Abstract
The photochemistry of cobalamins has recently been found to have biological importance, with the discovery of bacterial photoreceptor proteins, such as CarH and AerR. CarH and AerR, are involved in the light regulation of carotenoid biosynthesis and bacteriochlorophyll biosynthesis, respectively, in bacteria. Experimental transient absorption spectroscopic studies have indicated unusual photochemical behavior of 5′-deoxy-5′-adenosylcobalamin (AdoCbl) in CarH, with excited-state charge separation between cobalt and adenosyl and possible heterolytic cleavage of the Co-adenosyl bond, as opposed to the homolytic cleavage observed in aqueous solution and in many AdoCbl-based enzymes. We employ molecular dynamics and hybrid quantum mechanical/molecular mechanical calculations to obtain a microscopic understanding of the modulation of the excited electronic states of AdoCbl by the CarH protein environment, in contrast to aqueous solution and AdoCbl-based enzymes. Our results indicate a progressive stabilization of the electronic states involving charge transfer (CT) from cobalt/corrin to adenine on changing the environment from gas phase to water to solvated CarH. The solvent exposure of the adenosyl ligand in CarH, the π-stacking interaction between a tryptophan and the adenine moiety, and the hydrogen-bonding interaction between a glutamate and the lower axial ligand of cobalt are found to contribute to the stabilization of the states involving CT to adenine. The combination of these three factors, the latter two of which can be experimentally tested via mutagenesis studies, is absent in an aqueous solvent environment and in AdoCbl-based enzymes. The favored CT from metal and/or corrin to adenine in CarH may promote heterolytic cleavage of the cobalt-adenosyl bond proposed by experimental studies. Overall, this work provides novel, to our knowledge, physical insights into the mechanism of CarH function and directions for future experimental investigations. The fundamental understanding of the mechanism of CarH functioning will serve the development of optogenetic tools based on the new class of B12-dependent photoreceptors.
Significance
The discovery of B12 (cobalamin)-based bacterial photoreceptor proteins, such as CarH, has revealed the biological importance of cobalamin photochemistry, which was earlier unknown. CarH is involved in light regulation of bacterial biosynthesis of carotenoids that prevent photo-oxidative damage. Experiments have indicated that CarH modulates the photochemistry of 5′-deoxy-5′-adenosylcobalamin (AdoCbl) differently compared with bulk water and enzymes, promoting excited-state charge transfer and possibly heterolytic cleavage of the cobalt-adenosyl bond. Our molecular dynamics and quantum mechanical/molecular mechanical calculations reveal that the AdoCbl environment in CarH exhibits unique features contributing to stabilization of excited electronic states involving long range charge transfer. The physical insights obtained provide directions for future experimental investigations and contribute to the fundamental knowledge that will serve development of B12-dependent optogenetic tools.
Introduction
The discovery of cobalamin-binding photoreceptor proteins, such as CarH and AerR, that regulate transcription in bacteria has revealed the biological importance of cobalamin photochemistry, which was earlier unknown (1, 2, 3, 4, 5, 6, 7). For many years, the only known biological function of cobalamins or B12 derivatives was as cofactor to enzymes in humans, animals, and microbes, with the Co-C bond undergoing thermally-driven heterolytic (methylcobalamin) or homolytic (5′-deoxy-5′-adenosylcobalamin (AdoCbl)) cleavage to provide methyl cations or free radicals for enzymatic reactions (8,9). Excitation by light was also known to lead to cleavage of the Co-C bond in methylcobalamin and AdoCbl. This photolytic cleavage was, however, not believed to have biological relevance.
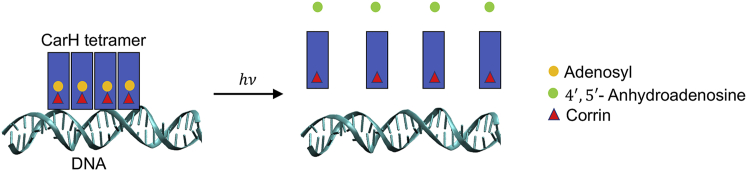
CarH and AerR are involved in the light regulation of carotenoid biosynthesis and bacteriochlorophyll biosynthesis, respectively, in bacteria (10). AerR (3,4) shows tight binding for hydroxocobalamin, whereas CarH uses AdoCbl as a chromophore. CarH is a bacterial transcriptional repressor that controls the biosynthesis of carotenoids in response to light (1,2,5, 6, 7). Carotenoids protect bacteria from photo-oxidative damage by quenching the singlet oxygen free radicals produced by light absorption. In the dark, upon AdoCbl binding, CarH forms tetramers that bind operator DNA and block transcription. Upon light absorption, the tetramers dissociate, releasing DNA and allowing transcription and the subsequent synthesis of carotenoids that protect the bacteria against damage by oxygen free radicals (Fig. 1) (5,6).
Figure 1.
Dissociation of DNA-bound CarH tetramer in the presence of light, leading to DNA release and synthesis of carotenoids. Light excitation leads to Co-Ado bond cleavage and release of Ado from the protein. Once released from the protein, Ado gets converted to 4′,5′-anhydroadenosine. To see this figure in color, go online.
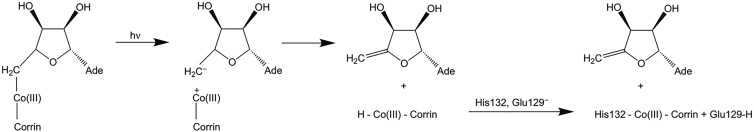
Experimental studies have indicated the modulation of the photochemistry of AdoCbl by the CarH protein environment (6). In contrast to AdoCbl photochemistry in aqueous solution that leads to the cob(II):Ado. radical pair through homolytic cleavage of the Co-C bond, photolysis of CarH-bound AdoCbl yields 4′,5′-anhydroadenosine, a species not observed before as a product of AdoCbl photolysis. Ultrafast spectroscopy experiments on CarH (6), which analyzed the photochemical mechanism on a timescale of femtoseconds to seconds, concluded that photoexcitation of CarH-bound AdoCbl is followed by the formation of a metal-to-ligand charge transfer (CT) state and, possibly, heterolytic cleavage of the Co-C bond. The cob(II):Ado. radical pair formed from a small percentage of homolytic cleavage was found to undergo quantitative recombination. Hydride and proton transfers have also been implicated in the proposed mechanism (Fig. 2).
Figure 2.
Proposed scheme of photochemical reactions in CarH (5, 6, 7). Postexcited-state charge transfer, the formation of 4′,5′-anhydroadenosine may be accompanied by hydride coordination to Co(III), with the resulting hydridocobalamin subsequently undergoing oxidation. X-ray crystallography has revealed His132 to be the final upper axial ligand of Co (5), which probably gets deprotonated by the nearby Glu129.
