Abstract
HOXC10 and mitochondrial fission regulator 2 (MTFR2) have been reported to be abnormally expressed in multiple types of cancer tissues. However, the effects of HOXC10 and MTFR2 on colorectal cancer (CRC) remain poorly understood. Therefore, the present study aimed to investigate the expression of HOXC10 and MTFR2 in CRC tissues and cells, and analyze their effects on CRC cell proliferation, invasion and migration. Reverse transcription-quantitative PCR and western blotting were used to detect the expression levels of MTFR2 and HOXC10 in tissues and cells. To investigate the association between MTFR2 and HOXC10, short hairpin RNA-MTFR2 and overexpression vector-HOXC10 were transfected into the cells, respectively. Furthermore, western blotting was performed to detect the expression levels of invasion-associated proteins. The proliferation, clone formation, invasion and migration of colorectal cancer cells were in turn analyzed by the Cell Counting Kit-8, clone formation, wound healing and Transwell assays. Japan Automotive Software Platform and Architecture software predicted the binding sites between HOXC10 and MTFR2, which was confirmed by the dual-luciferase reporter assay and chromatin immunoprecipitation. The present study demonstrated that HOXC10 and MTFR2 mRNA and protein expression levels were significantly upregulated in CRC tissues and cells. MTFR2 knockdown significantly inhibited CRC cell proliferation, clone formation, invasion and migration. Furthermore, HOXC10 was shown to interact with MTFR2. HOXC10 overexpression was able to significantly reverse the inhibitory effects of MTFR2 knockdown on CRC cells. In conclusion, HOXC10 overexpression activated MTFR2 expression to enhance the proliferation, clone formation, invasion and migration of CRC cells.
Keywords: HOXC10, mitochondrial fission regulator 2, proliferation, invasion, migration, colorectal cancer
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors. The incidence rate of CRC in women ranks third only to lung cancer and breast cancer, and ranks fourth only to lung cancer, stomach cancer and liver cancer in men (1). According to the World Health Organization's International Agency for Research on Cancer, in 2018 there were ~1.8 million new CRC diagnoses, accounting for 10% of all patients with cancer, and ~860,000 CRC related-deaths, accounting for 9.9% of all cancer-related mortalities (1). The prevalence of CRC has been increasing over the last 5–10 years, with an estimated 2.2 million new CRC cases and 1.1 million deaths worldwide predicted by 2030 (2). In 1990, CRC was ranked 21st among the leading causes of death in China, but in 2017 it rose to the 11th, with a CRC mortality rate of 13.24 per 100,000 people (3). In 2017, CRC was therefore accountable for 1.79% of all deaths, seriously threatening human health and imposing a huge economic burden on society (3). Thus, it is important to find a new, safe and effective treatment for CRC.
According to the Gene Expression Omnibus database (dataset, GSE10950), the expression of mitochondrial fission regulator (MTFR) 2 was significantly upregulated in patients with CRC (4). MTFR2 is a gene that encodes mitochondrial proteins and belongs to the MTFR1/family with sequence similarity 54 member A family. MTFR2 induces fission of mitochondria in eukaryotic cells and provides energy for aerobic respiration (5). Wang et al (6) demonstrated that MTFR2 is highly expressed in glioblastoma and is associated with a poor prognosis (6). MTFR2 has also been reported to be highly expressed in oral squamous cell carcinoma tissues and to be negatively related to the overall survival (OS) of patients (7). MTFR2 may also aggravate proliferation, migration and invasion of oral squamous carcinoma cells by switching oxidative phosphorylation to glycolysis (7). A previous study revealed that high MTFR2 expression is related to poor prognosis in patients with breast cancer (BC) and is more prevalent in patients with aggressive tumors (8). The invasion, migration and epithelial-mesenchymal transition of BC cells has also been shown to be suppressed by the knockdown of MTFR2 (9). However, the biological function of MTFR2 in CRC remains unclear.
A previous study revealed that HOXC10 expression is significantly higher in CRC samples than in normal adjacent tissues, and may be used as a biomarker for the diagnosis of CRC and as a potential therapeutic target (10). HOXC10 is also considered a key modulator in ovarian cancer (OC), which is associated with poor prognosis in patients with OC, actively regulating Slug transcription to promote OC metastasis (11). Furthermore, knockdown of HOXC10 has been reported to suppress the proliferation, migration and invasion of gastric cancer (GC) cells in vitro, and to suppress tumor growth and induce apoptosis in vivo (12). HOXC10 expression is also markedly upregulated in glioblastoma tissues and cells, resulting in a poor OS in patients with glioblastoma. Notably, inhibition of HOXC10 could inhibit the proliferation, migration and invasion of glioblastoma cells (13). Therefore, it is speculated that HOXC10 could also regulate CRC progression.
The aim of the present study was to investigate whether HOXC10 binding to MTFR2 could regulate the proliferation, invasion and migration of CRC cells.
Materials and methods
Human tumor samples
The present study was approved by the Clinical Research Ethics Committee of Nanjing Medical University (Nanjing, China; approval no. 2021–448) and written informed consent was obtained from each patient. In total, CRC tissues and paraneoplastic tissues (>2 cm away from the edge of the tumor) were obtained from 16 patients with CRC in the Jiangsu Cancer Hospital (Nanjing, China) between May 2019 and June 2020. None of the patients had received cancer treatment beforehand. The tumor samples were stored at −80°C until further use. The histopathological characteristics of the patients are shown in Table I.
