Summary
CYP2C9 encodes a cytochrome P450 enzyme responsible for metabolizing up to 15% of small molecule drugs, and CYP2C9 variants can alter the safety and efficacy of these therapeutics. In particular, the anti-coagulant warfarin is prescribed to over 15 million people annually and polymorphisms in CYP2C9 can affect individual drug response and lead to an increased risk of hemorrhage. We developed click-seq, a pooled yeast-based activity assay, to test thousands of variants. Using click-seq, we measured the activity of 6,142 missense variants in yeast. We also measured the steady-state cellular abundance of 6,370 missense variants in a human cell line by using variant abundance by massively parallel sequencing (VAMP-seq). These data revealed that almost two-thirds of CYP2C9 variants showed decreased activity and that protein abundance accounted for half of the variation in CYP2C9 function. We also measured activity scores for 319 previously unannotated human variants, many of which may have clinical relevance.
Keywords: CYP2C9, deep mutational scanning, pharmacogenomics, warfarin, humanized yeast
Introduction
Recent sequencing efforts have resulted in an avalanche of new variants, many of which are variants of uncertain significance (VUSs)—variants identified through genetic testing whose functional significance is unknown. VUSs hamper the implementation of precision medicine because they must be classified as pathogenic or benign before they can be used to inform clinical decisions. Over half of the missense variants in ClinVar1 are VUSs.2 VUSs are a particular problem in the field of pharmacogenomics, which seeks to understand the genetic sources of inter-individual variation in drug response. Functionally annotated pharmacogene variants can be used to guide dosing decisions and predict adverse drug reactions (ADRs), which cost U.S. hospitals up to 30 billion dollars annually and are a leading cause of hospitalization and death.3,4 30% of ADRs are predicted to be caused by inter-individual variability in drug metabolizing enzymes and other drug-related genes.5 Genetic variants predict drug response for a subset of important drugs, and implementing genotype-guided drug dosing can improve individual outcomes.6 However, the vast majority of pharmacogene variants discovered so far are of unclear functional effect.
One important group of pharmacogenes is the cytochromes P450 (CYPs). CYPs are a superfamily of monooxygenase enzymes that use heme as a cofactor, and there are 57 CYP genes in humans.7 CYP2C9 (MIM: 601130) in particular is the primary metabolic enzyme for a wide range of drugs, including drugs that must be dosed carefully, such as phenytoin (for seizures) and the widely prescribed oral anticoagulant warfarin.8,9 CYP2C9 polymorphisms contribute to an estimated 15% of the variation in warfarin dose,10 and some common coding variants have large effects. For example, the CYP2C9 p.Ile359Leu (c.42614A>C) missense variant results in substantially diminished S-warfarin clearance leading to warfarin sensitivity.11 Genotype-guided warfarin dosing based on CYP2C9 and VKORC1 (MIM: 608547) alleles can improve treatment in some situations12 but relies on knowing the function of alleles to guide dosing decisions.
Only a subset of CYP alleles have been studied adequately for genotype-guided dosing. As human CYP alleles are discovered, they are named according to the star (∗) system13 and curated by the PharmVar Consortium.14 There are 70 documented CYP2C9 star alleles in the PharmVar database. The Clinical Pharmacogenetics Implementation Consortium (CPIC) reviews in vitro and in vivo evidence and provides clinical functional recommendations for CYP2C9 and other pharmacogenes.15 CPIC has provided clinical allele functional annotations for 40 of the 70 CYP2C9 star alleles.16,17 However, there are many more CYP2C9 alleles than those documented in PharmVar. CYP2C9 has eight common alleles (MAF > 1%)18 and hundreds of documented rare alleles (MAF < 1%).19,20 In the population database gnomAD,20 there are 466 missense variants in CYP2C9, half of which are singletons. The vast majority of variation in CYP2C9 is unannotated, and so knowing the functional consequence of existing and yet-to-be discovered variants will help improve dosing of drugs cleared by CYP2C9. Thus, there is a need for a large-scale experimental effort to comprehensively characterize CYP2C9 variants.
We used deep mutational scanning (DMS) to measure the enzyme activity and steady-state cellular abundance of thousands of CYP2C9 missense variants. DMS is a high-throughput method for probing variant function via application of a functional selection, which enriches variants with high function and depletes variants with low function.21 High-throughput DNA sequencing is then used for the quantification of the change in each variant’s frequency during the selection, yielding a functional score for every variant in the library. Selections can take many forms but often couple variant function to cell growth or measure protein or ligand binding and rarely measure enzyme activity directly (e.g., Romero et al.22). DMS approaches have the potential to transform pharmacogenomic implementation,23,24 but so far they have been applied to only a handful of pharmacogenes, including TPMT, NUDT15, and VKORC1.25, 26, 27 Very few multiplexed methods for quantifying enzyme activity directly in cells currently exist, precluding the quantification of variant effects on human CYP enzymes despite the clear need for such comprehensive functional data.
To meet this challenge, we developed click-seq, a multiplexed, sequencing-based method for quantifying protein variant activity, and used it to measure the activity of 6,142 CYP2C9 missense variants in yeast cells. Additionally, we leveraged the variant abundance by massively parallel sequencing (VAMP-seq) assay we developed previously,25 which uses a fluorescent protein reporter coupled with FACS, to measure the abundance of 6,370 CYP2C9 missense variants in cultured human cells. Comparison of both activity and abundance revealed that the mechanism behind variant loss of function could be attributed to reduced abundance for at least 50% of variants. Additionally, these data highlighted key regions of CYP2C9 crucial to function, including many residues involved in heme binding. Finally, our experimental functional scores are concordant with existing CYP2C9 functional annotations. We used our activity scores to annotate 319 previously unannotated human CYP2C9 missense variants in gnomAD, over half of which had reduced activity. In addition to annotating these 319 variants, we provide activity scores for 5,797 additional variants. This information will be of great utility to clinicians as a key source of evidence when presented with VUSs and will aid in improving dosing efficacy of drugs metabolized by CYP2C9.
Material and methods
General reagents
Unless otherwise noted, all chemicals were obtained from Sigma Aldrich Chemical (St. Louis, MO) and all enzymes were obtained from New England Biolabs. Tienilic acid, 6-hydroxywarfarin-d5, 7-hydroxywarfarin-d5, and 4-hydroxyphenytoin-d5 were synthesized according to published protocols.28 Hex-5-yn-1-amine was purchased from GFS Chemicals (Powell, OH). All cell culture reagents were purchased from Thermo Fisher Scientific unless otherwise noted. All yeast strains are listed in Table S1. All plasmids and oligonucleotides are listed in Table S4.
Growth media and culturing techniques
E. coli were cultured at 37°C in Luria broth (LB). Yeast were cultured at 30°C. Yeast culture media was prepared according to the following recipes. Yeast peptone (YP) media: 1% yeast extract and 2% peptone. Yeast peptone dextrose (YPD) media: 1% yeast extract, 2% peptone, and 2% (w/v) glucose. Synthetic drop-out media lacking uracil (C-ura): 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 0.2% dropout mix lacking uracil, and 2% (w/v) glucose. Synthetic drop-out media lacking tryptophan (C-trp): 0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate, 0.2% dropout mix lacking tryptophan, and 2% (w/v) glucose. Unless otherwise specified, all yeast transformations were performed with the lithium acetate/single-stranded carrier DNA/PEG method.29
Yeast cells carrying CYP2C9 wild-type (WT) or variant plasmid were induced as follows: a single colony was inoculated into 5 mL YPD media supplemented with 200 μg/mL G418 and grown overnight with rotation. This culture was diluted 1:50 into fresh YP media containing 2% (w/v) raffinose and supplemented with 200 μg/mL G418 and grown for at least two cell doublings. Cultures were then inoculated to optical density at 600 nm (OD600) 0.0125 into fresh YP media containing 2% (w/v) galactose and 200 μg/mL G418 and collected after seven doublings the following day.
HEK293T cells (ATCC CRL-3216) and derivatives thereof were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 0.1 mg/mL streptomycin. Cells were induced with 2.5 μg/mL doxycycline. Cells were passaged by detachment with trypsin-EDTA 0.25%, and cells were prepared for sorting by detachment with versene. All cell lines tested negative for mycoplasma.
CYP2C9 library mutagenesis
The activity and abundance CYP2C9 variant libraries were generated via inverse PCR-based site-directed saturation mutagenesis30 modified as follows. Saturation mutagenesis primers were designed for each codon in CYP2C9 from positions 2 to 490 such that the forward primer contained an NNK at the 5′ end of the sequence. Primers were ordered resuspended from Integrated DNA Technologies (IDT). Forward and reverse primers for each position were mixed at 2.5 μM and used in a PCR reaction with 125 pg of template, 5% DMSO, and 5 μL of KAPA Hifi Hotstart 2X ReadyMix. We visualized PCR products on a 0.7% agarose gel to confirm amplification of the correct product, and we then quantified PCR products by using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen) or Qubit fluorometry (Life Technologies). Products were pooled at equimolar ratios and cleaned with the DNA Clean and Concentrator Kit (Zymo Research), and then products were gel extracted. The pooled libraries were phosphorylated with T4 polynucleotide kinase, incubated at 37°C for 30 min, and heat inactivated at 65°C for 20 min. 8.5 μL of this phosphorylated product was combined with 1 μL of 10× T4 ligase buffer and 0.5 μL of T4 DNA ligase (NEB) and incubated at 16°C overnight.
The overnight ligation was cleaned and concentrated (Zymo) and used for transformation of electrocompetent E. coli cells (NEB C2989K or C3020K) with the ligated products via electroporation (settings: 2 kV). Pre-warmed SOC media was added to each cuvette after electroporation and the culture was recovered at 37°C with shaking for 1 h. At 1 h, we took 1 and 10 μL samples from all cultures and plated them on LB + kanamycin, and we used the remaining 989 μL to inoculate a 50 mL LB + kanamycin culture (LB + kanamycin). After overnight growth, we midiprepped the liquid culture and counted the plates to calculate how many unique molecules were transformed to gauge coverage of the library. Libraries were then subcloned into their respective expression and recombination vectors and barcoded as detailed below.
CYP2C9 yeast codon-optimized variant library construction in S. cerevisiae
To generate the template vector for mutagenesis, we synthesized (IDT) CYP2C9 sequence (UniProt: P11712) codon optimized for S. cerevisiae expression and cloned it into the vector pHSG298 (Clontech) by using restriction sites SalI and XbaI. We used this vector to generate a site-saturated mutagenesis library, as described above. Next, we subcloned the library back into the low-copy p41KGAL1 vector by using restriction sites SpeI and SalI, and we used ligated products to transform electrocompetent E. coli cells (NEB C2989K) as described above, but this time we selected for growth on LB + ampicillin (antibiotic switching strategy).
