Summary
An important goal of clinical genomics is to be able to estimate the risk of adverse disease outcomes. Between 5% and 10% of individuals with ulcerative colitis (UC) require colectomy within 5 years of diagnosis, but polygenic risk scores (PRSs) utilizing findings from genome-wide association studies (GWASs) are unable to provide meaningful prediction of this adverse status. By contrast, in Crohn disease, gene expression profiling of GWAS-significant genes does provide some stratification of risk of progression to complicated disease in the form of a transcriptional risk score (TRS). Here, we demonstrate that a measured TRS based on bulk rectal gene expression in the PROTECT inception cohort study has a positive predictive value approaching 50% for colectomy. Single-cell profiling demonstrates that the genes are active in multiple diverse cell types from both the epithelial and immune compartments. Expression quantitative trait locus (QTL) analysis identifies genes with differential effects at baseline and week 52 follow-up, but for the most part, differential expression associated with colectomy risk is independent of local genetic regulation. Nevertheless, a predicted polygenic transcriptional risk score (PPTRS) derived by summation of transcriptome-wide association study (TWAS) effects identifies UC-affected individuals at 5-fold elevated risk of colectomy with data from the UK Biobank population cohort studies, independently replicated in an NIDDK-IBDGC dataset. Prediction of gene expression from relatively small transcriptome datasets can thus be used in conjunction with TWASs for stratification of risk of disease complications.
Keywords: prediction of disease progression, eQTLs, transcriptional risk scores, predicted polygenic transcriptional risk scores, transcriptome-wide association studies, cell-type-specific gene expression
Introduction
Genetic risk assessment in humans has to date focused mainly on prediction of disease onset,1 whereas arguably the greater clinical need is for prediction of disease progression.2,3 Polygenic risk scores (PRSs) may sometimes meet both needs, such as the ability of a PRS for coronary artery disease to stratify people with respect to the likely effectiveness of statins or PCSK9 inhibitors.4, 5, 6 This is not generally expected to be the case, however, and in the context of inflammatory bowel disease (IBD), there appears to be little influence of the heritability for disease on progression to complicated disease.7 Because genome-wide association studies (GWASs) sufficiently powered to develop accurate PRSs for progression or therapeutic response are not yet available, there is a need for alternative genomic strategies.
A promising approach is gene expression profiling, which very often discriminates disease-affected and unaffected groups. For both Crohn disease (MIM: 266600) and ulcerative colitis (UC [MIM: 619398]), RNA sequencing (RNA-seq) of ileal and rectal biopsies, respectively, generates discriminators of disease severity and progression to complications or remission that are at least as good as clinical indices.8, 9, 10 For example, the first principal component (PC1) of the rectal transcriptome in the PROTECT study embodies differential expression of genes related to mitochondrial function, mucosal healing, and antibacterial defense, and PC1 also stratifies affected individuals with respect to remission.11 Relatedly, calprotectin (S100A8/9 [MIM: 123885 and 123886]) expression is one of a handful of transcriptional markers of failure to respond to anti-tumor necrosis factor (anti-TNF) therapy.12 Because avoidance of colectomy is a compelling therapeutic objective in IBD, we set out to ask, again using the PROTECT study data, whether a gene expression signature predictive of progression to this adverse outcome could be developed.
Secondary to this clinical objective is the research question of to what extent genetic variation contributes to the differential expression and whether it alone might be used for development of a predictor that rivals, or even improves upon, standard PRSs. Considering that the vast majority of signals from GWASs localize to regulatory regions, it is generally accepted that polymorphisms often mediate disease risk through their impact on gene expression. Such variants, known as expression quantitative trait loci (eQTLs), are ubiquitous, yet in IBD there is surprisingly small correspondence between the fine-mapped identities of the variants that associate with disease risk and transcript abundance.13,14 Conversely, the majority of genes that contribute to signatures of disease do not appear to have GWAS signals and hence may not be causal in pathology. We have therefore argued that genes harboring joint eQTL and GWAS associations might be used for development of transcriptional risk scores (TRSs), namely weighted sums of polarized Z scores of transcript abundance, that link expression to pathology.15 Indeed, in the RISK pediatric Crohn disease study, TRSs derived from 26 genes predict stricturing or penetrating outcomes whereas PRSs do not.16 Complicating the picture, further analysis argued that incoherent associations where the sign of the eQTL effect is opposite that of the expected effect on case gene expression imply that some differential expression represents a protective response whose magnitude is modulated by eQTL effects. As profiling moves to the single-cell level, it is clear that gene expression will also define the identities of critical cell types in which pathogenic alleles act17, 18, 19 and most likely refine transcript-based risk assessment. The main limitation of this approach is the ability to obtain appropriate tissue biopsies.
In parallel, transcriptome-wide association studies (TWASs) have emerged as an alternative approach linking genetic variation to disease-related gene expression.20,21 These are analyses that essentially sum the cis-eQTL effects at a locus in a discovery cohort in order to predict gene expression in a case-control cohort where only genotypes are available. Rather than relying on marginal univariate effects, a variety of Bayesian methods have been developed that assign weights to all polymorphisms in an interval and explain more of the variance at each locus. Differential expression predictions have been shown to highlight candidate genes for a range of disease.22 Here, we demonstrate the further utility of TWASs to generate a predicted polygenic transcriptional risk score (PPTRS) for UC, which not only discriminates the UC from the non-IBD group but also progression to major disease complication requiring colectomy for up to 10% of affected individuals.23, 24, 25 Genomic analysis of just hundreds of individuals, projected onto the UK Biobank,26 supports polygenic risk assessment that performs at least as well as the current PRS for UC. Our analyses also provide insight into the cell-type specificity in both epithelial and immune compartments for IBD-GWAS loci.
Subjects and methods
The PROTECT cohort
428 participants aged 4 to 17 years were enrolled from 29 centers across North America into the PROTECT study27 upon clinical, histological, and endoscopic diagnosis of UC. Affected individuals with disease extent beyond the rectum, a pediatric ulcerative colitis activity index (PUCAI) score of ≥10,28 no prior therapy for colitis, and negative enteric bacterial stool culture were eligible to participate. All baseline assessments and sample collections were performed prior to the initiation of therapy. Initial treatment with mesalamine, oral corticosteroids, or intravenous corticosteroids was decided on the basis of mild, moderate, or severe PUCAI. Following the baseline assessment, follow-up assessments were performed at 4, 12, and 52 weeks and other therapeutic interventions were administered on the basis of guidelines for need for additional medical therapy. The study parameters are described in further detail in Hyams et al. (2017).27 Each site’s institutional review board approved the protocol and safety monitoring plan. Informed consent or assent was obtained for each participant.
RNA-seq data processing and differential expression analyses
RNA was isolated from 340 rectal biopsies taken at baseline and 92 rectal biopsies taken at week 52 follow-up. RNA-seq was performed with the Lexogen QuantSeq 3′ platform.29 Single-end 150 bp reads were trimmed, and adapters were removed with FastQC.30 Reads were mapped to human genome hg19 via hisat2,31 and the aligned reads were converted into read counts per gene with SAMtools and HTSeq in the default union mode.32,33 The raw read counts were normalized via trimmed mean of M-values normalization with the edgeR R package.34
We used expression of the sex-specific genes RPS4Y1, DDX3Y, EIF1AY, KDM5D, and XIST (MIM: 470000, 400010, 400014, 426000, and 314670, respectively) to validate the gender of each individual, resulting in the removal of two mismatches. Further adjustment and removal of batch effects was performed with surrogate variable analysis (SVA)35 combined with supervised normalization (SNM).36 Ancestry, gender, initial treatment group, time of sampling, and week 52 colectomy status were modeled with the SVA R package where initial treatment group, time of sampling, and week 52 colectomy status were protected variables, which resulted in the identification of 28 confounding factors. Of these, five variables significantly correlated with protected variables were preserved, while the remaining 23 were statistically removed with SNM. Because PROTECT was predominantly European (see genotypic PCA in Figure S1), ancestry did not have a significant impact on the eQTL analysis and very few gene models were affected by exclusion from the SNM. Two individuals that were outliers in a principal-component analysis of total gene expression were removed.
