Summary
Genome-wide association studies (GWASs) have identified a melanoma-associated locus on chromosome band 7p21.1 with rs117132860 as the lead SNP and a secondary independent signal marked by rs73069846. rs117132860 is also associated with tanning ability and cutaneous squamous cell carcinoma (cSCC). Because ultraviolet radiation (UVR) is a key environmental exposure for all three traits, we investigated the mechanisms by which this locus contributes to melanoma risk, focusing on cellular response to UVR. Fine-mapping of melanoma GWASs identified four independent sets of candidate causal variants. A GWAS region-focused Capture-C study of primary melanocytes identified physical interactions between two causal sets and the promoter of the aryl hydrocarbon receptor (AHR). Subsequent chromatin state annotation, eQTL, and luciferase assays identified rs117132860 as a functional variant and reinforced AHR as a likely causal gene. Because AHR plays critical roles in cellular response to dioxin and UVR, we explored links between this SNP and AHR expression after both 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and ultraviolet B (UVB) exposure. Allele-specific AHR binding to rs117132860-G was enhanced following both, consistent with predicted weakened AHR binding to the risk/poor-tanning rs117132860-A allele, and allele-preferential AHR expression driven from the protective rs117132860-G allele was observed following UVB exposure. Small deletions surrounding rs117132860 introduced via CRISPR abrogates AHR binding, reduces melanocyte cell growth, and prolongs growth arrest following UVB exposure. These data suggest AHR is a melanoma susceptibility gene at the 7p21.1 risk locus and rs117132860 is a functional variant within a UVB-responsive element, leading to allelic AHR expression and altering melanocyte growth phenotypes upon exposure.
Keywords: melanoma, tanning, ultraviolet radiation, UVR, genome-wide association study, GWAS, expression quantitative trait locus, eQTL, fine-mapping, aryl hydrocarbon receptor, AHR, melanocyte
Introduction
Cutaneous melanoma (CM) is the deadliest form of skin cancer,1 and it arises from melanocytes, pigment-producing cells in the skin. Ultraviolet radiation (UVR) exposure is a well-established environmental risk factor2 and genetics also plays a clear role; twin studies suggest melanoma to be the most heritable of solid tumors.3 Over the past decade, a series of progressively larger genome-wide association studies (GWASs) have identified 68 independent signals at 54 genomic loci associated with cutaneous melanoma at a level of genome-wide significance,4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14 highlighting the role of genetics in disease risk. Of these loci, many have also been implicated in risk-associated complex traits, including nevus count8,14, 15, 16 and multiple pigmentation phenotypes.14,17, 18, 19, 20, 21 Notably, an analysis by Landi and colleagues found that of these 54 loci, almost half (n = 26) were also significantly associated with ease of tanning in UK Biobank participants.14
Among these tanning- and melanoma-associated loci is a risk locus on chromosome band 7p21.1 between AGR3 and AHR, initially identified by Law and colleagues in a meta-analysis of GWAS data from Europe, Australia, and the United States.4 The association to this region has since been confirmed by a larger meta-analysis, which identified two independent genome-wide significant signals, as well as a third following conditional analyses,14 all of which lie closest to AGR3. The strongest association signal at this locus has been found to be the lead signal reported to be associated with both tanning response to sun exposure14,22 as well as cutaneous squamous cell carcinoma (cSCC),23 where the risk alleles for melanoma and cSCC were found to be associated with an inability to tan. Thus, these data suggest that response to UVR is the common underlying genetic etiology for these traits.
Among the potential candidate causal genes at this locus is the gene encoding the aryl hydrocarbon receptor (AHR), a ligand-activated transcription factor expressed in all skin cell types.24,25 It is well characterized as a pleiotropic sensor of environmental factors in response to UVR, dioxins, polycyclic aromatic hydrocarbon (PAH), tryptophan and its metabolites, and cigarette smoke.25, 26, 27, 28 Upon activation, AHR translocates to the nucleus and forms a complex with aryl hydrocarbon receptor nuclear translocator (ARNT), which binds to a DNA recognition sequence and initiates gene transcription.25,29 Notably, AHR signaling has been reported to modulate melanogenesis in mice via multiple mechanisms, inducing hyperpigmentation in response to benzanthrone,30 mediating ultraviolet B (UVB)-induced skin tanning in mice via changing expression of genes involved in melanocyte proliferation and differentiation including kitl,26 or regulating melanoma-stroma interaction.31 AHR has also been found to play a key role in melanoma cell behavior, where AHR expression was associated with MEK inhibitor efficacy in NRAS mutant cell lines.32 Sustained activation of AHR signaling by exogenous and endogenous signals, including UVR, leads to a transcription signature that is similar to that of metastatic and dedifferentiated melanoma cells, as well as that of BRAF inhibitor resistance. Chronic activation of the canonical AHR signaling pathway can switch BRAF inhibitor sensitive cells into persister/resistant cells, emphasizing the importance of AHR antagonists as potential sensitizers for melanoma target therapy.33
In this report, we establish the regulation of AHR by the melanoma susceptibility locus at 7p21.1 marked by SNP rs117132860. In melanocytes, we provide evidence for a chromatin interaction between the region surrounding rs117132860 and the promoter and gene body of AHR, as well as similar interactions between candidate causal variants from a second independent signal at this locus. Chromatin immunoprecipitation (ChIP) of AHR indicated allele-specific binding to the protective rs117132860-G allele, which was enhanced in response to UVB and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) exposure, accompanied by an increase of AHR expression. CRISPR knockout (KO) of rs117132860 abolished AHR binding to this region and notably alters melanocyte proliferation and cellular response to UVB exposure. In summary, the association of 7p21.1 with both melanoma risk and tanning response implies a mechanistic link between two processes, and we show this occurs through AHR, providing a direct link between germline genetics and environmental factors on cancer risk.
Material and methods
Fine-mapping
For fine-mapping, we used melanoma GWAS summary data derived from both confirmed, as well as self-reported melanoma cases from 23andMe and UK Biobank (UKBB), and controls as previously described;14 all participants provided informed consent and participation was IRB approved. Participants from 23andMe provided informed consent and participated in the research online under a protocol approved by the external AAHRPP-accredited IRB, Ethical and Independent Review Services (E and I Review). We performed conditional and joint analyses of summary GWAS meta-analysis data as previously described14 by using genome-wide complex trait analysis (GCTA, v.1.26.0)34 to identify independently associated variants. To ensure we were only detecting completely independent SNPs, we set the collinearity threshold (–cojo-collinear) to R2 = 0.05. Conditional analyses of summary data in GCTA were calculated with a reference population of 5,000 individuals selected randomly from the portion of the UKBB population determined to be European by PCA (LDEUR). Variants were converted to best-guess genotype (threshold 0.3). Best-guess data were cleaned for missingness > 3%, HWE p < 1 × 10−6, and MAF < 0.001. Individual credible sets for each of the three association signals in the AHR region were separately identified by conditioning on both of the lead SNPs marking the two other signals reported by Landi and colleagues across a 4 Mb region centered on rs117132860;14 for each isolated signal, we subsequently retained as potential causal variants those with a log-likelihood ratio less than 1:100 relative to the lead variant based on conditional p values. We further fine-mapped this region by using DAP-G.35,36 Briefly, DAP-G (v.1.0) analyses used a window size of 500 kb centered on rs117132860 and allowing for a maximum number of four causals. The pairwise linkage disequilibrium (LD) between all SNPs in each window was computed with the 1000 Genomes phase 3 EUR (1000G EUR) data via PLINK v.1.9 and R v.4.0.2.
Cell culture
Melanoma cell lines were grown in RPMI1640 medium containing 10% FBS, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), and amphotericin B/penicillin/streptomycin. We obtained frozen aliquots of melanocytes isolated from foreskin healthy newborn males, mainly of European descent, following an established protocol37 from the Specialized Programs of Research Excellence (SPORE) in Skin Cancer Specimen Resource Core at Yale University. Cells were either grown in Dermal Cell Basal Medium (American Type Culture Collection/ATCC PCS-200-030) supplemented with Melanocyte Growth Kit (ATCC PCS-200-041) and 1% amphotericin B/penicillin/streptomycin (120-096-711, Quality Biological) for expression quantitative trait locus (eQTL) and Capture-C analysis or alternatively in M254 (Invitrogen, M254500) supplemented with HMGS-2 (Invitrogen, S0165) for all other experiments, grown at 37°C with 5% CO2. All cells tested negative for mycoplasma contamination via MycoAlert PLUS Mycoplasma Detection Kit (LT07-710, Lonza).
Massively parallel reporter assay data
Data were generated and analyzed as previously described.38 Data are presented for massively parallel reporter assay (MPRA) runs with UACC903 and HEK293FT cells analyzed jointly, as well as for UACC903 cells alone.
Luciferase reporter assays
We generated luciferase constructs to include the DNaseI hypersensitivity (DHS) region encompassing rs117132860 (375 bp; chr7: 17,134,641–17,135,015). Sequences encompassing each variant were synthesized via GeneArt service from Invitrogen on the basis of the genomic sequence of HapMap CEU panel samples; sequences are listed in Table S1. Sequences were then cloned into the pGL4.23 vector with BglII and XhoI sequence overhangs. Sequence-verified pGL4.23 constructs were then co-transfected with pGL4.74 (Renilla luciferase) into the melanoma cell line UACC 903 and multiple human melanocyte cultures (C23, C197, and C87) with Lipofectamine 2000 reagent (Thermo Fisher Scientific) for UACC903 or electroporation with the Lonza Amaxa P2 Kit and the CA-137 protocol on the 4D-Nucleofector System for primary melanocytes. Cells were collected 24 h after transfection, and luciferase activity was measured with the Dual-Luciferase Reporter System (Promega) on the GLOMAX Multi Detection System (Promega). All experiments were performed in at least three biological replicates in sets of four replicates.
eQTL analysis
Melanocyte eQTL data were generated and analyzed as previously described39 (dbGaP: phs001500.v1.p1). Genes within the topologically associated domain (TAD) were considered expressed in melanocytes on the basis of RNA-seq by expectation maximization (RSEM) reading with subsequent Bonferroni correction based on the number of expressed genes in the TAD (n = 3; 0.05/3). Pre-analyzed cis-eQTL data from the Genotype-Tissue Expression (GTEx) project (v.8) were downloaded from the GTEx portal.