Table I.
Histopathological characteristics of the patients.
| No. | Age, years | Sex | Number of LNM | Grading | Tumor location | Differentiation degree | Distant metastasis |
|---|---|---|---|---|---|---|---|
| 1 | 63 | Female | 1 | II | Sigmoid colon | Middle-low | M1 |
| 2 | 16 | Male | 1 | IV | Middle segment of transverse colon | Low | M1 |
| 3 | 69 | Female | 0 | II | Rectum | Middle | M0 |
| 4 | 39 | Male | 0 | II | Transverse colon | Middle | M0 |
| 5 | 66 | Male | 1 | III | Junction of rectum and sigmoid colon | Middle | M0 |
| 6 | 43 | Female | 2 | III | Sigmoid colon | Middle | M0 |
| 7 | 60 | Male | 0 | I | Rectum | Middle | M0 |
| 8 | 75 | Female | 0 | II | Ascending colon | Middle | M0 |
| 9 | 67 | Female | 0 | I | Rectum | Middle | M0 |
| 10 | 63 | Female | 0 | II | Transverse colon (proximal to hepatic curvature) | Middle | M0 |
| 11 | 48 | Female | 0 | I | Rectum | Middle | M0 |
| 12 | 54 | Male | 2 | III | Sigmoid colon | Middle | M0 |
| 13 | 54 | Female | 1 | III | Colon | Middle | M0 |
| 14 | 51 | Male | 2 | IV | Sigmoid colon | Middle-low | M1 |
| 15 | 66 | Female | 0 | IV | Ileocecal junction | Middle-low | M1 |
| 16 | 55 | Male | 1 | III | Rectum | Middle | M0 |
LNM, lymph node metastasis (32); M0, no distant metastasis; M1, have distant metastasis.
Cell culture and transfection
The human intestinal epithelial cell line (HIEC-6) and colorectal cancer cell lines (Caco-2, HCT116, SW480 and LoVo) were all obtained from the American Type Culture Collection. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin solution at 37°C with 5% CO2. Short hairpin RNA (sh)-MTFR2-1/2 (its negative control was sh-NC) and overexpression (Oe)-HOXC10 (its negative control was Oe-NC) plasmids were provided by Shanghai GenePharma Co., Ltd. HCT116 cells were seeded at 1.5×105 cells/well in 6-well plates for 24 h at 37°C. When cell density reached 60% confluence, HCT116 cells were transfected with sh-NC (40 nM), sh-MTFR2-1/2 (40 nM), Oe-NC (60 nM) and Oe-HOXC10 (60 nM) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Cells were then cultured in normal medium at 37°C for 48 h and subsequent experiments were conducted.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells and tissue samples using TRIzol® reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Total RNA (1 µg) was reverse transcribed into cDNA at 42°C for 30 min, according to the manufacturer's protocol of the PrimeScript RT Reagent Kit (cat. no. RR037A; Takara Biotechnology Co., Ltd.). qPCR was performed using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) on an Applied Biosystems 7900 PCR system (Thermo Fisher Scientific, Inc.). The thermocycling conditions were used: Initial denaturation at 95°C for 10 min; followed by 40 cycles of denaturation at 95°C for 15 sec and annealing at 60°C for 1 min; and a final extension of 10 min at 72°C. The following primer pairs were used for qPCR: MTFR2 forward, 5′-AGGGCTACGGGCCAATTTGA-3′ and reverse, 5′-TTCCTAAATAAAGTTTGGTCCAC-3′; HOXC10 forward, 5′-CTCGGATAACGAAGCGAAAG-3′ and reverse, 5′-CGCTCTCGCGTCAAATACAT-3′; and GAPDH forward, 5′-TGTGGGCATCAATGGATTTGG−3′ and reverse, 5′-ACACCATGTATTCCGGGTCAAT-3′. MTFR2 and HOXC10 mRNA expression levels were quantified using the 2−ΔΔCq method and normalized to the internal reference gene GAPDH (14).
Western blotting
Total protein was extracted from cells using cold RIPA buffer (cat. no. P0013C; Beyotime Institute of Biotechnology), and were centrifuged at 3,000 × g for 15 min at 4°C. The non-transfected cells were used as the control group. Total protein was quantified using a BCA kit (Beyotime Institute of Biotechnology) and 20 µg protein/lane was separated by SDS-PAGE on a 12% gel. The separated proteins were transferred to PVDF membranes and blocked with 5% non-fat milk with TBS-0.1% Tween-20 (TBST) for 1 h at room temperature. The membranes were incubated overnight at 4°C with primary antibodies against the following: MTFR2 (1:500; cat. no. ab155678; Abcam), MMP2 (1:1,000; cat. no. ab92536; Abcam), MMP9 (1:1,000; cat. no. ab76003; Abcam), HOXC10 (1:500; cat. no. ab153904; Abcam) and GAPDH (1:2,500; cat. no. ab9485; Abcam). Following the primary incubation, membranes were washed with TBST and incubated with an appropriate HRP-conjugated secondary antibody (1:1,000; cat. no. 7074; Cell Signaling Technology, Inc.) for 1 h at room temperature. Protein bands were observed using a chemiluminescence reagent (ECL) kit (Beyotime Institute of Biotechnology). The gray values of bands were semi-quantified using ImageJ software (version 1.0; National Institutes of Health) with GAPDH as the loading control.
