Abstract
Automated quantitative and probabilistic medical image analysis has the potential to improve the accuracy and efficiency of the radiology workflow. We sought to determine whether AI systems for brain MRI diagnosis could be used as a clinical decision support tool to augment radiologist performance. We utilized previously developed AI systems that combine convolutional neural networks and expert-derived Bayesian networks to distinguish among 50 diagnostic entities on multimodal brain MRIs. We tested whether these systems could augment radiologist performance through an interactive clinical decision support tool known as Adaptive Radiology Interpretation and Education System (ARIES) in 194 test cases. Four radiology residents and three academic neuroradiologists viewed half of the cases unassisted and half with the results of the AI system displayed on ARIES. Diagnostic accuracy of radiologists for top diagnosis (TDx) and top three differential diagnosis (T3DDx) was compared with and without ARIES. Radiology resident performance was significantly better with ARIES for both TDx (55% vs 30%; P < .001) and T3DDx (79% vs 52%; P = 0.002), with the largest improvement for rare diseases (39% increase for T3DDx; P < 0.001). There was no significant difference between attending performance with and without ARIES for TDx (72% vs 69%; P = 0.48) or T3DDx (86% vs 89%; P = 0.39). These findings suggest that a hybrid deep learning and Bayesian inference clinical decision support system has the potential to augment diagnostic accuracy of non-specialists to approach the level of subspecialists for a large array of diseases on brain MRI.
Keywords: Deep learning, Convolutional neural networks, Neuroradiology, Brain MRI, U-Net, Artificial intelligence, Bayesian inference, Clinical decision support, Augmented performance
Introduction
Medical image interpretation currently relies almost entirely on visual review and analysis by radiologists or other subspecialized clinicians. Although these clinicians are highly trained and skilled, they face increasing volume demands [1] and are susceptible to certain cognitive biases, which result in predictable diagnostic errors [2, 3], such as “satisfaction of search,” availability bias [4], and confirmation bias [5]. Computational methods for quantitative image analysis and other forms of artificial intelligence (AI) hold potential to augment radiologists’ performance by decreasing the impact of these biases on clinical image interpretation. As both the complexity and volume of medical imaging increases, methods that can augment non-specialists’ ability to provide diagnoses on complex radiologic exams would also be advantageous, particularly in general practices and developing countries where there is a shortage of subspecialist radiologists [6].
Deep learning, a subclass of machine learning methods based on neural networks [7, 8], is a powerful data-driven approach that has shown promise in many biomedical imaging tasks [9–14]. However, construction of an AI system for classification on deep learning typically requires hundreds or thousands of examples of each category. Constraints of data collection and annotation therefore limit the potential for deep learning to classify rare and uncommon diseases, atypical presentations of common diseases, or the large spectrum of diseases commonly seen in clinical practice. Instead, deep learning excels at extracting features of medical images, such as hemorrhage types [12, 13], FLAIR signal abnormality [14], or brain tumor tissue volumes [11].
Bayesian networks integrate multiple variables to probabilistically model conditional independence [15]. They can be used to discriminate numerous disease possibilities given a set of imaging and clinical variables, driven by expert-derived conditional probabilities [16]. This feature allows Bayesian networks to model experts’ explicit knowledge of an array of possible diseases, and to perform the cognitive task of generating a differential diagnosis, without requiring large amounts of labeled data for each disease. Previous work utilizing Bayesian networks for probabilistic diagnosis in radiology includes a variety of modalities and organ systems [17–22]. We recently developed AI systems [25, 26] combining deep learning and Bayesian networks for generation of differential diagnosis on clinical brain MRIs, which performed as well as academic neuroradiologists across 50 diagnoses.
Successful integration of AI technologies into clinical practice requires real-time interactions with radiologists. At our institution, we developed a Bayesian network interface for assisting in radiology interpretation and education, known as Adaptive Radiology Interpretation and Education System (ARIES) [23, 24], which allows the user to select different imaging and clinical features and see estimated disease probabilities update in real time for different disease networks. For example, the predictions of the recently developed brain MRI diagnostic AI systems can be integrated with ARIES to allow radiologists’ real-time interaction with the systems by overriding AI-generated imaging descriptions and observing the resulting changes in predicted differential diagnoses.
In this study, we sought to determine whether these AI systems, instantiated in ARIES, could improve the performance of radiologists in generating differential diagnoses. A handful of recent studies have shown the ability of artificial intelligence methods to augment radiologist performance. This includes convolutional neural networks for the detection of fractures on radiographs [27, 28], cerebral aneurysms on computed tomographic angiograms [29], anterior cruciate ligament and meniscal tears on knee magnetic resonance imaging [30], and cancer screening mammograms [31, 32]. These prior studies all utilized end-to-end deep learning models and focused on just one or two diagnostic entities. While the findings of these studies are promising for these particular tasks, when compared to the spectrum of possible diagnoses that a radiologist must consider when interpreting medical imaging, they represent a narrow perspective. No prior studies have evaluated whether AI diagnostic systems encompassing 50 diagnostic entities, as we test here, could augment radiologists’ performance.