Despite the experimental studies, several details of the photochemical mechanism remain unknown and not easily accessible to experimental tools. Computational studies are indispensable for a fundamental detailed understanding of CarH photochemistry. CarH is inherently a very flexible biomolecule (5, 6, 7,10), and molecular dynamics (MD) simulations can provide information about the structural flexibility of CarH that is not available from the crystal structures. Quantum mechanical (QM) calculations are necessary to understand the photochemical bond breaking and formation reactions in CarH. To the best of our knowledge, this is the first computational study on CarH that involves a systematic investigation of the effect of the environment on the excited electronic states of AdoCbl with the protein (CarH) adequately solvated and its conformational flexibility accounted for (11). The goal of this study is to understand what features of the CarH protein environment contribute to the differences in AdoCbl photochemistry compared with aqueous solution and AdoCbl-based enzymes. The insights obtained will guide new experiments on CarH and contribute to the fundamental understanding of the mechanism of CarH functioning that will serve the development of optogenetic tools based on the new class of B12-dependent photoreceptors. B12 absorbs green light, making it suitable for tackling the “blind spot” in optogenetics. Green-light-absorbing B12-binding domains have been used to regulate signaling in human cells and development in zebrafish embryos (12). The development of a B12-based system for conditional expression of essential genes in Myxococcus xanthus has also been pursued (13).
Materials and methods
Molecular dynamics simulations
MD simulations were carried out using the Nanoscale Molecular Dynamics (NAMD) software package version 2.13. (14). The initial structure of an AdoCbl-bound CarH monomer was taken from Protein Data Bank (PDB): 5C8E (5). NAMD was used to simulate the CarH monomer bound to AdoCbl in a water box, using periodic boundary conditions. Based on the solvent exposure and the hydrogen-bonding environment, all His residues were treated as neutral, with the N site protonated, and all Lys, Arg, Asp, and Glu residues were treated as being in their standard protonation states at pH 7. Because of the closeness of His142 to Glu141 in the crystal structure, both neutral and protonated states of His142 were studied. The minimum distance between a protein atom and the edges of the water box was 15 Å. Na+ and Cl− ions were added to neutralize the charge of the protein and to bring the salt concentration to 0.15 M. These were added using the Visual Molecular Dynamics solvate and autoionize plugins (15). The cutoff, switch, and pairlist distance values used with the switch scheme for treatment of Van der Waals interactions were 14, 12, and 16 Å, respectively. The particle mesh Ewald method (16) was used for electrostatic interactions, with the default particle mesh Ewald parameters in NAMD (14). The CHARMM36 force field parameters for the protein (17) and modified TIP3P model for water (18) were used. Force field parameters for the ions and AdoCbl were taken from (19) and (20), respectively. The force field parameters used for AdoCbl in our MD simulations have been extensively verified by the developers (20). The quality of the parameters was judged based on gas-phase-optimized geometries, the corrin fold angle in MD simulations of cobalamins in aqueous and biomolecular environments, and the stability of cobalamins in the binding sites of proteins and RNA during MD simulations. The QM and molecular mechanical (MM) geometries showed excellent agreement. The corrin fold angle was also reproduced well in MD simulations, and cobalamin remained stable in the binding sites of proteins/RNA.
The initial structure of the wild-type (WT) monomer was energy minimized and equilibrated for 1 ns at 298 K in an NVT ensemble in which the volume and temperature were held constant. During the production simulations, an NPT ensemble was maintained in which the pressure and temperature were held constant at 1 atm and 298 K, respectively. For the E175Q and E175D mutants, the crystal structure of the AdoCbl-bound CarH monomer was mutated appropriately. Several independent simulations were carried out for both the WT protein and the mutants. Details of these simulations are provided in Table S1. The independent simulations for the WT protein, the E175Q mutant, and the E175D mutant were labeled traj. 1–4, traj. 5 and 6, and traj. 7 and 8, respectively. Unless noted otherwise, these simulations utilized the same protocol as outlined above. In cases where restraints on the N-terminal region, the Gln175-His177 distance or the Asp175-His177 distance were applied during equilibration, the NVT equilibration was followed by an additional 50 ns NPT equilibration.
Hybrid QM/MM calculations
Hybrid QM/MM calculations using a locally modified NAMD-Gaussian interface (21,22) were carried out to investigate the effect of the explicitly solvated protein environment on the excited electronic states of AdoCbl. 25 structures, at intervals of 4 ns, were selected from the first 100 ns of the traj. 1 WT simulation for these calculations. Additionally, six structures were also randomly selected after 100 ns of the traj. 3 WT simulation because of the sampling of shorter Glu141-sugar and Trp131-sugar distances compared with traj. 1. In the QM/MM calculation on each structure, the entire protein and primary water box along with the ions were retained. The water box was centered at AdoCbl, and its longest edge was aligned with the longest axis of the protein. The QM region included the central corrin ring without the substituents, Co, the upper axial adenosyl (Ado) ligand, and the side chain of the lower axial His177 ligand. The region of the corrin ring chosen to be studied quantum mechanically is depicted in Fig. S1. For His177, the boundary between the QM and MM regions was between the Cα and Cβ atoms, with all backbone atoms treated as point charges.
All QM atoms at the QM/MM boundary were capped using H link atoms to satisfy their valence and to mirror the nonpolarity of the C-C bond. The redistributed charge scheme (23) implemented in NAMD was used to adjust the charges on the MM atoms at the QM/MM boundary. To maintain a net charge of +1 for the QM region (Co being in the +3 oxidation state), a negligible charge redistribution on the MM atoms was carried out. Based on the partial charges assigned to cobalamin atoms by the force field (20), the net charge of the QM atoms belonging to the corrin ring and Co was 0.954. The protein structure file was edited so that the charges of the 23 heavy atoms in this set were increased by 0.002 each, resulting in a net charge of +1 for the QM region. The charges of the 23 MM atoms farthest away from the QM region (the dimethylbenzimidazole moiety of corrin) were decreased by 0.002 each to conserve the total charge of AdoCbl.
Time-dependent density functional theory (TDDFT) (24) calculation for the QM region was carried out in the presence of the MM point charges, mostly using the BP86 functional and the Def2SVP basis set. A comprehensive benchmarking of density functionals has shown the BP86 functional to best describe the Co-C (upper axial ligand) dissociation energy and structural and spectral features of cobalamins (25, 26, 27). Test calculations of AdoCbl excited states in the gas phase and in implicit water with TD-BP86/def2SVP (data provided in Supporting materials and methods, Data S1, Excel sheet) agree well with those in (25) using BP86 and a Pople basis set for C, N, O, and H.
