Summary
Transportin-2 (TNPO2) mediates multiple pathways including non-classical nucleocytoplasmic shuttling of >60 cargoes, such as developmental and neuronal proteins. We identified 15 individuals carrying de novo coding variants in TNPO2 who presented with global developmental delay (GDD), dysmorphic features, ophthalmologic abnormalities, and neurological features. To assess the nature of these variants, functional studies were performed in Drosophila. We found that fly dTnpo (orthologous to TNPO2) is expressed in a subset of neurons. dTnpo is critical for neuronal maintenance and function as downregulating dTnpo in mature neurons using RNAi disrupts neuronal activity and survival. Altering the activity and expression of dTnpo using mutant alleles or RNAi causes developmental defects, including eye and wing deformities and lethality. These effects are dosage dependent as more severe phenotypes are associated with stronger dTnpo loss. Interestingly, similar phenotypes are observed with dTnpo upregulation and ectopic expression of TNPO2, showing that loss and gain of Transportin activity causes developmental defects. Further, proband-associated variants can cause more or less severe developmental abnormalities compared to wild-type TNPO2 when ectopically expressed. The impact of the variants tested seems to correlate with their position within the protein. Specifically, those that fall within the RAN binding domain cause more severe toxicity and those in the acidic loop are less toxic. Variants within the cargo binding domain show tissue-dependent effects. In summary, dTnpo is an essential gene in flies during development and in neurons. Further, proband-associated de novo variants within TNPO2 disrupt the function of the encoded protein. Hence, TNPO2 variants are causative for neurodevelopmental abnormalities.
Keywords: Transportin, Drosophila, TNPO1, TNPO2, intellectual disability, global developmental delays, nucleocytoplasmic shuttling, rare disease, Karyopherin-β2b, Importin-3
Introduction
Genomic sequencing in combination with functional investigations in model organisms has led to the discovery of numerous novel Mendelian diseases.1,2 Functional investigations may be particularly impactful when considering contributions of potential disease-associated variants that occur in genes encoding pleiotropic proteins,3,4 defined as proteins that function in a diverse number of unrelated pathways.
Here, we identified Transportin-2 (TNPO2 [MIM: 603002]; Importin-3; Karyopherin-β2b) as a disease-associated gene. TNPO2 primarily mediates a non-classical nucleocytoplasmic shuttling pathway.5,6 TNPO2 activity is dependent on the Ras-related nuclear protein (RAN) GTP/GDP gradient.7 During nucleocytoplasmic shuttling, TNPO2 is bound by RAN-GDP at its N terminus, promoting interactions with cytoplasmic protein cargoes at its C terminus.5,6 Subsequently, the RAN-GDP:TNPO2:cargo complex is shuttled into the nucleus via the nuclear pore complex (NPC). Conversion of RAN-GDP to RAN-GTP in the nucleus causes a conformational change in TNPO2’s acidic loop—a flexible domain found between the RAN and cargo binding domains. This releases the cargo. RAN-GTP:TNPO2 is then shuttled back to the cytoplasm, destined to repeat the process.
TNPO2 is closely related to Transportin-1 (TNPO1 [MIM: 602901]; Importin-2; Karyopherin-β2)5 and neither gene has been associated with a Mendelian disease. TNPO2 is the less studied of the two as it was discovered later. Human TNPO2 and TNPO1 protein sequences are 84% identical and 92% similar.5 Differences primarily occur in their flexible acidic loops and, to a lesser extent, their cargo-binding domains.8 Current data support that TNPO2 and TNPO1 are functionally redundant.5 Although the two genes are expressed ubiquitously, they differ in their expression levels in different tissues. Expression profiling data in mice demonstrated that TNPO2 is more highly expressed in the brain than TNPO1.9 These results are consistent with other mammalian datasets.5 At the protein level, TNPO2 is more abundant in cultured neurons, astrocytes, and neural stem cells than TNPO1.10 TNPO2 may also be more critical in muscles as TNPO1 is not detected in cultured myoblasts.11 TNPO2 is required during myoblast differentiation into myotubes.11
More than 150 proteins are predicted to interact with TNPO1/2 based on high-throughput studies and more than 60 proteins have been confirmed as TNPO1 cargoes.5,12,13 Cargoes confirmed to be shuttled by TNPO2 include FUS (MIM: 137070),14 HuR/ElavL1 (MIM: 603466),8,11,15,16 hnRNPA1 (MIM: 164017),8,17 and NF-κB Essential Modulator (NEMO [MIM: 300248]).18 All of these are also TNPO1 cargoes. Recent high-throughput studies have detected rare proteins that uniquely interact with TNPO212,13 but direct investigations are needed to confirm them as TNPO2-specific cargoes.
The majority of TNPO1/2 cargoes carry a non-canonical nuclear-localization signal (NLS), a PY-NLS, defined as a C-terminal R/H/K-X2-5PY motif.17 However, a large number of cargoes do not have a PY-NLS and are simply described as being structurally disordered and having a hydrophobic or basic N-terminal sequence.5,19 RNA-binding proteins and transcription factors needing import into the nucleus to regulate expression of a diverse number of genes are common targets of TNPO1/25. Other nucleus-bound cargoes include histones, splicing factors, and ribosomal proteins.5,12,13 TNPO1/2 also interacts with ciliary proteins,20, 21, 22 spindle assembly factors,23,24 and nucleoporins,23,25,26 shuttling these cargoes to the appropriate region of the cell for them to function. This means TNPO1/2 activity directly impacts ciliogenesis, mitotic spindle assembly, and nuclear envelope and pore assembly. Last, TNPO1/2 has been implicated in mechanisms that promote aging and neurodegenerative diseases.14,27, 28, 29
Here we characterize a cohort that carry de novo variants within TNPO2, finding that common features include developmental and neurological abnormalities. Using Drosophila to perform functional studies, we provide evidence that de novo, pathogenic variants in TNPO2 are the genetic causes of individuals’ symptoms.
Material and methods
Recruitment and sequencing of individuals
Fifteen individuals were recruited through the Undiagnosed Diseases Network (UDN)30 and GeneMatcher.31 The procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national). Proper informed consent was obtained from family members for all probands in this study. All of the UDN work, including clinical and model organism work, and coordination for this publication, was performed under NIH IRB protocol 15-HG-0130.
Sequencing details for each proband can be found in Data S1. Briefly, trio (proband and both biological parents) whole-exome sequencing (WES) was done in 14 of 15 affected individuals as previously described.32, 33, 34 Trio whole-genome sequencing (WGS) used the Illumina Novaseq 6000 platform. Sequencing libraries were generated using the Truseq Nano DNA HT Sample Preparation Kit (Illumina). Alignment of 150 bp paired-end reads to the hg19 reference genome was performed using Burrows-Wheeler Aligner (BWA) software, before sorting with samtools and marking duplicates with Picard. Single-nucleotide variants (SNVs) and small insertions/deletions (indels) were labeled using Genome Analysis Tool Kit (GATK v.3.1), structural variants (SVs) were detected using DELLY (v0.7.3) software, and copy number variants (CNVs) were detected using the control-FREEC (v.9.9) tool. Following genomic variants detection, variants were annotated using ANNOVAR. Identification of genomic regions affected by each variant and possible changes in protein was performed using RefSeq and Gencode databases. The presence of the variants were assessed in dbSNP, GnomAD, 1000 Genomes Project, Exome Aggregation Consortium (ExAC), exome sequencing project (ESP), and Clinvar. Databases dbSNP, COSMIC, OMIM, GWAS Catalog, and HGMD were used to find reported information of variants. SIFT, PolyPhen, MutationAssessor, LRT, and CADD scores were used to predict the deleteriousness of mutations and GERP++ scores were used to access the conservation of mutations.
Drosophila husbandry and established fly lines
All fly lines were raised and maintained as described.35 Publicly available fly lines are detailed in Table S1 and were obtained from Vienna Drosophila Research Center (VDRC), Bloomington Drosophila Stock Center (BDSC), and Kyoto Stock Center (Kyoto). Rh1-GAL4 on II (w∗; Rh1-GAL4;), elav-GAL4 on II (y∗ w∗; elav-GAL4) and UAS-(empty) control (y w; PBac{UAS-empty}VK37/SM6a) were published previously.36,37 da-GAL4GS (w∗; P{da-GSGAL4.T};) was generously provided by H. Tricoire.38
dTnpo mutant alleles and genomic rescue line
dTnpoGly736Asp, Tnpo-RA (GenBank: NM_058020.4):c.2207G>A (p.Gly736Asp), was identified in a forward genetic screen of FRT80B isogenized flies.39 dTnpoΔ11 is an imprecise excision line derived from P{GawB}NP440840 (Kyoto #104668). A dTnpo genomic rescue construct, GRdTnpo, was cloned from the endogenous dTnpo loci using genomic DNA from isogenized FRT80B and inserted into the VK37 docking site using φC31-mediated transgenesis as described.41 The dTnpo CRIMIC (T2A-GAL4) allele was designed as part of the Gene Disruption Project (construct CR92235) as described42 using sgRNA 5′-CAAGCGTAATTTAAGAGTAATGG-3′.
UAS-hTNPO2 lines
UAS-hTNPO2 lines were developed as described.35 Q5 site directed mutagenesis (NEB # E00554S) was done on a pDONR223-hTNPO2 cDNA construct (GenBank: NM_001136196.1; Horizon Discovery # OHS1770-202312693) to introduce a stop codon and variants. Primers are detailed in Table S2. LR clonase II (ThermoFisher # 11791020) was used to transfer the cDNA sequence to a pGW-attB-3xHA destination vector,43 creating pGW-hTNPO2 constructs. All clones were PCR and sequence confirmed. Sequencing primers included ones specific to the hTNPO2 sequence (Table S2) and M13 primers. pGW-hTNPO2 constructs were inserted into the VK37 docking site using φC31-mediated transgenesis.41 Final genotype: w1118; PBac{UAS-hTNPO2}VK37/SM6a.
GeneSwitch-driven transgene expression and lifespan
da-GAL4GS and elav-GAL4GS assays were performed as previously described44 with the following changes. At 1–2 day post-eclosion, animals were placed onto 300 μM for elav-GAL4GS or 500 μM for da-GAL4GS RU486-containing food. RU486-food was prepared by mixing molten (60°C–65°C) food with 10 mM RU486 (Sigma #M8046; prepared using 200 pf ethanol) to the desired concentration at 2 mL per vial. Molten food was solidified for 1–24 h in a fume hood. For elav-GAL4GS lifespan assays, female flies were maintained at 29°C. For da-GAL4GS studies, female flies were maintained for 4 days on RU486 at 25°C.
Quantitative real-time polymerase chain reactions (qPCR)
qPCR was performed as previously described44 with the following changes. The iScript gDNA Clear cDNA Synthesis Kit (BioRad #1725034), iTaq Universal SYBR Green Master Mix (BioRad #1725120), and a BioRad C1000 Touch Cycler were used. Multiple housekeeping genes (RP49, RPS20, and Tubulin) were included for normalizing the data. qPCR primers are described in Table S2 and those for housekeeping genes were previously published.44
Immunofluorescence (IF) and confocal microscopy
IF for L3 larval CNS and adult brains was performed as described.45,46 Primary antibodies: anti-FasII (DSHB #7G10; 1:100), anti-Elav (DSHB #7E8A10; 1:500), anti-Repo (DSHB #8D12; 1:60), anti-mCherry (Genetex # GTX59788; 1:200; also targets RFP). Goat-derived secondary antibodies were used at 1:500 (Jackson ImmunoResearch Laboratories). A Leica Sp8x with lightning deconvolution was used for confocal microscopy. Images were taken with a 20× oil immersion Leica objective (HC PL APO 20x/0.75 IMM CORR CS2).
Western immunoblots (WBs)
The BioRad Mini-PROTEAN Electrophoresis System was used with 4%–20% Mini-PROTEAN TGX Precast Gel (BioRad #4561095), 1× Tris/Glycine/SDS running buffer, 1× Tris/Glycine transfer buffer with 10% methanol, and PVDF membrane. For lysates, whole frozen flies were homogenized as described44,47 into 1× SDS sample buffer at 50 μL per animal. For 1× SDS sample buffer, 60 μL of β-mercaptoethanol was added to 1 mL of diluted 6× SDS sample buffer (0.35M Tris-HCl [pH 6.8], 10% SDS, 30% glycerol, 30% β-mercaptoethanol, 1% Bromophenol Blue). 10 μL of lysate was loaded per lane. Membranes were stained and reprobed as described.44,47 Antibodies: anti-hTNPO1/2[A11] (1:1,000, Santa Cruz #sc-365179), anti-mouse-HRP (1:5,000), and anti-α-Tubulin[11H10]-HRP (1:2,000, Cell Signaling #9099). HRP activity was measured using SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Sci #34577) and a BioRad Chemidoc MP Imaging System.
Results
Coding variants in TNPO2 are associated with global developmental delay, dysmorphic features, ophthalmologic abnormalities, and neurological features
Fifteen individuals who primarily presented with feeding difficulties and developmental delays during infancy or in early childhood were evaluated clinically by their providers in the respective institution (Data S1). Trio (proband and both biological parents) sequencing, primarily whole-exome sequencing (WES), was performed by these clinical sites and results showed that these individuals carry a potentially pathogenic, heterozygous coding-variant in TNPO2 (GenBank: NM_001136196.1) (Table 1; extended data in Data S1). Based on the presence of this variant, individuals were recruited to this study through the Undiagnosed Diseases Network (UDN)44 and GeneMatcher,45 independent of their respective clinical features.
Table 1.