Materials and Methods
Patient Data
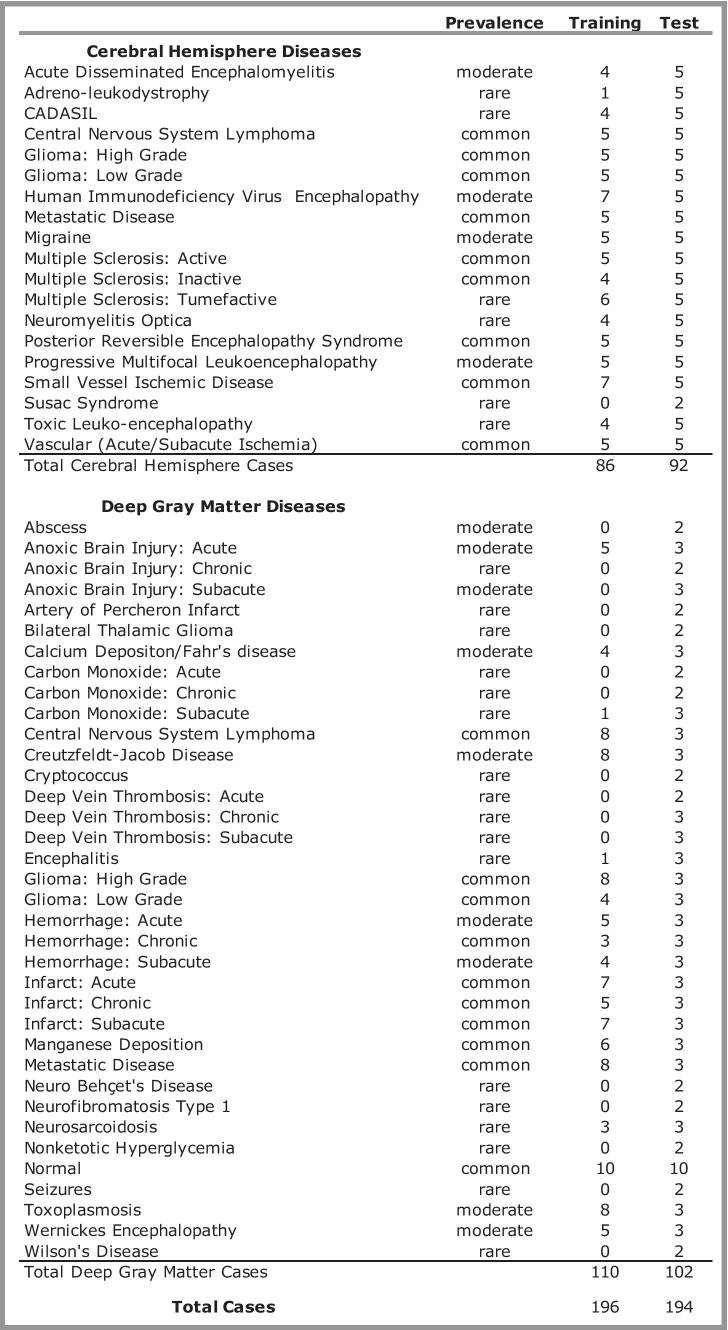
As part of an institutional review board approved study at our institution, multimodal brain MRIs from 390 patients (52 ± 17 years [standard deviation], 228 women) with diseases representing 50 different neurologic entities and normal were included in this study (Table 1). The brain MRIs were collected as part of two separate AI systems evaluating 19 diseases involving cerebral hemispheres [25] and (36) diseases involving deep gray matter [26]. These distinct systems were initially chosen to encompass a large array of brain diseases with notable exceptions including extra-axial diseases and diseases primarily involving the posterior fossa. There were five entities (high-grade glioma, low-grade glioma, central nervous system lymphoma, and metastatic disease) overlapping between the two systems. The details of the case selection and randomization process of MRIs into training (n = 196) and testing samples (n = 194) of the two studies are fully described elsewhere [25, 26], with no individual MRI overlapping between the two studies. The diseases were classified as “common,” “uncommon,” or “rare” according to the relative frequency with which they are diagnosed on brain MRIs at a tertiary care academic institution, based on the consensus of two academic neuroradiologists (7- and 12-year post fellowship experience).
Table 1.
Diagnostic entities included in the study. Prevalence ratings were determined by consensus of two neuroradiologists. CADASIL = cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy
Automated MRI Processing Pipeline
The automated MRI processing pipelines have been extensively described in prior publications [25, 26]. We utilized features derived from these previously developed image processing pipelines for diseases involving the cerebral hemispheres [25] and deep gray matter [26] to populate ARIES prior to radiologists’ review of images. Briefly, these pipelines perform segmentation of brain tissues, structures, and abnormal signal using a variety of CNNs and traditional atlas-based neuroimaging methods [25, 26, 33–35]. In addition to five signal features (T1-weighted, FLAIR, GRE, T1-post, diffusion), the pipelines also extracted anatomic, spatial pattern, and volumetric features. The deep gray pipeline included four anatomic subregion features (caudate, putamen, globus pallidus, and thalamus) and two spatial pattern features (bilateral and symmetric). The cerebral hemisphere pipeline included seven spatial pattern features (lobar distribution, periventricular, juxtacortical, symmetry, corpus callosum, anterior temporal lobe, cortical gray matter) and six volumetric features (lesion number, lesion volume, lesion extent, ventricular volume, mass effect, enhancement ratio) [25, 26].