Tests with the B3LYP and CAM-B3LYP functionals and the larger Def2TZVP basis set were also carried out. The B3LYP and CAM-B3LYP functionals were chosen as representatives of the class of hybrid and range-separated functionals, respectively. For each selected structure, TDDFT calculation for the QM region was also carried out in the presence of implicit water using the polarizable continuum model (PCM) (28) and in the gas phase. For calculations in implicit water, linear response nonequilibrium solvation was used for the calculation of vertical excitation energies at the ground state geometries. The default self-consistent reaction field method in Gaussian09 and Gaussian16, PCM using the integral equation formalism PCM, was employed. For some structures, the effect of the net charge of selected charged amino acid residues on the excited electronic states was investigated by setting the partial charges of the side-chain atoms of these residues to 0 and repeating the TDDFT/MM calculations. The procedure adopted for testing the effect of explicit solvent versus implicit solvent on the excited electronic states of AdoCbl is explained in the Supporting materials and methods.
The process of generation of isosurface plots of the natural transition orbitals (NTOs) was automated using Python and Bash scripts, provided as part of the Supporting materials and methods. An isovalue of 0.07 bohr−3/2 was used for most of the isosurface plots. In selected cases, an isovalue of 0.06 or 0.05 bohr−3/2 was used for better understanding of the electronic transition. The images were visually analyzed to determine the basic nature of the electronic transitions.
Data availability
Images of natural transition orbitals corresponding to all the calculated excited states in all the time-dependent density functional theory calculations performed are available in a Google Drive repository (https://drive.google.com/drive/folders/1cnldtIQksAtH0Lsts6Hn-JueJK3hbSfw?usp=sharing).
Results and discussion
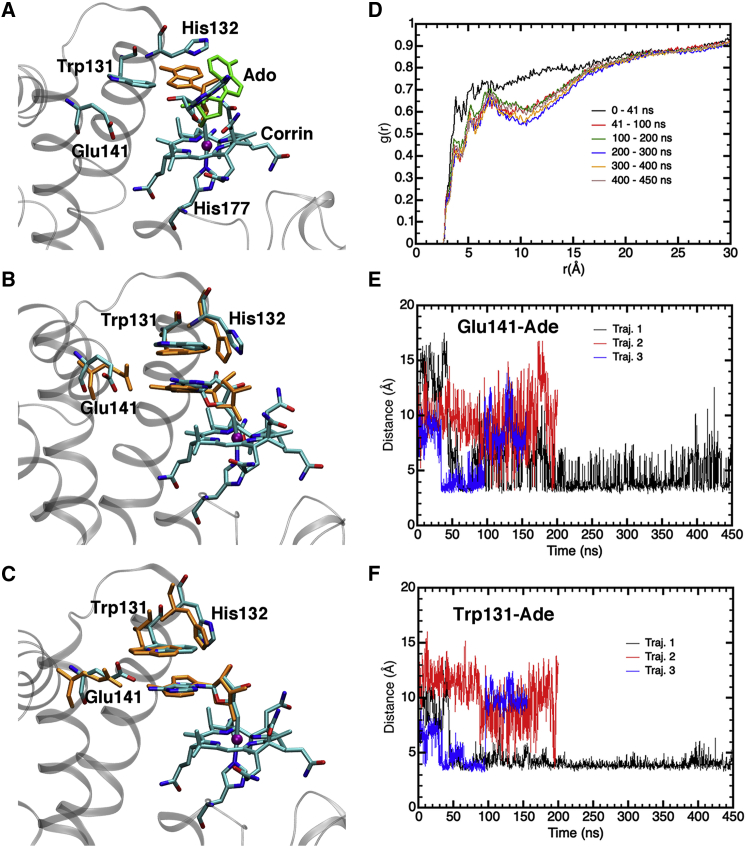
Four independent simulations for the WT CarH monomer ranging in length from 155 to 450 ns were carried out (Table S1). The root mean-square deviation (RMSD) of the backbone heavy atoms (Fig. S2, A and B) and the root mean-square fluctuation of the amino acid residues (Fig. S3, A and B) indicate that the protein is stable and that the N-terminal region exhibits larger flexibility compared with the C-terminal region. As a test, one of the trajectories (traj. 2) was equilibrated for an additional 50 ns in the NPT ensemble with weak restraints on the N-terminal region residues, after which the restraints were released. The RMSD and root mean-square fluctuation plots were found to have no significant dependence on the application of restraints in the N-terminal region during equilibration. Because Glu141 is close to His142 in the crystal structure, His142 was treated as protonated in traj. 4 to investigate the effect of its protonation on the conformational sampling of Glu141. Although Glu141 remained hydrogen bonded to His142 throughout most of the trajectory (Fig. S4 B), unlike the other three trajectories with a neutral His142 (Fig. S4 A), Glu141 still showed significant hydrogen-bonding to adenine (Ade), like the other trajectories (Fig. S5, A and B). The hydrogen bond between Glu141 and Trp131 was not observed often (Fig. S7, A and B), which is similar to the behavior of traj. 1 but in contrast with traj. 2 and 3.
In all four WT trajectories, the Ado ligand is found to sample distinct orientations, as shown using representative snapshots from traj. 1 (Fig. 3). In the first ∼40 ns of this trajectory, the Ado ligand maintains an orientation similar to that in the crystal structure, remaining highly solvent exposed and relatively distant from Glu141 and Trp131. In the last ∼410 ns, it resides closer to Glu141 and Trp131. For all the trajectories, it is observed that in correlation with Ado rotation from a solvent-exposed to a π-stacked conformation (Figs. S8, A and B and S9, A and B), there is a decrease in the distance between the Glu141 side chain and the NH2 group of Ade (Fig. S5, A and B). The formation of the π-stacked conformation between Trp131 and Ade is monitored by a dihedral angle involving Trp131 and Ade (Fig. S8, A and B) and the distance between the centers of mass of Trp131 and Ade (Fig. S9, A and B). The ribose moiety of Ado is also found to become more distant from Glu141 (Fig. S10, A and B) and Trp131 (Fig. S11, A and B), with the formation of the π-stacked conformation between Trp131 and Ade. The radial distribution function (29) plots in Figs. S12–S15 show that the solvent exposure of both the five- and six-membered rings of Ade decreases with formation of the π-stacking interaction between Ade and Trp131.
Figure 3.
(A–C) Selected structures from an MD simulation (traj. 1) of the WT CarH monomer to demonstrate the conformational flexibility of the Ado ligand. (A) The structure at 28 ns (colored by atom type) is overlaid with that at 4 ns (orange) and 44 ns (green). Despite its flexibility, Ado remains relatively far from Trp131 and Glu141 and highly solvent exposed. (B) The structure at 52 ns (colored by atom type) is overlaid with that at 68 ns (orange). The Ade moiety is roughly coplanar with Trp131, and Glu141 forms no or one hydrogen bond to Ade. The solvent exposure of Ade is lower than in the first ~40 ns. (C) The structure at 64 ns (colored by atom type) is overlaid with that at 80 ns (orange). The difference from (B) is the formation of hydrogen bonds between Glu141 and both Trp131 and Ade. (D) Radial distribution function for water O atoms around the heavy atoms (excluding the N atom of the −NH2 substituent) of the six-membered ring of the Ade moiety. (E) Time evolution of the distance between the Cδ atom (carboxylate C atom) of Glu141 and the N6 atom (N atom of −NH2 group, also see Fig. S6) of Ade in three independent WT simulations with deprotonated His142. (F) Time evolution of the distance between the center of mass of the heavy atoms of the Trp131 side chain and the heavy atoms of the five- and six-membered rings of Ade in three independent WT simulations with deprotonated His142. To see this figure in color, go online.
