Summary
Kainate receptors (KARs) are glutamate-gated cation channels with diverse roles in the central nervous system. Bi-allelic loss of function of the KAR-encoding gene GRIK2 causes a nonsyndromic neurodevelopmental disorder (NDD) with intellectual disability and developmental delay as core features. The extent to which mono-allelic variants in GRIK2 also underlie NDDs is less understood because only a single individual has been reported previously. Here, we describe an additional eleven individuals with heterozygous de novo variants in GRIK2 causative for neurodevelopmental deficits that include intellectual disability. Five children harbored recurrent de novo variants (three encoding p.Thr660Lys and two p.Thr660Arg), and four children and one adult were homozygous for a previously reported variant (c.1969G>A [p.Ala657Thr]). Individuals with shared variants had some overlapping behavioral and neurological dysfunction, suggesting that the GRIK2 variants are likely pathogenic. Analogous mutations introduced into recombinant GluK2 KAR subunits at sites within the M3 transmembrane domain (encoding p.Ala657Thr, p.Thr660Lys, and p.Thr660Arg) and the M3-S2 linker domain (encoding p.Ile668Thr) had complex effects on functional properties and membrane localization of homomeric and heteromeric KARs. Both p.Thr660Lys and p.Thr660Arg mutant KARs exhibited markedly slowed gating kinetics, similar to p.Ala657Thr-containing receptors. Moreover, we observed emerging genotype-phenotype correlations, including the presence of severe epilepsy in individuals with the p.Thr660Lys variant and hypomyelination in individuals with either the p.Thr660Lys or p.Thr660Arg variant. Collectively, these results demonstrate that human GRIK2 variants predicted to alter channel function are causative for early childhood development disorders and further emphasize the importance of clarifying the role of KARs in early nervous system development.
Keywords: glutamate receptor, intellectual disability, epilepsy, GluK2, ataxia, white matter abnormalities, electrophysiology, channel gating, whole-exome sequencing
Introduction
Fast glutamatergic signaling in the central nervous system (CNS) is transduced by three structurally related ionotropic receptors referred to as α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptors (AMPARs), N-methyl-d-aspartic acid receptors (NMDARs), and kainate receptors (KARs).1 Each family of ionotropic glutamate receptors (iGluRs) serves a variety of roles in the CNS that include phasic postsynaptic depolarization (AMPARs), induction of some forms of long-term synaptic plasticity (NMDARs), and modulation of neuronal excitability and the balance between circuit excitation and inhibition (KARs). In addition to their discrete functions in the adult CNS, iGluRs can play critical roles in neuronal and circuit maturation during the earliest periods of development. For example, NMDAR activation is necessary for the formation of topographic maps during axon pathfinding2 and in the establishment of mature synaptic contacts.3 KAR signaling also is required for development of appropriate circuit and synaptic function in the hippocampus.
KARs subserve diverse functions in the mature central and peripheral nervous systems that encompass modulation of presynaptic neurotransmitter release and intrinsic neuronal excitability and establishment of appropriate synaptic connectivity in early development.4,5 Five KAR genes, GRIK1–5 (MIM: 138243–138245, 600282, and 600283), encode the subunit proteins GluK1–GluK5.1 Like other iGluR subunit proteins, KAR subunits are modular structures that each contain an extracellular N-terminal domain (NTD), a ligand-binding domain (LBD), three transmembrane domains (TMDs) and a re-entrant P loop, and an intracellular C-terminal domain.6 The GluK1–3 subunits have a relatively low affinity for glutamate and can form homomeric cation channels, whereas the higher-affinity GluK4 and GluK5 subunits co-assemble with low-affinity subunits to generate receptors with a wide range of biophysical properties.4 Diversity in KAR function is increased through assembly with the neuropilin and tolloid-like proteins-1 and -2 (Neto1 and Neto2) auxiliary proteins, which alter fundamental aspects of receptor pharmacology, gating, and neuronal trafficking and targeting.7,8
Variants in genes encoding iGluRs give rise to neurodevelopmental disorders (NDDs) of diverse presentation and phenotype. Pathogenic missense variants in human NMDAR subunit genes now number in the hundreds and lead to a variety of neurological disorders of childhood that include autism spectrum disorders (ASDs), intellectual disability (ID), and epilepsies.9 Far fewer disease-causing missense variants in AMPAR and KAR subunit genes have been reported to date. Several genes in these two receptor families are under strong evolutionary constraint based on the paucity of loss-of-function (LoF) variants in gnomAD.10 Nonsynonymous variants in AMPA receptor subunit genes can underlie ASD and other neurodevelopmental abnormalities.11, 12, 13, 14 GRIK2, which encodes the GluK2 KAR subunit, is among the most intolerant to LoF (probability of intolerance to heterozygous predicted loss of function variation [pLI] of 0.998) in the iGluR family of subunit genes. Bi-allelic presumed LoF variants in GRIK2 cause nonsyndromic ID with additional clinical features.15,16 In a previous study, we identified a de novo missense variant in GRIK2 in a child with neurodevelopmental dysfunction that included ID, ataxia, and speech impairment.17
We report here a set of de novo, missense variants in GRIK2 in eleven individuals with neurodevelopmental abnormalities and additional comorbidities. The variants cluster in two critical functional domains: the pore-forming M3 transmembrane helix and the adjacent M3-S2 gating linker. Five of the probands have a GRIK2 variant identical to one reported previously that is predicted to cause an alanine to threonine change at position 657 in M3 (c.1969G>A [p.Ala657Thr]; GenBank: NM_021956.5).17 The other variants have both functional and biochemical consequences on GluK2-containing KAR signaling in vitro as described in this study. This work therefore provides insight into a growing body of NDDs arising from KAR dysfunction and establishes an autosomal-dominant GRIK2-related phenotype.
Subjects and methods
Subjects
All probands were identified by either clinical or research investigations for determination of the underlying etiology of their NDDs and were included in the study following direct communication by researchers or through GeneMatcher.18 Pediatricians, neurologists, and clinical geneticists at participating centers carried out standard neurological assessments, magnetic resonance imaging (MRI), and assessment of exome sequencing data. Trio exome sequencing of the probands and their parents was performed as follows: probands A657T.2 and A657T.6, exomes were captured by BGI-Europe (Copenhagen, Denmark) via Agilent SureSelect XT Human All Exon V5 (Agilent Technologies, Santa Clara, CA, US) and sequenced on an Illumina HiSeq4000 sequencer (Illumina, San Diego, CA, US); proband A657T.4, Agilent SureSelect Human All Exon V5 followed by sequencing on Hiseq2500 (Illumina) at Leiden Clinical Genetics, the Netherlands; proband A657T.5, exome was captured by the Exeter Genomics Laboratory, UK, with the Twist Biosciences Human Core Exome Kit and was sequenced on an Illumina NovaSeq6000; proband T660K.2, exome sequencing was performed in-house at Tartu University Hospital, Estonia; proband T660K.3, Agilent SureSelect QXT CREv1 Kit followed by sequencing on NextSeq500 (Illumina) at Victorian Clinical Genetics Services, Australia, as part of the Australian Genomics Acute Care study; proband T660R.1, Agilent SureSelect CREv1 Kit followed by sequencing on NextSeq at Greenwood Genetic Center, South Carolina; and proband I668T.1, a TruSight One Panel on a NextSeq 500 (Illumina) at Hôpital Pitié-Salpêtrière, Paris, France. Other exome sequencing was performed by GeneDx (Gaithersburg, MD, US) or Blueprint Genetics (Helsinki, Finland).19,20 Variant allele frequencies were assessed with the gnomAD dataset,10 and the variants are classified according to the American College of Medical Genetics and Genomics (ACMG) guidelines for variant interpretation.21 All parental or legal guardians provided consent for this study and publication of clinical and genetic data. Procedures followed were in accordance with the ethical standards of the national and institutional responsible committees on human experimentation.
Constructs and image preparation
The rat cDNA constructs used in this study were obtained from the sources described previously unless otherwise stated.17 The rat GluK2a isoform was used as a template for generating GluK2(T660K), GluK2(T660R), and GluK2(I668T) mutants with a PCR-based approach, and mutations were confirmed by sequencing of the open reading frames by the Northwestern University Sanger Sequencing Facility (Chicago, IL, US). Structural images were generated in Pymol 2.3.3.
Cell culture and electrophysiology
Human embryonic kidney cells expressing T-antigen, clone 17 (HEK293-T/17) from the American Culture Collection (Manassas, VA, US) were cultured in Dulbecco’s modified Eagle’s medium (Corning Cellgro, Manassas, VA, US) supplemented with 10% heat-inactivated fetal bovine serum (Gemini Bio-Products, West Sacramento, CA, US), 100 mg penicillin, and 100 mg/mL streptomycin (Corning Cellgro) at 37°C with 5% CO2. HEK293-T/17 cells were transfected with TransIT-LT1 (Mirus Bio, Madison, WI, US) according to the manufacturer’s protocol. Media was supplemented with 1-naphthyl acetyl spermine trihydrochloride (NASPM, 20 μM) following transfection. Receptor plasmid DNAs were co-transfected with an enhanced green fluorescent protein (eGFP) construct at ratios of 1:4 [GluK2(A657T)] or 1:6 [GluK2(T660K), GluK2(T660R), and GluK2(I668T)] for GluK2 and eGFP cDNAs, respectively. For all GluK2 variants, the transfection ratio was 1:3:0.5 for GluK2, Neto2, and eGFP and 1:3:0.5 for GluK2, GluK5, and eGFP cDNAs, respectively.
Whole-cell voltage-clamp recordings were performed 1–3 days after transfection. The external solution contained 150 mM NaCl, 2.8 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM glucose, and 10 mM HEPES (pH 7.3). The internal solution contained 110 mM CsF, 30 mM CsCl, 10 mM Cs-HEPES, 5 mM EGTA, 4 mM NaCl, and 0.5 mM CaCl2 (pH 7.3). Cells were voltage clamped at −70 mV. We fast-applied glutamate (10 mM) via three-barreled theta glass attached to a Siskiyou MXPZT-300 solution switcher (Siskiyou Corporation, Grants Pass, OR, US) for 1 s to determine peak current amplitude and desensitization and to generate concentration-response relationships. Cells were excluded from analysis if 10%–90% rise times were greater than 3 ms. Glutamate applications of 1–2 ms for deactivation were confirmed by recording of the junction potential after each experiment. We best-fit current decays in response to either 1–2 ms or 1 s applications with either an exponential function or the sum of two exponential functions to generate a weighted mean τdes by using the formula [(τ1 × amplitude1) + (τ2 × amplitude2)]/[amplitude1 + amplitude2]. The tau values are the time constants from the exponential fits, and the amplitudes are the estimated contributions of each component to the total peak current amplitude. The degree of desensitization was calculated as the inverse of the ratio between the current amplitude at the end of the 1 s application and the peak amplitude. The time courses of recovery from desensitization were fit with a one-phase association exponential function. Desensitization-concentration response curves were fitted with logistic functions with variable Hill slopes. Analyses of current amplitudes and kinetics were carried out with Clampfit10 (Molecular Devices, Sunnyvale, CA, US). The half-maximal effective concentration (EC50) values and recoveries from desensitization were calculated from fitting data to logistic curves in GraphPad (San Diego, CA, US).
