Summary
Although expression quantitative trait loci (eQTLs) have been powerful in identifying susceptibility genes from genome-wide association study (GWAS) findings, most trait-associated loci are not explained by eQTLs alone. Alternative QTLs, including DNA methylation QTLs (meQTLs), are emerging, but cell-type-specific meQTLs using cells of disease origin have been lacking. Here, we established an meQTL dataset by using primary melanocytes from 106 individuals and identified 1,497,502 significant cis-meQTLs. Multi-QTL colocalization with meQTLs, eQTLs, and mRNA splice-junction QTLs from the same individuals together with imputed methylome-wide and transcriptome-wide association studies identified candidate susceptibility genes at 63% of melanoma GWAS loci. Among the three molecular QTLs, meQTLs were the single largest contributor. To compare melanocyte meQTLs with those from malignant melanomas, we performed meQTL analysis on skin cutaneous melanomas from The Cancer Genome Atlas (n = 444). A substantial proportion of meQTL probes (45.9%) in primary melanocytes is preserved in melanomas, while a smaller fraction of eQTL genes is preserved (12.7%). Integration of melanocyte multi-QTLs and melanoma meQTLs identified candidate susceptibility genes at 72% of melanoma GWAS loci. Beyond GWAS annotation, meQTL-eQTL colocalization in melanocytes suggested that 841 unique genes potentially share a causal variant with a nearby methylation probe in melanocytes. Finally, melanocyte trans-meQTLs identified a hotspot for rs12203592, a cis-eQTL of a transcription factor, IRF4, with 131 candidate target CpGs. Motif enrichment and IRF4 ChIP-seq analysis demonstrated that these target CpGs are enriched in IRF4 binding sites, suggesting an IRF4-mediated regulatory network. Our study highlights the utility of cell-type-specific meQTLs.
Keywords: quantitative trait loci, QTL, genome-wide association study, GWAS, melanoma, melanocyte, colocalization, DNA methylation, trans-QTL, methylome-wide association study, MWAS, mediation analysis, IRF4
Introduction
Expression quantitative trait locus (eQTL) studies have been powerful for nominating candidate causal genes for loci identified via genome-wide association studies (GWASs) of many complex traits and diseases, including cancer susceptibility. Most prominently, the Genotype-Tissue Expression (GTEx) project has made eQTL data publicly available for more than 50 tissue types.1 Most eQTL datasets, including GTEx, however, are based on heterogeneous bulk tissues where cell-type-specific allelic regulation of gene expression in rarer cell types may be obscured by signals from other cell types and thus may go undetected. Colocalization analyses with the most recent GTEx dataset (v.8) demonstrated that a median of 21% of GWAS loci from 87 tested complex traits colocalized with a cis-eQTL when aggregated across 49 tissue types.1 While cell-type-interacting eQTLs by computational deconvolution of bulk tissue data improves colocalization compared to that by standard eQTLs only,2,3 most GWAS loci nonetheless lack colocalizing eQTLs.
A recent melanoma GWAS meta-analysis identified a total of 54 loci reaching genome-wide significance,4 increasing the total number of melanoma-risk-associated loci by more than 3-fold compared to the largest existing study.5 We previously demonstrated that eQTLs from cultured melanocytes,6 the cell type of origin for melanoma, efficiently identified candidate susceptibility genes for 25%5,6 and 16%4 of loci from two recent melanoma GWASs through colocalization. Notably, as melanocytes represent only a small fraction of typical skin biopsies, even this moderately sized melanocyte eQTL dataset (n = 106) was able to identify candidate causal genes that were not captured by GTEx skin tissue eQTLs from sample sets three times larger,6 some of which were functionally validated.7,8 These data highlighted the utility of cell-type-specific QTL resources, however, eQTLs alone were still not sufficient to explain the majority of GWAS loci.
DNA methylation of cytosine at CpG dinucleotides is an important mode of epigenetic gene regulation. While CpG methylation is interconnected with mRNA expression, their relationship is rather complex. In tumors, hypermethylation has been observed in the promoters of inactivated tumor suppressor genes.9 Gene body methylation, on the other hand, is usually correlated with higher mRNA expression and tends to be inversely correlated with promoter methylation.10 Further, it is not always clear whether methylation/demethylation actively initiates gene expression repression/activation or, instead, methylation levels reflect repressed/activated expression status.11 While DNA methylation has been more widely studied as a marker of epigenetic regulation in population studies (e.g., EWAS12), DNA methylation is also under tight genetic control where an individual’s heritable genotypes could influence DNA methylation levels. Methylation QTL (meQTL) studies have been performed to detect local (cis-meQTL) and distant (trans-meQTL) correlation between the genotype of SNPs and CpG methylation. In particular, trans-meQTL has been powerful in identifying transcription-factor-mediated regulation networks and large numbers of target CpGs,13, 14, 15 in contrast to relatively small numbers of trans-eQTL genes or trans-splice QTL (sQTL) genes when using gene expression data.1
meQTL studies to date have largely been limited to blood and blood-related cell types13,14,16, 17, 18, 19, 20, 21, 22 with a few exceptions of studies of normal bulk tissues15,23, 24, 25 and tumor tissues.26,27 Overall, cell-type-specific meQTL studies from non-blood samples have largely been lacking. Particularly in the context of cancer, understanding a heritable component of DNA methylation in the cell types where the tumor originates may help answer questions about how methylation and gene expression are co-regulated through genetic variants and how much of that genetic regulation is still observed during the malignant transformation where multiple genetic and non-genetic events could mask gene expression variance explained by germline variants.
In this study, we explore the roles of cell-type-specific meQTLs derived from human primary melanocytes in explaining melanoma-risk-associated genetic signals through multi-QTL colocalization as well as an imputed methylome-wide association study (MWAS).28 We further compare genetic control of DNA methylation in melanocytes with that of malignant melanoma tissues. We then investigate whether eQTLs and meQTLs are connected by common causal variants in melanocytes and further identify a melanocyte-specific transcriptional hub through trans-meQTL study.
Material and methods
Melanocyte samples
Primary cultures of melanocytes from 106 newborn males mainly of European descent were used in this study as previously described.6 Of 106 individuals, 77 (73%) are of >80% European ancestry (CEU) and 100 (94%) are at least 20% European on the basis of ADMIXTURE29 analysis.6 Non-European samples include three individuals of African (YRI) descent and three individuals of Asian (CHB) descent at >80% and 23 individuals displaying admixed ancestry (Figure S1).
DNA methylation profiling
Genome-wide DNA methylation was profiled on the Illumina HumanMethylation450 BeadChip (Illumina, San Diego, USA). Genomic DNA was extracted as previously described,6 and DNA methylation was measured according to Illumina's standard procedure at the Cancer Genomics Research Laboratory (CGR), National Cancer Institute. Basic intensity quality control (QC) was performed with the minfi R package.30 Briefly, we background corrected and dye-bias equalized raw methylated and unmethylated intensities to correct for technical variation in signal between arrays. We applied the following criteria to filter probes and samples. (1) Probes located on chrX and chrY were removed. (2) Probes including common SNPs with minor allele frequency (MAF) > 5% (1000 Genomes, phase 3, EUR) were removed. No melanoma GWAS loci were found within 1 Mbp of these SNPs. (3) Probes located in repetitive genomic regions (repeatmask hg19 database) were removed. (4) Probes with detection p value > 0.01 were marked as missing. Probes with a missing rate > 5% were removed and samples with a missing rate > 4% were removed. (5) Control samples and samples without matched genotyping data were removed. (6) For duplicate samples, the better one of the two was selected on the basis of probe intensity, SNP call rate, and the percentage of missing probes. No batch effects or plating issues were identified across plates, wells, and barcode IDs on the basis of the assessment of methylated and unmethylated intensities, failed samples, and beta distributions. We used functional normalization implemented in the minfi R package30 to calculate the final methylation levels (beta value) after normalization. In total, we retained 386,520 probes (average density 134.2 probes/Mb) and 106 samples for the downstream meQTL analysis. We also calculated the top ten probabilistic estimation of expression residuals (PEERs)31 as potential hidden covariates for QTL analysis.
Quantification of RNA splicing
We re-analyzed RNA-sequencing (RNA-seq) data of the same 106 melanocytes from our previous publication6 to quantify RNA splicing. We used the processed BAM files to create the junction files and intron clustering based on the instructions of LeafCutter.32 The normalized quantification of 117,570 junctions was generated as the phenotype and ten principal components (PCs) were included as covariates for splice QTL (sQTL) analysis.
meQTL and sQTL detection
cis-meQTL and cis-sQTL analyses were performed with the same cis-QTL pipeline and the same processed genotype data (variant call format [vcf]) as described in our previous cis-eQTL analysis.6 Briefly, we used FastQTL to perform cis-QTL mapping,33 and we generated nominal p values for genetic variants located within ±1 Mb of the target CpG site of each probe (cis-meQTL) or splice junction (cis-sQTL) tested. For covariates of QTL analyses, we included three PCs inferred on the basis of genotype data and independent methylation variables (Pearson correlation coefficient < 0.8) from ten PEER factors (meQTLs) or independent splice junction usage variables (Pearson correlation < 0.8) from ten PCs (sQTLs). We then used the beta-distribution-adjusted empirical p values from FastQTL to calculate q values,34 and we applied a false discovery rate (FDR) threshold of ≤0.05 to identify probes or junctions with a significant QTL (“meProbes” or “sJunctions”). We used a similar method as that used for the GTEx study1 (using FastQTL) to identify all significant variant-probe or junction pairs. In summary, a genome-wide empirical p value threshold, pt, was defined as the empirical p value of the probe or junction closest to the 0.05 FDR threshold. We then used pt to calculate a nominal p value threshold for each gene on the basis of the beta distribution model of the minimum p value distribution f(pmin) obtained from the permutations for the probe or junction. Specifically, the nominal threshold was calculated as F−1(pt), where F−1 is the inverse cumulative distribution. For each probe or junction, variants with a nominal p value below the probe- or junction-level threshold were considered significant and included in the final list of genome-wide significant cis-QTL variants. The effect (slope) of QTLs is relative to the alternative allele.
trans-meQTLs detection
For identification of trans-meQTLs, we followed the methods that have been described previously by Shi and colleagues.15 Prior to meQTL analysis, each methylation trait was regressed on batches and independent PEER factors based on methylation profiles. The regression residuals were then quantile-normalized to the standard normal distribution N(0,1) for QTL analysis. We performed the genetic association testing by using tensorQTL,35 adjusted for the top three PCs based on GWAS SNPs to control for potential population stratification. To identify the threshold for genome-wide-significant trans-meQTLs, we applied the following statistical steps. For each CpG probe, the trans region was defined as being more than 5 Mb from the target CpG site in the same chromosome or on different chromosomes. For the nth methylation trait with m SNPs in the trans region, let (qn1,⋯,qnm) be the p values for testing the marginal association between the trait and the m SNPs. Let pn = min(qn1,⋯,qnm) be the minimum p value for m SNPs. We performed one million permutations for one random methylation trait with k SNPs in the trans region. We calculated the minimal p value as pp among these k SNPs for each permutation and then sorted all permutation minimal p values as pp1 to pp1000000. We then converted pn into the genome-wide empirical p value, padjn, by ranking pn among pp1 to pp1000000. Because a cis region is very short compared with the whole genome, padjn computed based on SNPs in trans regions is very close to that based on permutations with genome-wide SNPs. Thus, we use the genome-wide p value computed based on all SNPs to approximate padjn. Furthermore, all quantile-normalized traits follow the same standard normal distribution N(0,1); thus, the permutation-based null distributions are the same for all traits. We then applied the Benjamini–Hochberg36 procedure to (padj1,⋯,padjN) to identify trans-meQTLs by controlling FDR at 1%, which corresponded to a nominal p value of 1.03E−11.