Cell Counting Kit-8 (CCK-8) assay
The proliferation of HCT116 cells was detected by performing the CCK-8 assay. Following transfection, cells (2×103 cells/well) were seeded into 96-well plates. The non-transfected cells were used as the control group. Cell proliferation was detected at 24, 48 and 72 h using the CCK-8 reagent (Beyotime Institute of Biotechnology) according to the manufacturer's instructions. Cells were incubated with 10 µl CCK-8 reagent for 2 h before the absorbance value at 450 nm was detected via a microplate reader (Bio-Rad Laboratories, Inc.).
Clone formation assay
Following transfection, HCT116 cells (1×103 cells/well) were seeded into a 6-well plate and cultured in DMEM at 37°C for 2 weeks. The non-transfected cells were used as the control group. Cell culture was stopped when clones were visible to the naked eye in the culture dish. The cells were fixed with 10% formaldehyde for 10 min at room temperature and stained with 0.5% crystal violet for 5 min at room temperature. The number of cloned cells was quantified by eye.
Wound healing assay
The migratory ability of HCT116 cells was detected by the wound healing assay. Following transfection, HCT116 cells (5×105 cells/well) were seeded into 6-well plates and cultured until cells reached 100% confluence. The non-transfected cells were used as the control group. Serum-free medium replaced normal medium and a sterile 100-µl pipette tip was used to scratch the cells (0 h). Unattached cells were removed and the remaining cells were cultured at 37°C for 24 h. The wound gap was observed under an inverted light microscope (magnification, ×100) and the cell migration rate was calculated using ImageJ (version 1.52r; National Institutes of Health).
Transwell assay
The invasive ability of HCT116 cells was detected via the Transwell assay. Following transfection, HCT116 cells (1×105 cells/well) in 200 µl DMEM were placedin the upper Transwell chamber with the 8-µm pore inserted membranes pre-coated with Matrigel (BD Biosciences) overnight at 37°C. The non-transfected cells were used as the control group. In the lower chamber, 600 µl DMEM containing 10% FBS was added. After cells were cultured at 37°C for 24 h, cells in the upper side of the chamber were removed and those in the lower side of the chamber were fixed with methanol for 20 min at room temperature and then stained with crystal violet for 10 min at room temperature. Cells were observed and imaged using an inverted light microscope (magnification, ×100), and the cell invasion rate was calculated using ImageJ (version 1.52r; National Institutes of Health). The number of invasive cells were counted from five randomly selected fields.
Bioinformatics analysis
Japan Automotive Software Platform and Architecture [JASPAR, 8th (2020); http://jaspar.genereg.net/] predicted that HOXC10 could bind to the promoter sequence of MTFR2.
Dual-luciferase reporter assay
The interaction between HOXC10 and the MTFR2 promoter in HCT116 cells was determined via the dual-luciferase reporter assay. The wild-type (WT) and mutant (MUT) sequences of MTFR2 untranslated region were amplified by Shanghai GenePharma Co., Ltd., cloned into a pGL3 luciferase vector (Promega Corporation) and respectively named MTFR2-WT and MTFR2-MUT. HCT116 cells at 80% confluence were co-transfected with MTFR2-WT (50 nM) or MTFR2-MUT (50 nM) and Oe-HOXC10 (50 nM) or Oe-NC (50 nM) using Lipofectamine 2000. Following incubation for 48 h at 37°C, cells were collected and firefly and Renilla luciferase activities were detected using a Dual-Luciferase Reporter Assay System (Promega Corporation) according to the manufacturer's protocol. Firefly luciferase was normalized to Renilla luciferase activity.
Chromatin immunoprecipitation (ChIP)
The interaction between HOXC10 and the MTFR2 promoter in HCT116 cells was also determined by ChIP. ChIP was conducted using the ChIP Assay Kit (Beyotime Institute of Biotechnology) according to the manufacturer's protocol. Protein and DNA in HCT116 cells were cross-linked in 1% formaldehyde for 10 min at room temperature, extracted by SDS lysis buffer, and separated by ultrasonication to obtain DNA fragments. The cell lysate was then immunoprecipitated using anti-HOXC10 (1:500; cat. no. ab153904; Abcam) or IgG (1:1,000; cat. no. 7074; Cell Signaling Technology, Inc.) antibodies. Immunoprecipitated DNA was analyzed via RT-qPCR.
Statistical analysis
All experiments were repeated independently in triplicate. All statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software, Inc.). Data are presented as the mean ± SD. Comparisons among two groups were analyzed using a paired or unpaired Student's t-test, whereas comparisons among multiple groups were analyzed using one-way ANOVA, followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
mRNA expression levels of HOXC10 and MTFR2 are upregulated in CRC tissues
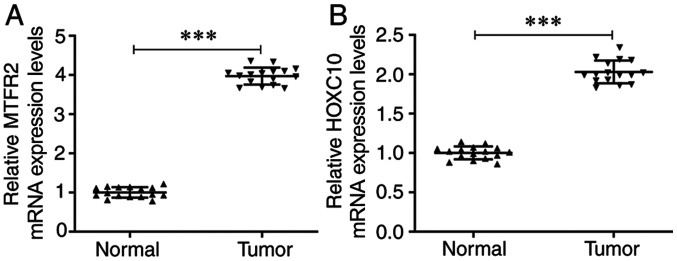
The mRNA expression levels of MTFR2 were significantly increased in CRC tissues compared with those in the matched normal adjacent tissues (Fig. 1A). Furthermore, the mRNA expression levels of HOXC10 were also significantly upregulated in CRC tissues compared with those in the matched adjacent tissues (Fig. 1B).