Individuals with TNPO2 variants present with developmental delays, intellectual disability, behavioral deficits, and strabismus
| Summary | Proband |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | ||
| Protein variant (p.) | SNV, del, delins | Gln28Arg | Gln32Arg | Pro61Arg | Lys118Asn | Lys152del, mosaic (16/21%) | Asp156Asn | Trp370Arg | Trp370Cys | Lys491_Arg492delinsGlnTrp | Pro514Leu | Ala546Val | Ser548Phe | Phe598Leu | Ala649_Leu652del | Trp727Cys |
| CADD score | 22.7–34.0 | 27.8 | 24.3 | 23.9 | 22.7 | – | 27.1 | 34.0 | 28.8 | – | 30.0 | 27.0 | 31.0 | 29.4 | – | 25.5 |
| Sanger confirmed? | 10 of 15 | yes | yes | – | yes | yes | – | yes | yes | yes | – | yes | – | yes | – | yes |
| Inheritance | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | de novo | mother, mosaic (1%) | de novo | de novo |
| Additional variants of uncertain significance | 6 of 15 | – | – | – | – | SETBP1:p.Leu1522Argfs∗59; CUX2: p.His1253Pro; 12q13.13_dup | ANKFY1: p.Thr1088Serfs∗9 | – | ARMC9: p.Asp330Asn | – | 1q21.1_ins522Kb | – | – | – | PDE4D: p.Arg237∗ | INA p.Leu376Pro, mosaic (20%) |
| Age at onset | neo-18mo | 1mo | 4mo | neonat. | 13mo | 3mo | 6mo | 4mo | neonatal | prenatal | 9mo | 8mo | neonat. | 15mo | 18mo | neonat. |
| Age at exam | 14mo-20y | 6y | 18mo | 6mo | 3y | 23mo | 4y | 10y | 8y | 14mo | 5y | 9y | 20y | 11y | 12y | 7y |
| Global developmental delays | 15 of 15 | ++ | +, regress. | + | ++, regress. | +++ | ++ | +++ | + | + | ++ | + | +++ | +, regress. | + | ++ |
| Speech impaired | 15 of 15 | ++ | + | + | ++ | +++ | +++ | +++ | + | + | ++ | + | +++ | + | + | ++ |
| Intellectual disability | 9 of 9 | +++ | ND | ND | +++ | ND | ++ | +++ | ++ | ND | ND | ++ | +++ | + | ++ | ND |
| Motor Impaired | 15 of 15 | + | + | + | ++ | +++ | ++ | +++ | + | + | + | + | +++ | + | + | + |
| Dysmorphic features | 11 of 15 | + | – | + | + | – | + | – | + | + | + | – | + | + | + | + |
| Behavioral deficits | 10 of 14 | + | + | + | + | – | + | – | + | – | – | + | ND | + | + | + |
| GI / feeding abnormalities | 11 of 15 | + | + | + | – | + | – | + | + | – | + | – | + | + | + | + |
| Ophthalmologic abnormalities | 10 of 15 | + | – | – | + | + | + | – | + | + | + | + | – | + | – | + |
| Muscle tone abnormalities | 11 of 15 | +, hypo | – | +, hypo | – | +, hypo | +, variable | +, hypo | – | +, hypo | +, variable | – | +, hypo | +, hypo | +, hypo | +, hyper |
| Movement /neurological disorder | 6 of 15 | + | – | – | – | + | + | – | – | – | – | + | + | + | – | - |
| Seizures | 6 of 15 | +, febrile | – | – | – | – | +, febrile to non-febrile | +, febrile to non-febrile | – | – | +, febrile to non-febrile | +, febrile to non-febrile | + | – | – | – |
| Microcephaly | 5 of 15 | + | – | – | + | + | + | – | – | + | – | – | – | – | – | – |
| MRI brain abnormalities | 7 of 13 | – | ND | + | – | + | + | – | + | + | + | – | + | – | ND | – |
misZ for TNPO2 loss is 5.88 (o/e = 0.28). pLI for TNPO2 loss is 1.00 (o/e = 0.04). TNPO2 coding DNA (GenBank: NM_001136196.1). All individuals are heterozygous for variants. No variants are found in control genetic databases. See Data S1 and Note S1 for additional details on probands’ features and additional variants of uncertain significance. ND, no data; CADD, combined annotation dependent depletion.
All variants are de novo except the one in proband 13, whose mother was low-level mosaic. The variant is in 1% of NGS reads in the mother by WES. To learn more about TNPO2 and potential impact of these variants, we used information accumulated into the Model organism Aggregated Resources or Rare Variant ExpLoration (MARRVEL) tool, v.2.48 MARRVEL is a valuable resource that brings together multiple sources of information for the investigation of human and model organism based disease research. Here, we found that TNPO2 is highly constrained, having a missense constraint (misZ) score49 of 5.88 (observed/expected (o/e) = 0.28) and a probability of loss-of-function intolerance (pLI) score49,50 of 1.00 (o/e = 0.04) based on gnomAD (genome Aggregation Database), v.2.1.1.51 Twelve probands carry single-nucleotide variants (SNVs) in TNPO2 that are predicted to be deleterious using combined annotation dependent depletion (CADD) scores (phred > 20), v.1.4.52 Proband 5 carries a mosaic, in-frame deletion of p.Lys152del (16% by Sanger, 21% by WGS of reads, DNA from blood). Proband 9 carries a deletion-insertion of p.Lys491_Arg492delinsGlnTrp. Proband 14 carries an in-frame deletion of p.Ala649_Leu652, removing four codons. None of these variants are found in genetic databases containing control populations, including information in gnomAD.51
To evaluate common features among probands, information was extracted from chart review as well as clinic visits at the respective institutions. We found that all probands present with global developmental delay (GDD), with either slow or plateaued developmental progress (Tables 1 and S1). Probands 2, 4, and 13 show regression of milestones, mostly transiently. All probands have delayed speech, with expressive language more severely impacted compared with receptive speech. Four individuals are nonverbal (+++) including proband 7 at age 10 years and proband 12 at age 20 years. Motor impairments appear to be comparatively less severe in our cohort compared with speech delays, although probands 5, 7, and 12 are non-ambulatory (+++). Intellectual disability (ID) was assessed and found in nine probands, ranging from mild (+) to severe (+++). ID is also suspected in another three individuals.
Behavioral deficits are observed in 10 of 14 probands with variable presentation (Tables 1 and S1). The most common neuropsychiatric concerns are inattention and autistic behaviors. Proband 12 is severely delayed so behavioral analysis was not done.
Gastrointestinal (GI) features appear to be shared within the cohort, impacting 11 of 15 probands (Tables 1 and S1). The most common features include neonatal feeding difficulties and poor weight gain.
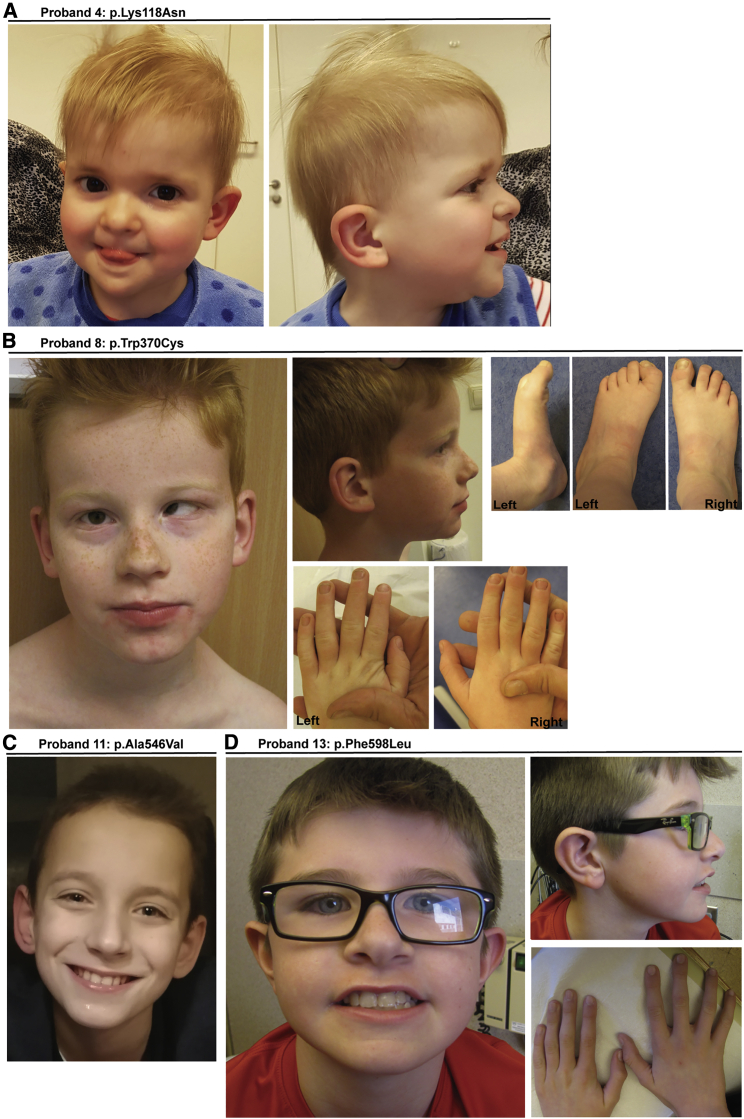
No single craniofacial dysmorphism is reported across the cohort, although dysmorphic features are noted in 11 of 15 individuals (Figure 1; Tables 1 and S1). The most common abnormalities include a broad or high nasal bridge, retrognathia, and a shortened philtrum. Skull and facial features include dolichocephaly, bitemporal narrowing or narrow face/high arched palate in four probands, and microcephaly (defined as less than –2 standard deviations; SD) in five probands. Noticeably, dysmorphisms surrounding the eye area are observed in five probands although presentation varies. This includes deep-set eyes and palpebral fissure length, spacing, or slant irregularities. Dysmorphic ears are also noted in six probands.
Figure 1.
TNPO2 variants are associated with varied dysmorphic features in individuals
(A) Proband 4 at age 3 years with short philtrum, broad nasal bridge, large fleshy ears, and coarse facial features.
(B) Proband 8 at age 8 years with strabismus, high nasal bridge, eversion of the lower lip, and clinodactyly.
(C) Proband 11 at age 9 years has no clear dysmorphism.
(D) Proband 13 at age 11 years with deep set eyes and large cupped ears.
Ophthalmologic abnormalities are reported within the cohort and impact 10 of 15 individuals (Tables 1 and S1). Strabismus is observed in seven probands. Saccadic and rapid eye movements are noted in three individuals, resolving in proband 5 by age 23 months. Myopia, hyperopia, or astigmatism are described in four probands.
Muscle tone abnormalities are described in 11 of 15 individuals, primarily hypotonia. Interestingly, probands 6 and 10 show signs of both hypertonia and hypotonia. In addition, movement and neurological disorders, primarily tremors and ataxia, were seen in 6 of 15 probands.
Neurologic impairments are detected in some probands (Tables 1 and S1). Of the 15 individuals, 6 had seizures starting between 1 and 2.5 years of age. Initial presentation in five of these individuals was febrile induced and in four of five, individuals developed non-febrile seizures. Electroencephalograms (EEGs) were abnormal in three of ten probands assessed, with proband 7 showing severe abnormalities consistent with epileptic encephalopathy. Magnetic resonance imaging (MRI) of the brain was done on 13 probands and cerebellar hypoplasia or dysplasia were seen in three probands. Tnpo2 is highly expressed in this region in mice.53 Other findings include white matter loss, mild ventricular dilation, hypoplastic caudate nuclei, thin corpus callosum, as well as minor anomalies such as cavum septum pellucidum, enlarged Virchow Robin spaces, and borderline delay in myelination.
Other, less common features for individuals are also observed. This includes renal abnormalities (bilateral pyeloureteral junction stenosis requiring surgery at age 3 months in proband 4, left kidney agenesis in proband 8, and kidney stones in proband 12), nipple abnormalities, cardiac abnormalities (patent ductus arteriosus requiring transcatheter closure in proband 7, mild dilation of the aortic root in proband 12), finger anomalies, hip dysplasia, (kypho)scoliosis, and pes planus (Data S1).
Of 15 probands, 6 carry additional heterozygous, de novo genomic alterations (variants of uncertain significance; VUSs) which were not the primary candidate for further investigation (detailed in Note S1). Briefly, probands 6, 8, and 15 carry SNVs in Rabankyrin-5 (ANKFY1 [MIM: 607927]; no disease association), Armadillo repeat containing 9 (ARMC9 [MIM: 617612]; associated with autosomal-recessive Joubert syndrome 30 [MIM: 617622]), and α-Internexin (INA [MIM: 605338]; no disease association), respectively. Importantly, these genes are less constrained than TNPO2 (see Note S1). Proband 5 carries three VUSs in addition to the one in TNPO2. Two are not predicted to be pathogenic based on CADD and other information (see Note S1), including a SNV in Cut-like Homeobox 2 (CUX2 [MIM: 610648]; associated with autosomal-dominant developmental and epileptic encephalopathy 67 [MIM: 618141]) and a duplication of 12q13.13. The third is a a deletion-insertion in SET binding protein 1 (SETBP1 [MIM: 611060]) that occurs considerably further down in the gene from known pathogenic variants associated with mental retardation, autosomal dominant 2954 and this individual has no suggestive features for Schinzel-Giedion syndrome55 ([MIM: 269150]; autosomal dominant). Proband 10 carries multiple VUSs (see Note S1), most notably a 522 Kb gain in 1q21.1. No impacted genes were thought to explain the individual’s features. Proband 14 carries a truncating SNV in the highly constrained gene Phosphodiesterase-4D (PDE4D [MIM: 600129]) and is diagnosed with acrodysostosis 256 (ACRDYS2 [MIM: 614613]).
In summary, 15 individuals were identified who carry potential disease-causing variants in TNPO2. All individuals present with global developmental delays. Speech abilities and intelligence are typically more impaired than motor abilities. Other common features between probands include variable dysmorphic features, ophthalmologic abnormalities (primarily strabismus), muscle tone abnormalities (primarily hypotonia), movement/neurological disorders, and neurological features.
Drosophila Tnpo is orthologous to human TNPO2 and most proband variants affect evolutionarily conserved residues
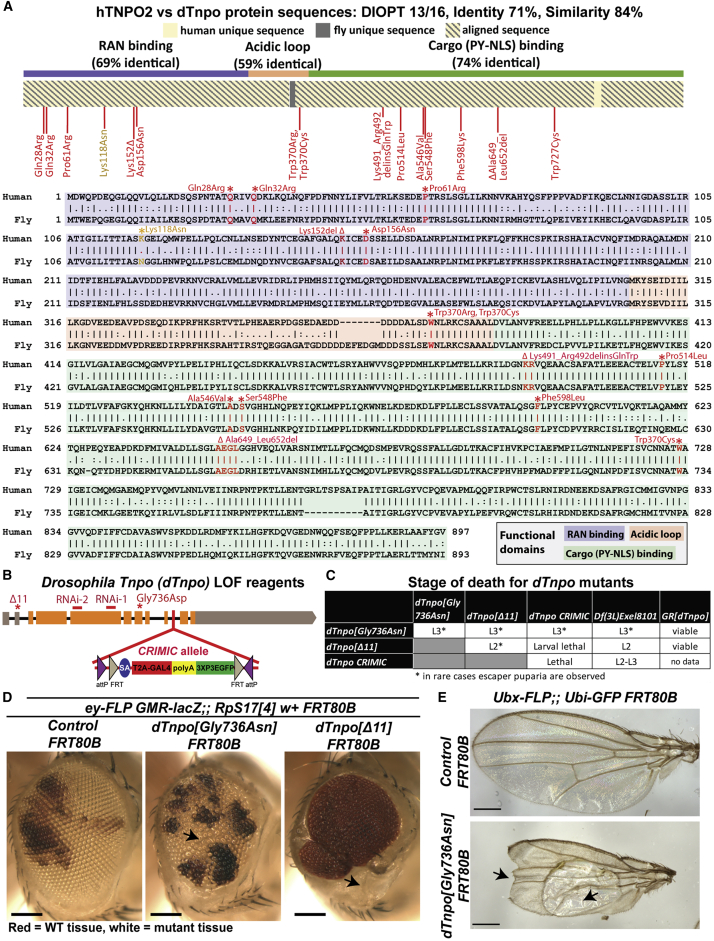
To investigate whether the TNPO2 variants identified in our cohort underlie individuals’ features, we utilized the model organism, Drosophila melanogaster. The fly ortholog to human TNPO2 (hTNPO2) is Drosophila Tnpo (dTnpo) and the encoded proteins from these two genes shuttle the same cargoes into the nucleus.57, 58, 59, 60, 61 The amino acid sequences encoded by these two genes are 71% identical and 84% similar (Figure 2A). The DRSC Integrative Ortholog Prediction Tool (DIOPT, v.7.1)62 score between these genes is 13 of 16, giving strong confidence that dTnpo is indeed orthologous to hTNPO2. The sequences of the RAN binding and cargo binding domains are more conserved than that of the acidic loop. Specifically, sequences encoding the acidic loops are 59% identical and 70% similar compared to the RAN binding domain (69% identical, 84% similar) and cargo binding domain (74% identical, 85% similar).8 dTnpo is also orthologous to human TNPO1 (DIOPT score = 13/16), so one fly gene corresponds to two human genes.