Differential gene expression testing was performed on the basis of colectomy status with the voom R package.37 Log fold change and Benjamini-Hochberg adjusted p values were obtained for all genes, and these are listed in Table S1. Because of high significance levels despite p value adjustment, significant genes were pared down to 150 following testing for robustness at multiple thresholds. Although 2,188 transcripts significantly differentiate colectomy status after Bonferroni adjustment, PC1 explains less of the variance of these genes than when we contrasted PC1 scores with 10, 100, 150, or 500 genes, and although these were all highly correlated (r > 0.9), we found that the top 150 robustly maximized discrimination of colectomy cases while including diverse genes and hence not being susceptible to a few outliers. The first PC of these top 150 genes differentially expressed at baseline between affected individuals who required colectomy by week 52 follow-up (n = 21) and affected individuals who did not (n = 310) formed the gene-expression-based risk score for colectomy (PC1col). This score is moderately correlated (r = 0.46) with PC1 of overall expression of genes differentiating UC-affected individuals and control individuals, reported by Haberman et al. (2019).11
We performed cross validation for PC1col by randomizing colectomy status among individuals prior to differential gene expression testing and calculation of PC1colRand, as in the calculation for PC1col. Analysis of variance (ANOVA) tests were performed between randomized colectomy and non-colectomy individuals, and results from 1,000 such tests are reported in Figure S2.
We compared expression of the genes comprising PC1col at baseline and week 52 with Mayo score as a marker for mucosal healing. PC1col was calculated as previously described in the subset of individuals with baseline gene expression. Additionally, we calculated a restricted PC1col-week52 calculated by finding PC1 of the 150 genes used in the calculation of PC1col within the subset of individuals with week 52 gene expression. Change in PC1 score was simply calculated as the difference between PC1col and PC1col-week52. All p values were generated with ANOVA tests.
TRSs, introduced by Marigorta et al.16 for discriminating IBD-affected individuals versus control individuals, capture the summation of polarized expression of genes incorporated on the basis of both proximity to IBD-GWAS hits and presence of eQTL in peripheral blood. We generated the TRSs with four different strategies, all of which gave similar highly significant differentiation between colectomy and no colectomy samples. Model 1 was a generalized linear model (GLM) with nine genes, RGS14, APEH, MRPL20, POP7, RORC, EDN3, PTK2B, STAT3, and CDC42SE2 (MIM: 602513, 102645, 611833, 606113, 602943, 131242, 601212, and 102582, respectively, CDC42SE2 does not have a MIM entry), that in forward stepwise regression most strongly differentiate affected individuals by colectomy status, essentially the sum of the Z scores weighted by their magnitude of differential expression. Model 2 was a GLM with the ten genes discussed in the text because of strong co-regulation and association with colectomy. Models 3 and 4 were based on all 26 genes, generated with a weighted GLM or simple PC1 score, respectively. All four scores are highly correlated, r > 0.8, indicating that they are capturing similar aspects of differential expression (Figure S3). We report model 4 in the text. This TRS is highly correlated with PC1col (r = 0.64).
Relative proportions of epithelial and immune contributions to total rectal gene expression reported in Figure S4 were evaluated by contrasting PC1 of epithelial- and immune-enriched transcripts. We first identified 200 genes upregulated (fold difference in expression, regardless of significance) specifically in either the total epithelial or immune components of the single-cell gene expression dataset reported by Smillie et al. (2019).19 Then we computed PC1 for these two gene sets in the bulk RNA-seq data and contrasted the mean scores at baseline and week 52. We checked each PC to ensure that positive values associate with elevated expression of the respective genes.
Replication of colectomy risk score and cell-type enrichment
Surgical specimens from 210 UC-affected individuals undergoing bowel resection for IBD at Mount Sinai Health System and affiliated clinicians were recruited to be part of the Mount Sinai Crohn’s and Colitis Registry (MSCCR) between December 2013 and September 2016 as described.38,39 The protocol required written informed consent that was approved by the Icahn School of Medicine at Mount Sinai Institutional Review Board (HSM#14-00210). Affected individuals who were enrolled in the study were asked to provide blood and/or biopsies, which were collected during a colonoscopy planned for regular care. Clinical and demographic information was obtained through a questionnaire. Affected individuals were treated with a range of medications, including corticosteroids, infliximab, azathioprine, and mesalamine. All macroscopically moderate-to-severely inflamed tissues were confirmed as active colitis by pathology examination provided by the Mount Sinai Hospital (MSH) Pathology Department. Freshly collected representative 0.5-cm-wide tissue fragments were isolated from surgical specimen samples, flash frozen, and stored at −80°C.
RNA was isolated from frozen tissue with QIAGEN QIAsymphony RNA Kit (cat. # 931636), and samples with RIN scores > 7 were retained. One microgram of total RNA depleted of ribosomal RNA via the Ribo-Zero Kit (Illumina cat. # MRZG12324) was used for the preparation of sequencing libraries with RNA TruSeq Kits (Illumina [cat. # RS-122-2001-48]). These were sequenced on the Illumina HiSeq 2500 platform with 100 bp paired-end protocol. Base calling from images and fluorescence intensities of the reads was done in situ on the HiSeq 2500 computer with Illumina software, aiming for 70,000 paired-end reads per sample. Short reads were mapped to the GRCh37/hg19 assembly (UCSC Genome Browser) with two-pass spliced transcripts alignment to a reference (STAR) and processed with RAPiD, which is an RNA-seq analysis framework developed and maintained by the technology development group at the Icahn Institute for Genomics and Multiscale Biology. Detailed quality control (QC) metrics were generated with the RNASeQC package. We pre-filtered raw count data to keep genes with CPM > 0.5 for at least 3% of the samples. After filtering, count data were normalized via the weighted trimmed mean of M-values and further variance stabilized with a logarithmic transformation. Normalized counts were further transformed into normally distributed expression values via the voom-transformation37 with a model that included technical covariates (processing batch, RIN, exonic rate, and ribosomal RNA rate) while accounting for the intra-individual correlation across regions.
We repeated the transcriptional risk assessment analysis in this external dataset after normalization for gender, age, exonic RNA ratio, and rRNA expression levels by using the prcomp function in R with the 150 genes from the PROTECT PC1col or the 26 gene TRS. We then used the R package ggplot240 to plot the distribution of PC1 for affected individuals who did (ten individuals) or did not (201 individuals) have follow-up colectomies (Figure S5). Additionally, we performed hierarchical clustering of single-cell gene expression data to identify cell types implicated by both the PC1 and TRS gene sets. Cell types enriched for PC1 genes included plasmacytoid dendritic cells, endothelial cells, group I innate lymphoid cells, fibroblasts, and macrophages.
SNP data processing and eQTL studies
We used the Affymetrix UK Biobank Axiom Array to perform genotyping of 424 individuals across 800,000 SNPs. We performed imputation by using IMPUTE2 software,41 after which we used quality control performed with PLINK42 to remove SNPs not in Hardy-Weinberg equilibrium at p < 10−3, SNPs with a minor allele frequency < 1%, or a rate of missing data across individuals > 5%. Approximately 7 million imputed SNPs passed these thresholds and were tested in the eQTL analysis. SNPs within 250 kb of the start and stop sites of a gene were considered to be cis to the gene and tested for a potential eQTL association. Mapping was performed with the mixed linear modeling method in GEMMA,43 which tested a set of approximately 12 million SNP-gene pairs for associations at a common p value threshold of 1 × 10−5. Two separate comparative analyses were performed: the initial set of eQTL mapping was performed on all 330 baseline samples and all 87 follow-up samples acquired at week 52, and the secondary analysis was performed on 78 samples matched between the two time points. The initial full analysis yielded 98,491 significant SNP-gene associations at baseline and 20,509 associations at week 52 follow-up, and the secondary matched analysis yielded 15,078 significant unique SNP-gene associations at baseline and 13,236 significant associations at week 52 follow-up. These were further refined to 1,432, 432, 402, and 322 peak SNP to unique gene associations, respectively, at FDR 5%.