Surface biotinylation
HEK293-T/17 cells were maintained and transfected with GluK2a cDNA as described above. The cells were washed with ice-cold PBS three times and incubated with 0.5 mg/mL biotin (Thermo Fisher, Waltham, MA, US; #21335) in cold PBS for 30 min at 4°C with gentle agitation. The biotin solution was aspirated and replaced with 100 mM glycine in cold PBS and agitated for 5 min at 4°C. Following biotin quenching, 550 mL TEEN-TX (50 mM Tris, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 0.1% Triton X-100) with protease inhibitor cocktail (Thermo Fisher, #78430) was added to each well. The cells were sonicated briefly three times and centrifuged at 21,000 g for 10 min. An input fraction (50 l) was saved from each well and added to 12.5 l Laemmli sample buffer (Bio-Rad, Hercules, CA, US) containing β-mercaptoethanol and boiled at 95°C for 5 min. Streptavidin beads were pre-blocked by incubation of 40 mL Streptavidin Sepharose bead slurry (Millipore Sigma, Burlington, MA, US; #17-5113-01) with PBS plus 1% Triton X-100 and 1% normal goat serum and rotated at 4°C for 30 min. The remainder of each cell lysate (450 l) was added to the pre-blocked streptavidin beads and rocked for 2 h at 4°C. Following incubation, 50 l of the supernatant fraction was saved and added to 12.5 l Laemmli sample buffer (Bio-Rad) and boiled at 95°C for 5 min. The beads were washed with ice-cold PBS + 1% Triton X-100 for 3 min with three repetitions and then boiled in 40 l Laemmli sample buffer (Bio-Rad) at 95°C for 5 min. Samples were resolved by SDS-PAGE as described below with 10 l from each input, supernatant, and bead fraction.
Immunoblotting
Protein extracts were obtained from HEK293-T/17 cells and resolved by SDS-PAGE. The proteins were transferred to Immobilon-P PVDF membranes (Millipore Sigma, Billerica, MA, US) and blocked for 1 h at room temperature (5% milk and 0.1% Tween 20 in TBS [TBS-T]). Primary antibodies (rabbit -GluR6/7, 1:1,000 [Thermo Fisher, #PA1-37780] and mouse -actin, 1:500 [Millipore Sigma, #A4700]) were diluted in blocking solution and incubated at room temperature for 1 h or overnight at 4°C. Secondary antibodies were diluted in blocking solution and incubated at room temperature for 1 h and imaged by the Pierce ECL substrate (Thermo Fisher) or the near-infrared detection system (Li-Cor Biosciences, Lincoln, NE, US; IRDye 800CW, 680RD). Individual band density was quantified with Li-Cor ImageStudio software.
Statistical analysis
All statistical analyses were performed in GraphPad Prism 8 and statistical significance was denoted as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. We corrected for familywise error rates in recordings used for multiple measurements when determining p values that met statistical significance. Summary data are represented as mean ± standard error of the mean (SEM) from n cells. One-way analysis of variance (ANOVA) with Tukey’s post hoc test was performed for comparisons of three or more groups in which the data were normally distributed; a Kruskal-Wallis multiple comparison test was used if the data were nonparametric.
Results
Genetic findings in individuals with heterozygous GRIK2 variants
The contribution of pathogenic alterations in GRIK2 to NDDs in the human population is unknown. In the current study, eleven probands with de novo, heterozygous variants in GRIK2 at or near the c.1969G>A (p.Ala657Thr) variant described in our earlier case study17 were identified through direct communication between research groups or the GeneMatcher database.18 All missense variants are clustered in a region of GRIK2 that encodes critical functional domains of the GluK2 KAR subunit (Table 1). Four children and one adult harbored the previously reported missense variant c.1969G>A (p.Ala657Thr);17 the individual with that variant described earlier is therefore referred to as “A657T.1” in this report. We identified six additional individuals with one of three GRIK2 variants implicated in their NDDs: five children had variants at residue threonine 660 (three with c.1979C>A [p.Thr660Lys] and two with c.1979C>G [p.Thr660Arg]) and one child had a variant at isoleucine 668 (c.2003T>C [p.Ile668Thr]). All of these variants arose de novo, were not present in gnomAD, and were predicted to be damaging by in silico tools combined annotation-dependent depletion (CADD)22 and PolyPhen (PP2).23 Bioinformatic analyses of other variants that are not predicted to be causative for principal features of NDDs in the probands reported here are available in the supplemental notes, as is clinical information not directly related to the neurodevelopmental, neurological, or behavioral features described below.
Table 1.
Genetic characteristics of GRIK2 missense variants in individuals with NDDs
| Variant | Number of probands | GRIK2 (GRCh38, chr6) | cDNA | CADD | PP2 |
|---|---|---|---|---|---|
| p.Ala657Thr | 6 | 101928516G>A | c.1969G>A | 32 | D; 1.0 |
| p.Thr660Lys | 3 | 101928526C>A | c.1979C>A | 24.6 | D; 1.0 |
| p.Thr660Arg | 2 | 102376401C>G | c.1979C>G | 28 | D; 1.0 |
| p.Ile668Thr | 1 | 102376425T>C | c.2003T>C | 29.9 | D; 1.0 |
The table shows the four GRIK2 variants identified by the resultant change in amino acid, the number of probands harboring that variant, the nucleotide change in GRIK2, and the site of the variants in cDNA encoding GluK2 protein. Note that the six individuals with p.Ala657Thr variants include a child described previously,17 who is referred to as proband A657T.1 in this study. CADD scores predicted that all four variants are among the most deleterious substitutions in the human genome.22 PP2 analysis predicted that the variants are most likely damaging (D).23 CADD, combined annotation-dependent depletion; NDD, neurodevelopmental disorder; PP2, PolyPhen2.
Clinical phenotypes of individuals with a GRIK2 c.1969G>A (p.Ala657Thr) variant
Four children and one adult harbored the p.Ala657Thr variant identical to a 15-year-old female reported previously17 (proband A657T.1, Table 2), who exhibits ID, speech and motor developmental delay (DD), ataxia, and a short attention span. At 2–3 years of age, she began using single words, and around 5 years, she was able to walk short distances (with an ataxic gait and poor balance).
Table 2.
Clinical features of individuals harboring GRIK2 missense variants
| Proband | Age (years) | Sex | Diagnosis | Motor dysfunction | Hypotonia | Hypertonia | Visual impairment | Seizure onset (age in months) | Myelination |
|---|---|---|---|---|---|---|---|---|---|
| A657T.1a | 15 | F | ID, DD | ataxia | resolved | N.O. | N.O. | N.O. | normal |
| A657T.2 | 8 | M | ID, DD | uncoordinated gait | N.O. | N.O. | N.O. | N.O. | N.R. |
| A657T.3 | 13 | M | ID, DD | uncoordinated gait | N.O. | N.O. | hypermetropia astigmatism | N.O. | normal |
| A657T.4 | 7 | M | ID, DD, ASD | imbalance | resolved | N.O. | N.O. | N.O. | normal |
| A657T.5 | 6 | F | ID, DD | ataxia | N.O. | N.O. | N.O. | N.O. | N.R. |
| A657T.6 | 37 | F | ID, DDb | imbalance | N.O. | truncal and limbs | N.O. | N.O. | normal |
| T660K.1 | 5 | M | global DD | not ambulatory | truncal | limbs | nystagmus | 8 | abnormal |
| T660K.2 | 2 | M | global DD | not ambulatory | N.R. | N.R. | nystagmus | 7 | abnormal |
| T660K.3 | 2 | F | global DD | not ambulatory | truncal | limbs | CVI | 8 | abnormal |
| T660R.1 | 4 | F | global DD | limited function (standing only) | truncal | N.O. | CVI | 27 | abnormal |
| T660R.2 | 12 | M | ID, DD | ataxia | resolved | N.O. | strabismus | N.O.c | abnormal |
| I668T.1 | 12 | M | ASD | N.R. | N.O. | N.O. | N.R. | N.O. | N.R. |
F, female; M, male; ASD, autism spectrum disorder; CVI, cortical visual impairment; DD, developmental motor and speech delay; ID, intellectual disability; N.O., not observed; N.R., not reported.
Proband A657T.1 was reported in an earlier study17 and is included for comparative purposes.
Proband A657T.6: autistiform behaviors were observed, but ASD was not formally diagnosed.
Proband T660R.2: definitive seizures have not been observed, but the child did exhibit a single event during which he became unresponsive and his eyes deviated upward.
Proband A657T.2 is an 8-year-old male who started walking without support at 18 months of age and speaking when he was 2 years old. At 5 years of age, he was diagnosed with moderate ID with severe speech and mild motor DD. He was able to speak single words and was assessed by his parents as having a developmental level roughly corresponding to a 2-year-old. His gait was unsteady without assistance and did not follow a linear path when prompted, but he could ride a bicycle with support wheels. No visual abnormalities were noted. The proband exhibited a happy and social demeanor at times that devolved into aggressive behavior when angry, which occurred when presented with unexpected changes to daily routines. His motor, speech, and social skills continue to develop slowly. For example, he is unable to draw or write but can hold a pencil between his thumb and index finger and recognizes symbols. His speech continues to improve, but he still cannot be understood by unfamiliar people. Finally, his social interactions with other children remain positive, consistent with the happy demeanor noted at younger ages. No seizures have been noted, and electroencephalograms (EEGs) and MRIs have not been acquired.
Proband A657T.3 is a 13-year-old male with ID and global DD. He exhibited early delays in gross and fine motor milestones, first sat up at 9 months of age, and began walking at 19 months. Walking and running continue to be uncoordinated and are accompanied by purposeful but disorganized hand movements. Language development was slow and included echolalia, and fewer than 10 words were used at 24 months of age. Currently his speech is poorly articulated, slurred, and perseverative, but he can communicate without devices. By 6th grade, he had plateaued at roughly a 1st grade academic level. Neurological assessments included attention deficit hyperactivity disorder (ADHD), anxiety, and impulsivity. A screening sleep study showed possible epileptiform activity, and a subsequent screening EEG and overnight long-term monitoring video EEG at ∼10 years of age revealed sharply contoured fronto-central epileptiform sleep features but no definitively abnormal epileptiform discharges and no clinical or electrographic seizures. He remains seizure free. An MRI at 9 years of age was normal.
The A657T.4 proband is a 7-year-old male who was referred to a pediatrician at the age of 18 months because of general hypotonia. He began physiotherapy at 11 months because of delayed motor development and began walking unaided at the age of 27 months. At the age of 3 years, he was diagnosed with ASD. His language development was slow; he began using words at 4 years of age and his current use of speech is equivalent to that of a 3-year-old. Articulation problems predominate and make communication challenging, particularly with people unfamiliar with his speech. His reading skills are appropriate for his age and his writing ability is progressing. Development of both gross and fine motor skills continues to be slow, and he has difficulty with balance. Socially, he functions at the level of a 2-year-old and has a happy demeanor. No seizures were reported, and no abnormalities were observed on an MRI at 18 months of age or an MRI with MR spectroscopy at 24 months of age. Metabolic assessments were normal.
Individual A657T.5 is a 6-year-old female who was diagnosed with global DD within 9 months of age. She was able to sit at around 18 to 20 months, crawl at around 18 to 24 months, and walk at the age of 5 years. She has an ataxic gait and holds her arms out when walking. Hand flapping occurs in response to sensory input. She tolerates changes to her routine well and is generally quiet with more vocalizations in familiar environments. Prolonged, idiopathic periods of screaming earlier in development have lessened in frequency. At the age of 5, she began to talk and now can string a few words together. Her vision and hearing are normal. No seizures have been noted and an MRI has not been acquired.
