol 128:109043, 2020 [DOI] [PubMed]
- 33.Fritz B, Müller DA, Sutter R, Wurnig MC, Wagner MW, Pfirrmann CWA, Fischer MA. Magnetic Resonance Imaging-Based Grading of Cartilaginous Bone Tumors. Invest Radiol. 2018;53:663–672. doi: 10.1097/RLI.0000000000000486. [DOI] [PubMed] [Google Scholar]
- 34.Gitto S, Cuocolo R, Annovazzi A, Anelli V, Acquasanta M, Cincotta A, Albano D, Chianca V, Ferraresi V, Messina C, Zoccali C, Armiraglio E, Parafioriti A, Sciuto R, Luzzati A, Biagini R, Imbriaco M, Sconfienza LM: CT radiomics-based machine learning classification of atypical cartilaginous tumours and appendicular chondrosarcomas. EBioMedicine 68:103407, 2021 [DOI] [PMC free article] [PubMed]
- 35.Lisson CS, Lisson CG, Flosdorf K, Mayer-Steinacker R, Schultheiss M, von Baer A, Barth TFE, Beer AJ, Baumhauer M, Meier R, Beer M, Schmidt SA. Diagnostic value of MRI-based 3D texture analysis for tissue characterisation and discrimination of low-grade chondrosarcoma from enchondroma: a pilot study. Eur Radiol. 2018;28:468–477. doi: 10.1007/s00330-017-5014-6. [DOI] [PubMed] [Google Scholar]
- 36.Li L, Wang K, Ma X, Liu Z, Wang S, Du J, Tian K, Zhou X, Wei W, Sun K, Lin Y, Wu Z, Tian J. Radiomic analysis of multiparametric magnetic resonance imaging for differentiating skull base chordoma and chondrosarcoma. Eur J Radiol. 2019;118:81–87. doi: 10.1016/j.ejrad.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 37.Yin P, Mao N, Liu X, Sun C, Wang S, Chen L, Hong N. Can clinical radiomics nomogram based on 3D multiparametric MRI features and clinical characteristics estimate early recurrence of pelvic chondrosarcoma? J Magn Reson Imaging. 2020;51:435–445. doi: 10.1002/jmri.26834. [DOI] [PubMed] [Google Scholar]
- 38.Mongan J, Moy L, Kahn CE: Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol Artif Intell 2:e200029, 2020 [DOI] [PMC free article] [PubMed]
- 39.De Coninck T, Jans L, Sys G, Huysse W, Verstraeten T, Forsyth R, Poffyn B, Verstraete K. Dynamic contrast-enhanced MR imaging for differentiation between enchondroma and chondrosarcoma. Eur Radiol. 2013;23:3140–3152. doi: 10.1007/s00330-013-2913-z. [DOI] [PubMed] [Google Scholar]
- 40.Geirnaerdt MJA, Hogendoorn PCW, Bloem JL, Taminiau AHM, van der Woude H-J. Cartilaginous Tumors: Fast Contrast-enhanced MR Imaging. Radiology. 2000;214:539–546. doi: 10.1148/radiology.214.2.r00fe12539. [DOI] [PubMed] [Google Scholar]
- 41.Yoo HJ, Hong SH, Choi J, Moon KC, Kim H, Choi J, Kang HS. Differentiating high-grade from low-grade chondrosarcoma with MR imaging. Eur Radiol. 2009;19:3008–3014. doi: 10.1007/s00330-009-1493-4. [DOI] [PubMed] [Google Scholar]