Figure 2.
Fly Transportin is essential for proper animal development and dTnpo loss in eyes and wings causes dysmorphisms
(A) Protein sequence comparison of human TNPO2 (hTNPO2) and Drosophila Tnpo (dTnpo) shown as a diagram and a detailed amino acid alignment. All variants are at conserved amino acids (red) except p.Lys118Asn (orange). Symbols in the protein alignment: identical (|), similar (:), different (.), absent (_).
(B) dTnpo mutants (red) created for loss-of-function (LoF) studies include dTnpoΔ11 (an imprecise excision of the P-element, NP4408), dTnpoGly736Asp (an EMS-induced mutation), and a CRIMIC allele. Two independent RNAi lines, RNAi-1 and RNAi-2, were also obtained.
(C) Animals homozygous for dTnpo mutant alleles demonstrate larval lethality due to dTnpo loss. None of the alleles or a large deficiency allele which lacks dTnpo, Df(3L)Exel8101, complement each other. Lethality caused by dTnpoΔ11 and dTnpoGly736Asp can be rescued using a genomic rescue construct, GRdTnpo.
(D) The FRT/FLP system was used to make mosaic tissue in the fly eye during development. dTnpoGly736Asp causes a rough eye phenotype. No homozygous dTnpoΔ11 mutant tissue is observed, indicating cell lethality. Scale bar = 100 μm.
(E) The FRT/FLP system was used to make mosaic tissue in the developing wing. dTnpoGly736Asp causes notch and blister phenotypes. Scale bar = 200 μm.
In (D) and (E), “Control” is yw;; FRT80B. Full fly genotypes for this and following figures are in Data S2. dTnpo-targeting RNAi produce consistent phenotypes (see Figure S1).
Of 15 variants found within our cohort, 14 occur at conserved amino acids between hTNPO2 and dTnpo (Figure 2A, red). Five variants are within the RAN binding domain. Two variants are at the same position within the acidic loop. Seven variants localize to the cargo binding domain. The p.Lys118Asn variant associated with proband 4 is not at a conserved amino acid (Figure 2A, orange) and the amino acid within the fly protein is an asparagine (Asn). This variant is at a conserved amino acid in vertebrate models (see data in MARRVEL).
Developmental loss of dTnpo causes lethality and morphologic defects
To gain an understanding of whether hTNPO2 may be essential during development, we assessed phenotypes associated with dTnpo loss in the fly. For this purpose, three dTnpo loss-of-function (LoF) mutant alleles were generated using different strategies (Figure 2B). First, we previously identified dTnpoGly736Asp in a genetic screen.39 Second, a truncated dTnpo mutant, dTnpoΔ11, was generated by an imprecise excision of a P-element. Last, a CRISPR-Mediated Integration Cassette (CRIMIC) allele was created by insertion of a Splice Acceptor-T2A-GAL4-polyA sequence into a shared intron of all dTnpo transcripts, effectively disrupting the gene’s expression by creating a truncated mRNA.42 We also obtained two available UAS-RNAi fly lines designed to target dTnpo63,64 (Figure 2B). These RNAi lines effectively reduce expression of dTnpo based on qPCR. dTnpo RNAi-1 causes an 81% ± 0.05% reduction and dTnpo RNAi-2 (previously used in Shi et al.60) causes a 58% ± 0.16% reduction of dTnpo mRNA compared to control RNAi expressing animals (Figure S1A).
The three dTnpo mutant alleles are homozygous lethal and no obvious phenotypes are observed in heterozygous animals. Notably, homozygous mutant animals do not survive beyond larval stages of development, shown in Figure 2C. In rare cases, escaper puparia could be observed in dTnpoGly736Asp cultures and, less commonly, in dTnpoΔ11 cultures. Based on a complementation test with a deletion line that lacks dTnpo, Df(3L)Exel8101, we conclude that dTnpoΔ11 is the most severe LoF allele, causing lethality at larval stage 2 (L2). dTnpoGly736Asp behaves as a hypomorph based on complementation failure with Df(3L)Exel8101, causing death in larval stage 3 (L3). Finally, the dTnpo CRIMIC allele also behaves as a hypomorph, causing lethality between L2 and L3. The dTnpoΔ11 and dTnpoGly736Asp alleles are rescuable by a genomic rescue line, GRdTnpo, which carries an independent copy of the dTnpo loci. Consistent with these data, ubiquitous expression of the strong UAS-dTnpo RNAi-1 using da-GAL4 causes lethality at L2, similar to dTnpoΔ11 mutants (Figure S1B). Further, da-GAL4 driven expression of the weaker UAS-dTnpo RNAi-2 causes lethality at L3, similar to dTnpoGly736Asp. Overall, these data show that dTnpo is essential during fly development.
Since hTNPO2 is likely required in multiple tissues and probands with hTNPO2 coding variants have diverse features, we assessed whether dTnpo loss impacts different tissues. Given that the majority of our cohort have ophthalmologic abnormalities, we first focused on the fly eye. The formation of this tissue is well studied and the developmental pathways required for proper eye formation are conserved.65 The mutant alleles dTnpoGly736Asp and dTnpoΔ11 were recombined onto FRT80B chromosomes. Using the FRT/FLP system,66 we crossed these flies to ey-FLP GMR-lacZ;; RpS174 w+ FRT80B to create mosaic eyes that include either homozygous mutant clonal tissue (white) or wild-type clonal tissue (red) (Figure 2D). Compared to FRT80B controls, dTnpoGly736Asp FRT80B causes eye deformities, including disorganized ommatidia consistent with a rough eye phenotype and smaller eyes. Interestingly, no homozygous mutant tissue is seen in animals carrying the stronger mutation, dTnpoΔ11 FRT80B, demonstrating that dTnpo is essential for eye development. Expression of dTnpo RNAi in the developing fly eye using ey-GAL4 shows consistent results, with the stronger UAS-dTnpo RNAi-1 causing developmental lethality and the weaker UAS-dTnpo RNAi-2 causing a rough eye phenotype and small eyes (Figure S1D). Thus, effects of dTnpo loss on eye development seem to be dosage dependent. Interestingly, expressing dTnpo RNAi with GMR-GAL4, which expresses later in eye development, did not cause significant alterations to the external fly eye (Figure S1E). These data argue that dTnpo is required during early eye imaginal disc development but do not rule out a requirement at later stages.
We next tested for dTnpo requirement during fly wing development, also a well-studied tissue that involves conserved signaling pathways for proper formation.67,68 We created mosaic tissue in the wing disc of dTnpoGly736Asp FRT80B larvae using Ubx-FLP;; Ubi-GFP FRT80B. Interestingly, wing notch phenotypes and large blisters can be observed in dTnpoGly736Asp mutant animals (Figure 2E). Taking an alternative approach, we used nub-GAL4 to express UAS-dTnpo RNAi in the developing wing disc and partially in the thorax. The stronger RNAi-1 causes lethality, consistent with dTnpo being required for development. The weaker RNAi-2 causes severe defects in wing morphology with hardly any wing forming (Figure S1C). Hence, dTnpo is required for wing development.
In sum, we found that dTnpo is required in multiple fly tissues for proper development using dTnpo LOF reagents. Interestingly, dTnpo loss was dosage dependent with the stronger mutant allele, dTnpoΔ11, and the stronger dTnpo RNAi, causing more severe defects than other hypomorphic reagents.
dTnpo is expressed primarily in neurons of the fly CNS
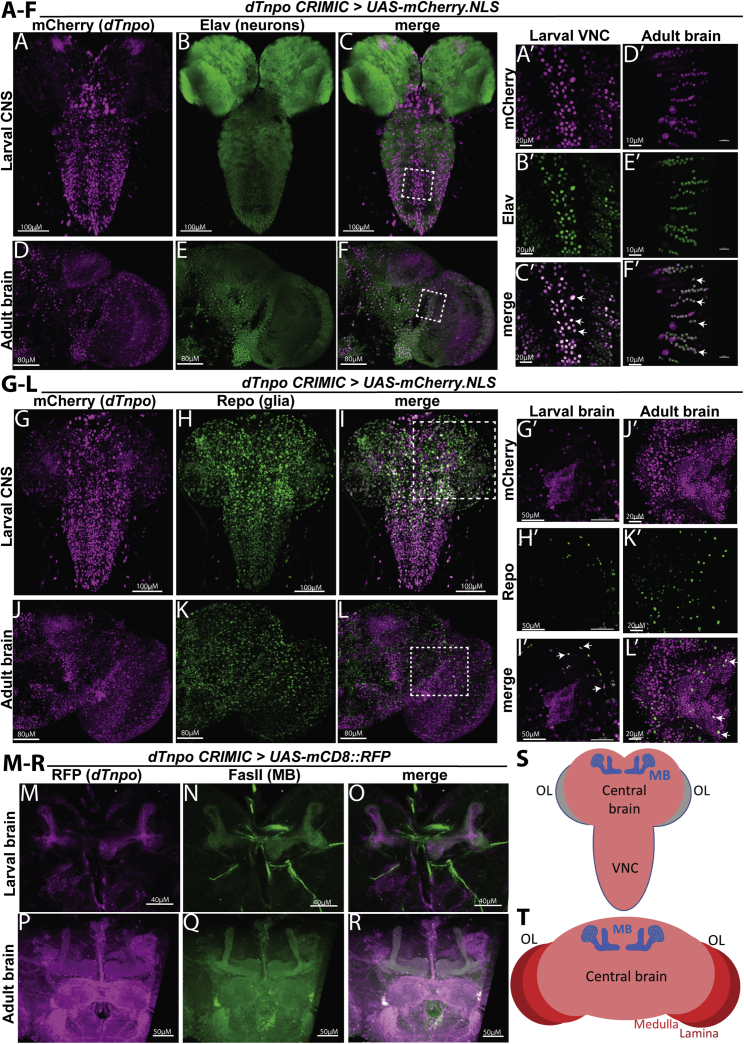
Given that the majority of the hTNPO2 cohort have features commonly associated with neurologic deficits and TNPO2 is highly expressed in the mammalian brain,9,10 we explored the importance of dTnpo in this tissue. First, we defined dTnpo’s expression pattern in the L3 larval central nervous system (CNS) and the adult fly brain (Figure 3). The dTnpo CRIMIC allele (see Figure 2B) carries a T2A-GAL4 sequence that expresses a GAL4 transcription factor under control of dTnpo’s regulatory elements.42 This GAL4 can drive expression of any UAS-transgene in the same spatial and temporal pattern as dTnpo.69 Thus, we used the dTnpo CRIMIC allele to express UAS-mCherry.NLS (mCherry fluorescent protein localized to the nucleus) (Figures 3A–3L). In larvae, mCherry (dTnpo) staining is most common in the central brain, including the cell bodies of mushroom body (MB) neurons, and ventral nerve cord (VNC; corresponding to the mammalian spinal cord) (Figures 3A and 3G); a schematic of the larval CNS is shown in Figure 3S for reference. These are areas that harbor a high density of active neurons at this stage.70 In adults, mCherry (dTnpo) staining shows the highest density in the optic lobe, MB cell bodies, and the central complex (Figures 3D and 3J); a schematic of the adult brain is shown in Figure 3T for reference.
Figure 3.
dTnpo is highly expressed in neurons, including mushroom body neurons
The dTnpo CRIMIC (T2A-GAL4) allele was used to drive expression of UAS-fluorescent reporter transgenes.
(A–L) UAS-mCherry.NLS (nuclear mCherry) was expressed and tissue were dissected from L3 larvae (CNS, includes central brain and VNC) or adults (brain). Shown is half of the adult brain. Tissue were counterstained with markers for neurons (Elav) or glia (Repo). Z stacked images showing dTnpo expression pattern compared to neurons (A–F) or glia (G–L). Dashed squares indicate regions used in (A′)–(L′).
(A′–L′) Single slice images were used to better visualize cellular co-localization of mCherry.NLS signal with neurons or glia. White arrows highlight co-localized nuclei with most neurons and some glia.
(M–R) dTnpo CRIMIC driven expression of UAS-mCD8::RFP (membrane-bound RFP) and FasII counter-staining confirmed overlap of dTnpo expression and mushroom body (MB) neurons in both larval and adult brains.
(S and T) Schematics of the larval CNS (S) and adult brain (T) highlighting MB neurons (blue), the ventral nerve cord (VNC), the central brain, and optic lobes (OL). The adult OL includes the medulla and lamina. The adult brain also includes the subesophageal ganglion (not shown in the schematic).
To define localization to specific cell types, the tissue were counterstained with Elav (predominantly marks neurons) or Repo (predominantly marks glia except midline glia).71, 72, 73 In both the larval CNS (Figures 3B and 3C) and adult brain (Figures 3E and 3F), not all Elav-positive cells stain positive for mCherry (dTnpo) in whole-mount, Z stacked images, supporting that dTnpo is expressed in a subset of neurons. These findings were consistent when using single-slice images of regions that show high mCherry staining in both the larval CNS (Figures 3A′–3C′) and adult brain (Figures 3D′–3F′). In whole mount with Z stacked images, there is no obvious overlap with glia and cells expressing mCherry (dTnpo) in larvae (Figures 3H and 3I) and adults (Figures 3K and 3L). However, in single-slice images of larval CNS (Figures 3G′–3I′) and adult brains (Figures 3J′–3L′), some Repo-positive cells show overlap with mCherry-positive cells, arguing that a small subset of glia express dTnpo.
The MB is of interest as this is the primary learning and memory center in Drosophila74 and the individuals in our cohort present with intellectual disability. To confirm dTnpo expression in these cells, UAS-mCD8::RFP (RFP fluorescent protein localized to the membrane) was expressed using the dTnpo CRIMIC allele and tissue were counterstained with an established MB marker, FasII,75 in larvae (Figures 3M–3O) and adults (Figures 3P–3R). Indeed, we see consistent overlap between RFP (dTnpo) and FasII signal, supporting that dTnpo is expressed in these neurons.
