Abstract
Monoderm bacteria utilize coproheme decarboxylases (ChdCs) to generate heme b by a stepwise decarboxylation of two propionate groups of iron coproporphyrin III (coproheme), forming two vinyl groups. This work focuses on actinobacterial ChdC from Corynebacterium diphtheriae (CdChdC) to elucidate the hydrogen peroxide-mediated decarboxylation of coproheme via monovinyl monopropionyl deuteroheme (MMD) to heme b, with the principal aim being to understand the reorientation mechanism of MMD during turnover. Wild-type CdChdC and variants, namely H118A, H118F, and A207E, were studied by resonance Raman and ultraviolet-visible spectroscopy, mass spectrometry, and molecular dynamics simulations. As actinobacterial ChdCs use a histidine (H118) as a distal base, we studied the H118A and H118F variants to elucidate the effect of 1) the elimination of the proton acceptor and 2) steric constraints within the active site. The A207E variant mimics the proximal H-bonding network found in chlorite dismutases. This mutation potentially increases the rigidity of the proximal site and might impair the rotation of the reaction intermediate MMD. We found that both wild-type CdChdC and the variant H118A convert coproheme mainly to heme b upon titration with H2O2. Interestingly, the variant A207E mostly accumulates MMD along with small amounts of heme b, whereas H118F is unable to produce heme b and accumulates only MMD. Together with molecular dynamics simulations, the spectroscopic data provide insight into the reaction mechanism and the mode of reorientation of MMD, i.e., a rotation in the active site versus a release and rebinding.
Significance
Coproheme decarboxylation by ChdC is the final step of the prokaryotic coproporphyrin-dependent heme b biosynthesis. Elucidation of the molecular mechanism of ChdCs is an ongoing process. The numerous steps of this complex reaction have been explained little by little in several studies. In this work, we focus on the reorientation of the transiently formed three-propionate reaction intermediate (monovinyl monopropionyl deuteroheme). A comprehensive study, including electronic absorption and resonance Raman spectroscopies, mass spectrometry, and molecular dynamics simulations of WT CdChdC and selected variants, allowed us to follow the decarboxylation steps. Based on the influence of distal and proximal heme mutations on the catalyzed reaction, we propose a mechanism in which the substrate remains bound in the heme pocket without being released.
Introduction
In monoderm bacteria, coproheme decarboxylases (ChdCs) catalyze the final reaction to generate heme b (1, 2, 3, 4). In the “coproporphyrin-dependent” heme biosynthesis pathway, heme b is produced by a stepwise decarboxylation of the propionate groups at position 2 (p2) and position 4 (p4) of the pyrrole rings A and B of iron coproporphyrin III (coproheme) to form vinyl groups (vinyl 2 (v2) and vinyl 4 (v4)). The hydrogen peroxide-dependent oxidative radical decarboxylation is carried out in two consecutive steps. After cleavage of p2, the transiently formed three-propionate intermediate, monovinyl monopropionyl deuteroheme (MMD), undergoes a reorientation before the decarboxylation of p4 and the resulting formation of heme b occur (5, 6, 7, 8).
In previous work, extensive electronic absorption and resonance Raman (RR) spectroscopic studies of different variants carrying mutations in the surroundings of the coproheme propionates, allowed 1) the vibrational assignment of all four propionate groups in the resting state of the firmicute ChdC of Listeria monocytogenes (LmChdC) and 2) monitoring the formation of the vinyl groups during turnover (7,9, 10, 11). Moreover, similarly to ChdC from Staphylococcus aureus (SaChdC) (8), a tyrosine (Y147 L. monocytogenes numbering) was identified as the catalytically relevant radical site that is involved in both decarboxylation reactions (7).
The reaction cycle of ChdCs (Fig. S1) is initiated by the H2O2-mediated oxidation of the ferric state of coproheme [Fe(III) Por-p2p4…Tyr] to Compound I, i.e., an oxoiron(IV) porphyryl radical [Fe(IV)=O Por•+-p2p4…Tyr] (Reaction 1). In firmicute ChdCs, a proton acceptor at the distal heme site is missing, and thus, the mechanism of heterolytic cleavage of hydrogen peroxide is still unclear. By contrast, in the actinobacterial ChdC from Corynebacterium diphtheriae (CdChdC), H118 has been demonstrated to act as a distal base that promotes Compound I formation. As a consequence of the more efficient formation of Compound I, the overall catalytic efficiency of actinobacterial ChdCs is higher compared with that of representatives from Firmicutes (5,12). In fact, Compound I is immediately converted to catalytically active Compound I∗ by an internal electron transfer and formation of a tyrosyl radical [Fe(IV)=O Por-p2p4… Tyr•] (Reaction 2). Compound I∗ initiates the decarboxylation of p2, thereby forming MMD [Fe(III) Por-v2p4…Tyr] (Reaction 3). After the reorientation of MMD, the next cycle is initiated by the H2O2-mediated oxidation to MMD Compound I [Fe(IV)=O Por•+-v2p4…Tyr] (Reaction 4). Finally, MMD Compound I∗ [Fe(IV)=O Por-v2p4… Tyr•] initiates the decarboxylation of p4, thereby forming the heme b protein [Fe(III) Por-v2v4…Tyr] (Reactions 5 and 6). Heme b is weakly associated with ChdC, thus facilitating its transfer to other apo-proteins with vacant heme binding sites. The reactions are as follows:
-
1)
[Fe(III) Por-p2p4…Tyr] + H-O-O-H → [Fe(IV)=O Por•+-p2p4…Tyr] + H2O Reaction 1;
-
2)
[Fe(IV)=O Por•+-p2p4…Tyr] → [Fe(IV)=O Por-p2p4… Tyr•] Reaction 2;
-
3)
[Fe(IV)-OH Por-p2p4…Tyr•] → [Fe(III) Por-v2p4…Tyr] + CO2 Reaction 3;
-
4)
[Fe(III) Por-v2p4…Tyr] + H-O-O-H → [Fe(IV)=O Por•+-v2p4…Tyr] + H2O Reaction 4;
-
5)
[Fe(IV)=O Por•+-v2p4…Tyr] → [Fe(IV)=O Por-v2p4… Tyr•] Reaction 5;
-
6)
[Fe(IV)-OH Por-v2p4… Tyr•] → [Fe(III) Por-v2v4…Tyr] + CO2 Reaction 6.
This work is focused on the actinobacterial CdChdC with the objective of elucidating the mechanism of the stepwise decarboxylation of coproheme to heme b (Reactions 1–6). Wild-type (WT) CdChdC and the variants H118A, H118F, and A207E (Fig. 1) were studied by RR and ultraviolet-visible (UV-vis) spectroscopy, mass spectrometry, and molecular dynamics (MD) simulations to probe the impact of the mutations on the two-step reaction and reorientation of MMD during turnover. The fully conserved alanine 207 (12) was targeted because of its proximity to the iron-coordinating H158, in which a conserved glutamate is present in chlorite dismutases (Clds). Clds and ChdCs belong to the same structural superfamily and share the same overall subunit and oligomeric structure (6). In Clds, a proximal H-bonding network spans from the proximal histidine via the conserved glutamate to the propionate groups of heme b, significantly defining and modulating the active site for efficient catalysis of chlorite degradation. In the CdChdC A207E variant, an H-bond between the inserted glutamate and the proximal histidine is introduced. By mimicking the situation in Clds, the rigidity of the proximal side is thus potentially increased. Therefore, we investigated the impact of this mutation on the rotation of MMD after it is formed (Reaction 3).
Figure 1.
Active site of WT coproheme-CdChdC (Protein Data Bank: 6XUC) and the variants H118A, H118F (A), and A207E (B) obtained by in silico mutations. Introduced amino acids are shown as black outlines. Expected H-bonds are shown as black dashed lines, and the distance between Y135 and the β-carbon of propionate p2 is indicated as red solid lines. To see this figure in color, go online.
Here, we demonstrate that 1) the electronic structure of ferric coproheme-CdChdC closely resembles that of ferric coproheme-LmChdC, being a mixture of five-coordinated (5c) quantum mixed-spin (QS) and high-spin (HS), and 2) the mutations affect the coordination and spin state(s) of the respective coproheme complexes. Both CdChdC WT and H118A convert coproheme mainly to heme b upon titration with H2O2, which is in agreement with previous data (5). Interestingly, the variant A207E accumulates MMD along with smaller amounts of heme b, whereas H118F is unable to produce heme b and accumulates only MMD. We provide the spectroscopic marker bands of coproheme-CdChdC, MMD-CdChdC, and heme b-CdChdC. Spectroscopic data, together with MD simulations, provide insight into the reaction mechanism and the mode of reorientation of MMD, i.e., a rotation in the active site versus a release and rebinding.
