Summary
Coatomer complexes function in the sorting and trafficking of proteins between subcellular organelles. Pathogenic variants in coatomer subunits or associated factors have been reported in multi-systemic disorders, i.e., coatopathies, that can affect the skeletal and central nervous systems. We have identified loss-of-function variants in COPB2, a component of the coatomer complex I (COPI), in individuals presenting with osteoporosis, fractures, and developmental delay of variable severity. Electron microscopy of COPB2-deficient subjects’ fibroblasts showed dilated endoplasmic reticulum (ER) with granular material, prominent rough ER, and vacuoles, consistent with an intracellular trafficking defect. We studied the effect of COPB2 deficiency on collagen trafficking because of the critical role of collagen secretion in bone biology. COPB2 siRNA-treated fibroblasts showed delayed collagen secretion with retention of type I collagen in the ER and Golgi and altered distribution of Golgi markers. copb2-null zebrafish embryos showed retention of type II collagen, disorganization of the ER and Golgi, and early larval lethality. Copb2+/− mice exhibited low bone mass, and consistent with the findings in human cells and zebrafish, studies in Copb2+/− mouse fibroblasts suggest ER stress and a Golgi defect. Interestingly, ascorbic acid treatment partially rescued the zebrafish developmental phenotype and the cellular phenotype in Copb2+/− mouse fibroblasts. This work identifies a form of coatopathy due to COPB2 haploinsufficiency, explores a potential therapeutic approach for this disorder, and highlights the role of the COPI complex as a regulator of skeletal homeostasis.
Keywords: coatopathy, COPI complex, COPB2, collagen trafficking, juvenile osteoporosis
Introduction
Vesicle coat proteins are an essential and evolutionarily conserved group of proteins that form the molecular machinery responsible for sorting and trafficking of proteins and lipids within the cell.1, 2, 3 Pathogenic variants in genes encoding subunits of coat complexes (coatomers), or in accessory and regulatory factors important for their function, have been implicated in a number of genetic disorders collectively termed coatopathies.2 Disruption of secretory pathways can cause retention of cargo proteins within the endoplasmic reticulum (ER) and Golgi compartments, ultimately leading to impaired biogenesis and function of these organelles, ER stress, and decreased cell viability. Abnormal vesicular transport also affects post-translational modifications (specifically glycosylation) of secreted proteins at the Golgi complex and may alter the extracellular matrix composition. Although this can affect multiple organ systems, the central nervous system is most frequently affected, and individuals typically manifest with microcephaly and developmental delay.2,4,5
Skeletal development and bone growth and strength are highly dependent on the proper biosynthesis, post-translational modifications, assembly, and cross-linking of collagen fibrils,6 including type I collagen within the bone and type II collagen within the cartilage. Procollagen (PC) molecules undergo multiple post-translational modifications while being transported through the ER, Golgi complex, and the plasma membrane.7 The ER export of fibrillar PC, such as type I and type II procollagen (PCI and PCII), requires the coordinated action of PC receptors such as transport and Golgi organization (TANGO)/cutaneous T-cell lymphoma-associated antigen 5 (cTAGE5) proteins, coat protein complex II (COPII) and transport protein particle (TRAPP) complex components, and the retrograde recruitment of coat protein complex I (COPI)-coated ERGIC53-containing vesicles.3,8, 9, 10 Abnormal export of PCI and PCII from the ER to the Golgi and plasma membrane due to defects in vesicular trafficking components has been associated with skeletal dysplasias.11, 12, 13, 14, 15
The coatomer complex COPI is essential for retrograde Golgi/ ER-Golgi intermediate compartment (ERGIC) to ER trafficking (which is required for recycling of membranes and proteins from the Golgi to ER) and within the Golgi cisternae and is also involved in anterograde trafficking between the ER to Golgi.16, 17, 18 COPI subunits are essential for notochord development in zebrafish.19 In humans, geroderma osteodysplasticum (MIM: 231070) is caused by loss of the COPI scaffold protein GORAB (MIM: 607983),20 and defects in ARCN1 (MIM: 600820), encoding COPI complex subunit, are associated with syndromic rhizomelic short stature (MIM: 617164).21
Here, we report loss-of-function variants in COPB2 (MIM: 606990), a component of the COPI coatomer complex, in six individuals from five unrelated families presenting with a clinical spectrum of osteoporosis or osteopenia, with or without fractures, and developmental delay of variable severity. A hypomorphic, homozygous missense variant in COPB2 was previously reported in two siblings with microcephaly, spasticity, and developmental delay (MIM: 617800)22 in whom we also here identified low bone mass. We focused on the effect of COPB2 haploinsufficiency on the integrity of ER-Golgi machinery and collagen trafficking and correlate it with the phenotypic spectrum. Our data demonstrate that pathogenic variants in COPB2 lead to early onset osteoporosis and variable developmental delay and that COPB2 and the COPI complex are essential regulators of skeletal homeostasis.
Material and methods
Study approval
All zebrafish care and experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Oregon. Mice were housed in the Baylor College of Medicine Animal Vivarium, and all studies were approved by the IACUC at Baylor College of Medicine. For human studies, informed consent was obtained as part of the Undiagnosed Diseases Network for subjects 1 and 2, according to the institutional review board of the National Human Genome Research Institute. The research sequencing study (exome and genome sequencing) and transfer of clinical and sequencing data were approved by the human subject ethics committees as part of the 100,000 Genomes Project for subject 3 and at Baylor College of Medicine for subjects 4, 5, and 6. All families provided informed consent prior to participation.
Exome or genome sequencing and identification of COPB2 variants
Subject 1 had research trio exome sequencing performed at the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC). Briefly, library was constructed as described in the BCM-HGSC protocol and hybridized to the HGSC VCRome 2.1 design (42 Mb NimbleGen, cat. no. 06266380001).23 Sequencing was performed in paired-end mode with the Illumina HiSeq 2000 platform with sequencing-by-synthesis reactions extended for 101 cycles from each end and an additional seven cycles for the index read. With a sequencing yield of 6.6 Gb, the samples achieved 92% of the targeted exome bases covered to a depth of 20× or greater. Illumina sequence analysis was performed with the HGSC Mercury analysis pipeline,24,25 which moves data through various analysis tools from the initial sequence generation on the instrument to annotated variant calls. In parallel to the exome workflow, an Illumina Infinium Exome-24 v.1.1 array was generated for a final quality assessment. This included orthogonal confirmation of sample identity and purity via the error rate in sequencing (ERIS) pipeline developed at the HGSC. A successfully sequenced sample met quality control metrics of ERIS SNP array concordance (>90%) and ERIS average contamination rate (<5%).
GeneMatcher26 assisted in the recruitment of subject 2, who had clinical exome sequencing at GeneDx. Subject 3 had genome sequencing through the UK 100,000 Genomes Project. In brief, DNA was extracted from lymphocytes at the North Thames Genomic Laboratory Hub, Great Ormond Street Hospital for Children, NHS Foundation Trust, and sent to Illumina for genome sequencing. Sequencing data were passed through Genomics England’s bioinformatics pipeline for alignment, annotation, and variant calling. On the basis of the Human Phenotype Ontology terms entered for this individual, the following gene panel was applied: osteogenesis imperfecta 1.13. The COPB2 c.1206−2A>G variant was not called by the PanelApp but was identified subsequently with the Exomiser tool.27 The variant was confirmed via bidirectional Sanger sequencing at the North Thames Genomic Laboratory Hub, Great Ormond Street Hospital for Children, NHS Foundation Trust. Subject 4 had clinical sequencing at GeneDx. The sequencing analysis for subjects 5 and 6 was reported in a prior publication.22
RNA sequencing
Messenger RNA from whole blood was extracted, quantified, and processed prior to library preparation via the Illumina TruSeq Kit (Illumina, SD) according to manufacturer’s instructions. Samples were subsequently multiplexed and subject to 150 bp paired-end sequencing on the Illumina HiSeq 2000 platform. Resulting raw base call (bcl) reads were converted to FASTQ prior to being processed with a pipeline adapted from the Genotype-Tissue Expression (GTEx) Consortium.28 Briefly, reads were aligned to the GRCh37 human reference via Spliced Transcript Alignment to a Reference (STAR)29 followed by gene-based quantification with RNA sequencing (RNA-seq) by expectation maximization (RSEM)30 and alignment visualization in the Integrative Genomics Viewer (IGV).31
RNA extraction and real-time qPCR
Total RNA was extracted from lymphoblastoid cells derived from subject 1, her parents, and sibling with Trizol reagent. Similarly, total RNA was extracted from fibroblasts derived from subject 2 and her mother. cDNA was synthesized with the Superscript III First Strand RT-PCR kit according to the manufacturer’s protocol (Invitrogen). Real-time qPCR was performed on a LightCycler instrument (Roche) with FastStart Essential DNA Master reagent (Roche) with GAPDH or B2m (beta-2 microglubulin) as internal control.
Electron microscopy
Cells were pelleted and fixed in 4% paraformaldehyde, rinsed, and then dehydrated in a graded ethanol series from 30%–100%. The samples were washed in propylene oxide and embedded in Spurrs epoxy. Ultrathin sections were stained in uranyl acetate followed by Reynolds lead citrate and examined via an FEI Tecnai G2 TEM.