In sum, we found that dTnpo is highly expressed in a subset of neurons, including those that mediate associative learning, in the larval CNS and adult fly brain.
dTnpo is required for neuron function and maintenance
We next examined whether dTnpo was essential in fly neurons. We first assessed whether dTnpo was required during neural development. When we express UAS-dTnpo RNAi-1 in neuroblasts (neural stem cells) using insc-GAL4, no significant reductions in L3 larval CNS size are seen (Figure S2). In contrast, expressing UAS-dTnpo RNAi-1 using the pan-neuronal driver, elav-GAL4, is lethal (Figure S1F).
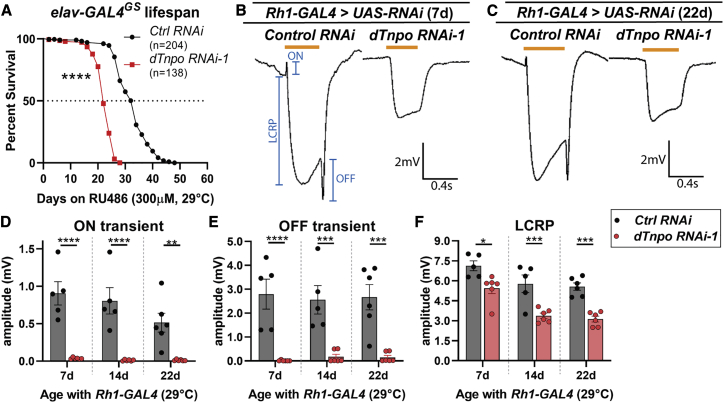
To avoid developmental lethality caused by expressing UAS-dTnpo RNAi-1 with elav-GAL4, we utilized a drug, RU486, inducible version of this neuronal driver, elav-GAL4[GeneSwitch] (elav-GAL4GS)76 to express the dTnpo RNAi-1. 1- to 2-day-old flies were transferred onto RU486-containing food, thus avoiding RNAi expression prior to adulthood. These animals were maintained on RU486 and survival curves were calculated for animals expressing UAS-dTnpo RNAi-1 compared to animals expressing UAS-control (Luciferase) RNAi. Interestingly, there is a significant decrease in survival when dTnpo is downregulated using RNAi expression in the adult fly neurons (Figure 4A). 50% of UAS-dTnpo RNAi-1-expressing animals die by 22 days compared to 32 days for control RNAi-expressing animals. The max survival is also decreased by 20 days with 100% of dTnpo RNAi-1 animals dying by 28 days, compared to 48 days for control RNAi animals.
Figure 4.
Fly Transportin is required in neurons for survival and eye function
(A) The drug-inducible elav-GAL4GS driver was used to express RNAi in adult fly neurons while avoiding RNAi expression during development. Expression of dTnpo RNAi-1 significantly impacts animal survival, indicating a progressive loss of neuron function due to dTnpo loss.
(B and C) Rh1-GAL4 was used to express RNAi in mature photoreceptor neurons and electroretinograms (ERGs) were used to measure neuronal function at 7 days, 14 days, and 22 days. Blue annotation shows where amplitudes are measured. Orange bars indicate the light pulses.
(D–F) dTnpo RNAi-1 nearly abolishes ON and OFF transients (D, E) and reduces the light coincident receptor potential (LCRP; F) compared to a control RNAi.
Statistics: (A) log-rank, (D–F) 2-way ANOVAs with Sidak’s multiple comparisons test. ∗p < 0.02, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Each dot represents the mean of 5 recorded ERGs per animal. The mean from 5–6 animals is shown. Error bars denote SEM; “Control (Ctrl) RNAi” is UAS-Luciferase RNAi (TRiP.JF01355). UAS-dTnpo RNAi-1 is TRiP.HMJ23009.
To examine whether dTnpo is required for neuronal activity, we performed electroretinograms (ERGs) on UAS-dTnpo RNAi-1 and UAS-control RNAi-expressing animals (Figures 4B–4F). ERGs are an established method for measuring neuron dysfunction in the synaptic circuit that makes up the fly optic system. ERGs quantify the light coincident receptor potentials (LCRP) and ON/OFF transients in the adult eye.77, 78, 79 LCRP amplitudes measure the phototransduction pathway that is dependent on light exposure.77,78 ON/OFF transients measure synaptic transmission between photoreceptor neurons and post-synaptic neurons in the lamina.77,78 At 7 days, the downregulation of dTnpo by expressing RNAi in mature photoreceptor neurons using Rh1-GAL4 causes significant changes to the ON and OFF transient amplitudes (Figures 4D and 4E), indicating a loss of synaptic activity. LCRP defects are also observed in dTnpo RNAi-1-expressing animals based on reductions in depolarization amplitude (Figure 4F). This impact seems to become stronger with age as there is a reduction of 24% in LCRP at 7 days when compared to control RNAi-expressing flies. This reduction is more robust by 14 days and 22 days at 42% and 44%, respectively.
In sum, dTnpo expression in neurons was found to be essential for animal survival. Further, neuronal function in the fly eye is disrupted by dTnpo loss, supporting that dTnpo is required in mature neurons.
Upregulation of dTnpo causes similar phenotypes to dTnpo LoF mutants
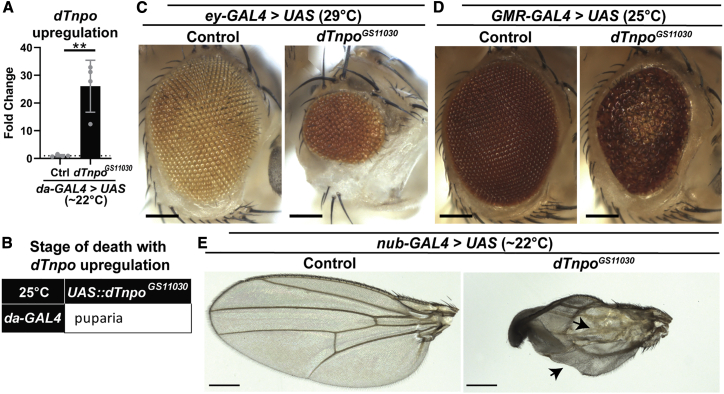
Thus far, we found that phenotypes associated with dTnpo loss are dosage dependent. We therefore considered whether dTnpo overexpression could also be detrimental. We obtained a fly line, UAS::dTnpoGS11030, that contains a P-element insertion with a UAS element upstream of the dTnpo gene.80 This allows us to upregulate dTnpo under control of the GAL4/UAS system69 by a 25- ± 8.1-fold increase in dTnpo mRNA levels (Figure 5A). Interestingly, upregulation of dTnpo using the ubiquitous driver, da-GAL4, causes lethality after pupariation (Figure 5B). Further, upregulation of dTnpo in both early (ey-GAL4) and late (GMR-GAL4) eye development causes rough eye phenotypes and reduced eye size (Figures 5B and 5C). dTnpo upregulation in the wing using nub-GAL4 causes wing notching and large blisters in 100% of animals (Figure 5D). In sum, we see that upregulating dTnpo causes similar phenotypes as dTnpo loss (see Figures 2 and S1).
Figure 5.
Upregulation of dTnpo disrupts morphology of eyes and wings
(A) Ubiquitous expression of UAS::dTnpoGS11030 using da-GAL4 in flies causes a 25-fold increase in dTnpo mRNA levels by qPCR. L3 larvae were analyzed at 22°C. Unpaired t test, ∗∗∗p = 0.0003. Each dot represents the mean from replicate wells per sample. The mean from 4 individual samples is shown. Error bars denote SD.
(B) da-GAL4>UAS::dTnpoGS11030 animals do not survive beyond pupariation at 25°C.
(C and D) Upregulation of dTnpo during eye development, using either ey-GAL4 (early development) or GMR-GAL4 (late development) driven expression of UAS::dTnpoGS11030, causes small eyes and rough eye phenotypes. Scale bar = 100 μm.
(E) nub-GAL4 driven expression of UAS::dTnpoGS11030 causes notch and blister phenotypes (arrows) in the fly wing. Scale bar = 200 μm. “Control (Ctrl)” is UAS-empty.
Ectopic expression of human TNPO2 in flies causes developmental toxicity
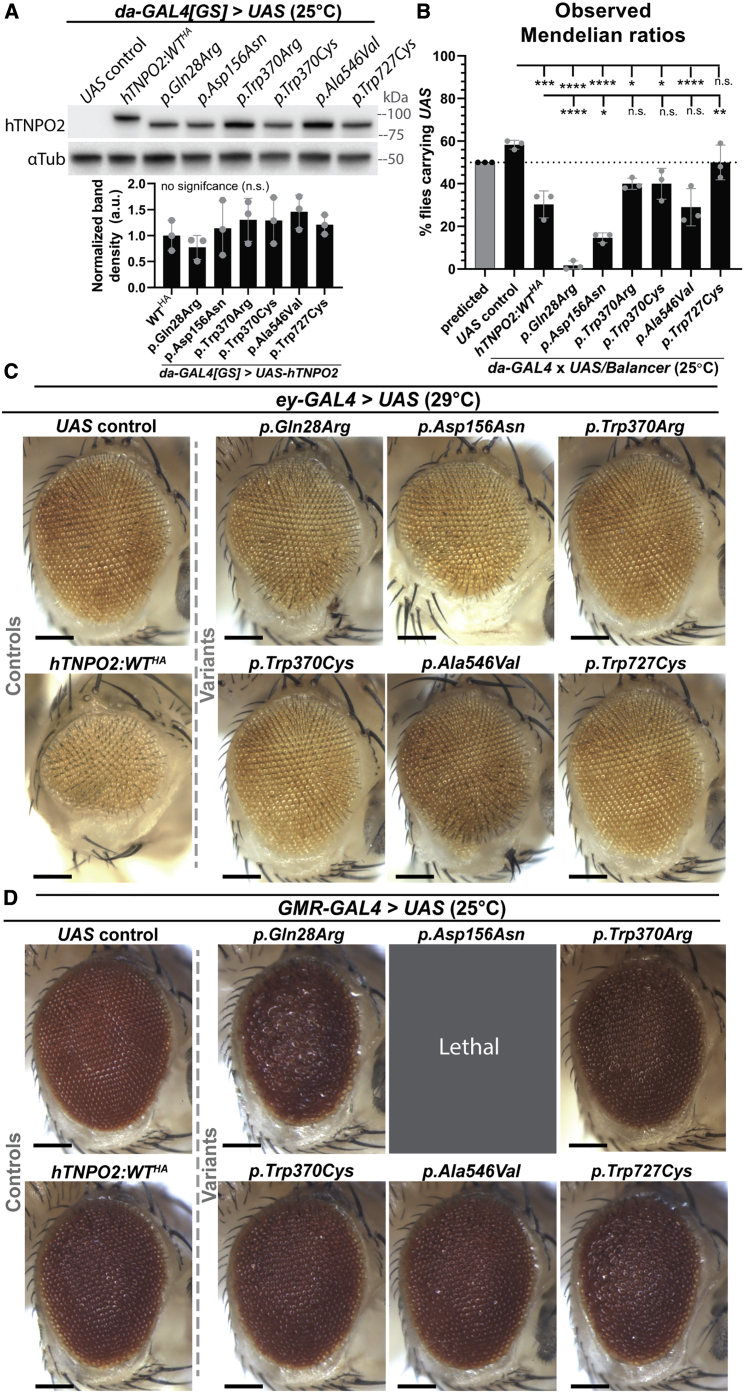
We next aimed to define whether proband-associated variants in hTNPO2 could alter the function of the encoded protein in vivo. For this purpose, we established a series of UAS-hTNPO2 fly lines expressing wild-type (WT; reference) or variant human TNPO2 cDNA (GenBank: NM_001136196.1) under control of the GAL4/UAS system. We selected 6 of the 14 variants that are at conserved amino acids for analysis, two from each protein domain. This included p.Gln28Arg, p.Asp156Asn, p.Trp370Arg, p.Trp370Cys, p.Ala546Val, and p.Trp727Cys. We confirmed that these lines properly express the UAS-hTNPO2 transgenes at comparable levels using western immunoblots and an antibody specific to human TNPO1/2 (Figure 6A). We expressed UAS transgenes in 1- to 2-day-old adult animals using the drug-inducible, ubiquitous driver, da-GAL4GS, to avoid any toxicity during development.
Figure 6.
Variants in hTNPO2 cause different amounts of toxicity compared to wild-type hTNPO2 during fly development and in the eye
(A) UAS-hTNPO2 fly lines were generated. Western immunoblots (WBs) confirmed hTNPO2 protein levels are similar between lines using a drug-inducible ubiquitous driver (da-GAL4GS) to express transgenes and a human TNPO1/2 antibody. Normalized hTNPO2 band density from three independent westerns were quantified. Each dot represents one independent sample. The mean from 3 individual samples is shown.
(B) da-GAL4 driven ectopic expression of UAS-hTNPO2:WTHA reduces Mendelian ratios compared to UAS control flies, demonstrating toxicity during development. Variants p.Gln28Arg and p.Asp156Asn are more toxic than hTNPO2:WT whereas p.Trp727Cys is less toxic. Each dot represents one independent cross with >100 animals scored. The mean from three independent crosses is shown.
(C and D) Ectopic expression of UAS-hTNPO2:WTHA disrupts eye development using either ey-GAL4 (early development) or GMR-GAL4 (late development). Scale bars = 100 μm.
(C) With ey-GAL4>hTNPO2:WTHA, eyes are smaller than controls and have a rough eye phenotype. p.Trp370Cys and p.Trp370Arg are less toxic.
(D) With GMR-GAL4>hTNPO2:WTHA, eyes are moderately smaller and there is a mild rough eye phenotype compared to controls. p.Gln28Arg and p.Asp156Asn are more toxic.
Statistics: 1-way ANOVAs with Dunnett’s (A) or Tukey’s (B) multiple comparisons test. no significance (n.s.) ≥ 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Error bars denote SD “UAS Control” is UAS-(empty).
Previously, we had found that animals trans-heterozygous for the hypomorph alleles, dTnpoGly736Asp and dTnpo CRIMIC, did not survive past larval stage 3 (see Figure 2C). Thus, we examined whether expression of hTNPO2 could rescue this phenotype. Using the dTnpo CRIMIC (T2A-GAL4) allele, we expressed UAS-hTNPO2:WTHA (wild-type hTNPO2 cDNA with a 3′ 3xHA-tag) or control UAS-(empty) in these dTnpo trans-heterozygous hypomorph animals. No rescue is observed with the expression of hTNPO2 cDNA in these mutant animals (Figure S3A). We noted that hTNPO2:WT expression causes death earlier, at L2, rather than L3 when compared to UAS-(empty) control expressing flies, demonstrating that the expression of hTNPO2:WT increased, rather than reduced, toxicity in these dTnpo mutant flies. We also tested whether five of the variants found in our cohort could rescue lethality in dTnpo trans-heterozygous animals. Interestingly, the p.Trp370Cys and p.Ala546Val variants result in death at L3 rather than L2 when compared to hTNPO2:WT. The other variants tested—p.Gln28Arg, p.Asp156Asn, and p.Trp727Cys—caused lethality at L2 like hTNPO2:WT.