Materials and methods
Expression and purification of WT CdChdC and variants
Cloning, site-directed mutagenesis, and expression of the CdChdC gene (DIP1394) and its variants were performed as described in detail previously (5). Site-directed mutagenesis was carried out using the QuikChange Lightning Kit (Agilent Technologies, Santa Clara, CA). The pD441-NH vector (ATUM, Newark, CA), containing a protease cleavage site in front of the gene of interest was transformed in Escherichia coli Tuner (DE3) (Merck/Novagen, Darmstadt, Germany) cells for heterologous expression. The cells were cultivated in Luria-Bertani medium, containing 100 μg mL−1 kanamycin, at 37°C and 180 rpm for 3 h before cooling to 16°C, induction with 0.5 mM isopropyl-β-D-thiogalactopyranoside, and further overnight cultivation.
The cells were then harvested by centrifugation, resuspended in lysis buffer (50 mM phosphate buffer (pH 7.4), with 500 mM NaCl, 5% glycerol, and 0.5% Triton X-100), and lysed by ultrasonication. The lysate was centrifuged, and the supernatant was filtered before loading it onto a 5 mL His-trap FF column (GE Healthcare, Chicago, IL). The column was washed with binding buffer (50 mM phosphate buffer (pH 7.4), with 500 mM NaCl) and equilibrated with cleavage buffer (50 mM Tris-HCl with 150 mM NaCl and 1 mM EDTA) before the target protein was eluted with three column volumes of cleavage buffer after overnight on-column cleavage at 4°C using a His-tagged HRV 3C PreScission Protease (Thermo Fischer Scientific, Waltham, MA). The eluate was further purified by size exclusion chromatography on a HiLoad 16/600 Superdex 200 pg column (GE Healthcare, Chicago, IL) with 100 mM phosphate buffer (pH 7.4), with 100 mM NaCl. The protein was concentrated to 750–1000 μM and stored at −80°C.
UV-vis Electronic absorption
UV-vis electronic absorption spectra were recorded using a 5 mm NMR tube (300 nm min−1 scan rate) or a 1 mm cuvette (600 nm min−1 scan rate) at 25°C by means of a Cary 60 spectrophotometer (Agilent Technologies, Santa Clara, CA) with a resolution of 1.5 nm. For calculation of the second derivative spectra, the Savitzky-Golay method was applied using 15 data points (LabCalc; Galactic Industries, Salem, NH). No changes in the wavelength or in the bandwidth were observed when the number of points was increased or decreased. For the sake of clarity, the 450–750 nm region in all the absorption spectra is magnified by a factor from 6- to 15-fold depending on the sample.
Resonance Raman
RR spectra were obtained using a 5 mm NMR tube by excitation with the 406.7 nm line of a Kr+ laser (Innova300 C; Coherent, Santa Clara, CA). Back-scattered light from a slowly rotating NMR tube was collected and focused into a triple spectrometer (consisting of two SpectraPro 2300i instruments working in the subtractive mode and a SpectraPro 2500i instrument in the final stage with a grating of 3600 grooves/mm by Acton Research, Acton, MA), equipped with a liquid nitrogen-cooled charge-coupled device detector. Spectral resolution, calibration, and details on the experiments in polarized light have been reported previously (13) To avoid sample denaturation, a stream of cooled nitrogen gas was directed toward the rotating sample tube (15°C).
All RR measurements were repeated several times under the same conditions to ensure reproducibility. To improve the signal/noise ratio, a number of spectra were accumulated and summed only if no spectral differences were noted. Table S1 summarizes the integration time and the number of averaged spectra reported in the figures. The electronic absorption spectra were measured both before and after RR measurements to cross-check that no degradation of the sample occurred under the experimental conditions used. All spectra were baseline corrected. The RR spectra were normalized to the intensity of the ν4 band at 1370–1373 cm−1 (data not shown) for the high frequency region and to the ν8 band at 341–345 cm−1 in the low frequency region, respectively. A 5.5 mW laser power at the sample for the coproheme complex was employed for the WT and all the variants, whereas a 6.5 mW laser power at the sample was employed for the complexes upon titration with hydrogen peroxide for the WT and all the variants. In the figures, the spectra were shifted along the ordinate axis to allow better visualization.
A spectral simulation program (LabCalc; Galactic Industries, Salem, NH) was used to determine the peak positions, bandwidth, and intensity, assuming Lorentzian line shapes. The frequencies of the bands were optimized to an accuracy of 1 cm−1 and the bandwidths to an accuracy of 0.5 cm−1.
Sample preparation for UV-vis and RR experiments
Ferric coproheme was purchased from Frontier Scientific (Logan, Utah) as lyophilized powder. Ferric coproheme-CdChdC samples were prepared by adding the coproheme solution (coproheme dissolved in 0.5 M NaOH) to apo-CdChdC dissolved in 50 mM phosphate buffer (pH 7.0). Typically, for both the variants and the WT protein, the coproheme/apoprotein ratio was 1:2.
Ferric heme b-CdChdC samples were prepared by adding small aliquots of a concentrated solution of H2O2 to coproheme-CdChdC. Hydrogen peroxide (Sigma-Aldrich, St. Louis, MO) was diluted in 50 mM phosphate buffer (pH 7.0), and its concentration was determined using an extinction coefficient of 39.4 M−1 cm−1 at 240 nm. In detail, in a 1 cm cuvette, 1 μL aliquots of a 1 mM solution of H2O2 were added to 500 μL of a 25–35 μM coproheme complex buffered solution under continuous magnetic stirring at room temperature. To ensure the complete reaction of the H2O2, an interval of 2–10 min was set before adding each H2O2 aliquot. Titration was monitored by UV-vis absorption and RR spectroscopies. The sample concentrations, in the range 25–35 μM for both coproheme- and heme b-CdChdC, were determined using an extinction coefficient of 68,000 M−1 cm−1 at 392 nm for both the WT and mutant coproheme-ChdCs.
Mass spectrometry and coproheme decarboxylase activity
Experimental conditions for the determination of the coproheme decarboxylation activity via titration with hydrogen peroxide monitored by UV-vis absorption and mass spectrometric analysis have been described in detail previously (5). Experiments were performed in 100 mM phosphate buffer (pH 7) using a Cary 60 spectrophotometer for UV-vis absorption. 10 μL samples were drawn from the 1200 μL reaction mix, deactivated by the addition of 10 mM cyanide and analyzed using a Dionex Ultimate 3000 system directly linked to a quadrupole time-of-flight mass spectrometer (maXis 4G ETD; Bruker, Billerica, MA) equipped with the standard electrospray ionization source in the positive ion mode. MS scans were recorded within a range from m/z 400 to 3800, and the instrument was tuned to detect both the rather small free heme derivatives and intact proteins in a single run. A Thermo ProSwift RP-4H analytical separation column (250 × 0.200 mm) (Thermo Fisher Scientific, Vienna, Austria) was used to separate the analytes. A gradient from 99% solvent A and 1% solvent B (solvent A, 0.05% trifluoroacetic acid; solvent B, 80.00% acetyl cyanide and 20% solvent A) to 65% B in 11 min was applied, followed by a 2 min gradient from 65% B to 95% B, at a flow rate of 8 μL min−1 and at 65°C. A blank run (5.0 μL of H2O) was performed after each sample to minimize carryover effects.
Cyanide binding to coproheme-CdChdC WT and variants
Steady-state kinetics of cyanide binding were determined using a Cary 60 spectrophotometer (Agilent Technologies, Santa Clara, CA). Typically, 5–10 μM holo-enzyme was titrated with NaCN using the DOSY titration device (DOSTAL, Halle/Saale, Germany) or by hand to a 5–20-fold excess of ligand. The absorbance change at the maximum of the resulting low-spin (LS) Soret band at 412/413 nm was plotted against ligand concentration and fitted in SigmaPlot with a single rectangular hyperbola and three parameters (), in which yo is the absorbance at zero ligand concentration, ΔAbs is the absorbance difference at maximal saturation, and KD is the dissociation constant.
Pre-steady-state spectroscopic changes upon the addition of the NaCN ligand were monitored using a stopped-flow apparatus equipped with a photodiode array detector (SX-18MV; Applied Photophysics, Leatherhead, UK). Path length and volume of the optical quartz cell were 10 mm and 20 μL. The first spectrum was recorded 1 ms after mixing. All measurements were performed in 100 mM phosphate buffer (pH 7) at 25°C. Solutions of 4 μM apo-CdChdC WT and variants were reconstituted with 2 μM coproheme directly before the measurements. Ligand concentrations varied from 100 μM to 2 mM.
Rate constants for cyanide binding were determined at a single wavelength by electronic absorption spectroscopy using a PiStar-180 spectrometer (Applied Photophysics, Leatherhead, UK). Time traces were taken at 407 nm to follow the formation of the LS complex. Typically, the enzyme concentration was 2 μM, and ligand concentrations varied from 0.1 to 5.0 mM. For the single wavelength measurements, a minimum of three time traces for each ligand concentration were averaged. kobs values for each ligand concentration were determined by fitting the respective average time traces with a single exponential equation ), where a is the absorbance at the beginning of ligand binding, kobs is the absorbance change per second, and c is the absorbance at the end of the ligand binding. The kobs values were fitted using the Pro-Data viewer software (Applied Photophysics, Leatherhead, UK) and plotted against the respective ligand concentration.