25 structures from the first 100 ns of traj. 1, at intervals of 4 ns, were selected for QM/MM calculations. Six structures with short Glu141/Trp131-sugar distances were also randomly selected from traj. 3. TDDFT calculations for the QM region (AdoCbl) were carried out in the presence of the point charges corresponding to the rest of the system. The entire protein and primary water box were retained in these calculations, with the water box centered at cobalamin and its longest edge aligned with the length of the protein. Table 1 shows the effect of the environment on the excitation wavelength of the lowest energy excited states involving CT from metal/corrin to Ade, CT from metal to the σ∗ orbital corresponding to the Co-C(Ado) bond, and CT from metal to the corrin ring (Fig. 4). These three categories of excited states were investigated because the first can possibly lead to heterolytic cleavage of the Co-C bond, especially when accompanied by population of the Co-C σ∗ orbital, and the latter two have been studied widely in previous investigations of homolytic cleavage of the Co-C bond in enzymes and in CarH (11,30, 31, 32, 33, 34, 35). Table S2 shows the presence of the metal/corrin to Ade/σ∗ transition in several structures selected from the MD simulation. This transition is absent in most calculations in only implicit solvent.
Table 1.
Effect of environment on the excitation wavelength in nanometers and index of the lowest energy states involving metal/corrin to Ade, metal to σ∗ and metal to corrin transitions
| Time (ns) | Type of transition | Solvent | Protein |
|---|---|---|---|
| 24a | metal/corrin to Ade | 387.2, S20 | 445.3, S8 |
| metal to σ∗ | 481.3, S7 | 491.0, S4 | |
| metal to corrin | 568.6, S2 | 540.7, S2 | |
| 44a | metal/corrin to Ade | 399.0, S20 | 519.8, S4 |
| metal to σ∗ | 493.9, S6 | 488.0, S7 | |
| metal to corrin | 621.8, S3 | 600.0, S2 | |
| 52a | metal/corrin to Ade | 423.4, S14 | 579.6, S3 |
| metal to σ∗ | 475.0, S9 | 486.5, S8 | |
| metal to corrin | 619.3, S2 | 595.2, S2 | |
| 96a | metal/corrin to Ade | 473.5, S10 | 596.9, S4 |
| metal to σ∗ | 601.2, S2 | 601.9, S3 | |
| metal to corrin | 593.6, S3 | 572.7, S6 | |
| 108b | metal/corrin to Ade | 453.8, S14 | 476.9, S10 |
| metal to σ∗ | 511.2, S8 | 516.4, S6 | |
| metal to corrin | 624.2, S2 | 604.8, S2 | |
| 115b | metal/corrin to Ade | 439.1, S14 | 627.0, S2 |
| metal to σ∗ | 544.7, S4 | 545.0, S5 | |
| metal to corrin | 590.8, S2 | 699.3, S1 | |
| 125b | metal/corrin to Ade | 483.2, S11 | 493.2, S8 |
| metal to σ∗ | 559.4, S5 | 559.9, S4 | |
| metal to corrin | 638.9, S3 | 640.5, S3 | |
| 137b | metal/corrin to Ade | 468.0, S14 | 661.6, S3 |
| metal to σ∗ | 703.8, S2 | 695.3, S2 | |
| metal to corrin | 612.7, S3 | 594.7, S4 | |
| 146b | metal/corrin to Ade | 458.6, S11 | 565.3, S5 |
| metal to σ∗ | 558.5, S4 | 567.3, S4 | |
| metal to corrin | 598.6, S2 | 681.9, S3 | |
| 154b | metal/corrin to Ade | 399.7, S23 | 475.9, S10 |
| metal to σ∗ | 562.1, S4 | 559.1, S3 | |
| metal to corrin | 712.1, S1 | 668.1, S1 |
The Def2SVP basis set was used for all calculations. Sn indicates the nth excited singlet state.
Structures from traj. 1 (Detailed data for the other structures are provided in the Supporting materials and methods).
Structures from traj. 3.
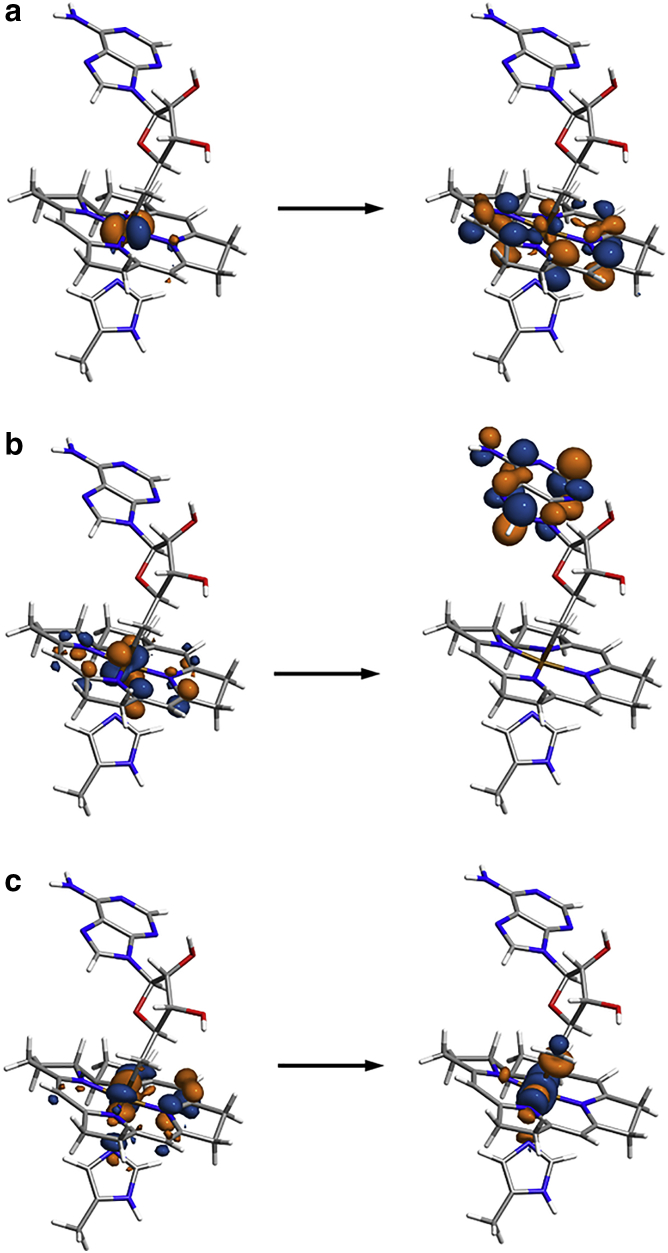
Figure 4.