Figure 1.
mRNA expression levels of HOXC10 and MTFR2 are upregulated in CRC tissues. mRNA expression levels of (A) MTFR2 and (B) HOXC10 in CRC tissues and matched normal adjacent tissues were detected by reverse transcription-quantitative PCR. ***P<0.001. MTFR2, mitochondrial fission regulator 2; CRC, colorectal cancer.
MTFR2 knockdown inhibits CRC cell proliferation
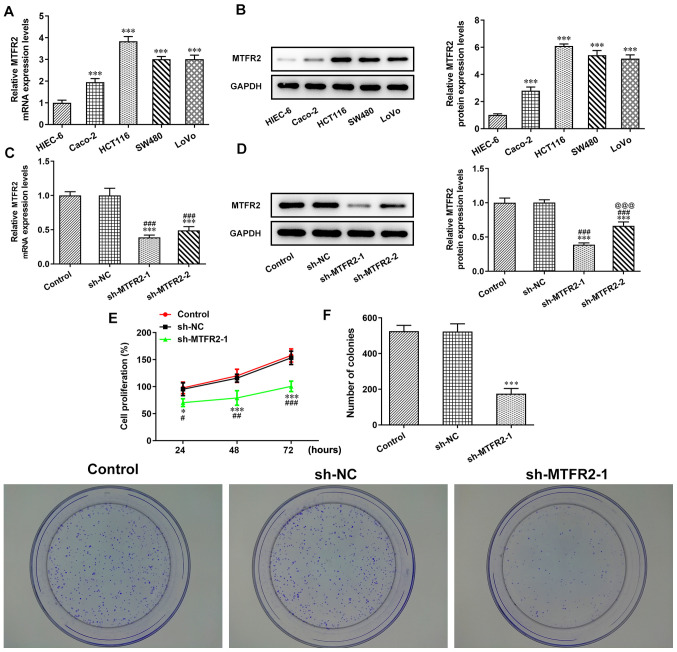
The mRNA and protein expression levels of MTFR2 in the different CRC cell lines were significantly higher compared with those in the HIEC-6 cells (Fig. 2A and B). Notably, the MTFR2 mRNA and protein expression levels were highest in HCT116 cells, which were therefore chosen for subsequent experiments. When HCT116 cells were transfected with sh-MTFR2-1/2, the mRNA and protein expression levels of MTFR2 were significantly downregulated compared with those in the control and sh-NC groups (Fig. 2C and D). Furthermore, MTFR2 protein expression levels in the sh-MTFR2-1 group were significantly lower compared with those in the sh-MTFR2-2 group, sh-MTFR2-1 was therefore used for subsequent experiments. Proliferation and clone formation were also shown to be significantly reduced in response to sh-MTFR2-1 compared with in the sh-NC group (Fig. 2E and F).
Figure 2.
MTFR2 knockdown inhibits CRC cell proliferation. (A) mRNA and (B) protein expression levels of MTFR2 in CRC and the HIEC-6 cell lines were analyzed by RT-qPCR and western blotting, respectively. ***P<0.001 vs. HIEC-6. (C) mRNA and (D) protein expression levels of MTFR2 in HCT116 cells transfected with sh-MTFR2-1/2 were analyzed by RT-qPCR and western blotting, respectively. (E) Cell proliferation and (F) clone formation ability of HCT116 cells transfected with sh-MTFR2-1 were determined by the Cell Counting Kit-8 assay and clone formation assay, respectively. *P<0.05 and ***P<0.001 vs. control group; #P<0.05, ##P<0.01 and ###P<0.001 vs. sh-NC group; @@@P<0.001 vs. sh-MTFR2-1 group. MTFR2, mitochondrial fission regulator 2; CRC, colorectal cancer; RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin RNA; NC, negative control.
MTFR2 knockdown inhibits invasion and migration of CRC cells
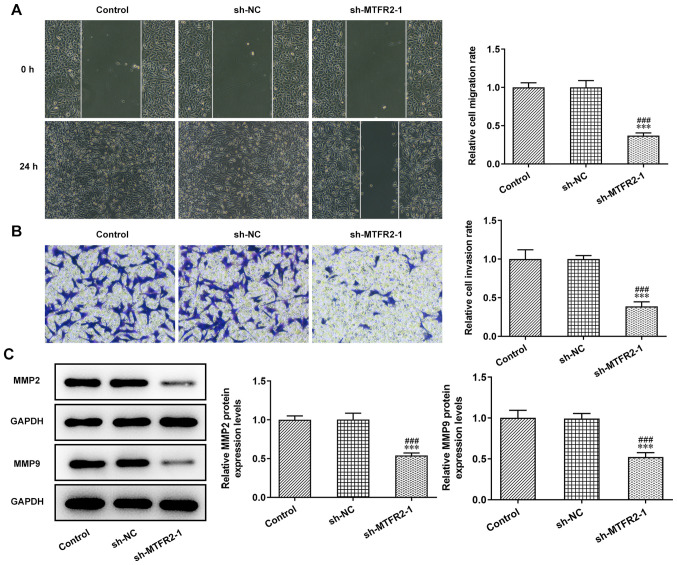
The invasion and migration of HCT116 cells were significantly inhibited by sh-MTFR2-1 compared with in the control and sh-NC groups (Fig. 3A and B). Furthermore, knockdown of MTFR2 significantly downregulated the protein expression levels of MMP2 and MMP9 in HCT116 cells compared with those in the control and sh-NC groups (Fig. 3C).