We next considered if the lack of rescue seen with expression of hTNPO2 in dTnpo trans-heterozygous hypomorph animals is due to the overexpression of hTNPO2 in flies being toxic. This is because we found that robust upregulation of dTnpo was toxic (see Figure 5) and the expression of UAS-hTNPO2 by the dTnpo CRIMIC allele will result in an overexpression of hTNPO2,42 albeit likely at significantly lower levels than that caused by the UAS::dTnpoGS11030 allele.69 Thus, we would have two sources of toxicity in rescue experiments, that from the mutations in dTnpo and that from overexpressing hTNPO2. To test this hypothesis, Mendelian ratios were calculated for progeny from crosses between da-GAL4 and UAS fly lines, including UAS-hTNPO2 lines and the control UAS-(empty) line (Figure 6B). As expected, 58% ± 2.1% of control progeny carry the UAS transgene. In contrast, only 28% ± 6.4% of progeny from UAS-hTNPO2:WTHA crosses carry the UAS transgene, showing that significant toxicity occurs during development (Figure 6B). The presence of the HA-tag on the hTNPO2:WT transgene does not alter this effect (Figure S3B). Overall, these data demonstrate that ubiquitous, ectopic expression of hTNPO2 is toxic.
We also investigated whether proband-associated variants could induce the same toxicity as wild-type hTNPO2. Notably, p.Gln28Arg and p.Asp156Asn are more toxic than hTNPO2:WT (Figure 6B). Further, variants p.Trp370Arg, p.Trp370Cys, and p.Ala546Val cause similar toxicity compared to that caused by hTNPO2:WT expression (Figure 6B). In contrast, the p.Trp727Cys variant is significantly less toxic than hTNPO2:WT, producing 50% ± 8.2% of UAS carrying progeny (Figure 6B).
In summary, ectopic expression of hTNPO2 in flies causes toxicity consistent with phenotypes observed when upregulating dTnpo (see Figure 5). As three of six proband-associated variants tested caused significant differences in the amount of toxicity than that caused by wild-type hTNPO2, these data suggest that these variants alter the function of the hTNPO2 protein. Specifically, p.Gln28Arg and p.Asp156Asn may cause gain-of-function (GoF) effects and p.Trp727Cys may cause LoF effects.
Toxicity caused by variants in the fly eye differ from that of wild-type hTNPO2
Next, we assessed whether ectopically expressing wild-type and variant hTNPO2 in the fly eye can cause morphologic disruptions similar to wild-type dTnpo upregulation (see Figure 5). Using ey-GAL4, expression of hTNPO2:WT causes a smaller eye and a rough eye phenotype compared to animals expressing a UAS control (Figure 6C). Expression of the p.Trp727Cys variant leads to eyes more similar to controls than hTNPO2:WT (Figure 6C), consistent with it being less toxic than hTNPO2:WT during animal development (see Figure 6B). Interestingly, variants p.Trp370Arg and p.Trp370Cys are also less toxic than hTNPO2:WT, suggesting that during early eye development these variants act as LoF-variants. Expression of variants p.Gln28Arg, p.Asp156Asn, and p.Ala546Val cause similar eye phenotypes as hTNPO2:WT.
To further explore the impacts of variants at later stages of the developing eye than those affected by ey-GAL4, we expressed UAS transgenes using GMR-GAL4. Ectopic expression of hTNPO2:WT causes a moderate rough-eye phenotype and smaller eyes compared to animals expressing UAS control (Figure 6D). Consistent with previous data using da-GAL4 (see Figure 6B), expression of p.Gln28Arg and p.Asp156Asn cause more toxicity than hTNPO2:WT expression (Figure 6D). Specifically, p.Gln28Arg causes a more robust rough eye phenotype and smaller eyes and p.Asp156Asn causes developmental lethality. GMR-GAL4 is expressed at low levels in the larval brain81,82 and has been reported to cause lethality in extremely toxic situations.44,83 In contrast, eye phenotypes caused by expression of p.Trp370Arg, p.Trp370Cys, and p.Ala546Val are similar to hTNPO2:WT (Figure 6D). In turn, expressing p.Trp727Cys causes a slightly more robust rough eye phenotype compared to hTNPO2:WT expressing animals.
In sum, ectopic expression of wild-type hTNPO2 in the fly eye causes morphologic defects. Interestingly, these defects are different when comparing animals expressing proband-associated variants versus hTNPO2:WT. The specific effects of each variant are dependent on the developmental stage during which the transgenes are expressed in the fly eye, fitting with TNPO2 encoding a pleiotropic protein that may play different roles at different stages of development.
Toxicity caused by variants in the developing wing differ from that of wild-type hTNPO2
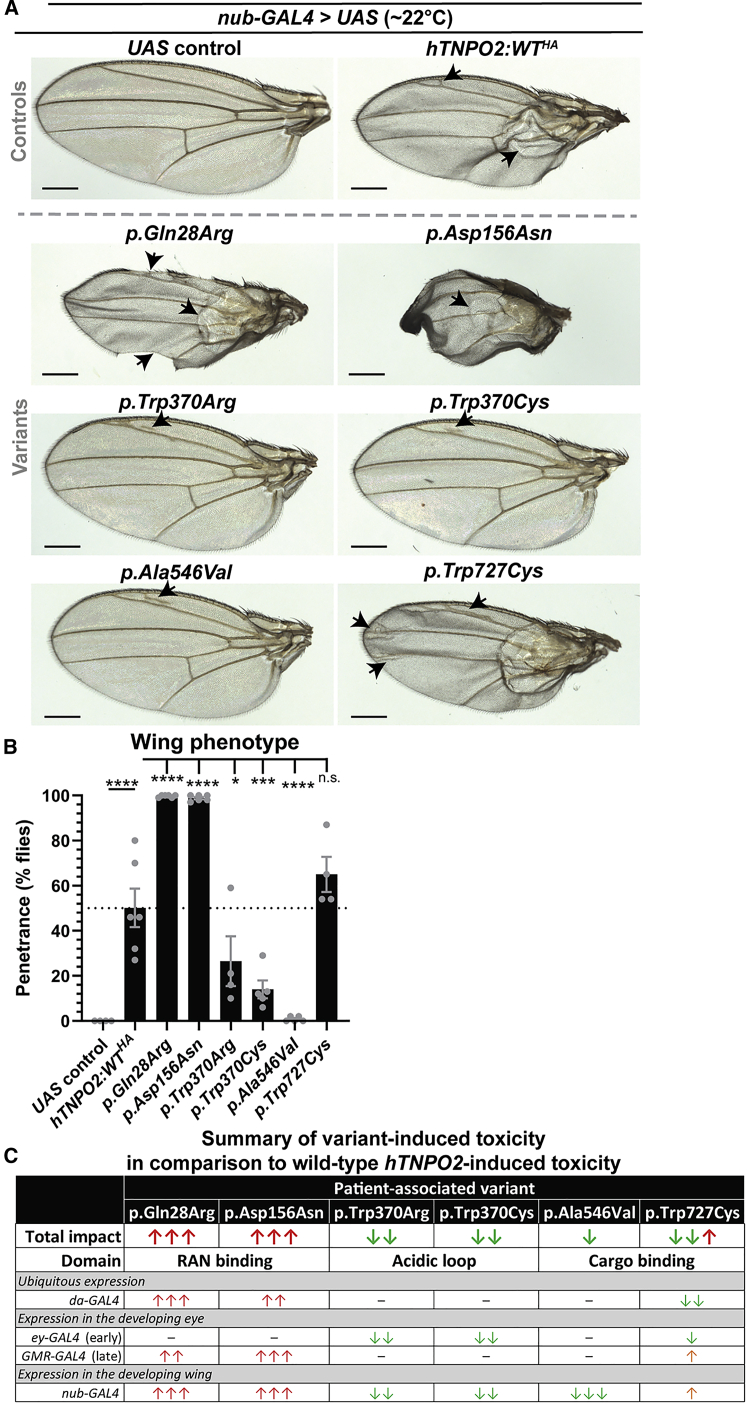
Thus far, we defined differences between toxicity induced by ectopically expressing wild-type and variant hTNPO2 by expressing them ubiquitously during development or in the developing fly eye (see Figure 6). Given that toxicity in response to expressing these proteins may differ depending on the tissue, we further analyzed impacts in the fly wing using nub-GAL4. When UAS-hTNPO2:WTHA was ectopically expressed in the developing wing, progeny had blister phenotypes and gain-of-vein phenotypes and the wings were smaller than control UAS wings (Figure 7A). hTNPO2:WT phenotypes are 50% ± 8.5% penetrant, allowing us to quantitatively analyze the effects of proband-associated variants in this tissue (Figure 7B). Consistent with previous data, expression of p.Gln28Arg and p.Asp156Asn cause more severe wing phenotypes than hTNPO2:WT expressing animals (Figure 7A). In addition to blisters and gain-of-vein phenotypes, wings from p.Gln28Arg and, particularly, p.Asp156Asn, commonly have notch phenotypes, are smaller, and have more disruptions to wing inflation than hTNPO2:WT animals. Further, blister and notch phenotypes are significantly more penetrant with these variants, at 99.6% ± 0.2% and 98.8% ± 0.5%, respectively (Figure 7B). In contrast, animals expressing variants p.Trp370Arg, p.Trp370Cys, and p.Ala546Val rarely have blister phenotypes (Figure 7A) with a concomitant and significant reduction in phenotype penetrance compared to hTNPO2:WT expressing animals (Figure 7B). Interestingly, animals expressing p.Ala546Val, which had minimal effects in the developing animal and eye (see Figure 6), showed few defects and phenotype penetrance was significantly lower than that for hTNPO2:WT expressing animals at 0.8% ± 0.5%. Last, expression of p.Trp727Cys, which had variable effects in other tissues, causes a more severe gain-of-vein phenotype compared to hTNPO2:WT expressing animals (Figure 7A). However, penetrance of the blister and notch phenotypes is similar between animals expressing p.Trp727Cys and hTNPO2:WT (Figure 7B).
Figure 7.
Variants alter hTNPO2-induced phenotypes and penetrance in the fly wing and impact of the variants corresponds with their location within the protein
(A) Ectopic expression of UAS-hTNPO2:WTHA using nub-GAL4 disrupts wing development, causing notching, blisters, and gain-of-vein phenotypes (arrows). p.Trp370Cys, p.Trp370Arg, and p.Ala546Val have less severe phenotypes whereas p.Gln28Arg and p.Asp156Asn are significantly more toxic. p.Trp727Cys has a moderately stronger gain-of-vein phenotype than hTNPO2:WT. Scale bar = 200 μm.
(B) Blister and notch phenotypes caused by hTNPO2 expression in the wing occurs in 50% of wings, representing penetrance. Penetrance is significantly different for all variants except p.Trp727Cys. Statistics: 1-way ANOVAs with Dunnett’s multiple comparisons test. No significance (n.s.) ≥ 0.05, ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Error bars denote SEM. Each dot represents the results from one cross with >50 animals scored. The mean from two independent experiments that included 2–3 individual crosses is shown.
(A and B) “UAS Control” is UAS-(empty).
(C) Table summarizing phenotype severity associated with variants when compared to hTNPO2:WT-associated phenotypes. Symbols: strong decrease in toxicity (green arrows), strong increase in toxicity (red arrows), mild increase in toxicity (orange arrows), no obvious difference in toxicity (dash). p.Trp727Cys strongly reduces toxicity in two situations and mildly increases toxicity in two situations, earning two green and one red arrow in the summary.
In sum, ectopic expression of hTNPO2 in the developing fly wing causes morphologic defects that are altered by proband-associated variants. Concomitantly, phenotype penetrance is significantly different for most variants.
Discussion
We identified 15 individuals who carry pathogenic coding variants in the pleiotropic protein, TNPO2. Probands uniformly present with global developmental delay (GDD), including speech/motor deficits and intellectual disability. The majority also have behavioral deficits, feeding difficulties, dysmorphic features, strabismus, and muscle tone abnormalities (primarily hypotonia). Movement and neurological disorders (e.g., tremors, ataxia) and neurological features, including seizures and abnormal MRIs, are also seen. Using Drosophila, we found that Transportin (dTnpo) is an essential gene during animal development and is required for proper eye and wing formation. Malformations caused by decreasing dTnpo activity are dosage dependent with greater reductions in dTnpo activity causing more severe defects. Interestingly, upregulation of dTnpo causes similar defects as dTnpo loss. We further found that dTnpo is required in the fly nervous system. It is expressed mostly in neurons and abundant in mushroom body (MB) neurons. Downregulating dTnpo in mature neurons disrupts their function and reduces animal survival. Ectopic expression of wild-type, human TNPO2 causes similar phenotypes in the fly as gain and loss of dTnpo, suggesting that its function is evolutionarily conserved. Interestingly, ectopic expression of UAS-hTNPO2 transgenes carrying proband-associated variants cause different levels of toxicity compared to animals expressing wild-type UAS-hTNPO2, supporting that these variants disrupt the encoded protein’s normal activity in vivo. Impacts seem to vary based on the variant’s location within the protein (discussed below). In conclusion, these data demonstrate that de novo coding variants within TNPO2 can alter the function of the encoded, essential protein and are associated with developmental phenotypes in individuals.
Potential roles of TNPO2 variants in disease
While de novo variants in TNPO2 had been recorded in individuals with intellectual disability, previous studies did not find that TNPO2 variants were a significant cause of GDD in large datasets that included hundreds to thousands of cases.84, 85, 86, 87 Our independent identification of 15 individuals with GDD that carry pathogenic TNPO2 variants and functional studies using Drosophila provide strong evidence of the important role this gene plays in human development and in the nervous system. Together, these data demonstrate that while TNPO2 variants are associated with GDD, this is a rare cause of disabilities.
