Abstract
The gut microbiome is a pivotal player in toxicological responses. We investigated the effects of maternal exposure to 3 human health-relevant toxicants (BDE-47, tetrabromobisphenol [TBBPA], and bisphenol S [BPS]) on the composition and metabolite levels (bile acids [BAs] and short-chain fatty acids [SCFAs]) of the gut microbiome in adult pups. CD-1 mouse dams were orally exposed to vehicle (corn oil, 10 ml/kg), BDE-47 (0.2 mg/kg), TBBPA (0.2 mg/kg), or BPS (0.2 mg/kg) once daily from gestational day 8 to the end of lactation (postnatal day 21). 16S rRNA sequencing and targeted metabolomics were performed in feces of 20-week-old adult male pups (n = 14 − 23/group). Host gene expression and BA levels were quantified in liver. BPS had the most prominent effect on the beta-diversity of the fecal microbiome compared with TBPPA and BDE-47 (QIIME). Seventy-three taxa were persistently altered by at least 1 chemical, and 12 taxa were commonly regulated by all chemicals (most of which were from the Clostridia class and were decreased). The most distinct microbial biomarkers were S24-7 for BDE-47, Rikenellaceae for TBBPA, and Lactobacillus for BPS (LefSe). The community-wide contributions to the shift in microbial pathways were predicted using FishTaco. Consistent with FishTaco predictions, BDE-47 persistently increased fecal and hepatic BAs within the 12α hydroxylation pathway, corresponding to an up-regulation with the hepatic BA-synthetic enzyme Cyp7a1. Fecal BAs were also persistently up-regulated by TBBPA and BPS (liquid chromatography-mass spectrometry). TBBPA increased propionic acid and succinate, whereas BPS decreased acetic acid (gas chromatography-mass spectrometry). There was a general trend in the hepatic down-regulation of proinflammatory cytokines and the oxidative stress sensor target gene (Nqo1), and a decrease in G6Pdx (the deficiency of which leads to dyslipidemia). In conclusion, maternal exposure to these toxicants persistently modified the gut-liver axis, which may produce an immune-suppressive and dyslipidemia-prone signature later in life.
Keywords: PBDE, BPS, TBBPA, microbiome, mice, bile acid, SCFA
The human microbiome is a highly dynamic and metabolically active “second genome” estimated to have over 3 million unique genes, varying in composition due to host genetics, diet, xenobiotic exposure, and location in the body. Emerging research suggests that the human microbiome is an important player in the developmental origins of human health and disease (Mandy and Nyirenda, 2018), playing key roles in metabolic and immune systems (Shreiner et al., 2008). In addition, there is growing concern towards alteration of gut microbiota in early life. Variations in the composition of early-life exposure have been associated with specific disease outcomes such as asthma, obesity, and neurodevelopmental disorders (Stiemsma and Michels, 2018). Gut dysbiosis has been linked to gastrointestinal conditions such as inflammatory bowel disease and irritable bowel syndrome, as well as obesity and diabetes (Bull and Plummer, 2014). The composition of gut microbiome also plays a key role in disease susceptibility, where enrichment of opportunistic bacteria has been linked to increased gut permeability and increased likelihood of infections (Tsolis and Baumler, 2020). Microbiome produces distinct metabolites such as secondary bile acids (BAs) (from liver-derived primary BAs) as well as short-chain fatty acids (SCFAs) (from fermentation of complex carbohydrates) (Baxter et al., 2019). Secondary BAs at exceedingly high concentrations, such as during cholestasis, are toxic to the host due to their hydrophobic nature and can induce inflammation and cytotoxicity (Woolbright and Jaeschke, 2019). However, under physiological conditions, secondary BAs protect against intestinal inflammation in a mouse model of colitis (Sinha et al., 2020), highlighting the context-specific roles of microbial derived BAs in host health and diseases. Short-chain fatty acids, such as acetic acid, propionic acid, and butyric acid, are considered beneficial to the host in a variety of physiological and pathophysiological models, due to their anti-inflammatory, antiobesity, neuroprotective, and epigenetic reprogramming capacities (Al-Lahham et al., 2010; Govindarajan et al., 2011; Olson et al., 2018; Sun et al., 2018).
Exposure to environmental toxicants has led to growing safety concerns. The formerly banned flame retardants polybrominated diphenyl ethers (PBDEs), the current-use PBDE-alternative tetrabromobisphenol (TBBPA), as well as the bisphenol A (BPA), an alternative to bisphenol S (BPS) are well-known environmental toxicants that are linked to human diseases. Polybrominated diphenyl ethers are a class of bio-accumulative persistent compounds that were used as flame retardants for a variety of electronics, furniture, plastics, and industrial buildings (Siddiqi et al., 2003). In 2006, the EPA committed to phase out these commercial mixtures of multiple congeners, mainly pentaBDE and OctaBDE. Among various PBDE congeners, BDE-47 is a diet-enriched PBDE congener that has been linked to thyroid toxicity (Richardson et al., 2008), neurotoxicity (Dong et al., 2020; Ji et al., 2019; Ta et al., 2011), as well as increased risk for metabolic syndrome (Zhang et al., 2016). BDE-47 made up 40% of many pentaBDE mixtures (technical fact—PBDEs). Due to the persistent and bio-accumulative nature, BDE-47 continues to be present in the environment and bio-accumulate in fatty bio-compartments of humans and other wildlife.
TBBPA is an environmental pollutant that exists as a currently approved flame-retardant present in common house furniture, sewage, and house dust. TBBPA is a well-known endocrine disruptor (Van der Ven et al., 2008). A study in 2008 revealed that brominated compounds like TBBPA were efficient in competing for the thyroxine (T4) binding site regulating thyroid hormone synthesis. TBBPA also plays a role in immunotoxicity. A study in 2017 showed that TBBPA increased the production of interleukin 1 beta (IL-1β) from natural killer cells, blood mononuclear cells, and peripheral blood mononuclear cells, in a concentration and time-dependent manner (Anisuzzaman and Whalen, 2016).
BPS is a BPA alternative and a plasticizer associated with endocrine-disrupting effects (Huang et al., 2020), commonly found in plastic bottles and electronics. A study in 2017 showed that rats that were fed 30, 60, and 120 mg/kg BW/day of BPS for 30 days had significantly decreased levels of blood cells, white blood cells, and hemoglobin in a dose-dependent manner (Pal et al., 2017). Aside from metabolic toxicity, BPS is known to induce neurotoxicity and halt behavioral growth. For example, a study found that Caenorhabditis elegans treated with 0.1 μM BPS noted inhibition of head thrashes had decreased body length and weakened bends in nematode models (Xiao et al., 2019).
The environmental levels of BDE-47, TBBPA, and BPS have been estimated through detecting levels of these chemicals in human bio-compartments. For example, as reported in a study on the North American human population, BDE-47 levels are approximately 399 ng/g lipids (Baxter et al., 2019). As we discussed previously (Kim et al., 2015), the doses used in the present study are relevant to the levels of these toxicants in human bio-compartments as a result of environmental exposures. For example, Dr Suvorov’s laboratory has previously shown that exposing pregnant rats to 0.2 mg/kg body weight of BDE-47 resulted in 234.3 ng BDE-47 per gram lipid in the adipose tissue of dams (Suvorov et al., 2009). In addition, because the half-life of BDE-47 is faster in mice than in rats (Staskal et al., 2004; von Meyerinck et al., 1990), the dose of BDE-47 is expected to represent the lower end of the environmentally levels that are relevant to the human exposure settings. There are limited data on BPS and TBBPA tissue burdens between mice and humans, but human tissue burdens are likely lower than the doses used in the present study. Regarding TBBPA, average TBBPA levels in human adipose tissue is approximately 0.048/g lipid (Johnson-Restrepo et al., 2008), whereas the daily exposure of toddlers to TBBPA is about 0.2 ng/kg body weight per day via dust (Abb et al., 2011). Regarding BPS, the median BPS daily intake is estimated to be 1.67 μg per person in Japan and 0.338 μg per person in the United States, based on urinary data extrapolations (Liao et al., 2012).
Although there has been extensive research on the acute exposure outcomes of all these chemicals, relatively less is known regarding to what extent early-life exposure to these chemicals modulate the gut microbiome during adulthood. Considering that the gut microbiome is important for health and diseases, it has been suggested to be a key player in developmental origins of health and diseases. Therefore, it is important to characterize the potential persistent effect of these chemicals on the microbiome long after the initial chemical exposure ceases. The goals of the present study were to: (1) identify the differentially regulated bacteria in adult male mice from early-life exposure, (2) identify which differentially regulated bacteria modulate different metabolic pathways within the gut-liver axis, and (3) assess which of these altered pathways may lead to increased risk of disease (Table 1).
Table 1.
Overall Summary of Major Findings
| Chemicals | No. of Uniquely Altered Microbiota | Most Representative Microbial Biomarkers | Fecal Metabolites | Liver BAs | Hepatic Gene Expression | Potential Outcomes Within Gut-Liver Axis |
|---|---|---|---|---|---|---|
| BDE-47 | 12 | S24-7 |
↑Total, secondary, and unconjugated BAs (all 3 chemicals) ↑Propionic acid and succinate (TBBPA) ↓Acetic acid (BPS) |
Trend in ↑ CA, T-DCA, and DCA |
↑Cyp7a1 (BA synthetic enzyme) ↓Cytokines ↓G6pdx (glucose and lipid regulator) |
-> Enhanced BA synthesis by host liver and gut bacteria -> Immune suppression -> Dyslipidemia |
| TBBPA | 15 | Rikenellaceae | ↓CDCA |
↓Cytokines ↓Nqo1 (prototypical target of the oxidative stress sensor Nrf2) |
||
| BPS | 9 | Lactobacillus | ↓UDCA |
↓Cytokines ↓Nqo1 |
MATERIALS AND METHODS
Animals and chemical exposure
CD-1 mice were obtained from Charles River Laboratories (Kingston, New York) and were housed in the animal facility at the University of Massachusetts Amherst as described before (Kim et al., 2015). From pregnancy day 8 through postnatal day (PND) 21, dams were fed daily either vehicle (corn oil) or 0.2 mg/ml solutions of TBPPA, BDE-47, or BPS at 1 μl/gram body weight from the tip of a pipette, resulting in 0.2 mg/kg body weight/day. In addition, a nonexposed group of CD-1 dams were included to control for any potential effect due to vehicle exposure. At PND 21, pups were weaned and allowed to mature naturally without any additional chemical exposure as described above. Livers and feces were collected from male pups only at 20 weeks of young adult age (n = 15 − 24 per group; specifically, n = 19 for the no treatment group; n = 18 for the vehicle exposure group; n = 24 for the BDE-47 exposure group, n = 17 for the TBBPA exposure group, and n = 15 for the BPS exposure group). Samples were stored at 2.0 μl microcentrifuge tubes and stored at −80° C until further analyses. All procedures met the guidelines of the National Institutes of Health Guide for Care and Use of Laboratory Animals, and this study was approved by the Institutional Animal Care and Use Committee at University of Massachusetts, Amherst (key procedures are summarized in Figure 1).
Figure 1.