Representative natural transition orbitals (NTOs) depicting (a) CT from metal to the corrin ring, (b) CT from metal/corrin to Ade, (c) CT from metal to the σ∗ orbital corresponding to the Co-C(Ado) bond. The Co atom is located at the center of the tetrapyrrole ring, the upper axial ligand of Co is the Ado group, and the lower axial ligand of Co is the His177 side chain. An isovalue of 0.05 bohr−3/2 was used for these plots. The orange and blue colors of the isosurfaces indicate positive and negative signs of the wavefunction, respectively. To see this figure in color, go online.
For several structures selected from the MD simulations (Table 1), the replacement of the explicitly solvated protein environment with implicit solvent (water in this case) leads to a significant blue shift in the excitation wavelength of the lowest lying excited state involving CT to Ade (see Table S2 for more details on other structures). In contrast, this change of environment has a small effect on the excitation wavelength of the lowest lying excited states involving CT from metal to the Co-C σ∗ orbital and CT from metal to the corrin ring. For the latter, a small red shift is observed upon changing the environment of the chromophore from explicitly solvated protein to implicit solvent in most of the structures. Tests with AdoCbl solvated in implicit versus explicit water show that the effect of treating the solvent implicitly versus explicitly is minimal (Table S3). Changing the basis set from Def2SVP to the larger Def2TZVP basis set has significant effect on the excitation wavelength only if the CT to Ade is accompanied by population of the σ∗ orbital corresponding to the Co-C(Ado) bond (Table S4).
The observed red shift for the lowest lying excited state involving CT to Ade upon changing the environment from implicit solvent to explicitly solvated protein was reproducible using a hybrid functional, B3LYP, and a range-separated functional, CAM-B3LYP (Table 2) (36). The magnitude of red shift, however, decreases in the order BP86, B3LYP, CAM-B3LYP. Previous studies have shown that for cobalamins, the BP86 functional best describes the Co-C(upper axial ligand) dissociation energy, and structural and spectral features (25, 26, 27). This was explained based on artificial widening of the energy gap between the occupied and virtual orbitals for cobalamins with range-separated functionals such as CAM-B3LYP. Because polarizability is related to the gap between the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO) gap (37,38), the large red shifts predicted by BP86 are likely more accurate than the smaller shifts predicted by the B3LYP and CAM-B3LYP functionals.
Table 2.
Effect of environment and DFT functional on the excitation wavelength in nanometers of the lowest energy state involving CT from metal/corrin to Ade
| Time |
BP86 |
B3LYP |
CAM-B3LYP |
||||||
|---|---|---|---|---|---|---|---|---|---|
| (ns) |
|||||||||
| Gas phase | Solventa | Proteinb | Gas phase | Solventa | Proteinb | Gas phase | Solventa | Proteinb | |
| 24 | NA | 387.2 | 445.3 (58.1) | 247.0 | 294.4 (47.4) | 319.7 (25.3) | NA | 214.3 | 226.3 (12.0) |
| 44 | NA | 399.0 | 519.8 (102.8) | NA | 297.8 | 357.0 (59.2) | NA | 218.1 | 248.4 (30.3) |
| 52 | 330.1 | 423.4 (93.3) | 579.6 (156.2) | 257.2 | 313.6 (56.4) | 385.8 (72.2) | NA | 223.9 | 258.0 (34.1) |
| 96 | 363.2 | 473.5 (110.3) | 596.9 (123.3) | 261.1 | 316.0 (54.9) | 447.3 (131.3) | NA | 220.4 | 277.5 (57.1) |
The Def2SVP basis set was used for all calculations. Structures were taken from traj. 1.
NA, not available indicates that the transition could not be located in the first 60 excited states.
Numbers in parentheses indicate the change in excitation wavelength on changing the environment from gas phase to solvent.
Numbers in parentheses indicate the change in excitation wavelength on changing the environment from solvent to protein.
Whereas the absolute excitation wavelengths of states involving long range CT, like those involving CT from metal/corrin to the Ade ring, may not be well described by a generalized gradient approximation functional, such as BP86 (39), the shift in the excitation wavelength upon change of environment is expected to be more reliable, especially because BP86 describes the HOMO-LUMO gap for cobalamins well. Table 2 shows that the red shift in the excitation wavelength of the lowest lying excited state involving CT to Ade occurs not only upon changing the environment from implicit water to explicitly solvated protein, but also upon changing the environment from gas phase to implicit water.
Additional QM/MM calculations with side-chain charges of selected amino acids zeroed out provided insight into the aforementioned red shift upon changing the environment from implicit water to explicitly solvated protein. The structures at 32 and 72 ns of traj. 1 were selected because they showed the largest number of transitions involving CT to Ade in the first 60 excited states. Two other randomly selected structures (at 24 and 96 ns) were also investigated. The residues whose side-chain charges were zeroed out one at a time were Arg, Lys, Glu, and Asp residues with any atom within 8 Å of a heavy atom in the corrin central ring or of a heavy atom in the Ado ligand. Tables S5–S8 show that Glu175 is responsible for a large red shift in the excitation wavelength of the lowest lying excited state involving CT to Ade. An examination of the MD trajectory and the crystal structure (PDB: 5C8E) (5) shows that Glu175 is hydrogen bonded to the lower axial ligand of Co, His177 (see Fig. S20). The presence of this negatively charged residue close to the lower axial ligand is expected to promote CT to the upper axial ligand, as is observed in our calculations. The role of Glu175 in modulating the excited electronic states can be experimentally investigated by mutating it to a Gln residue that conserves the hydrogen-bonding interaction with His177 but removes the negative charge, or an Asp residue that possibly removes the hydrogen-bonding interaction but retains the negative charge.
Two independent simulations each for the E175Q and E175D mutant were carried out, and the detailed analysis in Figs. S2–S11, C and D and Figs. S16–S19 show similar trends to those in the WT trajectories, except greater sampling of longer distances between Glu141/Trp131 and the ribose O2′ atom in one of the E175Q trajectories. The other E175Q trajectory samples larger RMSD-values, primarily due to greater flexibility of the N-terminal region. Fig. S21 shows that, unlike Glu175, which forms a stable hydrogen bond to the His177 side chain in all independent trajectories for the WT protein, Gln175 and Asp175 do not form stable hydrogen bonds with His177. The rotation of Gln175 and Asp175 away from His177, as well as the rotation of the Nδ atom of His177 away from these residues is observed. Some test QM/MM calculations on structures selected from the E175Q and E175D trajectories show that a very large number of calculations would be required to make a meaningful distinction between the WT and mutant spectra because of the large number of variables that can be different between snapshots selected from the WT and mutant trajectories. The loss of a stable hydrogen bond between residue 175 and His177 in the E175Q and E175D mutants, as observed in MD simulations, besides the removal of a negative charge in the E175Q mutant, provides further motivation for experimental investigations of the E175Q and E175D mutants to understand the role of Glu175 in modulating cobalamin photochemistry in CarH.