ATAC-seq library generation and peak calls
We lysed 30,000–50,000 primary melanocytes with cold lysis buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630) and centrifuged them to get nuclei. The nuclei were resuspended in the transposition reaction mix (2× TD buffer [Illumina Cat #FC-121–1030, Nextera], 2.5 μl Tn5 transposase [Illumina Cat #FC-121–1030, Nextera] and nuclease free H2O) on ice and then incubated for 30 min at 37°. We then purified the transposed DNA by using the MinElute Kit (QIAGEN) and PCR amplified the transposed DNA by using Nextera primers for 12 cycles to generate each library. The PCR reaction was subsequently cleaned up with AMPureXP beads (Agencourt) and libraries were paired-end sequenced on an Illumina HiSeq 4000 (100 bp read length) and the Illumina NovaSeq platform. 15 assay for transposase accessible chromatin sequencing (ATAC-seq) libraries were made from five independent primary melanocyte cultures (C56, C140, C205, C24, and C27) with three replicates for each. ATAC-seq data analysis was done with the ENCODE ATAC-seq pipeline v.1.6.1. ATAC-seq reads from independent replicates of each of the five melanocyte cultures (three replicates per culture) were merged together. The five melanocyte cultures were treated as biological replicates, and their sequencing reads were aligned to hg19 genome with bowtie240,41 and duplicate reads were removed. Two pseudo replicates were generated by random sampling of reads from pooled biological replicates. ATAC peaks were called with MACS2 peak caller (2.1.0)42 (p < 0.01), and the ENCODE blacklist regions were removed during the peak calling process.43 Peaks were called from individual biological replicates, pooled data of biological replicates, and pooled data of pseudo replicates. We calculated irreproducible discovery rate (IDR) to find peaks that are reproducible and rank consistently across individual replicates and pooled replicate data. Peaks that overlapped between individual replicates and pooled replicate data with IDR < 0.05 were selected. This peak set is also known as the conservative IDR set. The ATAC-seq peaks that overlapped between the pseudo replicates and pooled replicate data with IDR < 0.05 were selected and referred to as the pseudo IDR set (PIS). The number of peaks between the conservative IDR set and PIS were compared, and the maximum of the two were selected as the optimal IDR set. The conservative IDR and optimal IDR sets passed the reproducibility test because they met the conditions of self-consistency ratio < 2 and rescue ratio < 2. Furthermore, the peaks from both sets between the AHR TAD region (chr7: 16,720,000–17,720,000) were analyzed and visualized on the UCSC Genome Browser44 or WashU Epigenome Browser.45
Capture-C
Custom capture baits were designed by Arima Genomics (San Diego, CA) with an Agilent SureSelect library design targeting specific restriction fragments encompassing two genome-wide significant 7p21 signals from single-SNP analyses.14 The probe library was synthesized by Agilent. Regions for bait design were determined for each locus via identification of the set of SNPs with a log-likelihood ratio < 100 relative to the leading SNP of a given GWAS signal. For the AHR locus, we covered signals for both genome-wide significant signals defined by Landi and colleagues14 (chr7: 17,134,618–17,135,137 and chr7: 16,966,279–17,005,842); baited fragments are listed in Table S2. Capture-C libraries were made with the Arima HiC Kit (Arima Genomics) and the KAPA HyperPrep Kit (KAPA Biosystems) following the manufacturer’s protocol. Briefly, 2–4 million cells were crosslinked, enzyme digested, and ligated. The ligated DNA was reverse crosslinked, fragmented by sonication, and size-selected for adaptor ligation and library amplification. The HiC library was then hybridized with the custom capture baits and captured by the SureSelect XT HS and XT Low Input Library Preparation Kit for ILM (Agilent). 15 Capture-C libraries were made from five human primary melanocyte cultures (C56, C140, C205, C24, and C27) with three biological replicates for each. The captured libraries were pooled and sequenced via an Illumina Novaseq, with one run on an SP and a second run on an S1 flow cell, generating ∼5.7 billion paired-end reads with 150 bp read length. Paired-end sequencing reads from biological replicates were pre-processed with the HiCUP pipeline46 and aligned to human genome version 19 via bowtie2.40,41 Chromatin interaction loops were detected at one- and four-fragment resolutions via CHiCAGO pipeline v.1.16.0.47 Default parameters were used for one fragment analysis except for minFragLen, maxFragLen, binsize, and maxLBrownEst, which were set to 75, 1,200, 2,000, and 150,000, respectively. Four-fragment resolution was created with artificial .baitmap and .rmap files where four consecutive restriction digestion fragments were grouped into one fragment. Four-fragment analysis was conducted with default parameters except for minFragLen, maxFragLen, binsize, and maxLBrownEst, which were set to 150, 5,000, 8,000, and 600,000, respectively. Chromatin interactions with CHiCAGO scores ≥ 5 were considered high-confidence interactions and were further analyzed. The output file was generated with long-range interaction format and was used for visualization on the WashU Epigenome Browser.45
Chromatin conformation capture
Chromatin conformation capture (3C) assays were done on the basis of protocol from Dekker Lab.48 RP11-317K18 and RP11-594C23 bacterial artificial chromosome (BAC) clones were purchased from BACPAC Resources Center and purified with QIAGEN large-construct Maxi Kit (#12462) to cover the genomic region between rs117132860 and AHR to be mapped (RP11-317K18, chr7: 17,121,094–17,313,112; RP11-594C23, chr7: 17,306,808–17,483,385). BAC libraries were made by HindIII digestion of BAC plasmids, followed by ligation and DNA purification. To generate melanocyte 3C libraries, we fixed and lysed ∼20 million cells, followed by HindIII digestion and ligation. Both the melanocyte 3C library and BAC libraries were amplified with a Taqman assay containing a primer localized to various regions between rs117132860 and AHR and a fixed primer near rs117132860 plus a FAM-labeled probe annealing to 3′ of the fixed primer (primer sequences are listed in Table S3). PCR cycle conditions were as follows: 50°C, 2 min; 95°C, 10 min; [95°C, 15 s; 60°C 1 min]×40. Amplification for different primer pairs from the melanocyte 3C library was normalized to that of BAC libraries, reflecting the potential of chromatin interaction.
Chromatin immunoprecipitation
Primary melanocytes were fixed with 1% formaldehyde when ∼85% confluent, following the instructions of the Active Motif ChIP-IT High Sensitivity Kit. 7.5 × 106 cells were then homogenized and sheared by sonication with a Bioruptor (Diagenode) at high setting for 15 min with 20 s on and 30 s off cycles. 5 μg sheared chromatin was used for each immunoprecipitation with antibodies against AHR (Cat#83200s, Cell Signaling) or normal rabbit IgG (Cat# ab37415, Abcam) following the manufacturer’s instructions. Purified antibody pulled-down DNA or input DNA was assayed by SYBR Green qPCR for enrichment of target sites with primers listed in Table S4. Relative quantity of each sample was derived from a standard curve of each primer set and normalized to 1/1,000 input DNA. For rs117132860 genotyping, input DNA or genomic DNA from each cell line and DNA pulled down with an anti-AHR antibody from melanocytes heterozygous for rs117132860 (c87, c197, and c262) were used as template DNA for a Taqman genotyping assay (Assay ID: ANU699F, Thermo Fisher Scientific). All experiments were performed for at least three biological replicates.
UVB and TCDD treatment, AHR quantitative real-time PCR, and immunoblotting
Human primary melanocytes were treated with 10 nM TCDD dissolved in toluene (Sigma, Cat#48599) for 1–24 h. Toluene treated cells served as non-treated control. Cells were exposed with UVB (312 nm) for 10 s from a Spectronics ENB-280C Handheld UV Lamp. The UVB dosage was measured to be 13.2 mJ/cm2 by a radiometer. Cells were lysed with Trizol (Thermo Fisher Scientific) for RNA prep or with RIPA buffer (Thermo Fisher Scientific) for cell extract prep. For assessment of AHR mRNA levels, we purified total RNA by using the QIAGEN RNeasy Mini Kit (Cat#74106) and made cDNA by using the iScript cDNA Synthesis Kit from Biorad (Cat#1725036) followed by a Taqman assay to detect AHR transcription (Hs00169233-m1 for AHR expression and Hs0442063-g1 for GAPDH; Thermo Fisher Scientific). For protein analysis, whole-cell extracts were subjected to water bath sonication and samples were resolved by 4%–12% Bis-Tris ready gel (Invitrogen) electrophoresis. The primary antibodies used were rabbit antibody to AHR (Cat#83200s, Cell Signaling) and mouse antibody to β-actin (A5316, Sigma).
AHR allele-specific expression
Total RNA was isolated via a RNAeasy Mini Kit (217004, QIAGEN). cDNA was synthesized from total RNA via iScript Advanced cDNA Synthesis Kit (Bio-Rad). Genomic DNA and cDNA were then genotyped for rs17779352 with a custom Taqman genotyping assay for rs17779352 (ANCE9Z2, Thermo Fisher Scientific; primers and probes listed in Table S4) recognizing both genomic DNA and cDNA (ENST00000543692). The AHR expression ratio from the risk rs117132860-G/rs17779352-C allele relative to the risk rs117132860-A/rs17779352-T allele was calculated from dRn value on the basis of the linkage between rs117132860 and rs17779352. All experiments were performed in at least three biological replicates in sets of triplicates.
Gene-based CRISPR-Cas9 knockout screen
Target genes were predominantly selected from genome-wide significant melanoma-risk loci.14 Up to ten single guide RNAs (sgRNAs) per gene were designed against the list of target genes. First, sequences were drawn from pre-existing genome-wide libraries.49,50 The remaining sequences were obtained from a published computationally predicted list.51 200 negative control sequences were obtained from the GeCKO V2 library.52 Oligonucleotides, with flanking sequences for PCR sub-pool amplification and isothermal assembly, were synthesized by Twist Biosciences (Table S5). Primers for sub-pool amplification (Table S4) were obtained from a previously published list of sequences orthogonal to the human genome.53 We PCR amplified sub-pools by using quantitative PCR to minimize amplification bias and then gel extracted PCR products from a 2% E-gel EX. 5 ng of gel-extracted PCR product was then assembled with 50 ng of BsmBI digested lentiGuide-Puro (PMID: 25075903) via the NEB HiFi Builder. lentiGuide-Puro was a gift from Feng Zhang (Addgene plasmid # 52963; RRID:Addgene_52963). Assemblies were then purified with the Zymo Clean and Concentrate 5 and eluted in 10 μL of elution buffer. 5 μL of eluted product was then electroporated into Lucigen Endura electrocompetent cells with the BioRad Gene Pulser. Cells were recovered with super optimal broth with catabolite repression (SOC) and plated across 5–15 cm plate and grown at 30°C. ∼18 h later, colonies were immersed in LB and scraped and pelleted and plasmid DNA was extracted with the QIAGEN Plasmid Plus Maxiprep Kit. For production of pooled lentivirus expressing guide RNA (gRNA), the pooled plasmids were co-transfected into HEK293 cells with the pSPAX2, pMD2-G, and pCAG4-RTR2 packaging vectors. Virus was collected 2 days after transfection and concentrated by Vivaspin. Virus titer was measured by HIV-1 p24 Antigen Elisa Kit by ZaptoMetrix Corporation. The TERT-immortalized human melanocyte cell C283T was infected with pCW-Cas9-Blast from Addgene followed by introduction of lentiGuide-Puro (Addgene)-expressing pooled gRNAs. We controlled infection rate at ∼0.3 to allow no more than one copy of sgRNA in each cell. After drug selection and cell recovery (∼day 7 after infection; NoDOX), we collected a fraction of cells and treated the remaining cells with 0.5 μg/mL dox to induce Cas9 expression. Cells were then collected at roughly day 14 (DOX_T1), day 21 (DOX_T2), and day 28 (DOX_T3) after infection (Table S6) and DNA was purified for sequencing by MiSeq. We aligned the raw sequences conservatively (only perfect matches counted) via MAGeCK and compared the normalized read counts for each sgRNA from samples before and after dox treatment (three biological replicates; Exp1, Exp2, Exp3) and analyzed them by MAGeCK together to rank both positively and negatively selected genes. Genes and gRNAs are supplied in Table S5.
CRISPR-Cas9 editing of the rs117132860 AHR-binding motif
We designed a sgRNA targeted to rs117132860 (Table S4) to introduce small deletions into the AHR-binding motif encompassing rs117132860. The sgRNA was cloned and expressed in LentiGuide-Puro (Addgene, Cat#52963). A Cas9-expressing lentiviral vector pCW-Cas9-Blast was virally introduced into an TERT-immortalized human melanocyte culture, C283T, followed by further introduction of either the plasmid containing sgRNA targeted to rs117132860 or a non-targeting sgRNA (Addgene, Cat#80226). Following selection, we isolated monoclonal cell lines from the mixed population through limited dilution. For candidate clones grown from single cells, we sequenced the genomic region around rs117132860 and deconvoluted the sequences to identify potential cell clones with deletion/mutation of the AHR-binding motif containing rs117132860 (Mutation Surveyor, Soft Genetics). We identified two clones with deletion/mutation of both alleles of the AHR-binding motif around rs117132860 (KO1 and KO2), one clone with one copy of wild-type sequence and one copy with deletion of AHR-binding motif (heterozygous [HT]), and two clones with no change of the sequence around the SNP (wild-type [WT]).
Cell cycle and proliferation assays
Cell proliferation was assayed with a BrdU Flow Kit (BD PharMingen) according to the manufacturer’s protocol. Briefly, human melanocytes were labeled with 10 μM BrdU for 3 h before they were fixed, permeabilized, and subjected to DNase I treatment. Cells were then stained with FITC-conjugated antibody to BrdU and 7-AAD, followed by flow cytometry analysis with a BD Accuri instrument (BD PharMingen). For crystal violet staining, cells were seeded at equal numbers and subjected to UVB exposure before being fixed and stained with crystal violet.
Statistical analyses
All cell-based experiments were repeated at least three times with multiple cell cultures. When a representative set is shown, replicate experiments displayed similar but not identical patterns. For all plots, mean and SEM are shown, except for violin plots, where individual data points are shown with the median or mean, range (maximum and minimum), and 25th and 75th percentiles (where applicable). The statistical method, number of data points, and number and type of replicates are indicated in each figure legend.