Molecular dynamics simulation
MD simulations of WT CdChdC and the variants H118A, H118F, and A207E, in complex with either coproheme or MMD, as redox active substrates were performed to analyze the dynamics of their hydrogen bonding interactions within the respective active sites. MMD was additionally simulated in the pose of coproheme to investigate interactions directly after cleavage of p2 and before the 90° rotation. The simulation of one pentameric CdChdC is comparable to five independent subunit simulations.
Equilibrated structures of WT CdChdC with different binding poses of the cofactors were taken as initial structures from our previous work (5). The structures of the three variants were each based on the equilibrated WT structures. Mutations were introduced using the mutation wizard of PyMol (14) (version 2.3, 2019) by selecting the rotamer with the least conformational strain. Coordinates of the variants were then minimized by steepest descent energy minimization (15) using the GROMOS11 (16) software for biomolecular simulations together with the GROMOS 54a8 force field parameter set (17). Coproheme and MMD parameters were taken from previous work (5) and parameterized in analogy to heme b in its ferric form (18).
Water was modeled explicitly and described by means of the simple point charge (SPC) water model (19). Minimized mutant structures were solvated in preequilibrated periodic rectangular boxes of SPC water with box sizes of ∼10.2 nm × 10.4 nm × 11.0 nm. Minimal distances of solute-box edge and solute-solvent were set to 1.4 and 0.23 nm, respectively. The box was co-solvated with 25 mM sodium chloride, and additional sodium atoms were added to neutralize the boxes.
The GROMOS11 (16) (version 1.5.0) and GROMACS (20) (release 2020) packages for molecular simulations were used to perform MD simulations. Equilibrations of the variants were performed using the MD engine of GROMOS. Equations of motions were integrated using the leap-frog algorithm (21); the SHAKE algorithm (22) was applied for bond length constraints, allowing for integration time steps of 2 fs. Nonbonded interactions were treated using a twin range cutoff (short range 0.8 nm, long-range 1.4 nm). For long-range interactions, a reaction field contribution (23) with a relative dielectric permittivity of 61, as appropriate for the SPC water model, was added (24). Initial velocities were sampled from a Maxwell-Boltzmann distribution at 60 K. The systems were slowly heated up to 298 K in five discrete steps of 100 ps each. The positions of the solutes were restrained by a harmonic potential with a decreasing force constant factor of 10 for each equilibration step starting from an initial value of 2.5 × 104 kJ mol−1. The final equilibration step was prolonged to 150 ps, in which position restraints were replaced by rototranslational restraints.
After equilibration, production simulations of the CdChdC WT and variants were performed for 100 ns each using the GROMACS MD engine. Bond lengths were constrained using the LINCS algorithm with an order of 4. An atom-based, Verlet-buffered pair list (25) was used to treat nonbonded interactions. A reaction field contribution due to a homogenous medium with a dielectric permittivity of 61 was added to account for the influence of particles beyond the long-range cutoff (1.4 nm) (23,24). The temperature was maintained at 298 K by using a weak-coupling thermostat (26) of solute and solvent to two separate heat baths with a coupling time of 0.1 ps; pressure was kept constant at 1 atm with a coupling time of 0.5 ps and an isothermal compressibility of 4.575 × 10−4 kJ nm3. The center-of-mass motion was removed every 2 ps to avoid the flying ice cube problem (27). Trajectories were written every 2 ps.
Analyses of the coordinate trajectories were performed using the GROMOS++ programs (28). Hydrogen bonds were considered to be present if the distance between a hydrogen atom connected to a donor atom is within 0.25 nm from an acceptor and the donor-hydrogen-acceptor angle is larger than 135°. For the hydrogen bonding interaction between propionates of the substrate and the amino acid residues around the active sites, two types of hydrogen bonds were distinguished in this work: 1) direct hydrogen bonds (solute-solute bonds) and 2) bridging hydrogen bonds (solute-solvent-solute bridges), in which a water molecule facilitates the hydrogen bonding between solutes. The mean number of H-bonds over 100-ns simulation time averaged over the five subunits per propionate, and residues were calculated. They were considered for residues with an average occurrence of more than 0.01 H-bonds per residue. The dynamics of the selected residues were evaluated by analyzing distance-time series.
Results
Stepwise conversion of WT coproheme-CdChdC to heme b-CdChdC
The electronic configuration of the actinobacterial WT coproheme-CdChdC is very similar to that of the well-studied firmicute representative LmChdC (7,9,13) (Fig. S2). The UV-vis spectrum is characterized by a Soret band at 392 nm with a shoulder at 377 nm, as clearly observed in the second derivative (D2) spectra. In addition, the Q-bands (β, α) are at 496 and 537 nm and the charge transfer band (CT1) at 631 nm (Fig. S3, left). Accordingly, in the high frequency RR spectra, the core size marker bands characteristic of 5cQS and 5cHS species have been identified (ν3 at 1491 (5cHS) and 1505 cm−1 (5cQS), ν2 at 1580 cm−1 (5cQS), and ν10 at 1624 (5cHS) and 1636 cm−1 (5cQS)) (Fig. 2, right). The 5cQS species has been also identified in the firmicute representative LmChdC (7,9,13) (Fig. S2). The QS state results from a quantum mechanical admixture of intermediate (S = 3/2) and high (S = 5/2) spin states. It has been found to be a distinctive characteristic of several ferric model compounds and heme proteins, including class III of the peroxidase-catalase superfamily, but its structural origin and functional significance remain elusive (29, 30, 31). A 5cQS state is characterized by electronic absorption spectra similar to those of a 5cHS heme but with shorter wavelength π→π∗ transitions together and a CT1 at 630–635 nm, with the RR core size marker band wavenumbers typical of a LS heme (32).
Figure 2.
Tracing coproheme decarboxylation in WT CdChdC upon titration with H2O2. Low frequency (left) and high frequency RR (right) region spectra of WT coproheme complex with increasing amounts of H2O2 are shown. RR core size marker band wavenumbers are reported in black, propionate bending modes δ(CβCcCd) are indicated in purple, and vinyl bending modes δ(CβCaCb) and vinyl stretching modes (νC=C) are in orange. To see this figure in color, go online.
Only two δ(CβCcCd) propionate bending modes are observed in the low frequency RR spectra of the WT coproheme-CdChdC, giving rise to a strong band at 372 cm−1 and a weak shoulder at 384 cm−1, respectively (Fig. 2, left). This indicates a strong overlap of the expected four propionate bending modes.
The decarboxylation of the p2 and p4 to vinyl groups upon the addition of H2O2 is expected to cause an overall red shift of the electronic absorption spectra compared to the coproheme complex because of the conjugation of the vinyl double bonds with the porphyrin macrocycle. To gain a comprehensive understanding of the effect of the hydrogen peroxide titration, we followed the H2O2-mediated decarboxylation of coproheme-CdChdC to heme b-CdChdC by mass spectrometry, UV-vis absorption, and RR spectroscopies.
Conversion of WT coproheme (708.2 Da, green) to MMD (662.2 Da, cyan) and, finally, to heme b (616.2 Da, red), as traced by mass spectrometry (Fig. 3, left), shows that the formation of MMD (cyan) is detected after the addition of ∼0.5 equivalent(s) of (eq.) H2O2 to CdChdC, whereas heme b (red) formation requires ∼1 eq. H2O2. Full conversion to heme b is seen with a ∼3-fold excess of hydrogen peroxide. The theoretical stoichiometry of the reaction (2:1) (13) is most probably exceeded because of in vitro side reactions (e.g., oxidation of amino acids; cross-linking) observed in this experimental setup. In fact, in the highly sensitive mass spectrometric analysis, porphyrin species are separated from the whole protein in the course of the analysis. Therefore, unlike in the RR spectra (see below), in the presence of an excess of H2O2, oxidized porphyrin species, which are potentially cross-linked to the protein moiety, are not observed. The conversion of WT coproheme-CdChdC as studied by UV-vis absorption (and D2; see Fig. S3, right) displays a continuous red shifting of the Soret maximum to 404 nm (and 405 nm in D2) up to 2 eq. of H2O2. The shoulder at 392 nm disappears, and a new band at 586 nm evolves. The conversion of coproheme to heme b as followed by RR spectroscopy (Fig. 2) allowed us to monitor the changes in heme coordination and spin state(s) of the heme iron (see also Fig. S2 for a comparison with the firmicute LmChdC). Upon the addition of 0.25 eq. H2O2, the amount of 5cQS species decreases, whereas that of the six-coordinated (6c) HS and 5cHS forms grows. These species increase upon further addition of H2O2 (ν3 at 1482 (6cHS) and 1488 (5cHS), ν2 at 1567 (5 and 6cHS together), and ν10 at 1609 (6cHS) and 1628 (5cHS) cm−1). The presence of a 6cHS species is also confirmed by the appearance of the ν38 and ν37 bands at 1516 and 1579 cm−1, respectively. It is worth noting that the frequencies of the core size marker bands of the 5cHS form after titration with hydrogen peroxide differ from those found in the coproheme complex, thus suggesting a new species.
Figure 3.