Unlike the other individuals in this report, individual A657T.6 is a 37-year-old female adult assessed during childhood for ID and delayed speech and motor development when she first entered school. The c.1969G>A (p.Ala657Thr) variant was identified in exome sequencing in 2017 and then was established as de novo following trio sequencing in 2018. Her cognitive function was assessed recently to be at the level of an 18-to-36-month-old child. Although she is unable to read or write, her speech is well-developed and she communicates verbally with other people. She experiences mood swings and can become aggressive when her immediate environment changes or when she is frustrated in her intentions. Her suite of behaviors was characterized as autistiform, but the absence of a psychological assessment precluded a formal diagnosis of ASD. Motor development was delayed, but she is now able to walk, albeit with difficulties in balance that result in a lean to one side. She does not have seizures and her MRI at 7–8 years of age was normal.
Clinical phenotypes of children with a GRIK2 c.1979C>A (p.Thr660Lys) or c.1979C>G (p.Thr660Arg) variant
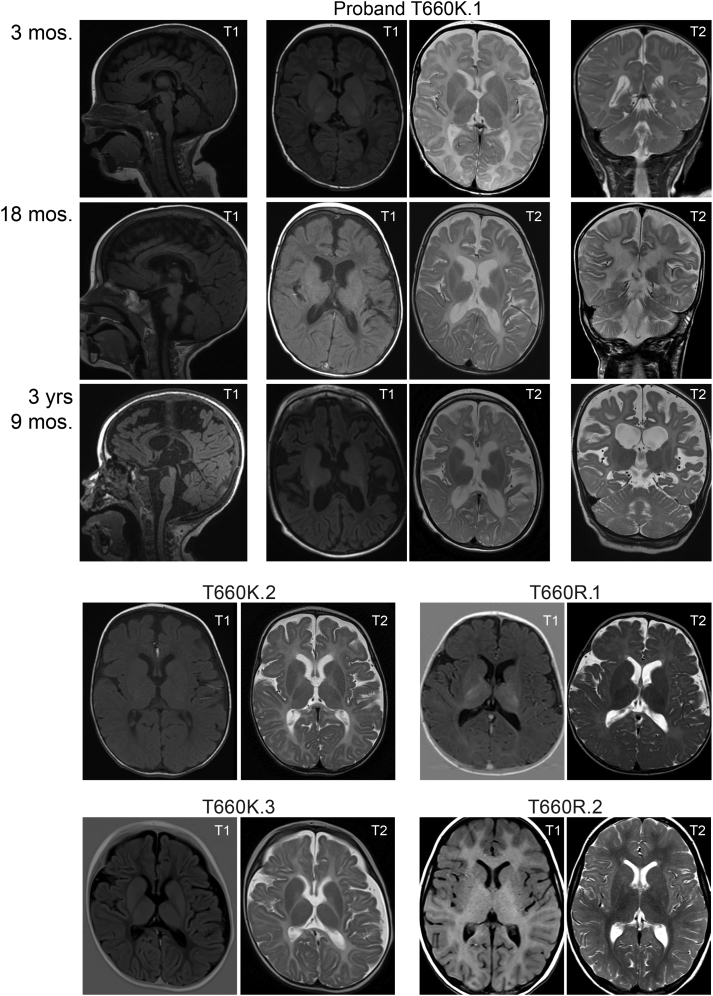
Five children with de novo variants of threonine 660 were identified (Table 2). The T660K.1 proband is a 5-year-old male born with global DD, congenital nystagmus, and hearing impairment. He is confined to a wheelchair, exhibits few movements, has truncal hypotonia with limb hypertonia, exhibits occasional tongue thrusting, and does not speak. At 8 months of age, he began experiencing daily seizures. An EEG at 27 months was poorly organized and lacked normal sleep architecture, consistent with severe encephalopathy. Multifocal interictal spikes, multiple tonic seizures, and clusters of epileptic spasms were observed in the EEG. Lennox-Gastaut syndrome was diagnosed at 4 years of age on the basis of his history of refractory epileptic spasms; bilateral tonic, tonic-clonic, and myoclonic seizures; severe encephalopathy; and global DD. His current treatment regimen consists of ketogenic diet with levetiracetam, vigabatrin, and valproic acid, but these are only partially effective at seizure reduction. MRIs were taken at the ages of 3 months, 18 months, and 3 years 9 months and are shown at the level of the midline (sagittal), lentiform nucleus (axial), and hippocampus (coronal) in Figure 1. The earliest MRIs showed normal ventricles, extra-axial spaces, and cerebral white matter volume. T2 hyperintensity was observed in the cerebrum and deep white matter tracts. At 18 months, both ventricles and extra-axial spaces had slightly increased in volume, which was particularly notable in the Sylvian fissure. White matter volume was decreased relative to the earlier images, and T2 prolongation suggested aberrant myelination. A last set of MRI images at 3 years and 9 months of age revealed progressive enlargement of cerebrospinal fluid spaces. The images also show reduced cerebral white matter relative to the earlier scans, continued white matter abnormalities, and a thin corpus callosum. Hippocampal and cerebellar atrophy was noted, but malformations were not present, and the global pallidus exhibited increased bilateral signal (Figure 1). The series of images underscore a failure of myelination, both in volume and in character, which are consistent with a neurodegenerative trajectory.
Figure 1.
MRIs reveal abnormal myelination in children with GRIK2 variants that alter threonine 660 in GluK2
A series of images from MRIs taken at the indicated ages are shown for proband T660K.1. Sagittal images are from T1-weighted pulse sequences; T1 and T2 axial images are shown, and coronal images are T2 weighted. MRIs for other probands are axial images from T1- and T2-weighted scans. Ages where not indicated on the figure are as follows: T660K.2, 10 months; T660K.3, 3 months; T660R.1, 11 months; T660R.2, 6 months. MRI, magnetic resonance imaging.
The T660K.2 proband is a 2-year-old male with global DD and horizontal congenital nystagmus. At 2 months of age, he adopted an opisthotonic position and spastic movements that gradually improved within several months. At 7 months of age, he was able to hold his head slightly and sometimes fixate his gaze. He remains non-ambulatory and does not speak. The child developed an intractable seizure disorder (MIM: 600669) that began with epileptic spasms and consisted of 6–10 bilateral tonic-clonic seizures per day at 7 months of age; in the subsequent year, the seizures reduced to 1–2 per day. His current course of treatment manages the seizures well and consists of vigabatrin, valproic acid, and clonazepam. An MRI at 10 months indicated that myelination had not progressed compared to images taken 3 months earlier and revealed mild loss of white matter on both sides and dilated gyri consistent with atrophy (Figure 1), similar to the T660K.1 proband. In addition, the child developed bilateral and symmetric T2 hyperintensities in the midbrain and dorsal pons.
The T660K.3 proband is a 2-year-old female with global DD, early-onset breath-holding episodes, cortical visual impairment (CVI), auditory neuropathy spectrum disorder, and a seizure disorder. This individual was reported previously in a study on the performance and feasibility of rapid exome sequencing in the context of people with suspected monogenic disorders.20 By 3 months of age, she experienced truncal hypotonia and limb hypertonia and was unable to fixate her gaze. Daily breath-holding episodes commenced at 5 months of age with a pattern of a period of distress in the evening, which could last for several hours, followed by a prolonged apnea with cyanosis. An EEG showed no evidence of seizure activity during the episodes. Breath-holding episodes ceased at 8 months old but were replaced by bilateral tonic seizures lasting 5–30 s; clusters lasted from a few minutes to a few hours each day. By 15 months, she was able to recognize family members, but her responses were limited. This assessment noted persistent clenched fists with some opening and purposeful hand movements, no definite understanding of speech, and simple monosyllabic vocalizations. She was not yet sitting unsupported and still exhibited truncal hypotonia and limb hypertonia. Currently, she remains unable to walk or talk and continues to have severe to profound DD. An MRI at 3 months highlighted mildly enlarged ventricles, extra-axial spaces, and aberrant myelination in T2-weighted images (Figure 1). The corpus callosum appeared complete but was generally thin and had not increased in thickness as would be expected with increasing age. There was an absence of normal myelin signal within the white matter on T2, T1, and true inversion recovery scans and no progression of myelination on MRI over a 12-month period. The appearances were consistent with hypomyelination rather than demyelination.
The T660R.1 proband is a 4-year-old female with global DD, CVI, truncal hypotonia, and right-sided weakness that were first noted at 4–5 months of age. She sat unassisted at 1 year of age and was crawling by 18 months. The individual now stands for approximately 20 s but cannot walk without assistance. At 26 months of age, she spoke her first word, but progress was very limited; currently, she employs only two words and remains largely non-verbal. She began having staring spells at about 20 months old; an EEG at that time was consistent with potential epileptogenicity in the mesial fronto-central and right temporal regions. Subsequently, at 27 months she was diagnosed with focal symptomatic epilepsy and seizures with impaired awareness that are managed with oxcarbazepine. She continues to receive early intervention services, including speech therapy, occupational therapy, and physical therapy. Like the children with Thr660Lys variants, an MRI at 11 months revealed aberrant myelination (Figure 1).
The T660R.2 proband is a 12-year-old boy who exhibits significant ID and speech and motor DD. He remains non-verbal with attempts at augmented communication in progress. Initial hypotonia resolved by the time of a neurological exam at 18 months. Walking with an unsteady, ataxic gait began at 35 months. After initial strabismus, which self-resolved, he does not have visual impairments other than a requirement for corrective lenses. A neurological exam at 2 years of age did not reveal definitive seizures while under observation but did exhibit a single event during which he became unresponsive and his eyes deviated upward. No EEG has been performed on the proband. Axial MRI revealed modest hypomyelination that did not appear as severe as that in the children with Thr660Lys variants (Figure 1).
Clinical phenotype of a child with a GRIK2 c.2003T>C (p.Ile668Thr) variant
The I668T.1 proband is a 12-year-old male diagnosed with autism, overgrowth, and macrocephaly. His healthy father has similar macrocephaly (+2.5 SD) and tall stature. The proband sat unaided at 5.5 months; he said his first words and walked independently at 12 months. His language skills subsequently plateaued, and he exhibited isolation, repetitive behaviors, and hand stereotypies consistent with the diagnosis of ASD.
Structural modeling of GluK2 amino acid substitutions
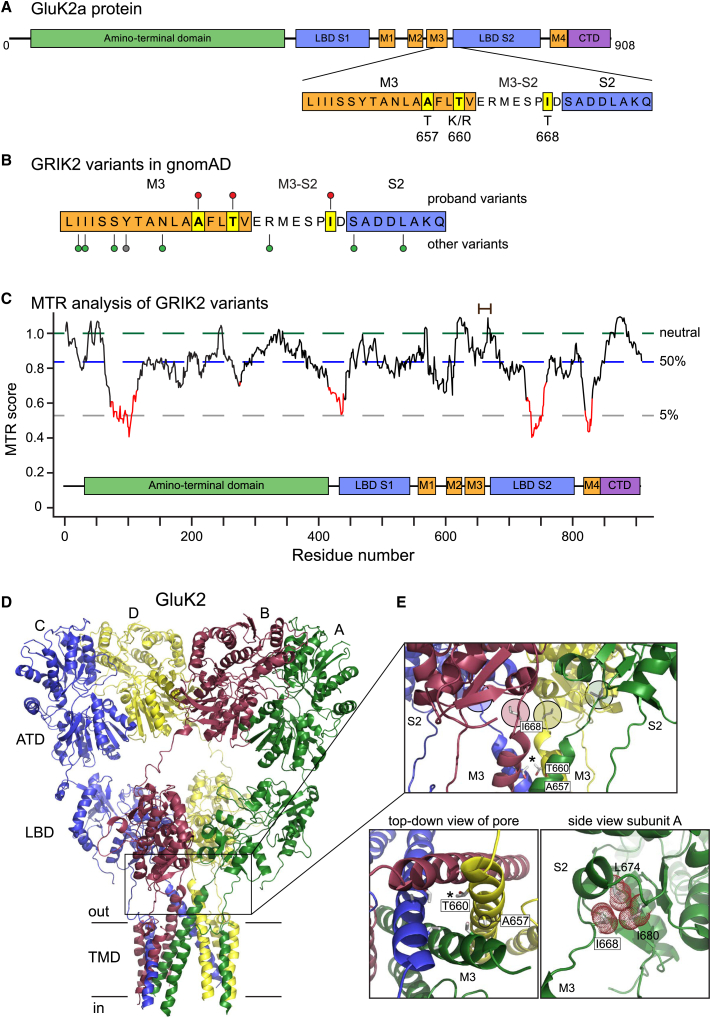
The GluK2 Ala657, Thr660, and Ile668 residues are located within the highly conserved M3 transmembrane domain and adjacent M3-S2 linker region, as shown in the cartoon representation of a single subunit protein with component domains in Figure 2A.1 No variants at each of these sites exist in gnomAD (Figure 2B).10,24 Although missense variants are abundant in the initial part of the M3 helix, relatively few occur in the distal M3 and the M3-S2 linker. Accordingly, a missense tolerance ratio (MTR) analysis did not reveal an unusual sensitivity to missense variants in this protein domain with a 31-residue sliding window (Figure 2C),25 underscoring the critical importance of discrete residues transducing key functional components of gating in these receptors.