To barcode the library, we digested library plasmid with SalI at 37°C for 1 h and heat inactivated it at 65°C for 20 min. We then ordered barcode oligos with 18 bp random sequences IDT, resuspended them at 100 μM, and then annealed them by combining 1 μL each of primer with 4 μL CutSmart Buffer and 34 μL ddH2O and running at 98°C for 3 min followed by ramping down to 25°C at −0.1°C/s. After annealing, we then combined 0.8 μL of Klenow polymerase (exonuclease negative, NEB) and 1.35 μL of 1 mM dNTPS with the 40 μL of product to fill in the barcode oligo (cycling conditions: 25°C for 15 min, 70°C for 20 min, ramp down to 37°C at −0.1°C/s). Digested vector and barcode oligo were then ligated overnight at 16°C. We used the barcoded library to transform electrocompetent E. coli cells (NEB C2989K) and midiprepped them (QIAGEN). The size of the barcoded library was estimated via colony counts to be 280,000. To reduce library size, we again used the barcoded library to transform electrocompetent E. coli cells (NEB C2989K) and bottlenecked and midiprepped it (QIAGEN). The size of the barcoded library was estimated via colony counts to be 42,000.
To determine more accurate library barcode counts, we amplified two PCR replicates each using 1.5 μg of plasmid-extracted library by using custom barseq primers CJA120/CJA138 via KAPA2G Robust HotStart ReadyMix (Sigma 2GRHSRMKB) with the following conditions: 95°C for 3 min, five cycles of 95°C for 15 s, 60°C for 15 s, 72°C for 15 s, and 72°C for 1 min. We then purified the products by using AMPure XP beads (Beckman Coulter A63880) at 1:1 ratio (beads:DNA). We amplified the purified products by using primers CJA135 and JS486 or JS487 via KAPA2G Robust HotStart ReadyMix with the following PCR conditions: 95°C for 3 min, ten cycles of 95°C for 15 s, 65°C for 15 s, 72°C for 15 s, and 72°C for 1 min. We then gel extracted them by using the QIAquick Gel Extraction Kit (QIAGEN) and quantified them by Qubit fluorometry (Life Technologies). We pooled PCR replicates at equimolar ratios and deep sequenced them on an Illumina NextSeq500 to determine the number of barcodes present. Briefly, we merged forward and reverse reads with Pear,31 counted barcodes with Enrich2,32 and removed barcodes with less than ten reads, resulting in a total of ∼160,000 unique barcodes in the CYP2C9 library for an average of 17× coverage.
We used the barcoded CYP2C9 library to transform the humanized yeast strain YMD4256 (a strain expressing human CYP accessory proteins CPR and b5, see supplemental material and methods) by using the standard high-efficiency lithium acetate procedure mentioned above. We pooled four independent transformations to generate a library stock of OD600 5.7, equivalent to an average of 11× coverage (independent transformants) for each of the 160,000 independent barcoded variants. The latter estimate assumes that each yeast cell harbors one CYP2C9 variant and that all growth rates are similar. Library stocks were stored at −80°C in 25% (v/v) glycerol.
CYP2C9 human library construction
CYP2C9 sequence (UniProt: P11712) codon-optimized for human expression was synthesized (IDT) and cloned into the vector pHSG298. As with the yeast activity library, we used this template to generate a CYP2C9 library by using inverse PCR as described above. This library was transferred from pHSG298 to the recombination vector (attB-CYP2C9-EGFP-IRES-mCherry) via restriction site MluI and SphI. As before, we used ligated products to transform electrocompetent E. coli cells (NEB C2989K), selecting for growth on LB + ampicillin (antibiotic switching strategy).
This library was barcoded via the same method as the yeast activity library but with the AgeI site for barcode insertion. We used the overnight barcode ligation to transform electrocompetent E. coli and inoculated and plated it onto LB + ampicillin media at several different dilutions to ensure proper library size.
PacBio sequencing of CYP2C9 libraries for barcode-variant mapping
PacBio libraries were generated with the SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences) according to manufacturer’s directions with the following modifications. Barcoded variant sequences were excised with SpeI-HF and PspXI (activity library) or NheI and SmaI (abundance library) restriction enzymes and purified with AMPure PB beads (Pacific Biosciences 100-265-900) at 1:1 ratio (beads:DNA). Following end-repair and blunt end adaptor ligation, according to manufacturer’s instructions, we subjected PacBio libraries to two additional rounds of restriction digestion to remove any backbone plasmid contamination present in the library. Finally, libraries were cleaned in three consecutive rounds of AMPure PB beads (Pacific Biosciences 100-265-900) at 0.6:1 ratio (beads:DNA). The purity and size of PacBio libraries were confirmed by Tapestation (Agilent) and Bioanalyzer 2100 (Agilent) before proceeding with the sequencing run. Samples were submitted to University of Washington PacBio Sequencing Services and sequenced on two SMRT cells per library in a sequel run. The yeast activity library was sequenced with two replicate library preparations from the same miniprep, while the human abundance library was sequenced from two replicate samplings of the same E. coli ligation transformation.
We filtered long reads for at least ten passes and analyzed them by using a custom analysis pipeline to identify and link gene and barcode regions, AssemblyByPacBio. The activity library contained 66,958 unique nucleotide variants (22,421 of these full length, i.e., without indels) tagged by 105,372 unique barcodes, while the abundance library contained 37,758 unique nucleotide variants (22,669 of these full length) tagged by 78,740 unique barcodes (Table S3).
FACS-based deep mutational scan of activity library (click-seq)
CYP2C9 enzymatic activity was probed via a flow-cytometry-based method with a click chemistry compatible probe tienilic acid hexynyl amide activity-based probe (TAHA-ABP, synthesis described in supplemental material and methods) that has specificity for CYP2C9 activity with minimum reactivity toward other yeast proteins. We grew yeast cultures as described above to induce CYP expression, and for each sample, we collected 1 OD600/mL of overnight yeast culture via centrifugation at 4,000 rpm for 2 min, washed it with 0.5 mL of PBS by resuspension and centrifugation at 4,000 rpm for 2 min, and resuspended it in 100 μL PBS:0.1% saponin (w/v). Each sample was pre-incubated with 2 mM NADPH (Sigma N1630) at 37°C for 20 min. We treated all samples except a “no probe” control with 10 μM TAHA-ABP and incubated them with rotation at 37°C for 20 h to form activity-dependent CYP2C9-probe adducts. Samples were collected via centrifugation at 4,000 rpm for 2 min and washed three times with 0.5 mL PBS as above. Samples were resuspended in 100 μL PBS:0.1% saponin (w/v) and incubated at room temperature for 20 min. We added 100 μL 2× copper-catalyzed azide-alkyne cycloaddition (CuAAC) reaction buffer to cells to append a fluorophore reporter (2× concentrations: 10 μM CF488A picolyl azide [Biotium #92187], 2 mM CuSO4 [Sigma C8027], 4 mM THPTA [Sigma 762342], 6 mM ascorbic acid [Sigma A7631] in PBS) and vortexed it vigorously to mix. Samples were incubated in the dark at room temperature for 30 min and collected by centrifugation as above. Cells were washed five times in 0.5 mL PBS, resuspended in 1 mL PBS, and stored at 4°C up to 1 day.
To label and sort the CYP2C9 yeast library, we induced isogenic humanized yeast strains carrying control CYP2C9 variants (WT, p.Arg144Cys, p.Ile359Leu, and p.Cys435His) in galactose as described above. The barcoded CYP2C9 variant library was thawed at room temperature and ∼8 OD600/mL of library was inoculated into 25 mL YPD media supplemented with 200 μg/mL G418 and grown overnight at 150 rpm. The rest of the induction was performed as described above, with 5× culture volumes and shaking instead of rotation. For each control variant, one sample was collected (1 OD600/mL), and for the library, four samples (1 OD600/mL each) were collected. A “no probe” sample was included as a control. All samples were labeled with the activity assay described above with CYP2C9-specific activity-based probe TAHA-ABP.
Labeled cells were run on a BD AriaIII sorter (BD Biosciences, San Jose, CA) and a standard yeast singlet gate was used. For this population, we collected data on the AF488A channel (488 nm excitation; 530/30 nm detection filter), and we drew gates to contain approximately 10%, 10%, 20%, and 60% of events from the library sample, from most fluorescent (AF488A channel) to least fluorescent. Gates were sorted into 5 mL tubes, harvested by centrifugation, and stored at −20°C before library preparation. Flow cytometry data were collected via FACSDiva v.8.0.1 (BD Biosciences). Table S2 contains details of numbers of cells collected. Four biological replicates of the FACS-based deep mutational scan were performed.
Sorted activity library amplification and sequencing
For the yeast activity library, sorted samples were harvested by centrifugation and stored at −20°C. Plasmids were extracted from sorted cell pellets via the Zymoprep Yeast Plasmid Miniprep I Kit (Zymo Research D2001). Each sorted sample was split into two for PCR replicates. For each sample, the barcode region was amplified and an 18 bp unique molecular identifier (UMI) sequence was added with primers CJA120/CJA124 via KAPA2G Robust HotStart ReadyMix with the following conditions: 95°C for 3 min, two cycles of 95°C for 20 s, 60°C for 15 s, 72°C for 30 s, and 72°C for 1 min. It was then purified with AMPure XP beads (Beckman Coulter A63880) at 1:1 ratio (beads:DNA). Purified products were amplified with various forward (CJA135, CJA139, or CJA144) and reverse indexing primers (JS409-412, JS470-477) via KAPA2G Robust HotStart ReadyMix with 0.5× SYBR green (Roche #04707516001) on a miniOpticon (Bio-Rad) with the following PCR conditions: 95°C for 3 min, up to 30 cycles of 95°C for 20 s, 65°C for 15 s, 72°C for 30 s. Products were then removed from the thermocycler when the relative fluorescence units (RFUs) were between 0.5 and 1. These products were again purified with AMPure XP beads (Beckman Coulter A63880) at 1:1 ratio (beads:DNA) and were then gel extracted via the QIAquick Gel Extraction Kit (QIAGEN) and quantified by Qubit fluorometry (Life Technologies). Samples were pooled at equimolar ratios and deep sequenced on an Illumina NextSeq500. Within each sort there was a good correlation of barcode frequencies from PCR replicates (mean Pearson’s r = 0.859, mean Spearman’s ρ = 0.694, Figure S2).
FACS-based deep mutational scan of abundance library (VAMP-seq)
HEK293T cells with a Bxb1 serine integrase landing pad integrated via lentivirus with a selectable inducible Caspase 9 cassette (HEK293T-LLP-iCasp9)33 were used for all human cell experiments, enabling expression of a single variant per cell. To recombine variants into HEK293T cells, we transfected cells in 10 cm plates, 3,500,000 cells per plate (four plates per replicate). 7.1 μg of barcoded library plasmid was mixed with 0.48 μg of Bxb1 plasmid in 710 μL of OptiMEM. In a separate tube, 28.5 μL of Fugene was diluted in 685 μL of OptiMEM. The tubes were then combined and incubated at room temperature for 15 min. After incubation period, Fugene/DNA mixture was added to cells dropwise, and plates were placed in incubator at 37°C. A minimum of 48 h after transfection, cells were induced with doxycycline at a final concentration of 2.5 μg/mL. 24 h after induction with doxycycline, we added small molecule AP1903 to select from recombinant cells, which causes inducible Caspase 9 in unrecombined landing pads to dimerize and activate.
Recombined HEK293T cells were run on a BD AriaIII sorter. Cells were gated for live, recombined singlets. For this population, a ratio of eGFP/mCherry was calculated, and the histogram of this ratio was divided into four quartiles. Each quartile was sorted into a 5 mL tube. We grew out sorted cells for 2–4 days after sorting to ensure enough DNA for sequencing. Three biological replicates of the FACS-based deep mutational scan were performed.