Bayesian Network Construction
In addition to the signal, anatomic, spatial pattern, and volumetric features derived from the image processing pipelines, the cerebral hemisphere Bayesian network included five clinical features (age, gender, chronicity, viral prodrome, immune status) and the deep gray Bayesian network included four clinical features (age, gender, chronicity, immune status), which were manually extracted from the electronic medical record system. These features represent all of the possible feature states on the ARIES website. The probabilities of each features for each of the 19 possible diagnoses for the cerebral hemispheres network and 36 possible diagnoses for the deep gray network were determined by the consensus of four neuroradiologists as previously detailed [25, 26]. These previously developed Bayesian networks have shown excellent performance relative to radiologists [25, 26].
ARIES Interface
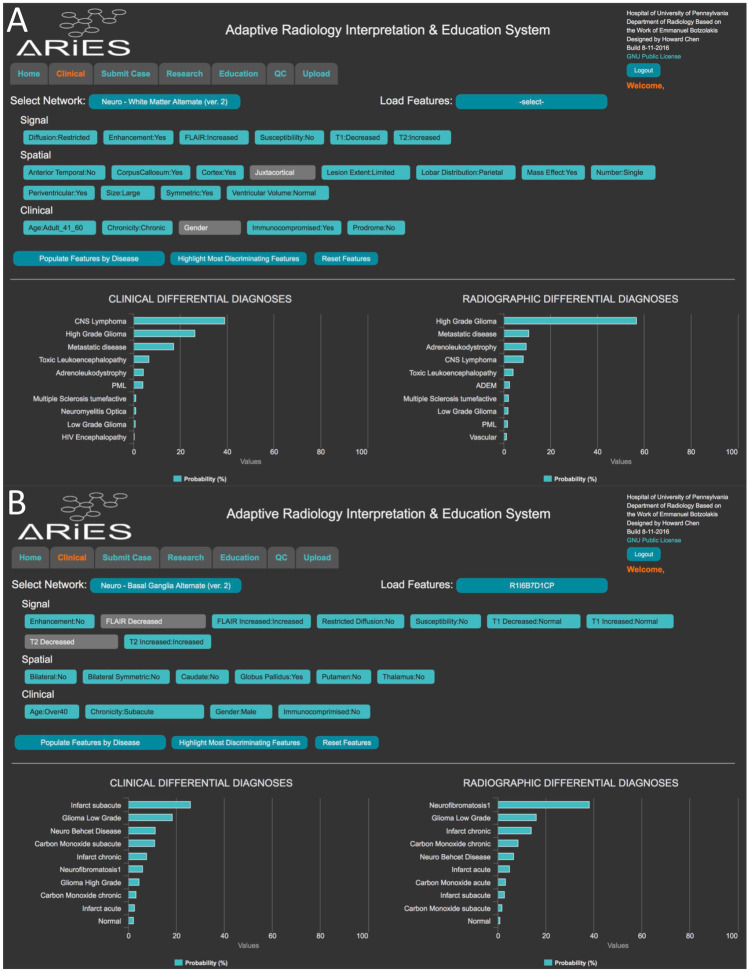
The feature inputs and disease probability outputs of the Bayesian networks were displayed on the web ARIES interface (Fig. 1) [23–34]. The ARIES web interface utilized a commercial Bayesian network backend (Netica, Norysys Software Company, Vancouver, Canada) to calculate naïve Bayes probabilities for each disease, depending on what feature states are selected [23, 24]. A web open-source version of the interface was also developed (https://aries-penn.appspot.com/) with all source code available at https://github.com/jeffduda/aries-app. ARIES allows for the radiologist to either enter individual feature states or utilize automatically derived feature states, adjust feature states, and visualize the probabilities of different diseases updated in real time. The system provides a “radiographic” differential diagnosis, which is based on imaging features and a “clinical” differential diagnosis, which adjusts the “radiographic” differential diagnosis based upon additional clinical features and disease prevalence/prior probabilities. For the purposes of these experiments, the prior probabilities of each disease were set to be equivalent. The clinical differential diagnosis was used for final results of the AI systems. ARIES was customized to preload each of the test cases with the feature states derived from the previously developed image processing pipelines [25, 26].
Fig. 1.
Adaptive Radiology Interpretation and Education System (ARIES). Example screen shots from the interactive ARIES web portal displaying results of sample anonymized test cases with features prefilled by the cerebral hemisphere (A) and deep gray matter (B) image processing pipelines
Clinical Validation for Augmented Performance
As part of the same institutional review board approved study at our institution, under signed written consent, four radiology residents (three PGY-3 radiology residents and one PGY-4 radiology resident, each with ~3–4 months of neuroradiology experience) and three academic neuroradiology attendings (with 7, 1, and 2 years of post-fellowship experience) reviewed the test cases, which were copied into anonymized accession numbers and then displayed in a standard fashion in the clinical picture archiving and communication system (PACS; Sectra AB, Linköping, Sweden).