Mass spectrometric analysis of WT (left), H118F (middle), and A207E (right) CdChdC titrated with H2O2. Coproheme (708.2 Da, green), the reaction intermediate MMD (662.2 Da, cyan), and final product heme b (616.2 Da, pink) are shown as elution profiles (left) and relative amounts (right). H118F: the pink asterisks indicate that detected heme b, which originates from recombinant production in E. coli, is constant throughout the titration and regarded as an artifact. A207E: The oxidized species are indicated as dashed boxes/lines (see text). To see this figure in color, go online.
The decarboxylation of the propionates and the formation of the vinyl groups is easily followed by RR spectroscopy because of an intense νC=C vinyl stretching band in the high frequency region and the corresponding δ(CβCaCb) bending vibrations in the low frequency region. After the addition of 0.25 eq. of H2O2, new bands at ∼1617 cm−1 (v2) (Fig. 2, right) and 416 cm−1 (Fig. 2, left) are clearly observed and grow in upon increasing the concentration of hydrogen peroxide. Moreover, starting from 1 eq. of hydrogen peroxide, in agreement with the formation of heme b observed by mass spectrometry, the vinyl stretching band broadens and grows in intensity, and its maximum shifts to 1623 cm−1. Corresponding changes are observed in the low frequency region as the intensity of the vinyl bending mode is enhanced. At the final point of the titration (2 eq. of hydrogen peroxide) together with the growth of the 6cHS heme population, the intensity of the vinyl stretching mode at 1623 cm−1 and of the bending modes shifted to 421 cm−1, increase. These overall changes suggest the formation of the second vinyl group (v4), as also confirmed by the RR spectra taken in polarized light (see below, Fig. 4 a). Concomitantly, the propionyl δ(CβCcCd) bending mode at 372 cm−1 decreases in intensity upon increasing hydrogen peroxide concentration, together with a broadening and frequency shift of the band. In fact, upon titration, the propionate bending modes are detected at 375 and 384 cm−1. A small amount of unconverted 5cQS remains up to the end of the titration, as shown by the weak ν3 at 1505 cm−1 and ν10 at 1636 cm−1, respectively (Fig. 2, right). We were unable to proceed with a further addition of H2O2 because an excess of hydrogen peroxide (>2 eq.) causes degradation of the protein, as reflected by the presence of a weak band at 1359 cm−1 and a shoulder at 1385 cm−1 in the RR spectrum, with no changes in the UV-vis spectra (data not shown). The intensity increase of these new bands is not correlated to the value of the laser power at the sample but only to the amount of hydrogen peroxide. Therefore, photoreduction can be excluded. This behavior is similar to the case of SaChdC, in which an excess of hydrogen peroxide was necessary for full conversion of the two propionates to the respective vinyl groups, instead of the 2 eq. of H2O2, which are, in principle, necessary. This excess of H2O2 suggested that some of the hydrogen peroxide reacts producing other species, with concurrent degradation of heme b evidenced by the decrease in intensity of the Soret band (11).
Figure 4.
Comparison of the RR spectra of WT coproheme-CdChdC and variants before and after titration with H2O2. Spectra were recorded in polarized light (parallel, blue lines, and perpendicular, red lines) in the 1535–1660 cm−1 frequency region. RR core size marker band wavenumbers are indicated in black, and vinyl stretching modes (νC=C) are indicated in orange. To see this figure in color, go online.
Fig. 4a shows the RR spectra taken in polarized light. These data together with the curve fitting of the RR spectrum of the WT coproheme titrated with 2 eq. of H2O2 (Fig. S4) allowed us to identify the depolarized bands because of the ν10 modes of the 6cHS, 5cHS, and 5cQS at 1609, 1628, and 1636 cm−1, respectively, and to confirm the presence of two polarized νC=C stretching modes at 1617 (v2) and 1623 (v4) cm−1.
Impairing the decarboxylation of propionate 4
It has been previously reported that actinobacterial CdChdC uses a distal histidine residue (H118) for deprotonation and subsequent heterolytic cleavage of hydrogen peroxide (5). H118 has been found to have a relatively low flexibility because of an H-bond interaction with the propionate 7 (p7). Fig. 1 shows the structures obtained by in silico mutation of WT CdChdC with PyMOL. No crystal structures of the three variants are available because the screening for crystallization conditions has failed so far. The exchange of H118 by either Phe or Ala is expected to disrupt the H-bond to p7 (5). Furthermore, the substitution of H118 by the bulkier and more hydrophobic phenylalanine might sterically hinder the MD within the active site, most probably by narrowing the substrate access channel.
The exchange of A207 by a glutamate is expected to lead to the formation of an H-bond to the proximal coproheme iron-coordinating H158, thereby reducing its mobility and increasing its imidazolate character. In the structurally related Clds, this H-bonding network has been established and is important for the heme integrity and stability (13). Moreover, as E207 interacts with R139, which in turn is H-bonded to p4, this mutation should also affect the H-bond interaction of this propionate group (Fig. 1).
As previously reported (5), the UV-vis spectrum of H118A differs from that of the WT enzyme (Fig. S3). Its Soret maximum is sharper and red shifted to 398 nm, whereas the CT1 band is blue shifted to 625 nm. These changes suggest the presence of a 6cHS heme, as confirmed by the RR core size marker bands (Fig. 5, right). In fact, the H118A coproheme complex is mainly a 6cHS species (ν3 at 1481 cm−1 and ν10 at 1611 cm−1) in equilibrium with a 5cHS (ν3 at 1490 cm−1 and ν10 at 1624 cm−1), with a minor amount of 5cQS species (ν3 at 1505 cm−1 and ν10 at 1636 cm−1). In the low frequency region (Fig. 5, left), the propionate bending modes are WT-like and observed at 371 (strong band) and 387 cm−1 (shoulder).
Figure 5.
Tracing of coproheme decarboxylation in H118A CdChdC upon titration with H2O2. Low frequency (left) and (right) high frequency RR spectral regions of the H118A-coproheme complex titrated with increasing amounts of H2O2 are shown. RR core size marker band wavenumbers are reported in black, propionate bending modes δ(CβCcCd) are indicated in purple, and vinyl bending modes δ(CβCaCb) and vinyl stretching modes (νC=C) are in orange. To see this figure in color, go online.
The titration of the variant H118A with hydrogen peroxide causes a red shift of the Soret band. After the addition of 2 eq. of hydrogen peroxide, the UV-vis spectrum resembles that of the WT heme b complex with a Soret maximum at 404 nm, a weak band at 586 nm, and the CT band at 631 nm (Fig. S3). The RR core size marker bands (Fig. 5, right) indicate that the heme b complex formed is mainly characterized by a 6cHS state. This is in agreement with the recently published mass spectrometry data of this variant (5). An intensity increase of the band at 1623 cm−1 (Fig. 5, right), together with a reduction in intensity of the propionate bending mode at 371 cm−1, is observed as the H2O2-mediated decarboxylation proceeds, starting from 0.2 eq. of H2O2 (Fig. 5, left). Moreover, a new band appears at 413 cm−1, which is assigned to the vinyl bending mode. Upon increasing the amount of hydrogen peroxide, these changes become more evident, and after the addition of 2.0 eq. of H2O2, both the bands at 1623 cm−1 (polarized; see Fig. 4 b) and 413 cm−1 gain intensity and broaden, suggesting the presence of two overlapped νC=C stretching and two overlapped vinyl bending modes, respectively.
Unlike H118A, H118F exhibits WT-like UV-vis spectral signatures with no alteration of the coordination and spin states of the coproheme complex (Fig. S3). Accordingly, the RR spectra are characteristic of a major 5cQS and a minor 5cHS species (Fig. 6, right). However, in the low frequency region, three-propionate bending modes are detected at 365, 372, and 386 cm−1 (Fig. 6, left). This suggests an alteration of the H-bonds, which was not observed in H118A. The titration of H118F with hydrogen peroxide does not induce significant spectral changes in the UV-vis spectrum. Only a new weak band at 586 nm appears, and the Soret band is upshifted by 1 nm upon titration with twofold H2O2 excess. Interestingly, the D2 spectrum shows also the presence of a shoulder at 401 nm (Fig. S3). It is worth noting that the difference spectra of both the absorption and second derivative spectra (Fig. S5) in the Soret region, obtained by subtracting the coproheme spectrum from that of the final form after the addition of H2O2, indicate the presence of a Soret band characterized by a maximum at ∼402 nm.
Figure 6.
Tracing of coproheme decarboxylation in H118F CdChdC upon titration with H2O2. Low frequency (left) and (right) high frequency RR spectral regions of the H118F-coproheme complex titrated with increasing amounts of H2O2 are shown. RR core size marker band wavenumbers are reported in black, propionate bending modes δ(CβCcCd) are indicated in purple, and vinyl bending modes δ(CβCaCb) and vinyl stretching modes (νC=C) are in orange. To see this figure in color, go online.