Sorted abundance library amplification and sequencing
For the abundance library, cells were collected, pelleted by centrifugation, and stored at −20°C. Genomic DNA was prepared with a DNEasy Kit, according to the manufacturer’s instructions (QIAGEN), with the addition of a 30 min incubation at 37 °C with RNase in the re-suspension step. Eight 50 μL first-round PCR reactions were each prepared with a final concentration of ∼50 ng/μL input genomic DNA, 1 × Q5 High-Fidelity Master Mix, and 0.25 μM of the KAM499/VKORampR 1.1 primers. The reaction conditions were 98°C for 30 s, 98°C for 10 s, 65°C for 20 s, 72°C for 60 s, repeat 5 times, 72°C for 2 min, 4°C hold. Eight 50 μL reactions were combined, bound to AMPure XP (Beckman Coulter), cleaned, and eluted with 21 μL water. 40% of the eluted volume was mixed with Q5 High-Fidelity Master Mix; VKOR_indexF_1.1 and one of the indexed reverse primers, JS385 through JS388, were added at 0.25 μM each. These reactions were run with Sybr Green I on a Bio-Rad MiniOpticon; reactions were denatured for 3 min at 95°C and cycled 20 times at 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s with a final 3 min extension at 72°C. The indexed amplicons were mixed based in RFUs and run on a 1% agarose gel with Sybr Safe and gel extracted via a freeze and squeeze column (Bio-Rad). The product was quantified with a KAPA Library Quant Kit (KAPA Biosystems).
Library sequence analysis
For the activity library, barcode and UMI sequences were trimmed and filtered for minimum base quality Q20 via FASTX-toolkit. We collapsed barcodes according to UMIs by pasting the UMI sequence after the barcode sequence for each read and then identifying unique combinations of barcode-UMI (sort | uniq -c). We used the barcode from each unique barcode-UMI pair to generate a FASTQ file that we then input into Enrich232 to count variants. Barcodes assigned to variants containing insertions, deletions, or multiple amino-acid alterations were removed from the analysis, and barcode counts were collapsed into variant counts. Variants were kept if they had a total (across bin) frequency greater than 1 × 10−5 in each replicate (Figure S11). For each replicate, we used a weighted average of variant frequency across bins to determine activity score. To determine optimal bin weights, we performed a linear regression on activity score (pool score) versus individual variant TAHA-ABP labeling with 14 variants. We varied bin weights to determine the best fit regression between pool score and individual score, resulting in the following bin weights: w1 = 0.05 (bin1), w2 = 0.2, w3 = 0.25, w4 = 1 (bin4), R2 = 0.986. Scores were normalized to the median synonymous weighted average (set to a score of 1) and the median nonsense weighted average of nonsense variants in the first 90% of the protein (set to score of 0), and scores were averaged across replicates. Variants with less than two replicates were removed. Scores for missense variants range from −0.046 to 1.305 and have a bimodal distribution with peaks approximately matching the synonymous and nonsense distributions.
For the abundance library, barcode sequences were trimmed and filtered for minimum base quality Q20 via FASTX-toolkit. As with the activity library, barcodes were counted with Enrich2. Barcodes assigned to variants containing insertions, deletions, or multiple amino-acid alterations were removed from the analysis, and barcode counts were collapsed into variant counts. Variants were kept if they had a total (across bin) frequency greater than 1 × 10−4 in each replicate (Figure S11). Abundance scores were calculated as above but with the following weights: w1 = 0.25 (bin1), w2 = 0.5, w3 = 0.75, w4 = 1 (bin4). Scores were normalized to the synonymous and nonsense distributions as above, but only normalizing to nonsense scores in the middle 80% of positions, excluding the first and last 10% of the protein. Variants with less than two replicates were removed. Scores for missense variants range from −0.29 to 1.59 and have a trimodal distribution with upper and lower peaks approximately matching the synonymous and nonsense distributions.
To calculate specific activity score, we normalized activity and abundance scores such that the lowest and highest scores in the dataset were set to 0 and 1, respectively, and we calculated a ratio of normalized activity score to normalized abundance score (specific activity). Specific activity scores were only calculated for variants that had both activity and abundance scores.
Activity and abundance classes were determined as follows, on the basis of a method modified from Matreyek et al., 201825 (Figure S4). We used a synonymous score threshold to discriminate between “WT-like” and “decreased” scores. This threshold was set at the 5th percentile of synonymous scores (0.879 for activity score and 0.77 for abundance score). We used an upper synonymous threshold to discriminate between “WT-like” and “increased” scores, set at the 95th percentile of synonymous scores (1.102 for activity score and 1.212 for abundance score). Additionally, we used a nonsense score threshold to discriminate between “decreased” and “nonsense-like” scores; this threshold was the 95th percentile of nonsense scores (0.093 for activity score and 0.282 for abundance score). Variants were classified as “nonsense-like” if their score and upper confidence interval were less than the nonsense threshold or “possibly nonsense-like” if just their score was less than the threshold.
Click-seq internal validation with individual CYP2C9 variants
14 individual CYP2C9 variants were generated via an inverse PCR site-directed mutagenesis. Oligonucleotide pairs for each of the 14 variants are listed in Table S4. With these, point mutations were generated with KAPA HiFi DNA Polymerase (KAPA Biosystems KK2601) and 500 pg of CYP2C9 template sequence p41KGAL1-hCYP2C9-HA. After performing inverse PCR for each variant, products were run on a 0.7% agarose gel, extracted with the QIAquick Gel Extraction Kit (QIAGEN), treated with T4 polynucleotide kinase (NEB M0201) at 37°C for 30 min, and ligated with T4 DNA ligase (NEB M0202) at 16°C overnight. We used ligated products to transform chemically competent E. coli cells (NEB C2987 or Bioline BIO-85027). Bacterial clones were prepared for plasmid extraction with the QIAprep Spin Miniprep Kit (QIAGEN), and variant sequences were confirmed with Sanger sequencing. Plasmids containing missense variants were individually transformed into YMD4256 via the one-step transformation protocol34 and selection for growth in YPD supplemented with 200 μg/mL G418. Individual clones from each transformation were stored at −80°C.
Individual CYP2C9-yeast-expressed variants were grown and induced in galactose as described above, and 1 OD600/mL of culture was collected for each variant. All samples were labeled with the CYP2C9 functional assay described above with CYP2C9-specific activity-based probe TAHA-ABP. Labeled cells were analyzed with a BD LSRII and a standard yeast singlet gate was used. For this population, data were collected on the FITC channel (488 nm excitation; 530/30 nm detection filter) for 20,000 events. Flow cytometry data were collected with FACSDiva v.8.0.1 (BD Biosciences) and analyzed with FlowJo v.10.7.1 (Ashland, OR). Fluorescence (FITC geometric mean of gated single cells) was normalized to background labeling (“no probe” control) and variant ABPP labeling relative to WT was calculated. Three biological replicates of CYP2C9 individual variant validation were performed.
VAMP-seq internal validation with individual CYP2C9 variants
12 of the 14 variants used for click-seq validation that also had VAMP-seq abundance score were cloned via the IVA cloning35 site-directed mutagenesis method into the VAMP-seq recombination vector (attB-CYP2C9-EGFP-IRES-mCherry) via primers in Table S4 (MAC379 through MAC403). Point mutations were generated with KAPA HiFi DNA Polymerase (KAPA Biosystems KK2601) and 40 ng of CYP2C9 template sequence attB-CYP2C9-EGFP-IRES-mCherry. After performing inverse PCR for each variant, we digested products with DpnI and used them to transform chemically competent E. coli cells (NEB C2987 or Bioline BIO-85027). Bacterial clones were prepped with a midiprep kit, validated by Sanger sequencing, and HEK293T-LLP-iCasp9 cells with landing pad were transfected with these preps. To recombine variants into HEK293T cells, we transfected cells in 6-well plates with 400,000 cells per well. 2.7 μg of library plasmid was mixed with 0.300 μg of Bxb1 plasmid in 125 μL of OptiMEM and 5 μL P3000 reagent. In a separate tube, 2.25 μL of Lipofectamine was diluted in 125 μL of OptiMEM. The tubes were then combined and incubated at room temperature for 15 min. After incubation period, Lipofectamine/DNA mixture was added to cells dropwise and plates were placed in incubator at 37°C. A minimum of 48 h after transfection, cells were induced with doxycycline at a final concentration of 2.5 μg/mL. 24 h after induction with doxycycline, we added small molecule AP1903 to select for recombinant cells, which causes inducible Caspase 9 in unrecombined landing pads to dimerize and activate.
Recombined HEK293T cells were analyzed with a BD LSRII flow cytometer. Cells were gated for live, recombined singlets. For this population, a ratio of eGFP/mCherry was calculated and the geometric mean of the histogram of this ratio was reported. Flow cytometry data were collected with FACSDiva v.8.0.1 (BD Biosciences) and analyzed with FlowJo v.10.7.1 (Ashland, OR). Two biological replicates of CYP2C9 individual variant validation were performed.
Warfarin metabolism validation assay
S-warfarin (50 μM) was mixed together with yeast lysate (microsomes), prepared from CYP2C9-variant-expressing cells, at 5 mg/mL total protein in 100 mM KPi buffer (pH 7.4) (100 μL final incubation volume). After 3 min pre-incubation at 37°C in a water bath, we added NADPH to initiate (to 1 mM final concentration). Reactions were incubated for 20 min and were then quenched with the addition of 5 μL of ice-cold 70% HClO4. We added an internal standard solution, containing 5 ng each of 6-hydroxywarfarin-d5 and 7-hydroxywarfarin-d5, and vortexed and centrifuged the reaction products to remove protein. Supernatants were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Three technical replicates were carried out for each CYP2C9 variant lysate. We prepared calibration curves by spiking variable amounts of unlabeled 6- and 7-hydroxywarfarins into 100 μL volumes of KPi buffer to generate standard mixtures with final concentrations ranging from 1 nM to 1 μM. These standard solutions were worked up and analyzed in an identical fashion to that described for the incubation samples.
Phenytoin metabolism validation assay
Phenytoin (100 μM) was mixed together with yeast lysate (microsomes), prepared from CYP2C9-variant-expressing cells, at 5 mg/mL total protein in 100 mM KPi buffer (pH 7.4) (200 μL final incubation volume). After 3 min pre-incubation at 37°C in a water bath, we added NADPH stock (to 1 mM final concentration) to initiate the reactions. Reactions were incubated for 20 min and were then quenched with the addition of 20 μL of ice-cold 15% ZnSO4. We added 4-hydroxyphenytoin-d5 (p-HPPH-d5, 10 ng) as the internal standard, vortexed the reactions and then centrifuged them to remove protein, and analyzed the supernatants by LC-MS/MS. Again, three technical replicates were carried out for each CYP2C9 variant lysate. We prepared calibration curves by spiking variable amounts of unlabeled 4-hydroxyphenytoin (p-HPPH) into 200 μL volumes of KPi buffer, generating standard mixtures with final concentrations ranging from 1 nM to 1 μM. These standard solutions were worked up and analyzed in an identical fashion to that described for the incubation samples.