TCGA SKCM meQTL analysis
444 skin cutaneous melanoma (SKCM) samples from The Cancer Genome Atlas (TCGA) with both genotype data and methylation data were included in our study. For genotype data, we collected our previously processed genotype data in vcf.6 The original raw intensity idat files from the HumanMethylation450 array with matched genotype data were downloaded from NCI Genomic Data Commons Data Portal (GDC Legacy Archive). The same DNA methylation processing pipelines for melanocytes described above were applied to TCGA methylation data, which included 384,273 high-quality probes for the downstream analysis. We selected the three PCs calculated from genotype data and uncorrected ten PEER factors from methylation data for the meQTL analysis. In addition, we adjusted for copy number alterations for each probe by including the segmentation’s logR value as a covariate for meQTL analysis. The segmentation CNV data was calculated from the SNP array as TCGA level 3 dataset, which was collected from the GDC portal. We followed the same melanocyte cis-meQTL analysis pipeline for the TCGA SKCM meQTL analysis. For trans-meQTL in TCGA SKCM, we only tested the association of significant melanocyte trans-meQTLs (FDR < 0.05) and applied a similar genome-wide p value threshold (1.03E−11) between SNPs and distant CpG probes.
Pairwise meQTL sharing between primary melanocytes and TCGA SKCM
To test the sharing of all significant SNP-CpG probe pairs of our melanocyte cis-meQTLs with those identified in TCGA SKCM, we calculated pairwise π1 statistics, where π1 is the proportion of all genome-wide significant meQTLs (using a threshold of FDR < 0.05) from one dataset found to also be genome-wide significant in the other. We used QVALUE34 to calculate π1, which indicates the proportion of true positives. A higher π1 value indicates an increased replication of meQTLs.
Multi-QTL colocalization
Melanoma GWAS summary statistics from a meta-analysis of 36,760 clinically confirmed and self-reported cutaneous melanoma cases were collected from a recent study,4 which included 54 significant loci with 68 independent SNPs. All study participants provided informed consent reviewed by IRBs, including 23andMe participants who gave online informed consent and participation, under a protocol approved by the external AAHRPP-accredited IRB, Ethical and Independent Review Services (E and I Review). We performed multi-QTL colocalization analyses among GWAS, eQTL, meQTL, and sQTL datasets. We used HyPrColoc37 to perform colocalization analysis with the following default parameters: prior.1 (1E−4) and prior.2 (0.980). We only considered genome-wide-significant QTL SNPs within ±250 kb of the GWAS lead SNP of each locus. Phased linkage disequilibrium (LD) matrices from 1000 Genomes, phase 3 (EUR), and sample overlap correction (as eQTL, meQTL, and sQTL datasets are coming from the same 106 individuals) were used for the colocalization analysis. We started with two-trait analyses comparing GWAS and each QTL one at a time: GWAS-eQTL, GWAS-meQTL, and GWAS-sQTL. Then, we performed three-trait (“G-e-m,” “G-s-e,” and “G-s-m”) and four-trait (“G-e-m-s”) analyses. For each matrix (trait × SNP), one gene/probe per trait is selected at a time. Any matrix (trait × SNP) from two-, three-, and four-trait analyses is dropped if there are fewer than 50 SNPs. The colocalization events showing the consistent number of tested traits and colocalizing traits were included as the final result. For sensitivity analysis, we performed a similar multi-QTL colocalization with the stricter prior.2 parameter in HyPrColoc: 0.990 and 0.995.
Imputed methylome-wide association study
We performed an imputed methylome-wide association study (MWAS) by predicting genetically regulated methylation levels of each CpG probe for the individuals from the GWAS dataset and performing an association analysis between predicted methylation levels and melanoma status. We used the same melanoma meta-analysis summary statistics as for the multi-QTL colocalization4 and methylation and genotype data from both TCGA SKCM and melanocyte data. We adapted TWAS FUSION,38 which was originally designed for transcriptome-wide association studies (TWASs), to perform the MWAS analysis. To summarize, we first collected the summary statistics without any significance thresholding. We then computed functional weights from our melanocyte methylation data one CpG probe at a time. Probes that failed to pass a heritability check (minimum heritability p value of 0.01) were excluded from further analysis. A cis-locus was restricted to 50 kb on either side of the CpG probe boundary. For melanocyte data, from 386,520 probes meeting basic quality control, 21,252 probes passed the heritability check and were included as MWAS weights for association analysis with the melanoma GWAS summary stats and 1000 Genomes, phase3 (EUR) LD reference. For the MWAS results, a genome-wide significance cutoff (MWAS p value < 0.05/number of probes tested) was applied.
eQTL/meQTL mediation analysis
We applied a workflow (Figure S2) to identify the potentially colocalized eQTL-meQTL pairs sharing a common causal variant, followed by mediation and partial correlation analysis as originally described by Pierce and colleagues.19 To identify candidate eQTL-meQTL pairs, we first restricted the meQTL analysis to 4,997 lead SNPs (eSNPs) for each eGene from eQTL results and 13,274 significant CpG probes (meProbes) from meQTL results to determine whether any of these eSNPs are significant cis-meQTLs for local meProbes (±1 Mb; FDR < 0.05). To reduce the redundant associations with the same SNP linking to a cluster of CpGs, we pruned our list of CpG probes by keeping only the CpGs whose lead meSNP had the highest LD with a lead eSNP. As a result, we identified each eGene paired with only one meCpG (eGene-meCpG pair), whose lead meSNP was in the strongest LD with the eSNP. In our melanocyte data, there were a total of 2,374 eGene-meCpG pairs showing association with a common SNP and available for colocalization analysis. We used HyPrColoc37 to perform colocalization analysis with the parameter prior.1 = 1E−4. A total of 841 potentially “colocalized” eQTL-meQTL pairs (including 296 common SNPs) were selected for downstream mediation analyses on the basis of the posterior probability of a common causal variant (CCV) above 0.8.
For mediation analysis, we used our melanocyte data on 106 genotyped individuals with both expression and methylation data to conduct tests of mediation for two hypothesized pathways: (1) SNP → methylation → expression, or “SME,” and (2) SNP → expression → methylation, or “SEM.” For all lead eSNPs, the cis-eQTL association was re-tested with adjustment for methylation of the CpG (and vice versa). Note that we cannot statistically exclude or account for a potential collider bias in both models. The difference between the beta coefficients before and after adjustment for the cis gene was expressed as the “proportion of the total effect that is mediated” (i.e., % mediation), calculated as |(βunadj – βadj)|/|βunadj| where βunadj and βadj represent the total effect and the direct effect of the variant, respectively.19,39 All regression analyses were adjusted for PCs inferred from expression or methylation data. The Sobel p value for mediation was calculated with the same formula as in previous publications.19,40
We also performed partial correlation analysis by using the colocalized eQTL-meQTL pairs in our 106 melanocyte datasets. The Pearson correlation coefficients between the gene expression and the methylation levels were calculated after adjusting for expression and methylation PCs, respectively. Both the gene expression and methylation levels were regressed on the lead eSNP, and the residuals from these regressions were obtained as the expression and methylation values that lack the phenotypic variance because of the effect of the SNP. We compared correlation coefficients before and after SNP adjustment to identify the eGene-meCpG pairs showing the partial correlation. To explore the extent to which partial correlation could be due to secondary, colocalized causal variants affecting both the expression trait and the CpG being analyzed, we also searched for secondary association signals for the eGene-meCpG pairs with partial correlation p < 0.05 and colocalization CCV > 0.8. For 73 pairs meeting these criteria, we adjusted for both the primary and secondary lead eSNP-meSNP. After this adjustment, 63 pairs were still significant (p > 0.05).
To explore the potential influence of CpG probe exclusion on methylation-expression mediation analysis, we surveyed the 5,575 methylation probes that were dropped from melanocyte meQTL analysis. These are probes with SNPs of MAF > 0.05 in EUR (minfi function dropLociWithSnps with SNPs parameters: “SBE” and “CpG”) were excluded to avoid technical artifacts’ affecting genotype effect on allelic methylation levels, as suggested by other studies.14,41 Among them, 583 unique methylation probes overlapped (within ±1 bp) with 594 unique melanocyte eQTL SNPs (595 unique probe-SNP pairs and 925 unique probe-SNP-gene trios). When overlaid with melanoma GWAS-melanocyte eQTL colocalization results (using HyPrColoc), none of the 594 eQTL SNPs overlapped with melanoma GWAS colocalized SNPs (posterior probability > 0.8) or their proxies (r2 > 0.8). Ten of the 594 eQTL SNPs were the strongest eQTL SNPs of an eGene (eSNP). Predicted allelic transcription factor binding for these ten SNPs was searched on Haploreg v.4.1.