Our fly data demonstrate that all variants tested can alter the anticipated function of hTNPO2 in vivo (see Figures 6 and 7). These data suggest that the impact of a variant depends on its position within the protein in addition to the tissue and developmental stage during which the transgenes are expressed. To better understand how these variants compare to wild-type TNPO2, we compiled a summary of our fly data that used ectopic expression of TNPO2 cDNA (Figure 7C; extended data in Figure S3C). Interestingly, expression of variants that fall within the RAN binding domain were significantly more toxic than hTNPO2:WT in almost all conditions. These data suggest these are GoF variants. In contrast, expression of variants that fall within the acidic loop of the protein tended to be less toxic than hTNPO2:WT expression, suggesting they are LoF variants. As these two variants fall at the same amino acid in the protein, future studies are needed to test additional variants from within this domain. Interestingly, we also observed that expression of variants in the cargo binding domain of hTNPO2 have more variable effects when compared to hTNPO2:WT expression. Generally, these have reduced toxicity compared to wild-type hTNPO2, but the tissue assayed seems more critical in defining their impact. This fits with the known function of this domain as cargoes are likely to differ between tissues and ages. Specifically, in our fly data UAS-hTNPO2:p.Ala546Val (associated with proband 11) has similar toxicity when ubiquitously expressed and when expressed in the fly eye as compared to UAS-hTNPO2:WT. However, expression of this variant is significantly less toxic than hTNPO2:WT in the fly wing. Consistent with these data, proband 11 shows fewer phenotypes than other persons within our cohort (see Table 1, Data S1, and Figure 1). Thus, this variant may be tissue specific. In addition to p.Ala546Val, the current fly data argue that the impact of p.Trp727Cys is also tissue dependent. Specifically, expression of this variant is significantly less toxic than wild-type TNPO2 with da-GAL4 (ubiquitous) and ey-GAL4 (early eye formation) and only mildly more toxic than wild-type TNPO2 with GMR-GAL4 (late eye formation) and nub-GAL4 (wing formation). In sum, these data suggest that it is more of a LoF variant than a GoF variant while mechanistic studies are needed in each tissue to strengthen these results. Interestingly, disease presentation in proband 15 (carries p.Trp727Cys) seems to diverge from the majority of the cohort as this individual presents with hypertonia and no signs of hypotonia, is the only individual with notable sleep deficits, and is the only individual with a rigid gait. Also, this person does not have any known neurological features despite undergoing an EEG and MRI testing. Overall, more directed investigation is needed to better understand the different roles of these cargo binding domain variants while considering their impacts in different tissues and at different developmental stages.
Interestingly, the p.Lys152del variant in proband 5 is only found at a 16% mosaicism by Sanger sequencing (21% by WGS) in blood. We believe that the deletion of p.Lys152 within this critical domain can explain this individual’s features given similarities of this person’s symptoms to others in the cohort, our fly data showing that the nearby variant of p.Asp156Asn significantly impacted the function of TNPO2 in multiple tissues (see Figure 7C), and the fact that this variant is at a conserved amino acid in multiple organisms, including mice (see data in MARRVEL48). It is also important to note that the amount of mosaicism in other tissues is not known.
Except for p.Ala546Val (proband 11), and potentially p.Trp727Cys (proband 15), no obvious association with the other variants tested in the fly and symptoms or severity of individual’s features are observed. We hypothesize that this is due to the findings that both up- or downregulation of Transportin in the fly cause similar phenotypes (see Figures 2, 5, and S1). It is likely that loss of dTnpo disrupts the shuttling of cargoes into the nucleus and this disrupts multiple pathways important during development and for neuron maintenance (see below). Speculatively, we predict that upregulation of dTnpo and ectopic expression of hTNPO2 causes similar phenotypes as dTnpo loss as these would cause an accumulation of the Transportin protein intracellularly. This could sequester the dTnpo cargos, making them unavailable to perform their normal functions. Thus, gain-of-function-associated toxicity would still result in similar phenotypes as loss-of-function mechanisms.
It is important to note that TNPO1/2 function in many pathways including ciliogenesis, mitotic spindle assembly, and nuclear envelope assembly. Further, it was recently shown that when TNPO1 binds the nuclear pore complex RAN GTP was retained in the nucleus.88 Thus, disrupting the function of TNPO2 could not only impact its cargoes but also the RAN GDP/GTP gradient used to drive the activity of this and other proteins.7 Notably, due to this pleiotropic nature of TNPO2, the impact of up/downregulating the fly gene, the impact of ectopically expressing the human gene in the fly, and the impact of variants on the endogenous function of the protein encoded by human TNPO2 in individuals is likely to be very complex. Interestingly, phenotypic variability of monogenic causes of neurologic disorders has been described for other genetic conditions, such as those associated with EEF1A2 (MIM: 602959).89 Overall, our fly data demonstrate that Transportin’s activity would likely need to be tightly regulated to prevent disease.
Supporting the hypothesis that both gain- or loss-of-function variants can contribute to TNPO2-associated disorder, it is notable that copy number variants (CNVs) that include TNPO2, both deletions and duplications, have both been reported as pathogenic in ClinVar90 by studies that evaluated CNVs in individuals with developmental delays, including accession numbers: VCV000059111.1 (SCV000080263.4; duplication)91 and VCV000153069.1 (SCV000182485.3; deletion).92 While the region of these CNVs includes other genes, it is intriguing to consider that the dosage sensitivity of TNPO2 could contribute to developmental phenotypes in these individuals. In particular, the whole gene deletion of TNPO2 in these cases are consistent with a potential haploinsufficiency as a part of the disease mechanism(s) associated with this gene.
In sum, our fly data support that any disruptions to TNPO2 activity (gain or loss) in individuals is likely to cause similar symptoms. Further, coding variants in TNPO2 can alter the function of the protein with ectopic expression of transgenes. Future mechanistic studies should focus on differences between the variants, considering the impact of variants as gain-of-function, loss-of-function, or dominant-negative mutations in multiple contexts and tissues. Further, additional variants will need to be tested to better understand the potential association between protein domain and a variant’s impact on TNPO2’s function. Last, as our studies depended on ectopic expression of human TNPO2, future studies should focus on defining the mechanisms underlying the impact of individual variants in an endogenous system.
TNPO2 during development
Consistent with TNPO2 being a pleiotropic protein, this gene is required in multiple tissues in the fly (see Figures 2, 4, and S1). Impacts of losing Transportin during development are dosage dependent with stronger dTnpo mutants and dTnpo-targeting RNAi causing more severe defects (see Figures 2 and S1). Upregulation of dTnpo and ectopic expression of its human ortholog, hTNPO2, causes similar defects (see Figures 5, 6, and 7), potentially by titering cargoes away from their normal function(s) as discussed above. We note that our dTnpo mutant and RNAi studies rely on robust depletion of dTnpo, either through the use of homozygous mutant animals or the significant downregulation of dTnpo mRNA, respectively (see Figures 2 and S1). Further, the UAS::dTnpoGS11030 allele causes a dramatic upregulation of the fly gene. Future studies should titer the expression of dTnpo to see when phenotypes occur and consider the possibility that heterozygous mutant animals may have minor anomalies. It is also important to note that the ectopic expression of the UAS-hTNPO2 lines is not expected to cause such robust overexpression of the Transportin protein as was seen with the UAS::dTnpoGS11030 allele.69 However, further investigations into the impacts of the variants in a system that does not rely on ectopic expression of genes would likely reveal additional information as to the role these variants play in disease.
Interestingly, TNPO1/2 interact with multiple, conserved factors important in developmental pathways, including NF-κB signaling,18 hedgehog signaling,60 insulin signaling,29 and Ras/ERK signaling.17,93,94 These pathways are involved in multiple aspects of fly eye and wing formation and disruptions can cause similar phenotypes to what we observe in our studies.65,67,68 Thus, data support that TNPO2 coding variants could impact multiple developmental pathways simultaneously and this could explain the varied features observed in our cohort (see Data S1). Further, TNPO2 was found to be critical for HuR/ElavL1-mediated muscle cell differentiation in cultured murine myoblasts.11 This may be related to muscle tone abnormalities in our cohort (see Table 1). Last, it is notable that most of the TNPO2 cohort present with gastrointestinal abnormalities while multiple TNPO1/2 cargoes are involved in stress pathways associated with chronic intestinal inflammation in mammals, including components of the Activator Protein-1 (AP1) transcription complex,95, 96, 97, 98 NEMO,18,99 ADAR1 (MIM: 146920),100,101 and HSP70 (MIM: 140550).16,102 While gastrointestinal abnormalities are commonly associated with GDD, symptoms may be exacerbated by TNPO2 variants. Overall, our findings in Drosophila support that hTNPO2 is an important developmental gene that can impact multiple systems.
TNPO2 in the nervous system
Accumulating data show that TNPO2 is an important neuronal gene. We found that dTnpo is primarily expressed in a subset of neurons, including MB neurons, in the fly CNS (see Figure 3). These findings are consistent with mammalian data as TNPO1/2 is highly expressed in the brain.9,10,53 In mouse brains, Tnpo2 seems to be more highly expressed overall than Tnpo19,10 but this may depend on the brain region.53 Interestingly, when considering regions associated with memory, Tnpo2 was shown to be more highly expressed in the cerebral cortex than Tnpo1 and both genes were highly expressed in the hippocampus and cerebellum.53 The hippocampus is considered most orthologous to the MB in flies as neurons of both are critical for associative learning and circadian rythms.74 Fittingly, Tnpo1/2 activity impacts circadian rhythms in mice53,103 and proband 15 has sleep disturbance. Attention deficit disorder (ADD) and autism spectrum disorder (ASD), which are seen in the majority of our cohort, are commonly associated with sleep deficits.104 Further, our data show that dTnpo reduction in mature neurons disrupts their function and survival of the flies (see Figure 4). This may be the result of many of its cargoes being important neuronal proteins including FET proteins105 (FUS,14 EWS [MIM: 133450],106 TAF15 [MIM: 601574]), HuR/ElavL1,8,11,15,16 hnRNPA1,8,17 and Huntington (HTT [MIM: 613004]).107 Thus, perturbations to the translocation of these and other ubiquitous cargoes into the nucleus due to disrupted TNPO1/2 activity are likely to contribute to neurotoxicity. In fact, HuR and FUS have been implicated in ASD108,109 and FET proteins in ADD.110 It is also notable that homozygous null Tnpo2 mice are viable but may have significant anxiety and locomotion abnormalities.111 Mouse phenotypes are likely weaker than fly phenotypes because of compensation from Tnpo1 for Tnpo2 loss.
Overall, current data support that disruptions to TNPO2 activity can contribute to GDD, intellectual disability, behavioral deficits, and neurologic features observed in our cohort (see Tables 1 and S1).
Potential genetic interactions
As TNPO2 is shown to be a dosage dependent and pleiotropic protein, an individual’s unique genetic profile may contribute to disease occurrence and presentation. Interestingly, probands 5, 6, and 15 carry heterozygous variants of uncertain significance (VUSs) in genes highly expressed in the brain and predicted to be involved in neurological pathways, including CUX2 (a transcription factor involved in neuron proliferation, differentiation, and synaptic plasticity112,113), ANKFY1 (likely involved in vesicle trafficking114 and required for murine brain development115), and INA (a class IV neuronal intermediate filament involved in neuron morphogenesis116) (see Note S1). Thus, there is potential for genetic interactions between heterozygous loss of these genes and TNPO2 variants. Further, proband 8 carries a VUS in ARCMC9 which is associated with Joubert syndrome 30 (JBTS30).117 Joubert syndrome is an autosomal-recessive disorder which also involves GDD, ophthalmologic abnormalities, dysmorphic features, and hypotonia. The protein encoded by ARCMC9 impacts ciliogenesis like TNPO1/2.20, 21, 22,117 Thus, heterozygous loss of this gene could contribute to TNPO2-associated phenotypes. Last, proband 14 carries a de novo SNV in PDE4D which is predicted to cause a truncation and nonsense-mediated decay. Accordingly, this individual is diagnosed with acrodysostosis 2.56 We hypothesize that the TNPO2 variant in this individual contributes to developmental delays.
Concluding remarks
Overall, our data show that TNPO2-associated disorder represents a rare genetic condition with global developmental delay and syndromic features. As both upregulation and downregulation of Transportin causes similar defects in the fly and coding variants may increase or decrease hTNPO2’s activity, it is difficult to differentiate symptoms associated with a gain- or loss-of-function variant in individuals within our cohort. We conclude that because of pleiotropic effects of TNPO2 variants, sequencing and phenotypic comparison to reported cases is the most valuable approach to diagnosing features related to TNPO2. Further examination of these and additional cases will likely delineate the genotype-phenotype correlation.
Acknowledgments
We thank all members of our cohort and their families for agreeing to participate in this study. We thank former and present Bellen, Wangler, and Yamamoto lab members for their input during investigations, particularly Akhila Rajan, David Li-Kroger, Paul C. Marcogliese, Yiming Zu, and Guang Lin. We thank Anthony (Ton) J. van Essen, who unfortunately passed away before this manuscript was compiled, for his initial work with proband 11. Thank you to Hervé Tricoire and Nancy M. Bonini for copies of the da-GAL4GS fly line. Drosophila stock centers have been instrumental to these studies and include Bloomington Drosophila Stock Center, Vienna Drosophila Research Center, and Kyoto Stock Center. Research reported in this manuscript was supported by the National Institutes of Health (NIH) Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under award numbers U54 NS093793 and U01 HG007672. Further support came from NIH award R24 OD02205, NIH award R01 GM067858, and HHMI to H.J.B. L.D.G. was supported by NIH training grant T32 NS043124-18. Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the NIH award P50 HD103555 for use of the Neurovisualization Core facility. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. W.-L.C. was supported by Taiwan Merit Scholarships Program sponsored by the National Science Council, award NSC-095-SAF-I-564-015-TMS. M.A.K. and D.S. were supported by a NIHR Professorship. M.A.K. and K.E.S.B. were supported by the Sir Jules Thorn Award for Biomedical Research and Rosetrees Trust. S.S.M. was supported by the Cerebral Palsy Alliance, Australia (award PG01217).
Declaration of interests
The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing completed at Baylor Genetics Laboratories. Y.S. and A.B. are employees of GeneDx, Inc.
Published: July 26, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.06.019.
Contributor Information
Hugo J. Bellen, Email: hbellen@bcm.edu.
Queenie K.-G. Tan, Email: khoon.tan@duke.edu.
Data and code availability
This study did not generate datasets. All reagents developed in this study are available upon request.