LC-MS/MS of warfarin and phenytoin metabolites
LC-MS/MS analyses of warfarin and phenytoin metabolic reactions were conducted on a Waters Xevo TQ-S Tandem Quadrupole Mass Spectrometer (Waters, Co., Milford, MA) coupled to an ACQUITY Ultra Performance LC (UPLC) System with integral autoinjector (Waters, Co.). The Xevo was operated in ESI+-MS/MS (selected reaction monitoring) mode at a source temperature of 150°C and a desolvation temperature of 350°C. The following mass transitions were monitored in separate ion channels for the various oxidative warfarin metabolites/standards: m/z 325 > 179 (6- and 7-hydroxywarfarins-d0) and m/z 330 > 179 (6- and 7-hydroxywarfarins-d5). The following mass transitions were monitored in separate ion channels for the phenytoin metabolite and standard: m/z 269 > 198 (p-HPPH-d0) and m/z 274 > 203 (p-HPPH-d5). Optimized cone voltages and collision energies were set to 25 V and 15 eV for all metabolites and standards of warfarin, while the cone voltage was set to 35 V with a collision energy of 15 eV for the phenytoin metabolite p-HPPH (both d0- and d5-labeled). Metabolic products from the warfarin incubations were separated on an Acquity BEH Phenyl, 1.7 μ, 2.1 × 150 mm UPLC column (Waters, Co.) via an isocratic gradient of 45% solvent A (0.1% aqueous formic acid) and 55% solvent B (methanol), with a constant flow rate of 0.35 mL/min. Phenytoin metabolites were separated with this same BEH Phenyl UPLC column with a solvent gradient of water (solvent A) and acetonitrile (solvent B), both of which contained 0.1% formic acid, running at a flow rate of 0.3 mL/min. Initially, solvent B was set to 28%, where it was maintained for 4.5 min, and then increased linearly to 95% over 0.5 min, where it was left for an additional 1.5 min. Metabolites were quantified through comparison of their peak area ratios (relative to either the 6- and 7-hydroxywarfarin-d5 or p-HPPH-d5 internal standard peak areas) to calibration curves via linear regression analysis. The limits of detection for all of the metabolites were below 5 fmol injected on column. Mass spectral data analyses for the Xevo TQ-S were performed on Windows XP-based Micromass MassLynxNT v.4.1 software (Waters, Co.).
Results
Click-seq: A multiplexed assay for CYP2C9 enzymatic activity
We developed a multiplexed assay of CYP activity, click-seq, that uses a CYP-selective, activity-based probe to modify CYP variant enzymes heterologously expressed in the budding yeast S. cerevisiae. Following probe attachment via mechanism-based adduction, we use click chemistry to label the enzyme-bound probe with a fluorophore, FACS to separate cells according to their degree of labeling, and high-throughput sequencing of the sorted cells to score each variant (Figure 1A). Click-seq directly measures enzyme activity by quantifying the amount of mechanism-based inhibitor covalently attached to the CYP enzymes in a cell after a period of incubation; thus, labeling is activity dependent. CYP-specific activity-based probes have been developed previously,36,37 but prior work has focused on in vitro assays (commonly CYP-rich microsomal preparations) rather than cell-based methods. We modified existing assays to work with intact yeast cells in a pooled format. We also synthesized an activity-based probe, tienilic acid hexynyl amide (TAHA-ABP), that is an analog of tienilic acid, a known covalent inhibitor of CYP2C9.38 TAHA-ABP showed better labeling than a generic P450 probe37 (Figure S1). Additionally, to improve recombinant CYP activity, we integrated human P450 accessory proteins cytochrome P450 reductase and cytochrome b5 into a modified laboratory strain (supplemental material and methods), resulting in a humanized yeast strain.
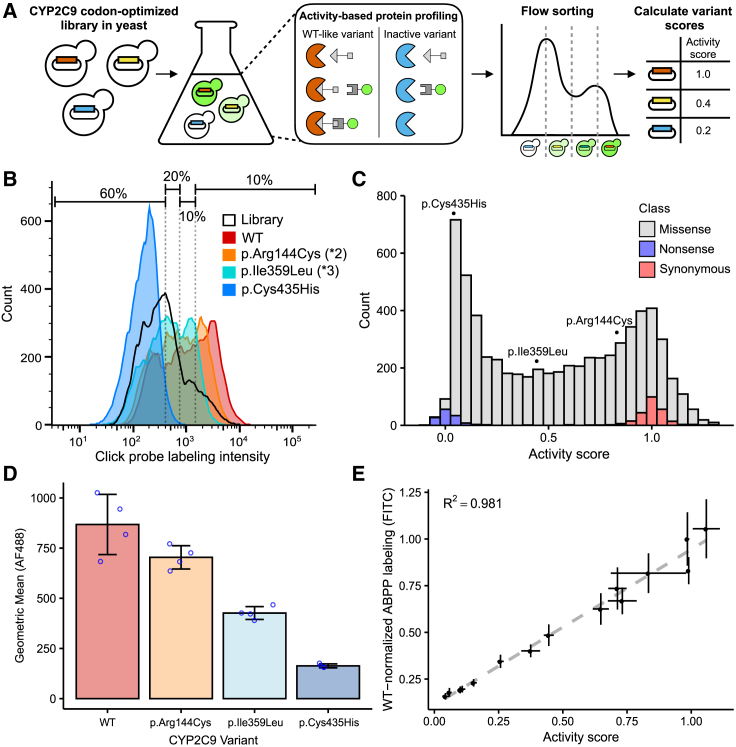
Figure 1.
Multiplexed measurement of CYP2C9 activity via click-seq
(A) A humanized yeast strain is transformed with a library of codon-optimized CYP2C9 variants, labeled with activity-based protein profiling (ABPP), resulting in a range of fluorescence levels, and sorted into four bins via fluorescence-activated cell sorting. Bins are sequenced for calculation of relative variant activity.
(B) Flow cytometry of ABPP-labeled yeast expressing CYP2C9 WT (red), reduced activity variants p.Arg144Cys (∗2, orange) and p.Ile359Leu (∗3, turquoise), null variant p.Cys435His (blue), and CYP2C9 variant library (black outline). Smoothed histograms are shown, and each sample represents ~20,000 cells. Note that some cells with low intensity are the result of plasmid loss and thus do not contribute to the downstream sequencing results. Histograms and binning shown are from one replicate and are representative of the other three replicates.
(C) Stacked histogram of activity score colored by type of variant. Individual scores of p.Cys435His, p.Ile359Leu (∗3), and p.Arg144Cys (∗2) are shown on top.
(D) Geometric mean of ABPP-labeled CYP2C9 variants. Individual replicates shown as blue points, and error bars show standard deviation.
(E) WT-normalized ABPP labeling (FITC-normalized fluorescence) for 14 CYP2C9 variants, expressed in the humanized yeast strain and labeled separately. Individual variants were labeled with the same ABPP protocol as the pooled assay. Scatterplot and linear regression of activity score (pool score) versus individual variant ABPP labeling (n = 3 replicates). Error bars show standard error for activity scores and standard error for ABPP labeling.
In order to demonstrate that click-seq accurately reflects enzyme activity, we cloned individual CYP2C9 variants of known activity and compared probe labeling levels to WT CYP2C9. We found that, as expected, CYP2C9 ∗2 (p.Arg144Cys) and ∗3 (p.Ile359Leu) had decreasing levels of labeling and a catalytically inactive variant, p.Cys435His, had labeling comparable to background levels (Figures 1B and 1D). We then constructed a barcoded, site-saturation mutagenesis library of CYP2C9 codon optimized for yeast expression and encompassing positions 2 to 490. This library covers 6,542 of the 9,780 possible single amino acid variants (67%) with 105,372 barcodes (mean of 5.8 and median of 3 for single amino acid variants; see Table S3 for details). We labeled the CYP2C9 activity library with the TAHA probe and flow sorted it into bins; we amplified, sequenced, and analyzed DNA collected from each bin to determine relative variant activity. We calculated activity scores for 6,524 single variants, of which 6,142 were missense, 131 were nonsense, and 250 were synonymous (Figure 1C). Activity scores were normalized to median nonsense and synonymous variant scores such that a score of 0 represented nonsense-like activity and a score of 1 represented WT-like activity. Variant activity scores correlated very well between the four replicate sorts we performed from four separate library outgrowths (mean Pearson’s r = 0.92, mean Spearman’s ρ = 0.919, Figure S3). We binned activity scores into activity classes (material and methods and Figure S4) and found that 64.9% (3,987) of missense variants showed significantly decreased activity compared to WT. As further confirmation that our classifications align with existing standards, the boundary between “WT-like” activity and “decreased” activity is very close to the activity score for CYP2C9 ∗2, a known decreased activity variant.
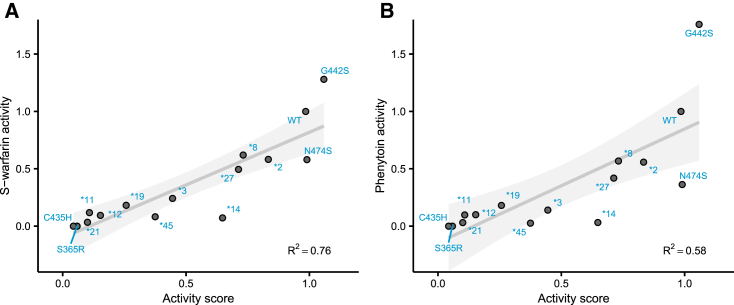
To internally validate our click-seq-derived activity data, we generated 14 CYP2C9 variants that spanned the full range of activity scores, labeled them individually, and found that individually tested and click-seq-derived activity scores were well correlated (Pearson’s r = 0.991, Figure 1E). To show that our large-scale activity scores determined with the TAHA probe were representative of CYP2C9 variant activity toward important CYP2C9 drug substrates, we performed gold-standard LC-MS assays of S-warfarin 7-hydroxylation and phenytoin 4-hydroxylation by using yeast microsomal preparations of the same 14 CYP2C9 variants generated for internal validation (Figure 2). Activity scores were well correlated with individual variant S-warfarin turnover (Pearson’s r = 0.874, Spearman’s ρ = 0.895) and phenytoin turnover (Pearson’s r = 0.764, Spearman’s ρ = 0.87). Both of these CYP2C9 drug substrates had highly similar activity levels across the variants tested (Pearson’s r = 0.965, Spearman’s ρ = 0.979, Figure S5). Additionally, we found that individual variant activity scores correlated well with an assay based on a fluorogenic substrate, BOMCC (Figure S6), indicating consistency across methods, as fluorogenic substrate assays are another standard method of measuring CYP activity.39
Figure 2.
Comparison of CYP2C9 activity scores with gold-standard activity assays on yeast microsomes
(A and B) Scatterplots of CYP2C9 activity scores plotted against individually tested CYP2C9 variants. Individual variants were expressed in the humanized yeast strain used in the pooled assay, and yeast microsomes were harvested from these individual strains. In (A), we used LC-MS to determine the rate of S-warfarin 7-hydroxylation. In (B), we used LC-MS to determine the rate of phenytoin 4-hydroxylation. The gray line is the regression line, and the shaded area shows the 95% confidence interval. All activities are shown normalized to wild-type rates. Variants shown are C435H (p.Cys435His), S365R (p.Ser365Arg), ∗21 (p.Pro30Leu), ∗11 (p.Arg335Trp), ∗12 (p.Pro489Ser), ∗19 (p.Gln454His), ∗45 (p.Arg132Trp), ∗3 (p.Ile359Leu), ∗14 (p.Arg125His), ∗27 (p.Arg150Leu), ∗8 (p.Arg150His), ∗2 (p.Arg144Cys), N474S (p.Asn474Ser), and G442S (p.Gly442Ser).