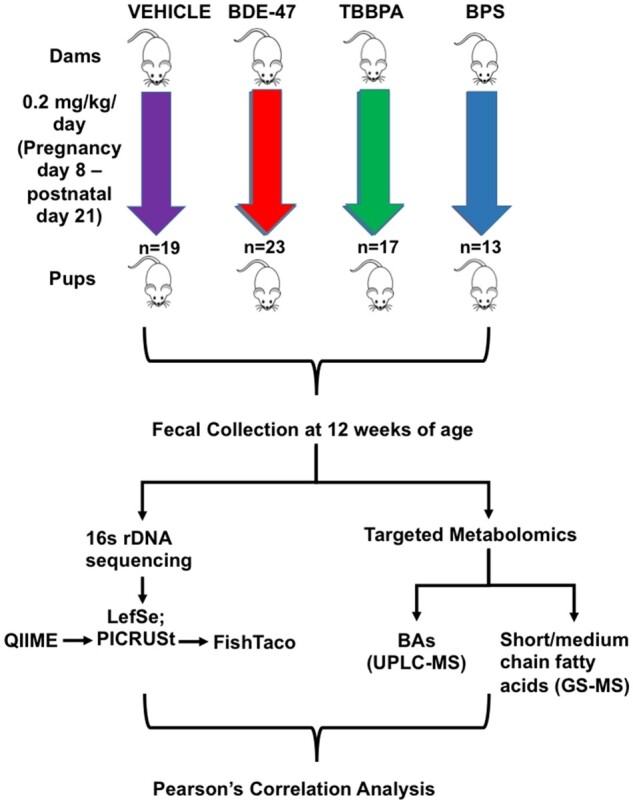
Experimental design that shows CD-1 mice treated with (Vehicle, BDE-47, TBBPA, BPS) (n = 13 − 23/group) daily for 21 days. Fecal samples from pups were collected at 20 weeks. Fecal samples were used for 16S rDNA sequencing for statistical analysis (QIIME-PiCRUSt-FishTaco) and targeted Metabolomics consisting of BA (LC-MS) and short-/medium-chain fatty acids (GS-MS). Pearson’s correlation analyses were performed between differentially regulated bacteria and metabolites.
Fecal DNA isolation and 16S rDNA sequencing
DNA was isolated and extracted using the E.Z.N.A. stool kit (OMEGA Bio-tek, Inc., Norcross, GA) following the manufacturer’s protocol as we described previously (Cheng et al., 2018; Dempsey et al., 2019; Li et al., 2018a,b; Scoville et al., 2019). DNA concentration was determined by a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, Massachusetts). Amplification and sequencing of the hypervariable V4 region of bacterial 16S rDNA was done using a HiSeq-2500 sequencing system (250 bp paired-end; n = 15 − 24 per group; Novogene, Sacramento, California).
Data analysis for the 16S rDNA sequencing data
The quality of raw reads from the demultiplexed FASTQ files was examined using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and all reads were kept for further analysis using Quantitative Insights Into Microbial Ecology (QIIME) version 1.9.1 (Caporaso et al., 2010). Specifically, the paired forward and reverse reads of the same sample were joined using “join_paired_ends.py.” The joined FASTQ files were then each uniquely labeled and then merged together into a FASTA file using “split_libraries.fastq.py.” Operational Taxonomic Units (OTUs) were assigned using “pick_open_reference_otus.py” against the 99_otus.fasta reference database (Version 13.8, Greengenes Database Consortium) (DeSantis et al., 2006), enabling both the forward and the reverse strand matches. The OTU tables were then sorted and summarized from the phylum (L2) to species (L7) levels. Alpha diversity was determined using “alpha_rarefaction.py” and beta diversity with “jackknifed_beta_diversity.py.” Functional profiles (KEGG pathways) of microbial communities were predicted using PICRUSt (Phylogenic Investigation of Communities by Reconstruction of Unobserved States) (Langille et al., 2013) using the following scripts: normalize by copy number.py, predict metagenomes.py, and categorize by function.py. FishTaco was used to predict the taxa that contribute to the functional shifts in the microbiome (http://elbo.gs.washington.edu/software_fishtaco.html) (Manor and Borenstein 2017) with taxonomic and functional abundance profiles as well as inferred genomic information. The 16S rDNA sequencing data are available for download at the Dryad database (https://zenodo.org/record/4069170#.X87Q5BNKjEY).
Preparation of solutions for BA standard curve internal standards
Cholic acid (CA), chenodeoxy CA (CDCA), deoxycholic acid (DCA), lithocholic acid (LCA), α muricholic acid (αMCA), βMCA, ωMCA, ursodeoxycholic acid (UDCA), hyodeoxycholic acid (HDCA), murideoxycholic acid (MDCA), taurine conjugated cholic acid (T-CA), T-CDCA, T-DCA, T-LCA, T-αMCA, T-βMCA, T-UDCA, and T-HDCA were purchased from Steraloids (Newport, Rhode Island). T-ωMCA was a kind gift from Dr Daniel Raftery’s laboratory at the University of Washington Northwest Metabolomics Research Center (Li et al., 2018a,b). A total of 5 deuterated (d4) ISs were used, among them the d4-CA was purchased from Toronto Research Chemicals Inc (Ontario, Canada), the d4-DCA and d4-chenodeoxycholic acid (d4-CDCA) were purchased from CDN Isotope Inc (Pointe-Claire, Quebec, Canada), the d4-glycine conjugated CDCA (d4-G-CDCA) was purchased from IsoSciences (Ambler, Pennsylvania), and d4-LCA was purchased from Steraloids (Newport, Rhode Island). One milligram per milliliter stock solutions of individual BAs (for standard curve) and IS were prepared in methanol and water (1:1). The 19 individual BA stock solutions were further diluted in 50% methanol to obtain 10 working standard solutions (0.05 − 10 000 ng/ml). The 5 ISs were mixed to obtain a working IS solution in which the concentration of each IS was 10 µmol/ml.
Extraction
All organic solvents (analytical grade) used in the procedure, unless noted otherwise, were purchased from Sigma-Aldrich (St Louis, Missouri). Fecal BAs were extracted from frozen fecal samples (n = 15 − 23/group) using a similar method as we described previously (Dempsey et al., 2019; Li et al., 2018a,b; Scoville et al., 2019) with modifications. Briefly, 75 − 85 mg of feces were used for each sample and were vortexed in sterile water (2.5 μl/mg of fecal mass) at 4° C. Afterwards, 200 μl of well-homogenized fecal suspension was taken out and mixed with 10 μl of internal standard (IS) mixture, and equilibrated on ice for 5 − 10 min; 1.5 ml of ice-cold alkaline acetonitrile (ACN) (5% ammonia in ACN) was added to the homogenate, which was then vortexed vigorously and shaken continuously for 1 h at room temperature. The mixture was then centrifuged at 12 000 × g for 15 min at 4° C, and the supernatant was collected into 5-ml glass tubes. The pellet was resuspended in 750 μl of 100% methanol, shaken for 20 min, and centrifuged at 12 000 × g for 20 min. The 2 supernatants obtained were combined, evaporated under vacuum (30°C) for 4 h, and reconstituted in 100 µl of 50% methanol. The suspension was transferred into a 0.2 µm Costar Spin-X HPLC microcentrifuge filter (Corning Inc., Corning, New York), and centrifuged at 12 000 × g for 10 min. Seventy microliters of this filtrate were loaded in the plate to inject into the ultraperformance liquid chromatography coupled with mass spectrometry in tandem (UPLC-MS/MS) for analysis. For liver BA extraction, as we described previously (Li et al., 2018b), approximately 50 − 60 mg of frozen liver was accurately weighed and homogenized in 5 volumes of water (eg, 300 μl for 60 mg liver tissue). For each 300-μl of homogenate (50 mg of liver in 250 μl of water), 1.5 ml of ACN with 5% ammonia hydroxide (NH4OH) and 10 μl of IS solution were added into the mixture. Tubes were vortexed and shaken for 1 h at room temperature. The mixtures were then centrifuged at 12 000 × g for 15 min at 4°C, and the supernatants were transferred to a separate set of tubes. A second BA extraction was performed using 750 μl analytical grade methanol, shaken for 20 min, sonicated for 5–10 min, and centrifuged at 15 000 × g for 20 min. The 2 extraction supernatants were then pooled together and dried under vacuum. The residue was then reconstituted by addition of 100 μl of 50% methanol water solution, vortexed, transferred into the 0.22-μm Costar Spin-X centrifuge tube filter (MilliporeSigma), and centrifuged at 20 000 × g for 10 min before injection. Calibrator and different quality control samples were prepared by adding the appropriate amount of the different standard stock solutions and were extracted using the similar sample preparation procedure described above for fecal sample extractions.
Ultraperformance liquid chromatography coupled with mass spectrometry in tandem method development
Liquid chromatographic separation and mass spectrometric detection were performed using an Agilent G6460 UPLC-MS/MS system (version A.00.07.32) combined with a triple quadrupole mass spectrometer via an electrospray ionization interface. The chromatographic separation was performed using a ZORBAX Eclipse Plus C18 analytical column (2.1×100mm; id: 1.8 µm). The gradient profile for the LC pump under the final chromatography conditions were as follows: 0 min, 95:5; 5 min, 95:5; 14 min, 86:14; 14.5 min, 75:25; 17.50 min, 75:25; 18 min, 50:50; 22 min, 50:50; 22.50 min, 20:80; 24.50 min, 20:80; 25 − 28 min, 95:5 (A:B, vol/vol). The injection volume of the samples was 5 µl. The column temperature was set at 45°C and the sample tray temperature was maintained at 9°C. MS/MS spectra were produced using the negative ionization mode.
Chemicals used for the quantification of short- and medium-chain fatty acids
Acetic acid was bought from Thermo Fisher Scientific (Fair Lawn, New Jersey). Propionic acid, isobutyric acid, butyric acid, 2-methylbutyric acid, isovaleric acid, valeric acid, 2-methylpentanoic acid, 3-methylpentanoic acid, isocaproic acid, caproic acid, 2-methylhexanoic acid, 4-methylhexanoic acid, heptanoic acid, hexanoic acid-6,6,6-d3 IS, N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide (MTBSTFA), and methoxyamine hydrochloride were purchased from Sigma-Aldrich.
Sample preparation for the quantification of fatty acids
Each fecal sample (∼20 mg) was mixed with 20 µl NaOH solution (0.5 M in water), 20 µl IS solution (hexanoic acid-6,6,6-d3 in water; 200 µM), and 460 µl MeOH in an Eppendorf tube (1.5 ml). Then stainless beads were added into the tube, and homogenization (2 min) was performed using a Bullet Blender homogenizer (Next Advance, Averill Park, New York). After homogenization, 400 µl 80% MeOH in water (vol:vol) were added into the sample. Then the sample was vortexed for 30 s, stored under −20°C for 30 min, and sonicated in an ice bath for 10 min. After centrifugation (14 000 rpm, 10 min), 700 µl supernatant was collected into a new Eppendorf tube. The sample was then dried under vacuum at 37°C for 240 min using a CentriVap Concentrator (Labconco, Fort Scott, Kansas).
Gas chromatography coupled with MS (GC-MS) for the quantification of short- and medium-chain fatty acids
Each sample was first derivatized with 40 µl of methoxyamine hydrochloride solution in pyridine (MeOX, 20 mg/ml) for 90 min under 60°C. Secondly, 60 µl of MTBSTFA was added, and the mixture was incubated for 30 min under 60°C. Then the sample was vortexed for 30 s, followed by centrifugation at 14 000 rpm for 10 min. Lastly, 70 µl of the supernatant was collected into a new glass vial for GC-MS analysis.
GC-MS experiments were performed using an Agilent 7820A gas chromatography system coupled to an Agilent 5977B mass spectrometer (Agilent Technologies, Santa Clara, California). Chemical derivatives in the samples were separated using an HP-5 ms capillary column coated with 5% phenyl-95% methylpolysiloxane (30 m × 250 µm i.d., 0.25 µm film thickness, Agilent Technologies). One microliter of each sample was injected, and the solvent delay time was set to 5 min. The initial oven temperature was held at 60°C for 1 min, shifted up to 325°C at a steady rate of 10°C/min, and finally held at 325°C for 10 min. Helium was used as the carrier gas at a constant flow rate of 20 ml/min through the column. The temperatures of the front inlet, transfer line, and electron impact ion source were set at 250°C, 290°C, and 230°C, respectively. The electron energy was −70 eV, and the mass spectral data were collected in the full scan mode (m/z 30 − 600).