Predicted transcription factor binding analysis
To predict the effects of candidate causal variants on potential transcription factor binding sites, we used motifbreakR54 v.2.4.0 on a total of 56 fine-mapped candidate causal variants (CCVs). Analysis used dbSNP144 for GRCh37, the UCSC genome build hg19 for the reference sequence, and 2,817 position frequency matrices from ENCODE-motif, FactorBook, HOCOMOCO, and HOMER (provided through the “MotifDb” package and using the default significance threshold of 1.0 × 10−4). Six variants were dropped from analysis (rs35785866, rs200020478, rs199662382, rs10532327, rs367629062, and rs10589929) because they were not included in dbSNP144/GRCh37. All motifbreakR analyses were run with R v.4.0.3.
Results
Fine-mapping identifies rs117132860 as the lone candidate causal variant representing the primary melanoma association signal at 7p21.1
Landi and colleagues14 recently reported the results from a meta-analysis of melanoma GWASs for 36,760 cases, providing evidence that multiple common sequence variants on chromosome band 7p21.1 confer melanoma susceptibility. The most significant association was for rs117132860 (pmeta = 3.83 × 10−21, odds ratio [OR]G-allele = 0.71), and there was an apparent second signal ∼138 kb away marked by rs73069846 (pmeta = 1.24 × 10−8, ORC-allele = 0.95; LD r2rs117132860 = 0.0004, D′rs117132860 = 0.13, 1000G EUR; Figure 1, Figure S1); this second signal appears to represent that locus/SNP previously identified by Law and colleagues in a smaller melanoma meta-analysis (rs1636744; r2rs73069846 = 0.83, D′rs73069846 = 0.94; r2rs117132860 = 0.0003, D′ rs117132860 = 0.11; 1000G EUR).4 Additionally, conditional and joint analysis identified a third independent signal of risk-associated variants marked by rs10487582 (pconditional = 2.30 × 10−8). Notably, rs117132860 had previously been reported to be the lead signal in this region associated with other UVR-associated traits, including tanning ability (p = 7.63 × 10−23; ORG-allele = 1.30)22 as well as cSCC (p = 3.6 × 10−8; ORG-allele = 0.68).23 Subsequent analyses by Landi and colleagues of nevus count15 as well as pigment trait data from UKBB14 found significant or marginal associations for both the primary and secondary melanoma signals (nevus count: prs117132860 = 6.53 × 10−7, prs73069846 = 0.00277; UKBB tanning: prs117132860 = 6.60 × 10−91, prs73069846 = 1.92 × 10−21; UKBB skin color: prs117132860 = 3.64 × 10−45, prs73069846 = 3.86 × 10−15; UKBB childhood sunburns: prs117132860 = 2.97 × 10−9, prs73069846 = 0.00261),14 suggesting a direct link at this locus between UV exposure, melanoma, and these melanoma-associated traits.
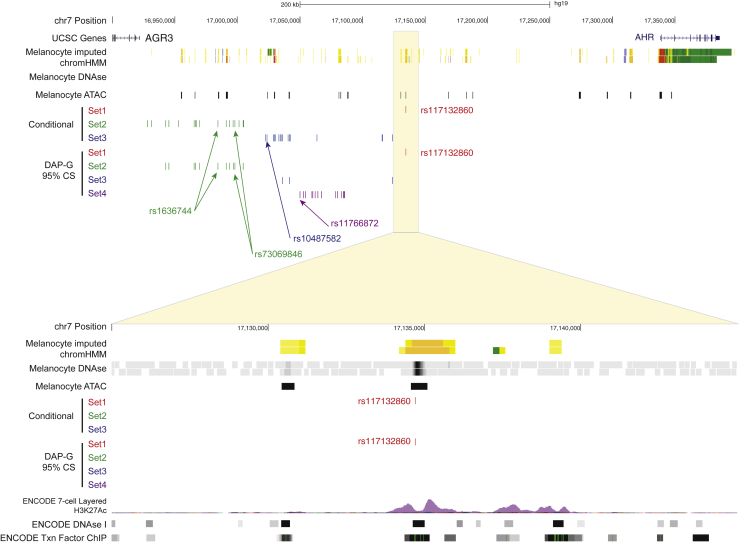
Figure 1.
Fine-mapping of multiple melanoma GWAS signals on chromosome band 7p21.1
(Top) A view of the 7p21.1 locus including UCSC genes, imputed ChromHMM and DNaseI hypersensitivity (DHS) data from two melanocyte cultures generated by the RoadMap Project, ATAC-seq data generated from five human primary melanocyte cultures, and candidate causal variant sets nominated by either conditional analysis or Bayesian fine-mapping (95% credible sets for each of four clusters) with DAP-G. Candidate causal variants mapping to signal/cluster 1 are in red, those mapping to signal/cluster 2 are in green, those mapping to signal/cluster 3 are in blue, and those mapping to signal/cluster 4 are in purple. (Bottom) A zoomed-in view of the region immediately surrounding rs117132860, showing rs117132860 mapping to open chromatin and annotated melanocyte enhancer. Genomic positions are based on hg19. For the zoomed-in region containing rs117132860 (bottom), layered 7-cell H3K27Ac, DHS clusters, and transcription factor CHIP tracks were generated by ENCODE and provided through the UCSC Genome Browser.
To identify credible causal melanoma-risk variants within this larger region, we performed both conditional association analyses as well as Bayesian fine-mapping. First, we performed conditional analysis iteratively for each of the three previously identified association signals by using a leave-one-out approach, conditioning on the lead SNP of the remaining two signals for each to obtain independent credible sets of variants representing each signal. The signal marked by rs117132860 had no other SNP within two orders of magnitude log likelihood relative to the lead p value following conditional analysis (rs117132860 pconditional = 2.99 × 10−20; conditioned on rs73069846 and rs10487582; Table S7, Figure 1, Figures S1 and S2A), suggesting rs117132860 as the lone candidate causal variant for this signal. The secondary signal consisted of a set of 17 variants within two orders of magnitude log-likelihood ratio relative to the lead SNP (rs73069846 pconditional = 1.44 × 10−10; conditioned on rs117132860 and rs10487582; Table S8, Figure 1, Figures S1 and S2B), while the third set consisted of 16 such variants relative to the lead (led by rs9638738, pconditional = 1.10 × 10−8; conditioned on rs117132860 and rs73069846; Table S9, Figure 1, Figures S1 and S2C). Lastly, conditioning on all three resulted in a marginal signal marked by rs11766872 (pconditional = 4.81 × 10−6; Table S10, Figure S2D). We also performed Bayesian fine-mapping of the 7p21.1 region (±250 kb centered on rs117132860) by using DAP-G,35,36 allowing for up to four causal variants given the results of the conditional analysis. Here, the analysis identified four clusters of candidate causal variants (Figure 1, Figure S1, Tables S11 and S12). The first consisted solely of rs117132860 (set 1; posterior inclusion probability [PIP] = 1), while a second cluster consisted of a 95% credible set of 11 variants (set 2; marked by rs73069846, PIP = 0.25). The third 95% credible set consisted of three variants marked by rs34585474 (set 3; PIP = 0.84), and the fourth cluster consisted of a set of 14 SNPs (set 4; led by rs12670784, PIP = 0.32). Finally, we assessed whether there may be other credible causal variants not successfully imputed in the meta-analysis or assessed in conditional or Bayesian analyses. Notably, the meta-analysis by Landi and colleagues performed imputation with a Haplotype Reference Consortium (HRC) reference panel that lacked small insertion/deletion (indel) variants. Thus, we also sought to identify additional variants not reported in the meta-analysis but in high LD with the lead SNP for each signal by using LDLink.55 Of variants from 1000G EUR that were not assessed in the meta-analysis, we identified no high-LD proxy variants (r2 > 0.6; 1000G EUR) for rs117132860, two indel variants in LD to rs73069846 (signal 2; rs35785866 and rs200020478), none for rs34585474, and five for rs12670784 (signal 4; rs13229759, rs199662382, rs10532327, rs367629062, and rs10589929). These data collectively suggest that there are multiple association signals within this locus, map to four sets of potential credible causal variants, and indicate that rs117132860 is the sole candidate causal variant for the signal most strongly associated with melanoma.
Functional annotation with human primary melanocyte ATAC-seq data we generated from a set of five genetically independent cultures (Table S13), as well as additional melanocyte histone-ChIPseq and DNaseI hypersensitivity (DHS) sequencing data from the Roadmap Epigenomics Project56 and ENCODE,57 indicated that rs117132860 lies in an open chromatin region marked as an active enhancer in primary cultured skin melanocytes (Figure 1, Figure S1). Furthermore, annotation of candidate causal variants in the region for predicted alteration of transcription factor binding via motifbreakR54 indicated that rs117132860 is within an AHR::ARNT-binding motif with the risk (A) allele weakening predicted binding (Figure 2A, Table S14). Notably, the gene encoding AHR itself lies 200 kb away from rs117132860 (Figure 1, Figure S1), suggesting a potential role for feedback regulation of AHR levels through rs117132860. Candidate causal variants for the other independent signals also overlap marks indicating potential melanocyte cis-regulatory regions, including rs847404 (ATAC-open), rs975603 (ATAC-open) and rs847377 (DNaseI open) for DAP-G set 2, rs847428 (ATAC-open) and rs847429 (ATAC-open) for DAP-G set 3, and rs4721562 (ATAC-open) for DAP-G set 4 (Table S15; set 1 highlighted in pink, set 2 in yellow, set 3 in green, and set 4 in blue). Altogether, these data identify candidate causal variants from multiple independent signals lying within putative cis-regulatory regions in melanocytes, including the sole variant within credible set 1, rs117132860.
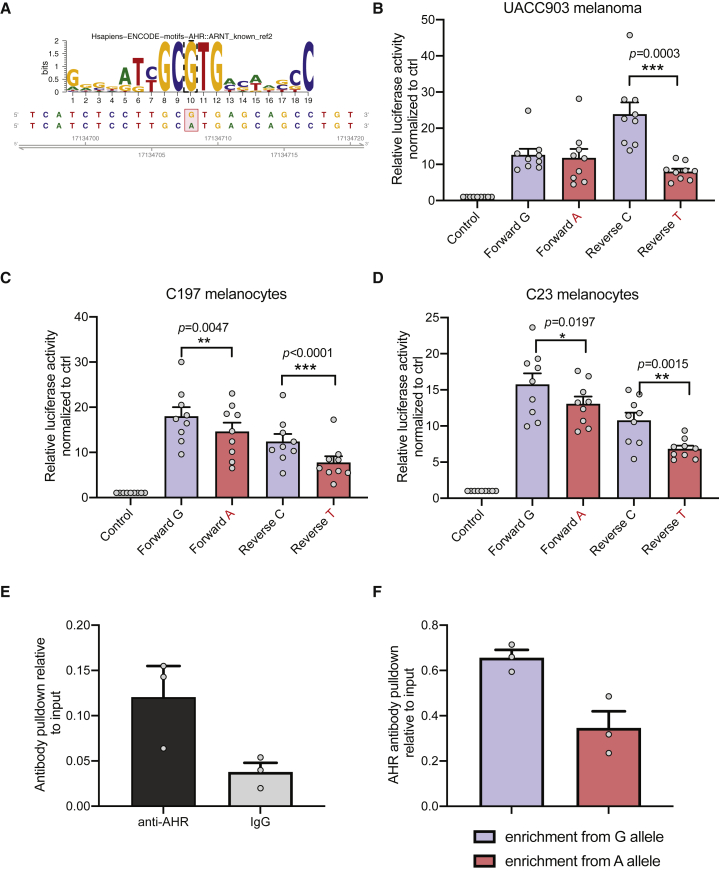
Figure 2.
Allele-specific transcriptional regulation and AHR binding via rs117132860
(A) motifbreakR analysis identified an AHR::ARNT binding motif as strongly altered, where the melanoma-protective rs117132860-G allele matches the motif, while rs117132860-A is predicted to weaken AHR binding.
(B–D) Luciferase reporter activity of a 375 bp fragment encompassing either allele of rs117132860 is measured in (B) the human melanoma cell line UACC903 and (C and D) two primary melanocyte cultures (C197 and C23; labeled). pGL4.23 is the control vector containing a minimal TATA promoter; risk (labeled in red) and protective alleles of rs117132860 are assayed in both forward and reverse directions relative to the promoter. Three biological replicates were combined for each cell (three technical replicates of each, mean, and SEM are plotted).
(E) rs117132860 displays allele-preferential binding to AHR in C87, melanocytes (heterozygous for rs117132860) by chromatin immunoprecipitation assay with anti-AHR antibody or normal IgG control. Relative quantities are shown as fold over input DNA (one representative experiment from three biological replicates is shown; individual datapoints, mean, and SEM are plotted).