In line with the UV-vis data, the high frequency RR spectrum of H118F upon titration with H2O2 indicates a mixture of 5cHS and 5cQS species (Fig. 6, left), similar to the H118F-coproheme complex, but with an increase of the 5cHS population in the titrated protein, as indicated by the shoulder at 1570 cm−1 (ν2, 5cHS). At 2 eq. of hydrogen peroxide, the variant H118F clearly shows also an upshifted frequency of the ν10 mode (5cHS) (depolarized band at 1628 cm−1) and a polarized band at ∼1624 cm−1 due to a νC=C stretching mode (Fig. 4 c; Fig. 6, right), similarly to the WT and the variant H118A. Concomitantly, in the low frequency region (Fig. 6, left), an intensity decrease of the propionate bending modes (now at 373 cm−1) is accompanied by the appearance of a new band at 415 cm−1 assigned to a vinyl bending mode. These data suggest the formation of the vinyl 2 at the expense of a propionate group upon titration with H2O2. The spectroscopic results completely agree with the mass spectrometric analysis, which indicates that H118F accumulates MMD (Fig. 3, middle, cyan). No heme b has been detected.
In comparison to the WT CdChdC, the variant A207E shows a red-shifted Soret at 397 nm and a blue-shifted CT1 at 627 nm (Fig. S3), similar to the H118A variant. Accordingly, its RR spectrum in the high frequency region (Fig. 7, right) resembles that of H118A, being a mixture of a 5c and 6cHS species (ν3 at 1485 cm−1) in equilibrium with a 5cQS population (ν3 at 1507 cm−1). In the low frequency region (Fig. 7, left), analogously to the other proteins, a very strong propionate bending mode at 372 cm−1 and a weak band at 387 cm−1 are observed. In addition, a new weak band at ∼397 cm−1 is present, which is tentatively assigned to propionate 4, because E207 is expected to interact with R139, which in turn is H-bonded to p4. In the UV-vis absorption spectra, upon the addition of H2O2, the variant A207E shows a very weak band at 586 nm, which increases slightly up to 1.5 eq. of H2O2, together with a red shift of the Soret band to 401 nm (403 nm in D2) (Fig. S3), similar to what is observed in the difference spectra of both the absorption and second derivative spectra of H118F (Fig. S5). Because of the high sensitivity of RR spectroscopy, upon the addition of 0.1 eq. of hydrogen peroxide, we already notice a decrease of the relative intensity of the bands of the 5cQS species as compared with the other species, together with the appearance of a new band at 1624 cm−1 and an intensity decrease of the propionate bending mode at 372 cm−1 in the low frequency region (data not shown). However, the appearance of a vinyl bending mode at 420 cm−1 is obvious only after the addition of 0.5 eq. of H2O2 (Fig. 7, left). Interestingly, after this addition, the band at 397 cm−1 disappears, whereas the intensity of the propionyl bending mode at 387 cm−1 increases. The final form obtained upon the addition of 1.5 eq. of H2O2, as for the WT and the H118A variant, is characterized by a mixture of a 5cHS (ν3 at 1490 and ν2 at 1570 cm−1) and a 6cHS species (ν3 at 1481 and ν10 at 1611 cm−1) with a minor 5cQS population. However, in contrast to that observed in the WT protein and the variant H118A, the broadening of the ν3 bands, together with the wavenumber upshift of the ν2 band (1570 cm−1 vs. 1566 cm−1), suggests that the 5cHS form is the preponderant species.
Figure 7.
Tracing of coproheme decarboxylation in A207E CdChdC upon titration with H2O2. Low frequency (left) and (right) high frequency RR spectral regions of the A207E-coproheme complex titrated with increasing amounts of H2O2 are shown. RR core size marker band wavenumbers are reported in black, propionate bending modes δ(CβCcCd) are indicated in purple, and vinyl bending modes δ(CβCaCb) and vinyl stretching modes (νC=C) are in orange. To see this figure in color, go online.
The decarboxylation of at least one propionate and the formation of at least one vinyl group is confirmed by 1) the polarized band at 1624 cm−1 (assigned to the νC=C stretching mode; Fig. 4 d), 2) the marked intensity decrease of the propionate bending mode at 372 cm−1 and its upshift to 375 cm−1, and 3) the new broad band at 420 cm−1 assigned to the vinyl bending modes. In line with the larger population of the 5cHS species (MMD) compared to the 6cHS (heme b), the vinyl stretching bands are much weaker than those observed for H118A and the WT protein, confirming that MMD is the dominating redox active reaction intermediate. This agrees with the mass spectrometry analysis: the H2O2-mediated conversion of coproheme (green lines) gives rise mainly to MMD (cyan lines) and partially to heme b (red lines) (Fig. 3, right). Moreover, significant protein denaturation occurs at H2O2 concentrations above 1.5 eq. (dashed boxes/lines), as also observed in the RR spectra (data not shown).
Table S2 summarizes the RR assignment of the core size marker bands and bands associated with the vinyl and propionate groups of WT CdChdC and variants in complex with coproheme, MMD, or heme b.
Active site accessibility
Steady-state and pre-steady-state cyanide binding kinetics at pH 7.0 were determined by UV-vis spectroscopy to assess the impact of the mutations on the accessibility of coproheme in the active site. H118, as the proposed distal base, deprotonates incoming hydrogen peroxide (5) as well as cyanide. As a consequence of its pKa value of 9.2, cyanide is mainly present in its protonated form at a neutral pH. Thus, formation of the LS cyanide complex mirrors Compound 0 formation of ferric CdChdC. Fig. S6 and Table 1 summarize the calculated dissociation constants (KD) and association rates (kon) of WT CdChdC and the variants H118A, H118F, and A207E. WT CdChdC and the A207E variant show similar cyanide binding kinetics and thermodynamics, whereas cyanide binding to H118F is significantly impaired. This suggests that this mutation influences the accessibility to the porphyrin iron by steric effects. Further, the hydrophobicity of the large phenylalanine side chain hinders the polar and negatively charged cyanide molecule from reaching the ferric heme iron. Interestingly, the exchange of A207 by glutamate improved the accessibility of coproheme for cyanide and its affinity to the central iron.
Table 1.
Kinetics of cyanide binding of WT CdChdCs and the variants H118A, H118F, and A207E at pH 7.0
| Protein | KD (μM) | kon (M−1 s−1) | koff (s−1) (=kon × KD) | Reference |
|---|---|---|---|---|
| WT | 29.1 ± 2.2 | 5.1 × 103 | 0.148 | (5) |
| H118A | 36.4 ± 1.1 | 1.8 × 103 | 0.066 | (5) |
| H118F | 285.1 ± 8.3 | 1.4 × 101 | 0.004 | this study |
| A207E | 5.4 ± 2.0 | 6.9 × 103 | 0.037 | this study |
Pre-steady-state cyanide binding kinetics was followed by stopped-flow UV-vis spectroscopy monitoring the change of absorbance in the Soret region. KD values were obtained from steady-state kinetics by using UV-vis spectroscopy to follow the titration of the enzymes with cyanide.
Molecular dynamics of coproheme- and MMD-CdChdC of the WT and the H118A, H118F, and A207E variants
MD simulations were performed to investigate the dynamics of the hydrogen bonding interactions of selected residues within the active site of CdChdC and the flexibility of the catalytically relevant H118 and Y135 and of the iron-coordinating proximal H158.
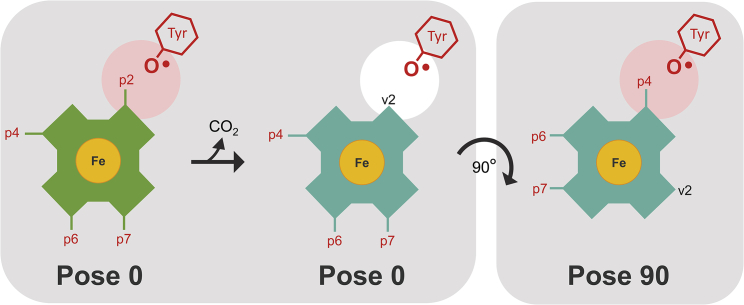
We analyzed the dynamics of the hydrogen bonding interactions of selected residues simulated in complex with coproheme (coproheme pose 0) and MMD in the catalytically active poses, namely MMD pose 90 and with MMD in the coproheme pose (MMD pose 0), i.e., before the 90° rotation (Fig. 8). Three variant structures, H118A, H118F, and A207E, were modeled on the basis of the equilibrated WT coordinates and simulated after an energy minimization of the mutated residues.
Figure 8.
Relevant binding orientations for coproheme (green) and MMD (cyan) to CdChdC used for MD simulations with respect to the catalytic Y135, which is the radical site during turnover. The propionate at p2 on the porphyrin ring is located in close proximity to Y135 in coproheme pose 0, whereas propionate at p4 is found in the position closest to Y135 in MMD pose 90. Not a propionate but a vinyl group at p2 (v2) is in close proximity to Y135 in MMD pose 0. To see this figure in color, go online.
MD simulations of the WT structures have been reported previously (5). Here, they were repeated for extended simulation times of 100 ns to be comparable to the variant simulations. The data are reported as the average over the five independent subunits constituting the protein. The analysis of the hydrogen bonding interactions between the propionates of the respective porphyrin and residues around the active sites is reported in Table S3 and summarized in Fig. 9 A. Overall, the results for WT CdChdC agree with the previous work: the total H-bond occurrence is the highest for coproheme (pose 0) and the lowest for MMD (pose 0). After the 90° rotation of MMD (pose 90), the number of H-bonds is higher than in pose 0. It is worth noting that the total number of H-bonds, averaged over 100-ns simulations and the five subunits, is lower compared to the 20-ns simulations of the previous work (by a factor of 1.2–1.4), as the total occurrences of direct hydrogen bonds and water-mediated (bridging) hydrogen bonds are more balanced when averaged over 100-ns simulations.