Examination of the QM/MM data with the side-chain charges of selected residues zeroed out shows that the small blue shift in the excitation wavelength of the CT from metal to the corrin ring upon changing the environment of the chromophore from implicit solvent to explicitly solvated protein (Table 1) has small contributions from the side-chain charges of residues Arg125, Arg133, and Glu175 (see Supporting materials and methods, Data S1, Excel sheet and NTO isosurface plots). Arg125 and Arg133 are located on the Ado side of the corrin ring. Whereas it is not straightforward to rationalize the effect of these residues on the stability of the excited states with CT from metal to the corrin ring, they are probably responsible for a shift of the α-band to higher energy in the protein environment (10). In the protein environment, the red shift in the wavelength of the excitations involving CT to Ade possibly has little effect on the spectrum because of the low oscillator strength associated with these excitations (Supporting materials and methods, Data S1, NTO isosurface plots).
In addition to Glu175, Trp131, which is often observed to be roughly π-stacked with the Ade ring of AdoCbl, is expected to further stabilize the excited states involving CT to Ade. Calculations with a larger QM region involving Trp131 were not carried out because of concerns with the description of CT to Trp131 by the BP86 functional. However, π-stacking between Ade and Trp131 will most likely lead to stabilization of CT to Ade. An examination of the crystal structures of several AdoCbl-based enzymes, ethanolamine ammonia-lyase (PDB: 3ABS) (40), diol dehydratase (PDB: 5YRV) (41), methylmalonyl-CoA mutase (PDB: 4REQ) (42), and glutamate mutase (PDB: 1I9C) (43) shows lack of a Trp residue close to the cobalamin. This indicates that CarH, in which Trp131 is a highly conserved residue (10), likely uses a special π-stacking mechanism to promote CT to Ade. This can be probed experimentally via mutation of Trp131 to a residue that cannot π-stack with Ade.
Overall, our calculations reveal that the CarH protein environment is associated with three key features that cause extra stabilization of excited states involving CT from metal/corrin to Ade. This extra stabilization of excited states involving CT to Ade in CarH is most likely related to the experimentally observed excited-state charge separation and possibly promotes subsequent heterolytic cleavage of the Co-Ado bond, especially when it is accompanied by population of the Co-C σ∗ orbital. Neither an aqueous solution environment nor the AdoCbl-based enzymes referred to above provide a combination of all these features. These three features are as follows: 1) solvent exposure of the Ado ligand, 2) hydrogen bond between a Glu and the lower axial His ligand of Co, and 3) π-stacking between the Ade ring and a Trp side chain. An aqueous solvent environment lacks features 2 and 3, methylmalonyl-CoA mutase (PDB: 4REQ) and glutamate mutase (PDB: 1I9C) lack features 1 and 3, and ethanolamine ammonia-lyase (PDB: 3ABS) and diol dehydratase (PDB: 5YRV) lack all three features. The cobalamin-dependent enzymes have limited conformational flexibility. Their crystal structures do not indicate significant conformational changes upon substrate binding (40,43, 44, 45, 46), thus justifying the above comparison between CarH and the enzymes based on the crystal structures of the latter. A comprehensive investigation, however, remains the subject of future studies.
In the crystal structure of the DNA-bound tetramer (Fig. 1) (5), 1) the small dimer interface in the vicinity of AdoCbl, 2) the lack of significant interactions between the monomers at the dimer interface in the vicinity of AdoCbl, and 3) the large solvent exposure of Ado, Trp131 and nearby residues indicate that Ado flexibility will be similar in the monomer and tetramer. Regardless, our observed stabilization of the Co/corrin to Ade CT states in CarH is independent of the protein conformation and Ado orientation. The three factors which promote this stabilization, =solvent exposure of Ado, π-stacking interactions between Ado and Trp131, and hydrogen bonding between His177 and Glu175, are independent of the protein oligomerization state or the rotation of Ado toward Glu141. Parallel displaced and T-shaped π-stacking interactions between Ado and Trp131 are formed in our simulations even without the rotation of the Ado ligand toward Glu141. Hence, this study, based on simulations of the CarH monomer, provides valuable insight into the modulation of AdoCbl excited states by the CarH protein environment. Simulations of the DNA-bound tetramer are a subject of future investigations.
A recent computational study of CarH (11) based on energy-minimized structures of the unsolvated protein monomer assumed the photochemistry to initiate from the Franck-Condon region of the S1 state, this region being associated with metal to corrin CT. Crossing of the energy barrier between the Franck-Condon region and the region of the S1 potential energy surface associated with metal to σ∗ transition was assumed to be related with homolytic cleavage of the Co-C bond. It should, however, be noted that the photochemical activity of CarH is experimentally observed for a broad range of excitation wavelengths, and the population decay from the excited states does not necessarily lead to population of the Franck-Condon region of the S1 state. In the absence of experimental evidence of nonreactivity on the higher excited states of CarH, occurrence of heterolytic cleavage of the Co-C bond correlated with stabilization of the excited states involving CT to Ade in CarH cannot be ruled out. Such heterolytic cleavage may be accompanied by abstraction of the H4′ atom of Ado as hydride by Co (Fig. 2) ; this atom is found to stay relatively close to Co throughout our MD simulation, despite the overall conformational flexibility of Ado (Fig. S22). The nonadiabatic dynamics associated with cobalamin photochemistry in CarH needs to be investigated using a combination of computational and experimental methods. Experimental studies of the wavelength dependence of AdoCbl photolysis in CarH can provide valuable insights in this regard.
Conclusions
In conclusion, we have investigated the effect of the CarH protein environment on the excited electronic states of AdoCbl, with adequate sampling of the protein conformations using MD simulation, QM/MM calculations with appropriate description of the protein and aqueous environment, and consideration of the role of CT states involving the Ade group in the photochemistry. Our results indicate a progressive red shift in the excitation wavelength and hence stabilization of the electronic states involving CT from metal/corrin to Ade on changing the environment from gas phase to water to solvated protein. The excited states with metal to σ∗ transition, which has been widely associated with homolytic cleavage of the Co-C bond (11,30, 31, 32, 33, 34, 35), do not exhibit such stabilization in CarH. The solvent exposure of AdoCbl in CarH, the π-stacking interaction between Trp131 and the Ade moiety, and the hydrogen-bonding interaction between Glu175 and the lower axial ligand of Co contribute to the stabilization of the states involving CT to Ade. The combination of these three factors may promote heterolytic cleavage of the cobalt-Ado bond as the protein evolves through excited electronic states, especially those with CT to Ade accompanied by population of the Co-C σ∗ orbital. A combination of all these factors is absent in an aqueous solvent environment and in common AdoCbl-based enzymes, which exhibit homolytic cleavage of the Co-C bond upon excitation by light. The role of Trp131 and Glu141 can be experimentally tested through site-directed mutagenesis studies. The involvement of higher excited electronic states in photochemistry can be investigated through studies of the wavelength dependence of AdoCbl photolysis in CarH. Overall, the insights obtained from this study contribute to the fundamental knowledge that will serve the development of optogenetic tools based on the new class of B12-dependent photoreceptors. Future studies using higher level electronic structure methods will allow greater insights into the influence of the CarH protein environment on the excited states of AdoCbl and an understanding of the evolution of the system on the excited states that determines the observed photochemistry.