Cell culture and PC transport assay
All cells were cultured at 37°C and 5% CO2 in a humid environment. HeLa (ATCC) and BJ-5ta hTERT-immortalized fibroblasts (ATCC), referred to as human fibroblasts (HFs), were cultured according to ATCC guidelines. The bone marrow-derived stromal cell line ST232 was provided by the laboratory of Fanxin Long (Department of Orthopedic Surgery, Washington University School of Medicine, St. Louis, Missouri, USA). ST2 cells were cultured in growth medium (α-MEM HyClone with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin). The following COPB2 siRNA sequences were used in this study: (1) CGAUGUAUCUCCUAGGCUA, (2) GGUCAAACAAUGUCGCUUU, and (3) CAGUAUCCACAGAUCCUGA in human cell lines and ON-TARGETplus Mouse Copb2 siRNA SMARTPool (Dharmacon) in ST2 cells. We treated HeLa and HFs for 36 h with Oligofectamine (Life Technologies) or RNAiMax (Life Technologies), respectively, by using different final concentrations of siRNAs (20 nM and 100 nM) to obtain different levels of silencing. ST2 cells were treated for 96 h with RNAiMax (Life Technologies) to a final concentration of 20 nM siRNA. We analyzed total cell lysates by immunoblot to quantify the COPB2 levels as described previously.12 Primary antibodies used in this study were mouse monoclonal anti-coatomer (CM1A1033), anti-GM130 (BD Transduction), anti-Giantin (Abcam), anti-GBF1 (BD Transduction), anti-KDEL receptor (Enzo LifeSciences), sheep polyclonal anti-TGN46 (SeroTech), rabbit polyclonal anti-PC-I (Rockland), anti-SAR1 (Sigma), anti-HSP47 (Novus Biologicals), anti-SEC24C (Sigma), anti-beta COP (Thermo Fisher Scientific), anti-GAPDH (Sigma), and anti-β-actin (Sigma). Immunofluorescence experiments were performed as previously described.12 All the images were acquired via a Zeiss confocal microscope LSM800. All the transport assays were performed as previously described.12 Briefly, to follow endogenous PC-I in fibroblasts, we incubated cells for 3 h at 40°C in DMEM supplemented with 1% serum and 20 mM HEPES (pH 7.2) and then shifted them to 32°C in the presence of cycloheximide (100 μg/mL) and ascorbate (50 μg/mL) for the indicated times. The retention using selective hook (RUSH) assay was performed as previously described.34 GPI-GFP-RUSH and CSF1-GFP-RUSH plasmids were kindly provided by Franck Perez and Gaelle Boncompain (Institut Curie, Paris, France). In brief, HeLa cells were transfected with GPI-GFP-RUSH or with CSF1-GFP-RUSH plasmids (0.5 μg/well) and cells were incubated for 16 h. After 16 h, cells were fixed (time 0 min) or treated with 40 μM biotin and 50 μg/mL of cycloheximide for promotion of the synchronized release of the cargo proteins from the ER. Quantification of GPI-positive and CSF-1-positive structures was performed via Fiji software with the same threshold for all the analyzed conditions. 50 cells/condition from three independent experiments were counted. We performed quantitative evaluation of PC-I transport by analyzing the immunofluorescence staining patterns of at least 100 cells in three independent experiments.
Zebrafish CRISPR-Cas9 model
Generation of mutant lines via CRISPR-Cas9
One single-guide RNA (sgRNA) was designed that contained targeting sequences in exon 2 (5′-GGACATCAAGCGCAAACTCA-3′) of copb2. sgRNA and Cas9 RNAs were co-injected at the one-cell stage. We used the following primers to identify introduced mutations: forward 5′-AGAGTACTGATGTATAATTTCTGC-3′ and reverse 5′-TCACTTGTGTTTCATGGTTC-3′. Wild-type (WT) ABCxTu and heterozygous mutant copb2b1327/+ adult zebrafish were maintained as previously described.35 Embryos and larvae were staged according to the staging series36 or by h post-fertilization (hpf) or days post-fertilization (dpf). Siblings (sibs) are defined as a mix of homozygous WT and heterozygous mutants from intercrosses of heterozygous mutant adults. Phenotypic assessment of heterozygous adult mutants was limited by the loss of this fish line because of lab shutdown during the COVID-19 pandemic.
Immunolabeling
Labeling of whole-mount larvae was performed as previously described37 with minor modifications. Thirty hpf embryos were fixed in BT fix35 overnight and permeabilized with proteinase K (10 μg/mL) for 10 min. The following antibodies were used: mouse anti-KDEL (Calbiochem, 1:500), mouse anti-GM-130 (BD Transduction Laboratories, 1:500), mouse anti-collagen type II (Developmental Studies Hybridoma Bank, II-II6B3; 1:200), and biotinylated horse anti-mouse (Vector Laboratories, 1:500). Images of immunolabeled embryos were acquired via a Zeiss LSM 5 confocal microscope and analyzed via ImageJ.
Ascorbic acid treatment
Homozygous copb2b1327/ b1327 embryos were treated from 5 hpf, when the notochord has not yet formed, to 30 hpf with 75 mM, 100 mM, and 200 mM ascorbic acid (Sigma). Ascorbic acid was diluted in embryo medium, and the pH of the solution was adjusted to 6.5. Control larvae were treated with an equal volume of water also diluted in embryo medium. At 30 hpf, each embryo was scored for its overall morphology and bisected at the end of the yolk extension. The posterior part was used for genotyping and the anterior part was fixed for immunolabeling. After genotyping and immunolabeling, the notochord was imaged.
In situ hybridization and histology
Assay was performed as previously described.38 Larvae were hybridized with a digoxigenin-labeled RNA probe spanning an 812 bp coding sequence between exons 8 and 14 of copb2. Stained larvae were embedded in 1% agarose, 0.5% agar, 5% sucrose medium and 16 μm cryosections were cut. Images were acquired via a Zeiss Axioplan2 compound microscope.
Mouse CRISPR-Cas9 model
Generation of mutant lines via CRISPR-Cas9
Single-guide RNAs (sgRNAs) were designed that contained targeting sequences to allow for exon 6 and 7 deletion [5′-CGGGTTTACCTGCCCAGCGG(TGG)-3′ and 5′-ACAGACCTCGGTTCAAAATC(AGG)-3′] of Copb2. sgRNA and Cas9 mRNA were co-injected into the cytoplasm of C57BL/6J mouse embryos at the one-cell stage. PCR cloning followed by Sanger sequencing with primers 5′-TCCAAGCATTATCCAAGGAAGT-3′, 5′-AACACCAGAGCCAAGAAGTG-3′, and 5′-GCCTTTTTCATGTCCTTCCA-3′ confirmed deletion of exon 6 through exon 7 in founder animals (Figure S6). Founders were backcrossed to generate N1 animals, inheritance of the deletion in N1 animals was confirmed by Sanger sequencing, and the N1 generation was backcrossed to generate N2 mice. N2 animals were intercrossed to generate the mouse line eventually used for phenotyping. Upon further breeding, no homozygous mice were detected, most likely because of early embryonic lethality of the Copb2−/− pups. Phenotypic analysis was therefore performed with mice that are heterozygous for the Copb2 deletion or their WT littermates. In parallel, the BCM component of KOMP2 generated a separate knockout mouse model of Copb2 [Copb2em1(IMPC)Bay] on the C57BL6/NJ background via the same sgRNAs targeting exon 6. Adult and embryo phenotyping data for this line are available on the International Mouse Phenotyping Consortium (IMPC) website.
Copb2+/− mice and wild-type (WT) littermates were euthanized at 8 weeks of age. The investigators were blinded to genotype in the micro computed tomography (micro-CT) and collagen mass-spectrometry analyses.
Micro-CT analysis
Spines and left femurs were scanned in 70% ethanol with a Scanco μCT-40 micro-CT system (55 kVp and 145 μA X-ray source), and scans were reconstructed at a 16 μm isotropic voxel size. We analyzed trabecular bone of L4 vertebrae and the distal metaphyses of left femurs with Scanco software by manually contouring trabecular bone. For vertebrae, the region of interest (ROI) was defined as the trabecular volume between the L4 vertebral endplates. At femoral trabecular ROI, 75 slides ( =1.2 mm) were analyzed proximal to the distal femoral growth plate. Quantification of trabecular parameters was performed via the Scanco software with a threshold value of 210. These parameters include bone volume/total volume (BV/TV), trabecular number (Tb.N) and trabecular thickness (Tb.Th), connectivity density (Conn.D), and tissue mineral density (TMD).39 Femur length was measured from the top of the femoral head to the bottom of the medial condyle. Cortical bone parameters of the femoral midshaft were measured at the exact center and at the distal 75% of femur length with the automated thresholding algorithm included in the Scanco software. Trabeculae in contact with cortical bone were manually removed from the ROI (11 slides analyzed per location, threshold 210). The cortical parameters include total cross-sectional area (Tt.Ar), cortical bone area (Ct.Ar), marrow area (Ma.Ar), cortical thickness (Ct.Th), cross-sectional moments of inertia (CSMI), anterior-posterior diameter, and TMD.39 Ten spines and femurs were scanned per group.