(F) Taqman genotyping of rs117132860 with AHR ChIP DNA from the same experiment was normalized to input DNA (one representative set from three biological replicates is shown; mean of PCR triplicates with SEM is plotted).
We also assessed whether any of these candidate causal variants demonstrated allele-specific reporter activity in a previously published MPRA analysis of the region.38 Because these MPRA data assessed allelic activity for SNPs in LD with the lead SNP (rs1636744) from the prior meta-analysis by Law and colleagues (2015),4 the SNPs tested largely corresponded to set 2, while rs117132860 was not assessed because this signal had not yet been found to be genome-wide significant. Four candidate causal variants, however, were false discovery rate (FDR) significant (FDR < 0.01) in a joint analysis of UACC903 and HEK cells, including two variants where the risk allele exhibited higher reporter activity (rs73069846 and rs847404) and two where the risk allele showed lower expression (rs847377 and rs9648229; Table S15). Two of these also overlap melanocyte regulatory marks: rs847404 (open chromatin in melanocyte ATAC-seq) and rs847377 (DNaseI accessible in melanocytes from RoadMap Epigenome Project).
rs117132860 is located within a melanocyte enhancer and displays allele-specific cis-regulatory activity
As described above, functional annotations indicate that rs117132860 is localized to an active melanocyte enhancer region (Figure 1), itself alters an AHR-binding motif (Figure 2A), and is the sole candidate causal variant for the lead melanoma-association signal. Thus, to assess the gene regulatory potential of this region as well as allelic differences in cis-regulatory activity, we cloned a 375 bp fragment encompassing the melanocyte DHS peak region harboring rs117132860 into a luciferase reporter plasmid and assessed luciferase activity in melanoma cells as well as human cultured primary melanocytes. Compared to control plasmid lacking this region, the fragment containing the genomic region surrounding rs117132860 displays strong transcriptional activity in both forward and reverse directions in UACC 903 melanoma cells (Figure 2B) and two independent melanocyte cultures (C197 and C23) (Figures 2C and 2D), consistent with this region’s acting as a transcriptional enhancer. Notably, in human primary melanocytes, we observed that the risk-associated A allele of rs117132860 exhibited significantly lower reporter activity than the protective G allele when cloned in both forward and reverse direction (Figures 2C and 2D; C197, p = 0.0047 and p = 8.09 × 10−6; C23, p = 0.0197 and p = 0.0015; forward and reverse, respectively; two-tailed Student’s t test). In melanoma cells, this allele-specific reporter activity was significant only when the fragment was cloned in reverse direction (Figure 2B, p = 0.0003; two-tailed Student’s t test). These data, together with melanocyte ATAC-seq and publicly available melanocyte DHS and histone-ChIPseq data, are consistent with the region surrounding rs117132860’s acting as an enhancer in cells of melanocytic lineage and suggest that rs117132860 alters the cis-regulatory activity of this enhancer in an allele-specific manner with lower expression associated with the risk allele.
The rs117132860 genotype is correlated with expression of AHR
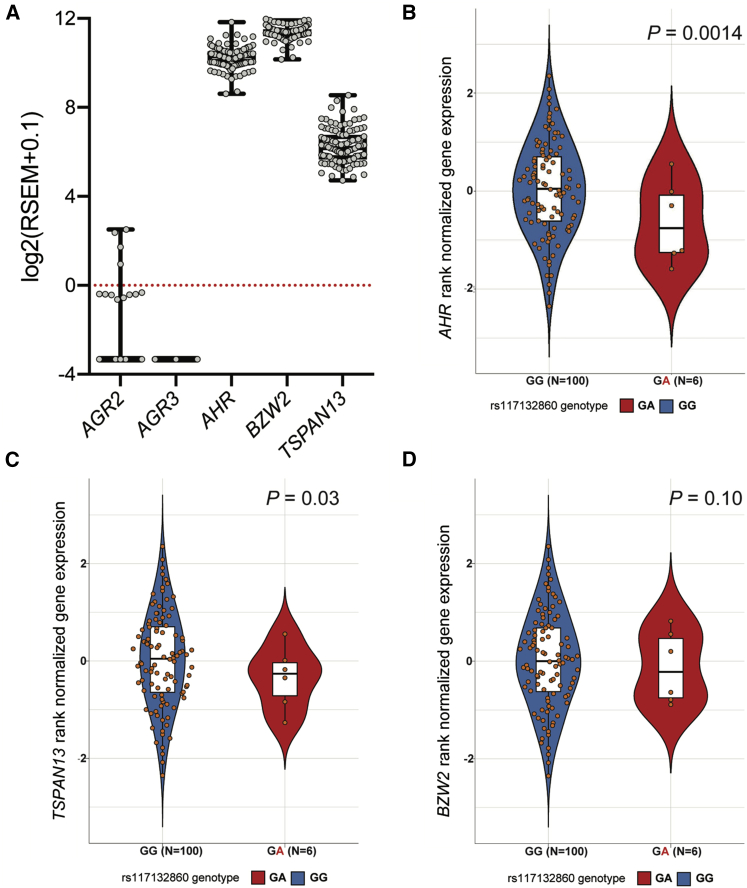
We next assessed expression quantitative trait locus (eQTL) data, both to assess rs117132860 as a potential cis-regulatory variant and to potentially link this variant to regulation of a candidate causal gene(s) within the topologically associated domain (TAD) harboring rs117132860. We turned to an eQTL dataset we previously generated from cultured primary human melanocytes (n = 106),39 where a genome-wide significant eQTL was not previously observed for rs117132860. Given low power due to the small sample size of this dataset and the relatively low minor allele frequency (MAF) for rs117132860 (MAF = 0.017, 1000G EUR, 6/106 samples in the melanocyte dataset are heterozygous; 0/106 are homozygous for the minor allele), we assessed whether rs117132860 might nonetheless be an eQTL for any of the five genes within the TAD. Of these, AGR2 and AGR3 were poorly expressed (detectable expression in <20% of melanocyte cultures, RSEM > 0; Figure 3A). Of the remaining, we found a strong correlation between the A-risk allele and lower levels of AHR expression (p = 0.00136, slope = 0.715; Figure 3B). The remaining genes exhibited weak correlations that were not significant after multiple-testing correction for the number of genes tested within the TAD (Figures 3C and 3D; TSPAN13, p = 0.03; BZW2, p = 0.10). Notably, the direction of correlation for AHR is consistent with reporter assay data where higher expression levels were associated with the protective rs117132860-G allele, providing further support that rs117132860 may be cis-regulatory and linking the risk allele to lower levels of AHR. Similar to that observed in our melanocyte eQTL dataset, there are no genome-wide significant eQTLs for rs117132860 in any tissue analyzed by the GTEx project, however there were three eQTLs with a nominal p value in a comparable range to that we observed for AHR in melanocytes (p < 0.002; Table S16): ANKMY2 in brain amygdala and AHR in non-sun-exposed skin, both of which had opposite direction of effect from the melanocyte eQTL, and HDAC9 in ovary, which had the same direction of effect as the melanocyte eQTL. Finally, although rs117132860 is not a genome-wide significant eQTL in any tissues assessed by the GTEx project, assessment of predicted AHR levels modeled from GTEx data via transcriptome-wide association study (TWAS) by the TWAS-Hub show a correlation between modeled AHR levels and tanning ability,58 further suggesting a role for common AHR-regulatory sequence variants and tanning.
Figure 3.
rs117132860 genotype is associated with AHR expression in human melanocytes
(A) Expression of the five genes located in the TAD containing rs117132860 in 106 human primary melanocyte cultures as assessed by RNA sequencing (box and whiskers with min. to max.).
(B–D) Rank-normalized gene expression of (B) AHR, (C) TSPAN13, and (D) BZW2 in melanocytes with GG or GA genotype at rs117132860. For violin plots, center lines show the medians, box limits indicate the 25th and 75th percentiles, whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, and all samples are represented by dots. n = 100 (GG) and 6 (GA) sample points.
The enhancer harboring rs117132860 physically interacts with the AHR promoter and gene body
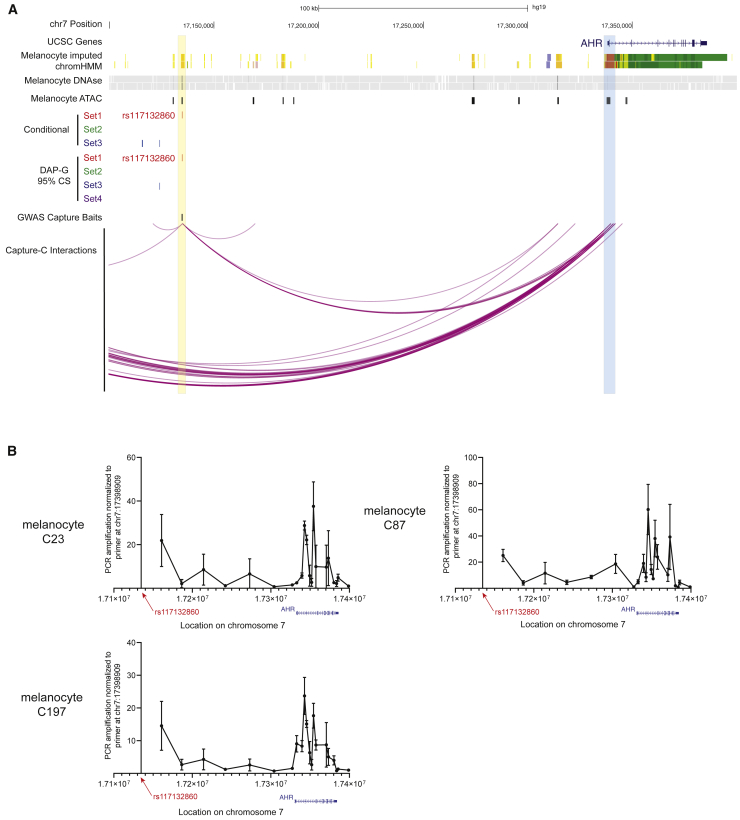
We next assessed physical interactions between regions of association and nearby genes by using multiple methods. As a part of a larger, region-specific Capture-C assay, we designed baits targeting the regions of association for the two signals at chromosome band 7p21.1 that were genome-wide significant in single-SNP analyses from the most recent meta-analysis by Landi and colleagues based on single-SNP data (set 1 and set 2; Figure 4A, Figure S3, Table S2).14 Using these baits, we performed Capture-C assays for a set of five genetically distinct primary human melanocyte cultures (three biological replicates per culture). Following loop calling by CHiCAGO47 with all replicates of all five melanocyte cultures grouped together, we assessed the targets of chromatin looping between our regions of association (and fine-mapped SNPs) to genes within the TAD harboring rs117132860 (Tables S17–S19). Notably, we observed a direct physical contact between the restriction fragment containing rs117132860 and the melanocyte-promoter (annotated by melanocyte imputed chromHMM data) region for AHR (Figure 4A; Figure S3; Tables S15, S18, and S19), suggesting that the enhancer containing rs117132860 may regulate AHR. We did not observe physical association between rs117132860 and any other gene within the TAD containing rs117132860, suggesting AHR is the primary target of this enhancer. We used the region harboring rs117132860 as probe and designed primers spanning from rs117132860 3′ to past the AHR gene body to subsequently confirm the physical association between rs117132860 and AHR by using chromatin conformation capture (3C). 3C analysis in three independent melanocyte cultures confirmed a physical association between rs117132860 and the AHR promoter, as well as the AHR gene body (Figure 4B), further implicating AHR as a target gene of the enhancer containing rs117132860.
Figure 4.
Region-specific Capture-C and chromatin conformation capture (3C) show a chromatin interaction between rs117132860 and the AHR promoter and gene body
(A) Significant chromatin interactions captured by Capture-C between rs117132860 and AHR. Region capture baits are labeled in black, and significant interactions are shown as purple arcs. UCSC genes, imputed ChromHMM and DNaseI hypersensitivity (DHS) data from two melanocyte cultures generated by the RoadMap Project, ATAC-seq data generated from five human primary melanocyte cultures, and candidate causal variant sets nominated by either conditional analysis or Bayesian fine-mapping (95% credible sets for each of four clusters) with DAP-G are also shown. Loops were called from data from five distinct melanocyte cultures (three biological replicates per culture) analyzed together for detection of the most reproducible interactions.