Figure 9.
MD simulations (100 ns) of WT CdChdC and the variants H118A, H118F, and A207E. Distances between the catalytic are shown. (A) Total sum of H-bonding interactions in each variant during the entire simulation time of every ensemble under investigation. (B) Y135 and propionate 2 in the unrotated coproheme pose (pose 0) or (C) Y135 and propionate 4 in the rotated MMD pose (pose 90). The normalized densities for the distances (in nanometers) between Y135 and p4 of the individual subunits are plotted on the right. To see this figure in color, go online.
In the H118A and H118F variants, hydrogen bonding is found to be the lowest in coproheme pose 0. In contrast to the WT protein, the rotated MMD intermediate (pose 90) seems to be stabilized by a higher number of H-bonds in H118A and H118F (Fig. 9 A). In A207E, MMD in the catalytically relevant position for p4 cleavage (pose 90) shows the lowest H-bond occurrence (3.75 H-bonds in total), whereas in the pose 0, the number of interactions is higher (4.40 H-bonds). Coproheme in pose 0 exhibits more H-bonds than MMD in pose 0 because of the additional propionate (p2).
The flexibility of catalytic residues was demonstrated by determining distance changes to reference points, like the heme iron or neighboring amino acid residues. We analyzed the flexibility of H118, i.e., the distance of the H118 δ-nitrogen to the iron centers of the respective porphyrins. The distance-time series for all five CdChdC subunits over 100-ns simulations of coproheme, in pose 0, and MMD, in both poses, were generated for the WT protein and the variant A207E. In H118F, the distance between the ζ-carbon of F118 to the respective iron center of coproheme or MMD was monitored instead (Fig. S7). In WT CdChdC and the A207E variant, pronounced density peaks are observed at a distance of 0.43 nm, particularly for coproheme in pose 0 and MMD in pose 0, which indicates a relatively low flexibility of the histidine residue. In the WT protein with MMD in pose 90, H118 fluctuates with maximal distances of up to 1.95 nm. In the variant A207E with MMD in pose 90, the distances between the δ-nitrogen and the iron exhibit a high fluctuation (maximal distances of up to 1.95 nm). In general, the normalized densities are broad and spread toward larger distances in the latter case. In H118F, the flexibility of F118 is comparable to that of H118 in the WT protein. Both coproheme (pose 0) and MMD (pose 0) show less flexibility (average distances around 0.6 nm) than MMD in pose 90, which exhibits a broad range of distances of up to 2 nm with high fluctuations (Fig. S7).
Additionally, the distances between the carboxy group of E207 and the δ-nitrogen of the iron-coordinating proximal H158 in the A207E variant were analyzed (Fig. S8). The time series agree with the hydrogen bonding analyses (Table S3). For simulation of MMD in pose 0, a pronounced density peak at 0.33 nm is observed. In all the subunits, the average distances were below 0.4 nm with small fluctuations, suggesting strong interactions.
Furthermore, the distances between Y135, which is the radical site for both decarboxylation steps, and the adjacent propionate (p2 in pose 0 and p4 in pose 90) over 100-ns simulations together with the normalized densities for the five subunits of both substrates are shown in Fig. 9. In the WT coproheme complex (pose 0), the distances between p2 and Y135 show variations with peaks up to 1 nm (Fig. 9 B). The distance-time series of the MMD (pose 90)-bound form shows fluctuating distances of around 0.4 nm between p4 and Y135 (Fig. 9 C). In H118A, the distances between p4 of MMD and Y135 are quite constant. Pronounced peaks in the density at 0.4 nm can be observed (average distances between 0.38 and 0.58 nm), similar to the WT protein. By contrast, in H118F, the distances between p4 of MMD and Y135 fluctuate with maximal distances of 1.42 nm. The average distances are above 0.58 nm, which is significantly higher than in the WT protein. In A207E, the distances between p4 of MMD and Y135 exhibit the biggest variations among the subunits. Three subunits (B, C, and E) are comparable to the WT protein with average distances between 0.41 and 0.56 nm. The other two subunits (A and D) show peaks above 0.9 nm in the density plot and far higher averages and fluctuations than the WT protein.
Discussion
Active site structure of WT actinobacterial CdChdC and the H118A, H118F, and A207E variants
Coproheme decarboxylases from Firmicutes and Actinobacteria exhibit structural differences in the coproheme cavity. Recently, it has been demonstrated that in the actinobacterial CdChdC, the distal H118 residue acts as a distal base that promotes deprotonation of hydrogen peroxide and Compound I formation, whereas coproheme decarboxylases from Firmicutes lack such a distal basic amino acid. The variants H118A and H118F were initially designed to probe the role of H118 in coproheme decarboxylation, but they have allowed us in this context to derive mechanistic conclusions because of the different size of the inserted residue (alanine versus phenylalanine). Furthermore, we mutated the proximal residue A207, which is fully conserved in ChdCs (12) (Fig. 1 B). In structurally related Clds, a conserved glutamate is found at the position of A207, which is H-bonded to the proximal histidine. Most interestingly, exchange of H118 by phenylalanine as well as A207 by glutamate significantly affected the two-step conversion of coproheme to heme b.
The electronic properties of WT actinobacterial coproheme-CdChdC resemble very closely those of WT coproheme-LmChdC from Firmicutes, i.e., being a mixture of (5c) QS and HS (Fig. S2). In the variants H118A, H118F, and A207E, the coordination and spin states of the respective ferric coproheme complexes were markedly affected. In fact, H118F exhibits WT-like coordination and spin states, whereas H118A and A207E show an increase of the 6cHS species with a concomitant decrease of the 5cQS population. These data suggest that upon replacing the distal H118 with the less-bulky residue alanine, a water molecule can enter the distal cavity and bind to the heme iron. In the case of A207E, the introduction of a polar residue might allow for the establishment of new H-bonds with the proximal H158 and with the R139 residue, which is also H-bonded to p4. The MD simulations suggest that the H-bond between E207 and the proximal H158 is established most frequently in the ensemble with MMD in pose 0 (Fig. S8). In the A207E variant with MMD in pose 90, this H-bond is not stably established and the distance between the catalytic Y135 and p4 is significantly larger than in the WT enzyme. This impairs the second radical decarboxylation and subsequent formation of v4 (Reaction 6) (Figs. 8 C and 9 B). Moreover, the flexibility of the distal H118 residue is increased in A207E, although the mutation was introduced on the proximal side (Fig. S7). This is likely responsible for the presence of a water molecule in the distal cavity, which is able to interact with the heme iron.
RR spectra clearly show that all the introduced mutations also affect the propionate surroundings and their H-bonding interactions in a different manner. The bending modes (δ(CβCcCd)) of the propionate substituents in WT coproheme-LmChdC were found in the low frequency region at 389 (p2), 381 (p4), 374 (p6), and 392 (p7) cm−1 (Supporting materials and methods; Fig. S2) (7). Their frequencies depend on the H-bond strength between the propionate and the nearby amino acids. An increase in the number and strength of the H-bonds leads to an upshift of the bending frequency and vice versa (7,33). Four propionate bending modes are expected for the WT coproheme-CdChdC. However, only two broad bands centered at 372 and 384 cm−1 and at 371 and 387 cm−1 are observed in the WT protein and in the variant H118A, respectively. Interestingly, in the H118F mutant, the band at 372 cm−1 is much narrower than in the WT and a shoulder appears at 365 cm−1, which remains unchanged upon the addition of H2O2 (see below). The x-ray crystal structure of WT CdChdC (PDB: 6XUC) indicates that H118 is strongly H-bonded to p7 (5). Therefore, we suggest that the shoulder at 365 cm−1 originates from propionate 7, whose frequency in the WT protein overlaps with other propionate bending modes. In A207E, another propionate bending mode is observed at 397 cm−1, which disappears upon titration with H2O2 (Fig. 7). According to the structures obtained by in silico mutation, the introduced E207 residue is expected to form an H-bond with the R139 residue, which is H-bonded to p4 (Fig. 1). Because a change in the H-bond interactions involving p4 is expected, we assign the new band observed at 397 cm−1 to p4 (7,33). This is consistent with the MD simulations, which show an increased H-bond interaction between p4 and the surrounding residues (Fig. 9 A; Table S3). Based on the frequency changes observed for the variants, we tentatively assign the propionate bending mode at 372 cm−1 of the WT to the overlapping contributions of p2 and p7 and the other bending mode at 384 cm−1 to p6 and p4.