Mass spectrometry of type I collagen
Collagen was prepared from minced femurs, collected from euthanized 8-week-old mice. Type I α chains were extracted by heat denaturation (90°C) in SDS-PAGE sample buffer, resolved on 6% SDS-PAGE gels, cut from gels, and digested with trypsin in-gel.40 Bone tissue was also digested with bacterial collagenase as described.41 Collagenase-generated peptides were separated by reverse phase HPLC (C8, Brownlee Aquapore RP-300, 4.6 mm × 25 cm) with a linear gradient of acetonitrile:n-propanol (3:1 v/v) in aqueous 0.1% (v/v) trifluoroacetic acid.42 Individual fractions were analyzed by liquid chromatography/mass spectrometry (LC/MS). Peptides were analyzed by electrospray LC/MS via an LTQ XL ion-trap mass spectrometer (Thermo Fisher Scientific) equipped with in-line liquid chromatography on a C4 5 μm capillary column (300 μm × 150 mm; Higgins Analytical RS-15M3-W045) and eluted at 4.5 μL/min. The LC mobile phase consisted of buffer A (0.1% formic acid in MilliQ water) and buffer B (0.1% formic acid in 3:1 acetonitrile:n-propanol v/v). An electrospray ionization source (ESI) introduced the LC sample stream into the mass spectrometer with a spray voltage of 3 kV. Proteome Discoverer search software (Thermo Fisher Scientific) was used for peptide identification with the NCBI protein database.
Golgi imaging in mouse primary fibroblasts
Quantitative imaging of Golgi structure and distribution was performed in skin fibroblasts derived from Copb+/− mice and WT littermates. Fibroblasts were cultured in chamber slides, and Golgi complex was labeled by CellLight Golgi-GFP BacMan 2.0 reagent per manufacturer’s protocol (Thermo Fisher Scientific). Nuclei were labeled with Hoechst staining. Acquisition and analysis of images was done with the assistance of the Baylor College of Medicine Integrated Microscopy Core with a Vala IC200 high-throughput microscope. For measurements, nuclei were segmented by watershed algorithm and cell mask was defined by tessellation. For each cell, Golgi were segmented by thresholding. Nuclear size and intensity, as well area, mean pixel intensity, and maximum length, were measured.
Ascorbic acid treatment
Copb2+/− female mice were provided with ascorbic-acid-enriched chow or the control chow for a period of 5 weeks. The ascorbic-acid-enriched chow (ENVIGO Teklad laboratory animal diet) contains 18% protein, 49.1% carbohydrate, and 6.3% fat, in addition to 1% ascorbic acid. The control chow (ENVIGO Teklad laboratory animal diet) contains 18.2% protein, 49.6% carbohydrate, and 6.4% fat, without ascorbic acid. Experimental diet was initiated at weaning age (P21), and mice were collected for analysis by micro-CT of spines at 8 weeks of age.
Statistical analysis
We used nonparametric two-tailed paired t test or one-way ANOVA as indicated to compare between samples. Statistical significance was determined as p value that is equal to or less than 0.05.
Results
Clinical presentation and molecular studies
We have identified six individuals (subjects 1–6) with overlapping phenotypes who carry heterozygous, putatively deleterious variants in COPB2. Their molecular and clinical findings are summarized below and in Table 1.
Table 1.
Clinical summary and COPB2 variants identified
| Subject 1 (family 1) | Subject 2 (family 2) | Subject 3 (family 3) | Subject 4 (family 4) | Subject 5 (family 5) | Subject 6 (family 5) | |
|---|---|---|---|---|---|---|
| Gender | female | female | male | female | male | female |
| Age at most recent evaluation (years) | 9 | 9 | 4 | 3 | 9 | 12 |
| Ethnicity | Caucasian/Native American | Caucasian/Native American | Caucasian | Asian | Caucasian/Native American | Caucasian/Native American |
| COPB2 variant (hg19, GenBank: NM_004766.2) | g.139088354_355delTT (c.1237_1238delAA; p.Lys413AspfsTer3) | g.139081338dupT (c.1906dupA; p.Thr636AsnfsTer7) | g.139088388T>C (c.1206−2A>G) | g.139097997dupC (c.247dupG; p.Val83GlyfsTer14) | g.139092642G>A (c.760C>T; p.Arg254Cys) | g.139092642G>A (c.760C>T; p.Arg254Cys) |
| Inheritance | de novo (heterozygous) | not maternal (heterozygous) | de novo (heterozygous) | de novo (heterozygous) | homozygous | homozygous |
| General | ||||||
| Prematurity | no (39 weeks) | yes (33 weeks) | no (stated as term) | no (40 weeks) | yes (36 weeks) | yes (35 weeks) |
| IUGR or small for age at birth | no | no | no | no | no | no |
| Growth parameters on most recent evaluation (percentile) | weight, 27th; height, 35th | weight, 43rd; height, 10th | weight, 5th; height, 1st | weight, 25th; height, 70th | weight, 63rd; height, 1st | weight, 1st; height, 30th |
| Head circumference (Z score) | 0.65 | −2.19 | −0.31 | −4 | −8 | −5.5 |
| Musculoskeletal features | ||||||
| Osteopenia | yes | yes | yes | unknown | yes | yes |
| Fractures | yes | no | yes | no | no | no |
| Neurological features | ||||||
| Developmental delay | yes | yes | yes | yes | yes | yes |
| Intellectual disability | no, but has learning disability and ADHD | yes, mild | N/A | N/A | yes, severe (nonverbal) | yes, severe (nonverbal) |
| Spasticity (ambulatory status) | no (normal gait) | yes (wide-based unsteady gait, progressed to require a walker and wheelchair) | no (wide-based gait) | possible in ankles bilaterally (mild, with normal gait) | yes (non- ambulatory) | yes (non- ambulatory) |
| Seizures | no | no | no | no | yes | yes |
| Brain MRI | focal cortical dysplasia | normal | normal | normal | microcephaly, simplified gyral pattern, thin corpus callosum, delayed myelination | microcephaly, simplified gyral pattern, thin corpus callosum, delayed myelination |
| Prior genetic testing | ||||||
| Copy number variant analysis | negative | copy number gain on chromosome 12p12.1 (24,692,382–25,353,987; hg19), maternally inherited | negative | negative | a 16.8 Mb region of absence of heterozygosity (AOH) on chromosome 3q22.1–3q24 (131,374,802–148,170,986; hg19) | a 16.8 Mb region of absence of heterozygosity (AOH) on chromosome 3q22.1–3q24 (131,374,802–148,170,986; hg19) |
| Other clinical testing | COL1A1/COL1A2 sequencing and recessive OI gene panel, negative; metabolic labs, normal; CDG, negative; clinical ES, negative | hereditary spastic paraplegia panel, negative; metabolic labs and CSF studies, normal; CDG, negative; clinical ES, negative | OI dominant gene panel (COL1A1/COL1A2/IFITM5), negative | autism/ID gene panel, negative | N/A | primary microcephaly gene panel, negative |
Abbreviations: CDG, congenital disorders of glycosylation; CSF, cerebrospinal fluid; ES, exome sequencing; ID, intellectual disability; N/A, not applicable; OI, osteogenesis imperfecta.
Subject 1 is a 9-year-old female who presented with her first fracture at 2 years of age, followed by more than ten fractures of long bones. Bone densitometry via dual energy X-ray absorptiometry (DXA) scan at 3 years of age indicated lumbar spine Z score of −3.2. Bisphosphonate therapy was initiated, and a repeat DXA scan at 5 years of age showed normal bone mineral density (BMD; lumbar spine Z score of 0.2; left hip Z score of 1.8; right hip Z score of 2.8). Over time, the frequency of her fractures decreased, but she continued to suffer chronic bone pain. Her growth was normal (height at the 60th percentile). She did not have dentinogenesis imperfecta, blue sclera, joint hypermobility, bone deformities, or scoliosis. There was no family history of osteopenia or recurrent fractures. Her development was significant for speech delay, attention deficit hyperactivity disorder (ADHD), and learning disabilities. Brain MRI showed focal cortical dysplasia of left frontal gyrus although she had no seizures. Previous genetic evaluation included normal COL1A1 (MIM: 120150)/COL1A2 (MIM: 120160), CRTAP (MIM: 605497), P3H1 (MIM: 610339), FKBP10 (MIM: 607063), PPIB (MIM: 123841), SERPINF1 (MIM: 172860), SERPINH1 (MIM: 600943), and SP7 (MIM: 606633) sequencing. SNP-based copy number variant (CNV) analysis did not detect pathogenic deletions or duplications. Metabolic evaluations, including plasma amino acids, urine amino acids, an acylcarnitine profile, a lipid panel, and a congenital disorders of glycosylation panel, were normal. Research trio exome sequencing did not detect variants in genes known to cause osteogenesis imperfecta or other low bone mass phenotypes. Further analysis of the data identified a heterozygous c.1237_1238delAA (p.Lys413Aspfs∗3) (GenBank: NM_004766.2) variant in COPB2. This variant has not been reported in gnomAD43 and is predicted to cause loss of function due to a shift in the reading frame. Sanger sequencing of DNA from the proband and her parents confirmed that the variant is de novo.