Author contributions
P.G. designed the research. All authors carried out calculations, analyzed the data, and wrote the article.
Acknowledgments
We thank Dr. Rafael da Silva and Dr. Amanda Morris for helpful discussions. All figures were made using version 1.9.4 of the Visual Molecular Dynamics program (15), Xmgrace (47), version 2.8 of Gimp (48), and ImageMagick (49).
This study was supported by funding from the Chemistry Department at the State University of New York at Binghamton and the Research Foundation of the State University of New York.
Editor: Alan Grossfield.
Footnotes
Courtney L. Cooper and Naftali Panitz contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.07.020.
Supporting citations
Reference (50) appears in the supporting material.
Supporting material
References
- 1.Pérez-Marín M.C., Padmanabhan S., Elías-Arnanz M. Vitamin B12 partners the CarH repressor to downregulate a photoinducible promoter in Myxococcus xanthus. Mol. Microbiol. 2008;67:804–819. doi: 10.1111/j.1365-2958.2007.06086.x. [DOI] [PubMed] [Google Scholar]
- 2.Ortiz-Guerrero J.M., Polanco M.C., Elías-Arnanz M. Light-dependent gene regulation by a coenzyme B12-based photoreceptor. Proc. Natl. Acad. Sci. USA. 2011;108:7565–7570. doi: 10.1073/pnas.1018972108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng Z., Li K., Bauer C.E. Vitamin B12 regulates photosystem gene expression via the CrtJ antirepressor AerR in Rhodobacter capsulatus. Mol. Microbiol. 2014;91:649–664. doi: 10.1111/mmi.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vermeulen A.J., Bauer C.E. Members of the PpaA/AerR antirepressor family bind cobalamin. J. Bacteriol. 2015;197:2694–2703. doi: 10.1128/JB.00374-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jost M., Fernández-Zapata J., Drennan C.L. Structural basis for gene regulation by a B12-dependent photoreceptor. Nature. 2015;526:536–541. doi: 10.1038/nature14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kutta R.J., Hardman S.J.O., Jones A.R. The photochemical mechanism of a B12-dependent photoreceptor protein. Nat. Commun. 2015;6:7907. doi: 10.1038/ncomms8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chemaly S.M. New light on vitamin B12: the adenosylcobalamin-dependent photoreceptor protein CarH. S. Afr. J. Sci. 2016;112:1–9. [Google Scholar]
- 8.Banerjee R., Ragsdale S.W. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu. Rev. Biochem. 2003;72:209–247. doi: 10.1146/annurev.biochem.72.121801.161828. [DOI] [PubMed] [Google Scholar]
- 9.Jones A.R. The photochemistry and photobiology of vitamin B12. Photochem. Photobiol. Sci. 2017;16:820–834. doi: 10.1039/c7pp00054e. [DOI] [PubMed] [Google Scholar]
- 10.Padmanabhan S., Jost M., Elías-Arnanz M. A new facet of vitamin B12: gene regulation by cobalamin-based photoreceptors. Annu. Rev. Biochem. 2017;86:485–514. doi: 10.1146/annurev-biochem-061516-044500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toda M.J., Mamun A.A., Kozlowski P.M. Why is CarH photolytically active in comparison to other B12-dependent enzymes? J. Photochem. Photobiol. B. 2020;209:111919. doi: 10.1016/j.jphotobiol.2020.111919. [DOI] [PubMed] [Google Scholar]
- 12.Kainrath S., Stadler M., Janovjak H. Green-light-induced inactivation of receptor signaling using cobalamin-binding domains. Angew. Chem. Int.Engl. 2017;56:4608–4611. doi: 10.1002/anie.201611998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Moreno D., Polanco M.C., Elías-Arnanz M. A vitamin B12-based system for conditional expression reveals dksA to be an essential gene in Myxococcus xanthus. J. Bacteriol. 2009;191:3108–3119. doi: 10.1128/JB.01737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips J.C., Hardy D.J., Tajkhorshid E. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020;153:044130. doi: 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38, 27–28.. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 16.Darden T., York D., Pedersen L. Particle Mesh Ewald: an N log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 17.Huang J., MacKerell A.D., Jr. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 19.Beglov D., Roux B. Finite representation of an infinite bulk system: solvent boundary potential for computer simulations. J. Chem. Phys. 1994;100:9050–9063. [Google Scholar]
- 20.Pavlova A., Parks J.M., Gumbart J.C. Development of CHARMM-compatible force-field parameters for cobalamin and related cofactors from quantum mechanical calculations. J. Chem. Theory Comput. 2018;14:784–798. doi: 10.1021/acs.jctc.7b01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melo M.C.R., Bernardi R.C., Luthey-Schulten Z. NAMD goes quantum: an integrative suite for hybrid simulations. Nat. Methods. 2018;15:351–354. doi: 10.1038/nmeth.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frisch M.J., Trucks G.W., Fox D.J. Gaussian Inc.; Wallingford, CT: 2009. Gaussian09 Revision E.01. [Google Scholar]
- 23.Lin H., Truhlar D.G. Redistributed charge and dipole schemes for combined quantum mechanical and molecular mechanical calculations. J. Phys. Chem. A. 2005;109:3991–4004. doi: 10.1021/jp0446332. [DOI] [PubMed] [Google Scholar]
- 24.Burke K., Werschnik J., Gross E.K.U. Time-dependent density functional theory: past, present, and future. J. Chem. Phys. 2005;123:62206. doi: 10.1063/1.1904586. [DOI] [PubMed] [Google Scholar]
- 25.Andruniów T., Jaworska M., Kozlowski P.M. Time-dependent density functional theory study of cobalt corrinoids: electronically excited states of coenzyme B12. J. Chem. Phys. 2009;131:105105. doi: 10.1063/1.2956836. [DOI] [PubMed] [Google Scholar]
- 26.Kornobis K., Kumar N., Kozlowski P.M. Electronically excited states of vitamin B12: benchmark calculations including time-dependent density functional theory and correlated ab initio methods. J. Phys. Chem. A. 2011;115:1280–1292. doi: 10.1021/jp110914y. [DOI] [PubMed] [Google Scholar]
- 27.Toda M.J., Lodowski P., Kozlowski P.M. Photolytic properties of the biologically active forms of vitamin B12. Coord. Chem. Rev. 2019;385:20–43. [Google Scholar]