Agilent MassHunter Workstation Software Quantitative Analysis (B.09.00) was used to process the GC-MS data for compound identification, peak picking, and quantification. The signal-to-noise ratio (S/N) was set to S/N = 3. The retention time and quantification mass for each SCFA were determined using its chemical standard. The concentrations of SCFAs in biological samples were calculated using the calibration curves constructed from the corresponding SCFAs standards.
Total RNA isolation
Total RNA was extracted from frozen livers using RNAzol Bee reagent (Tel-Test Inc., Friendswood, Texas) per the manufacturer’s protocol. RNA concentration was quantified using a NanoDrop Spectrophotometer (Thermo Scientific, Wilmington, Delaware) at 260 nm. The integrity of each RNA sample was evaluated by formaldehyde agarose gel electrophoresis to visualize the 18S and 28S rRNA bands.
RT-qPCR quantification of BA-processing genes and cytokines
The total RNAs of mouse livers from vehicle- and chemical-exposed groups were reverse-transcribed into cDNAs using the High Capacity cDNA Reverse Transcription Kit (Life Technologies, California). The resulting cDNA products were amplified by qPCR, using the Sso Advanced Universal SYBR Green Supermix in a Bio-Rad CFX384 Real-Time PCR Detection System (Bio-Rad, Hercules, California). The primers for all qPCR reactions were synthesized by Integrity DNA Technologies (Coralville, Iowa), and primer sequences are shown Supplementary Table S3. Data are expressed as % of the expression of the housekeeping gene Gapdh.
Statistics
Statistically significant differences among the 4 experimental groups were determined using 1-way analysis of variance (ANOVA) followed by Duncan’s post hoc test (p < .05). Two-way hierarchical clustering dendrograms were generated using JMP version 13 (SAS Institute Inc., Cary, North Carolina). Linear discriminant analysis effect size (LefSe) (SegaTa et al., 2011) was used for high-dimensional biomarker discovery for the 16S rDNA data at the species level. Pearson’s correlation analysis between metabolites and bacteria was performed using R.
RESULTS
Vehicle Effect on Adult Fecal Microbiome
Before investigating the effect of early-life exposure to environmental chemicals on the adult fecal microbiome, it is important to determine the effects of early-life exposure to vehicle (corn oil) on the microbiome in adult age. As shown in Supplementary Figures S1−S3, we compared the adult fecal microbiome between the no-treatment group and the vehicle-exposed group. Early life exposure to corn oil persistently decreased the overall richness (alpha diversity) of the fecal microbiome in adult age (Supplementary Figure S1A), and principal coordinate analysis showed that there was a distinct separation between the 2 groups (beta diversity) (Supplementary Figure S1B). Among the top 15 most enriched taxa, within the Firmicutes phylum, early life vehicle exposure persistently decreased a taxon in the S24-7 family and a taxon in the Lactobacillus genus, but persistently increased a taxon in the Clostridiales order, a taxon in the Lachnospiraceae family, and Ruminococcus gnavus. Within the Bacteroidetes phylum, early life vehicle exposure persistently decreased Bacteroides acidifaciens and another taxon of the same genus, but persistently increased a taxon in the Odoribacter genus. In addition, early life vehicle exposure persistently increased a taxon in the RF39 order in the Tenericutes phylum (Supplementary Figure S1C). The over-represented taxa in the adult fecal microbiome from the developmentally vehicle-exposed group versus the naïve no treatment group are shown in Supplementary Figures S2 (LefSe microbial markers) and S3 (a 2-way clustering dendrogram).
Effect of Early Life Exposure to Environmental Chemicals on Alpha and Beta Diversities of the Adult Fecal Microbiome
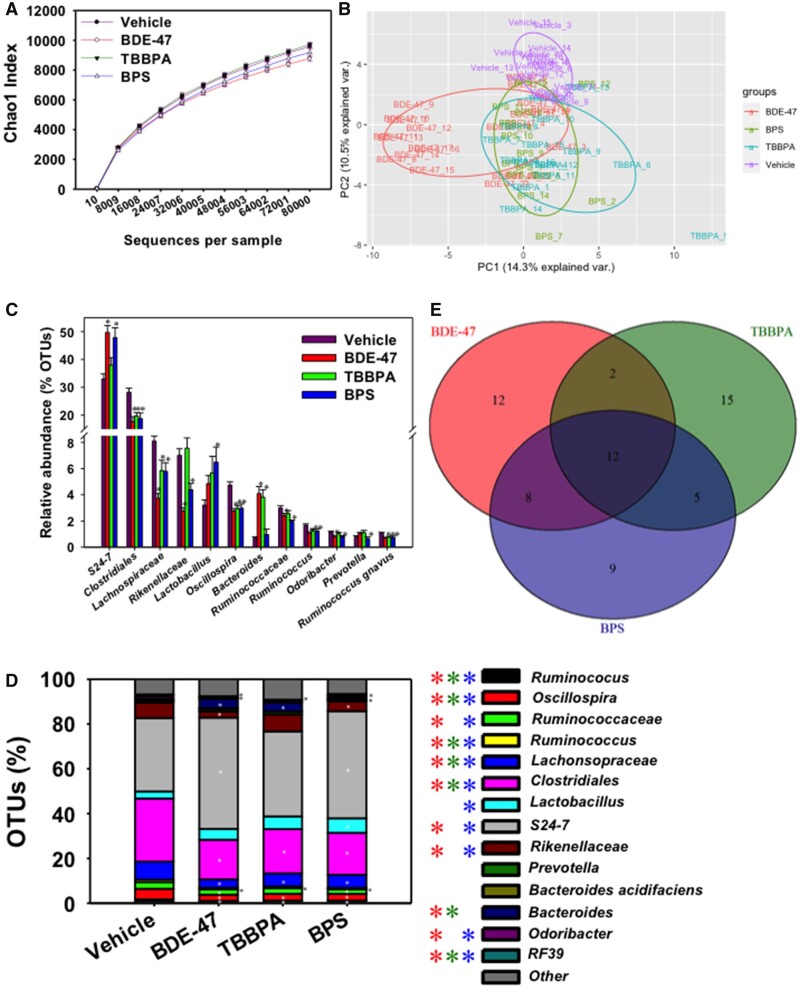
As shown in Figure 2A, early life exposure to BDE-47, TBBPA, or BPS did not markedly alter the overall richness (alpha diversity) of the fecal microbiome adult male pups compared with control groups. However, as shown in Figure 2B and Supplementary Figure S1D, beta diversity illustrated by principal coordinate analysis (weighted unifrac) showed that all 3 environmental chemical-exposed groups tended to separate from the vehicle-exposed group. In addition, the BDE-47-exposed group tended to show more distinct separation from the TBBPA- and BPS-exposed groups (Figure 2B; Supplementary Figure S1D).
Figure 2.
Composition and diversity of CD-1 mice that were maternally exposed to vehicle or one of the 3 chemicals (BDE-47, TBBPA, and BPS). A, Alpha diversity of vehicle and chemical exposure groups via chao1 index. B, Beta diversity of vehicle and exposure groups as shown by a PCA 2D plot. C, The top 12 most abundant and differentially regulated taxa (at L7 level). Asterisks represent statically significant changes as compared with vehicle-exposed group (1-way ANOVA followed by Duncan’s post hoc test, p < .05). D, Composition of L7 bacteria species in vehicle and chemical exposure groups. The top 15 most abundant taxa are shown (the remaining taxa are summed and shown in the “Other” category). Asterisks represent statically significant changes as compared with vehicle-exposed group (1-way ANOVA followed by Duncan’s post hoc test, p < .05). E, Venn diagram of numbers of commonly and uniquely regulated bacteria across all 4 exposure groups.
Effect of Early Life Chemical Exposure to Environmental Chemicals on the Relative Abundance of the Top 15 Taxa in the Adult Fecal Microbiome
The top 12 most abundant and differentially regulated taxa are shown in Figure 2C. Interestingly, most of the bacteria from the Firmicutes phylum were down-regulated by early life chemical exposure. For example, within the Firmicutes phylum, all 3 environmental chemicals persistently decreased the relative abundance of R. gnavus, and another taxon unknown at the species level a taxon in the Oscillospira genus, a taxon in the Lachnospiraceae family, and a taxon in the Clostridiales order; in addition, early life BDE-47 and BPS-exposure persistently decreased the relative abundance of a taxon in the Ruminococcaceae family. The only exception to the down-regulatory pattern within the abundant taxa of the Firmicutes phylum was a taxon in the Lactobacillus genus, which was persistently increased by early life BPS-exposure. Within the Bacteroidetes phylum, the relative abundance of Prevotella and B. acidifaciens remained unchanged following early life exposure to any of the 3 environmental chemicals. Conversely, a taxon within the S24-7 family, which was most relatively abundant within the entire fecal microbiome, was persistently increased following early life exposure to BDE-47 and BPS. In addition, a taxon in the Bacteroides genus was persistently increased following early life exposure to BDE-47 and TBPPA. A taxon in the Rikenellaceae and a taxon in the Odoribacter genus were persistently down-regulated following early life exposure to BDE-47 and BPS. In addition, a taxon in the RF-39 order of the Tenericutes phylum was persistently decreased by all 3 chemicals. There are 2 other abundant taxa that were not altered by early life chemical exposure, namely a taxon in the Prevotella genus, as well as B. acidifaciens (Figure 2D).
In summary, among the top 15 most abundant taxa in the adult fecal microbiome, early life environmental chemical exposure appeared to down-regulate most bacteria in the Firmicutes phylum; whereas there were less abundant taxa in the Bacteroidetes phylum that were persistently regulated by these chemicals. However, notably, the most abundant taxon in the adult fecal microbiome, namely S24-7 in the Bacteroidetes phylum, was persistently upregulated by early life exposure to BDE-47 and BPS.
Common and Unique Microbial Targets in Adult Fecal Microbiome Following Early Life Exposure to Environmental Chemicals
In the fecal microbiome of adult pups, there were 12 taxa that were commonly regulated by early life exposure to all 3 chemicals (Figure 2E), and most of them belong to the Firmicutes phylum and were persistently down-regulated, including a taxon in the Peptococcaceae family, a taxon in the Allobaculum genus (which is an antiobesity) microbial biomarker (Ravussin et al., 2012), a taxon in the order of RF39, a taxon in the Dorea genus, a taxon in the Bacillus genus, a taxon in the Clostridiales order, a taxon in the Lachnospiraceae family, and a taxon in the Enterococcus genus (Figure 3A). Bacteroidetes uniformis was the only taxon in the Bacteroidetes phylum that was persistently downregulated by early life exposure to all 3 environmental chemicals (Figure 3A). A previous study explored the effects of post-natal low-dose exposure to different human relevant environmental chemicals (DEP, MPB, TCS, and MIX) on the fecal microbiome that showed Prevotella genus and Bacilli class were reduced in all treated groups (Hu et al., 2016). These 12 commonly differentiated taxa from 3 different chemicals suggest that there may be common mechanisms in altering the fecal microbiome.
Figure 3.