Overall, click-seq yielded a map relating variant sequence to activity but did not provide information on the mechanisms underlying variant loss of function. Thus, we performed a second CYP2C9 DMS, scoring variants for their abundance in cells in order to determine to what degree decreases in variant activity could be explained by decreases in abundance.
A multiplexed assay for CYP2C9 abundance in cultured human cells
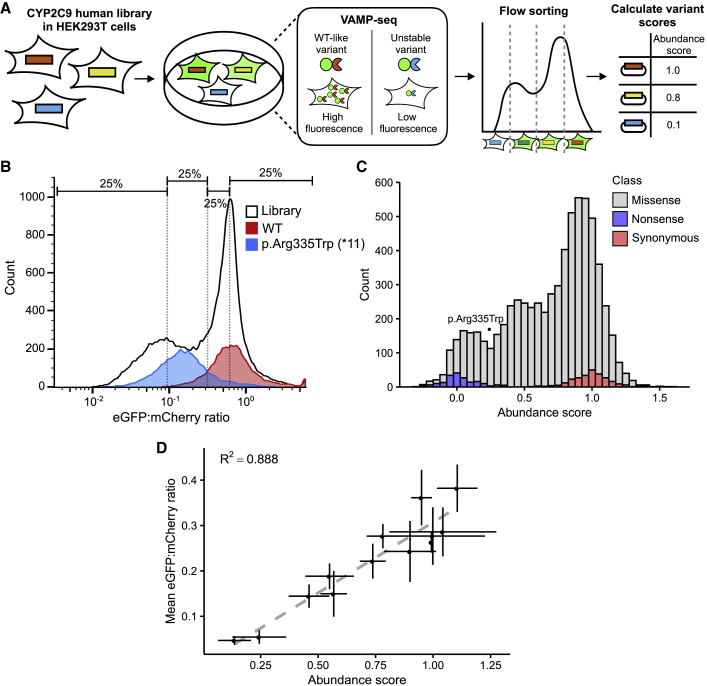
We recently developed a method, VAMP-seq,25 that enables measurement of steady-state protein abundance in cultured human cell lines via fluorescent reporters (Figure 3A). We applied VAMP-seq to CYP2C9, fusing eGFP C-terminally (Figure S7), and from the same construct expressing mCherry via an internal ribosomal entry site (IRES) to control for cell-to-cell differences in expression. The fluorescent reporters accurately quantified the loss of abundance of a known destabilized CYP2C9 variant,40 p.Arg335Trp (∗11), relative to WT as measured by the ratio of eGFP to mCherry (Figure 3B). We constructed a barcoded, site-saturation mutagenesis library of CYP2C9, encompassing positions 2 to 490. This library covered 8,310 of the 9,780 possible single amino acid variants (85%) with 78,740 barcodes (mean of 5.9 and median of 4 for single amino acid variants; see Table S3 for details).
Figure 3.
Multiplexed measurement of CYP2C9 activity via VAMP-seq
(A) Using VAMP-seq, we expressed a CYP2C9 library in HEK293T cells such that each variant was expressed as an eGFP fusion, resulting in a range of fluorescence according to variant stability. We then flow sorted cells into bins and sequenced them to determine relative variant abundance.
(B) Flow cytometry of CYP2C9 WT (red), destabilizing variant p.Arg335Trp (∗11, blue), and CYP2C9 eGFP fusion library expressed in HEK293T cells (black outline). Smoothed histograms of eGFP:mCherry ratios are shown. Approximate quartile bins for sorting shown are at the top.
(C) Stacked histogram of abundance score colored by type of variant. Abundance score of p.Arg335Trp (∗11) is shown as a point.
(D) Scatterplot and linear regression of individually measured cell eGFP:mCherry ratios for 12 CYP2C9 variants versus VAMP-seq-derived abundance scores for the same variants. Error bars show standard error for abundance scores and standard error for individually determined eGFP:mCherry ratio (n = 2 replicates).
We expressed this library in HEK293T cells by using a Bxb1 recombinase landing pad system previously used for other VAMP-seq deep mutational scans.25,33 Successfully recombined cells were selected with a small molecule, AP1903, and then sorted into quartile bins on the basis of eGFP:mCherry ratio (Figure 3A). We deeply sequenced bins and used the resulting sequencing reads to calculate frequencies across bins for each variant. Abundance scores were calculated with weighted averages of variant frequencies and normalized to the scores of synonymous and nonsense variants as for the activity scores (material and methods). Variant abundance scores showed distinct, separable distributions of synonymous and nonsense variants; missense variants spanned the range between them (Figure 3C). After filtering, we assigned variant scores to 6,821 single variants, of which 6,370 were missense, 189 were nonsense, and 261 were synonymous. Three replicate sorts from two separate transfections were performed on this library and the replicates correlated well (mean Pearson’s r = 0.789, mean Spearman’s ρ = 0.754, Figure S3). To internally validate our VAMP-seq-derived abundance data, we generated 12 CYP2C9 variants, recombined them individually, and found that individually measured and VAMP-seq-derived abundance scores were well correlated (Pearson’s r = 0.942, Figure 3D). In contrast to the activity classes, only 36.8% of missense variants (2,347 variants) had a significantly decreased abundance class. This fraction is similar to other VAMP-seq studies of pharmacogene abundance, as 34% of VKOR missense variants showed significantly decreased abundance.27
Mechanism of CYP2C9 variant loss of function
Between the click-seq and VAMP-seq datasets, 8,091 missense variants had at least one functional score and 4,421 variants had both activity and abundance scores (Figure 4). Among these variants, activity and abundance were strongly correlated (Pearson’s r = 0.748, Spearman’s ρ = 0.749) (Figure 4F). We observed an abundance threshold at a score of ∼0.5, below which variants had very low activity (median activity score ∼0.098), suggesting that for variants with abundance below this level, differences in click-seq signal are too small to detect. Conversely, variants with abundance scores greater than 0.5 had a wider range of activity scores. The overall positive trend between abundance and activity scores revealed that (1) using engineered yeast as a heterologous CYP expression system largely recapitulates protein behavior in human cells and (2) a substantial number of variants had low activity because they were less abundant. We estimated that approximately half of the variation in activity could be explained by abundance (R2 = 0.56, Figure 4F). Since there was no normalization to protein levels per cell, the yeast activity scores reported are each a combination of both variant activity and variant stability.
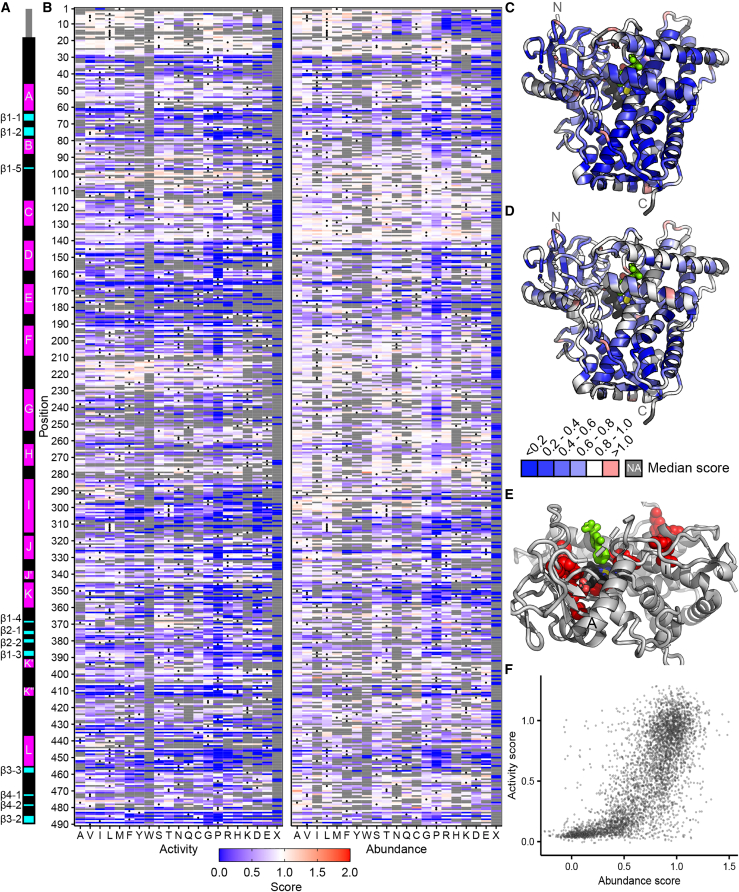
Figure 4.
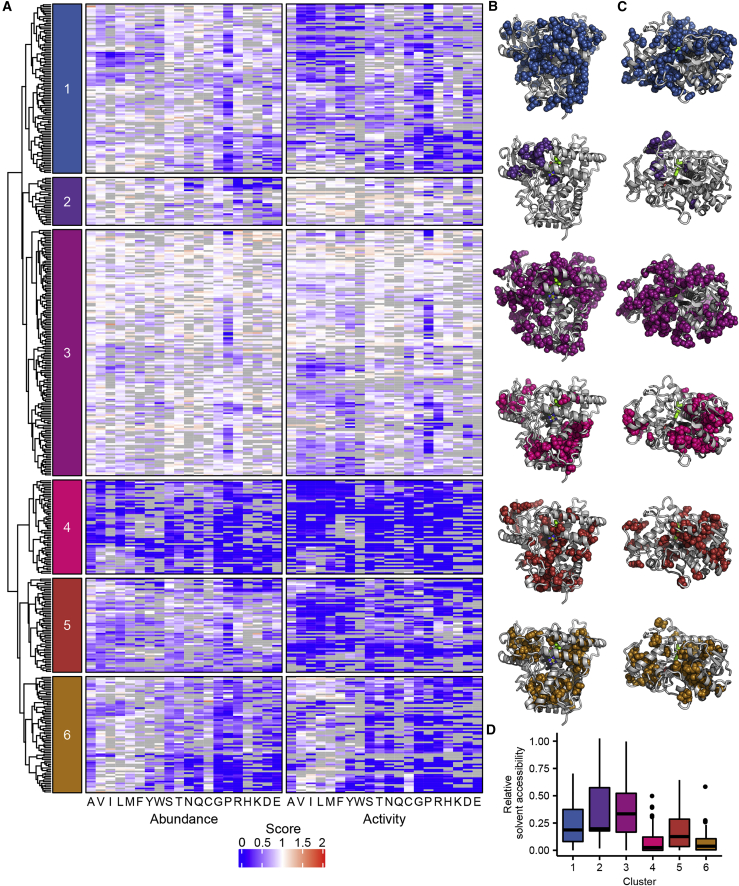
Click-seq activity scores and VAMP-seq abundance scores for CYP2C9
(A) Secondary structure of CYP2C9; alpha helices are shown in magenta, and beta sheets are shown in cyan. Helix and beta sheet names are labeled.
(B) Heatmaps of CYP2C9 activity (left) and abundance (right) scores. WT amino acids are denoted with a dot, and missing data are shown in gray. Scores range from nonfunctional (blue) to WT-like (white) to increased (red).