Single-cell sequence analysis of the lamina propria
We analyzed a total of 34,157 cells from paired inflamed rectum (n = 4) and uninflamed sigmoid colon (n = 5) from four UC-affected individuals undergoing treatment at MSH. Resected tissue biopsies were collected in ice-cold RPMI 1640 (Corning.) and processed within 1 h after termination of the surgery. To limit biased enrichment of specific cell populations related to local variations in the intestinal micro-organization, we pooled twenty mucosal biopsies sampled all along the resected specimens by using a biopsy forceps (EndoChoice). Epithelial cells were dissociated by incubation of the biopsies in a dissociation medium (HBSS without Ca2+ or Mg2+ with HEPES 10 mM [Life Technologies]) and enriched with 5 mM EDTA at 37°C with 100 rpm agitation for two cycles of 15 min. After each cycle, the biopsies were vortexed vigorously for 30 s and washed in complete RPMI media equilibrated at room temperature (RT). They were transferred to digestion medium (HBSS with Ca2+ Mg2+, FCS 2%, DNase I 0.5 mg/mL [Sigma-Aldrich] and collagenase IV 0.5 mg/mL [Sigma-Aldrich]) for 40 min at 37°C with 100 rpm agitation. After digestion, the cell suspension was filtered through a 70 mm cell strainer, washed in DBPS/2% FCS/1 mM EDTA and spun down at 400 g for 10 min. After red blood cell lysis (BioLegend), dead cells were depleted with the dead cell depletion kit (Miltenyi Biotec, Germany) following manufacturer’s recommendations. Viability of the final cell suspension was calculated with a Cellometer Auto 2000 (Nexcelom Biosciences) with AO/PI dye. The exclusion was routinely 70% or higher live cell rate.
Single cells were processed through the 10× Chromium platform via the Chromium Single Cell 3′ Library and Gel Bead Kit v.2 (10× Genomics, PN-120237) and the Chromium Single Cell A Chip Kit (10× Genomics, PN-120236) as per the manufacturer’s protocol. In brief, 10,000 cells from single-cell suspension were added to each lane of the 10× chip. The cells were partitioned into gel beads in emulsion in the Chromium instrument in which cell lysis and bar-coded reverse transcription of RNA occurred, followed by amplification, fragmentation, and 5′ adaptor and sample index attachment. Libraries were sequenced on an Illumina NextSeq 500.
We aligned reads to the GRCh38 reference with the Cell Ranger v.2.1.0 Single-Cell Software Suite from 10× Genomics. The unfiltered raw matrices were imported into R Studio as a Seurat object (Seurat v.3.0.1).44 Genes expressed in fewer than three cells in a sample were excluded, as were cells that expressed fewer than 500 genes and with unique molecular identifier (UMI) count less than 500 or greater than 60,000. We normalized by dividing the UMI count per gene by the total UMI count in the corresponding cell and log transforming. We used the Seurat integrated model44 to generate a combined UC model with cells from both inflamed and uninflamed samples retaining their group identity. We performed unsupervised clustering with shared nearest-neighbor graph-based clustering by using from 1 to 15 PCs of the highly variable genes; we also accordingly tuned the resolution parameter to determine the resulting number of clusters. Cell types were assigned with known markers previously described for the gut.17, 18, 19,45 Visualization of relative abundance of specific genes in each cell type was performed with Seurat functions in conjunction with the ggplot2.40
Gene expression imputation and prediction models
We performed a TWAS for association between the imputed cis-genetic component of gene expression with UC status. We used PROTECT as the prediction study with both genetic and transcriptomic data from which we estimated cis-eQTL effects,10 which we then used to impute gene expression in the UK Biobank validation dataset.26 Subsequently, these predicted gene expression models were associated with UC status in the UK Biobank, and the significant ones were combined into a weighted predicted polygenic transcriptional risk score (PPTRS), which was itself evaluated for association with UC, and secondarily with colectomy status, in PROTECT.27
Before building the gene expression imputation models, we ensured that the prediction and validation studies were harmonized such that the allele frequencies are correlated by requiring that the genotype matrix accounts correspond to the same allele in both datasets. Gene expression imputation models were built with a non-parametric Bayesian Dirichlet process regression (DPR) method46 in TIGAR,47 which assumes a Dirichlet process prior on the effect size variance to estimate cis-eQTL effect sizes. A linear regression model was assumed for estimating cis-eQTL effect sizes:
where Eg is the gene expression for a gene g, X is the genotype matrix for all cis-genotypes (SNPs within 1 Mb of the flanking 5′ and 3′ ends), w is the vector of cis-eQTL effect sizes, and ϵ is the error term assumed to be normally distributed with a mean of zero. The predicted (imputed) gene expression for gene g is computed as follows:
where Xnew is the cis-genotype matrix of the new genotype data or GWAS samples and Eg-pred is the predicted gene expression of the new data. The imputed gene expression is the cis-genetic component of the total gene expression derived from common cis-eQTLs and does not include the trans-component or environmental effects. TIGAR47 has been shown to generate a 2-fold improvement in variance explained by multi-SNP models relative to just capturing the top cis-eQTLs, more than with similar imputation methods such as Predixcan and FUSION.48, 49, 50As prediction datasets, we initially utilized the PROTECT cohort (rectal gene expression, n = 331),27 confirmed with GTEx transverse colon gene expression (n = 368),51 and contrasted with GTEx muscle gene expression (n = 706) and cortex gene expression (n = 205) negative controls. Sigmoid colon has fewer samples, so it was underpowered for these analyses despite its being closer to the rectum than transverse colon. We used threshold of 5% imputation R2 to select genes with valid imputation models that were taken forward for testing in the UK Biobank and PROTECT (Figure S6 shows boxplots of imputation R2 for all tissues and Table S2 shows the number and identity of genes in each tissue with imputation R2 > 5%). Note that colectomy status was not used in the modeling of either the cis gene expression or generation of the PPTRS, so prediction of colectomy in PROTECT from the UK Biobank score should not be circular. However, use of the GTEx colon expression to generate the imputation models ensures that prediction, validation, and testing are performed with three independent datasets (GTEx, UK Biobank, and PROTECT). Further, we also replicated these results on a larger and completely independent European subset of NIDDK IBD Genetics Consortium colectomy cohort,52 wherein the rectum- and colon-based PPTRS discriminated UC from colectomy, while the muscle- and cortex-based PPTRS were negative controls. Finally, we also generated the PPTRS on a subset of the UK Biobank, testing it on a held-out sample with similar results.
TWAS and PPTRS
For the validation dataset, the genotype data of UK Biobank was used, including 4,112 UC-affected individuals and 402,994 non-IBD control individuals. The gene expression of 407,106 white British individuals was predicted via gene expression imputation models for genes with imputation R2 > 5%. Subsequently, we performed a gene-based association test by fitting a logistic regression model of the predicted gene expression against UC versus non-IBD status to determine the weight (log odds ratio) and p value for each gene.
We then built a TWAS-based polygenic risk score, which we call a predicted polygenic transcriptional risk score (PPTRS). To assess the full polygenic architecture of the predicted cis-component of gene expression, we adopted an inclusive TWAS threshold for differentially expressed genes with TWAS p value < 0.05. Similar to PRS computation, inclusion of predictors that are not individually significant can nevertheless improve the score because false negatives contribute signal. We constructed the PPTRS by computing the weighted sum of the predicted gene expression, where the weights are the log of odds ratio from TWAS of UC in UK Biobank.26 This score, as expected, highly significantly differentiates affected individuals and control individuals in the UK Biobank and surprisingly also colectomy status. We then used the same weights to generate the PPTRS in PROTECT and NIDDK cohorts and to evaluate association with colectomy status. This procedure was repeated with the GTEx eQTL models.51 The contrasting PRS derived from GWAS weights, PRSUC, was constructed via 6,396 UC SNPs from summary statistics of the European UC GWAS meta-analysis53 (pruned with PLINK at p value < 0.001, LD r2 > 0.5 in 10 kb windows with a five-SNP sliding step).