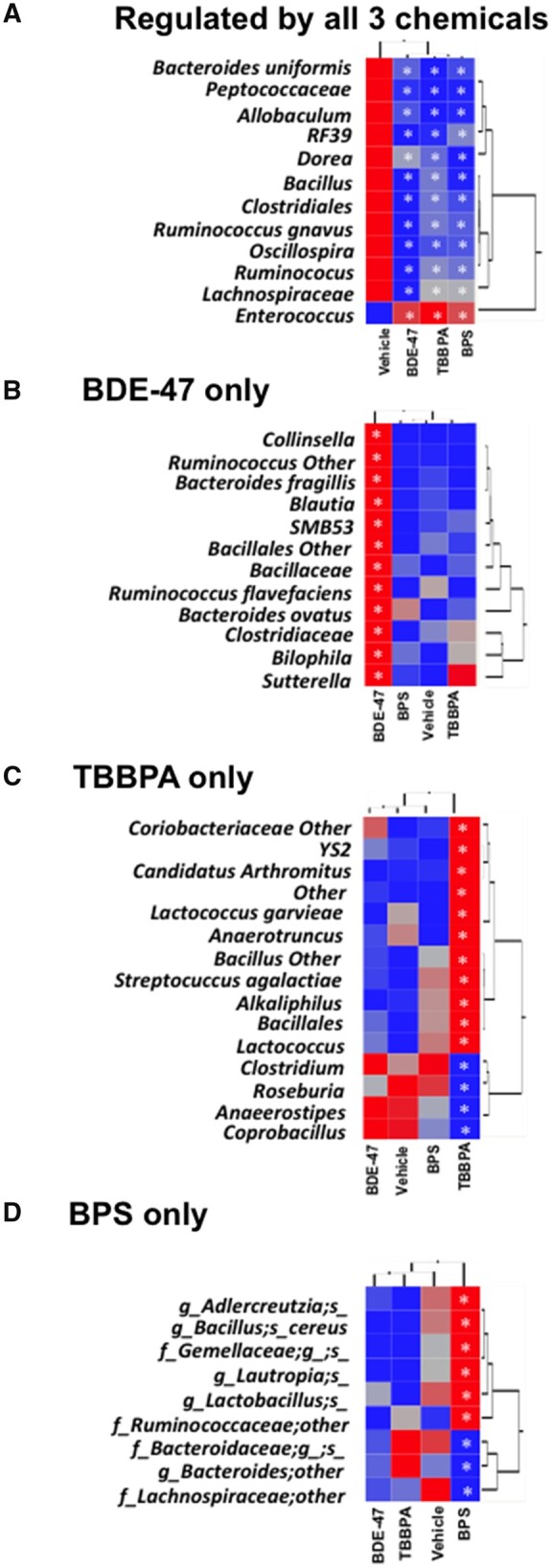
Two-way hierarchical clustering dendrograms showing commonly and uniquely regulated taxa by early life chemical exposures. A, Bacteria that were commonly regulated by all 3 chemicals. B, Bacteria uniquely regulated by BDE-47. C, Bacteria uniquely regulated by TBBPA. D, Bacteria uniquely regulated by BPS. Asterisks represent statically significant changes as compared with vehicle-exposed group (1-way ANOVA followed by Duncan’s post hoc test, p < .05).
There were 12 taxa that were persistently regulated by early life BDE-47 exposure (Figure 2D), and interestingly, all of these bacteria were upregulated, including a taxon in the Collinsella genus from the Actinobacteria phylum, 7 taxa in the Firmicutes phylum (a taxon in the Ruminococcus genus, the Blautia genus, the SMB53 genus, the Clostridiales order, the Bacillaceae family, Ruminococcus flavefaciens, and the Clostridiaceae family). In addition, 2 taxa in the Bacteroidetes phylum (Bacteroides fragilis and Bacteroides ovatus), and 2 taxa in the Proteobacteria phylum (a taxon in the Bilophila genus and a taxon in the Sutterella genus) were also upregulated by BDE-47 (Figure 3B).
Of the 15 taxa that were persistently regulated by early life TBBPA exposure (Figure 2D), 11 were persistently up-regulated, and 4 were persistently down-regulated (Figure 3C). The up-regulated bacteria include a taxon in the Coriobacteriaceae family, a taxon in the YS2 order, a taxon in the Candidatus arthromitus genus, Lactococcus garvieae, a taxon in the Anaerotruncus genus, a taxon in the Bacillus genus, Streptococcus agalactiae, a taxon in the Alkaliphilus genus, a taxon in the Bacillales order, and a taxon in the Lactococcus genus. The downregulated bacteria included a taxon in the Clostridium genus, a taxon in the Roseburia genus, a taxon in the Anaerostipes genus, and a taxon in the Coprobacillus genus (Figure 3C).
There were 9 taxa that were persistently regulated by early life BPS exposure (Figure 2D), 6 of which were persistently up-regulated, and 3 were persistently downregulated. The upregulated taxa include a taxon in the Adlercreutzia genus, Bacillus cereus, a taxon in the Gemellaceae family, a genus in the Lautropia genus, a taxon in the Lactobacillus genus, a taxon in the Ruminococcaceae family, a taxon in the Bacteroidaceae family, a taxon in the Bacteroides genus, and a taxon in the Lachnospiraceae family (Figure 3D). Meanwhile, the down-regulated taxa include a taxon in the Bacteroidaceae family, a taxon in the Bacteroides genus and a taxon in the Lachnospiraceae family.
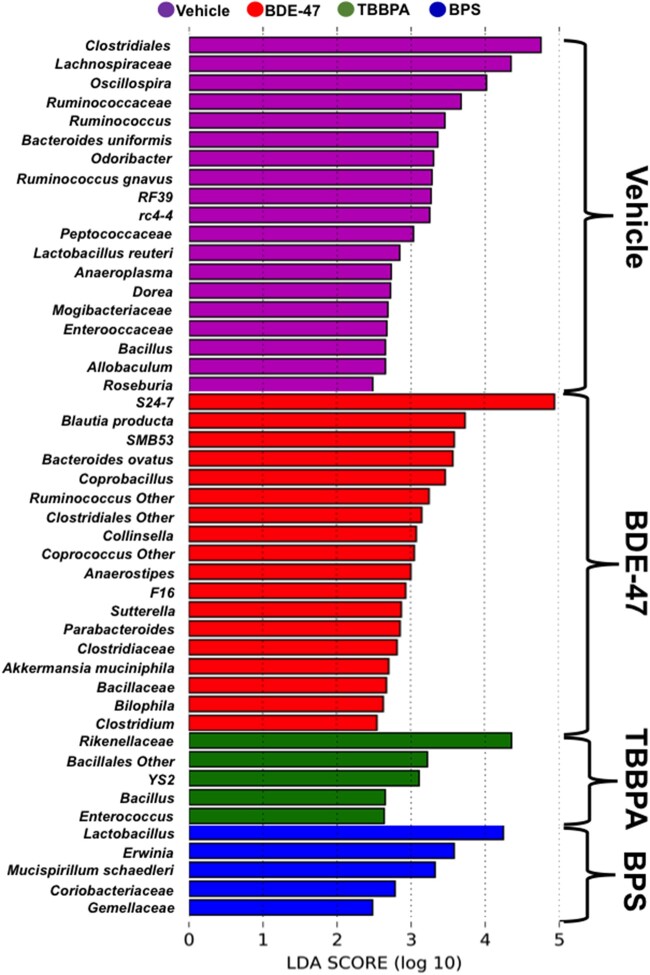
Persistent Unique Microbial Biomarkers Associated With Each Exposure
As shown in Figure 4, the most prominent adult microbial biomarkers from the early life vehicle-exposed groups include Clostridiales, Lachnospiraceae, Oscillospira, Ruminococcaceae, Ruminococcus, B. uniformis, Odoribacter, R. gnavus, RF39, rc-4-4, Peptococcaceae, Lactobacillus reuteri, Anaeroplasma, Dorea, Mogibacteriaceae, Enterococcaceae, Bacillus, Allobaculum, and Roseburia. The most prominent adult microbial biomarkers in the early life BDE-47-exposed group include S24-7, Blautia, SMB53, B. ovatus, Coprobacillus, Ruminococcus Other, Clostridiales Other, Anaerostipes, F16, Sutterella, Parabacteroides, Clostridiaceae, Akkermansia muciniphila, Bacillaceae, Bilophila, and Clostridium. The most prominent adult microbial biomarkers in the early life TBBPA-exposed group include Rikenellaceae, Bacillales Other, YS2, Bacillus, and Enterococcus; whereas the most prominent adult microbial biomarkers in the early life BPS-exposed group include Lactobacillus, Erwinia, Mucispirillum schaedleri, Coriobacteriaceae, and Gemellaceae.
Figure 4.
Microbial biomarkers associated with each exposure group using Lefse.
In summary, early life exposure to these three environmental contaminants resulted in distinct alterations in the adult fecal microbiome of chemically exposed groups. Although the overall richness of the microbiome was not markedly altered, early life exposure to all 3 chemicals resulted in a shift in microbiome structure as compared with the unexposed group. Most commonly regulated taxa by all 3 chemicals were persistently downregulated, whereas most uniquely regulated taxa for each chemical were persistently upregulated. These uniquely regulated taxa could be used as valuable biomarkers to assess the exposure history.
Bioinformatics Predictions of Key Taxa That Drive Persistent Microbial Functional Changes Using FishTaco
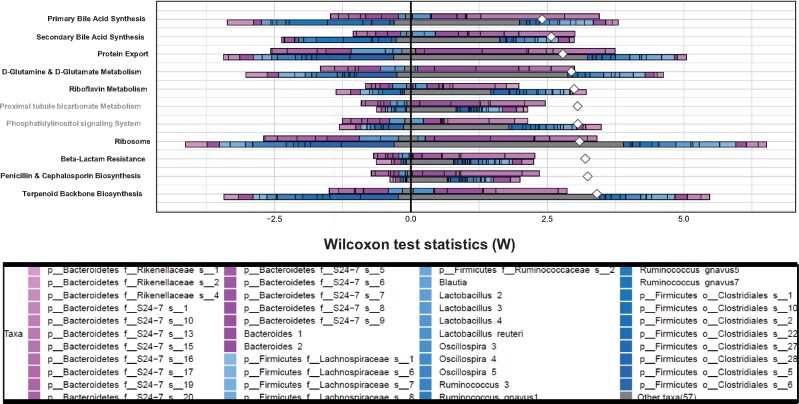
I. BDE-47. As shown in Figure 5, FishTaco (Manor and Borenstein, 2017) predicted that 9 relevant pathways were persistently altered in adult fecal microbiome following early life BDE-47 exposure. Although FishTaco predicted proximal tubule bicarbonate metabolism and phosphatidylinositol signaling system, they were removed from downstream analysis because they are not relevant to bacterial functions. The 9 pathways include BA metabolism (including primary and secondary BA synthesis), protein synthesis and transport (protein export, d-glutamine, and d-glutamate metabolism, ribosome), vitamin B2 (riboflavin) metabolism, membrane lipid and cell wall synthesis (phosphatidylinositol signaling system, beta-lactam resistance, penicillin and cephalosporin biosynthesis), as well as terpenoid backbone biosynthesis. Interestingly, the community-wide contribution to the shift in most basal microbial functions appeared to be weakened, and most notably for the following pathways: Terpenoid backbone synthesis, ribosome, phosphatidylinositol signaling system, riboflavin metabolism, d-glutamine and d-glutamate metabolism, and protein export. The taxa that contributed to the shift in these pathways in the BDE-47-exposed group were also much less diverse compared with the vehicle-exposed group (note: individual plots are shown in Supplementary Figs. 5 − 39).
Figure 5.
Predicted relative contributions of metabolic and regulatory bacteria to the shift in the pathway’s abundance by FishTaco for the early life BDE-47-exposed group (top bar of each pair) versus the vehicle-exposed group (bottom bar of each pair) and attenuators (left side).
Regarding BA metabolism, S24-7 appeared to contribute to this pathway under both control and BDE-47 exposed conditions; however, there were more taxa in this family that are predicted to drive the shift in BA metabolism under BDE-47 exposed conditions. Control group-enriched taxa such as Oscillospira and Lacnospiraceae (which lack the BA metabolizing enzymes), as well as BDE-47 exposure group-enriched taxa such as Lactobacillus and Bacteroides (which have BA-metabolizing enzymes), both contributed to the BDE-47-mediated functional shift toward BA metabolism.
Regarding protein-processing related pathways (protein export, d-glutamine and glutamate metabolism, and ribosome), control group-enriched taxa such as Oscillospira, Lachnospiraceae, and Lactobacillus (which lack the related enzymes), as well as BDE-47 exposure group-enriched taxa such as S24-7 and Bacteroides (which have the related enzymes), both contributed to the BDE-47-mediated functional shift of these protein-processing pathways.