Figure 2.
Mapping of GRIK2 variants onto protein structures and analysis of tolerance ratios
(A) The primary sequence and key structural domains of the GluK2 subunit are shown in cartoon form together with the predicted divergences from wild-type amino acids.
(B) Lollipop mapping of the sites of variants in this report (red lollipops), other missense variants (green), and a truncating variant (gray) in the “hotspot” region in the M3 helix and M3-S2 linker domains.24 Variant sites are from gnomAD.
(C) Missense tolerance ratio of GRIK2 variants analyzed with a 31 amino acid window. Red domains indicate highly intolerant regions of the gene mapped onto the primary protein sequence. The relative positions of the variants in this study are shown with a brown bar at the top.
(D) Tetrameric structure of the homomeric GluK2 KAR (PDB: 5kuf);26 each subunit is color-coded and denoted as A, B, C, or D according to convention.27 The boxed region harbors the protein variants and spans the bottom of the LBDs, the gating linkers, and external segments of the pore helices.
(E) The top box shows a side-on view of isoleucine 668 (I668) in the M3-S2 linker (circled, colored by chain) and threonine 660 (T660) in M3 (starred). Alanine 657 (A657) projects into neighboring M3 helices and is not visible in this view. The top-down view of the M3 helices reveals the projection of threonine side chains into the channel pore (asterisk marks threonine 660 in chain D). Alanine 657 stabilizes interactions between M3 domains (circled in chain D). A closer view of isoleucine 668 with the atomic surface shown as red dots illustrates the projection of its hydrophobic side chain into a pocket formed from residues in the S2 domain of the LBD. ATD, amino-terminal domain; CTD, carboxy-terminal domain; LBD, ligand-binding domain; MTR, missense tolerance ratio; TMD, transmembrane domain.
In tetrameric iGluRs, M3 domains form the ion channel pore and the M3-S2 linkers transduce the energy of agonist binding in the LBD to channel gating (boxed region, Figure 2D).28 The alanine at position 657 forms key inter-helical interactions at the tip of M3 that stabilize the closed structure of the pore and facilitate rapid deactivation and desensitization of iGluRs (Figure 2D).27,29 In the closed conformation of GluK2 KARs, threonine 660 also resides at the tip of the M3 domain in GluK2 KARs and projects into the channel pore, where it constricts the ion permeation pathway (indicated with an asterisk in the top panel and expanded in bottom left panel, Figure 2E).26 As with the p.Ala657Thr variant, the replacement of a threonine with a bulky and charged lysine or arginine at position 660 is predicted to destabilize the closed configuration of the M3 domains. Finally, isoleucine 668 is located at the distal end of the M3-S2 linker and forms hydrophobic interactions in a small pocket at the base of the D2 lobe of the LBD (highlighted with colored circles in the top panel and expanded in bottom right panel, Figure 2E). The inter-domain coupling between Ile668 and the S2 domain has been proposed to be necessary for transducing conformational changes associated with agonist binding into pore opening (Figure 2D).30 In summary, the four GRIK2 variants implicated in NDDs we describe here are positioned in critical structural domains in the GluK2 subunit protein.
Functional effects of variants on GluK2-containing KARs
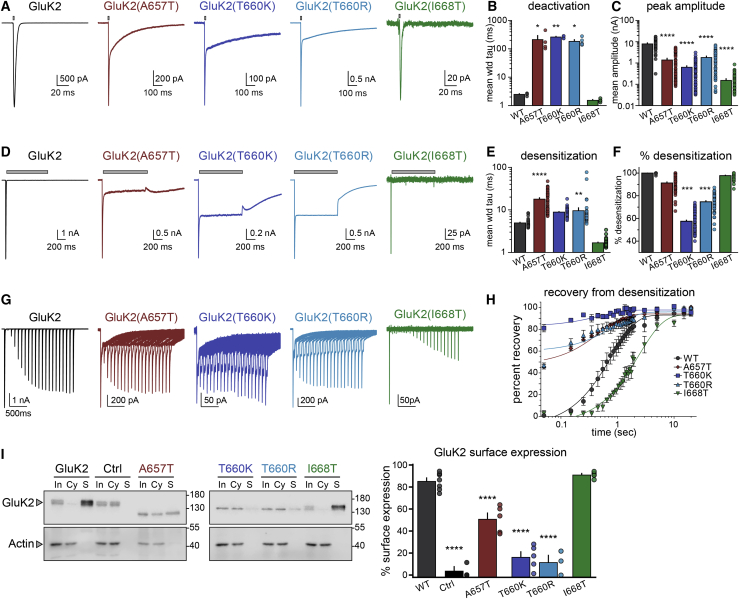
To determine whether the changes in protein structure introduced by the GRIK2 variants alter channel function, we generated recombinant GluK2 subunits containing the analogous mutations and performed electrophysiological analyses from homomeric and heteromeric KARs. We and others reported previously that the GluK2 p.Ala657Thr variant altered many functional parameters, which included slowing desensitization, increasing the relative steady state to peak current amplitude, decreasing the EC50 for activation by domoic acid, and apparent agonist-independent channel opening.17,29 In the current study, we compared these parameters and others in KARs containing the wild-type GluK2 subunit or those harboring p.Ala657Thr [GluK2(A657T)], p.Thr660Lys [GluK2(T660K)], p.Thr660Arg [GluK2(T660R)], or p.Ile668Thr [GluK2(I668T)] variants (Figure 3; Table 3).
Figure 3.
Functional and biochemical characterization of homomeric mutant GluK2 KARs
(A) Representative whole-cell currents evoked by a 1–2 ms application of glutamate (10 mM, gray bar) from recombinant receptors composed of the indicated subunits expressed in HEK293-T/17 cells. The holding potential was −70 mV in all recordings. Note that the GluK2(A657T) and GluK2(T660K/R) mutant receptors exhibited two temporally distinct phases to the current decay.
(B) Quantitation of the decay tau values fitted either to one exponential function (WT and I668T) or two components weighted by proportional contributions (A657T and T660K/R).
(C) Mean peak amplitude of glutamate-evoked currents. Note the semi-log y axis.
(D) Representative whole-cell currents evoked by a 1 s application of glutamate (10 mM, gray bar) from recombinant receptors composed of the indicated subunits.
(E) Quantitation of desensitization tau values fitted to two exponential decay functions weighted by proportional contributions.
(F) Quantitation of percent desensitization for glutamate-evoked currents.
(G) Representative traces in which two glutamate applications were made with variable intervals for generation of the time course of recovery from desensitization. Glutamate was applied for 50 ms in these experiments.
(H) Quantitation of recovery from desensitization data. Tau values from the fits are given in the text.
(I) Quantitation of GluK2 KAR membrane expression in HEK293-T/17 cells. Receptor-expressing cells were exposed to biotin followed by isolation on streptavidin-conjugated beads, separation on SDS-PAGE gels, and immunoblotting with anti-GluK2/3 or anti-actin antibodies. Samples shown are input (“In”), cytosol (“Cy”), and surface (“S”). Wild-type GluK2 and GluK2(I668T) proteins are predominantly found on plasma membrane (S) relative to intracellular compartments (Cy), whereas surface localization of A657T and T660K/R proteins is reduced. Biotin was omitted in the control (“Ctrl”) samples. The graph shows quantification of the data via densitometry normalized to actin staining. Molecular weight markers are shown in kDa. Statistical significance is denoted as ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. p values are also given in the text. WT, wild-type; wtd, weighted. Error bars represent SEM.
Table 3.
Summary of physiological parameters for homomeric wild-type and mutant GluK2 KARs
| GluK2 constructs | Deactivation (ms) | Desensitization (ms) | Degree of desensitization (%) | Recovery (s) | Glutamate EC50(μM) | Peak amplitude (−nA) | Surface localization (%) |
|---|---|---|---|---|---|---|---|
| Wild-type | 2.5 ± 0.1 | 4.9 ± 0.2 | 100 ± 0 | 0.8 (0.7–0.9) | 295 | 8.2 ± 0.6 | 85 ± 3 |
| p.Ala657Thr | 212 ± 82 (p = 0.011) | 18 ± 1 (p < 0.001) | 91 ± 1 (p < 0.0001) | 0.5 (0.4–0.6) | 47 (11–179) | 1.4 ± 0.2 (p < 0.0001) | 51 ± 5 (p < 0.0001) |
| p.Thr660Lys | 260 ± 14 (p = 0.002) | 8.9 ± 0.2 (NS) | 57 ± 1 (p < 0.0001) | 0.5 (0.2–0.9) | 19 (15–26) | 0.65 ± 0.08 (p < 0.0001) | 16 ± 5 (p < 0.0001) |
| p.Thr660Arg | 183 ± 27 (p = 0.023) | 9.6 ± 1.6 (p = 0.009) | 75 ± 1 (p < 0.0001) | 0.9 (0.4–1.4) | 52 (38–73) | 1.9 ± 0.2 (p < 0.0001) | 12 ± 6 (p < 0.0001) |
| p.Ile668Thr | 1.5 ± 0.1 (NS) | 1.7 ± 0.1 (NS) | 98 ± 0.5 (NS) | 2.4 (2.1–2.8) | 3700 (2,300–10,800) | 0.16 ± 0.02 (p < 0.0001) | 91 ± 1 (NS) |
τ values from fits to the data are given for deactivation, desensitization, and recovery from desensitization as described in the text. The data are expressed as mean ± SEM. Statistical information is given where significantly different from wild-type GluK2 receptors, except for 95% confidence intervals shown for recovery from desensitization and for the EC50 values. NS, not significant.
We first carried out electrophysiological recordings from homomeric receptors comprised of GluK2 wild-type or mutant subunits expressed in HEK293-T/17 cells. Deactivation time constants of the channels were determined by fitting exponential functions to currents following a 1–2 ms application of 10 mM glutamate. The representative traces in Figure 3A illustrate the profound slowing of deactivation in both the GluK2(A657T) and GluK2(T660K/R) mutants by ∼2 orders of magnitude relative to wild-type GluK2 receptors [GluK2 τdeact: 2.5 0.1 ms, n = 4; GluK2(A657T): 212 82 ms, n = 4, p = 0.011; GluK2(T660K): 260 14 ms, n = 4, p = 0.002; GluK2(T660R): 183 27 ms, n = 5, p = 0.023; GluK2(I668T): 1.5 0.1 ms, n = 4, p = 1.0; values are mean SEM and p values are compared to wild-type GluK2 via a one-way ANOVA with Tukey’s post hoc test] (Figures 3A and 3B; Table 3). The homomeric GluK2(A657T) and GluK2(T660K/R) mutant KARs were indistinguishable in their deactivation time constants.