The cases were split into four blocks with ARIES and four blocks without ARIES (24–26 cases per block, which allowed participants to complete each block in a ~1-h sitting). Each block contained half deep gray and half cerebral hemisphere cases. For blocks with ARIES, participants viewed the results of either the cerebral hemisphere or deep gray matter image processing pipelines preloaded onto the ARIES website with the same clinical information and provided their ranked top three differential diagnoses from an excel file dropdown menu of the 19 or 36 possible diagnostic entities. The order in which the cases were seen with or without prepopulated imaging pipeline results in ARIES was alternated for each subject and counterbalanced between subjects. All participants completed eight blocks, except for one of the radiology residents, who only completed four blocks. The same clinical information used in the Bayesian network was made available to each reader when viewing the cases with or without ARIES, except radiologists were provided the exact age of the patient and the Bayesian network received a thresholded age as input.
For the cases with ARIES, the participants provided two sets of their own ranked differential diagnoses. They first provided their top 3 differential diagnoses from the 19 or 36 possible diagnostic entities after reviewing both the prepopulated features and outputted disease probabilities/differential diagnoses, which was the result of the AI pipelines. The participants were not aware of the performance of the AI diagnostic pipelines and were instructed to use their best judgment when considering output presented by the Bayesian networks via ARIES. Then they were then allowed to interact with ARIES and change features, such as if they disagreed with one or more of the AI-provided features. Feature changes could result in updated disease probabilities, and the readers could then change their differential diagnoses if they thought different diagnoses were more likely based on new information provided by interacting with ARIES.
Statistics for Comparison to Human Performance
Chi-squared tests were used for comparing the fraction of cases correctly answered with and without ARIES for correct top diagnosis (TDx) and correct top three differential diagnosis (T3DDx). This analysis was done across all diagnoses and as a function of disease prevalence for residents and attendings. All reported p-values represent non-directional, two-tailed tests with significance defined as P < 0.05.
Results
Artificial Intelligence System Performance
As previously reported [25], the AI systems determined the correct top diagnosis (TDx) in 61% of the test cases (57% for cerebral hemisphere pipeline and 64% for deep gray matter pipeline) and the correct diagnosis within the top 3 of the DDx (T3DDx) in 88% of the test cases (91% for cerebral hemisphere pipeline and 85% for deep gray matter pipeline) for the 194 test cases, which in prior experiments was found to be significantly better than radiology residents, neuroradiology fellows, and general radiologists, and not different from academic neuroradiologists [25]. These autonomously derived features and diagnoses were presented to the radiologists in this experiment for their use via ARIES for subsequent analyses.
Radiology Resident Performance
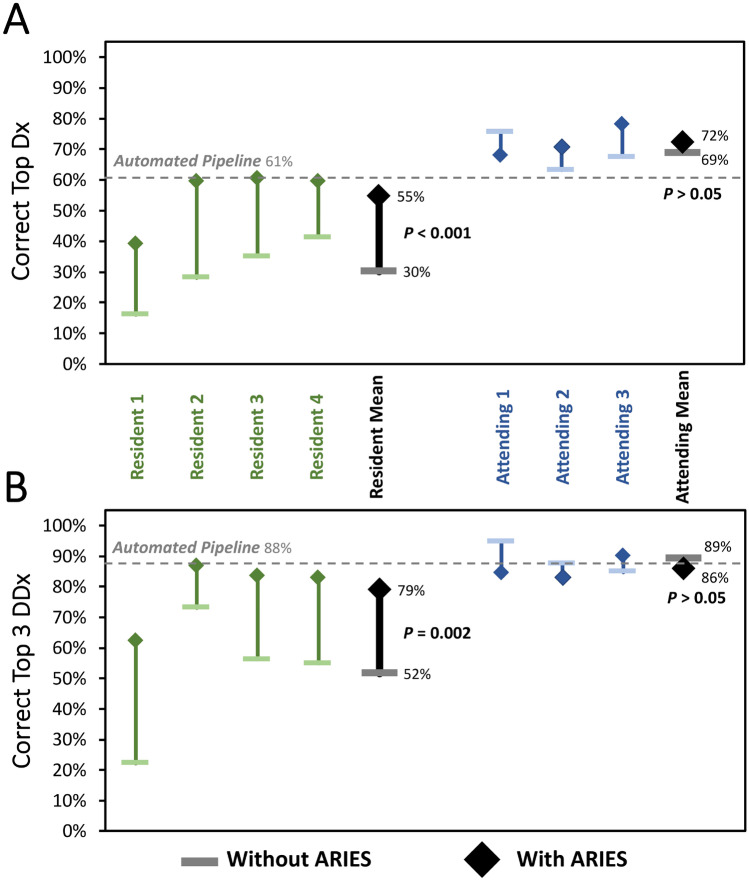
Without ARIES, the radiology residents determined the correct TDx in 30% of the test cases and the correct diagnosis within the T3DDx in 52% of the test cases. Radiology resident performance was significantly better when they were given access to the results of the AI pipelines and ARIES for both TDx (55% vs 30%; 83% improvement; P < 0.001; Fig. 2A) and T3DDx (79% vs 52%; 49% improvement; P = 0.002; Fig. 2B). Residents changed their differential diagnosis in 24% ± 12% of cases when allowed to manipulate the features with ARIES. Resident performance was not significantly further changed when allowed to manipulate the automatically derived feature states within ARIES for both TDx (55% vs 53%; P = 0.22) and T3DDx (79% vs 78%; P = 0.38).