Regarding vitamin B2 (riboflavin) metabolism, control group-enriched taxa such as Clostridiales, Oscillospira, Lachnospiraceae (which lack the related enzymes), as well as BDE-47 exposure group-enriched taxa such as Lactobacillus and Bacteroides (which have the related enzymes), both contributed to the BDE-47-mediated functional shift of this pathway.
Regarding bacterial cell wall functions (beta-lactam resistance, penicillin and cephalosporin biosynthesis), as well as terpenoid backbone synthesis, control group-enriched taxa such as Oscillospira and Lachnospiraceae (which lack the related enzymes), as well as BDE-47 exposure group-enriched L. reuteri and Bacteroides (which have the related enzymes), both contributed to the BDE-47-mediated functional shift of these pathways.
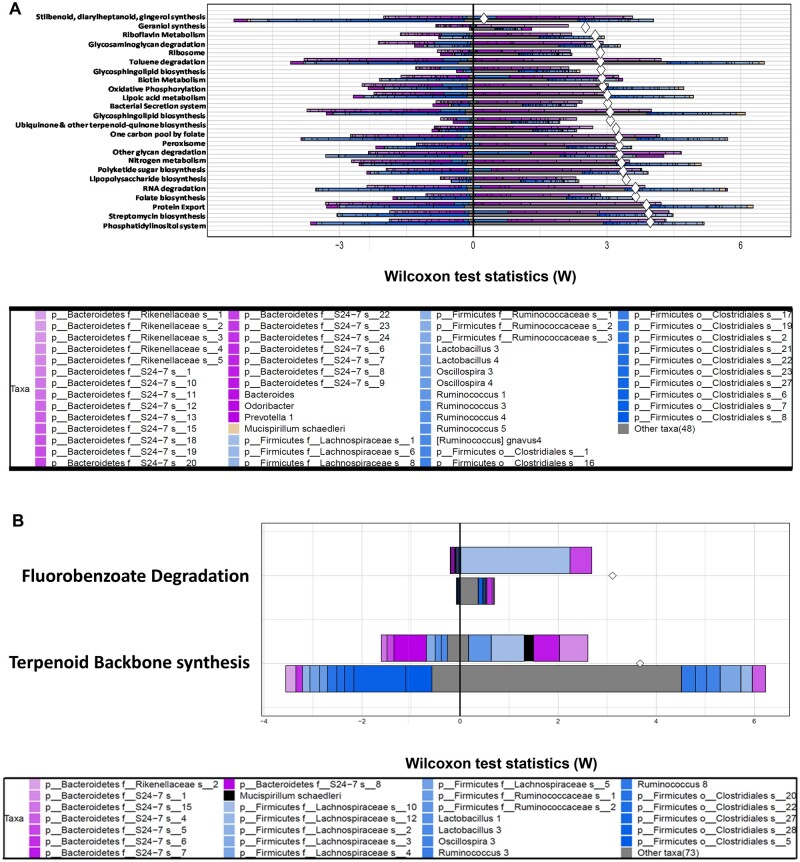
II. TBPPA. As shown in Figure 6A, FishTaco predicted that early life TBBPA exposure persistently altered the greatest number of pathways in the fecal microbiome (24 in total) of adult pups, as compared with the other chemicals.
Figure 6.
A, Predicted relative contributions of metabolic and regulatory bacteria to the shift in the pathway’s abundance by FishTaco for the early life TBPPA-exposed group (top bar of each pair) versus the vehicle-exposed group (bottom bar of each pair) and attenuators (left side). B, Predicted relative contributions of metabolic and regulatory bacteria to the shift in the pathway’s abundance by FishTaco for the early life BPS-exposed group (top bar of each pair) versus the vehicle-exposed group (bottom bar of each pair) and attenuators (left side).
Interestingly, similar to BDE-47 exposed group, the community-wide contribution to most basal microbial functions appeared to be weakened, and most notably for the following pathways: phosphatidylinositol system, protein export, folate biosynthesis, RNA degradation, nitrogen metabolism, one carbon pool by folate, bacterial secretion system, oxidative phosphorylation, biotin metabolism, and ribosome. Conversely, the community-wide contribution to most basal microbial functions appeared to be greater for the stilbenoid, diarylheptanoid, and gingerol synthesis pathway. In general, most of these pathways were contributed by multiple taxa labeled in the “Other” category, followed by many taxa in the Firmicutes phylum. Interestingly, following early life TBPPA exposure, much less diversity of the contributing microbes to the pathway’s shift was observed, and these functions were largely contributed by S24-7 (and to a lesser extent, Rikenellaceae) in the Bacteroides phylum.
III. BPS. As shown in Figure 6B, FishTaco predicted that early life BPS exposure persistently altered 2 pathways in the fecal microbiome of adult pups, namely fluorobenzoate degradation and terpenoid backbone synthesis. Fluorobenzoate belongs to a class of fluorinated aromatic compounds that are often used as pesticides (Murphy et al., 2009). Fluoroorganics pose a potential human health concern, and gut microbiome has been shown to be primarily responsible for fluorobenzoate degradation in vivo, whereas host only has minor contributions (Sridharan et al., 2014). In vehicle exposed control groups, a taxon in the Clostridiales order and a taxon in the S24-7 family were predicted to be the driving force for fluorobenzoate degradation; whereas following BPS early life exposure, these taxa were replaced by a taxon in the Lachnospiraceae, which became the major driving force for this pathway, as well as a different taxon in the S24-7 family.
Terpenoid backbone synthesis is important for many important biological functions such as membrane formation, glycoprotein synthesis, and intracellular electron transport (Goldstein and Brown, 1990). It has been shown that both the gut microbiome and the host contribute to terpenoid backbone synthesis in vivo, although host-specific contributions are minimal (Sridharan et al., 2014).
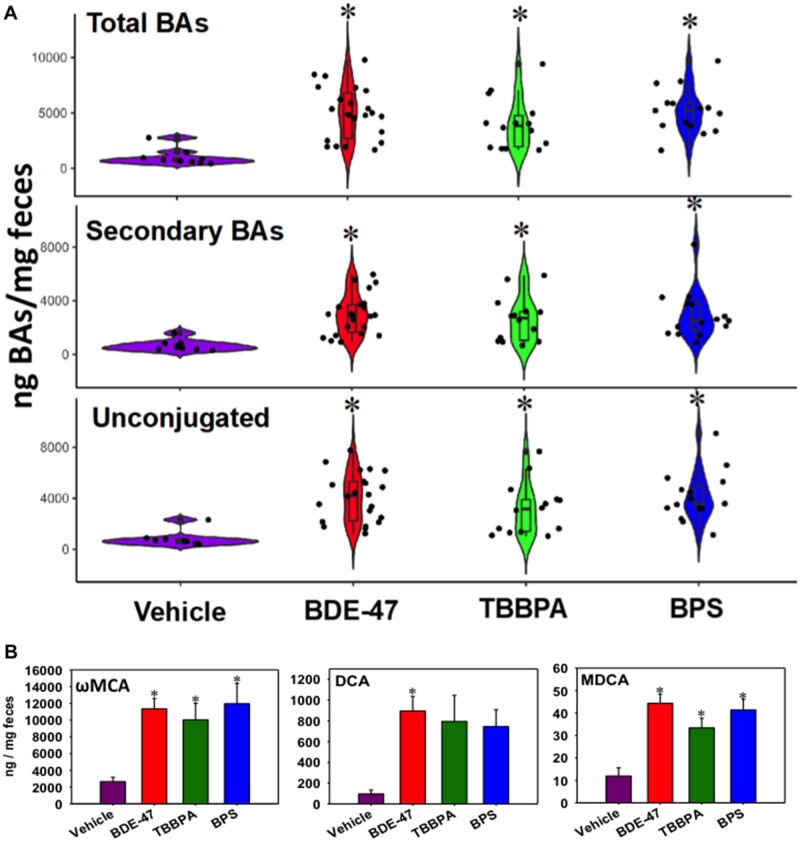
Fecal BA Quantification Using LC-MS
As shown in Figure 7A, early life exposure to all 3 chemicals markedly increased the fecal total, secondary, and unconjugated BAs. Specifically, regarding the secondary unconjugated BAs, which can only be synthesized by the gut microbiome, ωMCA (the most abundant) and MDCA were persistently up-regulated by early life exposure to all chemicals, whereas DCA was persistently up-regulated by BDE-47, and tended to be upregulated by the other 2 chemicals (although statistical significance was not achieved). Supplementary Figure S4A shows the correlation amongst all differentially regulated bacteria and all differentially regulated BAs. Most notably, Bacillales taxa and streptococcus genus positively associated with T-UDCA, T-CDCA, and T-CA; whereas the uniformis species negatively correlated with a number of BAs including MDCA, ωMCA, UDCA, CDCA, and βMCA. The fragilis species, Turicibacter genus, YS2 order, and the Clostridiales order had a positive correlation with αMCA. The Coprococcus genus had a strong positive correlation with T-CDCA.
Figure 7.
A, Fecal total, secondary, and unconjugated BAs in vehicle- and chemical-exposed groups as determined by LC-MS. B, Individual unconjugated secondary BAs that were differentially regulated by early life chemical exposures. Asterisks represent statically significant changes as compared with vehicle-exposed group (1-way ANOVA followed by Duncan’s post hoc test, p < .05).
Hepatic Expression of BA-Processing Genes and BA Quantification
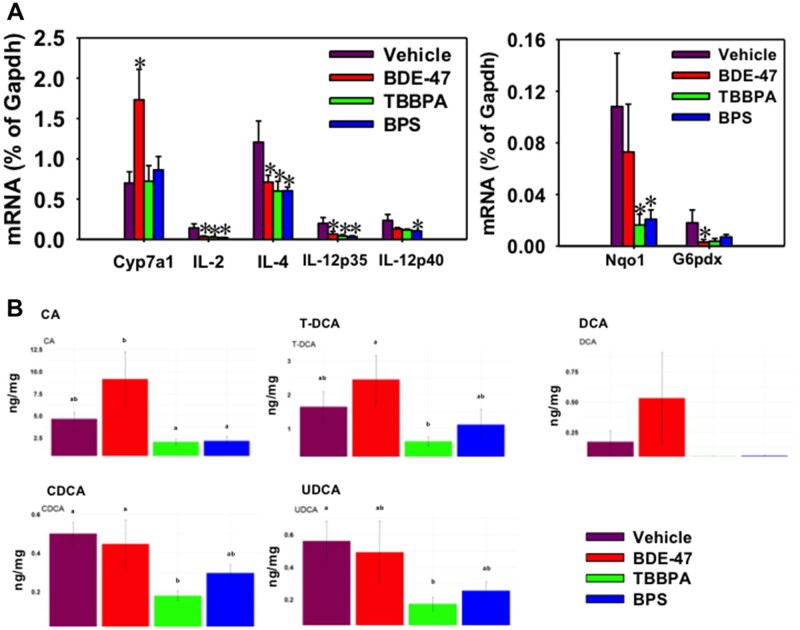
Because both bioinformatics predictions using Fishtaco and targeted BA metabolomics in feces indicated increased BA synthesis and metabolism capacities by BDE-47, we tested our hypothesis that the hepatic BA synthesis or transport may also contribute to the BA increase within the gut-liver axis. Indeed, the hepatic rate-limiting enzyme for BA synthesis, namely Cyp7a1, was persistently upregulated by early life BDE-47 exposure (Figure 8A), and this corresponds to a general trend in the increase in its downstream product CA and its taurine conjugated form (T-CA). The microbial metabolite of CA, namely DCA, also appeared to be increased, although with notable variations (Figure 8B). Interestingly, the hepatic levels of CDCA and UDCA were decreased by early life exposure to TBBPA. There were no changes in other BAs in liver by any of the 3 chemicals. In addition, there was no change in other BA synthetic enzymes (Cyp8b1, Cyp27a1, Cyp7b1) or transporters (Ntcp, Bsep, Oatp1b2) (data not shown), by any of the 3 chemicals.