Subject 2 is a 9-year-old female that presented with global developmental delay, microcephaly, and progressive spastic paraparesis. She was initially ambulatory but had wide-based gait and started developing spasticity at 2–3 years of age. She is able to walk a few steps independently but requires a walker or wheelchair for longer distances. She does not have a history of fractures, but a DXA scan performed at 9 years of age showed a total spine Z score of −1.9 and femoral neck Z score of −2.4 (Figure 1E). The family history is significant for a sibling who has severe developmental delay and is non-ambulatory due to a large, de novo 8q21.3–q24.3 duplication that is 52.42 Mb in size. There is no family history of osteopenia or recurrent fractures. MRI of brain and spine at 2 years of age showed scoliosis of the thoracolumbar spine but was otherwise normal. Electromyography and nerve conduction velocity studies were normal. Previous genetic evaluation included a normal hereditary spastic paraplegia gene panel. Array-based CNV analysis revealed a maternally inherited 12p12.1 duplication that is 662 kb in size (including SOX5 [MIM: 604975], BCAT1 [MIM: 113520], and LRMP [MIM: 602003]) that was considered an unlikely explanation for her phenotype. Clinical exome sequencing revealed a heterozygous c.1906dupA (p.Thr636Aspfs∗7) (GenBank: NM_004766.2) variant in COPB2 that was classified as a variant of uncertain significance. Subsequently, the family consented for transfer of those data to study in the Undiagnosed Diseases Network (UDN). The variant in COPB2 is novel and not reported in gnomAD.43 It is predicted to cause loss of function due to a shift in the reading frame. The mother and full siblings tested negative for this variant, but her deceased father was not available for testing.
Figure 1.
COPB2 variants lead to osteopenia and fractures
(A) Right knee radiograph of subject 3 at age 14 months showing fracture of distal femoral metadiaphysis and osteopenia.
(B) Left hand radiograph of subject 3 at age 20 months showing thin metacarpal cortices.
(C) Right foot radiograph of subject 5 at age 8 years showing diffuse osteopenia and hind-foot varus deformity.
(D) Right leg radiographs of subject 6 at age 12 years showing gracile, over-tubulated long bones.
(E) Bone densitometry scan in subject 2 showing low bone mineral density of the lumbar spine (left panel) and left hip (right panel).
(F) Schematic illustration of COPB2 variants identified in subjects with osteopenia and developmental delay (GenBank: NM_004766.2).
Subject 3 is a 4-year-old male who first presented with fracture of the right distal femoral metadiaphysis at 14 months of age (Figure 1A). He had another fracture of the right distal femur at 20 months due to minor trauma. A skeletal survey demonstrated recurrent insufficiency fractures of the distal right femur, generalized osteopenia (Figures 1A and 1B), varus bowing deformities of both ulnae, no rachitic changes, apparent cavo-varus deformities of the hind feet, and wormian bones at the lambdoid suture (the latter could be a normal variant). His growth was notable for short stature (height Z score at −2.3). His exam was significant for pale gray sclerae, normal dentition, low muscle tone, and mild joint hypermobility. A kyphotic posture was noted while sitting although the spine was straight. There was no family history of osteopenia or recurrent fractures. He is the seventh of eight children born to non-consanguineous Ashkenazi Jewish parents. His parents and siblings are all healthy. His development was significant for gross motor delay. A brain MRI was normal. An array-based CNV analysis and a dominant osteogenesis imperfecta gene panel, including COL1A1, COL1A2, and IFITM5 (MIM: 614757), were negative. This subject was recruited into the 100,000 Genomes Project (UK Department of Health funded genomes project) and genome sequencing identified a de novo heterozygous variant, c.1206−2A>G (GenBank: NM_004766.2), in COPB2 that is novel and not reported in gnomAD.43 The variant is predicted to affect splicing.
Subject 4 is a 3-year-old female who presented with global developmental delay, abnormal muscle tone, poor weight gain, and microcephaly. Prenatal history was unremarkable, and fetal growth measurements were within the normal limits. Head circumference growth began declining at 4 months of age from 2 SD below the mean to 2.86 SD below the mean at age 1 year 4 months, and to 4 SD below the mean at 3 years of age. Her medical history was also remarkable for atopic dermatitis, mild joint hypermobility, and feeding issues. She does not have a history of fractures and has not had skeletal radiographs for evaluation for osteopenia. A brain MRI performed at 2 years of age was normal. A SNP-based CNV analysis was negative. An Autism/Intellectual Disability XPanded Panel (GeneDx) revealed a de novo, heterozygous variant, c.247dupG (p.Val83Glyfs∗14), in COPB2 (GenBank: NM_004766.2) that is novel and not reported in gnomAD.43 This frameshift variant (located in exon 4 out of 22) is predicted to result in protein truncation or nonsense-mediated mRNA decay.
Subject 5, a 9-year-old male, and subject 6, a 12-year-old female, are siblings that were described in a prior publication.22 Their bone phenotype had not been evaluated at the time of the previous report. They presented with global developmental delay, spasticity, seizures, and severe microcephaly. They do not have a history of fractures, but skeletal radiographs showed gracile, over-tubulated bones and osteopenia (Figures 1C and 1D). Brain MRI findings in both siblings demonstrated microcephaly with simplified gyral pattern, thin corpus callosum, and delayed myelination. A primary microcephaly gene panel in subject 6, including ASPM (MIM: 605481), CDK5RAP2 (MIM: 608201), CENPJ (MIM: 609279), MCPH1 (MIM: 607117), and STIL (MIM: 181590), was negative. Array-based CNV analysis identified a 16.8 Mb region of absence of heterozygosity on chromosome 3q22.1–q24 (chr3: 131,374,802–148,170,986; hg19). A subsequent research exome sequencing analysis indicated a shared recessive homozygous missense variant, c.760C>T (p.Arg254Cys) (GenBank: NM_004766.2), in COPB2. This variant was reported three times in gnomAD43 in European (non-Finnish) population (allele frequency 0.00001063) but only in a heterozygous status.
COPB2 loss-of-function variants result in haploinsufficiency accompanied by abnormal cellular morphology
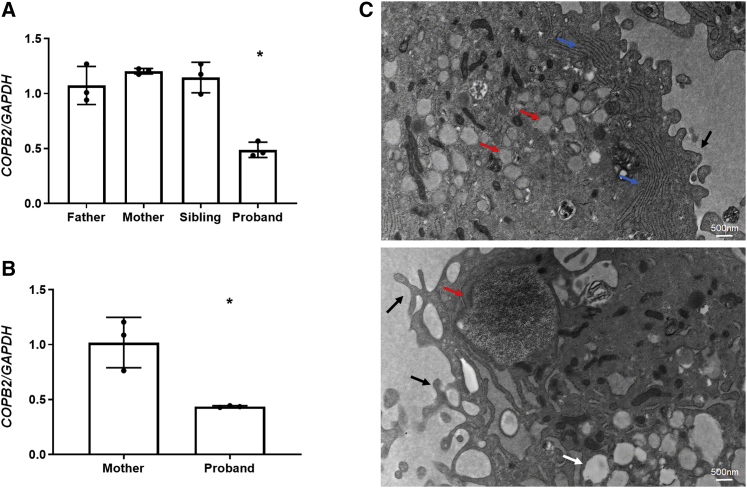
To better understand the molecular consequences of COPB2 variants, we studied the expression of COPB2 in cells from affected individuals. mRNA sequencing on whole blood from subject 1 demonstrated that the heterozygous c.1237_1238delAA variant leads to nonsense-mediated mRNA decay (Figure S1). Real-time qPCR analysis in lymphoblastoid cells derived from subject 1 supported this finding by showing significantly reduced expression of COPB2, as compared to her parents and sibling (Figure 2A). Real-time qPCR analysis in fibroblast cells derived from subject 2 showed significantly reduced expression of COPB2, as compared to her mother (Figure 2B). Electron microscopy of fibroblasts from subject 2 showed markedly dilated ER filled with granular material, prominent rough ER, multiple intracellular vacuoles, and prominent cell surface pseudopodia, collectively suggesting a disruption of vesicular trafficking in these cells (Figure 2C).
Figure 2.
COPB2 haploinsufficiency results in abnormal cellular morphology
(A) COPB2 qPCR in lymphoblastoid cells from subject 1 (proband) showing decreased COPB2 expression by about 50% compared to expression in parents and sibling.
(B) COPB2 qPCR in skin fibroblasts from subject 2 (proband) and her mother showing decreased COPB2 expression by about 50% compared to expression in parent. Results shown as COPB2/GAPDH relative mRNA expression, ∗p = 0.0002 (one-way ANOVA) for subject 1, and ∗p = 0.002 (t test) for subject 2.
(C) Electron microscopy of subject 2 fibroblasts showing dilated ER containing granular matrix (red arrows), prominent rough ER (blue arrows), vacuoles (white arrow), and dominant pseudopodia on the cytoplasmic membrane (black arrows).
COPB2 depletion is associated with delayed collagen trafficking and disorganization of the Golgi complex
In order to study the effect of COPB2 deficiency on intracellular trafficking, we assessed the organization of the ER-Golgi intermediate compartment (ERGIC) and Golgi complex and the trafficking of various classes of cargo including PCI by immunofluorescence microscopy in human fibroblasts or HeLa cells where the expression of COPB2 was titrated by siRNA treatment (Figure 3 and Figure S2). COPB2 siRNA-treated cells exhibited a disorganized distribution of Golgi markers that was proportional to the extent of the COPB2 depletion (Figure 3A and Figure S2). Additionally, COPB2 depletion resulted in marked reduction of the punctate peripheral pattern of the ERGIC, as assessed by COPI, GBF1, KDEL receptor, and ERGIC53 labeling, indicating impaired Golgi-to-ER retrograde trafficking (Figure 3B and Figure S2).
Figure 3.