(B) Interactions between rs117132860 and AHR were confirmed via chromatin conformation capture (3C) in three independent primary melanocyte cultures. Relative interaction frequencies of various genomic fragments to the rs117132860 region are shown according to their location in chromosome 7. For each experiment, the PCR amplification for each target primer from 3C libraries were first normalized to the PCR amplification of the target primer from BAC library DNA and subsequently normalized to the PCR amplification of the target primer HindIII17398909, which is immediately downstream of the AHR gene body. A total of four biological replicates were performed (one each for C23 and C87 and two for C197; mean and SEM are plotted for each primer pair).
In addition to the chromatin loop between AHR and rs117132860, we observed multiple chromatin loops between the secondary conditional/DAP-G signal at this locus and the promoter of AHR (Figure S3; Tables S15, S18, and S19), suggesting that candidate causal variants from multiple independent association signals in this region target AHR. Specifically, we see direct loops between fragments containing DAP-G set 2 SNPs rs847404, rs1721040, and rs12535242, as well as rs35785866, which was not fine-mapped but is in LD with rs73069846. Taken together, these data show a physical association between multiple association signals within the 7p21.1 melanoma-risk locus and the AHR promoter, strongly implicating AHR as the likely causal gene explaining the melanoma-risk association for this larger region.
Dynamic change of AHR expression and allele-specific binding of AHR to rs117132860-G in melanocytes upon dioxin and UVB exposure
As described previously, motif analysis indicates that rs117132860 is part of the AHR-binding motif “5′-GCGTG-3′,” with the risk-A allele weakening the binding relative to the protective-G allele (Figure 2A). AHR footprinting with data from multiple cell types, including epidermal melanocytes, also identified rs117132860 as within a region likely to be bound by AHR (RegulomeDB).59 We therefore assessed the potential binding of rs117132860 by AHR via chromatin immunoprecipitation (ChIP) assay in primary melanocyte cultures. As shown in Figure 2E, an AHR antibody specifically pulled down a fragment harboring rs117132860 compared to IgG control as assessed by quantitative real-time PCR, suggesting AHR indeed binds to this fragment. A quantitative real-time PCR genotyping assay was further designed with specific probes distinguishing between rs117132860-G and -A alleles. In a melanocyte culture heterozygous for rs117132860, enhanced IP by AHR antibody was detected for the G allele relative to the A allele (Figure 2F), consistent with the motif prediction. Together with the luciferase result, these data indicate that rs117132860 may regulate transcription in an allele-specific manner as part of an AHR transcription factor binding site.
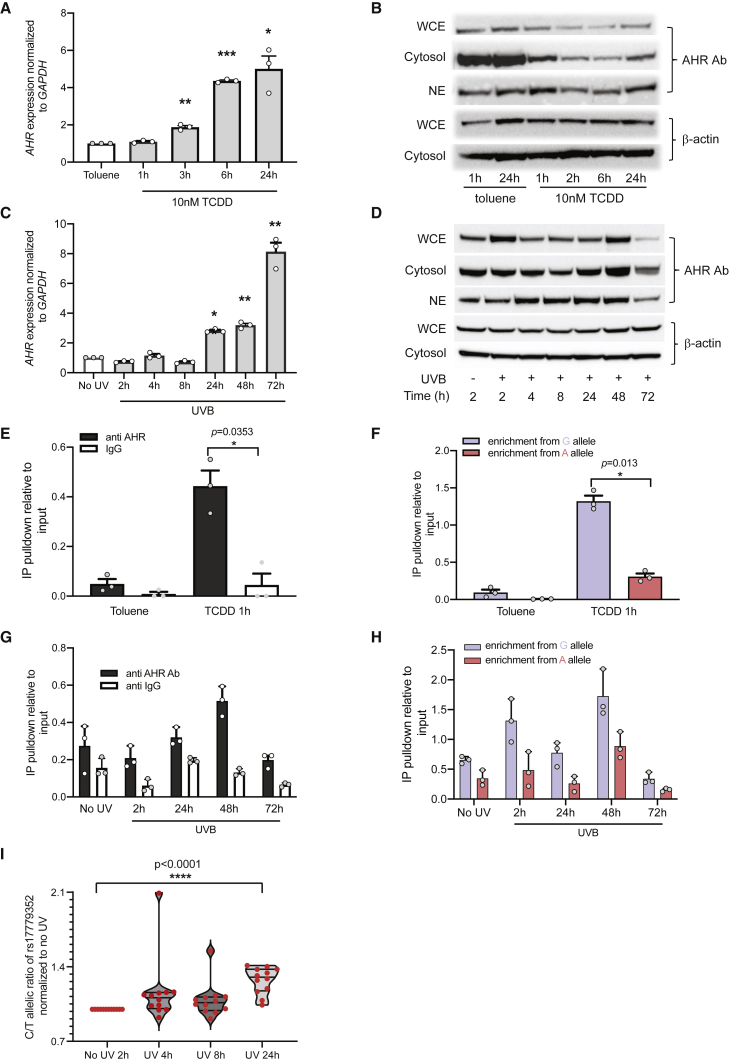
AHR is activated in response to a number of environmental factors, including dioxins and UVR, upon which it translocates to the nucleus and forms a complex with aryl hydrocarbon receptor nuclear translocator (ARNT), subsequently binding to the AHR DNA recognition sequence and initiating target gene transcription. Given this mode of activation, along with the relatively weak binding of AHR to rs117132860 observed in normal culturing conditions (Figure 2E), we assessed AHR expression and AHR binding to the enhancer harboring rs117132860 in the context of dioxin (TCDD) and UVB exposure in multiple human primary melanocyte cultures. We observe that both transcription of AHR as well as overall cellular and nuclear levels of AHR protein are changed dynamically following TCDD and UVB exposure. Following TCDD exposure, AHR mRNA expression is induced from 3 to 24 h after exposure (Figure 5A, Figure S4A), while we observe an increase in nuclear AHR via immunoblot at 1 h after exposure, followed by a reduction in both whole-cell and nuclear AHR from 2 to 6 h after exposure (Figure 5B); it has previously been reported that whole-cell AHR protein level decreases after TCDD treatment in human melanocytes, while AHR mRNA level increases.27 This difference between mRNA and protein level could be mediated by posttranslational ubiquitin-mediated protein degradation, similar to what has been reported following 6-formylindolo[3,2-b]carbazole (FICZ) treatment.60 Following UVB exposure (13.2 mJ/cm2), on the other hand, we observe an increase in the nuclear level of AHR starting at 4 h and a decrease of cytosolic level around 8–24 h following exposure, accompanied by increases in AHR mRNA from 24–72 h. (Figures 5C and 5D, Figure S4B). In summary, upon exposure with both TCDD and UVB in melanocytes, AHR protein translocates into the nucleus together with first a decrease and then an increase in AHR expression.
Figure 5.
Dynamic change of AHR expression and allele-preferential AHR binding to rs117132860-G in melanocytes upon dioxin and UVB exposure
(A) AHR expression normalized to GADPH was measured by Taqman assay in C87 human melanocytes before and after TCDD treatment (individual datapoints, mean, and SEM are plotted).
(B) AHR immunoblotting indicates a dynamic change of AHR protein nuclear localization after TCDD treatment.
(C) GAPDH-normalized AHR transcription is increased after UVB exposure in C87 melanocytes (individual datapoints, mean, and SEM are plotted).
(D) Nuclear translocation of AHR is observed after UVB exposure via immunoblotting in C87 melanocytes.
(E—H) AHR binding to rs117132860 measured by CHIP assay increased after (E) TCDD treatment in C197 melanocytes and (G) UVB exposure in C87 melanocytes. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 (individual datapoints, mean, and SEM are plotted). A genotyping assay with AHR CHIP DNA shows the enhanced binding of AHR to the rs117132860-G allele after (F) TCDD treatment and (H) UVB exposure in C87 melanocytes (individual datapoints, mean, and SEM are plotted).
(I) A genotyping assay of rs17779352, a proxy SNP for rs117132860 located in in the AHR coding sequence, indicates that the ratio of AHR expression from the melanoma-protective rs117132860-G/rs17779352-C allele significantly increased after UVB exposure relative to the rs117132860-A/rs17779352-T allele at 24 h.
All experiments except for those shown in (I) were done in both C87 and C197 human melanocytes with a total of four biological replicates each. The experiment set shown in the figure is a representative one with p value calculated from a two-way paired Student’s t test (A, C, E, and F) or two-way ANOVA (G and H), and the mean with SEM is plotted. The proxy SNP genotyping experiment (I) was only performed in C87 cells, which are heterozygous for both rs117132860 and rs17779352; the violin plot shows the combination of four experiments of three replicates each, and all points are shown.
Given these changes in AHR transcription as well as AHR nuclear localization, we assessed binding of AHR to the region harboring rs117132860 via AHR ChIP both with and without TCDD or UVB exposure in human primary melanocytes. Consistent with the data presented above, in the absence of TCDD or UVB treatment, we observe enriched binding of AHR to rs117132860 via enhanced pulldown by AHR antibody compared to control IgG (Figures 5E and 5G, Figures S5A and S5C). At 1 h after exposure, we observe significant increased binding of AHR to rs117132860 relative to untreated toluene control, consistent with the short-term changes in nuclear translocation of AHR in response to TCDD and the increased expression pattern reflected in latter time points (p = 0.035, Figure 5E; p = 0.018, Figure S5A). We further assessed allele-preferential binding of AHR following TCDD exposure in primary melanocytes heterozygous for rs117132860 via a Taqman-based allele discrimination assay with dye-labeled probes distinguishing between the G or A allele. Consistent with the predicted stronger AHR motif for rs117132860-G allele, we observe enhanced binding of AHR to rs117132860-G versus rs117132860-A both with and without TCDD treatment (p = 0.013, Figure 5F; p = 0.0014, Figure S5B; two-way paired Student’s t test), where the preferential binding to the G allele becomes significant after TCDD treatment. Likewise, following UVB exposure, we observe stronger binding of AHR to rs117132860, and the peak binding is detected at 48 h after exposure (Figure 5G, Figure S5C), and binding of AHR at all time points is enriched for the rs117132860-G allele relative to rs117132860-A (Figure 5H, Figure S5D), both of which are significant after UVB exposure for two melanocyte cultures (p = 0.0005, p = 2.62 × 10−6, respectively, for Figure 5H; p = 1.3 × 10−5, p = 7.2 × 10−12 for Figure S5D; two-way ANOVA). Thus, the dynamic changes of AHR binding to the SNP are consistent with the nuclear localization of AHR following both TCDD and UVB exposure and increased expression pattern reflected at later time points, suggesting that activated AHR binding to the SNP by those treatments may regulate AHR expression in an allelic manner.
Given the widespread effects of TCDD and, in particular, UVB on global gene expression, including many control genes, which may affect the quantification of transcription, we turned to assessing allelic expression in human primary melanocyte cultures heterozygous for rs117132860. Specifically, we identified an mRNA coding proxy SNP in LD with rs117132860 (rs17779352; r2 = 0.0014; D′ = 1, 1000G EUR), which allowed us to assess transcription of AHR in cis by rs117132860-A and rs117132860-G alleles. We treated a primary melanocyte culture heterozygous for both rs117132860 and rs17779352 (where the A-risk allele of rs117132860 is cis to rs17779352-T-allele and rs117132860-G is cis to rs17779352-C) with either TCDD or UVB (13.2 mJ/cm2) and collected RNA from cells at different time points. Notably, at 24 h following UVB treatment, we observed a significant increase in AHR transcribed from the protective (rs117132860-G, rs17779352-C) allele (p = 1.1 × 10−5; two-way paired Student’s t test) compared to the risk allele (Figure 5I). However, we did not observe the same allele-preferential changes in AHR expression in response to TCDD (Figure S6). These data are consistent in direction with both the luciferase reporter data and AHR eQTL with rs117132860. Thus, in response to UVB exposure, we observe an increase in transcription of AHR driven from the melanoma-protective rs117132860-G allele as compared to that driven from the rs117132860-A-risk allele.