Figure 8.
A, Hepatic mRNA expression of the rate-limiting BA-synthetic enzyme Cyp7a1, inflammatory cytokines (IL-2, IL-4, IL-12p35, and IL-12p40) (left panel), as well as Nqo1 (the oxidative stress sensor Nrf2-target gene) and G6pdx (glucose metabolism). The mRNAs were quantified using RT-qPCR as described in Materials and Methods section. Data are normalized to the house-keeping gene Gapdh. Asterisks represent statistically significant differences as compared with the vehicle group (p < .05, one-way ANOVA followed by Duncan’s post hoc test). B, Hepatic BAs that were differentially regulated in adult age following early life chemical exposure. BAs were quantified using ultraperformance liquid chromatography coupled with mass spectrometry in tandem as described in Materials and Methods section. The letters a and b represent different post hoc groups (1-way ANOVA followed by Duncan’s post hoc test).
Hepatic Expression of Genes Involved in Inflammation, Oxidative Stress, and Metabolic Syndrome
Because it is known that under physiological conditions, secondary BAs such as DCA are protective against intestinal inflammation during a mouse model of colitis (Sinha et al., 2020), we wanted to extend our investigation to liver, and determine to what extent the persistent increase of secondary BAs within the gut-liver axis contribute to the hepatic pathways involved in inflammation. And because inflammation is a key contributor to oxidative stress and metabolic syndrome (Fernandez-Garcia et al., 2014), we also investigated pathways involved in oxidative stress pathway and metabolic syndrome. Interestingly, the proinflammatory cytokines IL-2, IL-4, IL-12p35, and IL-12p40 were all persistently downregulated in liver by early life exposure to all 3 chemicals (Figure 8A). There was no change in the expression of other cytokines that we quantified (GM-CSF, IFNgamma, IL-1b, IL-10, MCP-1, or TNFα) (data not shown). The prototypical target gene of the major oxidative stress sensor Nrf2, namely Nqo1, was persistently downregulated by TBBPA and BPS, and tended to be downregulated by BDE-47. There was no change in the other Nrf2 target genes, namely Gclc and Gclm (data not shown). The mRNA of G6pdx, which is the rate-limiting enzyme involved in the oxidative branch within the pentose phosphate pathway, was persistently decreased by BDE-47 and also tended to decrease by TBBPA and BPS. The deficiency of G6pdx has been shown to produce dyslipidemia through elevating adipose and body mass, as well as serum free fatty acids and triglyceride during diet induced obesity (Hecker et al., 2012). There was no change in Fgf21, which is a master regulator of carbohydrate and lipid homeostasis (Emanuelli et al., 2014) (data not shown). Together these observations suggest that there is a general trend of immune suppression and oxidative stress, but a mild increase in the risk of dyslipidemia, following early life exposure to these chemicals.
Fecal Short- and Medium-Chain Fatty Acids Quantification Using GC-MS
As shown in Figure 9, early life exposure to BPS persistently decreased acetic acid, whereas early life exposure to TBBPA persistently increased propionic acid and succinate. Supplementary Figure S4B illustrates correlation between differentially regulated SCFAs acetic acid, propionic acid and succinate, and all differentially regulated taxa. Acetic acid was positively correlated most notably with the Coprococcus genus, Dorea genus, Blautia genus, and Collinsella genus. Propionic acid has shown a negative correlation with the Erwinia genus, as most notably. Succinate had a negative correlation with the genus rc4 and a slight positive correlation with the Ruminococcaceae family.
Figure 9.
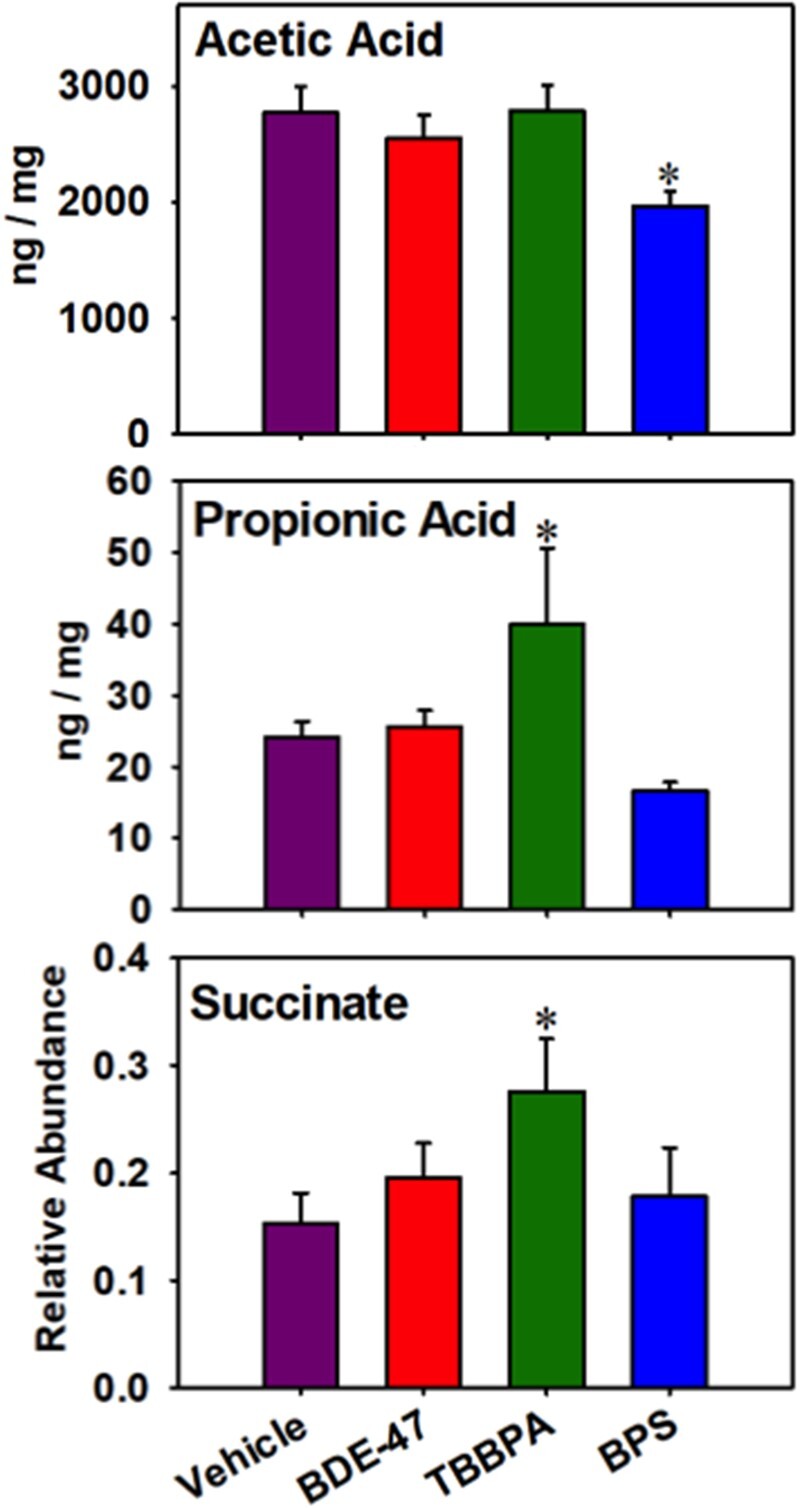
Fecal short- and medium-chain fatty acids that were differentially regulated by early life chemical exposures. Asterisks represent statically significant changes as compared with vehicle-exposed group (1-way ANOVA followed by Duncan’s post hoc test, p < .05).
DISCUSSION
The present study has demonstrated that early life exposure to the 3 chemicals persistently increased the fecal BA output, and most notably, multiple secondary unconjugated BAs that can only be produced from the gut microbiome. In addition, there appeared to be a functional shift in the types microbes that are responsible for BA metabolism following chemical exposure, with a lot less community-wide diversity and a focused cluster within the Firmicutes phylum especially S24-7, which is a hallmark for early life BDE-47 exposure and is predicted by FishTaco to contribute to both primary and secondary BA synthesis. To add, S24-7 has also been shown to be positively correlated with hepatic T-CDCA (a primary BA) and T-DCA (a secondary BA), and with cecum DCA (a secondary BA), as reported in a previous previously on a liver regeneration study (Liu et al., 2016). The predicted contribution of S24-7 in the shift in abundance of the primary BA synthesis pathway is likely due to an indirect role through the enhanced excretion of liver-derived primary BAs. This hypothesis is based on the previous observation that S24-7 positively associates with the hepatic efflux transporter Mdr1 (Liu et al., 2016), which can transport BAs as an adaptive response to increased levels of BAs (Kneuer et al., 2007). In our study, the major hepatic BA transporters remained unchanged at the mRNA level. However, post-transcriptional activities of these transporters may contribute to the increase in fecal BA output from liver. Although secondary BAs at exceedingly high concentrations are toxic to the host due to their hydrophobic nature where at high concentrations can induce inflammation and cytotoxicity (Woolbright and Jaeschke, 2019), under physiological conditions, secondary BAs protect against intestinal inflammation in a mouse model of colitis, suggesting that they may have immune-suppression effects in intestine. In addition, increased BAs may also lead to metabolic disorders (Sun et al., 2019), and the 3 chemicals (BDE-47, TBBPA, and BPS) have all been reported to dys-regulate intermediary metabolism pathways related to obesity and diabetes (Rezg et al., 2019; Suh et al., 2017; Zhang et al., 2016). This aligns with the hepatic decrease in G6pdx, the deficiency which is known to produce dyslipidemia in a mouse model of obesity (Hecker et al., 2012). Therefore, higher levels of secondary BAs and unconjugated BAs may contribute to environmental chemicals mediated toxicities such as immune suppression and metabolic syndrome.
A previous study done with BDE-47 showed increased expression of BA-processing genes such as Cyp7a1, which is the rate-limiting enzyme for host BA synthesis (Khalil et al., 2018). Our study aligns with the previous findings, and has provided further evidence that BDE-47 not only acutely but also persistently up-regulates Cyp7a1 gene expression. Cyp7a1 is a classic example of feedback inhibition whereby its end products directly inhibit the first enzyme (Chiang and Ferrell, 2018). Another study has shown that increased Cyp7a1 expression may be due to the reduced FXR signaling primarily induced by the gut microbiome (Li et al., 2018a). As such, decreased diversity and abundance of certain BA regulating bacteria may lead to reduced FXR signaling, due to lack of feedback inhibition, thus increasing the expression of Cyp7a1, that may contribute to an overall increase in BA synthesis.
Regarding the contribution of various taxa in BA metabolism among the 3 chemical exposure groups: (1) following BDE-47 exposure, within the Bacteroides genus, the species of fragilis and ovatus were persistently increased. The species of B. fragilis has been proved to have bile salt hydrolase activity, which is important for deconjugating primary BAs (Stellwag and Hylemon, 1976). The increase in deconjugated BAs for BDE-47 may be in part due to the persistent increase in the B. fragilis species. The dihydroxylation of primary BAs, such as CDCA and CA, are executed by a subset of genera of Clostridium and Eubacterium species (Gerard, 2013) into DCA and LCA. Additionally, it’s been shown that the dihydroxylation of primary BAs was enhanced when Eubacterium combined with strains of Bacteroides genus (Hirano and Masuda, 1982). This may contribute to the increase of unconjugated and secondary BAs for BDE-47 exposure. A study done in 2013 stated that advanced cirrhosis patients had higher primary BAs, but much lower secondary BAs. To add, cirrhosis patients also had lower Blautia bacterial concentrations, which positively correlated with LCA/CDCA and Ruminococcaceae with a positive correlation with DCA/CA and lower Lachnospiraceae concentrations (Kakiyama et al., 2013). This suggests that the genus Blautia along with Ruminococcaceae and other Lachnospiraceae taxa may play a pivotal role in the conversion of primary BAs into secondary BAs. The increased abundance of the genus Blautia, Ruminococcus genus, and Lachnospiraceae family by early life BDE-47 exposure may play a role in the increased secondary BA synthesis. (2) TBPPA also resulted in an overall increase in unconjugated and secondary BAs, and this positively associated with an increase in the genus of Anaerotruncus, from the Ruminococcaceae family. A recent study was performed using antibiotics to test childhood obesity and the results was that the overall concentration of secondary BAs decreased with an increase in primary BAs, conversely with a decrease in the genus of Anaerotruncus (Li et al., 2017). This suggests that Aneurotruncus plays a role in regulating secondary BA synthesis. (3) Following BPS exposure, Lactobacillus is the most prominent microbial biomarker that is persistently increased in feces of adult pups. Lactobacillus is known to have Bile salt hydrolase activity, which deconjugates primary BAs (more specifically glycine and taurine linked bile salts) (Kumar et al., 2006). Conversely, there was a persistent decrease in the Bacteroidaceae family, Bacteroides genus, and Lachnospiraceae family.