Partial depletion of COPB2 induces disorganization of the Golgi complex and impairs ER exit of PCI in human fibroblasts
(A) BJ-5ta human fibroblasts (HFs) were mock (CTRL) or COPB2-siRNA treated for 36 h for obtainment of different levels of COPB2 reduction (to 50% and 35% of control via 20 nM and 100 nM siRNAs, respectively). The gray bar indicates the residual COPB2 protein level after siRNA treatment. Cells were immunolabeled with an antibody against the cis-Golgi marker GM130 (upper panel), medial Golgi marker Giantin (middle panel), and trans-Golgi protein TGN46 (lower panel). Scale bars represent 10 μm.
(B) Immunofluorescence analyses of CTRL and COPB2-KD HFs for several proteins of the ER-Golgi intermediate compartment (ERGIC): GBF1, COPI (evaluated with an anti-coatomer mAbCM1A10), and KDEL receptor (all markers in gray). Scale bars represent 10 μm. Numbers represent quantification of GBF1-positive and COPI-positive spots per cell from one representative experiment. N = 3 experiments, n = 50 cells counted, ± SD.
(C) PCI transport in human fibroblasts. CTRL and COPB2-KD (50% of residual protein level) cells were shifted to 40°C for 3 h for accumulation of PCI (red) in the ER. Cells were then either fixed immediately (0 min) or incubated for different times (15 and 60 min) at 32°C in the presence of ascorbate and CHX. During the temperature block, both CTRL and COPB2-KD cells accumulate PCI into the ER. In control cells, PCI co-localizes with the Golgi marker Giantin (green) after 15 min but does not in COPB2-KD cells where it fails to exit the ER. PCI and HSP47 co-localized in control and COPB-KD cells at baseline (0 min) and continued to co-localize in COPB2-KD cells (but not in control cells) after 15 and 60 min because of ER retention of PCI. Scale bars represent 10 μm.
(D) Quantification of the ER exit of PCI in CTRL and COPB2-KD HFs expressed as percentage of cells with PCI in the ER. N = 3 experiments, n = 100 cells counted. Mean values ± SD (∗∗p < 0.001, ∗∗∗p < 0.0001).
(E) PCI transport assay in COPB2-KD human fibroblasts as in (C). Cells were immunolabeled for PCI and cTAGE5 as a marker of the ER exit sites. PCI co-localized with cTAGE5 15 and 60 min after the release. Bottom panels are enlargements of the boxed areas with white arrows indicating the co-localizing PCI with cTAGE5. Scale bars represent 10 μm.
To analyze the impact of COPB2 depletion on intracellular trafficking of newly synthesized cargos, we used a temperature-based synchronization protocol that depends on the temperature sensitivity of PCI folding, such that PCI is retained in the ER at 40°C.12 We found that COPB2 siRNA-treated fibroblasts had a marked delay in ER export of PCI, which was retained in the ER (Figures 3C and 3D) at the level of ER exit sites (as labeled by cTAGE5, Figure 3E) for up to 60 min after the release of the 40°C temperature block. As seen in Figure 3C, HSP47 is present and properly localized to the ER in both control and COPB2 siRNA-treated cells. Co-localization of HSP47 and PCI in the ER supports that PCI is properly folded in COPB2-depleted cells (Figure 3C). Levels of SEC24C and SAR1, essential members of the ER exit machinery, were not affected by COPB2 depletion (Figure S3). Moreover, COPB2 depletion did not affect the levels of COPI complex component beta COP (COPB, Figure S3). We then analyzed the anterograde trafficking of GPI-GFP and CSF1-GFP that served as control cargo proteins by using the RUSH (retention using selective hook) system.34 Transport of control cargo was not significantly different between COPB2-depleted and mock-transfected cells (Figure S2), suggesting a preferential requirement for COPB2 in the intracellular transport of PCI.
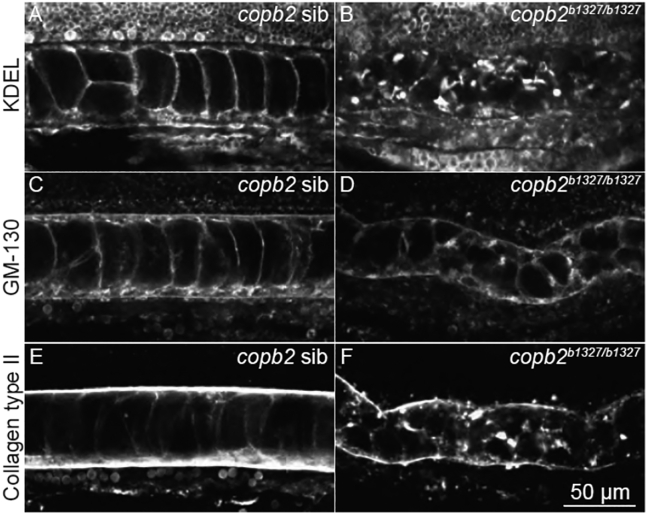
copb2 mutant zebrafish embryos show abnormal secretion of type II collagen and disruption of the Golgi and ER
To study the developmental consequences of COPB2 loss of function in vivo, we generated a mutant allele of the zebrafish ortholog, copb2, by using CRISPR-Cas9 technology to introduce a frameshift followed by a premature stop codon (copb2b1327, p.Lys10Thrfs∗11; Figure S4). Consistent with a previously analyzed zebrafish null allele of copb2,19,44 copb2b1327/b1327 homozygous mutant animals showed a severe phenotype at 24 hpf, including kinked notochord, pigmentation defect, hydrocephaly, thinner midbrain-hindbrain boundary, and embryonic lethality. In copb2b1327/b1327 homozygous mutant embryos, the ER and the Golgi apparatus appeared mislocalized and fragmented (Figures 4A–4D) as observed previously.19 Defect in type II collagen secretion with cytoplasmic aggregation in notochord cells was observed (Figures 4E and 4F), supporting the evolutionarily conserved role of the Golgi complex in collagen trafficking. Although copb2b1327/b1327 zebrafish exhibit hydrocephaly and a thinner midbrain-hindbrain boundary, sections through the brain did not reveal any other obvious abnormalities (Figure S5). Interestingly, developmental delay, with or without microcephaly, had been reported in coatopathies and other disorders related to vesicular transport dysfunction.2,4,5
Figure 4.
Structural integrity of the ER and Golgi apparatus and protein secretion are compromised in copb2b1327/b1327 zebrafish mutants
(A–F) Confocal sections of the notochord. Immunolabeling of KDEL in copb2 siblings (A) and copb2b1327/b1327 mutant (B) embryos. Immunolabeling of GM130 in copb2 siblings (C) and copb2b1327/b1327 mutant (D) embryos. Immunolabeling of type II collagen in copb2 siblings (E) and copb2b1327/b1327 mutant (F) embryos (anterior is to the left and dorsal to the top). Images were taken at the level of the yolk extension at 30 hpf.
Copb2+/− mice exhibit low bone mass
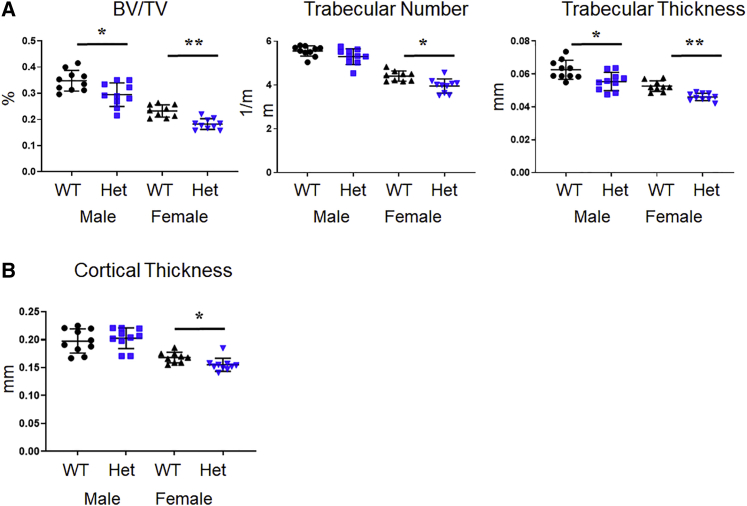
To better understand the clinical relevance of these findings, we generated a mouse carrying a Copb2-null allele by using CRISPR-Cas9 technology (Figure S6). In crosses of Copb2+/− mice, no Copb2−/− embryos were identified by screening at embryonic day 12.5 (E12.5)–E13.5. Homozygosity for a similar CRISPR-Cas9-generated allele (Copb2em1(IMPC)Bay) produced by the Baylor College of Medicine (BCM) component of the Knockout Mouse Phenotyping Program (KOMP2) project was found to be embryonic lethal, and no homozygous embryos were identified after E15.5. Therefore, phenotypic analysis was performed in Copb2+/− mice. Analysis of bone mass via micro-CT imaging showed a 15%–20% reduction in spine BV/TV in 2-month-old Copb+/− male and female mice compared to control WT littermates (Figure 5A). In females, the spinal trabecular number, trabecular thickness, and femur cortical thickness were significantly reduced, as compared to WT littermates (Figures 5A and 5B).
Figure 5.
Copb2 heterozygous mice exhibit low bone mass phenotype
(A) Micro-CT analysis of spine showed significant decrease in bone architectural parameters in Copb2+/− mice (Het), including the bone volume/total volume (BV/TV, p = 0.014 for males, p = 0.0006 for females) and trabecular thickness (p = 0.007 for males, p < 0.0001 for females). Females also showed decreased trabecular number (p = 0.002).