We next compared peak current amplitudes and desensitization elicited by 1 s applications of glutamate. Mean peak current amplitudes were lower in homomeric receptors composed of GluK2(T660K/R) or GluK2(I668T) mutant subunits compared to wild-type GluK2 receptors (Figure 3C; Table 3), and the reduced mean amplitude in GluK2(A657T) mutants in this new data is consistent with our earlier observations.17 At a holding potential of −70 mV, 10 mM glutamate evoked currents with a mean amplitude of 8.2 0.6 nA from wild-type GluK2 KARs (n = 47), 1.4 0.2 nA from GluK2(A657T) (n = 54), 0.65 0.08 nA from GluK2(T660K) (n = 70), 1.9 0.2 nA from GluK2(T660R) (n = 54), and 0.16 0.02 nA from GluK2(I668T) (n = 47) (p < 0.0001 for all comparisons to wild-type GluK2 KARs). Desensitization of GluK2(T660K)- and GluK2(T660R)-containing KARs was slower and relative steady-state currents were larger, similar to those observed with GluK2(A657T) KARs, as is shown in the traces in Figure 3D. In contrast, GluK2(I668T) receptors desensitized with a rapid time course indistinguishable from wild-type GluK2 KARs. The weighted mean time constants of desensitization derived from two-exponential fits to the currents were as follows: GluK2, 4.9 0.2 ms (n = 47); GluK2(A657T), 18 1 ms (n = 54, p < 0.0001); GluK2(T660K), 8.9 0.2 ms (n = 61, p = 0.03; not significantly different when corrected for familywise error rates); GluK2(T660R), 9.6 1.6 ms (n = 53, p = 0.009); and GluK2(I668T), 1.7 0.1 ms (n = 39, p = 0.19) (comparisons to mean weighted τdes of wild-type GluK2 KARs; Figure 3E; Table 3). Those mutant receptors with slower entry into desensitization also had a much greater likelihood of channel opening in the continued presence of glutamate, which we measured as the percent desensitization from peak to steady-state current amplitudes at the end of the one second application [GluK2: 100% 0%, n = 47; GluK2(A657T): 91% 1%, n = 54; GluK2(T660K): 57% 1%, n = 70; GluK2(T660R): 75% 1%, n = 54; GluK2(I668T): 98% 0.5%, n = 46] (Figure 3F). All mutant receptors except for GluK2(I668T) were statistically different from wild-type receptors at p < 0.0001 in a one-way ANOVA with Tukey’s post hoc test. These data demonstrate that all variants reduce peak current amplitude, but only the M3 variants—GluK2(A657T) and GluK2(T660K/R)—slowed channel kinetics and prevented full desensitization.
The rate of recovery from desensitization is a critical gating parameter that in part determines the efficacy of receptor gating over a range of stimulation frequencies. We measured the effect of GluK2 mutants on KAR recovery by comparing peak current amplitudes for pairs of applications at a range of intervals (Figures 3G and 3H). As noted in previous experiments, GluK2(A657T) and GluK2(T660K/R) currents did not desensitize completely; thus, a substantial current was evoked by glutamate even at the shortest interval of 50 ms. Fitting of the normalized amplitudes (test relative to control) revealed that recoveries for the GluK2(A657T) and GluK2(T660K) mutants were modestly faster than wild-type GluK2, whereas GluK2(T660R) was equivalent to wild-type GluK2 [τrec for GluK2: 0.8 s, 95% CI 0.7–0.9 s, n = 4; GluK2(A657T): 0.5 s, 95% CI 0.4–0.6 s, n = 4; GluK2(T660K): 0.5 s, 95% CI 0.2–0.9 s, n = 4; GluK2(T660R): 0.9 s, 95% CI 0.4–1.4 s, n = 4] (Table 3). Conversely, glutamate currents from GluK2(I668T) receptors exhibited a markedly slower recovery (τrec of 2.4 s, 95% CI 2.1–2.8 s, n = 4). The incomplete desensitization and slow deactivation of GluK2(T660K) receptors, in particular, would ensure that substantial currents are gated by the mutant KARs even at high frequencies of activation by glutamate.
Structural changes in key gating domains of iGluR subunits can impair biogenesis and plasma membrane expression in addition to altering channel function.31,32 Accordingly, we tested whether the reductions in mutant GluK2 receptor current amplitudes shown in Figure 3C resulted from attenuated surface expression of receptor protein. GluK2 receptors expressed in HEK293-T/17 cells were incubated with or without biotin, isolated with streptavidin, and then detected via immunoblotting with anti-GluK2/3 antibodies (Figure 3I). Band densities were quantified and expressed as the percent of receptors expressed on the cell surface (Figure 3I). Wild-type GluK2 subunits were predominantly expressed on the plasma membrane (85% 3%, n = 7), as has been reported previously.31 Surprisingly, the GluK2(I668T) mutant was expressed to an equivalent degree as wild-type receptors (91% 1%, n = 5) despite >90% functional attenuation (Figure 3C). KARs containing GluK2(A657T), GluK2(T660K), or GluK2(T660R) mutations had significantly reduced surface expression compared to wild-type GluK2 receptors [GluK2(A657T): 51% 5%, n = 5; GluK2(T660K): 16% 5%, n = 5; GluK2(T660R): 12% 6%, n = 3; p < 0.001 for each comparison] (Table 3). These data demonstrate that the mutations within the M3 domain reduce GluK2 receptor surface expression, whereas KARs containing the GluK2(I668T) mutant subunit traffic to the membrane efficiently but are nearly non-functional.
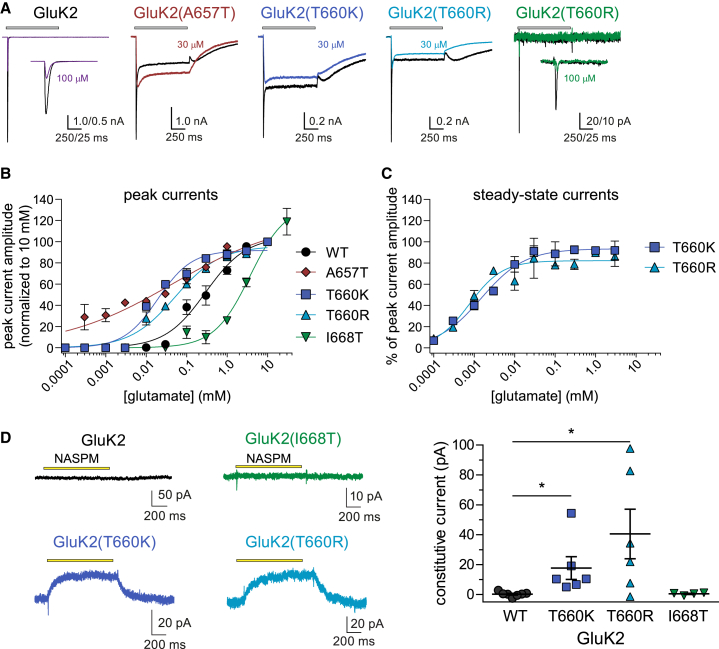
We next determined the EC50 for activation of mutant KARs by glutamate because the p.Ala657Thr variant and changes to other residues directly involved in iGluR gating also can alter the potency for channel activation (Table 3).29,33,34 Traces shown in Figure 4A are representative for currents evoked by saturating glutamate (10 mM) and a single test concentration [30 μM for M3 mutants and 100 μM for GluK2(I668T)]. At the lower concentration, GluK2 M3 mutants exhibit peak currents that modestly surpass the steady-state currents, whereas GluK2(I668T) was notably less responsive than wild-type GluK2 KARs to glutamate at concentrations between 100 nM and 30 μM. Normalized concentration-response data were fit to logistic functions (Figure 4B). GluK2 recordings yielded a peak EC50 value of 295 μM and a Hill slope of 0.9, which are close to the values reported previously.1 GluK2(A657T) receptors were notable in that both the mean EC50 and Hill slope were lower (47 μM, nH = 0.3) than wild-type, and consequently, the slope of the logistic function was shallow, suggesting that the receptors contain multiple binding sites with highly divergent affinities or cooperativity. These data are also consistent with an earlier proposal that analogous AMPAR mutant channels could be gated by extraordinarily low concentrations of glutamate present at contaminant levels in nominally glutamate-free extracellular solutions.33 Glutamate EC50 values for GluK2(T660K) and GluK2(T660R) currents were also lower than wild-type, but in contrast to GluK2(A657T), nH values did not differ from wild-type GluK2 currents [GluK2(T660K): EC50 = 19 μM, nH = 1.0; GluK2(T660R): 52 μM, nH = 0.8]. As noted, GluK2(I668T) receptors were less sensitive to activation by glutamate, and accordingly, the concentration-response curve was shifted to the right relative to wild-type GluK2 (EC50 = 3.7 mM, nH = 1.0). Fitting of the concentration-response data for steady-state currents (normalized to peak current amplitudes) yielded EC50 values of 1.5 and 0.7 μM for GluK2(T660K) and GluK2(T660R), respectively, which are greater than an order of magnitude lower than wild-type GluK2 receptors (Figure 4C).
Figure 4.
Concentration-response curves and constitutive activity of GluK2 mutants
(A) Representative whole-cell currents evoked by 1 s application of glutamate at two concentrations (10 mM, black traces; 30 μM, other colored traces) from recombinant receptors composed of the indicated subunits expressed in HEK293-T/17 cells.
(B) Peak current amplitudes at a range of glutamate concentrations were normalized to current amplitudes evoked by 10 mM glutamate. Logistic fits of the data yielded the EC50 data given in the text.
(C) Steady-state current amplitudes were normalized to peak currents over a range of glutamate concentrations before fitting with a logistic function.
(D) Representative traces showing the change in holding current after a 1 s application of 20 μM NASPM to cells expressing wild-type and mutant GluK2 KARs. Data are quantitated as the positive change in current upon application of NASPM in the graph. n = 4–6 recordings at each concentration. Error bars represent SEM. Statistical significance is denoted as ∗p < 0.05. p values are also given in the text. EC50, half-maximal effective concentration; KAR, kainate receptor; NASPM, 1-naphthyl acetyl spermine trihydrochloride; WT, wild-type.