A melanocyte gene-based CRISPR knockout screen identifies AHR as a gene hit mediating cell growth in an immortalized human melanocyte culture
We performed a gene-based pooled CRISPR-Cas9 cell proliferation knockout screen in an immortalized human melanocyte culture, C283T. Here, we designed 3,052 gRNAs (Table S5) covering a set of 288 genes, as well as ∼200 non-targeting gRNAs. The 288 genes focused primarily on gene candidates from genome-wide significant melanoma GWAS loci. Both Cas9 and gRNAs were expressed in lentiviral vectors, and Cas9 was induced by doxycycline (dox) treatment. DNA samples were collected from sgRNA infected cells before dox treatment (around day 7 after infection) and at three different time points after that (around day 14, day 21, and day 28 following infection). gRNAs were sequenced and normalized to counts of the integrated gRNA sequence from cells collected prior to dox treatment. We compared dox-treated cells from all three latter time points and prioritized gene hits critical for cell growth and/or survival by using MAGeCK.61 By combining three replicate multi-time-point experiments and comparing read counts (Table S6) from all samples treated with dox with untreated samples, MAGeCK ranked 55 genes as negatively selected genes and two genes as positively selected with significance score < 0.05 and FDR < 0.05 (Table S20). Consistent with established roles as tumor suppressors, guides targeting both TP53 and CDKN2A were found to be positively selected, while those targeting PARP1 and SETDB1, two genes previously reported to be important in melanogenesis,62,63 are among the top negatively selected gene group. Among the negatively selected genes, AHR was ranked eighth (log2(fold change) = −1.2515), thus suggesting that AHR plays a role in melanocyte growth and that risk-associated SNPs at 7p21.1 could potentially play a role in mediating melanocyte cellular phenotypes.
Deletion of the AHR-binding motif encompassing rs117132860 alters melanocyte response to UVB exposure
Finally, to further assess the contributions of the enhancer harboring rs117132860 on both expression of AHR and melanocyte cellular phenotypes, we utilized CRISPR-Cas9 editing to mutate/delete the conserved AHR-binding motif containing rs117132860. We designed a sgRNA targeted to rs117132860. A Cas9-expressing lentiviral vector pCW-Cas9-Blast was virally introduced into an TERT-immortalized human melanocyte culture, C283T, followed by further introduction of either an sgRNA targeted to rs117132860 or a non-target sgRNA. Following selection, we isolated monoclonal cell lines from the mixed population through limited dilution. For candidate clones grown from single cells, we sequenced the genomic region around rs117132860 and deconvoluted the sequences to identify potential cell clones with deletion/mutation regarding to the AHR-binding motif around rs117132860. We identified two clones with deletion/mutation of both alleles of the AHR-binding motif around rs117132860 (KO; deletion size in two knockout clones ranged from 2 to 9 bp), one clone with one copy of wild-type sequence and one copy with deletion of the AHR-binding motif (HT; deletion of 16 bp), and three clones with no change of the sequence around the SNP (WT) (Figure 6A, Figure S7). We first checked AHR binding to the SNP region in KO cells, and as expected, AHR does not bind to the deleted rs117132860 region both before and after UVB treatment (Figure S8). We assessed AHR expression from both WT and KO clones with and without UVB exposure. While we see a similar dynamic change of AHR expression pattern in WT and KO cells in response to UVB as observed in primary melanocytes, we did not observe a consistent difference in AHR expression between WT and KO cells both before and after UVB exposure, potentially because of the underlying complexity of AHR regulation (Figure S9). Nonetheless, we investigated melanocyte growth phenotypes in these clones. Notably, BrdU staining and flow cytometry of WT and KO clones indicated that WT clones had a larger proportion of BrdU-positive cells (range 42.8% to 48.6%) compared to KO clones (range 22.9% to 33.2%), and the heterozygous knockout clone was in between (range 33.4% to 39.3%; Figure S10A). This difference of proliferation between rs117132860-WT and -KO cells is confirmed by crystal violet staining of cells collected at different times after seeded at the same number (Figure S10B, WT versus KO clones, D4, p = 0.00031 for the representative experiment shown; p = 1.9 × 10−5 for three experiments combined, two-tailed paired Student’s t test). We also investigated cellular response to UVB exposure in these cells. At 72 h after being treated with 13 mJ/cm2 UVB, both WT and KO cells were cell cycle arrested, reflected by smaller cell numbers and the larger and flatter cell image via crystal violet staining (Figure 6B, Figure S11). At day 7 following UVB exposure, while many WT cells survived the UVB and went back to normal cell shape and cycle (“smaller cells” in Figure 6B and Figure S11), most KO cells remained arrested (large, flat cells), indicating a clearly different cellular response to UVB treatment between WT and KO cells. BrdU staining and flow cytometry confirmed a mixed population of growing and arrested cells at day 7 following UVB exposure, and significantly more cells remained arrested in KO lines (Figure 6C, Figure S12).
Figure 6.
Functional characterization of CRISPR-Cas9-edited monoclonal melanocyte cell lines
(A) Genomic DNA sequences of (multiple) monoclonal cells homozygous for rs117132860-G (WT), heterozygous monoclonal cells with one wild-type allele and second copy with a 16 bp deletion including the SNP (HT), and two monoclonal clones that each have both alleles harboring a short deletion encompassing the AHR-binding motif (KO1 and KO2).
(B) Crystal violet staining of rs117132860-WT1 and rs117132860-KO2 cells not treated with UVB (24 h), 72 h after UVB treatment, and at day 7 after UVB exposure, followed by a zoomed-in image of day 7.
(C) FACS analysis of the same WT and KO cells at day 7 following UVB treatment. Forward-scatter and side-scatter image indicates a mixed population, and BrdU and 7-AAD staining characterize the cell cycle status of each population. The images shown here are from a representative experiment from three biological replicates. Additional WT and KO clones were assessed similarly.
Discussion
Recent melanoma GWASs have successfully identified 68 independent signals at 54 genomic loci.14 Of these, almost half are also associated with ease of tanning, reflecting the well-established interplay between pigmentation traits, UV exposure, and melanoma risk and suggesting that environmental factors may work together with genetic variation leading to melanoma development. Here, we investigated a complex melanoma-risk signal on chromosome band 7p21, fine-mapping three to four independent genetic signals and demonstrating that candidate causal variants for at least two of these physically interact with the AHR promoter in melanocytes. We also show that in primary melanocytes, AHR levels correlate with genotype at the lone candidate causal variant marking the strongest signal, rs117132860, where lower AHR levels are associated with the risk/poor-tanning allele. We do not identify significant correlations between rs117132860 and other nearby genes, nor do we identify chromatin loops in melanocytes to the promoters of genes other than AHR. Taken together, these data strongly suggest that these signals most likely function via allele-specific cis-regulation of AHR.
We performed fine-mapping via conditional and joint analyses34 as well as with a Bayesian approach35,36 because it has been suggested that Bayesian fine-mapping methods may perform better than stepwise conditional regression methods.64 Conditional and joint analysis identified three sets of candidate causal variants between AGR3 and AHR (Figure 1, Figures S1 and S2, Tables S7–S9), which correlated to previously reported signals.4,14 Conditioning on all three signals revealed a fourth marginal signal, marked by rs11766872 (pconditional = 4.81 × 10−6; Figure S2D, Table S10) approximately 400 kb from the AHR promoter; SNX13 is the nearest gene. Bayesian analysis with DAP-G,35,36 on the other hand, identified three clusters of candidate causal variants largely overlapping the first three identified by conditional analysis, and a fourth was also located between AGR3 and AHR (Figure 1, Figure S1, Tables S11 and S12); note that the window size used for DAP-G did not include SNPs from the fourth conditional signal. Further functional studies, as well as perhaps fine-mapping with data from larger meta-analyses, will be required for better resolving of the spectrum of causal variants in this region.
Still, integration of melanocyte epigenomic, melanoma functional fine-mapping (MPRA), and melanocyte Capture-C data nominates at least one strong candidate for a causal variant marking the second most significant melanoma-risk signal (marked by rs73069846). Notably, of four candidate causal variants for this signal that directly loop to the promoter of AHR (rs847404, rs1721040, rs12535242, and rs35785866), only rs847404 both lies within open chromatin regions in melanocytes and exhibits significant allele-specific reporter activity (rs847404, p = 2.62 × 10−5 for UACC903/HEK combined, risk allele associated with higher expression; Table S15). MPRA data do nominate additional candidate causal variants for which we do not observe a direct loop between fragments containing the variant and AHR (rs73069846, p = 9.24 × 10−13 for UACC903/HEK cells combined; p = 0.0077 for UACC903 cells alone; risk allele associated with higher reporter activity; rs847377, p = 0.0095 for UACC903/HEK, risk allele associated with lower expression; Table S15). Given that many restriction fragments in the broader signal 2 region surrounding these variants were observed to associate with AHR in melanocytes via Capture-C, these data suggest that looping between these additional candidate causal variants and AHR is plausible. Finally of note, not all candidate causal variants were assessed via MPRA or baited for Capture-C analysis, and thus, we cannot rule out other variants as underlying risk in this region. Further work will be required for comprehensive assement of plausible functional variants in this region.
Because rs117132860 is the sole candidate causal variant associated with the lead signal at this locus, we adopted multiple approaches to characterize its potential functions in melanoma development. We used human primary melanocyte cultures as our model system to reflect tissue-specific effects. Region-focused Capture-C showed a clear association between the region encompassing rs117132860 and the promoter and gene body of AHR, which we further corroborated by 3C mapping in multiple melanocyte cultures. eQTL analysis of 106 primary melanocyte cultures similarly nominated AHR as the candidate causal gene for which cis-regulation through rs117132860 is likely in this cell type. Both luciferase reporter assay and ChIP assays demonstrated that AHR binds to its conserved binding motif containing the rs117132860-G allele and regulates expression in an allele-preferential manner. Because the signal marked by rs117132860 is also associated with tanning ability, cells were exposed to UVB, and we observed both increased allele-preferential binding of AHR to rs117132860-G as well as increased ratio of expression driven from the rs117132860-G-protective versus rs117132860-A-risk alleles. Thus, our work highlights interaction between a common genetic risk variant and a well-established environmental exposure.
AHR is a well-characterized pleiotropic sensor of environmental factors and has been reported to affect skin pigmentation and tanning ability after UVB exposure in a mouse knockout model where reduced pigmentation and tanning response in knockout mice functioned through melanocyte- rather than keratinocyte-specific expression of Ahr.26 Here, we were not able to test whether rs117132860 is directly involved in melanocyte pigmentation because of the limitation of our cell model, as the immortalized melanocytes we used had dramatically reduced pigmentation following immortalization by hTERT with diminished expression of TYPR1 and TYPR2 (data not shown) relative to parental cells. Still, CRISPR-Cas9 editing of melanocytes to interrupt the AHR-binding motif harboring rs117132860-G demonstrated a clear phenotypic impact of loss of this motif in human melanocytes, including reduced proliferation as assayed by both BrdU staining and assessed by flow cytometry (Figure S10A) and crystal violet staining in a growth assay (Figure S10B), consistent with the observed negative selection of AHR knockout itself on cell proliferation in our CRISPR screen (Table S20). This observation is consistent with a microarray analysis of cultured primary melanocytes from AhrWT and Ahr−/− mice, finding 3.6- to 6-fold lower expression of endothekin-1 (end1) and kit ligand (kitl) in Ahr−/− mice, two factors involved in melanocyte proliferation.26 We also observe a prolonged growth arrest following UVB irradiation in rs117132860-KO cells, and a heterozygous knockout clone showed an intermediate phenotype. This raises the question of whether these arrested/senescent cells can promote secretion of an array of cytokines, chemokines, and growth factors known as the senescence-associated secretory phenotype (SASP), which can create a permissive microenvironment to initiate carcinogenesis.65,66 Although we have not investigated the possible link between rs117132860 and AHR-activity-induced inflammation upon UVB because of the limitations of our system, multiple studies have been reported on the role of AHR in the regulation of the immune response in the context of inflammation and cancer,25,67,68 as well as the potential opportunities of developing AHR-targeted therapeutics. In particular, a recent report suggests a function of AHR activation on inflammation-induced melanoma cell dedifferentiation and metastases in a mouse model.60 Taken together, these data support the notion that rs117132860 may modulate tumor risk at least in part by regulating AHR function in melanocyte proliferation and/or growth arrest before and after UVB exposure, implying an additional mechanism associated with melanoma risk other than the reported effect through skin pigmentation.