Identifying cis-mediators for trans-meQTLs
To explore the mediation of trans-meQTLs by cis-eQTLs (e.g., of potential transcription factors), we performed mediation analysis by applying eQTLMAPT42 to the primary melanocyte meQTL data. Only trios with evidence of both cis-eQTL and trans-meQTL association were included. To detect the mediation effects, we derived 152 candidate trios from significant cis-eQTL and trans-meQTL associations (based on FDR < 0.05 and < 0.01, respectively). We performed the mediation analysis with an adaptive permutation scheme and generalized Pareto distribution approximation with parameters N = 10,000 and α = 0.05 for all candidate trios. All PEER factors included in eQTL and meQTL analyses and other covariates (top three genotype PCs) were adjusted and trios with suggestive mediation were reported with mediation p value threshold < 0.05.
Enrichment of melanoma GWAS variants in meQTLs
We generated quantile-quantile (QQ) plots to evaluate whether melanoma GWAS variants were enriched in meQTLs of melanocytes or TCGA SKCM. To minimize the impact of LD on the enrichment analysis, we performed LD pruning to identify independent SNPs among all the GWAS variants by using PLINK v.1.90 beta43 (r2 < 0.1 and window size 500 kb). QQ plots were made with p values (−log10) from the melanoma GWAS4 for non-meQTL SNPs versus meQTL SNPs after LD pruning. Deviation from the 45-degree line indicates that melanoma GWAS SNPs are enriched in meQTL SNPs.
Functional annotation of CpGs and meQTLs
Functional annotation of CpGs and meQTLs has been described previously.14 We annotated ten genomic features of CpGs, including CpGs located in CpG islands, low or high CpG regions, promoters, enhancers, gene bodies, 3 prime untranslated regions (3′ UTRs), 5 prime untranslated regions (5′ UTRs), 0–200 bases upstream of transcription start sites (TSS200), and 201–1,500 bases upstream of transcription start sites (TSS1,500). We used hypergeometric tests to evaluate whether the identified cis- and trans-meQTL CpGs showed enrichment for CpGs annotated with those genomic features. The significance threshold was defined by a fold change of >1.2 or <0.8 and a Bonferroni-corrected threshold p < 0.05/10 = 0.005.
In addition, we determined the distribution of genome-wide meCpG probes on the basis of their genomic position in relation to CpG islands and nearby genes. Enrichment fold change was calculated as the ratio of the fraction of meQTLs overlapping with genomic annotations versus the fraction of randomly selected SNPs overlapping with the genomic annotations; “epitools” was used for this analysis.
Motif enrichment analysis for trans-meQTLs
Enrichment of known sequence motifs among trans-CpGs was assessed with the PWMEnrich package in R. 131 CpG probes with trans-meQTL association with rs12203592 were selected for enrichment analysis. For PWMEnrich, the 101 bp sequence around each interrogated CpG site was used, similar to a previous study,13 and unique 2 kb promoters in humans were used as the pre-compiled background set. Although there is no minimum length of sequence required for the PWMEnrich analysis, it is recommended that the input sequences need to be longer than the length of the core sequence of the motif in the database to ensure the algorithm can properly compare them with a genomic background for score and p value calculation. Our detected top motifs are well within the range of 101 bp, indicating that our criteria sufficiently cover the necessary sequences. We also performed a sensitivity analysis to test whether varying lengths (51, 101, 201, 401, 1,001, 2,001, and 4,001 bp) of sequences could affect the resulting enriched motifs and found that all the different lengths of sequences, except 51 bp, identified IRF4 as one of the top three enriched motifs (data not shown).
IRF4 ChIP-sequencing in melanoma cells
To identify genome-wide binding sites of IRF4 in melanoma cells, we performed ChIP-sequencing against eGFP-tagged IRF4. We generated an inducible eGFP-tagged IRF4 cell line in 501Mel cells by cloning eGFP-tagged IRF4 downstream of the tetracycline response element in a PiggyBac transposon system.44 We used the Tetracycline-ON system where the expression of eGFP-IRF4 can be induced by adding doxycycline or tetracycline. For ChIP experiments, the eGFP-tagged-IRF4-expressing 501Mel cells were cultured on ten 10 cm dishes, and 1 μg/mL of doxycycline was added for the induction. Chromatin immunoprecipitation was performed according to Palomero and colleagues45 as follows: 20 million cells were crosslinked with 0.4% formaldehyde for 10 minutes at room temperature and quenched by 0.125 M glycine for 5 min at room temperature and chromatin was then sheared by 5 min sonication (25% amplitude, 30 sec off and 30 sec on) via a probe sonicator (Epishear, Active Motif). Immunoprecipitation was performed with Protein G Dynabeads (Life Technologies) with a total of 10 μg of anti-GFP antibody (3E6 from Molecular Probes, #A-11120). The bead-bound immune complexes were washed five times with wash buffer (50 M Hepes [pH 7.6], 1 mM EDTA, 0.7% Na-DOC, 1% NP-40, and 0.5 M LiCl) and once with Tris-EDTA. Crosslinking was reversed by washing the immune complexes and sonicated lysate input in elution buffer (50 mM Tris [pH 8], 10 mM EDTA, 1% SDS) overnight at 65ºC. Then the samples were treated with 0.2 μg/μL of RNase A for 1 h at 37ºC followed by treatment with 0.2 μg/μL proteinase K for 2 h at 55ºC. DNA was extracted from the samples using phenol:chloroform. ChIP-seq DNA libraries were prepared from the purified ChIP DNA and input DNA with the NEBNext ChIP-seq Library Prep Kit (E6200, NEB). Libraries were prepared from 8 to 15 ng of fragmented ChIP or input DNA, which were amplified with ten PCR cycles. The amplified libraries were purified with Agencourt AMPure XP beads (A63881, Beckman Coulter) and then were paired-end sequenced. Approximately 30 million raw reads were mapped of each sample to the human hg19 reference genome via Bowtie 2.46 The aligned reads were then used as an input for peak calling with MACS.47
IRF4 knockdown and RNA-seq
The human melanoma cell line 501Mel was cultured in RPMI-1640 cell culture medium (Gibco) supplemented with 10% FBS (Gibco) in a humid incubator at 5% CO2 and 37°C. IRF4 was knocked down in three biological replicates of 501Mel cells via transfection of the cells with Lipofectamine (RNAiMAX, Thermo Fisher) with siRNA (Silencer Select #AM16708, Thermo Fisher) for 48 h. Cells were harvested and RNA was extracted with the Quick-RNA Mini prep (#R1055, ZYMO Research). IRF4 knockdown was verified by RT-qPCR before generating sequencing libraries. RNA-seq was performed on the NovaSeq 6000 System, ∼150 million raw reads were mapped to human transcriptome GRCh38 with Kallisto,48 and differential expression analysis was performed with Sleuth.49
Results
Identification of cell-type-specific melanocyte meQTLs
To establish a melanocyte-specific meQTL dataset, we assessed DNA methylation levels in cultured melanocytes from 106 newborn males mainly of European descent by using Illumina 450K methylation arrays (material and methods; Figure S3). We then performed cis-meQTL analysis assessing variants within ±1 Mb of each CpG probe and identified 13,274 unique CpG probes (meProbes) with 1,497,502 significant cis-meQTLs (Table S1A). Most cis-meQTL variants are clustered near CpGs (<∼100 kb), where variants closer to the target CpGs tended to have lower p values and larger effect sizes (Figure S4). Among 13,274 meProbes, 29% were located in CpG islands and 34% in CpG-adjacent regions (shores and shelves), and the rest (38%) were away from CpG islands (open seas) (Figure S5). meProbes are also mainly located in or near the gene body (73% are within 1,500 bp of TSSs, UTRs, 1st exon, or gene body), and the rest (27%) are in intergenic regions. Compared to non-meProbes, meProbes are most enriched in open seas and intergenic regions, while most depleted in islands and 1st exons. At the variant level, cis-meQTLs are also significantly depleted in CpG islands and gene-promoter regions (Figure S6).
To supplement these meQTLs, as well as melanocyte-specific eQTLs we previously identified,6 we also performed mRNA splice junction QTL (sQTL) analysis by using previously generated RNA-seq data from the same melanocytes through which we identified 7,054 unique splice junctions with 887,233 cis-sQTLs (Table S1A). Together with our previous eQTL findings, we identified a total of 1,039,047 non-overlapping eQTL/meQTL/sQTL variants in melanocytes, a substantial proportion (40.4%) of which are only detected by meQTLs (Figure S7). Of meQTL variants, 27.4% and 21.8% were also detected as eQTLs and sQTLs, respectively, and 13.3% of meQTLs (n = 87,158) were significant for all three QTLs. Among eQTL variants, 42.3% and 36.7% were also detected as meQTLs and sQTLs, respectively. Among sQTLs, 44.2% and 40.5% displayed an overlap with eQTLs and meQTLs, respectively.
Multi-QTL colocalization improved melanoma GWAS annotation
To explore the contribution of cell-type-specific meQTLs and other QTLs to melanoma GWAS annotation, we first performed multi-trait colocalization by using HyPrColoc37 with summary data from a recent melanoma GWAS meta-analysis of 36,760 histologically confirmed and self-reported cases.4 Melanocyte meQTLs colocalized with melanoma GWAS signals (posterior probability > 0.8) at 13 of 54 loci, while sQTLs displayed colocalization at two loci (Figure 1, Tables S1B and S2). Together, at least one of three QTL types colocalized with melanoma GWAS signal at 21 of 54 melanoma loci (39%), which is a considerable improvement from the 12 loci (22%) explained by eQTLs alone via the same approach (HyPrColoc; note that this percentage differs slightly from the 16% reported in Landi et al.,4 where eCAVIAR50 was used for colocalization). Sensitivity analysis, adjusting the second prior from 0.98 to 0.99 and 0.995, indicated that 80% (61/76) and 64% (49/76) of colocalization events were still detected for the same traits at posterior probability > 0.8, respectively (Table S2). These data demonstrated that cell-type-specific multi-QTL colocalization could explain close to half of melanoma GWAS loci and that methylation QTL is the largest contributor colocalizing with 24% of the known loci.