Figure 3.
MTFR2 knockdown inhibits colorectal cancer cell invasion and migration. (A) Migration and (B) invasion of HCT116 cells transfected with sh-MTFR2-1 were analyzed by wound healing and Transwell assays, respectively (magnification, ×100). Relative migration and invasion rates were expressed normalized to the control group. (C) Expression of invasion- and migration-related proteins in HCT116 cells transfected with sh-MTFR2-1 were detected via western blotting. ***P<0.001 vs. control group; ###P<0.001 vs. sh-NC group. MTFR2, mitochondrial fission regulator 2; sh, short hairpin RNA; NC, negative control.
HOXC10 is upregulated in CRC cells and activates MTFR2 expression
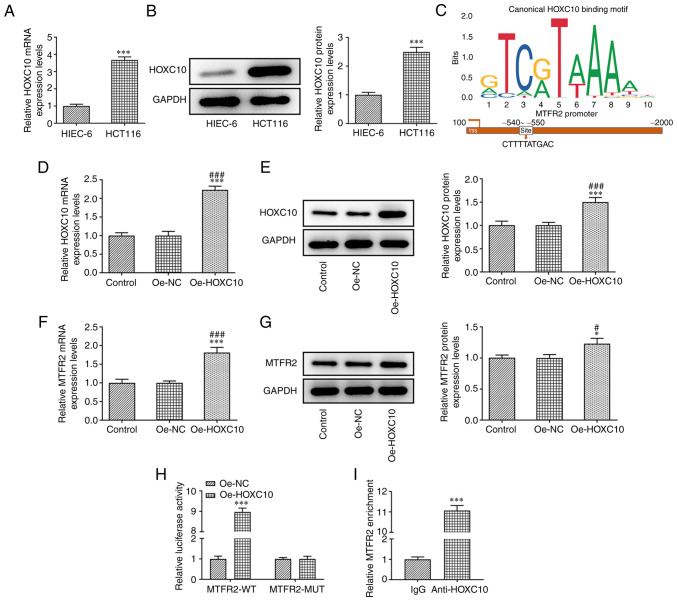
HOXC10 mRNA and protein expression levels in HCT116 cells were significantly higher compared with those in HIEC-6 cells (Fig. 4A and B). JASPAR predicted the binding sites between HOXC10 and MTFR2 (Fig. 4C). In HCT116 cells transfected with Oe-HOXC10, the mRNA and protein expression levels of HOXC10 (Fig. 4D and E) and MTFR2 (Fig. 4F and G) were significantly upregulated compared with those in the control and Oe-NC groups. Furthermore, the results demonstrated that relative luciferase activity was increased in HCT116 cells co-transfected with MTFR2-WT and Oe-HOXC10 compared with co-transfected with MTFR2-WT and Oe-NC, while there was no significant change in MTFR2-MUT (Fig. 4H). The ChIP results showed a significant increase in relative MTFR2 expression in anti-HOXC10 compared with IgG, which further confirmed that HOXC10 bound to MTFR2 (Fig. 4I).
Figure 4.
HOXC10 is upregulated in colorectal cancer cells, and binds to the MTFR2 promoter to activate MTFR2 expression. (A) mRNA and (B) protein expression levels of HOXC10 in HCT116 and HIEC-6 cell lines were analyzed by RT-qPCR and western blotting, respectively. ***P<0.001 vs. HIEC-6. (C) Binding sites between HOXC10 and MTFR2 were determined using the Japan Automotive Software Platform and Architecture program. (D) mRNA and (E) protein expression levels of HOXC10 in HCT116 cells transfected with Oe-HOXC10 were analyzed by RT-qPCR and western blotting, respectively. (F) mRNA and (G) protein expression of MTFR2 in HCT116 cells transfected with Oe-HOXC10 were analyzed by RT-qPCR and western blotting, respectively. *P<0.05 and ***P<0.001 vs. control group; #P<0.05 and ###P<0.001 vs. Oe-NC group. (H) Relative luciferase activity was determined using a dual-luciferase reporter assay in HCT116 cells co-transfected with Oe-HOXC10 or Oe-NC and MTFR2-WT or MTFR2-MUT. ***P<0.001 vs. Oe-NC group. (I) Chromatin immunoprecipitation confirmed the interaction between HOXC10 and MTFR2. ***P<0.001 vs. IgG group. MTFR2, mitochondrial fission regulator 2; RT-qPCR, reverse transcription-quantitative PCR; Oe, overexpression; NC, negative control; WT, wild-type; MUT, mutant.