The present study has identified that early life TBBPA exposure persistently increased propionic acid and succinate in feces of adult pups, whereas early life BPS decreased acetic acid in feces of adult pups. Propionic acid and acetic acid are mainly produced by the class of bacteria belonging in the Bacteroidetes phylum (Chakraborti, 2015). Propionic acid has been shown to lower fatty acid content in liver and plasma, reduce food intake, and have immunosuppressive effect through inhibiting NF-κB (Al-Lahham et al., 2010). This aligns with the hepatic decrease in several pro-inflammatory cytokines and the oxidative stress target gene.
Coincidentally, TBBPA has also been shown to have immunosuppressive effect in human natural killer cells (Kibakaya et al., 2009). Propionic acid may serve as a novel player in TBBPA-mediated immunosuppression.
However, it should also be noted that elevated propionic acid may have other important roles such as inducing satiety and stimulating glycogenolysis and hyperglycemia in mice (Tirosh et al., 2019). This coincides with the rise in succinate in feces of adult pups by TBBPA exposure. Succinate is a key component of TCA cycle, and TBBPA is known to disrupt the TCA cycle during development (Ye et al., 2016). Therefore, TBBPA may have multifaceted roles in the toxicity including alterations in immune functions and carbohydrate metabolism. Acetate constitutes about 60% of total SCFAs in humans, making it an important part of metabolic systems. A decrease in acetate by BPS may lead to decrease in immune-regulatory effects as acetate has been linked to the increase of interferon gamma level (Graham et al., 2015), and BPS is known to produce chronic inflammation (Qiu et al., 2019).
We have previously demonstrated the acute effects of PBDEs on the gut microbiome and BA homeostasis reported decreased alpha diversity of gut microbiome with regard to BDE-47 and BDE-99 (Li et al., 2018b). The mice were orally gavaged starting at 9 weeks daily with 100 µmol/kg for 4 days. The dams in the present study were fed with 0.2 mg/kg from gestational day 8 to the end of lactation (PND 21). Comparing the acute exposure model in adult mice and maternal exposure model in the present study, A. muciniphila is a biomarker for BDE-47 exposure in both studies. In the previous study by Li et al., Ruminoccoceea was down-regulated. In the present study, all Ruminoccoceae related taxa were also down-regulated persistently. No previous studies have shown the effects of the TBBPA or BPS on gut microbiome composition. Our study is among the first to show that early life exposure to these chemicals produced persistent dysbiosis of the fecal microbiome. Future studies are needed to determine the effect of these chemicals on female pups.
Fecal DNA has also been frequently used in many studies in the literature to recapitulate intestinal microbiome and have been shown to well correlate with metabolite milieu of the gut (Baxter et al., 2019). Therefore, we chose to use fecal compartments to recapitulate intestinal microbiome changes. The goal of the present study was to test our hypothesis that early life exposure to environmental toxicants lead to persistent alterations in adult age, therefore, the scope of this work was to focus this adult age time point (20 weeks of age), which was 17 weeks postchemical exposure. However, in future investigations, it is important to assess the full spectrum of the toxicokinetic curve of the microbiome changes involving multiple time points between weaning age and adulthood. It is also important to investigate the regulation of cecal and/or colonic microbiome, as well as the host intestinal tissues in response to early life toxicant exposure.
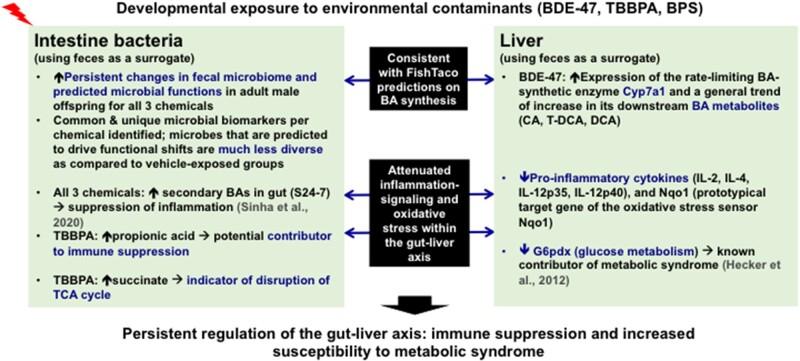
Taken together, as summarized in Figure 10, the present study has used fecal microbiome and host liver from adult male pups that were developmentally exposed to 3 human health-relevant environmental toxicants, namely BDE-47, TBBPA, and BPS, to demonstrate that the gut-liver axis is potentially reprogrammed long after the early life toxic exposure has stopped. Specifically, we have identified both common and unique microbial biomarkers per each chemical, and showed that the microbes predicted to drive various biological functions are much less diverse as compared with unexposed conditions. Furthermore, we validated the bioinformatics predictions of using targeted metabolomics. For BDE-47 exposed group, we showed that increased fecal BA output is a joint effort by increased hepatic synthesis of the primary BA CA as well as increased proportions of gut microbes that carry the BA-metabolizing enzymes. Overall, early life exposure to all 3 chemicals appeared to produce a general trend of immune suppression, evidenced by increased anti-inflammatory secondary BAs and the propionic acid (known immune suppressor) in feces, as well as decreased proinflammatory cytokines in liver. Early life exposure to all 3 chemicals may also increase the risk of metabolic syndrome, evidenced by decreased expression of genes involved in dyslipidemia (G6pdx) and elevated succinate (indicator of disruption of TCA cycle). Therefore, the potentially reprogrammed gut-liver axis following early life exposure to these toxicants may serve as an important mechanism for increased susceptibility to human diseases (such as immune suppression and metabolic syndrome).
Figure 10.
Overall summary of developmental exposure to BDE-47, TBPPA, and BPS producing persistent alterations in the gut-liver axis.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
National Institutes of Health (Grants R01 ES025708; R01 ES030197; R01 GM111381, R25 ES025503); the University of Washington Center for Exposures, Diseases, Genomics, and Environment (P30 ES0007033); the Sheldon Murphy Endowment.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the members of Dr Alexander Suvorov’s laboratory for the collection of liver and fecal compartments, as well as members from the Cui, Suvorov, Gu, and Mani laboratories for editing the manuscript. We would also like to thank Dr Elhanan Borenstein as well as Dr Alex Eng for their technical guidance in running FishTaco and data interpretation.
Dryad Digital Repository DOI: doi:10.5061/dryad.m905qftzn
REFERENCES
- Abb M., Stahl B., Lorenz W. (2011). Analysis of brominated flame retardants in house dust. Chemosphere 85, 1657–1663. [DOI] [PubMed] [Google Scholar]
- Al-Lahham S. H., Peppelenbosch M. P., Roelofsen H., Vonk R. J., Venema K. (2010). Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim. Biophys. Acta 1801, 1175–1183. [DOI] [PubMed] [Google Scholar]
- Anisuzzaman S., Whalen M. M. (2016). Tetrabromobisphenol A and hexabromocyclododecane alter secretion of IL-1beta from human immune cells. J. Immunotoxicol. 13, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter N. T., Schmidt A. W., Venkataraman A., Kim K. S., Waldron C., Schmidt T. M. (2019). Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio 10, e02566-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull M. J., Plummer N. T. (2014). Part 1: The human gut microbiome in health and disease. Integr. Med. (Encinitas) 13, 17–22. [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Pena A. G., Goodrich J. K., Gordon J. I., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborti C. K. (2015). New-found link between microbiota and obesity. World J. Gastrointest. Pathophysiol. 6, 110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. L., Li X., Lehmler H. J., Phillips B., Shen D., Cui J. Y. (2018). Gut microbiota modulates interactions between polychlorinated biphenyls and bile acid homeostasis. Toxicol. Sci. 166, 269–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang J. Y. L., Ferrell J. M. (2018). Bile acid metabolism in liver pathobiology. Gene Expr. 18, 71–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey J. L., Wang D., Siginir G., Fei Q., Raftery D., Gu H., Yue Cui J. (2019). Pharmacological activation of PXR and CAR downregulates distinct bile acid-metabolizing intestinal bacteria and alters bile acid homeostasis. Toxicol. Sci. 168, 40–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., Andersen G. L. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Li P., Yang K., Liu L., Gao H., Zhou G., Zhao Q., Xia T., Wang A., Zhang S. (2020). Promotion of mitochondrial fusion protects against developmental PBDE-47 neurotoxicity by restoring mitochondrial homeostasis and suppressing excessive apoptosis. Theranostics 10, 1245–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelli B., Vienberg S. G., Smyth G., Cheng C., Stanford K. I., Arumugam M., Michael M. D., Adams A. C., Kharitonenkov A., Kahn C. R. (2014). Interplay between FGF21 and insulin action in the liver regulates metabolism. J. Clin. Invest. 124, 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Garcia J. C., Cardona F., Tinahones F. J. (2014). Inflammation, oxidative stress and metabolic syndrome: dietary modulation. Curr. Vasc. Pharmacol. 11, 906–919. [DOI] [PubMed] [Google Scholar]
- Gerard P. (2013). Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 3, 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J. L., Brown M. S. (1990). Regulation of the mevalonate pathway. Nature 343, 425–430. [DOI] [PubMed] [Google Scholar]
- Govindarajan N., Agis-Balboa R. C., Walter J., Sananbenesi F., Fischer A. (2011). Sodium butyrate improves memory function in an Alzheimer's disease mouse model when administered at an advanced stage of disease progression. J. Alzheimers Dis. 26, 187–197. [DOI] [PubMed] [Google Scholar]
- Graham C., Mullen A., Whelan K. (2015). Obesity and the gastrointestinal microbiota: a review of associations and mechanisms. Nutr. Rev. 73, 376–385. [DOI] [PubMed] [Google Scholar]
- Hecker P. A., Mapanga R. F., Kimar C. P., Ribeiro R. F. Jr, Brown B. H., O'Connell K. A., Cox J. W., Shekar K. C., Asemu G., Essop M. F., et al. (2012). Effects of glucose-6-phosphate dehydrogenase deficiency on the metabolic and cardiac responses to obesogenic or high-fructose diets. Am. J. Physiol. Endocrinol. Metab. 303, E959–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S., Masuda N. (1982). Enhancement of the 7 alpha-dehydroxylase activity of a gram-positive intestinal anaerobe by bacteroides and its significance in the 7-dehydroxylation of ursodeoxycholic acid. J. Lipid Res. 23, 1152–1158. [PubMed] [Google Scholar]
- Hu J., Raikhel V., Gopalakrishnan K., Fernandez-Hernandez H., Lambertini L., Manservisi F., Falcioni L., Bua L., Belpoggi F., L.Teitelbaum S., et al. (2016). Effect of postnatal low-dose exposure to environmental chemicals on the gut microbiome in a rodent model. Microbiome 4, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Zhu L., Zhao C., Chen X., Cai Z. (2020). Integration of proteomics and metabolomics reveals promotion of proliferation by exposure of bisphenol S in human breast epithelial MCF-10A cells. Sci. Total Environ. 712, 136453. [DOI] [PubMed] [Google Scholar]