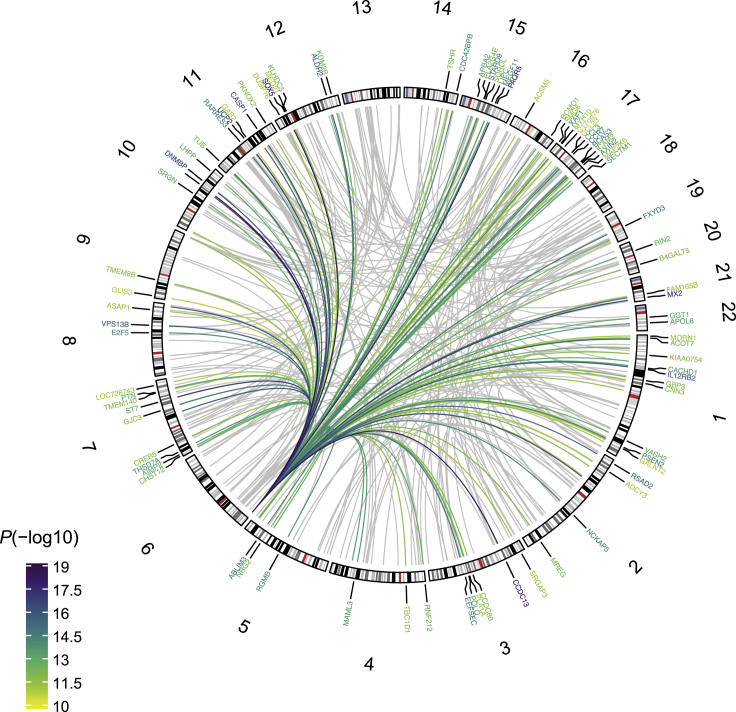
Figure 1.
Melanocyte meQTL and multi-QTL colocalization improved melanoma GWAS annotation
Circos plot shows significant colocalization of melanoma GWAS loci (top) with eQTLs (right), sQTLs (bottom), and meQTLs (left). Colocalization between individual GWAS loci with multiple QTL traits is depicted by thicker, colored lines. GWAS loci are sorted by genomic coordinate and labeled with GWAS lead SNPs with different colors; GWAS loci without any colocalizing QTLs are shown in black. QTL-associated gene symbols are also labeled with the same color as the GWAS loci they correspond to. Gene symbols are assigned on the basis of eQTLs/sQTLs for multi-QTL loci.
Further, multi-QTL colocalization identified four loci where more than one cell-type-specific QTL trait colocalizes with the melanoma GWAS signal (Figure 1, Table S2). For three loci, both eQTLs and meQTLs colocalized with the GWAS signal (MSC/RP11-383H13.1, OCA2/AC090696.2, and MX2; Figures S8A–S8C). At the fourth locus, all three QTL traits, including eQTL (CDH1), meQTL (meCpG near CDH1), and sQTL (splice junction in CDH3), were colocalized with the GWAS signal (Figure S8D). For the locus near MX2 (MIM: 147890), colocalization identified rs398206 as a common causal variant for eQTLs, meQTLs, and melanoma risk, validating our previous findings identifying this variant as a functional cis-regulatory variant regulating MX2.7 Here, meQTLs for two CpG probes in the gene body display the same allelic direction of effect as that of the MX2 eQTL, where higher methylation levels are correlated with the allele associated with increased MX2 expression, consistent with the observations that DNA methylation in the gene body is positively correlated with gene expression. OCA2 (MIM: 611409) is a known pigmentation gene and, within this locus, the lead GWAS SNP located in the HERC2 (MIM: 605837) gene, rs12913832, was identified as a common causal variant for eQTLs, meQTLs, and melanoma risk through the expression of both OCA2 and an antisense HERC2 transcript, AC090696.2. These results are consistent with the previous findings that a melanocyte-specific enhancer encompassing rs12913832 regulates OCA2 expression through an allele-preferential long-range chromatin interaction.51 The MSC (MIM: 603628)/RP11-383H13.1 locus was initially identified as a novel locus by melanoma TWAS with our melanocyte eQTL dataset6 and data from a prior melanoma GWAS meta-analysis,5 and this locus was subsequently identified as a genome-wide-significant GWAS locus by the larger melanoma GWAS.4 Our multi-QTL colocalization indicated that DNA methylation is also involved in this locus in mediating melanoma risk. Finally, for the CDH1/3 locus, rs4420522 in the intron of CDH1 (MIM: 602118) was identified as a common likely causal variant for an eQTL (CDH1), meQTL (CDH1 gene body open sea CpG), sQTL (CDH3; MIM: 602120), and melanoma risk. Notably, the eQTL (CDH1) and sQTL (CDH3) are for two different neighboring genes encoding E-cadherin and P-cadherin, respectively, that are located adjacent to each other. The same variants’ being an eQTL for one gene and a sQTL for another has been shown for a subset of GTEx sQTLs in a recent study,52 but whether they share candidate causal variants was not clear. Here, we show an example of a common candidate causal variant affecting gene expression or splicing of two different genes in the same cell type.
Imputed MWAS identified novel melanoma-associated loci
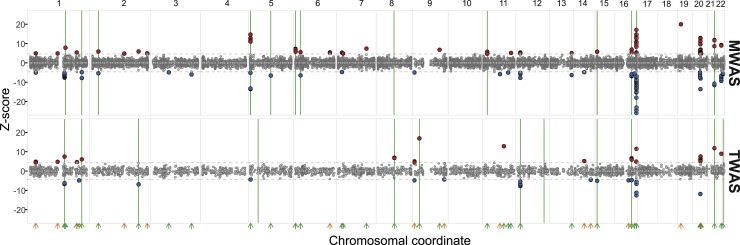
Given that meQTLs colocalize with a sizable proportion of melanoma GWAS signals, we further performed an imputed methylome-wide association study (MWAS)28 by using the melanocyte methylation data. Adopting the approach used for transcriptome-wide association studies (TWASs),38 we trained models of genetically regulated CpG methylation in our melanocyte dataset (material and methods) and tested the association of imputed methylation levels and melanoma risk by using the summary statistics from the melanoma GWAS. Significant MWAS was observed for 159 meCpGs (Bonferroni-corrected MWAS p < 0.05/21,252 tested probes), which overlapped 29 known genome-wide-significant melanoma GWAS loci and further nominated ten potentially “new” loci (Tables S3A and S4). Among these new loci, six overlapped with GWAS loci previously identified in a pleiotropic analysis between melanoma and nevus count and/or melanoma and hair color traits or loci identified by melanocyte TWAS (new loci 1, 2, 6, 7, 9, and 13; Table S4),4 suggesting that the MWAS approach effectively identifies bona fide susceptibility loci found via complementary approaches. Besides these six loci, the other four loci included CpG probes on or near SPOPL, NUMA1 (MIM: 164009)/LRTOMT (MIM: 612414), SNORD41/TNPO2 (MIM: 603002), EPB41L1 (MIM: 602879), and RPRD1B (MIM: 614694). These results demonstrated the potential of MWAS to nominate candidate susceptibility genes that are missed in the single-variant analysis.
Consistent with our comparisons between eQTL and meQTL colocalization, MWAS and TWAS together explained 54% of melanoma GWAS loci, which is a considerable improvement from 28% of GWAS loci by TWAS alone (Table S3A).4 Combined with the findings from colocalization analyses, melanocyte eQTLs and meQTLs together explained 63% of melanoma GWAS loci (Table S3B). TWAS, MWAS, and multi-QTL colocalization cross-validated each other in 18/54 (33%) of GWAS loci, where one or more approaches pointed to the same affected genes (Figure 2). Of the 16 genes that were supported by both TWAS and MWAS (gene assignment is based on CpG probes within 1.5 kb of the TSS, 5′ UTR, 1st exon, gene body, or 3′ UTR of a gene), six genes displayed the same direction of effect relative to melanoma risk (Z scores in the same direction), while five genes displayed the opposite direction of effect (Table S4). However, the other five genes (NIPAL3, CDH1, SPIRE2 [MIM: 609217], MX2, and MAFF [MIM: 604877]) were matched with CpG probes displaying the effect in both directions. These data suggest potential co-regulation of gene expression and promoter CpG methylation in these loci, contributing to melanoma risk.
Figure 2.
Manhattan plots of melanocyte TWAS and MWAS results combined with findings from eQTL and meQTL colocalization
Each circle represents the TWAS or MWAS Z score of a gene (TWAS) or a CpG probe (MWAS) reflecting significance and the direction of effect relative to melanoma risk (red, higher level correlates with melanoma risk; blue, lower level correlates with melanoma risk). Z scores are shown on the y-axis, and chromosomal positions are on the x-axis. Green arrows, overlapping melanoma GWAS loci; orange arrows, new loci detected by TWAS or MWAS; green lines, colocalization of eQTLs or meQTLs with melanoma GWAS loci; gray dashed horizontal lines, significance threshold defined by 0.05/number of probes or genes tested.
Through both colocalization and TWAS/MWAS, melanocyte eQTLs and meQTLs nominated a total of 107 unique candidate melanoma susceptibility genes. Ingenuity pathway analysis (Qiagen) identified biological pathways enriched by these genes, including those in melanin biosynthesis (L-dopachrome biosynthesis, L-dopa degradation, eumelanin biosynthesis), apoptosis (apoptosis signaling, Myc-mediated apoptosis signaling, retinoic acid-mediated apoptosis signaling), autophagy, adhesion junction signaling (epithelial adherens junction signaling, remodeling of epithelial adherens junctions), and melanoma-specific signaling (melanoma signaling, Wnt/beta-catenin signaling), among others (Table S5A). Of these, melanoma-specific signaling and apoptosis pathways are strengthened by adding meQTLs compared to a similar analysis using only eQTLs in melanocytes and skin tissues.4 Notably, upstream regulator analysis identified the transcription factor MITF (MIM: 156845) as the most significant regulator of these genes (Table S5B), which is consistent with its known role as the master regulator of melanocyte lineage53 and a melanoma susceptibility gene.54,55 Together, these data demonstrated that meQTL data are complementary to eQTL data and greatly increase the power to nominate candidate causal genes.