Overexpression of HOXC10 partially reverses the inhibitory effect of MTFR2 knockdown on CRC cell proliferation and migration
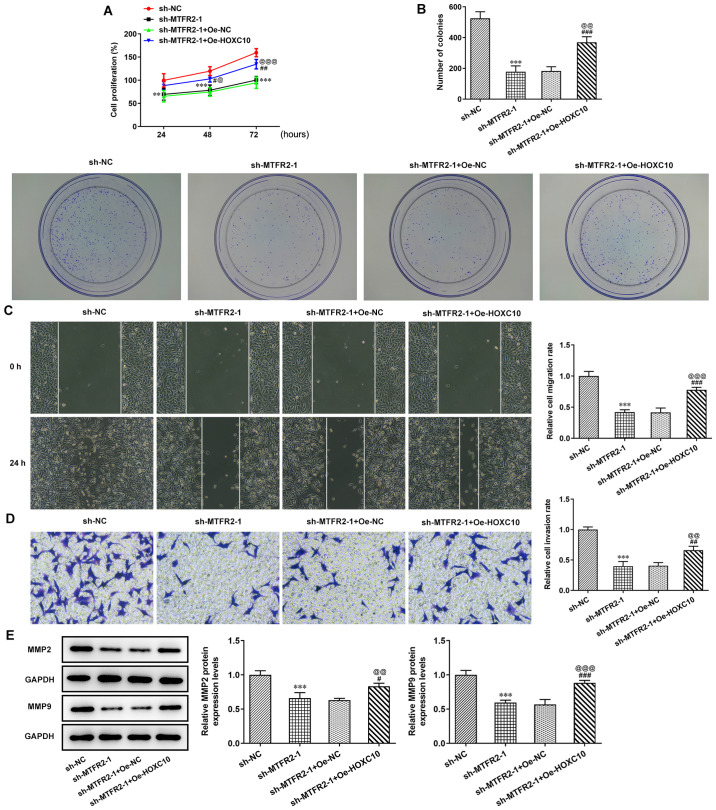
MTFR2 knockdown significantly suppressed the proliferation and clone formation of HCT116 cells compared with sh-NC; however, this effect was significantly reduced by HOXC10 overexpression compared with in the sh-MTFR2-1 + Oe-NC group (Fig. 5A and B). In the present study, MTFR2 knockdown significantly inhibited HCT116 cell invasion and migration (Fig. 5C and D), and significantly reduced MMP2 and MMP9 protein compression levels compared with in the sh-NC group (Fig. 5E); however, these effects were reversed by HOXC10 overexpression.
Figure 5.
Overexpression of HOXC10 partially reverses the inhibitory effect of MTFR2 knockdown on colorectal cancer cell proliferation and migration. (A) Proliferation, (B) clone formation, (C) migration and (D) invasion of HCT116 cells co-transfected with sh-MTFR2-1 and Oe-HOXC10 were in turn detected by the Cell Counting Kit-8, clone formation, wound healing and Transwell assays (magnification, ×100). Relative migration and invasion rates were expressed normalized to the control group. (E) Protein expression levels of invasion- and migration-related proteins in HCT116 cells co-transfected with sh-MTFR2-1 and Oe-HOXC10 were detected by western blotting. **P<0.01 and ***P<0.001 vs. sh-NC; #P<0.05, ##P<0.01 and ###P<0.001 vs. sh-MTFR2-1; @P<0.05, @@P<0.01 and @@@P<0.001 vs. sh-MTFR2-1 + Oe-NC. MTFR2, mitochondrial fission regulator 2; sh, short hairpin RNA; Oe, overexpression; NC, negative control.
Discussion
CRC is a malignancy of the digestive system with high morbidity and mortality rates; consequently, much focus has been given to CRC at both the medical and societal levels (15). If patients with CRC can be diagnosed and treated early there is a higher likelihood of a better prognosis; therefore, early diagnosis and treatment is extremely important (16). Colonoscopy is the international gold standard for CRC diagnosis; however, it is difficult to popularize this type of screening (17,18). Therefore, it is necessary to find more appropriate means and methods for the clinical diagnosis, prognosis, treatment effects and postoperative survival prediction of CRC.
MTFR2 is located on chromosome 6q23.3 and serves a key role in mitochondria, promoting mitochondrial division and aerobic respiration in eukaryotic cells (5,6). Previous studies have indicated that mitochondrial fission proteins could promote the cell cycle, proliferation, invasion and migration (19), and that mitochondrial fission was related to apoptosis (20). In addition, recent studies have demonstrated that MTFR2 expression was upregulated in BC and oral squamous cell carcinoma, which was related to the aggravation of invasion and migration (7–9). The present study indicated that MTFR2 expression was also significantly upregulated in CRC tissues and cells. Furthermore, MTFR2 knockdown could suppress the proliferation, invasion and migration of CRC cells.
In terms of the depth of infiltration, activation of MMP2 and MMP9 can promote the invasion of tumor cells, and are closely related to malignant phenotypes (21). Infiltration has been reported to be positively correlated with MMP2; with the increase in depth of infiltration, the positive expression intensity and expression rate of MMP2 were increased (22). It has also been reported that MMP9 was upregulated in colon cancer tumors and maintained high expression levels, suggesting that it may be involved in later invasion and metastasis (23). In the present study, MTFR2 knockdown significantly decreased MMP2 and MMP9 protein expression, but HOXC10 overexpression reversed these effects.