Fig. 2.
Comparison of radiologist performance with and without ARIES. Diagnostic performance of radiology residents (green) and academic neuroradiology attendings without ARIES (horizontal bar) and with ARIES (diamonds) for correct top diagnosis (A) and top 3 differential diagnosis (B)
Academic Attending Performance
For academic neuroradiology attendings, there was no significant difference between performance in cases with and without the use of ARIES for TDx (72% vs 69%; P = 0.48; Fig. 2A) or T3DDx (86% vs 89%; P = 0.39; Fig. 2B). In addition, academic neuroradiology attending performance was not significantly different when they were allowed to manipulate the automatically derived feature states within ARIES for both TDx (72% vs 70%; P = 0.58) and T3DDx (86% vs 86%; P = 0.58). Attendings changed their differential diagnosis in 21% ± 13% of cases when allowed to manipulate the features with ARIES, which was not significantly different from residents (21% vs 24%; P = 0.78).
Performance Relative to Disease Prevalence
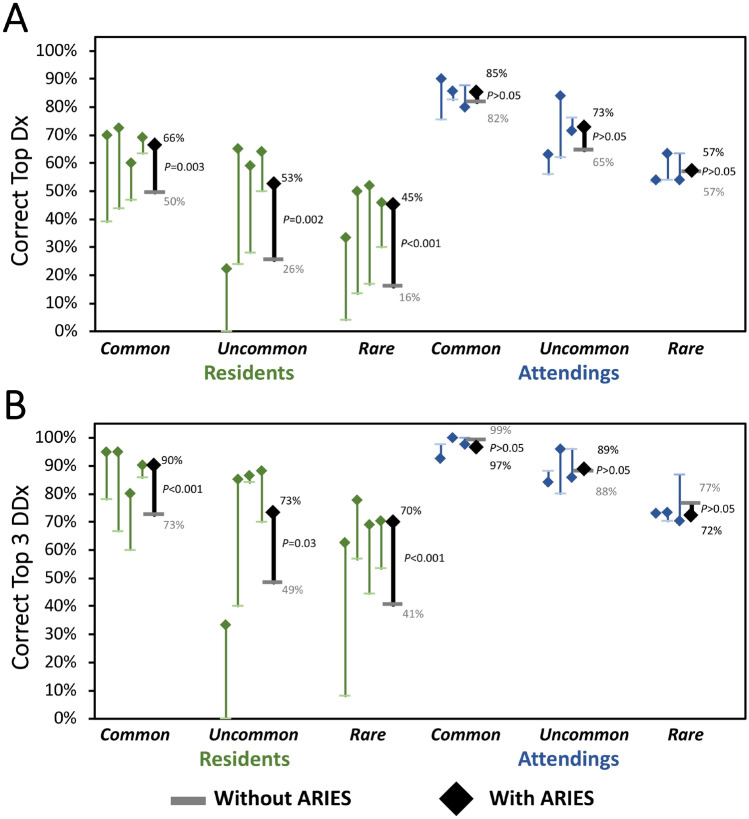
Next, we sought to assess how performance with and without ARIES varied as a function of disease prevalence (Fig. 3). For TDx, radiology residents were significantly better with ARIES for common (66% vs 50%; 32% improvement; P = 0.003), uncommon (53% vs 26%; 103% improvement; P = 0.002), and rare diseases (45% vs 16%; 181% improvement; P < 0.001). Radiology residents also performed significantly better with ARIES for T3DDx for common (90% vs 73%; 23% improvement; P < 0.001), uncommon (73% vs 49%; 49% improvement; P = 0.03) and rare diseases (70% vs 41%; 71% improvement; P < 0.001). Academic neuroradiology attendings did not perform significantly different with ARIES for TDx or T3DDx for common (85% vs 82% for TDx; 97% vs 99% for T3DDx; P’s > 0.19), uncommon (73% vs 65% for TDx; 89% vs 88% for T3DDx; P’s > 0.19), or rare diseases (57% vs 57% for TDx; 72% vs 77% for T3DDx; P’s > 0.21).
Fig. 3.
Comparison of radiologist performance with and without ARIES as a function of disease prevalence. Diagnostic performance of radiology residents (green) and academic neuroradiology attendings without ARIES (horizontal bar) and with ARIES (diamonds) as function of different disease prevalence for correct top diagnosis (A) and top 3 differential diagnosis (B)
Discussion
We previously developed two hybrid artificial intelligence systems for multimodal brain MRIs that performed at the level of academic neuroradiologists for distinguishing amongst 19 diagnoses involving the cerebral hemispheres [25] and 35 diagnoses involving deep gray matter and “normal” [26]. These systems automatically extract signal, anatomic, and quantitative imaging features, and then integrate them with clinical information to generate probabilistic differential diagnoses. Here we tested whether results from these systems displayed in ARIES, an interactive clinical decision support tool, could augment the performance of radiologists. We found that radiology residents had significantly better performance with ARIES, suggesting that such an AI-based clinical decision support system has the potential to improve the accuracy and efficiency of the radiology workflow for non-specialists.