Melanocyte meQTLs are substantially preserved in melanomas
Given the large contribution of melanocyte meQTLs underlying melanoma GWAS loci, we further investigated whether and, if so, to what extent the genetic control of CpG methylation in the melanocytic lineage is preserved in malignant melanomas. For this, we performed a meQTL analysis of 444 cutaneous melanomas from TCGA by using data generated from the same 450K methylation array platform and by using the same analytic approach, except for adding regional genomic copy number as a covariate (material and methods). First, we identified 3,794,446 genome-wide-significant cis-meQTLs for 15,308 unique meProbes from TCGA melanomas, which are higher numbers than those observed from melanocytes (15% more meProbes). When meProbes were compared between datasets, 45.9% of melanocyte meProbes were also significant in melanomas, while 39.8% of melanoma meProbes were observed in melanocytes (Figure S9A). Melanocyte meQTL preservation in melanoma is even higher at the gene level, showing 65% preservation when meProbes are assigned to genes on the basis of their position relative to gene bodies or promoters (Figure S9B). The effect sizes of the best meQTL for each meProbe were highly correlated for 6,087 common meProbes in both groups (p value < 2.2E−16; R = 0.74), and 88.4% of them displayed the same direction of effect (Figure S10). We further calculated the true positive rates (π1) of top cis-meQTLs (FDR < 0.05) from melanocytes by examining their p value distributions in melanoma meQTLs and vice versa. The true positive rates (π1) were 0.825 and 0.822 for melanocyte meQTLs in melanomas and melanoma meQTLs in melanocytes, respectively, displaying a high level of meQTL preservation between two datasets. Notably, the proportion of normal melanocyte QTLs preserved in melanomas was much smaller at the eGene level, where only 12.7% of melanocyte eQTL genes were preserved in melanomas (Figure S9A), in contrast to the high preservation rate of meQTLs (45.9%). Among 635 preserved eGenes, 230 (36%) were associated with one or more preserved eProbes.
We then investigated whether melanoma-specific meQTLs corroborate melanoma GWAS annotation through colocalization and MWAS. Melanoma meQTLs colocalized with the melanoma GWAS signal at 11 of 54 loci (20%) (Table S6), and melanoma MWAS overlapped with 19 GWAS loci (35%) and further identified six new loci (Table S7). Among these were loci only explained by melanoma meQTLs but not by melanocyte QTLs; melanoma meQTLs uniquely annotated five GWAS loci (CpG probes on or near C2orf58, PPARGC1B [MIM: 608886], STN1 [MIM: 613128], and SHANK3 [MIM: 606230] and cg07068045 in open sea) and identified four novel MWAS loci (Table S8). Through colocalization and MWAS, melanoma meQTLs explained 46% (25/54) of melanoma GWAS loci, which, despite the >4× larger sample size and an overall higher number of identified meProbes, is considerably less than that by melanocyte meQTLs (56%). Consistent with this observation, melanoma risk-associated variants are more enriched for melanocyte meQTLs than for melanoma meQTLs (Figure S11). Thus, these data demonstrate that genetically regulated CpG methylation observed in the melanocyte lineage is substantially preserved in tumors. Nevertheless, these data also show that cancer susceptibility reflected in GWAS signals is better explained by DNA methylation from normal homogeneous cells of disease origin than by that from heterogeneous tumor tissues, even with considerably larger sample size. Overall, melanocyte multi-QTLs and melanoma meQTLs collectively explain 39 melanoma-risk-associated loci (Figure 3), representing 72% (39/54) of all known genome-wide-significant loci.
Figure 3.
Summary of melanoma GWAS annotation with melanocyte multi-QTLs and TCGA-melanoma meQTLs
Known melanoma-associated loci (green circles) are defined by the findings from the newest melanoma meta-analysis. The new melanoma-associated loci (orange circles) are identified on the basis of TWAS or MWAS analysis. Known and new GWAS loci are sorted by genomic coordinate. The top bar plot shows the total number of annotations per locus by multi-QTL colocalization (shown by QTL types) or TWAS/MWAS from melanocyte and TCGA datasets. The right marginal bar plot shows the percentage of GWAS loci annotated by each approach (the percentage of the known loci is labeled in green).
Genetic control of DNA methylation and gene expression in melanocytes
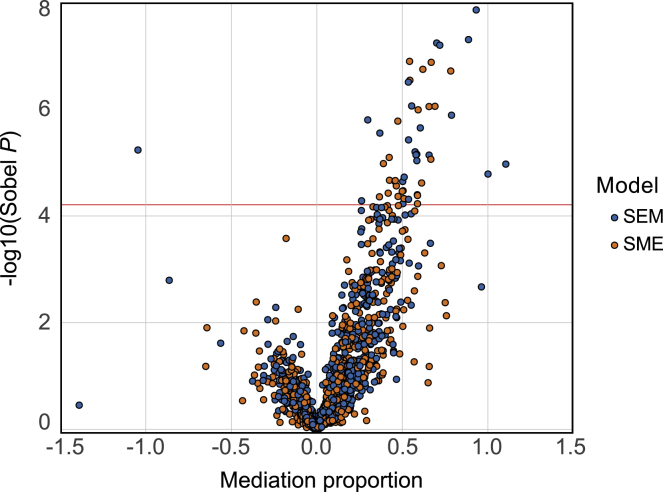
To investigate the genetic control of gene expression and DNA methylation in primary melanocytes beyond their contribution to melanoma risk, we sought to determine whether eQTLs and meQTLs more broadly share the same causal variants and whether one has a causal effect on the other. For this, we performed colocalization of eQTLs and meQTLs followed by mediation and partial correlation analysis as previously described by Pierce and colleagues19 (Figure S2). We first took 4,886 unique eSNPs (strongest eQTL SNP for each eGene) from eQTL data and re-identified cis-meQTLs, limiting to these 4,886 SNPs and 13,274 meCpG probes (meProbes). After pruning overlapping meProbes, we identified 2,374 unique eGene-meProbe pairs linked by the same eSNP, 841 of which (35%) were colocalized at a posterior probability > 0.8 via HyPrColoc (prior1 = 1E−4; prior2 = 0.95; material and methods). We then performed partial correlation analysis for those 841 eGene-meProbe pairs, of which 197 (23%) displayed correlation at a relaxed cutoff (p < 0.05) when conditioning on the primary variant and 50 (6%) displayed significant partial correlation (FDR < 0.05). Of 197, 73 pairs also had a significantly colocalizing secondary SNP and 63 of them remained significant (p < 0.05) when conditioning on both the primary and the secondary variants. These data suggested a link between DNA methylation and gene expression beyond that expected through common causal variants (Figure S12A; Table S9; material and methods). Next, we performed mediation analysis for 841 eGene-meProbe pairs to estimate the effect of SNPs on gene expression mediated by DNA methylation and vice versa. The results indicated that 32 unique eGene-meProbe pairs (4%) displayed significant mediation either of methylation on expression (25 pairs; FDR < 0.05 and % mediation > 0) or of expression on methylation (25 pairs; FDR < 0.05 and % mediation > 0), where 18 pairs were significant under both hypotheses (Figure 4; Table S10). All 32 significantly mediated pairs were included in 197 pairs displaying a marginal partial correlation (p < 0.05) (Figure S12B). Among 197 SNP-gene-probe trios, 69% (135 trios) displayed an opposite allelic direction of effect between meQTLs and eQTLs, while 31% (62 trios) displayed the same allelic direction of effect.
Figure 4.
Mediation analysis of potentially colocalizing SNP-eGene-meProbes
The volcano plot shows the mediation analysis results for both the SEM (blue) and SME (orange) models. Sobel p indicates the significance of the mediation analyses, where the red horizontal line indicates FDR = 0.05 cutoff. The mediation proportion shows the proportion of the total effect (cis-meQTL) mediated by a cis-gene (SEM) or the proportion of the total effect (cis-eQTL) mediated by cis-probes (SME). Mediation proportion can go in either direction depending on the directions of the effects of the confounders with the cis-mediator, the confounder on the cis-gene or cis-probes, and the non-reference allele on the cis-probes or cis-gene.
Our data suggest that a considerable proportion (∼35%; 841 of 2,374) of eQTLs and meQTLs for eGene-meProbe pairs may arise from the same causal variant in melanocytes. A subset (up to 23%; 197 of 841) of those displayed some evidence of methylation/expression co-regulation, where a majority displays an opposite directional effect. Notably, 841 potentially colocalizing eGene-meProbe pairs were significantly enriched in melanocyte eGenes that are preserved in malignant melanomas compared to non-preserved eGenes (Fisher's exact, p = 9.44E−7; OR = 1.68). We do not observe the same type of enrichment in preserved meProbes compared to non-preserved meProbes (p = 0.608; OR = 1.04). However, colocalizing eGene-meProbe pairs are significantly enriched in genes on or near the preserved meProbes compared to those with non-preserved meProbes (p = 3.36E−5; OR = 1.58). These data suggest that genetic influence on potentially co-regulated DNA methylation and gene expression in primary melanocytes tend to be well maintained during malignant transformation.
Although conventional meQTL analyses using array-based methylation measurement exclude SNPs overlapping CpGs themselves, SNPs on CpG sites could potentially have a high impact on allelic methylation and target gene expression. Among all the SNPs in assayed CpGs, 10.6% were significant eQTLs in melanocytes. Of these, we focused on ten CpG SNPs that are the strongest eQTL for an eGene (eSNPs) in our melanocyte dataset (Table S11). A majority of these CpG probes were located in promoter or enhancer regions near TSSs. While the allelic changes from C or G to A or T are considered to abolish the CpG sites preventing methylation, some of them were predicted to create transcription-factor-binding sites in exchange. In an example of cg16139068, a CpG probe near the TSS of OGDHL (MIM: 617513), an allelic change from CpG to CpA (rs61846889) dramatically increases predicted binding affinity for the Ahr::Arnt::HIF1 complex (Haploreg v.4.1 position weight matrix from Ward and Kellis56). rs61846889 is also a significant eQTL for OGDHL across multiple tissues as well as melanocytes, including sun-exposed skin (p = 1.2E−20, normalized effect size relative to A allele = 0.62; GTEx v.8). These data hint at a hypothesis that CpG SNPs could lead to allelic gene expression by directly affecting DNA methylation while simultaneously affecting transcription factor binding. Together our data provide insights into an intersection of eQTLs and meQTLs in the genetic control of gene expression and DNA methylation in melanocyte biology.