NIDDK-IBDGC colectomy cohort
Samples were genotyped on the Illumina Global Screening Array at Feinstein Institute for Medical Research (Manhasset, NY) or at the Broad Institute (Boston, MA) as a part of the National Institute of Diabetes and Digestive and Kidney Diseases Inflammatory Bowel Disease Genetics Consortium (NIDDK-IBDGC). Following stringent pre-imputation QC metrics as previously described,52 genotypes were phased with Eagle254 and imputation was performed with the Michigan Imputation Server and HRC r1.1 reference panel.55,56 Variants with estimated imputation accuracy (R2 < 0.3) and minor allele frequency > 0.1% were excluded after imputation, leaving 21.9 million variants available for analysis. Of the total 16,024 NIDDK-IBDGC samples available after QC, 14,659 were of European ancestry (defined as EUR Admixture proportion ≥ 0.70).57 These included 2,838 non-IBD control individuals, 2,298 UC-diagnosed individuals (1,325 established non-colectomy), and 753 individuals with known colectomy. The predicted polygenic risk score for colectomy was computed on these samples with predicted gene expression from the cis-eQTL weights calculated with DPR on the rectal gene expression from PROTECT or, alternatively, colon, cortex, and muscle gene expression from GTEx. The TWAS weights for inclusion in the PPTRScol from the UK Biobank are reported in Table S2; code was provided by S.N. to T.H.
Results
PROTECT is a multicenter pediatric inception cohort study of response to standardized colitis therapy.27 We have previously shown that a signature of rectal mucosal gene expression at diagnosis, prior to therapeutic intervention, associates with corticosteroid-free remission with mesalamine alone observed in 38% of 400 affected individuals by week 52 of follow-up.10 A signature of rectal mucosal gene expression associated with week 4 corticosteroid response in PROTECT is related to one indicative of response to anti-TNFa and anti-a4b7 integrin therapy in adults,11 and reciprocally, active pediatric UC was associated with suppression of mitochondrial gene expression and increasing disease severity with elevated innate immune function. In order to more explicitly model progression to colectomy observed in 6% (25 of 400) of the UC-affected individuals within 1 year of diagnosis, we performed differential expression analysis between baseline rectal RNA-seq biopsies of 21 affected individuals who progressed to colectomy and 310 who did not. The volcano plot in Figure 1A shows downregulation of 783 transcripts in the individuals who underwent colectomy and upregulation of 1,405 transcripts at the experiment-wide threshold of p < 4 × 10−6.
Figure 1.
Differential expression associated with colectomy in the PROTECT study
(A) Volcano plot of significance (negative log10 of the p value) against difference in expression on log2 scale; genes upregulated in colectomy are in blue.
(B) Six pathways highlighted by gene set enrichment analysis as upregulated in colectomy. Each bar represents a gene in the indicated pathway, and position along the axis is representative of rank order of differential expression. From left to right and top to bottom FDR < 10−4, < 10−4, < 10−4, < 10−4, < 2.4 × 10−4, and < 2.0 × 10−4. A full list of pathways can be found in Table S5.
(C–E) PC1 of the differentially expressed genes as a function of (C) colectomy status at week 52, p = 2 × 10−45; (D) initial treatment, p = 5 × 10−20; and (E) baseline or week 52 follow-up biopsy profile, p = 2 × 10−7. All boxplots indicate 1st and 3rd quartile as box ends, and center median line and whiskers extend to farthest point within 1.5 times the interquartile range.
Gene set enrichment analysis58 summarized in Figure 1B highlights engagement of multiple pathways previously implicated in adverse outcomes in IBD, including TNF and interferon signaling, and various signatures of inflammation and immune response.59,60 We recently showed in an admixed population that African ancestry also upwardly biases gene expression in several of these pathways61 but was not found to be driving the rectal profiles in this largely European ancestry dataset.
The first principal component (PC1col) of the top 150 of these differentially expressed genes has a weak negative correlation with our previously reported signature of remission detected in a subset of 206 affected individuals via a different RNA-seq protocol.11 With very high significance, it distinguishes the affected individuals who progressed to colectomy from non-progressors, as all but one individual have PC1 scores greater than 12, a value exceeded by only 20 of the 317 non-colectomy affected individuals (Figure 1C). This PC1col predictor is orders of magnitude more significant than observed with similar scores derived by 1,000 permutations of the data (Figure S2). All of the high PC1col individuals were placed initially on corticosteroids, the majority intravenously (Figure 1D); the score also correlates with a gradient of disease severity indicated by baseline PUCAI28 and initial treatment. We also obtained rectal biopsy RNA-seq data for 92 affected individuals at week 52 and observed significant depression of the score (Figure 1E), indicative of mucosal healing even in the affected individuals with elevated initial gene activity (none of the follow-up cases were colectomy because the surgical procedure had been performed earlier than week 52). Figure S7 shows that PC1 remains associated with Mayo endoscopic score62 even at week 52 and that the change in PC1 molecular score over time correlates with the degree of mucosal healing.
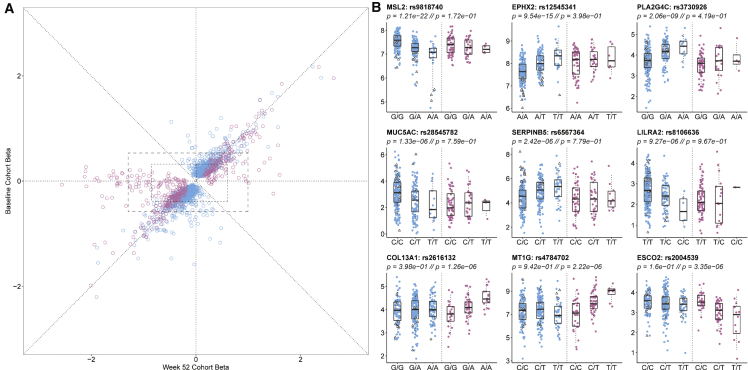
Given the marked shift in gene expression at follow-up, we next asked whether local regulation of the gene expression might contribute by performing comparative eQTL analysis. Figure 2A indicates generally high concordance in the effect sizes (betas) at both time points, with slight inflation of the estimates at baseline (1,416 blue effects) or week 52 (421 magenta effects), most likely due to winner’s curse. There were 72 eSNPs significantly regulating 308 genes at both time points, and the smaller number of eQTLs at week 52 was attributable to the smaller sample size. One quarter of the baseline eQTLs are at least 2-fold greater than at week 52, and one third of the follow-up eQTLs are at least 2-fold greater than at baseline. Examples of baseline- and follow-up-specific eQTLs affecting a variety of gene functions in immunity and epithelial cell biology are shown in Figure 2B. Some of the change in eQTL profiles is most likely attributable to an increase in the proportion of epithelial relative to immune cells at week 52 (Figure S4).
Figure 2.
eQTL contrast between baseline and week 52 follow-up in the PROTECT study
(A) Comparison of effect sizes (betas) for the effect of the minor allele on gene expression. Blue eQTLs were discovered at baseline and magenta only at week 52. All points along the diagonal have baseline point estimates within the 95% confidence interval of the week 52 estimate (~0.2 to 0.4 units). Dashed boxes represent 95% and 80% empirical ranges of effect estimates for the reciprocal time point for non-significant estimates at the other time point.
(B) Examples of nine genes with differential eQTL effects at the two time points showing observed transcript abundance as a function of genotype at baseline or week 52 follow-up. The bottom row are genes with eQTLs only at follow-up. All boxplots indicate 1st and 3rd quartile as box ends, and center median line and whiskers extend to farthest point within 1.5 times the interquartile range. Note that many of the genes with large negative follow-up betas in (A) have relatively small minor allele frequencies and hence insufficient homozygous minor allele genotypes to plot. A full list of peak eQTLs can be found in Table S3.