(C and D) CYP2C9 structure (PDB: 1R9O) colored by median activity (C) and abundance (D) at each position. Median scores are binned as depicted in the legend, and missing positions are shown in gray. Heme is colored by element (carbon:black, nitrogen:blue, oxygen:red, iron:yellow), and substrate (flurbiprofen) is colored bright green. Median activity scores shown in (C), and median abundance scores shown in (D).
(E) Zoomed view of partial CYP2C9 structure. Positions with the lowest 2.5% specific activity scores are shown as red spheres. F and G helices are hidden, A and I helices are labeled, and heme and substrate are colored as in (C) and (D).
(F) Scatterplot of CYP2C9 activity and abundance scores from a total of 4,421 missense variants.
By comparing activity and abundance, we were able to identify variants that abolish activity but not abundance. We hypothesized that functionally important regions, such as the active site and binding pocket of CYP2C9, would be enriched for such low activity, high abundance variants. To find such variants, we calculated variant specific activity by dividing the activity score by the abundance score (material and methods). We found that the positions with the lowest median-specific activity were not active site positions but instead mainly positions most likely involved in heme binding (Figure 4E). This finding implies that these positions are crucial for activity but did not strongly destabilize the protein when mutated. The positions with the lowest 2.5% median-specific activity scores included the heme-binding motif residues Gly431, Arg433, Cys435, and Gly43741 (Figure S8) as well as Arg97, which is important for heme propionate binding in CYP2C9.42 Finally, we found that variants in the active site43 had median activity scores of 0.61 and median abundance scores of 0.91, indicating that these variants were generally not destabilizing and also only had moderate effects on activity. This active site mutational tolerance is surprising, at least in contrast to VKOR, where active site positions had the lowest specific activity scores.27 However, CYPs have well-documented conformational flexibility, especially in the active site.44 Moreover, in CYP2C9, the BC loop which frames the substrate access channel is also highly flexible45 and has median activity and abundance scores of 0.81 and 0.88, respectively. Thus, CYP2C9 is apparently able to tolerate active site substitutions without loss of abundance.
Structural insights from CYP2C9 functional scores
Endoplasmic reticulum (ER)-localized CYP enzymes are composed of an N-terminal ER-transmembrane domain and a large, cytoplasmic catalytic domain.46 The CYP enzyme superfamily is diverse at the sequence level but members share a common structure including 12 major helices, labeled A through L, and four beta sheets, labeled β1 through β447 (Figure 4A). CYP2C9 has been crystallized with warfarin48 and flurbiprofen49 as well as with other substrates50 and also without a ligand.48 These structures are generally comparable and show small differences in substrate-interacting regions. We used the flurbiprofen-bound CYP2C9 structure for our analysis because the substrate is bound in a catalytically favorable orientation in this structure. Three positions are almost completely conserved across all CYPs: Glu354 and Arg357 in the ExxR motif involved in heme binding and core stabilizing51 and also the invariant heme-coordinating cysteine, Cys435.52 In our activity assay, the 36 missense variants at these three positions all had activity scores of <0.1. In our abundance assay, Arg357 was also extremely intolerant to substitution and had a median abundance score of 0.077, indicating that Arg357 is crucial for both activity and abundance. In addition to these highly conserved residues, our activity scores recapitulated the importance of the heme-binding motif at positions 428–43741 and the proline-rich PPGP motif in the linker (or hinge) region after the transmembrane domain,53 which is necessary for proper folding54 (Figure S8).
We mapped median positional activity and abundance scores onto the CYP2C9 structure (Figures 4C and 4D) to identify key regions important for activity and abundance. To determine the characteristic substitution patterns in different regions of CYP2C9, we performed hierarchical clustering of positions on the basis of both activity and abundance scores and identified six main clusters of positions (Figure 5). We found that substitutions in cluster 4 were universally not tolerated, and positions in this cluster generally grouped into two distinct regions: core-facing positions in helices D, E, I, J, K, and L comprising the highly conserved heme-binding structural core of the protein and positions in and directly abutting β sheet 1, located near the N terminus. Both of these regions are highly conserved across CYPs and are composed of buried, hydrophobic residues51,55 in which substitution leads to destabilization and degradation. In addition, substitutions in β sheet 1 may disrupt distal side chains that coordinate with the central heme iron.56 Clusters 5 and 6 were slightly more tolerant to substitution than cluster 4 and are also found in the core of the protein.
Figure 5.
Hierarchical clustering of activity and abundance scores and cluster accessibility
(A–D) In (A), dendrogram and heatmaps of CYP2C9 activity and abundance score clustered by position. Heatmaps colored as in Figure 4. Only positions that had at least 26 total substitutions were included in this analysis. Colored boxes on the left indicate the six major clusters and correspond to the colors shown in (B), (C), and (D). In (B) and (C), the positions that correspond to each of the six clusters are shown as spheres in the corresponding color on the CYP2C9 crystal structure (PDB: 1R9O). Alternate viewpoint is shown in (C). In (D), relative solvent accessibility of each cluster is shown as a boxplot. Bold black line shows median, box shows 25th and 75th percentile, vertical line shows 1.5 interquartile range above and below percentiles, and outliers are shown as black points.
Conversely, positions comprising clusters 1 and 3 were tolerant to substitution and were located on the surface of the protein, although cluster 1 was more sensitive to charged and proline amino acid substitutions. Cluster 2 contained many positions in the transmembrane domain not shown in the crystal structure. Cluster 2 also included part of the F-G loop, which defines a portion of the CYP2C9 substrate access channel and interacts with the membrane.57 Substitutions in the transmembrane domain (positions 1–20) had little effect on activity but had a larger effect on abundance. The largest effects in the transmembrane domain were from charged substitutions, which rarely occur in transmembrane domains. CYP2C9 is co-translationally inserted into the ER and the N-terminal transmembrane domain is involved in ER retention,58 so substitutions in the transmembrane domain that impacted abundance may have caused mislocalization.
Predicting the clinical impact of human CYP2C9 variants
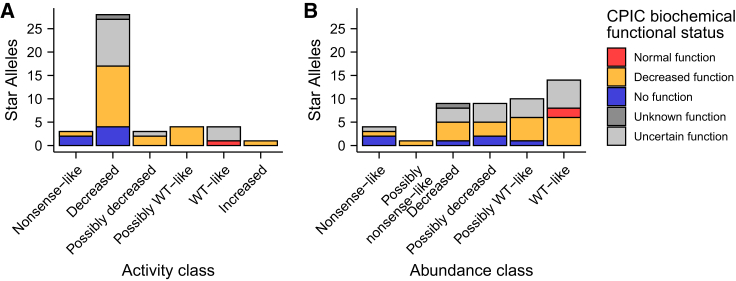
Genetic variation in CYP2C9 can drive variable drug response, but most CYP2C9 variants documented in humans so far have unknown functional consequences. The best-studied set of CYP2C9 variants are the 70 star alleles in PharmVar, some of which have been functionally characterized. The Clinical Pharmacogenetics Implementation Consortium (CPIC) reviews functional evidence and has made clinical recommendations for 34 of the 63 CYP2C9 single amino acid star alleles.16,17 We compared CPIC recommendations to our activity classes and found that CPIC allele function classes were largely concordant with our CYP2C9 activity classes (Figure 6). The few cases where our activity classes did not match CPIC classes were generally due to alleles with limited or inadequate functional evidence, as determined by CPIC (Table S7).
Figure 6.
Comparison of activity and abundance scores with clinical pharmacogenomic recommendations
(A and B) Stacked bar plot of number of CYP2C9 star alleles versus activity (A) or abundance (B) class, colored by clinical pharmacogenomic recommendation (CPIC biochemical functional class status). CPIC classes are taken from NSAID clinical functional status recommendations.16
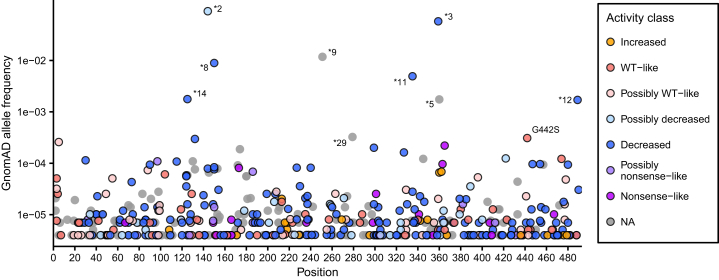
We additionally curated 629 synonymous, missense, and nonsense CYP2C9 variants from gnomAD,20 and 559 of these had at least one functional score from our datasets. Most of these variants lack functional annotations because only 27 of them are star alleles with an associated CPIC functional recommendation. All eight nonsense variants had very low activity and/or abundance scores, all 119 synonymous variants had high activity and/or abundance scores, and missense variants spanned the range of activity and abundance scores (Figure S9). Of the 466 total missense variants in gnomAD, 340 had an activity score (319 of these lack a CPIC functional recommendation) and a majority of these had significantly decreased activity. 48.8% of missense variants (166 variants) had “decreased” activity, and 9.7% (33 variants) had “nonsense-like” or “possibly nonsense-like” activity (Figure 7). 168 of the missense variants were singletons in gnomAD, and these had the same activity score pattern as the other missense variants where 49.4% (83 variants) had “decreased” or “possibly decreased” activity and 11.9% (20 variants) had “nonsense-like” or “possibly nonsense-like” activity. Finally, we compared our scores to several widely used computational predictors and found only moderate correlation between predicted functional status and experimentally derived activity scores (mean absolute Pearson’s r = 0.494, Figure S10). The fact that many human CYP2C9 variants have significantly decreased function is striking, highlighting that a large proportion of all possible CYP2C9 variants have the potential to impact the metabolism of warfarin and other drugs.
Figure 7.
Classification of human CYP2C9 variants via activity data
Frequency and protein position of CYP2C9 missense variants in human population database gnomAD, colored by click-seq activity class. Allele frequencies were calculated from combined v.2 and v.3 gnomAD allele frequencies. Variants at population frequency greater than 3 × 10−4 are labeled by star allele (if applicable) or amino acid change. Labeled variants are ∗2 (p.Arg144Cys), ∗3 (p.Ile359Leu), ∗5 (p.Asp350Glu), ∗8 (p.Arg150His), ∗9 (p.His251Arg), ∗11 (p.Arg335Trp), ∗12 (p.Pro489Ser), ∗14 (p.Arg125His), ∗29 (p.Pro279Thr), and G442S (p.Gly442Ser). Human variants lacking an activity class are shown in gray.
Discussion
CYP2C9 is a well-studied metabolic enzyme, and many small-scale functional characterizations of CYP2C9 have been performed;59, 60, 61 the largest of these comprises 109 CYP2C9 variants profiled for abundance via a VAMP-seq style assay.62 Abundance scores from this study correlate well with our abundance scores (Pearson’s r = 0.74). Collectively, previous studies of CYP2C9 variant function have tested a small fraction of the possible single mutations, focusing on already observed variants. Therefore, we developed a high-throughput yeast activity assay, applied VAMP-seq, and generated activity and abundance scores for a combined total of 8,091 missense variants, or 87% of the possible missense variants in CYP2C9. Our results were highly reproducible across biological replicates and validated well when tested against individual variants and clinical substrates of CYP2C9. Additionally, our activity scores were concordant with CYP2C9 variant functional status recommendations from CPIC.16 In addition to CYP2C9 variants of known function, we generated functional scores for over 300 CYP2C9 missense variants present in gnomAD that currently lack functional annotation.