(B) Micro-CT analysis of femurs showed mild reduction in cortical thickness in the females (p = 0.003).
Mean values ± SD are indicated for both (A) and (B) (∗p < 0.05, ∗∗p < 0.001).
Additionally, we analyzed mouse femurs for possible collagen over-modification to check for abnormal processing of type I collagen, which is characteristic of osteogenesis imperfecta caused by qualitative defects in type I collagen or by pathogenic variants in components of the collagen 3-hydroxylation complex.45 This analysis did not reveal significant changes in lysine or proline modification (Figure S7).
Ascorbic acid rescues the phenotype of copb2-deficient zebrafish and Copb2+/− mouse fibroblasts
Ascorbic acid (vitamin C) serves as a cofactor for PC hydroxylases and induces the intracellular trafficking and secretion of collagen molecules.7,16,46 In addition, ascorbic acid has been implicated in protein homeostasis in the ER and in regulating the ER stress response.47, 48, 49 Given the essential role of ascorbic acid in PC metabolism, and its potential role as a modulator of proteostasis and ER stress, the effect of ascorbic acid supplementation was studied in the COPB2-deficient animal models. Ascorbic acid treatment rescued type II collagen secretion in copb2b1327/b1327 homozygous zebrafish mutant embryos in a dose-dependent manner (Figures 6B, 6D, and 6F and Figure S8). A partial rescue of collagen type II secretion was observed in some copb2b1327/b1327 homozygous mutant embryos treated with 100 mM ascorbic acid (29% of mutants; Figures 6B and 6D and Figure S8). A full rescue (33% of mutants) was observed when copb2b1327/b1327 homozygous mutant embryos were treated with 200 mM ascorbic acid (Figure 5F and Figure S8). In vivo, the morphological defect of the notochord was rescued in embryos in which secretion of type II collagen was improved (Figure S9). Additionally, ascorbic acid treatment rescued the cellular phenotype in Copb2+/− mouse fibroblasts that exhibited altered Golgi distribution and increased expression of the ER stress marker "binding immunoglobulin protein" (BiP) (Figures 6G–6I), suggesting a beneficial effect on ER and Golgi dysfunction in COPB2 deficiency. A trend of improvement in bone mass was detected by micro-CT analysis of BV/TV in Copb2+/− mice that received ascorbic-acid-enriched diet; however, this effect was blunted by the fact that mice, unlike humans and zebrafish, have de novo endogenous synthesis of ascorbic acid (Figure S9).
Figure 6.
Secretion of collagen type II in copb2b1327/b1327 zebrafish mutants and ER/Golgi defects in Copb2+/− mouse fibroblasts can be rescued by treatment with ascorbic acid
(A–F) Confocal sections of zebrafish notochords (anterior is to the left and dorsal to the top, images were taken at the level of the yolk extension at 30 hpf). Immunolabeling of type II collagen in untreated copb2 siblings (A) and copb2b1327/b1327 (B) embryos, 100 mM ascorbic acid-treated copb2 siblings (C) and copb2b1327/b1327 embryos (D), and 200 mM ascorbic acid-treated copb2 siblings (E) and copb2b1327/b1327 embryos (F). See Figure S9 for quantification of the distribution of phenotypes in zebrafish.
(G) Representative image of Copb2+/− mouse fibroblasts (Het) and wild-type littermate control cells (WT) labeled by CellLight Golgi-GFP reagent for visualization of Golgi structure.
(H) Graph summarizing multiple single-cell measurements of Golgi area with or without ascorbic acid (n = 1,631/1,723 Het cells −/+ ascorbic acid treatment and n = 1,021/1,152 WT cells −/+ treatment).
(I) BiP/mHspa5 expression in Copb2+/− mouse fibroblasts (Het, n = 3) and wild-type littermate control cells (WT, n = 2) with or without ascorbic acid. Results are presented as mean values ± SD, summary of three independent experiments. ∗∗p = 0.002.
Discussion
Coatomer complexes and their interacting proteins play an essential role in the anterograde trafficking of cargo lipids and proteins and retrograde transport of proteins from the Golgi complex to the ER, maintaining the structure of the Golgi complex and preventing ER and Golgi stress.4,17,50,51 As such, their function is critical in development. Defects in various subunits have been implicated in both animal models and human genetic disorders.2,4,5 Here, we report a coatopathy due to pathogenic variants in COPB2 in five unrelated families. The subjects in this cohort present with osteopenia and fractures, thus expanding the phenotypic spectrum of previously reported neurodevelopmental features.22 Our data support a role for COPB2 in the intracellular trafficking of collagen as a mechanism for the early-onset osteoporosis.
Type I collagen is the major component of bone extracellular matrix. The biosynthesis of collagen involves multiple steps of post-translational modifications, including hydroxylation of lysine and proline residues in the ER and glycosylation in the Golgi complex.7,46 Abnormal intracellular trafficking of PC may result in altered post-translational modifications, abnormal folding of the triple helix of type I collagen, and ultimately instability of collagen fibers. Therefore, several defects in vesicular components associated with intracellular collagen trafficking have been reported to cause severe skeletal dysplasias.11, 12, 13, 14,21,52,53 The export of PC fibrils is a challenging task for the ER that requires a dedicated set of proteins.3,8, 9, 10,54 The ER-to-Golgi trafficking of PC has been the subject of study in different labs in the recent years and different and independent lines of evidence converge on a role of COPI in the release of newly synthesized ER-derived PC to the Golgi complex. There are essentially two potential mechanisms whereby COPB2 deficiency, and thus COPI deficiency, could lead to ER retention of PC. The first relates to the disruption of the input to the ER of ERGIC membranes through COPI-dependent retrograde trafficking, input that would feed the growth of large PC-containing carriers.55 We show, in fact, altered distribution of ERGIC components in COPB2-depleted cells, which is consistent with a derangement of retrograde trafficking (Figure 3 and Figure S2). The second mechanism by which COPI defects may impinge specifically on PC trafficking to the Golgi has been highlighted by two recent publications showing that large carriers/tunnels forming at the ER and containing PC acquire COPI before releasing their content into the Golgi complex.56,57 Our results demonstrate that COPB2 deficiency selectively affects the export of PCI in fibroblasts, providing evidence for the physiological relevance of COPI-mediated trafficking in the exit of PC from the ER. Curiously, despite the delayed transport causing retention of PC within the ER, mass spectrometry analysis of mouse bones did not detect significant changes in lysine and proline hydroxylation. Moreover, PC and HSP47 co-localized in the ER in COPB2-depleted cells. This suggests that although COPB2 deficiency delays secretion of type I collagen, it does not appear to affect its biosynthesis or post-translational processing in the ER. Dysfunction of the Golgi may result in altered protein glycosylation, as had been reported for defects in other components of the COPI complex,20 and this may be further contributing to the bone fragility.
Previous studies in Copb2R254C/R254C mice that are homozygous for the p.Arg254Cys variant (family 5, subjects 5 and 6) showed normal growth and no obvious brain abnormalities, as opposed to reduced brain size and cortical malformations in the Copb2R254C/Zfn mouse model.22 Interestingly, neurospheres derived from Copb2R254C/Zfn and Copb2R254C/wt mouse models showed significantly reduced cell viability, suggesting that the p.Arg254Cys is a hypomorphic variant.22 Although the molecular mechanism of the neurodevelopmental phenotype in our subjects is not well understood, this study supports a role for COPB2 in brain development. These findings are in agreement with the existence of other vesicular trafficking defects that are characterized by microcephaly and developmental delay.2,4
The anatomical and functional abnormalities seen in homozygous copb2 zebrafish and Copb2+/− mice were at least partially corrected by ascorbic acid supplementation. Ascorbic acid is known to stimulate collagen synthesis and secretion,16,46 enhance healing of fractured bones in preclinical studies,58 and significantly decrease the risk of osteoporosis and hip fracture in adults.59,60 Interestingly, it is also implicated in protein homeostasis in the ER,47, 48, 49 and our data suggest that the ER and Golgi dysfunction may be rescued by treatment with ascorbic acid. These experiments support the use of ascorbic acid as a therapeutic modality for improvement of bone fragility in individuals with COPB2 deficiency and provide a mechanistic rationale for its potential positive effects. Of note, the use of mouse models for studying of ascorbic acid supplementation is limited because of de novo synthesis of ascorbic acid in the mouse, while the Gulo knockout mice that have a block in the synthesis of ascorbic acid exhibit a strong skeletal phenotype.61 Testing the utility of ascorbic acid supplementation in COPB2-deficient subjects was beyond the scope and time frame of this work; however, this remains an important question to address in future studies.
One limitation of our study is the diversity of phenotypes noted within a small cohort of affected individuals. Although our subjects had overlapping skeletal and neurological features, the severity was variable, which may not be surprising for a heterozygous condition due to loss of function. Moreover, the neurological disability that is more severe in subjects 2, 5, and 6 may have contributed to the osteopenia, as would be frequently seen in individuals affected with neuromuscular disorders. Yet, it is important to note that other subjects who present with milder neurological phenotype, such as subjects 1 and 3, still showed significant osteopenia and fractures. It is also noteworthy that 4/6 subjects had heterozygous, de novo loss-of-function variants, whereas in one family a homozygous missense presumably hypomorphic variant was detected. Given the early embryonic lethality that was observed in a previously published mouse model,22 and in zebrafish model in this study, complete loss of function of COPB2 is most likely not tolerated in humans. The probability of loss-of-function intolerance (pLI) score reported in gnomAD43 for COPB2 is 1 and supports this prediction. It is difficult to discuss genotype-phenotype correlation in such a small cohort, and additional individuals must be identified before the spectrum of COPB2-related disorder can be fully characterized.