Still, the response of gene targets regulated by AHR is complex, and further work will be required for better characterization of the dynamics of transcriptional response to UVB irradiation. First, our fine-mapping clearly shows multiple melanoma-risk signals beyond rs117132860, which itself has a relatively low minor allele frequency. These data suggest multiple enhancers under genetic regulation combine to regulate AHR. Beyond this, cis-regulation via the enhancer harboring rs117132860 is likely to be more complex. Specifically, while the AHR/ARNT complex is generally a positive regulator of gene expression, AHR itself induces a transcriptional repressor (AHRR), which like AHR, forms a complex with ARNT and binds to the consensus AHR motif but negatively regulates expression. In addition, given the dependency of both AHR and AHRR activity on availability of ARNT, ARNT levels and competition by other ARNT-binding transcription factors (HIF) may influence this response. Finally, our motif analysis also suggested that rs117132860-A may create a MYC binding site, and ChIP sequencing for MYC by ENCODE identified this region as bound by MYC in some cell types. Indeed, consistent with this complexity, we observe several results suggesting that a simple model whereby AHR binding to rs117132860-G only upregulates AHR cannot fully explain short-term and long-term transcriptional dynamics upon UV exposure. Although genotyping of a rs117132860 proxy SNP within the AHR mRNA sequence (rs17779352) after UVB exposure demonstrated higher AHR expression driven from the protective rs117132860-G/rs17779352-C allele in cells heterozygous for both SNPs, we failed to observe such a difference following TCDD treatment at multiple time points. Further, we did not observe a consistent difference in AHR transcriptional dynamics following UVB in rs117132860-KO (or -HT) cells. Further work will be required for characterization of the dynamics of various rs117132860 allele-specific binding proteins both under normal growth conditions as well as in response to UVB.
Our data do not provide strong evidence as to the role or roles of the enhancer sequence harboring rs117132860 in other cell or tissue types. This region is annotated as enhancer across a large proportion of cell types characterized by the RoadMap Epigenomics Project, suggesting a broader role of this enhancer in regulation of AHR as well as potentially other genes in other cell types. Still, we speculate the activity of this element is likely to be highly dependent on cell type and cell state (e.g., AHR-activating exposures), both in terms of chromatin conformation and the availability of transcription factors that can bind to this element. As described above, transcriptional regulation via AHR response elements can be complex where nuclear localization of AHR is dependent on cell state and exposures and regulation of the repressor complex AHRR:ARNT is at least in part highly dependent on transcriptional activation by AHR itself. Furthermore, given that AHR and HIF share a common binding partner, ARNT, HIF signaling under some circumstances has been shown to antagonize gene regulation via AHR.69 Consistent with this complexity, the marginal eQTL observed in GTEx non-sun-exposed skin shows the opposite allelic effect observed in cultured melanocytes, suggesting that this region may be cis-regulatory in keratinocytes and/or fibroblasts but that activity of this element is indeed cell-type and cell-state dependent.
Finally, our findings linking the 7p21 melanoma susceptibility region to AHR suggests that signaling through the AHR pathway could be more broadly important in melanoma risk. Notably, multiple additional melanoma-risk loci harbor genes within this pathway. Specifically, the risk locus on chromosome band 1q21.3 (marked by rs8444) harbors the gene encoding ARNT, the binding partner of AHR itself on which the activity of AHR and AHRR depend. Notably, a TWAS with the same GWAS summary statistics found imputed melanocyte-specific ARNT expression to be negatively correlated with melanoma risk,14 while a TWAS of tanning found a positive correlation between predicted ARNT levels and ability to tan.58 Furthermore, a missense variant in CYP1B1 (rs1800440), a direct transcriptional target of AHR, has been reported to be associated with both melanoma risk4,14 and keratinocyte skin cancers.70 Thus, our work highlights a potential role for the larger AHR pathway in melanoma risk.
Acknowledgments
We would like to thank members at the National Cancer Institute Cancer Genomics Research Laboratory (CGR) for help with sequencing efforts and L. Amundadottir from the National Cancer Institute, Laboratory of Translational Genomics for helpful discussions. This work has been supported by the Intramural Research Program (IRP) of The National Cancer Institute, US National Institutes of Health, including through an NCI FLEX award to G.M. and K.M.B. M.M.I. is supported by Cancer Research UK (C588/A19167) and by the National Institutes of Health (R01 CA83115), and GWAS genotyping services were provided by the Center for Inherited Disease Research (CIDR); CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (HHSN268201100011I). M.H.L. acknowledges funding support from the Australian National Health and Medical Research Council (NHMRC; APP1129822 and APP1123248) and Worldwide Cancer Research (WCR16-101). We also thank all the cohorts, funders, and investigators who contributed to the melanoma GWAS, as originally acknowledged by Landi, 23andMe, and colleagues;14 data from this GWAS were used toward fine-mapping. S.F.A.G. is supported by NIH R01 HG010067 and the Daniel B. Burke Endowed Chair for Diabetes Research. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Declaration of interests
The authors declare no competing interests.
Published: August 2, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.07.002.
Data and code availability
Conditional fine-mapping data are available in Tables S7–S10; Bayesian fine-mapping results are available in Tables S11 and S12. Melanocyte ATAC-seq peaks for the region are available in Table S13. MotifbreakR analysis is provided in Table S14. Data from the 2020 melanoma GWAS meta-analysis performed by Landi and colleagues was obtained from dbGaP (dbGaP: phs001868.v1.p1), with the exclusion of self-reported data from 23andMe and UK Biobank. The full GWAS summary statistics for the 23andMe discovery dataset will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please visit the 23andMe website for more information and to apply to access the data. Summary data from the remaining self-reported cases are available from the corresponding authors of that manuscript (Matthew Law, matthew.law@qimrberghofer.edu.au; Mark Iles, m.m.iles@leeds.ac.uk; and Maria Teresa Landi, landim@mail.nih.gov). Melanocyte eQTL data are available from dbGaP (dbGaP: phs001500.v1.p1), and pre-analyzed cis-eQTL data from GTEx (v.8) were downloaded from the GTEx portal. MPRA data are available in the NCBI Gene Expression Omnibus as a SuperSeries under the accession number GEO: GSE129250. Restriction fragments baited for region-specific Capture-C assays are provided in Table S2, and loops called by ChICAGO are in Tables S17 and S18. gRNAs for the pooled gene-based CRISPR screen are provided in Table S5, and screen read counts for each and MaGeCK output are available in Tables S6 and S20, respectively. Luciferase assay fragments and other primers and gRNAs are listed in Tables S1, S3, and S4, respectively.
Web resources
23andMe, https://research.23andme.com/collaborate/#dataset-access/
ENCODE ATAC-seq pipeline v.1.6.1, https://www.encodeproject.org/atac-seq/
GTEx portal, https://www.gtexportal.org/home/
LDLink, https://ldlink.nci.nih.gov/
MotifDb, https://bioconductor.org/packages/release/bioc/html/MotifDb.html
NCBI GEO, https://www.ncbi.nlm.nih.gov/geo/
RegulomeDB, https://regulomedb.org/regulome-search/
TWAS-Hub, http://twas-hub.org/genes/AHR/
Supplemental information
References
- 1.Henley S.J., Ward E.M., Scott S., Ma J., Anderson R.N., Firth A.U., Thomas C.C., Islami F., Weir H.K., Lewis D.R. Annual report to the nation on the status of cancer, part I: National cancer statistics. Cancer. 2020;126:2225–2249. doi: 10.1002/cncr.32802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elwood J.M. Recent developments in melanoma epidemiology, 1993. Melanoma Res. 1993;3:149–156. doi: 10.1097/00008390-199306000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Mucci L.A., Hjelmborg J.B., Harris J.R., Czene K., Havelick D.J., Scheike T., Graff R.E., Holst K., Möller S., Unger R.H. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries. JAMA. 2016;315:68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Law M.H., Bishop D.T., Lee J.E., Brossard M., Martin N.G., Moses E.K., Song F., Barrett J.H., Kumar R., Easton D.F. Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nat. Genet. 2015;47:987–995. doi: 10.1038/ng.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gudbjartsson D.F., Sulem P., Stacey S.N., Goldstein A.M., Rafnar T., Sigurgeirsson B., Benediktsdottir K.R., Thorisdottir K., Ragnarsson R., Sveinsdottir S.G. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat. Genet. 2008;40:886–891. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 6.Brown K.M., Macgregor S., Montgomery G.W., Craig D.W., Zhao Z.Z., Iyadurai K., Henders A.K., Homer N., Campbell M.J., Stark M. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat. Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop D.T., Demenais F., Iles M.M., Harland M., Taylor J.C., Corda E., Randerson-Moor J., Aitken J.F., Avril M.F., Azizi E. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falchi M., Bataille V., Hayward N.K., Duffy D.L., Bishop J.A., Pastinen T., Cervino A., Zhao Z.Z., Deloukas P., Soranzo N. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat. Genet. 2009;41:915–919. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iles M.M., Law M.H., Stacey S.N., Han J., Fang S., Pfeiffer R., Harland M., Macgregor S., Taylor J.C., Aben K.K. A variant in FTO shows association with melanoma risk not due to BMI. Nat. Genet. 2013;45:428–432. doi: 10.1038/ng.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rafnar T., Sulem P., Stacey S.N., Geller F., Gudmundsson J., Sigurdsson A., Jakobsdottir M., Helgadottir H., Thorlacius S., Aben K.K. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macgregor S., Montgomery G.W., Liu J.Z., Zhao Z.Z., Henders A.K., Stark M., Schmid H., Holland E.A., Duffy D.L., Zhang M. Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat. Genet. 2011;43:1114–1118. doi: 10.1038/ng.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett J.H., Iles M.M., Harland M., Taylor J.C., Aitken J.F., Andresen P.A., Akslen L.A., Armstrong B.K., Avril M.F., Azizi E. Genome-wide association study identifies three new melanoma susceptibility loci. Nat. Genet. 2011;43:1108–1113. doi: 10.1038/ng.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ransohoff K.J., Wu W., Cho H.G., Chahal H.C., Lin Y., Dai H.J., Amos C.I., Lee J.E., Tang J.Y., Hinds D.A. Two-stage genome-wide association study identifies a novel susceptibility locus associated with melanoma. Oncotarget. 2017;8:17586–17592. doi: 10.18632/oncotarget.15230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landi M.T., Bishop D.T., MacGregor S., Machiela M.J., Stratigos A.J., Ghiorzo P., Brossard M., Calista D., Choi J., Fargnoli M.C. Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility. Nat. Genet. 2020;52:494–504. doi: 10.1038/s41588-020-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duffy D.L., Zhu G., Li X., Sanna M., Iles M.M., Jacobs L.C., Evans D.M., Yazar S., Beesley J., Law M.H. Novel pleiotropic risk loci for melanoma and nevus density implicate multiple biological pathways. Nat. Commun. 2018;9:4774. doi: 10.1038/s41467-018-06649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duffy D.L., Iles M.M., Glass D., Zhu G., Barrett J.H., Höiom V., Zhao Z.Z., Sturm R.A., Soranzo N., Hammond C. IRF4 variants have age-specific effects on nevus count and predispose to melanoma. Am. J. Hum. Genet. 2010;87:6–16. doi: 10.1016/j.ajhg.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duffy D.L., Zhao Z.Z., Sturm R.A., Hayward N.K., Martin N.G., Montgomery G.W. Multiple pigmentation gene polymorphisms account for a substantial proportion of risk of cutaneous malignant melanoma. J. Invest. Dermatol. 2010;130:520–528. doi: 10.1038/jid.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beaumont K.A., Shekar S.N., Newton R.A., James M.R., Stow J.L., Duffy D.L., Sturm R.A. Receptor function, dominant negative activity and phenotype correlations for MC1R variant alleles. Hum. Mol. Genet. 2007;16:2249–2260. doi: 10.1093/hmg/ddm177. [DOI] [PubMed] [Google Scholar]