Clearly visible in Figure 2A is a set of ∼50 apparently week-52-specific effects that have beta estimates less than −0.5 but lie close to the x axis, indicating little or no baseline effect. Given the reduced sample size at week 52, there was little power for estimation of significance of the difference, but in most of these cases, the baseline estimate lies outside 95% confidence limits given the week 52 effect estimate. Because these limits vary by gene, to visualize this approximately, the boxes in Figure 2A show the 90% and 95% empirical ranges of week 52 and baseline estimates for genes that had truly null (p > 0.05) eQTL estimates at the alternate time point as listed in Table S3. Of the 9,799 week 52 eQTL peaks that were non-significant at baseline, 80% had week 52 effect size estimates in the range −0.883 to 0.583, but of the 104 of these peaks that were also highly significant at week 52 (p < 10−5), 52% of the estimates were outside this range, a 2.5× enrichment. Similarly, 27% were outside the 95% range of −1.325 to 0.986, a more than 5× enrichment. The reciprocal analysis did not yield any enrichment (just 110 of 552 baseline-specific peaks are outside the 80% range [−0.412 to 0.321], and 35 [6%] outside the 95% range [−0.622 to 0.540]), implying that most baseline eQTLs have comparable effect estimates at week 52, and failure to replicate is often due to the reduction of power in the smaller sample.
Next, we asked whether the intersection of GWASs and eQTLs could be used for generation of a TRS that associates differential expression with colectomy, analogous to the one we recently developed for prediction of risk of progression to complicated Crohn disease.16 The heatmap in Figure 3A showing the abundance of 26 transcripts included in the TRSIBD derived with coloc63 overlap of IBD-GWAS and peripheral blood eQTL signals indicates striking enrichment for elevated or reduced expression of a dozen transcripts in the baseline rectal biopsies of PROTECT individuals destined for colectomy. The strongest clusters include RGS14, MRPL20, PTK2B, TNFRSF4 (MIM: 600315), TNFRSF18 (MIM: 603905), and CDC42SE2 upregulation and CISD1 (MIM: 611932), EDN3, RORC, and PLA2R1 (MIM: 604939) downregulation. PC1 of the entire set of 26 genes results in a TRSUC that discriminates colectomy from non-progressors at p = 1 × 10−28 (Figure 3B), noting that only four of 26 genes overlap with the 150 used for deriving of PC1col. A score above 3.24 has a sensitivity of 90% and specificity of 95% (Figure 3C), generating a positive predictive value of 55%, which is nine times the prevalence of the rate of progression in the study. Corresponding likelihood ratios for positive and negative prediction are 18 and 10, respectively. TRSUC also performs as well as the composite PC1col.
Figure 3.
Development of a transcriptional risk score for colectomy
(A) Heatmap of baseline rectal expression of 26 genes with evidence that the GWAS peak is the same as a blood eQTL (coloc H4 > 0.8), red representing high expression and blue low. The bar at the top indicates non-colectomy (gray) and colectomy (red) clinical status, highlighting a cluster of affected individuals for whom most of the genes are differentially expressed.
(B) PC1 of the genes generates a TRS that is highly discriminatory between colectomy and non-colectomy at baseline; p = 1 × 10−28. Boxplots indicate 1st and 3rd quartile as box ends, and center median line and whiskers extend to farthest point within 1.5 times the interquartile range.
(C) Receiver operating characteristic curve contrasting sensitivity and specificity for colectomy, showing that both the TRS (green) and PC1 of all differentially expressed genes (red) have high accuracy (area under the curve, AUC > 0.95) compared with PUCAI, a commonly used clinical disease severity index.
We replicated these findings in an independent adult UC cohort from Mount Sinai Medical School in New York.38,39 PC1 of the rectal expression of 146 of the 150 PROTECT PC1col genes detected in the Mount Sinai dataset highly significantly (p = 0.0015) distinguished ten individuals who have had colectomy from the remaining 201 (Figure S5), and the majority of genes were differentially expressed in the same direction. Similarly, a TRS derived from the GWAS-associated 26 transcripts showed a strong trend toward differentiation of colectomy status in the adult cohort, which was also significant (p = 0.010) after removal of two outliers characterized by aberrant expression of CDC42SE2, the only transcript in the list above that disagreed in direction of effect between the two studies.
Examination of the expression of colectomy-associated genes in a single-cell RNA-seq dataset obtained from rectal biopsies provides strong evidence that both epithelial and immune cells contribute to the risk of disease progression (Figure 4). Most of the genes are strongly expressed in just one or two of the 22 identified cell types, seven of which are notable for an excess of colectomy-associated genes: plasmacytoid dendritic cells, immunoregulatory T cells, ILC1/3 innate immune cells, and inflammatory macrophages from the immune compartment and fibroblasts, secretory epithelial cells, and endothelial cells from the gut itself. Because each of these cell types is also represented in the single-cell profiles of the TRS genes, which were selected on the basis of joint eQTL and GWAS associations, it is quite likely that cis-regulatory effects partly explain their relationship to the pathology. In both panels, three-quarters of the indicated genes are among the top FindMarker annotations for the observed cell type, which in almost all cases, implies expression bias in that cell type at p < 10−50 (Table S4). Prospective scRNA-seq studies will most likely reveal more insight into the cellular and genetic basis of the transcriptional risk of adverse disease progression.
Figure 4.
Cell-type-specific expression of colectomy-associated genes
(A) Heatmap showing upregulation (red) of each gene contributing to PC1 in a rectal scRNA-seq dataset. Dozens of genes are enriched in seven cell types.
(B) Similar analysis but for the TRSUC genes. Note the similarity of the cell types showing enrichment and the absence of B cell or plasma cell signals in both. In (A), 99 of the 136 genes are highly significant FindMarker transcripts for the cell type highlighted in the heatmap; exceptions are the fibroblast cluster and some of the endothelial/smooth muscle markers, which mark both subsets. In (B), 20 of 26 genes are similarly peak FindMarkers.
Much of the TRS reflects covariance of the expression of the genes, most likely because of a combination of environmental influences, variation in the proportions of contributing cell types, and trans-acting genetic influences. Cis-acting genetic effects will impact each gene independently yet cumulatively contribute, and the conjunction of GWAS and eQTL signals suggests that it may be possible to also predict disease progression from genotypes alone. To evaluate this, we performed a TWAS20,21 by using DPR implemented in TIGAR47 to capture the effects of all polymorphisms within 1 Mb of each transcript expressed in the PROTECT rectal biopsies and then used the weights to predict gene expression in the white British subset of the UK Biobank.26 We tested for differential predicted gene expression in 70% of the samples and discovered ∼800 genes either up- or downregulated in UC-affected individuals relative to non-IBD control individuals. A predicted polygenic transcriptional risk score (PPTRSUC) was then derived as a weighted sum of the effect sizes of the minor alleles (which polarizes effects of alleles that increase or decrease expression in affected individuals) and applied to the held-out 30% validation sample, as well as to the PROTECT genotypes. Figure 5A shows that the PPTRS efficiently discriminates UC-affected individuals from non-IBD control individuals in UK Biobank (p < 10−219) and remarkably that it also discriminates the indivduals who underwent colectomy in both UK Biobank and PROTECT (p = 0.002 and 0.006, respectively, p values computed with Kruskal-Wallis test in R). That is to say, as with the observed gene expression, individuals who underwent colectomy are distinguished by a trend toward yet more extreme predicted gene expression. The same trend was replicated in a larger and completely independent NIDDK-IBDGC colectomy cohort,52 consisting of 2,838 non-IBD control individuals, 2,298 individuals diagnosed as UC, and 753 individuals with known colectomy. The rectum-based PPTRS in this cohort discriminates UC-affected individuals from non-IBD control individuals (p = 8.5 × 10−7) as well as UC from colectomy (p = 0.0025) (Figure 5A).
Figure 5.
Properties of a predicted polygenic transcriptional risk score (PPTRS)
(A) PPTRS developed from predicted gene expression in PROTECT used for identification of predicted differentially expressed genes in the UK Biobank. The weighted sum of 820 predicted gene expression values clearly separates control individuals from UC-affected individuals in the UK Biobank, PROTECT, and NIDDK studies, while individuals with colectomy have even more highly elevated scores. In standard deviation units, the effect sizes are as follows: UKB controls versus UC, 0.54; UKB UC versus colectomy, 0.16; PROTECT UC versus colectomy, 0.58; NIDDK controls versus UC, 0.14; NIDDK UC versus colectomy, 0.13; p values derived from Kruskal-Wallis tests.