Melanocyte trans-meQTLs highlight an IRF4 transcriptional regulatory network
Next, we performed trans-meQTL analysis of melanocytes, testing SNPs outside the ±5 Mb boundary of each CpG probe or on a different chromosome. We observed 332 unique CpG probes with one or more significant trans-meQTL at FDR < 0.01 (Table S12; Figure 5). For 65% (215 of 332) of those CpG probes, the best trans-meQTL variant was also a significant cis-eQTL in melanocytes. Among all the significant trans-meQTL variants, only one variant was a hot spot trans-meQTL for more than 10 CpGs across the genome. Namely, rs12203592, a cis-eQTL for IRF4 (MIM: 601900) gene expression,6 was a trans-meQTL for 131 CpGs (40%). rs12203592 was previously shown as a functional variant in the melanocyte lineage that regulates the expression of the IRF4 transcription factor.57 In our previous study of melanocyte eQTLs, we identified rs12203592 as a significant cis-eQTL of IRF4 as well as a genome-wide-significant trans-eQTL for four different genes, TMEM140, MIR3681HG, PLA1A (MIM: 607460), and NEO1 (MIM: 601907),6 a subset of which displayed significant mediation by IRF4 cis-eQTLs. In the current study, rs12203592 was identified as a trans-meQTL for two CpG probes (cg14710552 and cg07972322) located in TMEM140 and one CpG probe (cg04330122) located in PLA1A, consistent with our findings in trans-eQTLs. Furthermore, 95.4% (125 of 131) of rs12203592 trans-meQTL-CpG pairs displayed a positive effect size relative to the alternative T allele, where lower IRF4 expression level is associated with higher methylation levels at the target CpGs (Table S13). These results are similar to the observation in blood samples, where trans-meQTL hotspots displayed consistent allelic directions.13,14 Our findings are consistent with the hypothesis that altered expression of IRF4 by the cis-eQTL SNP, rs12203592, affects allelic methylation changes of those CpGs on or near multiple downstream target genes in melanocytes.
Figure 5.
Melanocyte trans-meQTLs
Circos plot shows genome-wide significant trans-meQTLs at FDR < 0.01. The yellow-green gradient spikes show a hotspot trans-meQTL SNP, rs12203592, located at 6p25.3 that is associated with 131 CpG sites. Nearby genes of trans-meQTL-associated CpG sites are labeled outside of the circos plot.
We then asked whether any cis-eQTL variant is driving trans-meQTLs (i.e., via allelic expression of transcription factors and the subsequent effect on methylation of downstream targets) by performing mediation analysis with eQTLMAPT.42 For this, we tested 152 cis-eQTL variant:cis-eQTL gene:trans-meQTL probe trios (FDR < 0.05 for cis-meQTLs and < 0.01 for trans-meQTLs), of which 24 trios displayed significant mediation at p < 0.05 (Table S14; Figure S13). An overwhelming majority of the significant trios (92%; 22 of 24) included rs12203592, where a cis-eQTL of IRF4 expression mediates the trans-meQTL effect of 18 putative target genes, further supporting IRF4-mediated target gene regulation in melanocytes. Notably, among 18 putative IRF4 target genes was a melanoma-risk-associated gene, MX2 (MX dynamin-like GTPase 2). MX2 is an interferon-alpha-stimulated gene (ISG) with conventional roles in the innate immune response against HIV infection but was previously shown to have a melanocyte-lineage-specific function in promoting melanoma formation.7 Similarly, IRF4 was originally known as one of the IFN-regulatory factors with roles in B and T lymphocytes58, 59, 60 but also has melanocyte-lineage specific roles in pigmentation traits,57 which is consistent with its association with pigmentation traits,61,62 nevus counts,63 and melanoma risk.4,63 These data suggest a melanocyte-specific functional interaction between two melanoma-risk associated genes, IRF4 and MX2.
To further investigate whether the targets of rs12203592 trans-meQTLs are regulated by direct IRF4 binding, we performed IRF4 ChIP-seq by using 501Mel melanoma cells ectopically expressing IRF4. Among 131 significant trans-meQTL target CpGs (FDR < 0.01) of IRF4 cis-meQTL SNP rs12203592, 54 (41.2%) CpGs overlapped within ±100 bp of IRF4 ChIP-seq peaks (peaks detected at p < 1E−5 in at least one replicate) (Table S13). We also performed a motif enrichment analysis for the target CpGs of rs12203592 trans-meQTLs by using PWMEnrich, which showed that the motifs for IRF family proteins ranked at the top and the IRF4 motif was the second most significantly enriched motif (p = 3.09E−14) (Table S15; Figure S14). We further examined differentially expressed genes in 501Mel cells with IRF4 knockdown. Among 804 differentially expressed genes upon IRF4 knockdown (p < 0.01 and |log2(fold change)| > 1), seven genes overlapped with eight target CpG probes of rs12203592 trans-meQTLs (VPS13B [MIM: 607817], NCKAP5 [MIM: 608789], E2F5 [MIM: 600967], RGMB [MIM: 612687], SMG6 [MIM: 610963], MYH10 [MIM: 160776], and MAP2K6 [MIM: 601254]) (enrichment OR = 2.8, p value = 0.1), while none of them are near ChIP-seq peaks. These results provide support for IRF4 as a melanocyte-specific transcriptional regulator of multiple target genes. The data also support the hypothesis that allelic methylation changes in trans reflect altered gene expression driven by transcription factor binding rather than methylation changes themselves driving expression changes.
Finally, we tested whether significant melanocyte trans-meQTLs were also present in melanomas. Among 15,179 trans-meQTL variant-meProbe pairs found in melanocytes (FDR < 0.01), 11,714 were present in the TCGA SKCM dataset. rs12203592 was not present in the TCGA dataset and could not be tested. Of the tested variant-meProbe pairs, 9,868 (65% of 15,179 or 84% of 11,714), including all 332 melanocyte trans-meProbes, were significant in melanomas (p < 1E−11; equivalent to FDR < 0.01). A strong correlation of trans-meQTL effect sizes was observed between melanocyte and melanoma datasets (Pearson R = 0.71; p < 2.2E−16) (Figure S15). These data indicated that melanocyte trans-meQTLs are highly preserved in malignant melanomas.
Discussion
To date, meQTL studies have mainly been performed in blood and blood cell types,13,14,16, 17, 18, 19, 20, 21, 22 tumor tissues,26,27 and/or normal bulk tissues.15,23, 24, 25,64 However, cell-type-specific meQTL studies using the cell of origin for many diseases and traits have been largely lacking. Our study presents a rare example of a single cell type meQTL dataset accompanied by matching eQTL data. In this study, we explored the roles of cell-type-specific meQTLs in characterizing disease-associated genomic variants as well as understanding their roles in gene expression regulation. Using multi-trait colocalization and MWAS, we demonstrated that melanocyte meQTL data generated from a dataset of moderate sample size (n = 106) provides substantial power to detect melanoma-associated CpG probes. Comparison of meQTLs between melanocytes and malignant melanomas revealed that melanocyte meQTLs are far better preserved than eQTLs in melanomas. Together, melanocyte multi-QTL and melanoma meQTL data nominated molecular phenotypes underlying 72% of known genome-wide-significant melanoma GWAS loci (and identified multiple novel loci), which is higher than conventional eQTL-colocalization-based findings.1 Pathway analyses of these genes highlighted melanoma- and melanocyte-lineage-specific signaling, as well as a master regulator of melanocyte lineage, MITF, which was not apparent from the analyses using only eQTLs. Melanocyte meQTLs also extended our knowledge on genetic regulation of gene expression involving DNA methylation. eQTL-meQTL colocalization/mediation analyses and trans-meQTL hotspot analysis highlighted the roles of transcription factors in allelic methylation patterns, including those through lineage-specific transcription factors and target genes.
Melanocyte trans-meQTL analysis identified a melanocyte-specific regulatory network involving a transcription factor, IRF4. Previous studies suggested that trans-meQTL hotspots could affect the expression of nearby transcription factors (i.e., cis-eQTLs), which might be reflected on the allelic methylation of their potential binding sites across the genome.13, 14, 15 In our study, a trans-meQTL hotspot SNP, rs12203592, displayed multiple lines of support for regulation by the IRF4 transcription factor. IRF4 is primarily known as an interferon regulatory factor highly expressed in lymphocytes and blood cells, but rs12203592 is located in a melanocyte-specific enhancer element and seems to be regulated through a melanocyte-lineage-specific transcriptional program affecting pigmentation phenotypes.57 Consistent with this observation, two large blood trans-meQTL studies using thousands of samples did not identify trans-meQTL hotspots through rs12203592.13,14 Among the target CpGs of rs12203592 trans-meQTLs is the recently identified melanoma susceptibility gene, MX2, which also has pleiotropic roles in both melanoma promotion and immune response, hinting at potential functional interaction between IRF4 and MX2 in melanomagenesis. By combining eQTLs, meQTLs, and mediation analysis as well as ChIP-seq and knockdown analyses, our study presents a unique example of a cell-type-specific transcriptional network mediated by a multi-function transcription factor. Notably, the IRF4-mediated regulatory network in melanocytes was marginally detectable by trans-eQTLs,6 but trans-meQTL analysis in the current study revealed orders of magnitude larger plausible downstream targets (four genes at FDR < 0.1 versus 131 CpGs at FDR < 0.01). These data suggest that CpG methylation might better represent the dynamic status of transcription-factor-binding-related chromatin changes than gross gene expression changes do.
Our study provides a formal comparison of meQTLs and eQTLs between tumor tissues and cells of tumor origin. We show that a substantial proportion (45.9%) of genome-wide-significant meCpG probes in melanocytes are preserved in melanomas. This is a much larger overlap compared to that of eGenes observed in our previous eQTL study using the same datasets, where only 12.7% of melanocyte eGenes were preserved in TCGA melanomas. One can speculate that the proportion of gene expression variance explained by genotypes could become relatively smaller and undetectable in malignantly transformed cells, where multiple factors, including alterations of DNA methylation, chromatin modifications, genomic copy number, genomic structures, as well as mutations in somatic driver genes, could collectively influence local and global gene expression levels. Loss of the majority of normal tissue eQTLs in tumors has been observed in prostate tumors, although this was not examined genome wide.23 Our comparisons of eQTLs and meQTLs from the same samples suggest that genetic control of lineage-specific CpG methylation is still largely detectable even in the presence of presumably high variation of methylation in tumor genomes. Our eQTL-meQTL colocalization analysis also indicated that a substantial portion of tested genes in melanocytes are potentially co-regulated with DNA methylation through common genetic variants. Importantly, these co-regulated genes and CpG probes are likely to remain under genetic control during malignant transformation even in the presence of somatic events. Consistent with this idea, melanocyte trans-meQTLs (presumably regulated through transcription factor binding) were preserved in melanomas at an even higher level (65%) than cis-meQTLs. These data provide an insight into our understanding of gene expression regulation in tumors, where both heritable and tumor-specific events contribute to the total transcriptome profile.