Hydrogen peroxide-mediated two-step decarboxylation of coproheme—spectral signatures of MMD and heme b
All coproheme decarboxylases of monoderm bacteria utilize hydrogen peroxide to generate heme b via a stepwise decarboxylation of the propionate groups p2 and p4 of the coproheme pyrrole rings A and B (1, 2, 3, 4). Recently, we have demonstrated that in both L. monocytogenes from Firmicutes (7) and in actinobacterial C. diphtheriae (5), heme b production includes 1) the transient formation of the intermediate monovinyl (v2) monopropionate deuteroheme (MMD) by decarboxylation of p2, 2) its 90° reorientation, and 3) the subsequent decarboxylation of p4. Both decarboxylation reactions strongly depend on the H2O2-mediated formation of Compound I∗ with a catalytic tyrosyl radical (5, 6, 7).
Mass spectrometry clearly indicates that WT CdChdC and the variants H118A and A207E convert coproheme to a mixture of MMD and heme b upon titration with H2O2 with different final populations of each species. Whereas A207E mainly accumulates MMD together with a small amount of heme b, in the WT protein and H118A, the amount of heme b is higher than MMD. Interestingly, the H118F variant, whose coproheme complex is spectroscopically identical to the WT protein, is unable to produce heme b. Mass spectrometry demonstrated the formation of MMD only. The spectroscopic characterization clearly indicates that upon titration with H2O2, an equilibrium between the unreacted coproheme species (5cQS) and a new 5cHS heme is observed, which is, therefore, assigned to MMD. The UV-vis absorption spectra, together with their second derivative and difference spectra, between the nontitrated and titrated variant H118F with H2O2 suggest an MMD spectrum characterized by a Soret maximum at around 401–402 nm and a band at 586 nm. This agrees with the results on A207E at 1.5 eq. of H2O2, which mainly accumulates MMD (Fig. S3). It also has to be noted that Celis and co-workers (11) previously reported Soret bands at 396 and 398 nm, with a CT1 at 610 nm for MMD (III) SaChdC and MMD (IV) SaChdC, respectively. The formation of this MMD species is also confirmed by the presence of a νC=C vinyl stretching at 1624 cm−1 (v2), a δ(CβCaCb) vinyl bending mode at 415 cm−1, and three δ(CβCcCd) propionate bending modes at 365, 373, and 386 cm−1 in the RR spectra of H118F.
Even though the H118F active site is poorly accessible and the reaction efficiency with hydrogen peroxide is impaired, MMD formation is performed efficiently in this mutant. MD simulations of H118F show that MMD after the reorientation from the coproheme pose (pose 90) establishes the highest number of H-bonding interactions within its active site compared with all the other variants investigated in this study (Fig. 9 A; Table S3). This should lead to a higher efficiency of the second step of the reaction, assuming that a stably bound redox active substrate is oxidized more easily. However, the decarboxylation of p4 and formation of heme b are not observed so it can be assumed that MMD never reaches position 90 in H118F. In contrast, when H118 is exchanged with a small alanine, heme b is formed. This suggests that the bulky and hydrophobic phenylalanine sterically hinders the rotation of MMD, which is essential for bringing p4 in close proximity to the catalytic residue and, consequently, for its decarboxylation by the Y135 radical.
The variant A207E, which mainly accumulates MMD (5cHS), is characterized by a narrow νC=C vinyl stretching mode at 1624 cm−1 and a weak, but broad, vinyl bending mode at 420 cm−1 (Fig. 7). The rapid intensity decrease of the band at 397 cm−1, tentatively assigned to p4, at low H2O2 concentrations is intriguing because this band should disappear only after the second decarboxylation (p4 to v4) (8). However, the concomitant increase in intensity of the propionate at 387 cm−1 suggests a frequency downshift of the p4 bending mode after the decarboxylation of p2 and rotation of the MMD, possibly because of altered H-bonding interactions. MD simulations of A207E support the observation that in this variant, MMD is accumulated during catalysis, and only a small amount of heme b is produced. H-bonding analysis shows that MMD in pose 90 has the least number of H-bonding interactions with the protein moiety, and therefore, reorientation of MMD is most probably hampered (Table S3). Furthermore, once MMD is in pose 90, the catalytic Y135 is not close enough to perform the decarboxylation of p4 efficiently (Fig. 9).
The titration of the WT protein and H118A variant with H2O2 ultimately results in a mixture of MMD and heme b found in the active site. The RR spectra indicate a predominant 6cHS population with core size marker bands typical of a water-ligated heme and of a minor population of 5cHS. However, the concomitant presence of the MMD compound makes it difficult to conclude whether heme b is only a 6cHS species or a mixture with a 5cHS species in equilibrium with a 5cHS MMD minor population(s).
Proposed reaction mechanism
Recent extensive spectroscopic and structural studies have demonstrated that MMD reorientates after decarboxylation of p2 (Reaction 3) to bring p4 in close proximity to the catalytic tyrosine before the predicted MMD Compound I species is formed (Reaction 4) (5,7). However, the mechanism of reorientation is under discussion, as there are basically two possibilities: 1) a rotation within the active site or 2) a release and rebinding mechanism. The fact that the variant H118F is unable to produce heme b and accumulates MMD strongly suggests that MMD is arrested in the noncatalytical pose 0. As a consequence, the distance between p4 and the catalytic Y135 is too large for initiation of the radical decarboxylation reaction. A long-range electron transfer from Y135 to p4 is highly unlikely (7). Therefore, the neutral tyrosyl radical (formed in Reaction 5) cannot attack the β-carbon of p4, which is essential for formation of the vinyl group and release of carbon dioxide (Reaction 6). Nevertheless, CdChdC H118F is able to perform the first decarboxylation reaction of p2 of coproheme (Reaction 3) and form the vinyl group (v2) at this position (Fig. 6). Consequently, the Compound I formation (Reaction 1) and the subsequent Compound I∗ formation (Reaction 2) are possible. The Compound I formation in H118F must be slow as can be deduced from the cyanide binding studies, in which the kon rate is found to be lower by two orders of magnitude compared to the WT protein (Table 1). This is in agreement with previously published data, which demonstrated that H118 is necessary to deprotonate the incoming oxidant or ligand (5). Furthermore, the overall accessibility of the active site in H118F is reduced by the presence of the bulky and hydrophobic F118 residue. Our conclusions are consistent with the fact that the variant H118A also exhibits a significantly reduced cyanide binding rate but is able to perform both decarboxylation steps (Fig. 5) (5). In H118A, MMD is able to rotate to the catalytically active pose 90 for decarboxylation of p4. The inability of the H118F variant to perform the second decarboxylation reaction together with the steric effects within the active site led us to the conclusion that MMD must be able to rotate within the active site to complete decarboxylation.
In addition, the introduction of the hydrogen bonding E207 residue on the proximal heme side (A207E) (Fig. 1) also leads to accumulation of MMD (Fig. 7), with only some formation of heme b. MD simulations clearly indicate that an H-bond between E207 and the proximal H158 is most frequently established in the unrotated MMD form (pose 0) (Fig. S8). Additionally, once MMD is in the less-stabilized reoriented position, the catalytic Y135 is not close enough to p4 to guarantee efficient decarboxylation by the neutral tyrosyl radical (MMD Compound I∗) (Reaction 5, Fig. 9). This observation strongly suggests that the lack of the proximal H-bonding network in ChdCs is not only necessary to deliver the product heme b (34,35) to other apo-proteins but also to facilitate the flexibility of MMD during turnover while maintaining the interaction with H158. The introduction of the proximal H-bonding network with H158 increases its imidazolate character and modifies the active site, which is reflected by the increased affinity of cyanide and its faster kon (Table 1). In addition, the E207-H158 interaction should modify the redox properties of the respective cofactors, which might explain the observed oxidation of coproheme, MMD, and heme b in the A207E variant (Fig. 3, right). MD simulations are consistent with the hypothesis that the lack of the H-bonding network is necessary to provide the freedom needed for MMD to rotate within the active site between Reaction 3 and Reaction 4. Clearly, MMD in pose 90 is not the favored orientation in A207E.
Available data on lysine variants in SaChdC and LmChdC, involved in p4 coordination of coproheme in the initial orientation, also agree with a rotation mechanism of MMD within the active site. In these lysine variants, the second decarboxylation (Reaction 6) is significantly hampered, resulting in accumulation of MMD (10,36). The surroundings and H-bonding of the p2 and p4 groups of coproheme are identical to that of p4 and p6 of MMD. As a consequence, the p4 and p6 binding interactions of MMD in pose 90 are identical to those of p2 and p4 of coproheme in pose 0. Therefore, after a complete release from the active site, MMD would most likely rebind in pose 90. In this case, decarboxylation of p4 would be catalyzed and no accumulation of MMD would be observed.
Conclusions
A comprehensive spectroscopic characterization and marker bands assignment of all the ferric iron porphyrin species taking part in the stepwise decarboxylation reaction of coproheme catalyzed by actinobacterial coproheme decarboxylases have been obtained. Additionally, important insight into the mechanistically relevant reorientation step of the transiently formed intermediate MMD is provided, favoring a rotation mechanism within the active site and not a release and rebinding mechanism.
Author contributions
G.S., S.H., and C. Obinger designed research. F.S., H.M., B.L., M.B., P.G.F., and C. Oostenbrink performed research. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Acknowledgments
We thank Dr. B. D. Howes for helpful discussions.