Previous work indicates that cognitive and perceptual errors made by humans differ from errors made by automated systems [3, 36]. This was borne out by our prior studies [25, 26], showing that errors produced by different humans were more alike than those produced by an AI system, thereby suggesting that these AI systems could improve the performance of radiologists. Here we found that the performance of radiology residents improved by 83% for TDx and 49% for T3DDx when using ARIES. This increase in performance was significant across all disease prevalences, with the highest increase in performance for rare diseases. There were no significant differences between performance with and without ARIES for attending neuroradiologists. The lack of differences for attendings may be due to ceiling level performance of the current imaging processing pipeline. In addition, like other educational resources, it is perhaps not surprising that ARIES had the greatest impact on trainees. Although general radiologists were not included in this study, it is likely that this system could also augment their performance to the level of academic subspecialists, as suggested by their similar performance to radiology residents in our prior studies [25]. However, this claim needs to be tested with further clinical validation studies which could include general radiologists as well as other specialized clinicians, such as neurologists. Increasing performance of non-specialist radiologists may prove particularly useful in countries where there is a shortage of subspecialists [6].
There are several limitations of the current AI diagnostic systems. The cases were preselected to focus on a limited set of pathologies separated into two previously developed pipelines compared hundreds to thousands in the real world. In addition, cases with multiple diagnoses or lacking a T1 sequence necessary for the image processing pipeline were excluded. Thus, these AI diagnostic systems and clinical decision support tools can currently be considered a proof-of-concept on a subset of diseases encountered on brain MRIs. Future improvements to a unified image processing pipeline could address these limitations, which might include additional features, diseases, improved segmentation methods, and updated Bayesian probabilities. A larger study that evaluates the efficacy of a more comprehensive unified pipeline on more cases, across different sites and on a larger number of radiologists of varying experience would also better support its ability to improve clinical decision making. In addition, it may also be possible to account for multiple diseases processes within individual subjects, such as has been used to distinguish white matter hyperintensities in patients with glioblastoma [11]. It is possible that further improvements to the system could ultimately prove synergistic with academic neuroradiologists, but this remains to be demonstrated. In addition, given the benefit for residents, ARIES may prove useful as a tool for precision radiology education [37] as it could provide diagnostic rationale about what features are heavily weighted and how confident it is about the differential diagnosis. Further studies could be performed to evaluate if and how a tool might improve radiology education in addition to compared more traditional forms of teaching and educational resources. The ARIES tool for these networks (https://aries-penn.appspot.com/) and ARIES source code (https://github.com/jeffduda/aries-app) has been made available for other individuals and institutions to adapt for their own applications and populations.
Along with improvements to the pipeline, an integration across PACS and dictation software could allow “real-time decision support” and workflow augmentation by pre-populating radiology reports, in an “end-to-end” solution in neuroradiology [38]. It has been shown that draft reports from trainees can provide 25%-time savings to academic neuroradiologists, suggesting that a real-time version of this system has the potential to accelerate image interpretation efficiency [39]. Importantly, a system such as this one, where intermediate features are made available to the radiologist and can be manipulated, keeps the human radiologist in the loop and renders the entire diagnostic process explainable to other humans. These features could allow a future version of this AI system to be seamlessly integrated into the radiologist’s workflow.
Conclusions
We found that results from AI systems displayed with a clinical decision support tool could augment radiologist performance on a classification task of multiple common and rare diseases on multimodal brain MRI, all while allowing for modification of the high-level features from the AI system to see disease probabilities updated in real time. Ultimately, this modular approach could be expanded across other modalities and organ systems, paving the way for comprehensive, automated, decision support tools that may improve the capacity of non-specialists across radiology.
Abbreviations
- AI
Artificial intelligence
- CNN
Convolutional neural network
- T3DDx
Top three differential diagnosis
- TDx
Top diagnosis
- ARIES
Adaptive Radiology Interpretation and Education System
Funding
Financial support for this project was provided by an RSNA Resident Research grant (AMR; RR1778). AMR and JDR were also supported by institutional T-32 Training Grants (Penn T32-EB004311-10 and UCSF T32-EB001631-14). The NVIDIA corporation donated two Titan Xp GPUs as part of the NVIDIA GPU grant program (JDR, AMR).