In summary, we have identified heterozygous COPB2 loss-of-function variants as a cause of coatopathy with variable developmental delay and juvenile osteoporosis. Early-onset osteopenia increases the risk for osteoporosis and fractures in adulthood. Identification of individuals with low bone mineral density early in life may enable preventive intervention to reduce the risk of fractures and adult osteoporosis. Our findings highlight the important role of the COPI complex and COPB2 in skeletal homeostasis and demonstrate that collagen trafficking is a biological determinant of bone mass. Our work on this rare disorder suggests that decreased collagen flux may contribute to more common forms of osteoporosis.
Acknowledgments
The authors would like to thank the subjects and their families for participation in the study. We thank Paula Patricia Hernandez, Judy Peirce, and Poh Kheng Loi for their technical assistance. We would like to acknowledge Gaelle Boncompain and Franck Perez for kindly sharing the GPI-GFP-RUSH and CSF1-GFP-RUSH plasmids. R.M. was supported by the Geisman- Osteogenesis Imperfecta Foundation fellowship award, the Lawrence Bone Disease Program of Texas Research Award, the NIH/NIGMS (T32GM07526-43), and a BCM Chao Physician-Scientist Award. This work was also supported by NIH U01HG007709 (B.L.), NIH P01 HD070394 (B.L.), NIH U54 AR068069 (B.L. and V.R.S.), NIH UM1HG006348 (M.E.D. and J.D.H.), NIH U54NS093793 (M. Westerfield), NIH K08 DK106453 (L.C.B.), NIH/NIGMS T32GM007526 (M.J.), NIH/NIDCR R03DE026233 (I.G.), NIH 5UM1HG006542 (V.R.S.), and NIH/NICHD U54HD083092 for the Baylor College of Medicine Intellectual and Developmental Disabilities Research Center (IDDRC). L.C.B. was also supported by a Career Award for Medical Scientists from the Burroughs Wellcome Fund. M.A.D.M. acknowledges the support of Telethon grant TGM11CB1, Associazione Italiana per la Ricerca sul Cancro grant IG2013_14761, and European Research Council Advanced Investigator grant 670881 (SYS MET project). R.V. acknowledges the support of The University of Naples Federico II (grant STAR2017 Linea1) and Associazione Italiana per la Ricerca sul Cancro MFAG2020-25174. G.M. was supported by the National Institute of Neurological Disorders and Stroke (NIH/NINDS) under award number K08NS092898, Jordan’s Guardian Angels, and the Brotman Baty Institute. See supplemental information for additional acknowledgments and consortium details.
Declaration of interests
The Department of Molecular and Human Genetics at Baylor College of Medicine receives financial support from Baylor Genetics. Dr. Brendan Lee serves on the Board of Directors of Baylor Genetics and chairs its Scientific Advisory Board but receives no personal income from these positions.
Published: August 26, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.08.002.
Web resources
BCM-HGSC protocols, https://www.hgsc.bcm.edu/content/protocols-sequencing-library-construction
GeneMatcher, https://genematcher.org/
Genomics England PanelApp, https://panelapp.genomicsengland.co.uk/
HGSC Mercury Analysis Pipeline, https://www.hgsc.bcm.edu/software/mercury
OMIM, https://www.omim.org/
Supplemental information
References
- 1.Brandizzi F., Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 2013;14:382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dell’Angelica E.C., Bonifacino J.S. Coatopathies: Genetic Disorders of Protein Coats. Annu. Rev. Cell Dev. Biol. 2019;35:131–168. doi: 10.1146/annurev-cellbio-100818-125234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venditti R., Wilson C., De Matteis M.A. Exiting the ER: what we know and what we don’t. Trends Cell Biol. 2014;24:9–18. doi: 10.1016/j.tcb.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Passemard S., Perez F., Colin-Lemesre E., Rasika S., Gressens P., El Ghouzzi V. Golgi trafficking defects in postnatal microcephaly: The evidence for “Golgipathies”. Prog. Neurobiol. 2017;153:46–63. doi: 10.1016/j.pneurobio.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Rasika S., Passemard S., Verloes A., Gressens P., El Ghouzzi V. Golgipathies in Neurodevelopment: A New View of Old Defects. Dev. Neurosci. 2018;40:396–416. doi: 10.1159/000497035. [DOI] [PubMed] [Google Scholar]
- 6.Garnero P. The Role of Collagen Organization on the Properties of Bone. Calcif. Tissue Int. 2015;97:229–240. doi: 10.1007/s00223-015-9996-2. [DOI] [PubMed] [Google Scholar]
- 7.Gelse K., Pöschl E., Aigner T. Collagens--structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003;55:1531–1546. doi: 10.1016/j.addr.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Malhotra V., Erlmann P. The pathway of collagen secretion. Annu. Rev. Cell Dev. Biol. 2015;31:109–124. doi: 10.1146/annurev-cellbio-100913-013002. [DOI] [PubMed] [Google Scholar]
- 9.Raote I., Ortega-Bellido M., Santos A.J., Foresti O., Zhang C., Garcia-Parajo M.F., Campelo F., Malhotra V. TANGO1 builds a machine for collagen export by recruiting and spatially organizing COPII, tethers and membranes. eLife. 2018;7:e32723. doi: 10.7554/eLife.32723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCaughey J., Stephens D.J. COPII-dependent ER export in animal cells: adaptation and control for diverse cargo. Histochem. Cell Biol. 2018;150:119–131. doi: 10.1007/s00418-018-1689-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garbes L., Kim K., Rieß A., Hoyer-Kuhn H., Beleggia F., Bevot A., Kim M.J., Huh Y.H., Kweon H.S., Savarirayan R. Mutations in SEC24D, encoding a component of the COPII machinery, cause a syndromic form of osteogenesis imperfecta. Am. J. Hum. Genet. 2015;96:432–439. doi: 10.1016/j.ajhg.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venditti R., Scanu T., Santoro M., Di Tullio G., Spaar A., Gaibisso R., Beznoussenko G.V., Mironov A.A., Mironov A., Jr., Zelante L. Sedlin controls the ER export of procollagen by regulating the Sar1 cycle. Science. 2012;337:1668–1672. doi: 10.1126/science.1224947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyadjiev S.A., Fromme J.C., Ben J., Chong S.S., Nauta C., Hur D.J., Zhang G., Hamamoto S., Schekman R., Ravazzola M. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat. Genet. 2006;38:1192–1197. doi: 10.1038/ng1876. [DOI] [PubMed] [Google Scholar]
- 14.Lekszas C., Foresti O., Raote I., Liedtke D., König E.M., Nanda I., Vona B., De Coster P., Cauwels R., Malhotra V., Haaf T. Biallelic TANGO1 mutations cause a novel syndromal disease due to hampered cellular collagen secretion. eLife. 2020;9:e51319. doi: 10.7554/eLife.51319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Unlu G., Qi X., Gamazon E.R., Melville D.B., Patel N., Rushing A.R., Hashem M., Al-Faifi A., Chen R., Li B. Phenome-based approach identifies RIC1-linked Mendelian syndrome through zebrafish models, biobank associations and clinical studies. Nat. Med. 2020;26:98–109. doi: 10.1038/s41591-019-0705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephens D.J., Pepperkok R. Imaging of procollagen transport reveals COPI-dependent cargo sorting during ER-to-Golgi transport in mammalian cells. J. Cell Sci. 2002;115:1149–1160. doi: 10.1242/jcs.115.6.1149. [DOI] [PubMed] [Google Scholar]
- 17.Szul T., Sztul E. COPII and COPI traffic at the ER-Golgi interface. Physiology (Bethesda) 2011;26:348–364. doi: 10.1152/physiol.00017.2011. [DOI] [PubMed] [Google Scholar]
- 18.Arakel E.C., Schwappach B. Formation of COPI-coated vesicles at a glance. J. Cell Sci. 2018;131:jcs209890. doi: 10.1242/jcs.209890. [DOI] [PubMed] [Google Scholar]