While a formal comparison of melanocyte meQTLs with those from other tissue types is warranted as more of them become available, we performed an initial comparison with bulk skin tissue meQTLs in the context of melanoma GWAS annotation. Roos and colleagues64 reported a meQTL analysis of skin tissues (n = 283, European ancestry) for 22 melanoma-risk-associated variants. At the CpG probe level, among 21 of 22 SNP-CpG probe pairs that passed our QC, only two SNPs were genome-wide-significant meQTLs in melanocytes, while ten SNPs were significant meQTLs in skin tissues on the basis of the cutoff defined by each study (Table S16). However, meQTL effect sizes between melanocytes and skin displayed a significant correlation (Pearson R = 0.517, p = 0.028, with absolute values of effect sizes). Further, at the gene level, when we inspected the local CpG probes (±100 kb of the SNP) with the best meQTL p values for 21 SNPs in melanocytes, 11 of them were associated with the same genes as the best CpG probes in skin. While an in-depth comparison with the same regression model is warranted, these data suggest that cell-type-specific melanocyte meQTL data may share some similarities but are complementary to skin meQTLs in annotating melanoma-risk-associated loci.
Although meQTL is powerful, sensitive, and reliable, we acknowledge a few limitations of the current study. First, our dataset has a relatively small sample size and limited genome-wide meProbe coverage (i.e., ∼450,000 meCpGs), which might have compromised statistical power for QTL detection, especially for mediation analysis. Second, our dataset is not of 100% European ancestry (73% European and 94% European or European-admixed ancestry; Figure S1), and although we adjusted genetic ancestry in our QTL analyses to account for this heterogeneity, we recognize a minor discrepancy in ancestry might have affected QTL-based analyses relying on matched LD structures between GWAS and QTL populations such as TWAS and MWAS. A sensitivity analysis using a subset of strictly European individuals (n = 77) demonstrated a significant and strong correlation of meQTL effect sizes (Pearson R = 0.91) between the European subset and the full dataset, indicating the inclusion of individuals with higher non-European ancestry was not adversely impacting our analyses. Finally, we recognize that assigning the effector genes to significant meCpG is still challenging in the absence of colocalizing eQTL support. Colocalization approaches with an improved detection power might help identify those left undetected with the current approaches. Additionally, some of the GWAS-colocalizing meQTLs without concurrent eQTL support might reflect loci poised to be connected with allelic differences in gene expression upon appropriate stimulations (e.g., UV exposure), which actively proliferating cultured melanocytes cannot recapitulate.
In conclusion, our study demonstrated the utility of cell-type-specific meQTLs in GWAS annotation and provided insights into melanocyte-specific gene expression regulation involving DNA methylation.
Acknowledgments
This work has been supported by the Intramural Research Program (IRP) of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, US National Institutes of Health. This work was also supported by the Research Fund of Iceland (E.S., 184861 and 207067), the University of Iceland Doctoral Grants Fund (R.D.), the University of Iceland Research Fund (B.O.E., postdoctoral grant), Cancer Research UK (Program Award C588/A19167), and NIH (R01-CA83115). This work utilized the Biowulf cluster computing system at the NIH. The results appearing here are in part based on data generated by the TCGA Research Network. We would like to thank members at the National Cancer Institute Cancer Genomics Research Laboratory (CGR) for help with sequencing efforts, Stacie Loftus and William Pavan from the National Human Genome Research Institute for the help with the melanocyte eQTL study, and Christopher Foley from the University of Cambridge for the help with HyPrColoc analysis. We thank Erping Long, Alyssa Klein, and Alyxandra Golden for proofreading the manuscript. We also thank all the cohorts, funders, and investigators who contributed to the melanoma GWAS, acknowledged by Landi et al.,4 23andMe, and colleagues, from which data was used towards fine-mapping. We would like to thank the research participants and employees of 23andMe for making this work possible. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Declaration of interests
The authors declare no competing interests.
Published: July 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.06.018.
Data and code availability
The raw data of Illumina HumanMethylation450 BeadChips from 106 primary human melanocytes have been submitted to the Gene Expression Omnibus (GEO) database under accession code GEO: GSE166069; melanocyte genotype data, RNA-seq expression data, and all meQTL association results are deposited in Genotypes and Phenotypes (dbGaP) under accession dbGaP: phs001500.v1.p1. IRF4 ChIP-seq and RNA-seq data are deposited in GEO under accession code GEO: GSE167945. Data from the 2020 melanoma GWAS meta-analysis performed by Landi and colleagues were obtained from dbGaP (dbGaP: phs001868.v1.p1), with the exclusion of self-reported data from 23andMe and UK Biobank. The full GWAS summary statistics for the 23andMe discovery dataset will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please visit https://research.23andme.com/collaborate/#dataset-access/ for more information and to apply to access the data. Summary data from the remaining self-reported cases are available from the corresponding authors of that manuscript4 (Matthew Law, matthew.law@qimrberghofer.edu.au; Mark Iles, m.m.iles@leeds.ac.uk; and Maria Teresa Landi, landim@mail.nih.gov).
Web resources
epitools, https://cran.r-project.org/web/packages/epitools/index.html
eQTLMAPT, https://github.com/QidiPeng/eQTLMAPT
FastQTL, http://fastqtl.sourceforge.net/
GDC Data Portal, https://portal.gdc.cancer.gov
Haploreg v.4.1, https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php
HyPrColoc, https://github.com/jrs95/hyprcoloc
Kallisto, https://pachterlab.github.io/kallisto/
LeafCutter, https://davidaknowles.github.io/leafcutter/
minfi, https://bioconductor.org/packages/release/bioc/html/minfi.html
NIH Biowulf Cluster, http://hpc.nih.gov
OMIM, http://www.omim.org
PWMEnrich, https://bioconductor.org/packages/release/bioc/html/PWMEnrich.html
QVALUE, https://bioconductor.org/packages/release/bioc/html/qvalue.html
tensorQTL, https://github.com/broadinstitute/tensorqtl
The Cancer Genome Atlas (TCGA) Research Network, http://cancergenome.nih.gov/
TWAS FUSION, http://gusevlab.org/projects/fusion/
Supplemental information
References
- 1.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim-Hellmuth S., Aguet F., Oliva M., Muñoz-Aguirre M., Kasela S., Wucher V., Castel S.E., Hamel A.R., Viñuela A., Roberts A.L. Cell type–specific genetic regulation of gene expression across human tissues. Science. 2020;369:eaaz8528. doi: 10.1126/science.aaz8528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donovan M.K.R., D’Antonio-Chronowska A., D’Antonio M., Frazer K.A. Cellular deconvolution of GTEx tissues powers discovery of disease and cell-type associated regulatory variants. Nat. Commun. 2020;11:955. doi: 10.1038/s41467-020-14561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landi M.T., Bishop D.T., MacGregor S., Machiela M.J., Stratigos A.J., Ghiorzo P., Brossard M., Calista D., Choi J., Fargnoli M.C. Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility. Nat. Genet. 2020;52:494–504. doi: 10.1038/s41588-020-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Law M.H., Bishop D.T., Lee J.E., Brossard M., Martin N.G., Moses E.K., Song F., Barrett J.H., Kumar R., Easton D.F. Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nat. Genet. 2015;47:987–995. doi: 10.1038/ng.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang T., Choi J., Kovacs M.A., Shi J., Xu M., Goldstein A.M., Trower A.J., Bishop D.T., Iles M.M., Duffy D.L. Cell-type-specific eQTL of primary melanocytes facilitates identification of melanoma susceptibility genes. Genome Res. 2018;28:1621–1635. doi: 10.1101/gr.233304.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi J., Zhang T., Vu A., Ablain J., Makowski M.M., Colli L.M., Xu M., Hennessey R.C., Yin J., Rothschild H. Massively parallel reporter assays of melanoma risk variants identify MX2 as a gene promoting melanoma. Nat. Commun. 2020;11:2718. doi: 10.1038/s41467-020-16590-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi J., Xu M., Makowski M.M., Zhang T., Law M.H., Kovacs M.A., Granzhan A., Kim W.J., Parikh H., Gartside M. A common intronic variant of PARP1 confers melanoma risk and mediates melanocyte growth via regulation of MITF. Nat. Genet. 2017;49:1326–1335. doi: 10.1038/ng.3927. [DOI] [PubMed] [Google Scholar]
- 9.Baylin S.B., Jones P.A. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulis M., Heath S., Bibikova M., Queirós A.C., Navarro A., Clot G., Martínez-Trillos A., Castellano G., Brun-Heath I., Pinyol M. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat. Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 11.Husquin L.T., Rotival M., Fagny M., Quach H., Zidane N., McEwen L.M., MacIsaac J.L., Kobor M.S., Aschard H., Patin E., Quintana-Murci L. Exploring the genetic basis of human population differences in DNA methylation and their causal impact on immune gene regulation. Genome Biol. 2018;19:222. doi: 10.1186/s13059-018-1601-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birney E., Smith G.D., Greally J.M. Epigenome-wide Association Studies and the Interpretation of Disease -Omics. PLoS Genet. 2016;12:e1006105. doi: 10.1371/journal.pgen.1006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonder M.J., Luijk R., Zhernakova D.V., Moed M., Deelen P., Vermaat M., van Iterson M., van Dijk F., van Galen M., Bot J. Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 2017;49:131–138. doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 14.Huan T., Joehanes R., Song C., Peng F., Guo Y., Mendelson M., Yao C., Liu C., Ma J., Richard M. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat. Commun. 2019;10:4267. doi: 10.1038/s41467-019-12228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi J., Marconett C.N., Duan J., Hyland P.L., Li P., Wang Z., Wheeler W., Zhou B., Campan M., Lee D.S. Characterizing the genetic basis of methylome diversity in histologically normal human lung tissue. Nat. Commun. 2014;5:3365. doi: 10.1038/ncomms4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson T.G., Haycock P.C., Zheng J., Timpson N.J., Gaunt T.R., Davey Smith G., Relton C.L., Hemani G. Systematic Mendelian randomization framework elucidates hundreds of CpG sites which may mediate the influence of genetic variants on disease. Hum. Mol. Genet. 