(B) Prevalence versus percentile plots for a PRS based on 6,396 genotypes for UC (red) and the PPTRS (green), showing enhanced prevalence for the upper deciles of the PPTRS. Whiskers show standard error of mean from 5-fold cross-validation.
Furthermore, PPTRSUC provides enhanced discrimination of affected individuals and control individuals in the UK Biobank, as shown in the prevalence versus risk score percentile plots in Figure 5B. Whereas the top percentile has 3-fold higher prevalence than the median via a PRS with 6,396 UC SNPs from summary statistics of the European UC GWAS meta-analysis53 (pruned with PLINK42 at p value < 0.001, LD r2 > 0.5), the top percentile of PPTRSUC is 4-fold higher, and higher prevalence is inferred for the top 20% of the entire cohort. Negative predictive values are similar for both scores. Comparison of the percent variance explained by PPTRSs for disease outcome (evaluated by computing the out of sample Nagelkerke R2 of the logistic regression between UC status and score at increasing inclusion thresholds) was similar to the 0.005 observed for the PRS when only highly significant predicted transcripts were included but jumped to 0.021 with all predicted transcripts at p < 0.05 (Figure S8).
Although colectomy status was not incorporated into either the DPR-based prediction of gene expression or the computation of PPTRSUC, the fact that the prediction and testing datasets are both from PROTECT could confound the interpretation with an element of circularity. We thus used the GTEx study51 transverse colon samples (n = 368) to generate independent prediction models, which we then ran through the same pipeline to generate a confirmatory PPTRSUC. Table 1 shows that this score was almost as good as the PROTECT-derived one in predicting colectomy in the UK Biobank, PROTECT, and NIDDK studies (p = 0.011, p = 0.007, and p = 0.006, respectively). Furthermore, neither cortex- nor muscle-derived PPTRS from GTEx significantly predicts progression to colectomy. Note that discrimination of cases and controls at very high significance levels is expected of any cumulative TWAS score in this setting, regardless of the tissue, because the genes were included precisely because of the differential expression. The observation that only the rectum and colon sample scores (not muscle or cortex) associate with colectomy (which was not a term in the prediction model) confirms that disease-relevant tissue-specific cis-eQTL effects are more informative for studying of disease progression.
Table 1.
Summary of PPTRS results
| Training data for transcriptomic imputation | Reference transcriptome | Number of genes with gene expression imputation R2> 5% | Number of genes with TWAS p < 0.05 in UKBB UC versus non-IBD association and used in PPTRS |
PPTRS p values |
||||
|---|---|---|---|---|---|---|---|---|
| UK Biobank UC versus non-IBD | UK Biobank UC versus colectomy | PROTECT UC versus colectomy | NIDDK-IBDGC UC versus non-IBD | NIDDK-IBDGC UC versus colectomy | ||||
| PROTECT | rectum (n = 331) | 9,392 | 820 | 2.94e−210∗∗ | 0.0023∗ | 0.0062∗ | 8.5e−07∗∗ | 0.0025∗ |
| GTEx | colon-transverse(n = 368) | 13,410 | 1,097 | 4.71e−170∗∗ | 0.011∗ | 0.0073∗ | 7.83e−12∗∗ | 0.006∗ |
| Negative controls for UC versus colectomy | ||||||||
| GTEx | muscle (n = 706) | 9,963 | 777 | 1.57e−181∗∗ | 0.089 | 0.220 | 1.69e−19∗∗ | 0.290 |
| GTEx | cortex (n = 205) | 13,486 | 1,075 | 5.54e−215∗∗ | 0.071 | 0.065 | 3.73e−19∗∗ | 0.110 |
All p values derived by Kruskal-Wallis tests.
Discussion
Our results highlight the potential of transcriptional profiling for prediction of colectomy in UC. Direct measurement of rectal biopsy RNA provides a replicated, highly discriminatory signature observed in almost all children who will need surgery. This signature has a positive predictive value for the adverse outcome approaching 50% (18 of 38 individuals with a score greater than 12), yet the expression profile reverts to a healthier state regardless of immunological therapy within 1 year. Although much of the mis-expression is thus associated with disease status and due to trans-regulation, we nevertheless show that prediction of gene expression from cis-linked SNPs is sufficient for generation of a PRS that outperforms one based purely on GWAS associations.
The two types of TRS that we describe, namely measured and predicted, are somewhat independent predictors of risk because their correlation is just 0.08. This is unsurprising because the predicted score is the summation of hundreds of independent models, one per transcript, whereas the measured score largely reflects co-regulation of the contributing transcript. This observation does, however, raise the question of to what extent cis-acting eQTLs contribute to the observed disease-associated expression signatures. Several lines of evidence suggest that this component of genetic risk, which is also the one typically detected by GWASs, is modest. First, the discrimination of affected individuals who underwent colectomy is much greater for the measured score, as the median score is 4 standard deviations greater than the median of all other affected individuals (Figure 3B), whereas the predicted score difference is just 0.2 standard deviations (Figure 5A). Second, the percent variance explained by case status for the abundance of transcripts that are differentially expressed at baseline in the colectomy subset is typically three times greater than that explained by peak eQTL genotype (Figure S9; conversely, genes with eQTL effects that explain more than 5% of the transcript variance tend not to be differentially expressed in the colectomy subset). Third, the overlap in identities of the 2,188 genes contributing to the measured PC1col and 820 to the PPTRScol is just 138 genes, consistent with the notion that many of the dysregulated genes are correlated with disease progression without having a measurable genetic contribution to it.
Given this modest contribution of eQTL effects to transcriptionally assessed risk, it is particularly remarkable that the PPTRScol outperforms the PRS based on more than 6,000 prune+threshold selected SNPs significant for disease status at p < 0.001 in cross-validated prediction of UC status. The most likely reason for this is that the DPR used in our TWAS captures much more of the locally acting regulatory variation than summation of marginal effects and hence more efficiently combines the genetic signal at each locus. Indeed, implementation of a Bayesian method for polygenic risk scoring using continuous shrinkage priors on SNP effect sizes (PRS-CS)64 gives rise to stronger PRS that has similar performance to the PPTRS (Figure S10). It remains noteworthy that the score built from just 331 PROTECT RNA-seq profiles and computed in 4,112 UC-affected individuals performs as well as the PRS with weights from meta-analysis of 20,000 affected individuals. Also notably, gut gene expression models from a completely different study (GTEx) have similar performance, and replication of risk stratification was observed in an independent adult colectomy cohort study. These data underscore the interpretation that it is the sum total of regulatory effects at a locus that contribute to genetic risk and that this can be evaluated even without understanding the direct correspondence between eQTL and GWAS association.
An additional component of our study is the demonstration that multiple cell types in the rectal mucosa contribute to pathology. In both the expanded set of 138 differentially expressed genes represented in the single-cell dataset and the focused set of TRS genes, immunoregulatory T cells, innate lymphoid cells, plasmacytoid dendritic cells, endothelial cells, secretory epithelial cells, and activated fibroblast cells are implicated by enrichment for expression of subsets of the genes that are among the defining markers of these cell types. That is not to say the genes are uniquely expressed in the cell types (most appear in two or three types, sometimes including both immune and epithelial cells), but it does indicate that the signature of pathology involves dysregulation in multiple cell layers. More extensive single-cell profiling, combined with cell-type-specific genetic analysis of gene expression, is likely to lead to the development of even better transcriptional risk signatures. It is also likely that such focused and personalized analysis may highlight specific pathological mechanisms active in particular affected individuals.
Our results are limited by the relatively small sample size of colectomies in the PROTECT study, which is nevertheless the largest treatment-naive inception cohort to date. The clinical significance of the PPTRScol is limited at this time because the precision remains low, but in the absence of gene expression profiles, it should be further evaluated as a component of total evidence models, supplementing histology-based indices such as PUCAI. Validation of cross-ancestry assessments should be a high priority, and it will be interesting to evaluate to what extent gene expression prediction is consistent across populations. It is likely that more widespread sampling of this and other forms of IBD will yield even more accurate predictors of disease progression, influencing personalized therapeutic decisions. Similar strategies might also be developed for other complex diseases for which sampling of the relevant tissue is impractical.