- 19.Coutinho P., Parsons M.J., Thomas K.A., Hirst E.M., Saúde L., Campos I., Williams P.H., Stemple D.L. Differential requirements for COPI transport during vertebrate early development. Dev. Cell. 2004;7:547–558. doi: 10.1016/j.devcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 20.Witkos T.M., Chan W.L., Joensuu M., Rhiel M., Pallister E., Thomas-Oates J., Mould A.P., Mironov A.A., Biot C., Guerardel Y. GORAB scaffolds COPI at the trans-Golgi for efficient enzyme recycling and correct protein glycosylation. Nat. Commun. 2019;10:127. doi: 10.1038/s41467-018-08044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izumi K., Brett M., Nishi E., Drunat S., Tan E.S., Fujiki K., Lebon S., Cham B., Masuda K., Arakawa M. ARCN1 Mutations Cause a Recognizable Craniofacial Syndrome Due to COPI-Mediated Transport Defects. Am. J. Hum. Genet. 2016;99:451–459. doi: 10.1016/j.ajhg.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DiStasio A., Driver A., Sund K., Donlin M., Muraleedharan R.M., Pooya S., Kline-Fath B., Kaufman K.M., Prows C.A., Schorry E. Copb2 is essential for embryogenesis and hypomorphic mutations cause human microcephaly. Hum. Mol. Genet. 2017;26:4836–4848. doi: 10.1093/hmg/ddx362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bainbridge M.N., Wang M., Wu Y., Newsham I., Muzny D.M., Jefferies J.L., Albert T.J., Burgess D.L., Gibbs R.A. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 2011;12:R68. doi: 10.1186/gb-2011-12-7-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Challis D., Yu J., Evani U.S., Jackson A.R., Paithankar S., Coarfa C., Milosavljevic A., Gibbs R.A., Yu F. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid J.G., Carroll A., Veeraraghavan N., Dahdouli M., Sundquist A., English A., Bainbridge M., White S., Salerno W., Buhay C. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson P.N., Köhler S., Oellrich A., Wang K., Mungall C.J., Lewis S.E., Washington N., Bauer S., Seelow D., Krawitz P. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res. 2014;24:340–348. doi: 10.1101/gr.160325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tong J., Kishi H., Matsuda T., Muraguchi A. A bone marrow-derived stroma cell line, ST2, can support the differentiation of fetal thymocytes from the CD4+ CD8+ double negative to the CD4+ CD8+ double positive differentiation stage in vitro. Immunology. 1999;97:672–678. doi: 10.1046/j.1365-2567.1999.00822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer D.J., Helms J.B., Beckers C.J., Orci L., Rothman J.E. Binding of coatomer to Golgi membranes requires ADP-ribosylation factor. J. Biol. Chem. 1993;268:12083–12089. [PubMed] [Google Scholar]
- 34.Boncompain G., Divoux S., Gareil N., de Forges H., Lescure A., Latreche L., Mercanti V., Jollivet F., Raposo G., Perez F. Synchronization of secretory protein traffic in populations of cells. Nat. Methods. 2012;9:493–498. doi: 10.1038/nmeth.1928. [DOI] [PubMed] [Google Scholar]
- 35.Westerfield M. University of Oregon Press; Eugene: 2007. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- 36.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 37.Blanco-Sánchez B., Clément A., Fierro J., Jr., Washbourne P., Westerfield M. Complexes of Usher proteins preassemble at the endoplasmic reticulum and are required for trafficking and ER homeostasis. Dis. Model. Mech. 2014;7:547–559. doi: 10.1242/dmm.014068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thisse C., Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 39.Bouxsein M.L., Boyd S.K., Christiansen B.A., Guldberg R.E., Jepsen K.J., Müller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res. 2010;25:1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 40.Hanna S.L., Sherman N.E., Kinter M.T., Goldberg J.B. Comparison of proteins expressed by Pseudomonas aeruginosa strains representing initial and chronic isolates from a cystic fibrosis patient: an analysis by 2-D gel electrophoresis and capillary column liquid chromatography-tandem mass spectrometry. Microbiology (Reading) 2000;146:2495–2508. doi: 10.1099/00221287-146-10-2495. [DOI] [PubMed] [Google Scholar]
- 41.Hanson D.A., Eyre D.R. Molecular site specificity of pyridinoline and pyrrole cross-links in type I collagen of human bone. J. Biol. Chem. 1996;271:26508–26516. doi: 10.1074/jbc.271.43.26508. [DOI] [PubMed] [Google Scholar]
- 42.Wu J.J., Woods P.E., Eyre D.R. Identification of cross-linking sites in bovine cartilage type IX collagen reveals an antiparallel type II-type IX molecular relationship and type IX to type IX bonding. J. Biol. Chem. 1992;267:23007–23014. [PubMed] [Google Scholar]
- 43.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stemple D.L., Solnica-Krezel L., Zwartkruis F., Neuhauss S.C., Schier A.F., Malicki J., Stainier D.Y., Abdelilah S., Rangini Z., Mountcastle-Shah E., Driever W. Mutations affecting development of the notochord in zebrafish. Development. 1996;123:117–128. doi: 10.1242/dev.123.1.117. [DOI] [PubMed] [Google Scholar]
- 45.Marom R., Lee Y.C., Grafe I., Lee B. Pharmacological and biological therapeutic strategies for osteogenesis imperfecta. Am. J. Med. Genet. C. Semin. Med. Genet. 2016;172:367–383. doi: 10.1002/ajmg.c.31532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nabavi N., Pustylnik S., Harrison R.E. Rab GTPase mediated procollagen trafficking in ascorbic acid stimulated osteoblasts. PLoS ONE. 2012;7:e46265. doi: 10.1371/journal.pone.0046265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pozzer D., Invernizzi R.W., Blaauw B., Cantoni O., Zito E. Ascorbic Acid Route to the Endoplasmic Reticulum: Function and Role in Disease. Antioxid. Redox Signal. 2021;34:845–855. doi: 10.1089/ars.2019.7912. [DOI] [PubMed] [Google Scholar]
- 48.Aumailley L., Dubois M.J., Brennan T.A., Garand C., Paquet E.R., Pignolo R.J., Marette A., Lebel M. Serum vitamin C levels modulate the lifespan and endoplasmic reticulum stress response pathways in mice synthesizing a nonfunctional mutant WRN protein. FASEB J. 2018;32:3623–3640. doi: 10.1096/fj.201701176R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bober P., Tomková Z., Alexovič M., Ropovik I., Sabo J. The unfolded protein response controls endoplasmic reticulum stress-induced apoptosis of MCF-7 cells via a high dose of vitamin C treatment. Mol. Biol. Rep. 2019;46:1275–1284. doi: 10.1007/s11033-019-04598-w. [DOI] [PubMed] [Google Scholar]
- 50.Liu L., Doray B., Kornfeld S. Recycling of Golgi glycosyltransferases requires direct binding to coatomer. Proc. Natl. Acad. Sci. USA. 2018;115:8984–8989. doi: 10.1073/pnas.1810291115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brunet S., Sacher M. In sickness and in health: the role of TRAPP and associated proteins in disease. Traffic. 2014;15:803–818. doi: 10.1111/tra.12183. [DOI] [PubMed] [Google Scholar]
- 52.Ferreira C.R., Zein W.M., Huryn L.A., Merker A., Berger S.I., Wilson W.G., Tiller G.E., Wolfe L.A., Merideth M., Carvalho D.R. Defining the clinical phenotype of Saul-Wilson syndrome. Genet. Med. 2020;22:857–866. doi: 10.1038/s41436-019-0737-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Dijk F.S., Semler O., Etich J., Köhler A., Jimenez-Estrada J.A., Bravenboer N., Claeys L., Riesebos E., Gegic S., Piersma S.R. Interaction between KDELR2 and HSP47 as a Key Determinant in Osteogenesis Imperfecta Caused by Bi-allelic Variants in KDELR2. Am. J. Hum. Genet. 2020;107:989–999. doi: 10.1016/j.ajhg.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Omari S., Makareeva E., Gorrell L., Jarnik M., Lippincott-Schwartz J., Leikin S. Mechanisms of procollagen and HSP47 sorting during ER-to-Golgi trafficking. Matrix Biol. 2020;93:79–94. doi: 10.1016/j.matbio.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santos A.J., Raote I., Scarpa M., Brouwers N., Malhotra V. TANGO1 recruits ERGIC membranes to the endoplasmic reticulum for procollagen export. eLife. 2015;4:e10982. doi: 10.7554/eLife.10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shomron O., Nevo-Yassaf I., Aviad T., Yaffe Y., Zahavi E.E., Dukhovny A., Perlson E., Brodsky I., Yeheskel A., Pasmanik-Chor M. COPII collar defines the boundary between ER and ER exit site and does not coat cargo containers. J. Cell Biol. 2021;220:e201907224. doi: 10.1083/jcb.201907224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weigel A.V., Chang C.L., Shtengel G., Xu C.S., Hoffman D.P., Freeman M., Iyer N., Aaron J., Khuon S., Bogovic J. ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell. 2021;184:2412–2429.e16. doi: 10.1016/j.cell.2021.03.035. [DOI] [PubMed] [Google Scholar]
- 58.DePhillipo N.N., Aman Z.S., Kennedy M.I., Begley J.P., Moatshe G., LaPrade R.F. Efficacy of Vitamin C Supplementation on Collagen Synthesis and Oxidative Stress After Musculoskeletal Injuries: A Systematic Review. Orthop. J. Sports Med. 2018;6 doi: 10.1177/2325967118804544. 2325967118804544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malmir H., Shab-Bidar S., Djafarian K. Vitamin C intake in relation to bone mineral density and risk of hip fracture and osteoporosis: a systematic review and meta-analysis of observational studies. Br. J. Nutr. 2018;119:847–858. doi: 10.1017/S0007114518000430. [DOI] [PubMed] [Google Scholar]
- 60.Sun Y., Liu C., Bo Y., You J., Zhu Y., Duan D., Cui H., Lu Q. Dietary vitamin C intake and the risk of hip fracture: a dose-response meta-analysis. Osteoporos. Int. 2018;29:79–87. doi: 10.1007/s00198-017-4284-9. [DOI] [PubMed] [Google Scholar]
- 61.Mohan S., Kapoor A., Singgih A., Zhang Z., Taylor T., Yu H., Chadwick R.B., Chung Y.S., Donahue L.R., Rosen C. Spontaneous fractures in the mouse mutant sfx are caused by deletion of the gulonolactone oxidase gene, causing vitamin C deficiency. J. Bone Miner. Res. 2005;20:1597–1610. doi: 10.1359/JBMR.050406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.