As noted above, homomeric GluK2(A657T) receptors had a very high affinity for glutamate; moreover, they lacked the profound desensitization observed with wild-type GluK2 receptors, which resulted in “constitutive” channel gating in nominally glutamate-free extracellular solution.17 We therefore predicted that the GluK2(T660K/R) mutants would demonstrate analogous agonist-independent currents in voltage-clamp recordings. Agonist-free activation can be characterized by positive shifts in holding current upon application of a polyamine pore blocker, which was NASPM in this case (Figure 4D). Application of 20 μM NASPM for 1 s did not alter the holding current in recordings from cells expressing wild-type GluK2 or GluK2(I668T) (Figures 4D and 4E). In contrast, a positive jump in holding current was observed when NASPM was applied to receptors containing the GluK2(T660K/R) mutant subunits (Figure 4D), which averaged 17 ± 8 pA (T660K) and 40 ± 17 pA (T660R) (n = 6 for both groups, p = 0.022 for T660K and 0.018 for T660R via a Kruskal-Wallis test). The data therefore provide supporting evidence that the GluK2(T660K/R) mutants were activated tonically in nominally glutamate-free extracellular solution, like GluK2(A657T) receptors.17
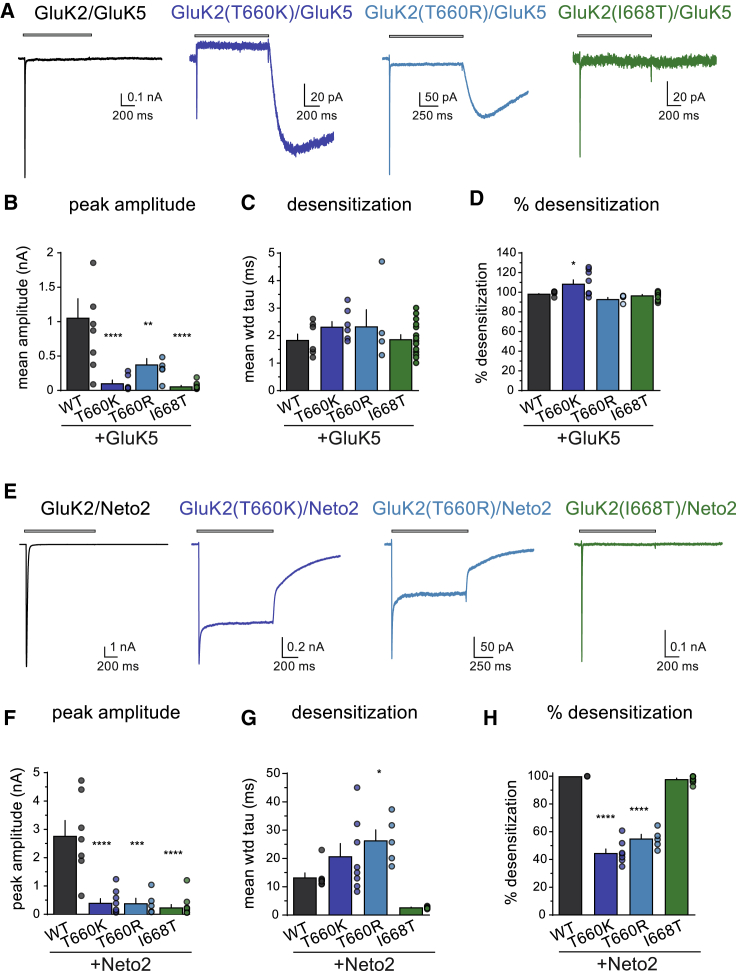
Many neuronal KARs are heteromeric assemblies of subunits that include the GluK5 subunit, which is expressed by a large proportion of neurons in the central and peripheral nervous systems35,36 and therefore most likely represents a constituent of many KARs in the brain.37 To determine whether the GRIK2 variants also influence properties of heteromeric, GluK5-containing receptors, we carried out voltage-clamp recordings from transfected cells expressing wild-type or mutant GluK2 subunits and GluK5 (Figures 5A–5D). [The properties of GluK2(A657T)/GluK5 receptors were reported previously.17] Co-assembly of the mutant GluK2 subunits with GluK5 altered current characteristics and introduced unusual features to the gating of a subset of the receptors (Figure 5A). Application of saturating glutamate to GluK2/GluK5 receptors evoked a current that desensitizes more rapidly (1.8 ± 0.2 ms, n = 7) than homomeric GluK2 receptors (4.9 ± 0.2 ms, n = 47). The GluK2(T660K/R) mutants exhibited large tail currents upon removal of glutamate. This unusual gating behavior has been described previously17,38,39 and results from incomplete desensitization during partial agonist occupancy of heteromeric KARs. GluK2(T660K)/GluK5 receptors also displayed a steady-state current amplitude lower than the control current, which arises from desensitization of the tonic current associated with the nominally agonist-independent activation described in the NASPM experiments. In contrast, GluK2(T660R)/GluK5 receptor currents did not desensitize to the same degree and therefore did not exhibit a positive displacement during glutamate application, despite the presence of the tonic current noted previously. Peak amplitudes for all three of the mutant heteromeric receptors were reduced by 3- to 18-fold compared to wild-type GluK2/GluK5 receptors [GluK2/GluK5: 1.1 ± 0.3 nA, n = 7; GluK2(T660K)/GluK5: 0.11 ± 0.05 nA, n = 8, p < 0.0001; GluK2(T660R)/GluK5: 0.38 ± 0.09 nA, n = 5, p = 0.009; GluK2(I668T)/GluK5: 0.06 ± 0.01 nA, n = 15, p < 0.0001] (Figure 5B). Desensitization rates for GluK2 mutants with GluK5 were not different from wild-type GluK2/GluK5 receptors (Figure 5C), and the percent desensitization from peak current varied as shown in Figure 5D (GluK2(T660K) had >100% because of desensitization of the tonic current).
Figure 5.
Functional characterization of wild-type and mutant heteromeric GluK2 KARs co-assembled with either GluK5 or Neto2
(A) Representative whole-cell currents evoked by a 1 s application of glutamate (10 mM, gray bar) from heteromeric GluK5-containing receptors assembled with the indicated GluK2 subunits expressed in HEK293-T/17 cells.
(B–D) Quantitation of the peak amplitude, desensitization, and percent desensitization for GluK2/GluK5 receptors.
(E) Representative whole-cell currents evoked by a 1 s application of glutamate (10 mM, gray bar) from wild-type and mutant GluK2/Neto2 receptors expressed in HEK293-T/17 cells.
(F–H) Quantitation of the peak amplitude, desensitization, and percent desensitization for GluK2/Neto2 receptors. n = 7 [GluK2/Neto2], 8 [GluK2(T660K)/Neto2], 5 [GluK2(T660R)/Neto2], and 11 [GluK2(I668T)/Neto2]. KAR, kainate receptor; Neto2, Neuropilin- and tolloid-like protein 2; WT, wild-type; wtd, weighted. Statistical significance is denoted as ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Error bars represent SEM.
Lastly, we determined how the auxiliary protein Neto2 altered gating of KARs composed of GluK2 wild-type and mutant subunits (Figures 5E–5H). The most striking feature of the glutamate-evoked currents from GluK2(T660K/R) receptors with Neto2 was their relatively large steady-state current due to incomplete desensitization (Figure 5E). Peak amplitudes were reduced by >80% for each of the mutant GluK2/Neto2 combinations (p < 0.0001 compared to wild-type GluK2/Neto2, Figure 5F). Neto2 slowed desensitization when assembled with wild-type GluK2 receptors (τdes 13 ± 2 ms, n = 7). The auxiliary subunit had a similar effect on GluK2(T660K)/Neto2 receptors (21 ± 5 ms, n = 8; p = 0.15), but GluK2(T660R)/Neto2 KARs (26 ± 4 ms, n = 5; p = 0.015) desensitized slower than wild-type GluK2-Neto2 receptors (Figure 5G). Unexpectedly, desensitization of GluK2(I668T)/Neto2 receptors was more rapid than wild-type (2.7 ± 0.1 ms, n = 11; p = 0.018), although this parameter was not significantly different when corrected for familywise error rate. Quantification of the degree of desensitization underscored the observation that steady-state currents were nearly half the amplitude of peak currents—roughly 50-fold larger than wild-type GluK2/Neto2 steady-state currents (Figure 5H). These data indicate that Neto2 modulates disease-causing variants of GluK2 receptors by altering the entry into the desensitized state and, in the GluK2(T660K/R) mutants, increasing the likelihood of channel opening in the presence of glutamate.
Discussion
Complex variants that potentially cause bi-allelic LoF of GRIK2 were the first in iGluR genes to be identified as co-segregating with ID in individuals from a consanguineous family,16 and missense and LoF variants in other GRIK genes have been associated with neurological disorders in genetic studies.40 However, the extent to which mono-allelic missense GRIK2 variants underlie NDDs is not clear, and missense variants have not been studied in cellular or animal models. In an earlier case study, we described a child whose NDD resulted from a de novo GRIK2 p.Ala657Thr variant.17 Here, we report eleven additional cases of individuals with likely pathogenic variants at three positions in GRIK2 who exhibit a diverse set of developmental and behavioral phenotypes. The GRIK2 variants occur within a domain characterized as a “hotspot” for disease-associated variants in several other iGluR genes.11,12 Each of the four GRIK2 variants profoundly alter GluK2-containing KAR function, and thus, our study underscores the critical importance of appropriate KAR signaling during early development of the CNS.
Genetic variants at positions encoding orthologs of alanine 657 occur in at least one member of all four iGluR gene families,11,41,42 including the GRIK2 variant reported previously (A657T.1) and in the five additional probands in the current study.17 The individuals with identical GRIK2 substitutions at this site have similar neurodevelopmental phenotypes but vary in their severity. All individuals with the p.Ala657Thr variant have ID; the five children show mild to prolonged delays in motor and speech development, whereas the single adult with this variant learned and employs speech but cannot read or write. Probands A657T.1 and A657T.5 have congenital gait ataxia, and the other four individuals exhibit poor balance or uncoordinated walking. Disorganized hand movements or flapping also was observed in several children. Three of the five children are reported to have happy demeanors, but the adult, A657T.6, has mood swings that can manifest as aggression, a behavior also exhibited by individual A657T.2. Development of the brain appears normal in the five individuals for whom MRIs are available, which most notably, distinguishes them from all the children with Thr660 variants. Other features of their disorders appear in only a few individuals; for example, proband A657T.3 was diagnosed with ADHD and only probands A657T.4 and A657T.6 were diagnosed with ASD or exhibit autistiform behaviors.