This project was supported by Fondazione Cassa Risparmio di Firenze, grant 2020.1397 (G.S.) and the Austrian Science Fund stand-alone project P29099 (S.H.) and FWF doctoral program “Biomolecular Technology of Proteins” W1224 (H.M., B.L., C. Oostenbrink, and C. Obinger). F.S. was the recipient of a fellowship partially funded by Italian Ministry of Education, University and Research (“Progetto Dipartimenti di Eccellenza 2018–2022” allocated to the Department of Chemistry “Ugo Schiff”).
Editor: Elsa Yan.
Footnotes
Federico Sebastiani and Hanna Michlits contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.06.042.
Contributor Information
Stefan Hofbauer, Email: stefan.hofbauer@boku.ac.at.
Giulietta Smulevich, Email: giulietta.smulevich@unifi.it.
Supporting material
References
- 1.Dailey H.A., Dailey T.A., Warren M.J. Prokaryotic heme biosynthesis: multiple pathways to a common essential product. Microbiol. Mol. Biol. Rev. 2017;81:e00048-16. doi: 10.1128/MMBR.00048-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dailey H.A., Gerdes S. HemQ: an iron-coproporphyrin oxidative decarboxylase for protoheme synthesis in Firmicutes and Actinobacteria. Arch. Biochem. Biophys. 2015;574:27–35. doi: 10.1016/j.abb.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lobo S.A., Scott A., Saraiva L.M. Staphylococcus aureus haem biosynthesis: characterisation of the enzymes involved in final steps of the pathway. Mol. Microbiol. 2015;97:472–487. doi: 10.1111/mmi.13041. [DOI] [PubMed] [Google Scholar]
- 4.Dailey H.A., Gerdes S., Phillips J.D. Noncanonical coproporphyrin-dependent bacterial heme biosynthesis pathway that does not use protoporphyrin. Proc. Natl. Acad. Sci. USA. 2015;112:2210–2215. doi: 10.1073/pnas.1416285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michlits H., Lier B., Hofbauer S. Actinobacterial coproheme decarboxylases use histidine as a distal base to promote Compound I formation. ACS Catal. 2020;10:5405–5418. doi: 10.1021/acscatal.0c00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hofbauer S., Pfanzagl V., Furtmüller P.G. Understanding molecular enzymology of porphyrin-binding α + β barrel proteins - one fold, multiple functions. Biochim. Biophys. Acta. Proteins Proteom. 2021;1869:140536. doi: 10.1016/j.bbapap.2020.140536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milazzo L., Gabler T., Hofbauer S. Redox cofactor rotates during its stepwise decarboxylation: molecular mechanism of conversion of coproheme to heme b. ACS Catal. 2019;9:6766–6782. doi: 10.1021/acscatal.9b00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Streit B.R., Celis A.I., DuBois J.L. Decarboxylation involving a ferryl, propionate, and a tyrosyl group in a radical relay yields heme b. J. Biol. Chem. 2018;293:3989–3999. doi: 10.1074/jbc.RA117.000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milazzo L., Hofbauer S., Smulevich G. Insights into the active site of coproheme decarboxylase from Listeria monocytogenes. Biochemistry. 2018;57:2044–2057. doi: 10.1021/acs.biochem.8b00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milazzo L., Gabler T., Smulevich G. The hydrogen bonding network of coproheme in coproheme decarboxylase from Listeria monocytogenes: effect on structure and catalysis. J. Inorg. Biochem. 2019;195:61–70. doi: 10.1016/j.jinorgbio.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celis A.I., Streit B.R., DuBois J.L. Unusual peroxide-dependent, heme-transforming reaction catalyzed by HemQ. Biochemistry. 2015;54:4022–4032. doi: 10.1021/acs.biochem.5b00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfanzagl V., Holcik L., Hofbauer S. Coproheme decarboxylases - phylogenetic prediction versus biochemical experiments. Arch. Biochem. Biophys. 2018;640:27–36. doi: 10.1016/j.abb.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofbauer S., Mlynek G., Obinger C. Hydrogen peroxide-mediated conversion of coproheme to heme b by HemQ-lessons from the first crystal structure and kinetic studies. FEBS J. 2016;283:4386–4401. doi: 10.1111/febs.13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLano W.L. Schrodinger LLC; 2010. The PyMOL Molecular Graphics System, version 1.3r1. Available online: http://www.pymol.org. [Google Scholar]
- 15.Vangunsteren W.F., Karplus M. A method for constrained energy minimization of macromolecules. J. Comput. Chem. 1980;1:266–274. [Google Scholar]
- 16.Schmid N., Christ C., van Gunsteren W. Architecture, implementation and parallelisation of the GROMOS software for biomolecular simulation. Comput. Phys. Commun. 2012;183:890–903. [Google Scholar]
- 17.Reif M.M., Hünenberger P.H., Oostenbrink C. New interaction parameters for charged amino acid side chains in the GROMOS force field. J. Chem. Theory Comput. 2012;8:3705–3723. doi: 10.1021/ct300156h. [DOI] [PubMed] [Google Scholar]
- 18.Zou C., Larisika M., Nowak C. Two-dimensional heterospectral correlation analysis of the redox-induced conformational transition in cytochrome c using surface-enhanced Raman and infrared absorption spectroscopies on a two-layer gold surface. J. Phys. Chem. B. 2013;117:9606–9614. doi: 10.1021/jp404573q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berendsen H.J.C., Postma J.P.M., Hermans J. Reidel; Dordrecht, the Netherlands: 1981. Interaction Models for Water in Relation to Protein Hydration. [Google Scholar]
- 20.Abraham M.J., Murtola T., Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 21.Hockney R.W. The potential calculation and some applications. Methods Comput. Phys. 1970;9:136–211. [Google Scholar]
- 22.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:324–341. [Google Scholar]
- 23.Tironi I.G., Sperb R., van Gunsteren W.F. A generalized reaction field method for molecular-dynamics simulations. J. Chem. Phys. 1995;102:5451–5459. [Google Scholar]
- 24.Heinz T.N., van Gunsteren W.F., Hünenberger P.H. Comparison of four methods to compute the dielectric permittivity of liquids from molecular dynamics simulations. J. Chem. Phys. 2001;115:1125–1136. [Google Scholar]
- 25.Pall S., Hess B. A flexible algorithm for calculating pair interactions on SIMD architectures. Comput. Phys. Commun. 2013;184:2641–2650. [Google Scholar]
- 26.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 27.Harvey S.C., Tan R.K.Z., Cheatham T.E., III The flying ice cube: velocity rescaling in molecular dynamics leads to violation of energy equipartition. J. Comput. Chem. 1998;19:726–740. [Google Scholar]
- 28.Eichenberger A.P., Allison J.R., van Gunsteren W.F. GROMOS++ software for the analysis of biomolecular simulation trajectories. J. Chem. Theory Comput. 2011;7:3379–3390. doi: 10.1021/ct2003622. [DOI] [PubMed] [Google Scholar]
- 29.Howes B.D., Schiodt C.B., Smulevich G. The quantum mixed-spin heme state of barley peroxidase: a paradigm for class III peroxidases. Biophys. J. 1999;77:478–492. doi: 10.1016/S0006-3495(99)76905-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smulevich G., Feis A., Howes B.D. Fifteen years of Raman spectroscopy of engineered heme containing peroxidases: what have we learned? Acc. Chem. Res. 2005;38:433–440. doi: 10.1021/ar020112q. [DOI] [PubMed] [Google Scholar]
- 31.Smulevich G., Howes B.D., Droghetti E. In: Heme Peroxidases, RSC Metallobiology Series. Raven E., Dunford B., editors. Vol. 4. The Royal Society of Chemistry; 2016. Structural and functional properties of heme-containing peroxidases: a resonance Raman perspective for the superfamily of plant, fungal and bacterial peroxidases; pp. 61–98. [Google Scholar]
- 32.Indiani C., Feis A., Smulevich G. Benzohydroxamic acid-peroxidase complexes: spectroscopic characterization of a novel heme spin species. J. Am. Chem. Soc. 2000;122:7368–7376. [Google Scholar]
- 33.Cerda-Colon J., Silfa E., Lopez-Garriga J. Unusual rocking freedom of the heme in the hydrogen sulfide-binding hemoglobin from Lucina pectinata. J. Am. Chem. Soc. 1998;120:9312–9317. doi: 10.1002/(sici)1520-6343(1998)4:5<311::aid-bspy3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 34.Hofbauer S., Howes B.D., Obinger C. From chlorite dismutase towards HemQ - the role of the proximal H-bonding network in haeme binding. Biosci. Rep. 2016;36:e00312. doi: 10.1042/BSR20150330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofbauer S., Hagmüller A., Furtmüller P.G. Structure and heme-binding properties of HemQ (chlorite dismutase-like protein) from Listeria monocytogenes. Arch. Biochem. Biophys. 2015;574:36–48. doi: 10.1016/j.abb.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Celis A.I., Gauss G.H., DuBois J.L. Structure-based mechanism for oxidative decarboxylation reactions mediated by amino acids and heme propionates in coproheme decarboxylase (HemQ) J. Am. Chem. Soc. 2017;139:1900–1911. doi: 10.1021/jacs.6b11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.