Acknowledgments
Support for this study was provided by NIDDK through grants U01DK095745, R01DK119991, P01DK046763, U01DK062413, U24DK062429, and U01DK062422 as well as the Leona M. and Harry B. Helmsley Charitable Trust. The authors thank Urko Marigorta for his counsel in development of TRS and Frank Hamilton, Dana Anderson, James Everhart, Jose Serrano, and Stephen James from NIDDK for their guidance. This research has been conducted with the UK Biobank resource under application number 17984 to G.G. The authors thank PROTECT site investigators for patient recruitment and data gathering, the research coordinators at the investigative sites for their tireless attention, and the affected individuals and families who graciously agreed to participate.
Declaration of interests
J.S.H. has served on an advisory board for Janssen and is acting as a consultant for AbbVie, Takeda, Lilly, Boehringer-Ingelheim, Allergan, Pfizer, Receptos, and AstraZeneca. S.D.T. has been a member of an independent data monitoring committee for Lycera Corporation. A.M.G. has received research support from AbbVie; been a consultant for AbbVie, Celgene, Janssen, Lilly, Pfizer, and Takeda; and been a speaker for AbbVie, Janssen, and Shire. N.S.L. has been a consultant for AbbVie. C.G.S. has been a consultant for AbbVie. J.M. has been a consultant for Janssen, Celgene, and Lilly. J.R.R. has been a consultant for AbbVie, Celgene, Janssen, Luitpold, and Pfizer and received grant funding from Janssen and AbbVie. A.S.P. has participated in speakers bureaus for AbbVie and Janssen. M.B.H. has received research grants from Genentech, AbbVie, Shire, Takeda, Mallinkrodt, Janssen, and Gilead. P.A.R. has been a consultant for Shire and Leutpold; been a speaker for AbbVie; and received research support from TechLab. S.K. has been a consultant for Janssen and UCB. L.A.D. has received grant support from AbbVie and Janssen. D.P.B.M. and T.H. are faculty members at Cedars-Sinai Medical Center. E.M. is an employee at Cedars-Sinai. Cedars-Sinai has financial interests in Prometheus Biosciences, a company that has access to the data and specimens in Cedars-Sinai’s MIRIAD Biobank. Prometheus Biosciences seeks to develop commercial products. D.M. is a paid consultant and shareholder of Prometheus Biosciences. D.M. has consulted for Pfizer, Gilead, Palatin Technologies, Bridge Biotherapeutics, and Takeda. All other authors declare no competing interests.
Published: August 26, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.07.013.
Data and code availability
The bulk RNA-seq data from PROTECT for this study has been deposited to the NCBI GEO database as GEO: GSE150961, and the single-cell RNA-seq data are available as GEO: GSE150516. No custom algorithms or software were utilized for this study, but the corresponding author will gladly share parameters used upon request. Code for computation of the PPTRS is available at the following GitHub link: https://github.com/sn-GT/Measured-and-predicted-TRS.
Supplemental information
References
- 1.Lambert S.A., Abraham G., Inouye M. Towards clinical utility of polygenic risk scores. Hum. Mol. Genet. 2019;28(R2):R133–R142. doi: 10.1093/hmg/ddz187. [DOI] [PubMed] [Google Scholar]
- 2.Lewis C.M., Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med. 2020;12:44. doi: 10.1186/s13073-020-00742-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson G. On the utilization of polygenic risk scores for therapeutic targeting. PLoS Genet. 2019;15:e1008060. doi: 10.1371/journal.pgen.1008060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Damask A., Steg P.G., Schwartz G.G., Szarek M., Hagström E., Badimon L., Chapman M.J., Boileau C., Tsimikas S., Ginsberg H.N., Regeneron Genetics Center and the ODYSSEY OUTCOMES Investigators Patients with high genome-wide polygenic risk scores for coronary artery disease may receive greater clinical benefit from alirocumab treatment in the ODYSSEY OUTCOMES Trial. Circulation. 2020;141:624–636. doi: 10.1161/CIRCULATIONAHA.119.044434. [DOI] [PubMed] [Google Scholar]
- 5.Natarajan P., Young R., Stitziel N.O., Padmanabhan S., Baber U., Mehran R., Sartori S., Fuster V., Reilly D.F., Butterworth A. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135:2091–2101. doi: 10.1161/CIRCULATIONAHA.116.024436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aragam K.G., Dobbyn A., Judy R., Chaffin M., Chaudhary K., Hindy G., Cagan A., Finneran P., Weng L.-C., Loos R.J.F. Limitations of contemporary guidelines for managing patients at high genetic risk of coronary artery disease. J. Am. Coll. Cardiol. 2020;75:2769–2780. doi: 10.1016/j.jacc.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J.C., Biasci D., Roberts R., Gearry R.B., Mansfield J.C., Ahmad T., Prescott N.J., Satsangi J., Wilson D.C., Jostins L., UK IBD Genetics Consortium Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn’s disease. Nat. Genet. 2017;49:262–268. doi: 10.1038/ng.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kugathasan S., Denson L.A., Walters T.D., Kim M.-O., Marigorta U.M., Schirmer M., Mondal K., Liu C., Griffiths A., Noe J.D. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet. 2017;389:1710–1718. doi: 10.1016/S0140-6736(17)30317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters L.A., Perrigoue J., Mortha A., Iuga A., Song W.-M., Neiman E.M., Llewellyn S.R., Di Narzo A., Kidd B.A., Telesco S.E. A functional genomics predictive network model identifies regulators of inflammatory bowel disease. Nat. Genet. 2017;49:1437–1449. doi: 10.1038/ng.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyams J.S., Davis Thomas S., Gotman N., Haberman Y., Karns R., Schirmer M., Mo A., Mack D.R., Boyle B., Griffiths A.M. Clinical and biological predictors of response to standardised paediatric colitis therapy (PROTECT): a multicentre inception cohort study. Lancet. 2019;393:1708–1720. doi: 10.1016/S0140-6736(18)32592-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haberman Y., Karns R., Dexheimer P.J., Schirmer M., Somekh J., Jurickova I., Braun T., Novak E., Bauman L., Collins M.H. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat. Commun. 2019;10:38. doi: 10.1038/s41467-018-07841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West N.R., Hegazy A.N., Owens B.M.J., Bullers S.J., Linggi B., Buonocore S., Coccia M., This S., Stockenhuber K., Pott J. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat. Med. 2017;23:579–589. doi: 10.1038/nm.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang H., Fang M., Jostins L., Umićević Mirkov M., Boucher G., Anderson C.A., Andersen V., Cleynen I., Cortes A., Crins F., International Inflammatory Bowel Disease Genetics Consortium Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173–178. doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Momozawa Y., Dmitrieva J., Théâtre E., Deffontaine V., Rahmouni S., Charloteaux B., Crins F., Docampo E., Elansary M., Gori A.-S., International IBD Genetics Consortium IBD risk loci are enriched in multigenic regulatory modules encompassing putative causative genes. Nat. Commun. 2018;9:2427. doi: 10.1038/s41467-018-04365-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson G., Powell J.E., Marigorta U.M. Expression quantitative trait locus analysis for translational medicine. Genome Med. 2015;7:60. doi: 10.1186/s13073-015-0186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marigorta U.M., Denson L.A., Hyams J.S., Mondal K., Prince J., Walters T.D., Griffiths A., Noe J.D., Crandall W.V., Rosh J.R. Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn’s disease. Nat. Genet. 2017;49:1517–1521. doi: 10.1038/ng.3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin J.C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J.A., Gettler K., Chuang L.-S., Nayar S., Greenstein A.J. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 2019;178:1493–1508.e20. doi: 10.1016/j.cell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parikh K., Antanaviciute A., Fawkner-Corbett D., Jagielowicz M., Aulicino A., Lagerholm C., Davis S., Kinchen J., Chen H.H., Alham N.K. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 2019;567:49–55. doi: 10.1038/s41586-019-0992-y. [DOI] [PubMed] [Google Scholar]