Declarations
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McDonald RJ, Schwartz KM, Eckel LJ, Diehn FE, Hunt CH, Bartholmai BJ, Erickson BJ, Kallmes DF. The effects of changes in utilization and technological advancements of cross-sectional imaging on radiologist workload. Acad Radiol. 2015;22:1191–1198. doi: 10.1016/j.acra.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Gunderman RB. Biases in radiologic reasoning. AJR Am J Roentgenol. 2009;192:561–564. doi: 10.2214/AJR.08.1220. [DOI] [PubMed] [Google Scholar]
- 3.Bruno MA, Walker EA, Abujudeh HH. Understanding and confronting our mistakes: the epidemiology of error in radiology and strategies for error reduction. Radiographics. 2015;35:1668–1676. doi: 10.1148/rg.2015150023. [DOI] [PubMed] [Google Scholar]
- 4.Tversky A, Kahneman D. Availability: a heuristic for judging frequency and probability. Cognitive Psychology. 1973;5:207–232. doi: 10.1016/0010-0285(73)90033-9. [DOI] [Google Scholar]
- 5.Busby LP, Courtier JL, Glastonbury CM. Bias in radiology: the how and why of misses and misinterpretations. Radiographics. 2018;38:236–247. doi: 10.1148/rg.2018170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.European Society of Radiology (ESR). Summary of the proceedings of the International Summit general and subspecialty radiology. Insights Imaging. 2015;2016(7):1–5. doi: 10.1007/s13244-015-0453-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 8.Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, Kadoury S, Tang A. Deep learning: a primer for radiologists. Radiographics. 2017;37:2113–2131. doi: 10.1148/rg.2017170077. [DOI] [PubMed] [Google Scholar]
- 9.Chang PJ, Grinband BD, Weinberg M, Bardis M, Khy G, Cadena MY Su, et al. Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas.” AJNR. American Journal of Neuroradiology 2019;39(7):1201–1207. 10.3174/ajnr.A5667. [DOI] [PMC free article] [PubMed]
- 10.Rudie JD, Rauschecker AM, Bryan RN, Davatzikos C, and Mohan S. Emerging applications of artificial intelligence in neuro-oncology. Radiology 10.1148/radiol.2018181928. [DOI] [PMC free article] [PubMed]
- 11.Rudie JD, Weiss DA, Saluja R, Rauschecker AM, Wang J, Sugrue L, Bakas S, Colby JB. Multi-disease segmentation of gliomas and white matter hyperintensities in the BraTS data using a 3D convolutional neural network. Front Comput Neurosci. 2019;13:84. doi: 10.3389/fncom.2019.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee H, Yune S, Mansouri M, Kim M, Tajmir SH, Guerrier CE, Ebert SA, Pomerantz SR, Romero JM, Kamalian S, Gonzalez RG, Lev MH, and Do S. An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat Biomed Eng Nature Biomedical Engineering 2018. [[Epub ahead of print]]. [DOI] [PubMed]
- 13.Kuo W, Hӓne C, Mukherjee P, Malik J, Yuh EL. Expert-level detection of acute intracranial hemorrhage on head computed tomography using deep learning. Proc Natl Acad Sci U S A. 2019;116:22737–22745. doi: 10.1073/pnas.1908021116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duong MT, Rudie JD, Wang J, et al. Convolutional Neural Network for Automated FLAIR Lesion Segmentation on Clinical Brain MR Imaging. AJNR Am J Neuroradiol 2019;40:1282–90. [DOI] [PMC free article] [PubMed]
- 15.Pearl J. Probabilistic Reasoning in Intelligent Systems. San Mateo, CA: Morgan Kaufmann; 1988. [Google Scholar]
- 16.Bielza C, Larrañaga P. Bayesian networks in neuroscience: a survey. Front Comput Neurosci. 2014;8:131. doi: 10.3389/fncom.2014.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tombropoulos R, Shiffman S, and Davidson C. A decision aid for diagnosis of liver lesions on MRI. Proc Annu Symp Comput Appl Med Care 1993. [PMC free article] [PubMed]
- 18.Kahn CE, Roberts LM, Shaffer KA, Haddawy P. Construction of a Bayesian network for mammographic diagnosis of breast cancer. Comput Biol Med. 1997;27:19–29. doi: 10.1016/S0010-4825(96)00039-X. [DOI] [PubMed] [Google Scholar]
- 19.Do BH, Langlotz C, Beaulieu CF. Bone tumor diagnosis using a naïve Bayesian model of demographic and radiographic features. J Digit Imaging. 2017;30:640–647. doi: 10.1007/s10278-017-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kahn CE, Laur JJ, Carrera GF. A Bayesian network for diagnosis of primary bone tumors. J Digit Imaging. 2001;14:56–57. doi: 10.1007/BF03190296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu J, Wu W, Zhu B, Wang H, Liu R, Zhang X, Li M, Yang Y, Yan J, Niu F, Tian C, Wang K, Yu H, Chen W, Wan S, Sun Y, Zhang B. Correction: cerebral glioma grading using bayesian network with features extracted from multiple modalities of magnetic resonance imaging. PLoS ONE. 2016;11:e0157095. doi: 10.1371/journal.pone.0157095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herskovits EH, Bryan RN, Yang F. Automated Bayesian segmentation of microvascular white-matter lesions in the ACCORD-MIND study. Adv Med Sci. 2008;53:182–190. doi: 10.2478/v10039-008-0039-3. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Elenee Argentinis JD, Weber G. IBM Watson: how cognitive computing can be applied to big data challenges in life sciences research. Clin Ther. 2016;38:688–701. doi: 10.1016/j.clinthera.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Duda J, Botzolakis E, Bryan RN, Chen P-H, Cook T, Gee J, Mohan S, Nasrallah I, Rauschecker A, and Rudie J. Bayesian network interface for assisting radiology interpretation and education. Proceedings of SPIE 2018;10579:105790S-105790S-10.