Children with variants in the GRIK2 codon for threonine 660 exhibited a spectrum of severe neurological and anatomical phenotypes that included intractable daily seizures (predominantly in children with the p.Thr660Lys variant), visual dysfunction, and the marked aberrant myelination observed in MRIs (in all five probands with Thr660 variants). Collectively, these observations highlight that there is a relatively homogeneous NDD phenotype associated with alteration of threonine 660. The abnormalities in MRIs in children with either p.Thr660Lys or p.Thr660Arg were not observed in other probands with GRIK2 variants and most likely underlie at least some of the neurological dysfunction in these children. The series of images at three developmental time points for proband T660K.1 are suggestive of a neurodegenerative trajectory on the basis of the reductions in white matter volume, failure of white matter to mature appropriately, and the atrophy of some brain regions at the 3 years and 9 months time points. Notably, a variant at the analogous site in the AMPAR gene GRIA2 (MIM: 13847) alters threonine 646 to asparagine (GluA2 p.Thr646Asn) and results in a phenotype reminiscent of the MRI changes observed in children with GluK2 Thr660Lys variants—that is, white matter abnormalities consistent with hypomyelination.12 The GluA2 p.Thr646Asn proband also suffered from cerebellar degeneration12 reminiscent of the appearance of cerebellar atrophy in the T660K.1 proband. It remains unclear why myelination fails to mature appropriately in children with either the GRIK2 or GRIA2 variants that lead to alternate residues at threonine 660. KARs are expressed by progenitor cells and some mature oligodendrocytes,43 and thus, a direct alteration of oligodendrocyte maturation is one possibility—especially given that over-activation of the receptors can result in excitotoxicity of glial cells and white matter damage.44,45
Each of the three probands with p.Thr660Lys variants developed epilepsies characterized by daily seizures and poor response to treatment. The severity of their epilepsies distinguishes them from children with the structurally similar p.Thr660Arg variant, who developed treatment-responsive epilepsy syndromes (T660R.1) or did not exhibit epilepsy (T660R.2). In addition to delayed myelination and epilepsy or electrographic abnormalities, most of the children with T660 variants have visual deficits that presented as nystagmus (T660K.1 and T660K.2) and CVI (T660K.3 and T660R.1). The GluK2 subunit is expressed in all areas of the visual system, including in the visual cortex and retina, where it is are localized to photoreceptor synapses46 and is expressed by amacrine and a small subset of ganglion cells.47 What role these receptors play in vision is unknown, however. In the T660K.3 proband, the rare combination of CVI paired with auditory neuropathy spectrum disorder is consistent with a central myelin disorder. These observations and the neuroimaging features of delayed or hypomyelination suggest that GRIK2 variants should be added to the diagnostic etiologic list of disorders of white matter and epilepsy syndromes.48
Individuals with variants at alanine 657 and threonine 660 exhibit surprising differences in disease severity and presentation despite the proximity of the two residues and similar effect of the subunit mutations on channel biophysical parameters. Substitution of a threonine in the p.Ala657Thr variants is poorly tolerated by the hydrophobic M3 domains and significantly increases the likelihood of opening upon agonist binding,27 and it seems likely that Thr660 substitutions similarly alter coupling between binding and gating. Resolution of tetrameric GluA2 structures containing mutations in the residues analogous to Ala657 (to a threonine) and Thr660 (to a glycine) predict that the variants induce structural changes in the M3-S2 gating linker rather than the M3 helix itself.30
The function and biogenesis of neuronal GluK2-containing KARs are most likely altered in diverse ways that produce the distinct phenotypes observed in individuals with the respective GRIK2 variants. Activation of neuronal KARs containing either Ala657 or Thr660 variant subunits will most likely occur at much lower concentrations of the neurotransmitter—possibly even in the 25–100 nM range estimated to be the ambient extracellular glutamate concentration in the CNS.49,50 As well, the slow deactivation rates of both GluK2(A657T)- and GluK2(T660K/R)-containing receptors (Figure 3) predicts that the time course of postsynaptic KAR-mediated depolarizations will be prolonged. Alteration of KAR signaling due to incorporation of the variant subunits will be also be neuron specific because functional properties are shaped by inclusion of other subunits (e.g., GluK5, Figures 5A–5D) or auxiliary proteins (Neto2, Figures 5E–5H). Subtle differences exist in receptor function and biogenesis, though, which we speculate could underlie the distinct neurodevelopmental outcomes of variants at the two sites. Although deactivation and desensitization rates were similarly slow for homomeric GluK2(A657T) and GluK2(T660K/R) mutant receptors, steady-state currents were much larger in proportion to peak currents for both GluK2(T660K) (43%) and GluK2(T660R) (25%) compared to either wild-type GluK2 (∼1%) and GluK2(A657T) (9%) (Figure 3). This phenomenon was even more pronounced when the receptors were co-expressed with the Neto2 auxiliary protein (Figure 5).17 Peak currents evoked from GluK2(T660K/R) mutants were also significantly reduced, and this functional attenuation was as least partially attributable to low receptor expression on the plasma membrane (Figure 3I). Thus, on the basis of the channel properties we measured, the salient differences between GluK2(A657T) and GluK2(T660K/R) mutant subunits were that the latter exhibited larger equilibrium currents but reduced membrane expression. To the extent that it is possible to predict from studies of recombinant KAR function, individuals with p.Thr660 variants might have a partial loss of GluK2-dependent KAR signaling while also gating equilibrium currents through those receptors during development. In contrast, p.Ala657Thr could result in a gain of function in GluK2-dependent KAR signaling in that peak conductances would not be affected while their decay kinetics would be significantly prolonged. It remains unknown how these differences in KAR biophysical function are ameliorated by the expression of GluK2 subunits derived from the wild-type GRIK2 allele in heterozygous individuals. Moreover, expression of the GluK2 isoforms most likely occurs during early stages of development and has the potential to alter axon outgrowth,51, 52, 53 maturation of neurons and circuits,54, 55, 56, 57, 58 and other critical processes that may be relevant to children harboring these variants.
The single individual with a variant at the codon for isoleucine 668 was diagnosed with ASD with no other neurological symptomology. GRIK2 has been identified as a susceptibility gene for ASD,59, 60, 61 and LoF has been implicated as causative for autism-like behavioral phenotypes in animal models.62 The functional consequence of the p.Ile668Thr variant was distinct from both wild-type GluK2 and the other GluK2 mutant subunits in this study: homomeric and heteromeric GluK2(I668T) KARs exhibited markedly reduced peak current amplitudes and very fast gating kinetics but undiminished homomeric receptor localization to the plasma membrane when expressed in heterologous cells. These altered channel properties are consistent with the central role that the residue is thought to play in coupling agonist binding and associated conformational changes with pore opening. Crystallographic resolution of a tetrameric GluA2 AMPA receptor demonstrated that the side chain of isoleucine 668 (Ile633 in GluA2) projects from its position in the M3-S2 linker into a complementary hydrophobic pocket formed from residues at the base of the D2 lobe, thereby bridging the binding and gating domains.30 Elimination of the hydrophobic connection between the LBD and M3-S2 linker through mutagenesis of isoleucine 633 in GluA2 resulted in an “uncoupled” phenotype in which the efficacy of agonism was greatly reduced.30 If Ile668 plays an analogous role in the GluK2 subunit, it is likely that individuals harboring this GRIK2 variant will have a partial LoF in those KARs containing the GluK2 subunit.
In summary, we report eleven individuals with disease-causing variants in GRIK2 and characterize the consequences of the variants on receptor function. Given the recurrent nature and de novo occurrence in individuals with complex NDDs, as well as our comprehensive functional analysis, we conclude that these missense GRIK2 variants are likely pathogenic and that GRIK2 is an NDD-associated gene. The pathogenic mechanisms that underlie the spectrum of phenotypes in individuals with missense GRIK2 variants are likely complex; however, some genotype-phenotype correlations are emerging, including the striking finding of severe epilepsies in the three individuals with the Thr660Lys variant. These results complement a recent genetic analysis that found a strong association between variants in several KAR and auxiliary subunit genes with neurological and NDDs.40 Further mechanistic studies and analysis of GRIK2 variants in model system organisms will be required for more clear understanding of KAR contributions to NDDs and for aiding in the development of precision therapies.
Acknowledgments
This work was supported by grants from the National Institute for Neurological Disorders and Stroke to G.T.S. (R01NS105502) and to G.L.C. (R00NS089858). G.T.S. thanks the Murphy family for additional support for these studies. The Acute Care Flagship of the Australian Genomics Health Alliances is supported by grants from the Sydney Children’s Hospital Network and the National Health and Medical Research Council (GNT1113531). K.Õ. and S.P. were supported by Estonian Research Council grants PRG471, MOBTP175, and PUTJD827. The research conducted at the Murdoch Children’s Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program. We are grateful to the families of the individuals for their willingness to participate in this effort.
Declaration of interests
The authors declare no competing interests.
Published: August 9, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.07.007.
Data and code availability
Additional clinical data supporting the current study have not been deposited in a public repository because they are assessments of individuals. The ClinVar accession numbers for the GRIK2 variants are c.1969G>A (p.Ala657Thr): SCV001754562, c.1979C>A (p.Thr660Lys): SCV001754563, c.1979C>G (p.Thr660Arg): SCV001754564, and c.2003T>C (p.Ile668Thr): SCV001754565. Other data are available from the corresponding author on request.
Web resources
Supplemental information
References
- 1.Traynelis S.F., Wollmuth L.P., McBain C.J., Menniti F.S., Vance K.M., Ogden K.K., Hansen K.B., Yuan H., Myers S.J., Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cline H.T., Constantine-Paton M. NMDA receptor antagonists disrupt the retinotectal topographic map. Neuron. 1989;3:413–426. doi: 10.1016/0896-6273(89)90201-8. [DOI] [PubMed] [Google Scholar]
- 3.Isaac J.T., Nicoll R.A., Malenka R.C. Evidence for silent synapses: implications for the expression of LTP. Neuron. 1995;15:427–434. doi: 10.1016/0896-6273(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 4.Contractor A., Mulle C., Swanson G.T. Kainate receptors coming of age: milestones of two decades of research. Trends Neurosci. 2011;34:154–163. doi: 10.1016/j.tins.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valbuena S., Lerma J. Kainate Receptors, Homeostatic Gatekeepers of Synaptic Plasticity. Neuroscience. 2021;456:17–26. doi: 10.1016/j.neuroscience.2019.11.050. [DOI] [PubMed] [Google Scholar]