- 19.Smillie C.S., Biton M., Ordovas-Montanes J., Sullivan K.M., Burgin G., Graham D.B., Herbst R.H., Rogel N., Slyper M., Waldman J. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 2019;178:714–730.e22. doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., Nicolae D.L., Cox N.J., Im H.K., GTEx Consortium A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W.J.H., Jansen R., de Geus E.J.C., Boomsma D.I., Wright F.A. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wainberg M., Sinnott-Armstrong N., Mancuso N., Barbeira A.N., Knowles D.A., Golan D., Ermel R., Ruusalepp A., Quertermous T., Hao K. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 2019;51:592–599. doi: 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leijonmarck C.E., Persson P.G., Hellers G. Factors affecting colectomy rate in ulcerative colitis: an epidemiologic study. Gut. 1990;31:329–333. doi: 10.1136/gut.31.3.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandborn W.J., Rutgeerts P., Feagan B.G., Reinisch W., Olson A., Johanns J., Lu J., Horgan K., Rachmilewitz D., Hanauer S.B. Colectomy rate comparison after treatment of ulcerative colitis with placebo or infliximab. Gastroenterology. 2009;137:1250–1260, quiz 1520. doi: 10.1053/j.gastro.2009.06.061. [DOI] [PubMed] [Google Scholar]
- 25.Ungaro R., Mehandru S., Allen P.B., Peyrin-Biroulet L., Colombel J.-F. Ulcerative colitis. Lancet. 2017;389:1756–1770. doi: 10.1016/S0140-6736(16)32126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bycroft C., Freeman C., Petkova D., Band G., Elliott L.T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyams J.S., Davis S., Mack D.R., Boyle B., Griffiths A.M., LeLeiko N.S., Sauer C.G., Keljo D.J., Markowitz J., Baker S.S. Factors associated with early outcomes following standardised therapy in children with ulcerative colitis (PROTECT): a multicentre inception cohort study. Lancet Gastroenterol. Hepatol. 2017;2:855–868. doi: 10.1016/S2468-1253(17)30252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner D., Hyams J., Markowitz J., Lerer T., Mack D.R., Evans J., Pfefferkorn M., Rosh J., Kay M., Crandall W., Pediatric IBD Collaborative Research Group Appraisal of the pediatric ulcerative colitis activity index (PUCAI) Inflamm. Bowel Dis. 2009;15:1218–1223. doi: 10.1002/ibd.20867. [DOI] [PubMed] [Google Scholar]
- 29.Moll P., Ante M., Seitz A., Reda T. QuantSeq 3′ mRNA sequencing for RNA quantification. Nat. Methods. 2014;11 i–iii. [Google Scholar]
- 30.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data.https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 31.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mecham B.H., Nelson P.S., Storey J.D. Supervised normalization of microarrays. Bioinformatics. 2010;26:1308–1315. doi: 10.1093/bioinformatics/btq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law C.W., Chen Y., Shi W., Smyth G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suárez-Fariñas M., Tokuyama M., Wei G., Huang R., Livanos A., Jha D., Levescot A., Irizar H., Kosoy R., Cording S. Intestinal inflammation modulates the expression of ACE2 and TMPRSS2 and potentially overlaps with the pathogenesis of SARS-CoV-2 related disease. Gastroenterology. 2021;160:287–301.e20. doi: 10.1053/j.gastro.2020.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gettler K., Levantovsky R., Moscati A., Giri M., Wu Y., Hsu N.-Y., Chuang L.-S., Sazonovs A., Venkateswaran S., Korie U., UK IBD Genetics Consortium, National Institute of Diabetes, Digestive and Kidney Diseases Inflammatory Bowel Disease Genetics Consortium Common and rare variant prediction and penetrance of IBD in a large, multi-ethnic, health system-based biobank cohort. Gastroenterology. 2021;160:1546–1557. doi: 10.1053/j.gastro.2020.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wickham H. 2nd ed. Springer; 2016. ggplot2: elegant graphics for data analysis. [Google Scholar]
- 41.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X., Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Song W., Wang J., Wang T., Xiong X., Qi Z., Fu W., Yang X., Chen Y.-G. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J. Exp. Med. 2020;217:e20191130. doi: 10.1084/jem.20191130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng P., Zhou X. Non-parametric genetic prediction of complex traits with latent Dirichlet process regression models. Nat. Commun. 2017;8:456. doi: 10.1038/s41467-017-00470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagpal S., Meng X., Epstein M.P., Tsoi L.C., Patrick M., Gibson G., De Jager P.L., Bennett D.A., Wingo A.P., Wingo T.S., Yang J. TIGAR: An improved Bayesian tool for transcriptomic data imputation enhances gene mapping of complex traits. Am. J. Hum. Genet. 2019;105:258–266. doi: 10.1016/j.ajhg.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ndungu A., Payne A., Torres J.M., van de Bunt M., McCarthy M.I. A multi-tissue transcriptome analysis of human metabolites guides interpretability of associations based on multi-SNP models for gene expression. Am. J. Hum. Genet. 2020;106:188–201. doi: 10.1016/j.ajhg.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gusev A., Mancuso N., Won H., Kousi M., Finucane H.K., Reshef Y., Song L., Safi A., McCarroll S., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 2018;50:538–548. doi: 10.1038/s41588-018-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mancuso N., Freund M.K., Johnson R., Shi H., Kichaev G., Gusev A., Pasaniuc B. Probabilistic fine-mapping of transcriptome-wide association studies. Nat. Genet. 2019;51:675–682. doi: 10.1038/s41588-019-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haritunians T., Taylor K.D., Targan S.R., Dubinsky M., Ippoliti A., Kwon S., Guo X., Melmed G.Y., Berel D., Mengesha E. Genetic predictors of medically refractory ulcerative colitis. Inflamm. Bowel Dis. 2010;16:1830–1840. doi: 10.1002/ibd.21293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., International Multiple Sclerosis Genetics Consortium. International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loh P.-R., Danecek P., Palamara P.F., Fuchsberger C., A Reshef Y., K Finucane H., Schoenherr S., Forer L., McCarthy S., Abecasis G.R. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Das S., Forer L., Schönherr S., Sidore C., Locke A.E., Kwong A., Vrieze S.I., Chew E.Y., Levy S., McGue M. Next-generation genotype imputation service and methods. Nat. Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K., Haplotype Reference Consortium A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Graham D.B., Xavier R.J. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature. 2020;578:527–539. doi: 10.1038/s41586-020-2025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naito T., Botwin G.J., Haritunians T., Li D., Yang S., Khrom M., Braun J., Abbou L., Mengesha E., Stevens C., NIDDK IBD Genetics Consortium Prevalence and effect of genetic risk of thromboembolic disease in inflammatory bowel disease. Gastroenterology. 2021;160:771–780.e4. doi: 10.1053/j.gastro.2020.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mo A., Krishnakumar C., Arafat D., Dhere T., Iskandar H., Dodd A., Prince J., Kugathasan S., Gibson G. African ancestry proportion influences ileal gene expression in inflammatory bowel disease. Cell. Mol. Gastroenterol. Hepatol. 2020;10:203–205. doi: 10.1016/j.jcmgh.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schroeder K.W., Tremaine W.J., Ilstrup D.M. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N. Engl. J. Med. 1987;317:1625–1629. doi: 10.1056/NEJM198712243172603. [DOI] [PubMed] [Google Scholar]
- 63.Giambartolomei C., Zhenli Liu J., Zhang W., Hauberg M., Shi H., Boocock J., Pickrell J., Jaffe A.E., Pasaniuc B., Roussos P., CommonMind Consortium A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics. 2018;34:2538–2545. doi: 10.1093/bioinformatics/bty147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ge T., Chen C.Y., Ni Y., Feng Y.A., Smoller J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019;10:1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The bulk RNA-seq data from PROTECT for this study has been deposited to the NCBI GEO database as GEO: GSE150961, and the single-cell RNA-seq data are available as GEO: GSE150516. No custom algorithms or software were utilized for this study, but the corresponding author will gladly share parameters used upon request. Code for computation of the PPTRS is available at the following GitHub link: https://github.com/sn-GT/Measured-and-predicted-TRS.