- Ji F., Sreenivasmurthy S. G., Wei J., Shao X., Luan H., Zhu L., Song J., Liu L., Li M., Cai Z. (2019). Study of BDE-47 induced Parkinson's disease-like metabolic changes in C57BL/6 mice by integrated metabolomic, lipidomic and proteomic analysis. J. Hazard Mater. 378, 120738. [DOI] [PubMed] [Google Scholar]
- Johnson-Restrepo B., Adams D. H., Kannan K. (2008). Tetrabromobisphenol A (TBBPA) and hexabromocyclododecanes (HBCDs) in tissues of humans, dolphins, and sharks from the United States. Chemosphere 70, 1935–1944. [DOI] [PubMed] [Google Scholar]
- Kakiyama G., Pandak W. M., Gillevet P. M., Hylemon P. B., Heuman D. M., Daita K., Takei H., Muto A., Nittono H., Ridlon J. M., et al. (2013). Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J. Hepatol. 58, 949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A., Cevik S. E., Hung S., Kolla S., Roy M. A., Suvorov A. (2018). Developmental exposure to 2,2',4,4'-tetrabromodiphenyl ether permanently alters blood-liver balance of lipids in male mice. Front. Endocrinol. (Lausanne) 9, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibakaya E. C., Stephen K., Whalen M. M. (2009). Tetrabromobisphenol A has immunosuppressive effects on human natural killer cells. J. Immunotoxicol. 6, 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B., Colon E., Chawla S., Vandenberg L. N., Suvorov A. (2015). Endocrine disruptors alter social behaviors and indirectly influence social hierarchies via changes in body weight. Environ Health 14, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneuer C., Honscha W., Gabel G., Honscha K. U. (2007). Adaptive response to increased bile acids: Induction of MDR1 gene expression and P-glycoprotein activity in renal epithelial cells. Pflugers Arch 454, 587–594. [DOI] [PubMed] [Google Scholar]
- Kumar R. S., Brannigan J. A., Prabhune A. A., Pundle A. V., Dodson G. G., Dodson E. J., Suresh C. G. (2006). Structural and functional analysis of a conjugated bile salt hydrolase from bifidobacterium longum reveals an evolutionary relationship with penicillin V acylase. J. Biol. Chem. 281, 32516–32525. [DOI] [PubMed] [Google Scholar]
- Langille M. G., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., Clemente J. C., Burkepile D. E., Vega Thurber R. L., Knight R., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C., Liu F., Alomirah H., Loi V. D., Mohd M. A., Moon H. B., Nakata H., Kannan K. (2012). Bisphenol S in urine from the United States and seven Asian countries: Occurrence and human exposures. Environ. Sci. Technol. 46, 6860–6866. [DOI] [PubMed] [Google Scholar]
- Li C. Y., Dempsey J. L., Wang D., Lee S., Weigel K. M., Fei Q., Bhatt D. K., Prasad B., Raftery D., Gu H., et al. (2018. b). PBDEs altered gut microbiome and bile acid homeostasis in male C57BL/6 mice. Drug Metab. Dispos. 46, 1226–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Selmi C., Tang R., Gershwin M. E., Ma X. (2018. a). The microbiome and autoimmunity: a paradigm from the gut-liver axis. Cell. Mol. Immunol. 15, 595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. X., Rocha C. S., Dandekar S., Wan Y. J. (2016). Functional analysis of the relationship between intestinal microbiota and the expression of hepatic genes and pathways during the course of liver regeneration. J. Hepatol. 64, 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Wang H., Shi Q., Wang N., Zhang Z., Xiong C., Liu J., Chen Y., Jiang L., Jiang Q. (2017). Effects of oral florfenicol and azithromycin on gut microbiota and adipogenesis in mice. PLoS One 12, e0181690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandy M., Nyirenda M. (2018). Developmental origins of health and disease: The relevance to developing nations. Int. Health 10, 66–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor O., Borenstein E. (2017). Systematic characterization and analysis of the taxonomic drivers of functional shifts in the human microbiome. Cell Host Microbe 21, 254–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. D., Clark B. R., Amadio J. (2009). Metabolism of fluoroorganic compounds in microorganisms: Impacts for the environment and the production of fine chemicals. Appl. Microbiol. Biotechnol. 84, 617–629. [DOI] [PubMed] [Google Scholar]
- Olson C. A., Vuong H. E., Yano J. M., Liang Q. Y., Nusbaum D. J., Hsiao E. Y. (2018). The gut microbiota mediates the anti-seizure effects of the ketogenic diet. Cell 173, 1728–1741.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S., Sarkar K., Nath P. P., Mondal M., Khatun A., Paul G. (2017). Bisphenol S impairs blood functions and induces cardiovascular risks in rats. Toxicol. Rep. 4, 560–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W., Yang M., Liu J., Xu H., Luo S., Wong M., Zheng C. (2019). Bisphenol S-induced chronic inflammatory stress in liver via peroxisome proliferator-activated receptor gamma using fish in vivo and in vitro models. Environ. Pollut. 246, 963–971. [DOI] [PubMed] [Google Scholar]
- Ravussin Y., Koren O., Spor A., LeDuc C., Gutman R., Stombaugh J., Knight R., Ley R. E., Leibel R. L. (2012). Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity (Silver Spring) 20, 738–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezg R., Abot A., Mornagui B., Knauf C. (2019). Bisphenol S exposure affects gene expression related to intestinal glucose absorption and glucose metabolism in mice. Environ. Sci. Pollut. Res. Int. 26, 3636–3642. [DOI] [PubMed] [Google Scholar]
- Richardson V. M., Staskal D. F., Ross D. G., Diliberto J. J., DeVito M. J., Birnbaum L. S. (2008). Possible mechanisms of thyroid hormone disruption in mice by BDE 47, a major polybrominated diphenyl ether congener. Toxicol. Appl. Pharmacol. 226, 244–250. [DOI] [PubMed] [Google Scholar]
- Scoville D. K., Li C. Y., Wang D., Dempsey J. L., Raftery D., Mani S., Gu H., Cui J. Y. (2019). Polybrominated diphenyl ethers and gut microbiome modulate metabolic syndrome-related aqueous metabolites in mice. Drug Metab. Dispos. 47, 928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreiner A., Huffnagle G. B., Noverr M. C. (2008). The “Microflora Hypothesis” of allergic disease. Adv. Exp. Med. Biol. 635, 113–134. [DOI] [PubMed] [Google Scholar]
- Siddiqi M. A., Laessig R. H., Reed K. D. (2003). Polybrominated diphenyl ethers (PBDEs): new pollutants-old diseases. Clin. Med. Res. 1, 281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S. R., Haileselassie Y., Nguyen L. P., Tropini C., Wang M., Becker L. S., Sim D., Jarr K., Spear E. T., Singh G., et al. (2020). Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe 27, 659–670.e655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan G. V., Choi K., Klemashevich C., Wu C., Prabakaran D., Pan L. B., Steinmeyer S., Mueller C., Yousofshahi M., Alaniz R. C., et al. (2014). Prediction and quantification of bioactive microbiota metabolites in the mouse gut. Nat. Commun. 5, 5492. [DOI] [PubMed] [Google Scholar]
- Staskal D. F., Diliberto J. J., DeVito M. J., Birnbaum L. S. (2004). Toxicokinetics of BDE 47 in female mice: effect of dose, route of exposure, and time. Toxicol. Sci. 83, 215–223. [DOI] [PubMed] [Google Scholar]
- Stellwag E. J., Hylemon P. B. (1976). Purification and characterization of bile salt hydrolase from Bacteroides fragilis subsp. fragilis. Biochim. Biophys. Acta 452, 165–176. [DOI] [PubMed] [Google Scholar]
- Stiemsma L. T., Michels K. B. (2018). The role of the microbiome in the developmental origins of health and disease. Pediatrics 141, e20172437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh K. S., Choi E. M., Rhee S. Y., Oh S., Kim S. W., Pak Y. K., Choe W., Ha J., Chon S. (2017). Tetrabromobisphenol A induces cellular damages in pancreatic beta-cells in vitro. J. Environ. Sci. Health A Tox. Hazard Subst. Environ. Eng. 52, 624–631. [DOI] [PubMed] [Google Scholar]
- Sun L., Pang Y., Wang X., Wu Q., Liu H., Liu B., Liu G., Ye M., Kong W., Jiang C. (2019). Ablation of gut microbiota alleviates obesity-induced hepatic steatosis and glucose intolerance by modulating bile acid metabolism in hamsters. Acta Pharm. Sin. B 9, 702–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Wu W., Chen L., Yang W., Huang X., Ma C., Chen F., Xiao Y., Zhao Y., Ma C., et al. (2018). Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat. Commun. 9, 3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorov A., Girard S., Lachapelle S., Abdelouahab N., Sebire G., Takser L. (2009). Perinatal exposure to low-dose BDE-47, an emergent environmental contaminant, causes hyperactivity in rat offspring. Neonatology 95, 203–209. [DOI] [PubMed] [Google Scholar]
- Ta T. A., Koenig C. M., Golub M. S., Pessah I. N., Qi L., Aronov P. A., Berman R. F. (2011). Bioaccumulation and behavioral effects of 2,2',4,4'-tetrabromodiphenyl ether (BDE-47) in perinatally exposed mice. Neurotoxicol. Teratol. 33, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh A., Calay E. S., Tuncman G., Claiborn K. C., Inouye K. E., Eguchi K., Alcala M., Rathaus M., Hollander K. S., Ron I., et al. (2019). The short-chain fatty acid propionate increases glucagon and FABP4 production, impairing insulin action in mice and humans. Sci. Transl. Med. 11, eaav0120. [DOI] [PubMed] [Google Scholar]
- Tsolis R. M., Baumler A. J. (2020). Gastrointestinal host-pathogen interaction in the age of microbiome research. Curr. Opin. Microbiol. 53, 78–89. [DOI] [PubMed] [Google Scholar]
- Van der Ven L. T., Van de Kuil T., Verhoef A., Verwer C. M., Lilienthal H., Leonards P. E., Schauer U. M., Canton R. F., Litens S., De Jong F. H., et al. (2008). Endocrine effects of tetrabromobisphenol-A (TBBPA) in Wistar rats as tested in a one-generation reproduction study and a subacute toxicity study. Toxicology 245, 76–89. [DOI] [PubMed] [Google Scholar]
- von Meyerinck L., Hufnagel B., Schmoldt A., Benthe H. F. (1990). Induction of rat liver microsomal cytochrome P-450 by the pentabromo diphenyl ether Bromkal 70 and half-lives of its components in the adipose tissue. Toxicology 61, 259–274. [DOI] [PubMed] [Google Scholar]
- Woolbright B. L., Jaeschke H. (2019). Inflammation and cell death during cholestasis: The evolving role of bile acids. Gene Expr. 19, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Zhang X., Zhang C., Li J., Zhao Y., Zhu Y., Zhang J., Zhou X. (2019). Toxicity and multigenerational effects of bisphenol S exposure to Caenorhabditis elegans on developmental, biochemical, reproductive and oxidative stress. Toxicol. Res. (Camb.) 8, 630–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye G., Chen Y., Wang H-o., Ye T., Lin Y., Huang Q., Chi Y., Dong S. (2016). Metabolomics approach reveals metabolic disorders and potential biomarkers associated with the developmental toxicity of tetrabromobisphenol A and tetrachlorobisphenol A. Sci. Rep. 6, 35257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li S., Liu L., Wang L., Xiao X., Sun Z., Wang X., Wang C., Wang M., Li L. (2016). Environmental exposure to BDE47 is associated with increased diabetes prevalence: Evidence from community-based case-control studies and an animal experiment. Sci. Rep. 6, 27854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.