- 25.Rauschecker AM, Rudie JD, Xie L, Wang J, Duong MT, Botzolakis EJ, Kovalovich AM, Egan J, Cook TC, Bryan RN, Nasrallah IM, Mohan S, and Gee JC. Artificial intelligence system approaching neuroradiologist-level differential diagnosis accuracy at brain MRI. Radiology10.1148/radiol.2020190283. [DOI] [PMC free article] [PubMed]
- 26.Rudie JD, Rauschecker AM, Xie L, Wang J, Botzolakis EJ, Kovalovich A, Egan J, Cook T, Bryan RN, Nasrallah IM, Mohan S, Gee J. Subspecialty-level deep gray matter differential diagnoses with deep learning and bayesian networks on clinical brain MRI: a pilot study. Radiology: Artificial Intelligence. In Press. [DOI] [PMC free article] [PubMed]
- 27.Lindsey R, Daluiski A, Chopra S, Lachapelle A, Mozer M, Sicular S, Hanel D, Gardner M, Gupta A, Hotchkiss R, Potter H. Deep neural network improves fracture detection by clinicians. Proc Natl Acad Sci USA. 2018;115:11591–11596. doi: 10.1073/pnas.1806905115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogue JD, Cheng KV, Hwang KM, Toogood P, Meinberg EG, Geiger EJ, Zaid M, McGill KC, Patel R, Sohn JH, Wright A, Darger BF, Padrez KA, Ozhinsky E, Majumdar S, and Pedoia V. Automatic hip fracture identification and functional subclassification with deep learning. Radiology: Artificial Intelligence. 2020;2:e190023-. [DOI] [PMC free article] [PubMed]
- 29.Park A, Chute C, Rajpurkar P, Lou J, Ball RL, Shpanskaya K, Jabarkheel R, Kim LH, McKenna E, Tseng J, Ni J, Wishah F, Wittber F, Hong DS, Wilson TJ, Halabi S, Basu S, Patel BN, Lungren MP, Ng AY, Yeom KW. Deep learning-assisted diagnosis of cerebral aneurysms using the HeadXNet model. JAMA Netw Open. 2019;2:e195600. doi: 10.1001/jamanetworkopen.2019.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bien N, Rajpurkar P, Ball RL, Irvin J, Park A, Jones E, Bereket M, Patel BN, Yeom KW, Shpanskaya K, Halabi S, Zucker E, Fanton G, Amanatullah DF, Beaulieu CF, Riley GM, Stewart RJ, Blankenberg FG, Larson DB, Jones RH, Langlotz CP, Ng AY, Lungren MP. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: development and retrospective validation of MRNet. PLoS Med. 2018;15:e1002699. doi: 10.1371/journal.pmed.1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conant EF, Toledano AY, Periaswamy S, Fotin SV, Go J, Boatsman JE, Hoffmeister JW. Improving accuracy and efficiency with concurrent use of artificial intelligence for digital breast tomosynthesis. Radiol Artif Intell. 2019;1:e180096. doi: 10.1148/ryai.2019180096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaffter T, Buist DSM, Lee CI, Nikulin Y, Ribli D, Guan Y, Lotter W, Jie Z, Du H, Wang S, Feng J, Feng M, Kim HE, Albiol F, Albiol A, Morrell S, Wojna Z, Ahsen ME, Asif U, Jimeno Yepes A, Yohanandan S, Rabinovici-Cohen S, Yi D, Hoff B, Yu T, Chaibub Neto E, Rubin DL, Lindholm P, Margolies LR, McBride RB, Rothstein JH, Sieh W, Ben-Ari R, Harrer S, Trister A, Friend S, Norman T, Sahiner B, Strand F, Guinney J, Stolovitzky G, Mackey L, Cahoon J, Shen L, Sohn JH, Trivedi H, Shen Y, Buturovic L, Pereira JC, Cardoso JS, Castro E, Kalleberg KT, Pelka O, Nedjar I, Geras KJ, Nensa F, Goan E, Koitka S, Caballero L, Cox DD, Krishnaswamy P, Pandey G, Friedrich CM, Perrin D, Fookes C, Shi B, Cardoso Negrie G, Kawczynski M, Cho K, Khoo CS, Lo JY, Sorensen AG, Jung H. Evaluation of combined artificial intelligence and radiologist assessment to interpret screening mammograms. JAMA Netw Open. 2020;3:e200265. doi: 10.1001/jamanetworkopen.2020.0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das SR, Avants BB, Grossman M, Gee JC. Registration based cortical thickness measurement. Neuroimage. 2009;45:867–879. doi: 10.1016/j.neuroimage.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(Suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 36.Parasuraman R, Sheridan TB, Wickens CD. A model for types and levels of human interaction with automation. IEEE Trans Syst Man Cybern A Syst Hum. 2000;30:286–297. doi: 10.1109/3468.844354. [DOI] [PubMed] [Google Scholar]
- 37.Duong MT, Rauschecker AM, Rudie JD, Chen PH, Cook TS, Bryan RN, Mohan S. Artificial intelligence for precision education in radiology. Br J Radiol. 2019;92:20190389. doi: 10.1259/bjr.20190389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zaharchuk G, Gong E, Wintermark M, Rubin D, Langlotz CP. Deep learning in neuroradiology. AJNR Am J Neuroradiol. 2018;39:1–9. doi: 10.3174/ajnr.A5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al Yassin A, Salehi Sadaghiani M, Mohan S, Bryan RN, Nasrallah I. It is about “time”: academic neuroradiologist time distribution for interpreting brain MRIs. Acad Radiol. 2018;25:1521–1525. doi: 10.1016/j.acra.2018.04.014. [DOI] [PubMed] [Google Scholar]