- 19.Tsetskhladze Z.R., Canfield V.A., Ang K.C., Wentzel S.M., Reid K.P., Berg A.S., Johnson S.L., Kawakami K., Cheng K.C. Functional assessment of human coding mutations affecting skin pigmentation using zebrafish. PLoS ONE. 2012;7:e47398. doi: 10.1371/journal.pone.0047398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halaban R., Svedine S., Cheng E., Smicun Y., Aron R., Hebert D.N. Endoplasmic reticulum retention is a common defect associated with tyrosinase-negative albinism. Proc. Natl. Acad. Sci. USA. 2000;97:5889–5894. doi: 10.1073/pnas.97.11.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu F., Visser M., Duffy D.L., Hysi P.G., Jacobs L.C., Lao O., Zhong K., Walsh S., Chaitanya L., Wollstein A. Genetics of skin color variation in Europeans: genome-wide association studies with functional follow-up. Hum. Genet. 2015;134:823–835. doi: 10.1007/s00439-015-1559-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Visconti A., Duffy D.L., Liu F., Zhu G., Wu W., Chen Y., Hysi P.G., Zeng C., Sanna M., Iles M.M. Genome-wide association study in 176,678 Europeans reveals genetic loci for tanning response to sun exposure. Nat. Commun. 2018;9:1684. doi: 10.1038/s41467-018-04086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chahal H.S., Lin Y., Ransohoff K.J., Hinds D.A., Wu W., Dai H.J., Qureshi A.A., Li W.Q., Kraft P., Tang J.Y. Genome-wide association study identifies novel susceptibility loci for cutaneous squamous cell carcinoma. Nat. Commun. 2016;7:12048. doi: 10.1038/ncomms12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haarmann-Stemmann T., Esser C., Krutmann J. The Janus-Faced Role of Aryl Hydrocarbon Receptor Signaling in the Skin: Consequences for Prevention and Treatment of Skin Disorders. J. Invest. Dermatol. 2015;135:2572–2576. doi: 10.1038/jid.2015.285. [DOI] [PubMed] [Google Scholar]
- 25.Vogeley C., Esser C., Tüting T., Krutmann J., Haarmann-Stemmann T. Role of the Aryl Hydrocarbon Receptor in Environmentally Induced Skin Aging and Skin Carcinogenesis. Int. J. Mol. Sci. 2019;20:6005. doi: 10.3390/ijms20236005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jux B., Kadow S., Luecke S., Rannug A., Krutmann J., Esser C. The aryl hydrocarbon receptor mediates UVB radiation-induced skin tanning. J. Invest. Dermatol. 2011;131:203–210. doi: 10.1038/jid.2010.269. [DOI] [PubMed] [Google Scholar]
- 27.Luecke S., Backlund M., Jux B., Esser C., Krutmann J., Rannug A. The aryl hydrocarbon receptor (AHR), a novel regulator of human melanogenesis. Pigment Cell Melanoma Res. 2010;23:828–833. doi: 10.1111/j.1755-148X.2010.00762.x. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura M., Ueda Y., Hayashi M., Kato H., Furuhashi T., Morita A. Tobacco smoke-induced skin pigmentation is mediated by the aryl hydrocarbon receptor. Exp. Dermatol. 2013;22:556–558. doi: 10.1111/exd.12170. [DOI] [PubMed] [Google Scholar]
- 29.Beischlag T.V., Luis Morales J., Hollingshead B.D., Perdew G.H. The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abbas S., Alam S., Singh K.P., Kumar M., Gupta S.K., Ansari K.M. Aryl Hydrocarbon Receptor Activation Contributes to Benzanthrone-Induced Hyperpigmentation via Modulation of Melanogenic Signaling Pathways. Chem. Res. Toxicol. 2017;30:625–634. doi: 10.1021/acs.chemrestox.6b00364. [DOI] [PubMed] [Google Scholar]
- 31.Contador-Troca M., Alvarez-Barrientos A., Barrasa E., Rico-Leo E.M., Catalina-Fernández I., Menacho-Márquez M., Bustelo X.R., García-Borrón J.C., Gómez-Durán A., Sáenz-Santamaría J., Fernández-Salguero P.M. The dioxin receptor has tumor suppressor activity in melanoma growth and metastasis. Carcinogenesis. 2013;34:2683–2693. doi: 10.1093/carcin/bgt248. [DOI] [PubMed] [Google Scholar]
- 32.Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehár J., Kryukov G.V., Sonkin D. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corre S., Tardif N., Mouchet N., Leclair H.M., Boussemart L., Gautron A., Bachelot L., Perrot A., Soshilov A., Rogiers A. Sustained activation of the Aryl hydrocarbon Receptor transcription factor promotes resistance to BRAF-inhibitors in melanoma. Nat. Commun. 2018;9:4775. doi: 10.1038/s41467-018-06951-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang J., Ferreira T., Morris A.P., Medland S.E., Madden P.A.F., Heath A.C., Martin N.G., Montgomery G.W., Weedon M.N., Loos R.J. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen X., Lee Y., Luca F., Pique-Regi R. Efficient Integrative Multi-SNP Association Analysis via Deterministic Approximation of Posteriors. Am. J. Hum. Genet. 2016;98:1114–1129. doi: 10.1016/j.ajhg.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee Y., Luca F., Pique-Regi R., Wen X. Bayesian Multi-SNP Genetic Association Analysis: Control of FDR and Use of Summary Statistics. bioRxiv. 2018 doi: 10.1101/316471. [DOI] [Google Scholar]
- 37.Halaban R., Cheng E., Smicun Y., Germino J. Deregulated E2F transcriptional activity in autonomously growing melanoma cells. J. Exp. Med. 2000;191:1005–1016. doi: 10.1084/jem.191.6.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi J., Zhang T., Vu A., Ablain J., Makowski M.M., Colli L.M., Xu M., Hennessey R.C., Yin J., Rothschild H. Massively parallel reporter assays of melanoma risk variants identify MX2 as a gene promoting melanoma. Nat. Commun. 2020;11:2718. doi: 10.1038/s41467-020-16590-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang T., Choi J., Kovacs M.A., Shi J., Xu M., Goldstein A.M., Trower A.J., Bishop D.T., Iles M.M., Duffy D.L. Cell-type-specific eQTL of primary melanocytes facilitates identification of melanoma susceptibility genes. Genome Res. 2018;28:1621–1635. doi: 10.1101/gr.233304.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langmead B., Wilks C., Antonescu V., Charles R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics. 2019;35:421–432. doi: 10.1093/bioinformatics/bty648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amemiya H.M., Kundaje A., Boyle A.P. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci. Rep. 2019;9:9354. doi: 10.1038/s41598-019-45839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li D., Hsu S., Purushotham D., Sears R.L., Wang T. WashU Epigenome Browser update 2019. Nucleic Acids Res. 2019;47(W1):W158–W165. doi: 10.1093/nar/gkz348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wingett S., Ewels P., Furlan-Magaril M., Nagano T., Schoenfelder S., Fraser P., Andrews S. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res. 2015;4:1310. doi: 10.12688/f1000research.7334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cairns J., Freire-Pritchett P., Wingett S.W., Várnai C., Dimond A., Plagnol V., Zerbino D., Schoenfelder S., Javierre B.M., Osborne C. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 2016;17:127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naumova N., Smith E.M., Zhan Y., Dekker J. Analysis of long-range chromatin interactions using Chromosome Conformation Capture. Methods. 2012;58:192–203. doi: 10.1016/j.ymeth.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hart T., Tong A.H.Y., Chan K., Van Leeuwen J., Seetharaman A., Aregger M., Chandrashekhar M., Hustedt N., Seth S., Noonan A. Evaluation and Design of Genome-Wide CRISPR/SpCas9 Knockout Screens. G3 (Bethesda) 2017;7:2719–2727. doi: 10.1534/g3.117.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chari R., Mali P., Moosburner M., Church G.M. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods. 2015;12:823–826. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Q., Schlabach M.R., Hannon G.J., Elledge S.J. Design of 240,000 orthogonal 25mer DNA barcode probes. Proc. Natl. Acad. Sci. USA. 2009;106:2289–2294. doi: 10.1073/pnas.0812506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coetzee S.G., Coetzee G.A., Hazelett D.J. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics. 2015;31:3847–3849. doi: 10.1093/bioinformatics/btv470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Machiela M.J., Chanock S.J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Consortium E.P., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mancuso N., Shi H., Goddard P., Kichaev G., Gusev A., Pasaniuc B. Integrating Gene Expression with Summary Association Statistics to Identify Genes Associated with 30 Complex Traits. Am. J. Hum. Genet. 2017;100:473–487. doi: 10.1016/j.ajhg.2017.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mengoni M., Braun A.D., Gaffal E., Tüting T. The aryl hydrocarbon receptor promotes inflammation-induced dedifferentiation and systemic metastatic spread of melanoma cells. Int. J. Cancer. 2020;147:2902–2913. doi: 10.1002/ijc.33252. [DOI] [PubMed] [Google Scholar]
- 61.Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., Irizarry R.A., Liu J.S., Brown M., Liu X.S. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi J., Xu M., Makowski M.M., Zhang T., Law M.H., Kovacs M.A., Granzhan A., Kim W.J., Parikh H., Gartside M. A common intronic variant of PARP1 confers melanoma risk and mediates melanocyte growth via regulation of MITF. Nat. Genet. 2017;49:1326–1335. doi: 10.1038/ng.3927. [DOI] [PubMed] [Google Scholar]
- 63.Ceol C.J., Houvras Y., Jane-Valbuena J., Bilodeau S., Orlando D.A., Battisti V., Fritsch L., Lin W.M., Hollmann T.J., Ferré F. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hormozdiari F., Kostem E., Kang E.Y., Pasaniuc B., Eskin E. Identifying causal variants at loci with multiple signals of association. Genetics. 2014;198:497–508. doi: 10.1534/genetics.114.167908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghosh K., Capell B.C. The Senescence-Associated Secretory Phenotype: Critical Effector in Skin Cancer and Aging. J. Invest. Dermatol. 2016;136:2133–2139. doi: 10.1016/j.jid.2016.06.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Acosta J.C., Banito A., Wuestefeld T., Georgilis A., Janich P., Morton J.P., Athineos D., Kang T.W., Lasitschka F., Andrulis M. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol. 2013;15:978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sadik A., Somarribas Patterson L.F., Ozturk S., Mohapatra S.R., Panitz V., Secker P.F., Pfander P., Loth S., Salem H., Prentzell M.T. IL4I1 Is a Metabolic Immune Checkpoint that Activates the AHR and Promotes Tumor Progression. Cell. 2020;182:1252–1270.e34. doi: 10.1016/j.cell.2020.07.038. [DOI] [PubMed] [Google Scholar]
- 68.Stockinger B., Di Meglio P., Gialitakis M., Duarte J.H. The aryl hydrocarbon receptor: multitasking in the immune system. Annu. Rev. Immunol. 2014;32:403–432. doi: 10.1146/annurev-immunol-032713-120245. [DOI] [PubMed] [Google Scholar]
- 69.Vorrink S.U., Domann F.E. Regulatory crosstalk and interference between the xenobiotic and hypoxia sensing pathways at the AhR-ARNT-HIF1α signaling node. Chem. Biol. Interact. 2014;218:82–88. doi: 10.1016/j.cbi.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liyanage U.E., Law M.H., Han X., An J., Ong J.S., Gharahkhani P., Gordon S., Neale R.E., Olsen C.M., MacGregor S., Whiteman D.C., 23andMe Research Team Combined analysis of keratinocyte cancers identifies novel genome-wide loci. Hum. Mol. Genet. 2019;28:3148–3160. doi: 10.1093/hmg/ddz121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Conditional fine-mapping data are available in Tables S7–S10; Bayesian fine-mapping results are available in Tables S11 and S12. Melanocyte ATAC-seq peaks for the region are available in Table S13. MotifbreakR analysis is provided in Table S14. Data from the 2020 melanoma GWAS meta-analysis performed by Landi and colleagues was obtained from dbGaP (dbGaP: phs001868.v1.p1), with the exclusion of self-reported data from 23andMe and UK Biobank. The full GWAS summary statistics for the 23andMe discovery dataset will be made available through 23andMe to qualified researchers under an agreement with 23andMe that protects the privacy of the 23andMe participants. Please visit the 23andMe website for more information and to apply to access the data. Summary data from the remaining self-reported cases are available from the corresponding authors of that manuscript (Matthew Law, matthew.law@qimrberghofer.edu.au; Mark Iles, m.m.iles@leeds.ac.uk; and Maria Teresa Landi, landim@mail.nih.gov). Melanocyte eQTL data are available from dbGaP (dbGaP: phs001500.v1.p1), and pre-analyzed cis-eQTL data from GTEx (v.8) were downloaded from the GTEx portal. MPRA data are available in the NCBI Gene Expression Omnibus as a SuperSeries under the accession number GEO: GSE129250. Restriction fragments baited for region-specific Capture-C assays are provided in Table S2, and loops called by ChICAGO are in Tables S17 and S18. gRNAs for the pooled gene-based CRISPR screen are provided in Table S5, and screen read counts for each and MaGeCK output are available in Tables S6 and S20, respectively. Luciferase assay fragments and other primers and gRNAs are listed in Tables S1, S3, and S4, respectively.