- 6.Mayer M.L. Structural biology of glutamate receptor ion channel complexes. Curr. Opin. Struct. Biol. 2016;41:119–127. doi: 10.1016/j.sbi.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Copits B.A., Swanson G.T. Dancing partners at the synapse: auxiliary subunits that shape kainate receptor function. Nat. Rev. Neurosci. 2012;13:675–686. doi: 10.1038/nrn3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomita S., Castillo P.E. Neto1 and Neto2: auxiliary subunits that determine key properties of native kainate receptors. J. Physiol. 2012;590:2217–2223. doi: 10.1113/jphysiol.2011.221101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myers S.J., Yuan H., Kang J.Q., Tan F.C.K., Traynelis S.F., Low C.M. Distinct roles of GRIN2A and GRIN2B variants in neurological conditions. F1000Res. 2019;8:F1000. doi: 10.12688/f1000research.18949.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geisheker M.R., Heymann G., Wang T., Coe B.P., Turner T.N., Stessman H.A.F., Hoekzema K., Kvarnung M., Shaw M., Friend K. Hotspots of missense mutation identify neurodevelopmental disorder genes and functional domains. Nat. Neurosci. 2017;20:1043–1051. doi: 10.1038/nn.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salpietro V., Dixon C.L., Guo H., Bello O.D., Vandrovcova J., Efthymiou S., Maroofian R., Heimer G., Burglen L., Valence S. AMPA receptor GluA2 subunit defects are a cause of neurodevelopmental disorders. Nat. Commun. 2019;10:3094. doi: 10.1038/s41467-019-10910-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin S., Chamberlin A., Shinde D.N., Hempel M., Strom T.M., Schreiber A., Johannsen J., Ousager L.B., Larsen M.J., Hansen L.K. De Novo Variants in GRIA4 Lead to Intellectual Disability with or without Seizures and Gait Abnormalities. Am. J. Hum. Genet. 2017;101:1013–1020. doi: 10.1016/j.ajhg.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies B., Brown L.A., Cais O., Watson J., Clayton A.J., Chang V.T., Biggs D., Preece C., Hernandez-Pliego P., Krohn J. A point mutation in the ion conduction pore of AMPA receptor GRIA3 causes dramatically perturbed sleep patterns as well as intellectual disability. Hum. Mol. Genet. 2017;26:3869–3882. doi: 10.1093/hmg/ddx270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Córdoba M., Rodriguez S., González Morón D., Medina N., Kauffman M.A. Expanding the spectrum of Grik2 mutations: intellectual disability, behavioural disorder, epilepsy and dystonia. Clin. Genet. 2015;87:293–295. doi: 10.1111/cge.12423. [DOI] [PubMed] [Google Scholar]
- 16.Motazacker M.M., Rost B.R., Hucho T., Garshasbi M., Kahrizi K., Ullmann R., Abedini S.S., Nieh S.E., Amini S.H., Goswami C. A defect in the ionotropic glutamate receptor 6 gene (GRIK2) is associated with autosomal recessive mental retardation. Am. J. Hum. Genet. 2007;81:792–798. doi: 10.1086/521275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzmán Y.F., Ramsey K., Stolz J.R., Craig D.W., Huentelman M.J., Narayanan V., Swanson G.T. A gain-of-function mutation in the GRIK2 gene causes neurodevelopmental deficits. Neurol. Genet. 2017;3:e129. doi: 10.1212/NXG.0000000000000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 20.Lunke S., Eggers S., Wilson M., Patel C., Barnett C.P., Pinner J., Sandaradura S.A., Buckley M.F., Krzesinski E.I., de Silva M.G. Feasibility of Ultra-Rapid Exome Sequencing in Critically Ill Infants and Children With Suspected Monogenic Conditions in the Australian Public Health Care System. JAMA. 2020;323:2503–2511. doi: 10.1001/jama.2020.7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jay J.J., Brouwer C. Lollipops in the Clinic: Information Dense Mutation Plots for Precision Medicine. PLoS ONE. 2016;11:e0160519. doi: 10.1371/journal.pone.0160519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyerson J.R., Chittori S., Merk A., Rao P., Han T.H., Serpe M., Mayer M.L., Subramaniam S. Structural basis of kainate subtype glutamate receptor desensitization. Nature. 2016;537:567–571. doi: 10.1038/nature19352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobolevsky A.I., Rosconi M.P., Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Twomey E.C., Sobolevsky A.I. Structural Mechanisms of Gating in Ionotropic Glutamate Receptors. Biochemistry. 2018;57:267–276. doi: 10.1021/acs.biochem.7b00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohda K., Wang Y., Yuzaki M. Mutation of a glutamate receptor motif reveals its role in gating and delta2 receptor channel properties. Nat. Neurosci. 2000;3:315–322. doi: 10.1038/73877. [DOI] [PubMed] [Google Scholar]
- 30.Chen L., Dürr K.L., Gouaux E. X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism. Science. 2014;345:1021–1026. doi: 10.1126/science.1258409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vivithanaporn P., Lash L.L., Marszalec W., Swanson G.T. Critical roles for the M3-S2 transduction linker domain in kainate receptor assembly and postassembly trafficking. J. Neurosci. 2007;27:10423–10433. doi: 10.1523/JNEUROSCI.2674-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greger I.H., Esteban J.A. AMPA receptor biogenesis and trafficking. Curr. Opin. Neurobiol. 2007;17:289–297. doi: 10.1016/j.conb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Klein R.M., Howe J.R. Effects of the lurcher mutation on GluR1 desensitization and activation kinetics. J. Neurosci. 2004;24:4941–4951. doi: 10.1523/JNEUROSCI.0660-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taverna F., Xiong Z.G., Brandes L., Roder J.C., Salter M.W., MacDonald J.F. The Lurcher mutation of an alpha-amino-3-hydroxy-5-methyl- 4-isoxazolepropionic acid receptor subunit enhances potency of glutamate and converts an antagonist to an agonist. J. Biol. Chem. 2000;275:8475–8479. doi: 10.1074/jbc.275.12.8475. [DOI] [PubMed] [Google Scholar]
- 35.Herb A., Burnashev N., Werner P., Sakmann B., Wisden W., Seeburg P.H. The KA-2 subunit of excitatory amino acid receptors shows widespread expression in brain and forms ion channels with distantly related subunits. Neuron. 1992;8:775–785. doi: 10.1016/0896-6273(92)90098-x. [DOI] [PubMed] [Google Scholar]
- 36.Wisden W., Seeburg P.H. A complex mosaic of high-affinity kainate receptors in rat brain. J. Neurosci. 1993;13:3582–3598. doi: 10.1523/JNEUROSCI.13-08-03582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Contractor A., Sailer A.W., Darstein M., Maron C., Xu J., Swanson G.T., Heinemann S.F. Loss of kainate receptor-mediated heterosynaptic facilitation of mossy-fiber synapses in KA2-/- mice. J. Neurosci. 2003;23:422–429. doi: 10.1523/JNEUROSCI.23-02-00422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanson G.T., Green T., Sakai R., Contractor A., Che W., Kamiya H., Heinemann S.F. Differential activation of individual subunits in heteromeric kainate receptors. Neuron. 2002;34:589–598. doi: 10.1016/s0896-6273(02)00676-1. [DOI] [PubMed] [Google Scholar]
- 39.Fisher J.L., Mott D.D. Distinct functional roles of subunits within the heteromeric kainate receptor. J. Neurosci. 2011;31:17113–17122. doi: 10.1523/JNEUROSCI.3685-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koromina M., Flitton M., Blockley A., Mellor I.R., Knight H.M. Damaging coding variants within kainate receptor channel genes are enriched in individuals with schizophrenia, autism and intellectual disabilities. Sci. Rep. 2019;9:19215. doi: 10.1038/s41598-019-55635-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fry A.E., Fawcett K.A., Zelnik N., Yuan H., Thompson B.A.N., Shemer-Meiri L., Cushion T.D., Mugalaasi H., Sims D., Stoodley N. De novo mutations in GRIN1 cause extensive bilateral polymicrogyria. Brain. 2018;141:698–712. doi: 10.1093/brain/awx358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuo J., De Jager P.L., Takahashi K.A., Jiang W., Linden D.J., Heintz N. Neurodegeneration in Lurcher mice caused by mutation in delta2 glutamate receptor gene. Nature. 1997;388:769–773. doi: 10.1038/42009. [DOI] [PubMed] [Google Scholar]
- 43.Puchalski R.B., Louis J.C., Brose N., Traynelis S.F., Egebjerg J., Kukekov V., Wenthold R.J., Rogers S.W., Lin F., Moran T. Selective RNA editing and subunit assembly of native glutamate receptors. Neuron. 1994;13:131–147. doi: 10.1016/0896-6273(94)90464-2. [DOI] [PubMed] [Google Scholar]
- 44.Matute C., Alberdi E., Domercq M., Sánchez-Gómez M.V., Pérez-Samartín A., Rodríguez-Antigüedad A., Pérez-Cerdá F. Excitotoxic damage to white matter. J. Anat. 2007;210:693–702. doi: 10.1111/j.1469-7580.2007.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alberdi E., Sánchez-Gómez M.V., Torre I., Domercq M., Pérez-Samartín A., Pérez-Cerdá F., Matute C. Activation of kainate receptors sensitizes oligodendrocytes to complement attack. J. Neurosci. 2006;26:3220–3228. doi: 10.1523/JNEUROSCI.3780-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harvey D.M., Calkins D.J. Localization of kainate receptors to the presynaptic active zone of the rod photoreceptor in primate retina. Vis. Neurosci. 2002;19:681–692. doi: 10.1017/s0952523802195137. [DOI] [PubMed] [Google Scholar]
- 47.Lindstrom S.H., Ryan D.G., Shi J., DeVries S.H. Kainate receptor subunit diversity underlying response diversity in retinal off bipolar cells. J. Physiol. 2014;592:1457–1477. doi: 10.1113/jphysiol.2013.265033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parikh S., Bernard G., Leventer R.J., van der Knaap M.S., van Hove J., Pizzino A., McNeill N.H., Helman G., Simons C., Schmidt J.L. A clinical approach to the diagnosis of patients with leukodystrophies and genetic leukoencephelopathies. Mol. Genet. Metab. 2015;114:501–515. doi: 10.1016/j.ymgme.2014.12.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herman M.A., Jahr C.E. Extracellular glutamate concentration in hippocampal slice. J. Neurosci. 2007;27:9736–9741. doi: 10.1523/JNEUROSCI.3009-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanson E., Armbruster M., Lau L.A., Sommer M.E., Klaft Z.J., Swanger S.A., Traynelis S.F., Moss S.J., Noubary F., Chadchankar J., Dulla C.G. Tonic Activation of GluN2C/GluN2D-Containing NMDA Receptors by Ambient Glutamate Facilitates Cortical Interneuron Maturation. J. Neurosci. 2019;39:3611–3626. doi: 10.1523/JNEUROSCI.1392-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marques J.M., Rodrigues R.J., Valbuena S., Rozas J.L., Selak S., Marin P., Aller M.I., Lerma J. CRMP2 tethers kainate receptor activity to cytoskeleton dynamics during neuronal maturation. J. Neurosci. 2013;33:18298–18310. doi: 10.1523/JNEUROSCI.3136-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ibarretxe G., Perrais D., Jaskolski F., Vimeney A., Mulle C. Fast regulation of axonal growth cone motility by electrical activity. J. Neurosci. 2007;27:7684–7695. doi: 10.1523/JNEUROSCI.1070-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vernon C.G., Swanson G.T. Neto2 Assembles with Kainate Receptors in DRG Neurons during Development and Modulates Neurite Outgrowth in Adult Sensory Neurons. J. Neurosci. 2017;37:3352–3363. doi: 10.1523/JNEUROSCI.2978-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lauri S.E., Segerstråle M., Vesikansa A., Maingret F., Mulle C., Collingridge G.L., Isaac J.T., Taira T. Endogenous activation of kainate receptors regulates glutamate release and network activity in the developing hippocampus. J. Neurosci. 2005;25:4473–4484. doi: 10.1523/JNEUROSCI.4050-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lauri S.E., Vesikansa A., Segerstråle M., Collingridge G.L., Isaac J.T., Taira T. Functional maturation of CA1 synapses involves activity-dependent loss of tonic kainate receptor-mediated inhibition of glutamate release. Neuron. 2006;50:415–429. doi: 10.1016/j.neuron.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 56.Lanore F., Labrousse V.F., Szabo Z., Normand E., Blanchet C., Mulle C. Deficits in morphofunctional maturation of hippocampal mossy fiber synapses in a mouse model of intellectual disability. J. Neurosci. 2012;32:17882–17893. doi: 10.1523/JNEUROSCI.2049-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marchal C., Mulle C. Postnatal maturation of mossy fibre excitatory transmission in mouse CA3 pyramidal cells: a potential role for kainate receptors. J. Physiol. 2004;561:27–37. doi: 10.1113/jphysiol.2004.069922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryazantseva M., Englund J., Shintyapina A., Huupponen J., Shteinikov V., Pitkänen A., Partanen J.M., Lauri S.E. Kainate receptors regulate development of glutamatergic synaptic circuitry in the rodent amygdala. eLife. 2020;9:e52798. doi: 10.7554/eLife.52798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jamain S., Betancur C., Quach H., Philippe A., Fellous M., Giros B., Gillberg C., Leboyer M., Bourgeron T., Paris Autism Research International Sibpair (PARIS) Study Linkage and association of the glutamate receptor 6 gene with autism. Mol. Psychiatry. 2002;7:302–310. doi: 10.1038/sj.mp.4000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shuang M., Liu J., Jia M.X., Yang J.Z., Wu S.P., Gong X.H., Ling Y.S., Ruan Y., Yang X.L., Zhang D. Family-based association study between autism and glutamate receptor 6 gene in Chinese Han trios. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2004;131B:48–50. doi: 10.1002/ajmg.b.30025. [DOI] [PubMed] [Google Scholar]
- 61.Casey J.P., Magalhaes T., Conroy J.M., Regan R., Shah N., Anney R., Shields D.C., Abrahams B.S., Almeida J., Bacchelli E. A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum. Genet. 2012;131:565–579. doi: 10.1007/s00439-011-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Micheau J., Vimeney A., Normand E., Mulle C., Riedel G. Impaired hippocampus-dependent spatial flexibility and sociability represent autism-like phenotypes in GluK2 mice. Hippocampus. 2014;24:1059–1069. doi: 10.1002/hipo.22290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional clinical data supporting the current study have not been deposited in a public repository because they are assessments of individuals. The ClinVar accession numbers for the GRIK2 variants are c.1969G>A (p.Ala657Thr): SCV001754562, c.1979C>A (p.Thr660Lys): SCV001754563, c.1979C>G (p.Thr660Arg): SCV001754564, and c.2003T>C (p.Ile668Thr): SCV001754565. Other data are available from the corresponding author on request.