- 28.Miertuš S., Scrocco E., Tomasi J. Electrostatic interaction of a solute with a continuum. A direct utilizaion of ab initio molecular potentials for the prevision of solvent effects. Chem. Phys. 1981;55:117–129. [Google Scholar]
- 29.Levine B.G., Stone J.E., Kohlmeyer A. Fast analysis of molecular dynamics trajectories with graphics processing units-radial distribution function histogramming. J. Comput. Phys. 2011;230:3556–3569. doi: 10.1016/j.jcp.2011.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mamun A.A., Toda M.J., Kozlowski P.M. Mechanism of light induced radical pair formation in coenzyme B12-dependent ethanolamine ammonia-lyase. ACS Catal. 2018;8:7164–7178. [Google Scholar]
- 31.Ghosh A.P., Mamun A.A., Kozlowski P.M. Mechanism of the photo-induced activation of CoC bond in methylcobalamin-dependent methionine synthase. J. Photochem. Photobiol. B. 2018;189:306–317. doi: 10.1016/j.jphotobiol.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh A.P., Mamun A.A., Kozlowski P.M. How does the mutation in the cap domain of methylcobalamin-dependent methionine synthase influence the photoactivation of the Co-C bond? Phys. Chem. Chem. Phys. 2019;21:20628–20640. doi: 10.1039/c9cp01849b. [DOI] [PubMed] [Google Scholar]
- 33.Mamun A.A., Toda M.J., Kozlowski P.M. Can photolysis of the CoC bond in coenzyme B12-dependent enzymes be used to mimic the native reaction? J. Photochem. Photobiol. B. 2019;191:175–184. doi: 10.1016/j.jphotobiol.2018.12.018. [DOI] [PubMed] [Google Scholar]
- 34.Kumar N., Bucher D., Kozlowski P.M. Mechanistic implications of reductive Co-C bond cleavage in B12-dependent methylmalonyl CoA mutase. J. Phys. Chem. B. 2019;123:2210–2216. doi: 10.1021/acs.jpcb.8b10820. [DOI] [PubMed] [Google Scholar]
- 35.Mamun A.A., Toda M.J., Kozlowski P.M. Photolytic cleavage of Co-C bond in coenzyme B12-dependent glutamate mutase. J. Phys. Chem. B. 2019;123:2585–2598. doi: 10.1021/acs.jpcb.8b07547. [DOI] [PubMed] [Google Scholar]
- 36.Mardirossian N., Head-Gordon M. Thirty years of density functional theory in computational chemistry: an overview and extensive assessment of 200 density functionals. Mol. Phys. 2017;115:2315–2372. [Google Scholar]
- 37.Pearson R.G. Absolute electronegativity and hardness correlated with molecular orbital theory. Proc. Natl. Acad. Sci. USA. 1986;83:8440–8441. doi: 10.1073/pnas.83.22.8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janjua M.R.S.A., Mahmood A., Pan S. Electronic absorption spectra and nonlinear optical properties of ruthenium acetylide complexes: a DFT study toward the designing of new high NLO response compounds. Acta Chim. Slov. 2014;61:382–390. [PubMed] [Google Scholar]
- 39.Kümmel S. Charge-transfer excitations: a challenge for time-dependent density functional theory that has been met. Adv. Energy Mater. 2017;7:1700440. [Google Scholar]
- 40.Shibata N., Tamagaki H., Toraya T. Crystal structures of ethanolamine ammonia-lyase complexed with coenzyme B12 analogs and substrates. J. Biol. Chem. 2010;285:26484–26493. doi: 10.1074/jbc.M110.125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shibata N., Sueyoshi Y., Toraya T. Direct participation of a peripheral side chain of a corrin ring in coenzyme B12 catalysis. Angew. Chem. Int.Engl. 2018;57:7830–7835. doi: 10.1002/anie.201803591. [DOI] [PubMed] [Google Scholar]
- 42.Mancia F., Evans P.R. Conformational changes on substrate binding to methylmalonyl CoA mutase and new insights into the free radical mechanism. Structure. 1998;6:711–720. doi: 10.1016/s0969-2126(98)00073-2. [DOI] [PubMed] [Google Scholar]
- 43.Gruber K., Reitzer R., Kratky C. Radical shuttling in a protein: ribose pseudorotation controls alkyl-radical transfer in the coenzyme B(12) dependent enzyme glutamate mutase This work was supported by the Österreichische Akademie der Wissenschaften (APART fellowship 614), the Österreichische Fonds zur Förderung der wissenschaftlichen Forschung (FWF-project 11599), and the European Commission (TMR project number ERB 4061 PL 95-0307). Crystallographic data were collected at the EMBL-beamline BW7B at DESY in Hamburg, Germany. We thank the beamline scientists for their assistance, and Ingrid Dreveny, Günter Gartler, Gerwald Jogl, and Oliver Sauer for their help during data collection. This research emerged from a collaboration with Prof. W. Buckel (Marburg) who supplied us with clones of the glutamate mutase proteins. Angew. Chem. Int. Ed. Engl. 2001;40:3377–3380. doi: 10.1002/1521-3773(20010917)40:18<3377::aid-anie3377>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 44.Yamanishi M., Yunoki M., Toraya T. The crystal structure of coenzyme B12-dependent glycerol dehydratase in complex with cobalamin and propane-1,2-diol. Eur. J. Biochem. 2002;269:4484–4494. doi: 10.1046/j.1432-1033.2002.03151.x. [DOI] [PubMed] [Google Scholar]
- 45.Gruber K., Kratky C. Coenzyme B(12) dependent glutamate mutase. Curr. Opin. Chem. Biol. 2002;6:598–603. doi: 10.1016/s1367-5931(02)00368-x. [DOI] [PubMed] [Google Scholar]
- 46.Toraya T., Honda S., Mori K. Coenzyme B12-dependent diol dehydratase is a potassium ion-requiring calcium metalloenzyme: evidence that the substrate-coordinated metal ion is calcium. Biochemistry. 2010;49:7210–7217. doi: 10.1021/bi100561m. [DOI] [PubMed] [Google Scholar]
- 47.Stambulchik E. Grace. https://plasma-gate.weizmann.ac.il/Grace/
- 48.The GIMP Development Team GIMP. https://www.gimp.org
- 49.The ImageMagick Development Team ImageMagick. https://imagemagick.org
- 50.Miertuš S., Scrocco E., Tomasi J. Electrostatic interaction of a solute with a continuum. A direct utilization of Ab initio molecular potentials for the prevision of solvent effects. Chem. Phys. 1981;55:117–129. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Images of natural transition orbitals corresponding to all the calculated excited states in all the time-dependent density functional theory calculations performed are available in a Google Drive repository (https://drive.google.com/drive/folders/1cnldtIQksAtH0Lsts6Hn-JueJK3hbSfw?usp=sharing).