HOXC10 is an important member of the HOX family, a group of evolutionarily conserved genes that control cell proliferation, differentiation and embryo development (24). It has previously been reported that the HOX gene family serves an important role in tumorigenesis and development. In cervical squamous cell carcinoma, the elevation of HOXC10 was determined to be correlated with increased invasion (25). Data from The Cancer Genome Atlas demonstrated that HOXC10 expression was significantly increased in human thyroid cancer tissues compared with in healthy thyroid tissues (26). Furthermore, HOXC10 promoted the migration and invasion of thyroid cancer cells, suggesting that HOXC10 may be a novel biomarker for the prognosis of human thyroid cancer (26). HOXC10 also promoted metastasis of human lung adenocarcinoma and indicated poor survival outcomes (27). Migration and invasion are important for tumor cell circulation and establishment of distant metastases (28). Previous studies demonstrated that HOXC10 overexpression promoted proliferation, inhibited apoptosis, and promoted metastasis and invasion of GC cells (12,29). The present study was consistent with previous research, whereby HOXC10 was highly expressed in CRC cancer cells (HCT116) compared with in healthy cells (HIEC-6). Moreover, HOXC10 overexpression attenuated the effects of MTFR2 knockdown to promote the proliferation, invasion and migration of CRC cells.
A recent study reported that increased expression of HOXC10 conferred resistance to radiotherapy and chemotherapy in esophageal squamous cell carcinoma (ESCC) tumor cells, and predicted the poor prognosis of patients with ESCC (30). In GC, knockdown of HOXC10 made GC cells more sensitive to apatinib, solving problems with apatinib resistance in the treatment of GC (31). Tang et al (27) indicated that HOXC10 expression was upregulated in lung cancer tissues, especially in tissues with metastatic potential, and predicted a poor prognosis of patients with lung cancer. HOXC10 expression has also been reported to be overexpressed in human thyroid cancer tissues, which may be positively related to a poor prognosis (26). In future work, the correlation analyses of HOXC10 expression with the prognosis of patients with CRC, especially those receiving adjuvant chemoradiotherapy, will be investigated. Furthermore, the results obtained in the present study will be confirmed in animal studies.
In conclusion, the present study demonstrated that HOXC10 overexpression activated the expression of MTFR2 to enhance the proliferation, clone formation, invasion and migration of CRC cells. HOXC10 may therefore be considered an ideal therapeutic target for CRC. However, the study did not perform an animal model, which was a limitation of this study.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YX, QW and GL conceived and designed the study. RC, LY and ZJ were responsible for the acquisition, analysis and interpretation of data. YX, RC and GL was responsible for manuscript preparation, writing and critical revisions. YX and RC confirm the authenticity of all the raw data. All authors have read and approved the manuscript.
Ethics approval and consent to participate
The present study was approved by the Clinical Research Ethics Committee of Nanjing Medical University (Nanjing, China; approval no. 2021-448) and informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 3.Yin J, Bai Z, Zhang J, Zheng Z, Yao H, Ye P, Li J, Gao X, Zhang Z. Burden of colorectal cancer in China, 1990–2017: Findings from the Global Burden of disease study 2017. Chin J Cancer Res. 2019;31:489–498. doi: 10.21147/j.issn.1000-9604.2019.03.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monticone M, Panfoli I, Ravera S, Puglisi R, Jiang MM, Morello R, Candiani S, Tonachini L, Biticchi R, Fabiano A, et al. The nuclear genes Mtfr1 and Dufd1 regulate mitochondrial dynamic and cellular respiration. J Cell Physiol. 2010;225:767–776. doi: 10.1002/jcp.22279. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Xie Y, Bai X, Wang N, Yu H, Deng Z, Lian M, Yu S, Liu H, Xie W, Wang M. Targeting dual specificity protein kinase TTK attenuates tumorigenesis of glioblastoma. Oncotarget. 2018;9:3081–3088. doi: 10.18632/oncotarget.23152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, Xiong M, Jiang L, Chen Z, Shao Y. MTFR2 promotes the proliferation, migration, and invasion of oral squamous carcinoma by switching OXPHOS to glycolysis. Front Oncol. 2020;10:858. doi: 10.3389/fonc.2020.00858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu W, Zang R, Du Y, Li X, Li H, Liu C, Song Y, Li Y, Wang Y. Overexpression of MTFR2 predicts poor prognosis of breast cancer. Cancer Manag Res. 2020;12:11095–11102. doi: 10.2147/CMAR.S272088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu G, Lai Y, Wang T, Lin W, Lu J, Ma Y, Chen Y, Ma H, Liu R, Li J. Mitochondrial fission regulator 2 (MTFR2) promotes growth, migration, invasion and tumour progression in breast cancer cells. Aging (Albany NY) 2019;11:10203–10219. doi: 10.18632/aging.102442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enteghami M, Ghorbani M, Zamani M, Galehdari H. HOXC10 is significantly overexpressed in colorectal cancer. Biomed Rep. 2020;13:18. doi: 10.3892/br.2020.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng Y, Li Y, Li Y, Wu A, Fan L, Huang W, Fu C, Deng Z, Wang K, Zhang Y, et al. HOXC10 promotes tumour metastasis by regulating the EMT-related gene Slug in ovarian cancer. Aging (Albany NY) 2020;12:19375–19398. doi: 10.18632/aging.103824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Tong G, Huang C, Luo Y, Wang S, Zhang Y, Cheng B, Zhang Z, Wu X, Liu Q, et al. HOXC10 promotes cell migration, invasion, and tumor growth in gastric carcinoma cells through upregulating proinflammatory cytokines. J Cell Physiol. 2020;235:3579–3591. doi: 10.1002/jcp.29246. [DOI] [PubMed] [Google Scholar]