Web resources
GeneMatcher (accessed December 2020), https://genematcher.org/
gnomAD Browser, v.2.1.1, https://gnomad.broadinstitute.org/
MARRVEL, http://marrvel.org/
OMIM, https://www.omim.org/
Supplemental information
References
- 1.Wangler M.F., Yamamoto S., Chao H.-T., Posey J.E., Westerfield M., Postlethwait J., Hieter P., Boycott K.M., Campeau P.M., Bellen H.J., Members of the Undiagnosed Diseases Network (UDN) Model Organisms Facilitate Rare Disease Diagnosis and Therapeutic Research. Genetics. 2017;207:9–27. doi: 10.1534/genetics.117.203067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellen H.J., Wangler M.F., Yamamoto S. The fruit fly at the interface of diagnosis and pathogenic mechanisms of rare and common human diseases. Hum. Mol. Genet. 2019;28(R2):R207–R214. doi: 10.1093/hmg/ddz135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ittisoponpisan S., Alhuzimi E., Sternberg M.J.E., David A. Landscape of Pleiotropic Proteins Causing Human Disease: Structural and System Biology Insights. Hum. Mutat. 2017;38:289–296. doi: 10.1002/humu.23155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown S.D.M., Lad H.V. The dark genome and pleiotropy: challenges for precision medicine. Mamm. Genome. 2019;30:212–216. doi: 10.1007/s00335-019-09813-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Twyffels L., Gueydan C., Kruys V. Transportin-1 and Transportin-2: protein nuclear import and beyond. FEBS Lett. 2014;588:1857–1868. doi: 10.1016/j.febslet.2014.04.023. [DOI] [PubMed] [Google Scholar]
- 6.Prpar Mihevc S., Darovic S., Kovanda A., Bajc Česnik A., Župunski V., Rogelj B. Nuclear trafficking in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Brain. 2017;140:13–26. doi: 10.1093/brain/aww197. [DOI] [PubMed] [Google Scholar]
- 7.Cavazza T., Vernos I. The RanGTP Pathway: From Nucleo-Cytoplasmic Transport to Spindle Assembly and Beyond. Front. Cell Dev. Biol. 2016;3:82. doi: 10.3389/fcell.2015.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rebane A., Aab A., Steitz J.A. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA. 2004;10:590–599. doi: 10.1261/rna.5224304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quan Y., Ji Z.-L., Wang X., Tartakoff A.M., Tao T. Evolutionary and transcriptional analysis of karyopherin β superfamily proteins. Mol. Cell. Proteomics. 2008;7:1254–1269. doi: 10.1074/mcp.M700511-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hock E.-M., Maniecka Z., Hruska-Plochan M., Reber S., Laferrière F., Sahadevan M K S., Ederle H., Gittings L., Pelkmans L., Dupuis L. Hypertonic Stress Causes Cytoplasmic Translocation of Neuronal, but Not Astrocytic, FUS due to Impaired Transportin Function. Cell Rep. 2018;24:987–1000.e7. doi: 10.1016/j.celrep.2018.06.094. [DOI] [PubMed] [Google Scholar]
- 11.van der Giessen K., Gallouzi I.-E. Involvement of transportin 2-mediated HuR import in muscle cell differentiation. Mol. Biol. Cell. 2007;18:2619–2629. doi: 10.1091/mbc.E07-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura M., Morinaka Y., Imai K., Kose S., Horton P., Imamoto N. Extensive cargo identification reveals distinct biological roles of the 12 importin pathways. eLife. 2017;6:e21184. doi: 10.7554/eLife.21184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackmull M.T., Klaus B., Heinze I., Chokkalingam M., Beyer A., Russell R.B., Ori A., Beck M. Landscape of nuclear transport receptor cargo specificity. Mol. Syst. Biol. 2017;13:962. doi: 10.15252/msb.20177608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dormann D., Rodde R., Edbauer D., Bentmann E., Fischer I., Hruscha A., Than M.E., Mackenzie I.R.A., Capell A., Schmid B. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Roretz C., Macri A.M., Gallouzi I.-E. Transportin 2 regulates apoptosis through the RNA-binding protein HuR. J. Biol. Chem. 2011;286:25983–25991. doi: 10.1074/jbc.M110.216184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Güttinger S., Mühlhäusser P., Koller-Eichhorn R., Brennecke J., Kutay U. Transportin2 functions as importin and mediates nuclear import of HuR. Proc. Natl. Acad. Sci. USA. 2004;101:2918–2923. doi: 10.1073/pnas.0400342101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee B.J., Cansizoglu A.E., Süel K.E., Louis T.H., Zhang Z., Chook Y.M. Rules for nuclear localization sequence recognition by karyopherin β 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang B., McCool K., Wan J., Wuerzberger-Davis S.M., Young E.W.K., Choi E.Y., Cingolani G., Weaver B.A., Miyamoto S. IPO3-mediated Nonclassical Nuclear Import of NF-κB Essential Modulator (NEMO) Drives DNA Damage-dependent NF-κB Activation. J. Biol. Chem. 2015;290:17967–17984. doi: 10.1074/jbc.M115.645960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cansizoglu A.E., Lee B.J., Zhang Z.C., Fontoura B.M.A., Chook Y.M. Structure-based design of a pathway-specific nuclear import inhibitor. Nat. Struct. Mol. Biol. 2007;14:452–454. doi: 10.1038/nsmb1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madugula V., Lu L. A ternary complex comprising transportin1, Rab8 and the ciliary targeting signal directs proteins to ciliary membranes. J. Cell Sci. 2016;129:3922–3934. doi: 10.1242/jcs.194019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dishinger J.F., Kee H.L., Jenkins P.M., Fan S., Hurd T.W., Hammond J.W., Truong Y.N.-T., Margolis B., Martens J.R., Verhey K.J. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-β2 and RanGTP. Nat. Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurd T.W., Fan S., Margolis B.L. Localization of retinitis pigmentosa 2 to cilia is regulated by Importin β2. J. Cell Sci. 2011;124:718–726. doi: 10.1242/jcs.070839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernis C., Swift-Taylor B., Nord M., Carmona S., Chook Y.M., Forbes D.J. Transportin acts to regulate mitotic assembly events by target binding rather than Ran sequestration. Mol. Biol. Cell. 2014;25:992–1009. doi: 10.1091/mbc.E13-08-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalab P., Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J. Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walther T.C., Askjaer P., Gentzel M., Habermann A., Griffiths G., Wilm M., Mattaj I.W., Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- 26.D’Angelo M.A., Anderson D.J., Richard E., Hetzer M.W. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- 27.Larrieu D., Viré E., Robson S., Breusegem S.Y., Kouzarides T., Jackson S.P. Inhibition of the acetyltransferase NAT10 normalizes progeric and aging cells by rebalancing the Transportin-1 nuclear import pathway. Sci. Signal. 2018;11:11. doi: 10.1126/scisignal.aar5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teratani T., Tomita K., Toma-Fukai S., Nakamura Y., Itoh T., Shimizu H., Shiraishi Y., Sugihara N., Higashiyama M., Shimizu T. Redox-dependent PPARγ/Tnpo1 complex formation enhances PPARγ nuclear localization and signaling. Free Radic. Biol. Med. 2020;156:45–56. doi: 10.1016/j.freeradbiomed.2020.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Putker M., Madl T., Vos H.R., de Ruiter H., Visscher M., van den Berg M.C.W., Kaplan M., Korswagen H.C., Boelens R., Vermeulen M. Redox-dependent control of FOXO/DAF-16 by transportin-1. Mol. Cell. 2013;49:730–742. doi: 10.1016/j.molcel.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 30.Splinter K., Adams D.R., Bacino C.A., Bellen H.J., Bernstein J.A., Cheatle-Jarvela A.M., Eng C.M., Esteves C., Gahl W.A., Hamid R., Undiagnosed Diseases Network Effect of Genetic Diagnosis on Patients with Previously Undiagnosed Disease. N. Engl. J. Med. 2018;379:2131–2139. doi: 10.1056/NEJMoa1714458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barish S., Barakat T.S., Michel B.C., Mashtalir N., Phillips J.B., Valencia A.M., Ugur B., Wegner J., Scott T.M., Bostwick B., Undiagnosed Diseases Network BICRA, a SWI/SNF Complex Member, Is Associated with BAF-Disorder Related Phenotypes in Humans and Model Organisms. Am. J. Hum. Genet. 2020;107:1096–1112. doi: 10.1016/j.ajhg.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao D., Reuter C.M., Ruzhnikov M.R.Z., Beck A.E., Farrow E.G., Emrick L.T., Rosenfeld J.A., Mackenzie K.M., Robak L., Wheeler M.T., Undiagnosed Diseases Network De novo EIF2AK1 and EIF2AK2 Variants Are Associated with Developmental Delay, Leukoencephalopathy, and Neurologic Decompensation. Am. J. Hum. Genet. 2020;106:570–583. doi: 10.1016/j.ajhg.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guillen Sacoto M.J., Tchasovnikarova I.A., Torti E., Forster C., Andrew E.H., Anselm I., Baranano K.W., Briere L.C., Cohen J.S., Craigen W.J., Undiagnosed Diseases Network De Novo Variants in the ATPase Module of MORC2 Cause a Neurodevelopmental Disorder with Growth Retardation and Variable Craniofacial Dysmorphism. Am. J. Hum. Genet. 2020;107:352–363. doi: 10.1016/j.ajhg.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanca O., Andrews J.C., Lee P.-T., Patel C., Braddock S.R., Slavotinek A.M., Cohen J.S., Gubbels C.S., Aldinger K.A., Williams J., Undiagnosed Diseases Network De Novo Variants in WDR37 Are Associated with Epilepsy, Colobomas, Dysmorphism, Developmental Delay, Intellectual Disability, and Cerebellar Hypoplasia. Am. J. Hum. Genet. 2019;105:413–424. doi: 10.1016/j.ajhg.2019.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon W.H., Sandoval H., Nagarkar-Jaiswal S., Jaiswal M., Yamamoto S., Haelterman N.A., Putluri N., Putluri V., Sreekumar A., Tos T. Loss of Nardilysin, a Mitochondrial Co-chaperone for α-Ketoglutarate Dehydrogenase, Promotes mTORC1 Activation and Neurodegeneration. Neuron. 2017;93:115–131. doi: 10.1016/j.neuron.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu L., Zhang K., Sandoval H., Yamamoto S., Jaiswal M., Sanz E., Li Z., Hui J., Graham B.H., Quintana A., Bellen H.J. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell. 2015;160:177–190. doi: 10.1016/j.cell.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tricoire H., Battisti V., Trannoy S., Lasbleiz C., Pret A.M., Monnier V. The steroid hormone receptor EcR finely modulates Drosophila lifespan during adulthood in a sex-specific manner. Mech. Ageing Dev. 2009;130:547–552. doi: 10.1016/j.mad.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Rajan A., Tien A.-C., Haueter C.M., Schulze K.L., Bellen H.J. The Arp2/3 complex and WASp are required for apical trafficking of Delta into microvilli during cell fate specification of sensory organ precursors. Nat. Cell Biol. 2009;11:815–824. doi: 10.1038/ncb1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayashi S., Ito K., Sado Y., Taniguchi M., Akimoto A., Takeuchi H., Aigaki T., Matsuzaki F., Nakagoshi H., Tanimura T. GETDB, a database compiling expression patterns and molecular locations of a collection of Gal4 enhancer traps. Genesis. 2002;34:58–61. doi: 10.1002/gene.10137. [DOI] [PubMed] [Google Scholar]
- 41.Venken K.J.T., He Y., Hoskins R.A., Bellen H.J. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 42.Lee P.-T., Zirin J., Kanca O., Lin W.-W., Schulze K.L., Li-Kroeger D., Tao R., Devereaux C., Hu Y., Chung V. A gene-specific T2A-GAL4 library for Drosophila. eLife. 2018;7:e35574. doi: 10.7554/eLife.35574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bischof J., Maeda R.K., Hediger M., Karch F., Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodman L.D., Prudencio M., Kramer N.J., Martinez-Ramirez L.F., Srinivasan A.R., Lan M., Parisi M.J., Zhu Y., Chew J., Cook C.N. Toxic expanded GGGGCC repeat transcription is mediated by the PAF1 complex in C9orf72-associated FTD. Nat. Neurosci. 2019;22:863–874. doi: 10.1038/s41593-019-0396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravenscroft T.A., Janssens J., Lee P.-T., Tepe B., Marcogliese P.C., Makhzami S., Holmes T.C., Aerts S., Bellen H.J. Drosophila Voltage-Gated Sodium Channels Are Only Expressed in Active Neurons and Are Localized to Distal Axonal Initial Segment-like Domains. J. Neurosci. 2020;40:7999–8024. doi: 10.1523/JNEUROSCI.0142-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Link N., Chung H., Jolly A., Withers M., Tepe B., Arenkiel B.R., Shah P.S., Krogan N.J., Aydin H., Geckinli B.B. Mutations in ANKLE2, a ZIKA Virus Target, Disrupt an Asymmetric Cell Division Pathway in Drosophila Neuroblasts to Cause Microcephaly. Dev. Cell. 2019;51:713–729.e6. doi: 10.1016/j.devcel.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodman L.D., Prudencio M., Srinivasan A.R., Rifai O.M., Lee V.M.-Y., Petrucelli L., Bonini N.M. eIF4B and eIF4H mediate GR production from expanded G4C2 in a Drosophila model for C9orf72-associated ALS. Acta Neuropathol. Commun. 2019;7:62. doi: 10.1186/s40478-019-0711-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J., Al-Ouran R., Hu Y., Kim S.-Y., Wan Y.-W., Wangler M.F., Yamamoto S., Chao H.-T., Comjean A., Mohr S.E., UDN MARRVEL: Integration of Human and Model Organism Genetic Resources to Facilitate Functional Annotation of the Human Genome. Am. J. Hum. Genet. 2017;100:843–853. doi: 10.1016/j.ajhg.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samocha K.E., Robinson E.B., Sanders S.J., Stevens C., Sabo A., McGrath L.M., Kosmicki J.A., Rehnström K., Mallick S., Kirby A. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuller Z.L., Berg J.J., Mostafavi H., Sella G., Przeworski M. Measuring intolerance to mutation in human genetics. Nat. Genet. 2019;51:772–776. doi: 10.1038/s41588-019-0383-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato M., Mizoro Y., Atobe Y., Fujimoto Y., Yamaguchi Y., Fustin J.-M., Doi M., Okamura H. Transportin 1 in the mouse brain: appearance in regions of neurogenesis, cerebrospinal fluid production/sensing, and circadian clock. J. Comp. Neurol. 2011;519:1770–1780. doi: 10.1002/cne.22600. [DOI] [PubMed] [Google Scholar]
- 54.Coe B.P., Witherspoon K., Rosenfeld J.A., van Bon B.W.M., Vulto-van Silfhout A.T., Bosco P., Friend K.L., Baker C., Buono S., Vissers L.E.L.M. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lehman A.M., McFadden D., Pugash D., Sangha K., Gibson W.T., Patel M.S. Schinzel-Giedion syndrome: report of splenopancreatic fusion and proposed diagnostic criteria. Am. J. Med. Genet. A. 2008;146A:1299–1306. doi: 10.1002/ajmg.a.32277. [DOI] [PubMed] [Google Scholar]
- 56.Lindstrand A., Grigelioniene G., Nilsson D., Pettersson M., Hofmeister W., Anderlid B.-M., Kant S.G., Ruivenkamp C.A.L., Gustavsson P., Valta H. Different mutations in PDE4D associated with developmental disorders with mirror phenotypes. J. Med. Genet. 2014;51:45–54. doi: 10.1136/jmedgenet-2013-101937. [DOI] [PubMed] [Google Scholar]