Our functional scores reflected known structural features of CYPs, including heme-binding residues and the highly conserved core regions of the protein. In general, residues involved in heme coordination and binding were crucial for activity but were less important to protein abundance, indicating that heme insertion, a process that is not fully understood, is not necessarily stabilizing.63 Somewhat surprisingly, residues in the active site were fairly tolerant to substitution and largely did not result in large decreases in activity. Instead, we found that substitutions in the hydrophobic core of the protein comprising helices D, E, I, J, K, and L were crucial for protein abundance and activity and substitutions in these regions probably most affect protein stability Additionally, we noticed that charged and polar substitutions in the transmembrane domain were less tolerated in the abundance assay than the activity assay, which may indicate that mislocalized CYP2C9 could have a more severe effect in human cells than in yeast. It is possible that mislocalized CYP2C9 variants in yeast could still be labeled in the click-seq assay but that in human cells these variants are degraded because of mislocalization.
We observed a strong correlation between activity and abundance scores, which is in contrast to other deep mutational scans. Paired activity and abundance data has been collected for VKOR and NUDT15,26,27 and for these proteins, activity and abundance scores were much less well correlated (VKOR, Pearson’s r = 0.261, Spearman’s ρ = 0.25; NUDT15, Pearson’s r = 0.384, Spearman’s ρ = 0.34). The strong correlation of CYP2C9 activity and abundance scores is partially due to the design of the click-seq vector, which does not include an expression control. Overall, we estimated that protein abundance could explain about 50% of the variation in CYP2C9 variant activity.
The variant functional datasets we generated are a resource for improving genotype-based dosing and also for improving our understanding of CYP biology. However, there are limitations to keep in mind. First, in both systems we expressed CYP2C9 as a cDNA by using an inducible promoter, so our assays did not capture splicing defects or transcriptional regulation. CYP2C9 is constitutively expressed in the liver but is also inducible via a number of substrates.9 We also cannot discern the impact of protein interactions, such as with CYP accessory proteins cytochrome P450 reductase (CPR) and cytochrome b5 or with other CYP enzymes. Certain protein variants might behave differently in yeast than in animal liver cells because of these interacting effects. Additionally, because of our flow cytometry binning strategy, we suspect that click-seq labeling was saturated at increased activity levels, so we would most likely need to re-sort our library with a modified binning strategy to detect variants with significantly increased activity. An example of this is the p.Gly442Ser variant, which had a “WT-like” activity score but showed 130% and 180% WT activity in individual tests (Figure 2), although this could also be explained by a substrate-dependent effect. Despite our binning strategy, we observed 240 variants with increased function, and 20 of these were also present in gnomAD. One increased activity variant, p.Ile434Phe (∗59), was also present in the PharmVar database, warranting further investigation, especially because there are no documented CYP2C9 increased activity variants,14 so the clinical impact of an increased activity variant is unknown.
Finally, click-seq measures CYP2C9 activity via a single substrate, so we cannot say how many variants may exhibit substrate-dependent effects, for which there is some evidence.64 As an example, Arg108 has been shown to be critical for binding negatively charged substrates42 and for binding flurbiprofen in particular,49 but substitutions at this position had little effect on activity as measured by click-seq or abundance. This is most likely because the TAHA probe is an amide and not acidic, so it is able to bind regardless of a strong electrostatic interaction with Arg108. Despite this, our activity scores correlate well with S-warfarin and phenytoin activity overall, indicating that they are generally informative of a larger set of substrates. In the future, we plan to re-test our library with a range of activity-based probes to identify variants that result in substrate-dependent changes in function.
We also anticipate that click-seq can be leveraged to examine other CYPs important to human drug metabolism, such as CYP2D6 and CYP2C19, both of which have also been successfully expressed in yeast previously and for which activity-based click probes have been designed.37 Expanding the repertoire of CYP deep mutational scans will allow us to investigate differences between CYP isoforms that are key to human drug metabolism and pharmacogenomics. Moreover, activity-based click probes are available for a variety of enzyme activities, so click-seq is likely to be useful beyond CYPs.
In addition to revealing details of how CYP2C9 sequence relates to its structure and function, we hope that the variant functional data presented here will be useful for informing drug dosing. In particular, we hope that the data will empower CPIC to provide functional classifications for additional variants, perhaps preemptively. Such preemptive classification could be extremely powerful because clinical genotyping efforts are increasing, guaranteeing we will continue to find new variants that may have consequences for drug dosing.
Acknowledgments
We thank D. Prunkard of the UW Pathology Flow Cytometry Core Facility for assistance with cell sorting; K. Munson of the UW PacBio Sequencing Services for assistance with long-read sequencing; A. Wright, C. Whidbey, and E. Stoddard for reagents and helpful discussion; J. Stephany for assistance with amplicon design and sequencing analysis; D. Nickerson for valuable advice in analyzing the data; B. Dunn for useful comments in writing the manuscript; and all members of the Dunham lab for helpful feedback on figures. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers R24GM115277 and R01GM132162 to D.M.F., M.J.D., and A.E.R. C.J.A. was supported by the National Human Genome Research Institute of the NIH under award T32 HG00035. A.E.R. was also supported by the National Institute of General Medical Sciences of the NIH under award number P01GM11669. The research of M.J.D. was supported in part by a Faculty Scholar grant from the Howard Hughes Medical Institute. M.J.D. acknowledges prior support as a Senior Fellow in the Genetic Networks program at the Canadian Institute for Advanced Research.
Declaration of interests
The authors declare no competing interests.
Published: July 26, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.07.001.
Contributor Information
Douglas M. Fowler, Email: dfowler@uw.edu.
Maitreya J. Dunham, Email: maitreya@uw.edu.
Data and code availability
The accession number for the sequencing data reported in this paper is NCBI GEO: GSE165412. Code and processed variant scores generated during this study are available at GitHub: https://github.com/dunhamlab/CYP2C9.
Web resources
AssemblyByPacBio, https://github.com/shendurelab/AssemblyByPacBio/
FASTX-toolkit, http://hannonlab.cshl.edu/fastx_toolkit/
PharmVar, https://www.pharmvar.org/
RCSB Protein Data Bank, https://www.rcsb.org/
UniProt, https://www.uniprot.org/
Supplemental information
References
- 1.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Starita L.M., Ahituv N., Dunham M.J., Kitzman J.O., Roth F.P., Seelig G., Shendure J., Fowler D.M. Variant Interpretation: Functional Assays to the Rescue. Am. J. Hum. Genet. 2017;101:315–325. doi: 10.1016/j.ajhg.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sultana J., Cutroneo P., Trifirò G. Clinical and economic burden of adverse drug reactions. J. Pharmacol. Pharmacother. 2013;4(Suppl 1):S73–S77. doi: 10.4103/0976-500X.120957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lazarou J., Pomeranz B.H., Corey P.N. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. JAMA. 1998;279:1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 5.Budnitz D.S., Pollock D.A., Weidenbach K.N., Mendelsohn A.B., Schroeder T.J., Annest J.L. National surveillance of emergency department visits for outpatient adverse drug events. JAMA. 2006;296:1858–1866. doi: 10.1001/jama.296.15.1858. [DOI] [PubMed] [Google Scholar]
- 6.Goulding R., Dawes D., Price M., Wilkie S., Dawes M. Genotype-guided drug prescribing: a systematic review and meta-analysis of randomized control trials. Br. J. Clin. Pharmacol. 2015;80:868–877. doi: 10.1111/bcp.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zanger U.M., Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Rettie A.E., Jones J.P. Clinical and toxicological relevance of CYP2C9: drug-drug interactions and pharmacogenetics. Annu. Rev. Pharmacol. Toxicol. 2005;45:477–494. doi: 10.1146/annurev.pharmtox.45.120403.095821. [DOI] [PubMed] [Google Scholar]
- 9.Daly A.K., Rettie A.E., Fowler D.M., Miners J.O. Pharmacogenomics of CYP2C9: Functional and clinical considerations. J. Pers. Med. 2017;8:1. doi: 10.3390/jpm8010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X., Li D., Wu J.C., Liu Z.Q., Zhou H.H., Yin J.Y. Precision dosing of warfarin: open questions and strategies. Pharmacogenomics J. 2019;19:219–229. doi: 10.1038/s41397-019-0083-3. [DOI] [PubMed] [Google Scholar]
- 11.Steward D.J., Haining R.L., Henne K.R., Davis G., Rushmore T.H., Trager W.F., Rettie A.E. Genetic association between sensitivity to warfarin and expression of CYP2C9∗3. Pharmacogenetics. 1997;7:361–367. doi: 10.1097/00008571-199710000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Pirmohamed M., Kamali F., Daly A.K., Wadelius M. Oral anticoagulation: a critique of recent advances and controversies. Trends Pharmacol. Sci. 2015;36:153–163. doi: 10.1016/j.tips.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Sim S.C., Ingelman-Sundberg M. The Human Cytochrome P450 (CYP) Allele Nomenclature website: a peer-reviewed database of CYP variants and their associated effects. Hum. Genomics. 2010;4:278–281. doi: 10.1186/1479-7364-4-4-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaedigk A., Ingelman-Sundberg M., Miller N.A., Leeder J.S., Whirl-Carrillo M., Klein T.E., PharmVar Steering Committee The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin. Pharmacol. Ther. 2018;103:399–401. doi: 10.1002/cpt.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Relling M.V., Klein T.E. CPIC: Clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin. Pharmacol. Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Theken K.N., Lee C.R., Gong L., Caudle K.E., Formea C.M., Gaedigk A., Klein T.E., Agúndez J.A.G., Grosser T. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and Nonsteroidal Anti-Inflammatory Drugs. Clin. Pharmacol. Ther. 2020;108:191–200. doi: 10.1002/cpt.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karnes J.H., Rettie A.E., Somogyi A.A., Huddart R., Fohner A.E., Formea C.M., Ta Michael Lee M., Llerena A., Whirl-Carrillo M., Klein T.E. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C9 and HLA-B Genotypes and Phenytoin Dosing: 2020 Update. Clin. Pharmacol. Ther. 2020;109:302–309. doi: 10.1002/cpt.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Y., Ingelman-Sundberg M., Lauschke V.M. Worldwide Distribution of Cytochrome P450 Alleles: A Meta-analysis of Population-scale Sequencing Projects. Clin. Pharmacol. Ther. 2017;102:688–700. doi: 10.1002/cpt.