- 13.Guan Y, He Y, Lv S, Hou X, Li L, Song J. Overexpression of HOXC10 promotes glioblastoma cell progression to a poor prognosis via the PI3K/AKT signalling pathway. J Drug Target. 2019;27:60–66. doi: 10.1080/1061186X.2018.1473408. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Liang PS, Shaukat A. Assessing the impact of lowering the colorectal cancer screening age to 45 years. Lancet Gastroenterol Hepatol. 2020;5:523–524. doi: 10.1016/S2468-1253(20)30054-6. [DOI] [PubMed] [Google Scholar]
- 16.Song WY, Zhang X, Zhang Q, Zhang PJ, Zhang R. Clinical value evaluation of serum markers for early diagnosis of colorectal cancer. World J Gastrointest Oncol. 2020;12:219–227. doi: 10.4251/wjgo.v12.i2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang K, Xu H, Li HT. Safety and efficacy of propofol alone or in combination with other agents for sedation of patients undergoing colonoscopy: An updated meta-analysis. Eur Rev Med Pharmacol Sci. 2020;24:4506–4518. doi: 10.26355/eurrev_202004_21033. [DOI] [PubMed] [Google Scholar]
- 18.Park SW, Shin SP, Hong JT. Efficacy and tolerability of prucalopride in bowel preparation for colonoscopy: A systematic review and Meta-analysis. Adv Ther. 2020;37:2507–2519. doi: 10.1007/s12325-020-01333-x. [DOI] [PubMed] [Google Scholar]
- 19.Lima AR, Santos L, Correia M, Soares P, Sobrinho-Simões M, Melo M, Máximo V. Dynamin-related protein 1 at the crossroads of cancer. Genes (Basel) 2018;9:115. doi: 10.3390/genes9020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheridan C, Martin SJ. Mitochondrial fission/fusion dynamics and apoptosis. Mitochondrion. 2010;10:640–648. doi: 10.1016/j.mito.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, Duan W, Guo H, Xu X, Bai Y. Meta-analysis of associations between polymorphisms in the promoter regions of matrix metalloproteinases and the risk of colorectal cancer. Int J Colorectal Dis. 2011;26:1099–1105. doi: 10.1007/s00384-011-1198-4. [DOI] [PubMed] [Google Scholar]
- 22.Dong W, Li H, Zhang Y, Yang H, Guo M, Li L, Liu T. Matrix metalloproteinase 2 promotes cell growth and invasion in colorectal cancer. Acta Biochim Biophys Sin (Shanghai) 2011;43:840–848. doi: 10.1093/abbs/gmr085. [DOI] [PubMed] [Google Scholar]
- 23.Morán A, Iniesta P, García-Aranda C, De Juan C, Díaz-López A, Sánchez-Pernaute A, Torres AJ, Díaz-Rubio E, Balibrea JL, Benito M. Clinical relevance of MMP-9, MMP-2, TIMP-1 and TIMP-2 in colorectal cancer. Oncol Rep. 2005;13:115–120. [PubMed] [Google Scholar]
- 24.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-N. [DOI] [PubMed] [Google Scholar]
- 25.Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ, Trimble CL, Fearon ER, Cho KR. Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Res. 2007;67:10163–10172. doi: 10.1158/0008-5472.CAN-07-2056. [DOI] [PubMed] [Google Scholar]
- 26.Feng X, Li T, Liu Z, Shi Y, Peng Y. HOXC10 up-regulation contributes to human thyroid cancer and indicates poor survival outcome. Mol Biosyst. 2015;11:2946–2954. doi: 10.1039/C5MB00253B. [DOI] [PubMed] [Google Scholar]
- 27.Tang XL, Ding BX, Hua Y, Chen H, Wu T, Chen ZQ, Yuan CH. HOXC10 promotes the metastasis of human lung adenocarcinoma and indicates poor survival outcome. Front Physiol. 2017;8:557. doi: 10.3389/fphys.2017.00557. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Li S, Zhang W, Wu C, Gao H, Yu J, Wang X, Li B, Jun Z, Zhang W, Zhou P, et al. HOXC10 promotes proliferation and invasion and induces immunosuppressive gene expression in glioma. FEBS J. 2018;285:2278–2291. doi: 10.1111/febs.14476. [DOI] [PubMed] [Google Scholar]
- 29.Zheng J, Ge P, Liu X, Wei J, Wu G, Li X. miR-136 inhibits gastric cancer-specific peritoneal metastasis by targeting HOXC10. Tumour Biol. 2017;39:1010428317706207. doi: 10.1177/1010428317706207. [DOI] [PubMed] [Google Scholar]
- 30.Suo D, Wang Z, Li L, Chen Q, Zeng T, Liu R, Yun J, Guan XY, Li Y. HOXC10 upregulation confers resistance to chemoradiotherapy in ESCC tumor cells and predicts poor prognosis. Oncogene. 2020;39:5441–5454. doi: 10.1038/s41388-020-1375-4. [DOI] [PubMed] [Google Scholar]
- 31.Yu J, Zhang X, Ma Y, Li Z, Tao R, Chen W, Xiong S, Han X. miR-129-5p restrains apatinib resistance in human gastric cancer cells via downregulating HOXC10. Cancer Biother Radiopharm. 2021;36:95–105. doi: 10.1089/cbr.2019.3107. [DOI] [PubMed] [Google Scholar]
- 32.Jin M, Frankel WL. Lymph node metastasis in colorectal cancer. Surg Oncol Clin N Am. 2018;27:401–412. doi: 10.1016/j.soc.2017.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.