- 57.Allemand E., Dokudovskaya S., Bordonné R., Tazi J. A conserved Drosophila transportin-serine/arginine-rich (SR) protein permits nuclear import of Drosophila SR protein splicing factors and their antagonist repressor splicing factor 1. Mol. Biol. Cell. 2002;13:2436–2447. doi: 10.1091/mbc.E02-02-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo L., Kim H.J., Wang H., Monaghan J., Freyermuth F., Sung J.C., O’Donovan K., Fare C.M., Diaz Z., Singh N. Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell. 2018;173:677–692.e20. doi: 10.1016/j.cell.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jäckel S., Summerer A.K., Thömmes C.M., Pan X., Voigt A., Schulz J.B., Rasse T.M., Dormann D., Haass C., Kahle P.J. Nuclear import factor transportin and arginine methyltransferase 1 modify FUS neurotoxicity in Drosophila. Neurobiol. Dis. 2015;74:76–88. doi: 10.1016/j.nbd.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 60.Shi Q., Han Y., Jiang J. Suppressor of fused impedes Ci/Gli nuclear import by opposing Trn/Kapβ2 in Hedgehog signaling. J. Cell Sci. 2014;127:1092–1103. doi: 10.1242/jcs.142828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siomi M.C., Fromont M., Rain J.-C., Wan L., Wang F., Legrain P., Dreyfuss G. Functional conservation of the transportin nuclear import pathway in divergent organisms. Mol. Cell. Biol. 1998;18:4141–4148. doi: 10.1128/mcb.18.7.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S.E. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perkins L.A., Holderbaum L., Tao R., Hu Y., Sopko R., McCall K., Yang-Zhou D., Flockhart I., Binari R., Shim H.-S. The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics. 2015;201:843–852. doi: 10.1534/genetics.115.180208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 65.Singh A., Kango-Singh M. Springer International Publishing; Cham: 2020. Molecular Genetics of Axial Patterning, Growth and Disease in Drosophila Eye. [Google Scholar]
- 66.Golic M.M., Rong Y.S., Petersen R.B., Lindquist S.L., Golic K.G. FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res. 1997;25:3665–3671. doi: 10.1093/nar/25.18.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beira J.V., Paro R. The legacy of Drosophila imaginal discs. Chromosoma. 2016;125:573–592. doi: 10.1007/s00412-016-0595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diaz de la Loza M.C., Thompson B.J. Forces shaping the Drosophila wing. Mech. Dev. 2017;144(Pt A):23–32. doi: 10.1016/j.mod.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 69.Duffy J.B. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- 70.Lemon W.C., Pulver S.R., Höckendorf B., McDole K., Branson K., Freeman J., Keller P.J. Whole-central nervous system functional imaging in larval Drosophila. Nat. Commun. 2015;6:7924. doi: 10.1038/ncomms8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sepp K.J., Schulte J., Auld V.J. Peripheral glia direct axon guidance across the CNS/PNS transition zone. Dev. Biol. 2001;238:47–63. doi: 10.1006/dbio.2001.0411. [DOI] [PubMed] [Google Scholar]
- 72.Spokony R.F., Restifo L.L. Broad Complex isoforms have unique distributions during central nervous system metamorphosis in Drosophila melanogaster. J. Comp. Neurol. 2009;517:15–36. doi: 10.1002/cne.22119. [DOI] [PubMed] [Google Scholar]
- 73.Robinow S., White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- 74.Modi M.N., Shuai Y., Turner G.C. The Drosophila Mushroom Body: From Architecture to Algorithm in a Learning Circuit. Annu. Rev. Neurosci. 2020;43:465–484. doi: 10.1146/annurev-neuro-080317-0621333. [DOI] [PubMed] [Google Scholar]
- 75.Fushima K., Tsujimura H. Precise control of fasciclin II expression is required for adult mushroom body development in Drosophila. Dev. Growth Differ. 2007;49:215–227. doi: 10.1111/j.1440-169X.2007.00922.x. [DOI] [PubMed] [Google Scholar]
- 76.Nicholson L., Singh G.K., Osterwalder T., Roman G.W., Davis R.L., Keshishian H. Spatial and temporal control of gene expression in Drosophila using the inducible GeneSwitch GAL4 system. I. Screen for larval nervous system drivers. Genetics. 2008;178:215–234. doi: 10.1534/genetics.107.081968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vilinsky I., Johnson K.G. Electroretinograms in Drosophila: a robust and genetically accessible electrophysiological system for the undergraduate laboratory. J. Undergrad. Neurosci. Educ. 2012;11:A149–A157. [PMC free article] [PubMed] [Google Scholar]
- 78.Wang T., Montell C. Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 2007;454:821–847. doi: 10.1007/s00424-007-0251-1. [DOI] [PubMed] [Google Scholar]
- 79.Harnish J.M., Deal S.L., Chao H.-T., Wangler M.F., Yamamoto S. In Vivo Functional Study of Disease-associated Rare Human Variants Using Drosophila. J. Vis. Exp. 2019 doi: 10.3791/59658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakayama M., Ishibashi T., Ishikawa H.O., Sato H., Usui T., Okuda T., Yashiro H., Ishikawa H., Taikou Y., Minami A. A gain-of-function screen to identify genes that reduce lifespan in the adult of Drosophila melanogaster. BMC Genet. 2014;15:46. doi: 10.1186/1471-2156-15-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li W.-Z., Li S.-L., Zheng H.Y., Zhang S.-P., Xue L. A broad expression profile of the GMR-GAL4 driver in Drosophila melanogaster. Genet. Mol. Res. 2012;11:1997–2002. doi: 10.4238/2012.August.6.4. [DOI] [PubMed] [Google Scholar]
- 82.Ray M., Lakhotia S.C. The commonly used eye-specific sev-GAL4 and GMR-GAL4 drivers in Drosophila melanogaster are expressed in tissues other than eyes also. J. Genet. 2015;94:407–416. doi: 10.1007/s12041-015-0535-8. [DOI] [PubMed] [Google Scholar]
- 83.Zhang P., Wang Q., Hughes H., Intrieri G. Synthetic Lethality Induced by a Strong Drosophila Enhancer of Expanded Polyglutamine Tract. OJGen. 2014;04:300–315. [Google Scholar]
- 84.de Ligt J., Willemsen M.H., van Bon B.W., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 85.Lelieveld S.H., Reijnders M.R.F., Pfundt R., Yntema H.G., Kamsteeg E.-J., de Vries P., de Vries B.B.A., Willemsen M.H., Kleefstra T., Löhner K. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 86.McRae J.F., Clayton S., Fitzgerald T.W., Kaplanis J., Prigmore E., Rajan D., Sifrim A., Aitken S., Akawi N., Deciphering Developmental Disorders Study Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Grozeva D., Carss K., Spasic-Boskovic O., Tejada M.-I., Gecz J., Shaw M., Corbett M., Haan E., Thompson E., Friend K., Italian X-linked Mental Retardation Project; UK10K Consortium; GOLD Consortium Targeted Next-Generation Sequencing Analysis of 1,000 Individuals with Intellectual Disability. Hum. Mutat. 2015;36:1197–1204. doi: 10.1002/humu.22901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barbato S., Kapinos L.E., Rencurel C., Lim R.Y.H. Karyopherin enrichment at the nuclear pore complex attenuates Ran permeability. J. Cell Sci. 2020;133:133. doi: 10.1242/jcs.238121. [DOI] [PubMed] [Google Scholar]
- 89.Lam W.W.K., Millichap J.J., Soares D.C., Chin R., McLellan A., FitzPatrick D.R., Elmslie F., Lees M.M., Schaefer G.B., Abbott C.M., DDD study Novel de novo EEF1A2 missense mutations causing epilepsy and intellectual disability. Mol. Genet. Genomic Med. 2016;4:465–474. doi: 10.1002/mgg3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kaminsky E.B., Kaul V., Paschall J., Church D.M., Bunke B., Kunig D., Moreno-De-Luca D., Moreno-De-Luca A., Mulle J.G., Warren S.T. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet. Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Miller D.T., Adam M.P., Aradhya S., Biesecker L.G., Brothman A.R., Carter N.P., Church D.M., Crolla J.A., Eichler E.E., Epstein C.J. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Janiszewska M., De Vito C., Le Bitoux M.-A., Fusco C., Stamenkovic I. Transportin regulates nuclear import of CD44. J. Biol. Chem. 2010;285:30548–30557. doi: 10.1074/jbc.M109.075838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kimura M., Kose S., Okumura N., Imai K., Furuta M., Sakiyama N., Tomii K., Horton P., Takao T., Imamoto N. Identification of cargo proteins specific for the nucleocytoplasmic transport carrier transportin by combination of an in vitro transport system and stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics. Mol. Cell. Proteomics. 2013;12:145–157. doi: 10.1074/mcp.M112.019414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Arnold M., Nath A., Wohlwend D., Kehlenbach R.H. Transportin is a major nuclear import receptor for c-Fos: a novel mode of cargo interaction. J. Biol. Chem. 2006;281:5492–5499. doi: 10.1074/jbc.M513281200. [DOI] [PubMed] [Google Scholar]
- 96.Malnou C.E., Salem T., Brockly F., Wodrich H., Piechaczyk M., Jariel-Encontre I. Heterodimerization with Jun family members regulates c-Fos nucleocytoplasmic traffic. J. Biol. Chem. 2007;282:31046–31059. doi: 10.1074/jbc.M702833200. [DOI] [PubMed] [Google Scholar]
- 97.Waldmann I., Wälde S., Kehlenbach R.H. Nuclear import of c-Jun is mediated by multiple transport receptors. J. Biol. Chem. 2007;282:27685–27692. doi: 10.1074/jbc.M703301200. [DOI] [PubMed] [Google Scholar]
- 98.Hasselblatt P., Gresh L., Kudo H., Guinea-Viniegra J., Wagner E.F. The role of the transcription factor AP-1 in colitis-associated and β-catenin-dependent intestinal tumorigenesis in mice. Oncogene. 2008;27:6102–6109. doi: 10.1038/onc.2008.211. [DOI] [PubMed] [Google Scholar]
- 99.Nenci A., Becker C., Wullaert A., Gareus R., van Loo G., Danese S., Huth M., Nikolaev A., Neufert C., Madison B. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557–561. doi: 10.1038/nature05698. [DOI] [PubMed] [Google Scholar]
- 100.Qiu W., Wang X., Buchanan M., He K., Sharma R., Zhang L., Wang Q., Yu J. ADAR1 is essential for intestinal homeostasis and stem cell maintenance. Cell Death Dis. 2013;4 doi: 10.1038/cddis.2013.125. e599–e599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fritz J., Strehblow A., Taschner A., Schopoff S., Pasierbek P., Jantsch M.F. RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol. Cell. Biol. 2009;29:1487–1497. doi: 10.1128/MCB.01519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Samborski P., Grzymisławski M. The Role of HSP70 Heat Shock Proteins in the Pathogenesis and Treatment of Inflammatory Bowel Diseases. Adv. Clin. Exp. Med. 2015;24:525–530. doi: 10.17219/acem/44144. [DOI] [PubMed] [Google Scholar]
- 103.Korge S., Maier B., Brüning F., Ehrhardt L., Korte T., Mann M., Herrmann A., Robles M.S., Kramer A. The non-classical nuclear import carrier Transportin 1 modulates circadian rhythms through its effect on PER1 nuclear localization. PLoS Genet. 2018;14:e1007189. doi: 10.1371/journal.pgen.1007189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Singh K., Zimmerman A.W. Sleep in Autism Spectrum Disorder and Attention Deficit Hyperactivity Disorder. Semin. Pediatr. Neurol. 2015;22:113–125. doi: 10.1016/j.spen.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 105.Dormann D., Madl T., Valori C.F., Bentmann E., Tahirovic S., Abou-Ajram C., Kremmer E., Ansorge O., Mackenzie I.R.A., Neumann M., Haass C. Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J. 2012;31:4258–4275. doi: 10.1038/emboj.2012.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Leemann-Zakaryan R.P., Pahlich S., Grossenbacher D., Gehring H. Tyrosine Phosphorylation in the C-Terminal Nuclear Localization and Retention Signal (C-NLS) of the EWS Protein. Sarcoma. 2011;2011:218483. doi: 10.1155/2011/218483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Desmond C.R., Atwal R.S., Xia J., Truant R. Identification of a karyopherin β1/β2 proline-tyrosine nuclear localization signal in huntingtin protein. J. Biol. Chem. 2012;287:39626–39633. doi: 10.1074/jbc.M112.412379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Popovitchenko T., Thompson K., Viljetic B., Jiao X., Kontonyiannis D.L., Kiledjian M., Hart R.P., Rasin M.R. The RNA binding protein HuR determines the differential translation of autism-associated FoxP subfamily members in the developing neocortex. Sci. Rep. 2016;6:28998. doi: 10.1038/srep28998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ho W.Y., Chang J.-C., Tyan S.-H., Yen Y.-C., Lim K., Tan B.S.Y., Ong J., Tucker-Kellogg G., Wong P., Koo E., Ling S.C. FUS-mediated dysregulation of Sema5a, an autism-related gene, in FUS mice with hippocampus-dependent cognitive deficits. Hum. Mol. Genet. 2019;28:3777–3791. doi: 10.1093/hmg/ddz217. [DOI] [PubMed] [Google Scholar]
- 110.Kino Y., Washizu C., Kurosawa M., Yamada M., Miyazaki H., Akagi T., Hashikawa T., Doi H., Takumi T., Hicks G.G. FUS/TLS deficiency causes behavioral and pathological abnormalities distinct from amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 2015;3:24. doi: 10.1186/s40478-015-0202-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Koscielny G., Yaikhom G., Iyer V., Meehan T.F., Morgan H., Atienza-Herrero J., Blake A., Chen C.-K., Easty R., Di Fenza A. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 2014;42:D802–D809. doi: 10.1093/nar/gkt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barington M., Risom L., Ek J., Uldall P., Ostergaard E. A recurrent de novo CUX2 missense variant associated with intellectual disability, seizures, and autism spectrum disorder. Eur. J. Hum. Genet. 2018;26:1388–1391. doi: 10.1038/s41431-018-0184-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cubelos B., Sebastián-Serrano A., Beccari L., Calcagnotto M.E., Cisneros E., Kim S., Dopazo A., Alvarez-Dolado M., Redondo J.M., Bovolenta P. Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron. 2010;66:523–535. doi: 10.1016/j.neuron.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kuriyama H., Asakawa S., Minoshima S., Maruyama H., Ishii N., Ito K., Gejyo F., Arakawa M., Shimizu N., Kuwano R. Characterization and chromosomal mapping of a novel human gene, ANKHZN. Gene. 2000;253:151–160. doi: 10.1016/s0378-1119(00)00247-x. [DOI] [PubMed] [Google Scholar]
- 115.Ding M., Weng C., Fan S., Cao Q., Lu Z. Purkinje Cell Degeneration and Motor Coordination Deficits in a New Mouse Model of Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay. Front. Mol. Neurosci. 2017;10:121. doi: 10.3389/fnmol.2017.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bott C.J., Winckler B. Intermediate filaments in developing neurons: Beyond structure. Cytoskeleton (Hoboken) 2020;77:110–128. doi: 10.1002/cm.21597. [DOI] [PubMed] [Google Scholar]
- 117.Van De Weghe J.C., Rusterholz T.D.S., Latour B., Grout M.E., Aldinger K.A., Shaheen R., Dempsey J.C., Maddirevula S., Cheng Y.-H.H., Phelps I.G., University of Washington Center for Mendelian Genomics Mutations in ARMC9, which Encodes a Basal Body Protein, Cause Joubert Syndrome in Humans and Ciliopathy Phenotypes in Zebrafish. Am. J. Hum. Genet. 2017;101:23–36. doi: 10.1016/j.ajhg.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets. All reagents developed in this study are available upon request.