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon A.S., Tabor H.K., Johnson A.D., Snively B.M., Assimes T.L., Auer P.L., Ioannidis J.P.A., Peters U., Robinson J.G., Sucheston L.E., NHLBI GO Exome Sequencing Project Quantifying rare, deleterious variation in 12 human cytochrome P450 drug-metabolism genes in a large-scale exome dataset. Hum. Mol. Genet. 2014;23:1957–1963. doi: 10.1093/hmg/ddt588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fowler D.M., Fields S. Deep mutational scanning: a new style of protein science. Nat. Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romero P.A., Tran T.M., Abate A.R. Dissecting enzyme function with microfluidic-based deep mutational scanning. Proc. Natl. Acad. Sci. USA. 2015;112:7159–7164. doi: 10.1073/pnas.1422285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Relling M.V., Krauss R.M., Roden D.M., Klein T.E., Fowler D.M., Terada N., Lin L., Riel-Mehan M., Do T.P., Kubo M. New Pharmacogenomics Research Network: An Open Community Catalyzing Research and Translation in Precision Medicine. Clin. Pharmacol. Ther. 2017;102:897–902. doi: 10.1002/cpt.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiasson M., Dunham M.J., Rettie A.E., Fowler D.M. Applying Multiplex Assays to Understand Variation in Pharmacogenes. Clin. Pharmacol. Ther. 2019;106:290–294. doi: 10.1002/cpt.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matreyek K.A., Starita L.M., Stephany J.J., Martin B., Chiasson M.A., Gray V.E., Kircher M., Khechaduri A., Dines J.N., Hause R.J. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat. Genet. 2018;50:874–882. doi: 10.1038/s41588-018-0122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suiter C.C., Moriyama T., Matreyek K.A., Yang W., Scaletti E.R., Nishii R., Yang W., Hoshitsuki K., Singh M., Trehan A. Massively parallel variant characterization identifies NUDT15 alleles associated with thiopurine toxicity. Proc. Natl. Acad. Sci. USA. 2020;117:5394–5401. doi: 10.1073/pnas.1915680117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiasson M.A., Rollins N.J., Stephany J.J., Sitko K.A., Matreyek K.A., Verby M., Sun S., Roth F.P., DeSloover D., Marks D.S. Multiplexed measurement of variant abundance and activity reveals VKOR topology, active site and human variant impact. eLife. 2020;9:1–25. doi: 10.7554/eLife.58026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rademacher P.M., Woods C.M., Huang Q., Szklarz G.D., Nelson S.D. Differential oxidation of two thiophene-containing regioisomers to reactive metabolites by cytochrome P450 2C9. Chem. Res. Toxicol. 2012;25:895–903. doi: 10.1021/tx200519d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gietz R.D., Schiestl R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 30.Jain P.C., Varadarajan R. A rapid, efficient, and economical inverse polymerase chain reaction-based method for generating a site saturation mutant library. Anal. Biochem. 2014;449:90–98. doi: 10.1016/j.ab.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J., Kobert K., Flouri T., Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubin A.F., Gelman H., Lucas N., Bajjalieh S.M., Papenfuss A.T., Speed T.P., Fowler D.M. A statistical framework for analyzing deep mutational scanning data. Genome Biol. 2017;18:150. doi: 10.1186/s13059-017-1272-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matreyek K.A., Stephany J.J., Chiasson M.A., Hasle N., Fowler D.M. An improved platform for functional assessment of large protein libraries in mammalian cells. Nucleic Acids Res. 2020;48:e1. doi: 10.1093/nar/gkz910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen D.C., Yang B.C., Kuo T.T. One-step transformation of yeast in stationary phase. Curr. Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 35.García-Nafría J., Watson J.F., Greger I.H. IVA cloning: A single-tube universal cloning system exploiting bacterial In Vivo Assembly. Sci. Rep. 2016;6:27459. doi: 10.1038/srep27459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright A.T., Cravatt B.F. Chemical proteomic probes for profiling cytochrome p450 activities and drug interactions in vivo. Chem. Biol. 2007;14:1043–1051. doi: 10.1016/j.chembiol.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright A.T., Song J.D., Cravatt B.F. A suite of activity-based probes for human cytochrome P450 enzymes. J. Am. Chem. Soc. 2009;131:10692–10700. doi: 10.1021/ja9037609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jean P., Lopez-Garcia P., Dansette P., Mansuy D., Goldstein J.L. Oxidation of tienilic acid by human yeast-expressed cytochromes P-450 2C8, 2C9, 2C18 and 2C19. Evidence that this drug is a mechanism-based inhibitor specific for cytochrome P-450 2C9. Eur. J. Biochem. 1996;241:797–804. doi: 10.1111/j.1432-1033.1996.00797.x. [DOI] [PubMed] [Google Scholar]
- 39.Donato M.T., Jiménez N., Castell J.V., Gómez-Lechón M.J. Fluorescence-based assays for screening nine cytochrome P450 (P450) activities in intact cells expressing individual human P450 enzymes. Drug Metab. Dispos. 2004;32:699–706. doi: 10.1124/dmd.32.7.699. [DOI] [PubMed] [Google Scholar]
- 40.Tai G., Farin F., Rieder M.J., Dreisbach A.W., Veenstra D.L., Verlinde C.L.M.J., Rettie A.E. In-vitro and in-vivo effects of the CYP2C9∗11 polymorphism on warfarin metabolism and dose. Pharmacogenet. Genomics. 2005;15:475–481. doi: 10.1097/01.fpc.0000162005.80857.98. [DOI] [PubMed] [Google Scholar]
- 41.Danielson P.B. The cytochrome P450 superfamily: biochemistry, evolution and drug metabolism in humans. Curr. Drug Metab. 2002;3:561–597. doi: 10.2174/1389200023337054. [DOI] [PubMed] [Google Scholar]
- 42.Dickmann L.J., Locuson C.W., Jones J.P., Rettie A.E. Differential roles of Arg97, Asp293, and Arg108 in enzyme stability and substrate specificity of CYP2C9. Mol. Pharmacol. 2004;65:842–850. doi: 10.1124/mol.65.4.842. [DOI] [PubMed] [Google Scholar]
- 43.Reynald R.L., Sansen S., Stout C.D., Johnson E.F. Structural characterization of human cytochrome P450 2C19: active site differences between P450s 2C8, 2C9, and 2C19. J. Biol. Chem. 2012;287:44581–44591. doi: 10.1074/jbc.M112.424895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nair P.C., McKinnon R.A., Miners J.O. Cytochrome P450 structure-function: insights from molecular dynamics simulations. Drug Metab. Rev. 2016;48:434–452. doi: 10.1080/03602532.2016.1178771. [DOI] [PubMed] [Google Scholar]
- 45.Roberts A.G., Cheesman M.J., Primak A., Bowman M.K., Atkins W.M., Rettie A.E. Intramolecular heme ligation of the cytochrome P450 2C9 R108H mutant demonstrates pronounced conformational flexibility of the B-C loop region: implications for substrate binding. Biochemistry. 2010;49:8700–8708. doi: 10.1021/bi100911q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson E.F., Stout C.D. Structural diversity of eukaryotic membrane cytochrome p450s. J. Biol. Chem. 2013;288:17082–17090. doi: 10.1074/jbc.R113.452805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gay S.C., Roberts A.G., Halpert J.R. Structural features of cytochromes P450 and ligands that affect drug metabolism as revealed by X-ray crystallography and NMR. Future Med. Chem. 2010;2:1451–1468. doi: 10.4155/fmc.10.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams P.A., Cosme J., Ward A., Angove H.C., Matak Vinković D., Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 49.Wester M.R., Yano J.K., Schoch G.A., Yang C., Griffin K.J., Stout C.D., Johnson E.F. The structure of human cytochrome P450 2C9 complexed with flurbiprofen at 2.0-A resolution. J. Biol. Chem. 2004;279:35630–35637. doi: 10.1074/jbc.M405427200. [DOI] [PubMed] [Google Scholar]
- 50.Maekawa K., Adachi M., Matsuzawa Y., Zhang Q., Kuroki R., Saito Y., Shah M.B. Structural Basis of Single-Nucleotide Polymorphisms in Cytochrome P450 2C9. Biochemistry. 2017;56:5476–5480. doi: 10.1021/acs.biochem.7b00795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hasemann C.A., Kurumbail R.G., Boddupalli S.S., Peterson J.A., Deisenhofer J. Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure. 1995;3:41–62. doi: 10.1016/s0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- 52.Sirim D., Widmann M., Wagner F., Pleiss J. Prediction and analysis of the modular structure of cytochrome P450 monooxygenases. BMC Struct. Biol. 2010;10:34. doi: 10.1186/1472-6807-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kemper B. Structural basis for the role in protein folding of conserved proline-rich regions in cytochromes P450. Toxicol. Appl. Pharmacol. 2004;199:305–315. doi: 10.1016/j.taap.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 54.Chen C.D., Doray B., Kemper B. A conserved proline-rich sequence between the N-terminal signal-anchor and catalytic domains is required for assembly of functional cytochrome P450 2C2. Arch. Biochem. Biophys. 1998;350:233–238. doi: 10.1006/abbi.1997.0524. [DOI] [PubMed] [Google Scholar]
- 55.Mustafa G., Yu X., Wade R.C. Drug Metabolism Prediction. Wiley Blackwell; 2014. Structure and Dynamics of Human Drug-Metabolizing Cytochrome P450 Enzymes; pp. 75–102. [Google Scholar]
- 56.Arendse L.B., Blackburn J.M. Effects of polymorphic variation on the thermostability of heterogenous populations of CYP3A4 and CYP2C9 enzymes in solution. Sci. Rep. 2018;8:11876. doi: 10.1038/s41598-018-30195-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cojocaru V., Winn P.J., Wade R.C. Multiple, ligand-dependent routes from the active site of cytochrome P450 2C9. Curr. Drug Metab. 2012;13:143–154. doi: 10.2174/138920012798918462. [DOI] [PubMed] [Google Scholar]
- 58.Szczesna-Skorupa E., Chen C.D., Rogers S., Kemper B. Mobility of cytochrome P450 in the endoplasmic reticulum membrane. Proc. Natl. Acad. Sci. USA. 1998;95:14793–14798. doi: 10.1073/pnas.95.25.14793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niinuma Y., Saito T., Takahashi M., Tsukada C., Ito M., Hirasawa N., Hiratsuka M. Functional characterization of 32 CYP2C9 allelic variants. Pharmacogenomics J. 2014;14:107–114. doi: 10.1038/tpj.2013.22. [DOI] [PubMed] [Google Scholar]
- 60.Wang Y.H., Pan P.P., Dai D.P., Wang S.H., Geng P.W., Cai J.P., Hu G.X. Effect of 36 CYP2C9 variants found in the Chinese population on losartan metabolism in vitro. Xenobiotica. 2014;44:270–275. doi: 10.3109/00498254.2013.820007. [DOI] [PubMed] [Google Scholar]
- 61.Dai D.P., Wang Y.H., Wang S.H., Geng P.W., Hu L.M., Hu G.X., Cai J.P. In vitro functional characterization of 37 CYP2C9 allelic isoforms found in Chinese Han population. Acta Pharmacol. Sin. 2013;34:1449–1456. doi: 10.1038/aps.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L., Sarangi V., Moon I., Yu J., Liu D., Devarajan S., Reid J.M., Kalari K.R., Wang L., Weinshilboum R. CYP2C9 and CYP2C19: Deep Mutational Scanning and Functional Characterization of Genomic Missense Variants. Clin. Transl. Sci. 2020;13:727–742. doi: 10.1111/cts.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Correia M.A., Sinclair P.R., De Matteis F. Cytochrome P450 regulation: the interplay between its heme and apoprotein moieties in synthesis, assembly, repair, and disposal. Drug Metab. Rev. 2011;43:1–26. doi: 10.3109/03602532.2010.515222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee C.R., Goldstein J.A., Pieper J.A. Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002;12:251–263. doi: 10.1097/00008571-200204000-00010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the sequencing data reported in this paper is NCBI GEO: GSE165412. Code and processed variant scores generated during this study are available at GitHub: https://github.com/dunhamlab/CYP2C9.