2018;27:3293–3304. doi: 10.1093/hmg/ddy210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McClay J.L., Shabalin A.A., Dozmorov M.G., Adkins D.E., Kumar G., Nerella S., Clark S.L., Bergen S.E., Hultman C.M., Magnusson P.K.E. High density methylation QTL analysis in human blood via next-generation sequencing of the methylated genomic DNA fraction. Genome Biol. 2015;16:291. doi: 10.1186/s13059-015-0842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark A.D., Nair N., Anderson A.E., Thalayasingam N., Naamane N., Skelton A.J., Diboll J., Barton A., Eyre S., Isaacs J.D. Lymphocyte DNA methylation mediates genetic risk at shared immune-mediated disease loci. J. Allergy Clin. Immunol. 2020;145:1438–1451. doi: 10.1016/j.jaci.2019.12.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierce B.L., Tong L., Argos M., Demanelis K., Jasmine F., Rakibuz-Zaman M., Sarwar G., Islam M.T., Shahriar H., Islam T. Co-occurring expression and methylation QTLs allow detection of common causal variants and shared biological mechanisms. Nat. Commun. 2018;9:804. doi: 10.1038/s41467-018-03209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banovich N.E., Lan X., McVicker G., van de Geijn B., Degner J.F., Blischak J.D., Roux J., Pritchard J.K., Gilad Y. Methylation QTLs are associated with coordinated changes in transcription factor binding, histone modifications, and gene expression levels. PLoS Genet. 2014;10:e1004663. doi: 10.1371/journal.pgen.1004663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaunt T.R., Shihab H.A., Hemani G., Min J.L., Woodward G., Lyttleton O., Zheng J., Duggirala A., McArdle W.L., Ho K. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 2016;17:61. doi: 10.1186/s13059-016-0926-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yan Y., Kundu K., Ecker S. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell. 2016;167:1398–1414.e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai J.Y., Wang X., Wang B., Sun W., Jordahl K.M., Kolb S., Nyame Y.A., Wright J.L., Ostrander E.A., Feng Z., Stanford J.L. DNA methylation and cis-regulation of gene expression by prostate cancer risk SNPs. PLoS Genet. 2020;16:e1008667. doi: 10.1371/journal.pgen.1008667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulz H., Ruppert A.-K., Herms S., Wolf C., Mirza-Schreiber N., Stegle O., Czamara D., Forstner A.J., Sivalingam S., Schoch S. Genome-wide mapping of genetic determinants influencing DNA methylation and gene expression in human hippocampus. Nat. Commun. 2017;8:1511. doi: 10.1038/s41467-017-01818-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grundberg E., Meduri E., Sandling J.K., Hedman A.K., Keildson S., Buil A., Busche S., Yuan W., Nisbet J., Sekowska M. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am. J. Hum. Genet. 2013;93:876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyn H., Sayols S., Moutinho C., Vidal E., Sanchez-Mut J.V., Stefansson O.A., Nadal E., Moran S., Eyfjord J.E., Gonzalez-Suarez E. Linkage of DNA methylation quantitative trait loci to human cancer risk. Cell Rep. 2014;7:331–338. doi: 10.1016/j.celrep.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 27.Gong J., Wan H., Mei S., Ruan H., Zhang Z., Liu C., Guo A.-Y., Diao L., Miao X., Han L. Pancan-meQTL: a database to systematically evaluate the effects of genetic variants on methylation in human cancer. Nucleic Acids Res. 2019;47(D1):D1066–D1072. doi: 10.1093/nar/gky814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baselmans B.M.L., Jansen R., Ip H.F., van Dongen J., Abdellaoui A., van de Weijer M.P., Bao Y., Smart M., Kumari M., Willemsen G. Multivariate genome-wide analyses of the well-being spectrum. Nat. Genet. 2019;51:445–451. doi: 10.1038/s41588-018-0320-8. [DOI] [PubMed] [Google Scholar]
- 29.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stegle O., Parts L., Piipari M., Winn J., Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y.I., Knowles D.A., Humphrey J., Barbeira A.N., Dickinson S.P., Im H.K., Pritchard J.K. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 2018;50:151–158. doi: 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ongen H., Buil A., Brown A.A., Dermitzakis E.T., Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32:1479–1485. doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor-Weiner A., Aguet F., Haradhvala N.J., Gosai S., Anand S., Kim J., Ardlie K., Van Allen E.M., Getz G. Scaling computational genomics to millions of individuals with GPUs. Genome Biol. 2019;20:228. doi: 10.1186/s13059-019-1836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamini Y., Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Series B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- 37.Foley C.N., Staley J.R., Breen P.G., Sun B.B., Kirk P.D.W., Burgess S., Howson J.M.M. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat. Commun. 2021;12:764. doi: 10.1038/s41467-020-20885-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W.J.H., Jansen R., de Geus E.J.C., Boomsma D.I., Wright F.A. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baron R.M., Kenny D.A. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J. Pers. Soc. Psychol. 1986;51:1173–1182. doi: 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- 40.Sobel M.E. Direct and Indirect Effects in Linear Structural Equation Models. Sociol. Methods Res. 1987;16:155–176. [Google Scholar]
- 41.Smith A.K., Kilaru V., Kocak M., Almli L.M., Mercer K.B., Ressler K.J., Tylavsky F.A., Conneely K.N. Methylation quantitative trait loci (meQTLs) are consistently detected across ancestry, developmental stage, and tissue type. BMC Genomics. 2014;15:145. doi: 10.1186/1471-2164-15-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang T., Peng Q., Liu B., Liu X., Liu Y., Peng J., Wang Y. eQTLMAPT: Fast and Accurate eQTL Mediation Analysis With Efficient Permutation Testing Approaches. Front. Genet. 2020;10:1309. doi: 10.3389/fgene.2019.01309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magnúsdóttir E., Dietmann S., Murakami K., Günesdogan U., Tang F., Bao S., Diamanti E., Lao K., Gottgens B., Azim Surani M. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat. Cell Biol. 2013;15:905–915. doi: 10.1038/ncb2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palomero T., Lim W.K., Odom D.T., Sulis M.L., Real P.J., Margolin A., Barnes K.C., O’Neil J., Neuberg D., Weng A.P. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 49.Pimentel H., Bray N.L., Puente S., Melsted P., Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods. 2017;14:687–690. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 50.Hormozdiari F., van de Bunt M., Segrè A.V., Li X., Joo J.W.J., Bilow M., Sul J.H., Sankararaman S., Pasaniuc B., Eskin E. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am. J. Hum. Genet. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Visser M., Kayser M., Palstra R.-J. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 2012;22:446–455. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garrido-Martín D., Borsari B., Calvo M., Reverter F., Guigó R. Identification and analysis of splicing quantitative trait loci across multiple tissues in the human genome. Nat. Commun. 2021;12:727. doi: 10.1038/s41467-020-20578-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levy C., Khaled M., Fisher D.E. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 54.Yokoyama S., Woods S.L., Boyle G.M., Aoude L.G., MacGregor S., Zismann V., Gartside M., Cust A.E., Haq R., Harland M. A novel recurrent mutation in MITF predisposes to familial and sporadic melanoma. Nature. 2011;480:99–103. doi: 10.1038/nature10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bertolotto C., Lesueur F., Giuliano S., Strub T., de Lichy M., Bille K., Dessen P., d’Hayer B., Mohamdi H., Remenieras A. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature. 2011;480:94–98. doi: 10.1038/nature10539. [DOI] [PubMed] [Google Scholar]
- 56.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gkr917. D930–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Praetorius C., Grill C., Stacey S.N., Metcalf A.M., Gorkin D.U., Robinson K.C., Van Otterloo E., Kim R.S.Q., Bergsteinsdottir K., Ogmundsdottir M.H. A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway. Cell. 2013;155:1022–1033. doi: 10.1016/j.cell.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hagman J. Critical Functions of IRF4 in B and T Lymphocytes. J. Immunol. 2017;199:3715–3716. doi: 10.4049/jimmunol.1701385. [DOI] [PubMed] [Google Scholar]
- 59.Shaffer A.L., Emre N.C.T., Romesser P.B., Staudt L.M. IRF4: Immunity. Malignancy! Therapy? Clin. Cancer Res. 2009;15:2954–2961. doi: 10.1158/1078-0432.CCR-08-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Man K., Miasari M., Shi W., Xin A., Henstridge D.C., Preston S., Pellegrini M., Belz G.T., Smyth G.K., Febbraio M.A. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol. 2013;14:1155–1165. doi: 10.1038/ni.2710. [DOI] [PubMed] [Google Scholar]
- 61.Morgan M.D., Pairo-Castineira E., Rawlik K., Canela-Xandri O., Rees J., Sims D., Tenesa A., Jackson I.J. Genome-wide study of hair colour in UK Biobank explains most of the SNP heritability. Nat. Commun. 2018;9:5271. doi: 10.1038/s41467-018-07691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hysi P.G., Valdes A.M., Liu F., Furlotte N.A., Evans D.M., Bataille V., Visconti A., Hemani G., McMahon G., Ring S.M. Genome-wide association meta-analysis of individuals of European ancestry identifies new loci explaining a substantial fraction of hair color variation and heritability. Nat. Genet. 2018;50:652–656. doi: 10.1038/s41588-018-0100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duffy D.L., Zhu G., Li X., Sanna M., Iles M.M., Jacobs L.C., Evans D.M., Yazar S., Beesley J., Law M.H., Melanoma GWAS Consortium Novel pleiotropic risk loci for melanoma and nevus density implicate multiple biological pathways. Nat. Commun. 2018;9:4774. doi: 10.1038/s41467-018-06649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roos L., Sandling J.K., Bell C.G., Glass D., Mangino M., Spector T.D., Deloukas P., Bataille V., Bell J.T. Higher Nevus Count Exhibits a Distinct DNA Methylation Signature in Healthy Human Skin: Implications for Melanoma. J. Invest. Dermatol. 2017;137:910–920. doi: 10.1016/j.jid.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of Illumina HumanMethylation450 BeadChips from 106 primary human melanocytes have been submitted to the Gene Expression Omnibus (GEO) database under accession code GEO: GSE166069; melanocyte genotype data, RNA-seq expression data, and all meQTL association results are deposited in Genotypes and Phenotypes (dbGaP) under accession dbGaP: phs001500.v1.p1. IRF4 ChIP-seq and RNA-seq data are deposited in GEO under accession code GEO: GSE167945. Data from the 2020 melanoma GWAS meta-analysis performed by Landi and colleagues were obtained from dbGaP (dbGaP: phs001868.v1.p1), with the exclusion of self-reported data from 23andMe and UK Biobank. The full GWAS summary statistics for the 23andMe discovery dataset will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please visit https://research.23andme.com/collaborate/#dataset-access/ for more information and to apply to access the data. Summary data from the remaining self-reported cases are available from the corresponding authors of that manuscript4 (Matthew Law, matthew.law@qimrberghofer.edu.au; Mark Iles, m.m.iles@leeds.ac.uk; and Maria Teresa Landi, landim@mail.nih.gov).