Abstract
Poor maternal nutrition in pregnancy affects fetal development, predisposing offspring to cardiometabolic diseases. The role of mitochondria during fetal development on later-life cardiac dysfunction caused by maternal nutrient reduction (MNR) remains unexplored.
We hypothesized that MNR during gestation causes fetal cardiac bioenergetic deficits, compromising cardiac mitochondrial metabolism and reserve capacity.
To enable human translation, we developed a primate baboon model (Papio spp) of moderate MNR in which mothers receive 70% of control nutrition during pregnancy, resulting in intrauterine growth restriction (IUGR) offspring and later exhibiting myocardial remodeling and heart failure at human equivalent ~25years. Term control and MNR baboon offspring were necropsied following cesarean-section, and left ventricle (LV) samples were collected. MNR adversely impacted fetal cardiac LV mitochondria in a sex-dependent fashion. Increased maternal plasma aspartate aminotransferase, creatine phosphokinase, and elevated cortisol levels in MNR concomitant with decreased blood insulin in male fetal MNR were measured. MNR resulted in a two-fold increase in fetal LV mtDNA. NMR resulted in increased transcripts for several respiratory chain (NDUFB8, UQCRC1, and cytochrome c) and ATP synthase proteins However, MNR fetal LV mitochondrial complex I and complex II/III activities were significantly decreased, possibly contributing to the 73% decreased ATP content and increased lipid peroxidation. MNR fetal LV showed mitochondria with sparse and disarranged cristae dysmorphology.
Conclusions:
MNR disruption of fetal cardiac mitochondrial fitness likely contributes to the documented developmental programming of adult cardiac dysfunction, indicating a programmed mitochondrial inability to deliver sufficient energy to cardiac tissues as a chronic mechanism for later-life heart failure.
Keywords: cardiometabolic disease, cardiac metabolic flexibility, sexual dimorphism, heart, maternal nutrition, fetal development
1. Introduction
Cardiovascular disease (CVD) is the leading cause of mortality worldwide (1). Human epidemiologic studies have demonstrated that developmental programming by a suboptimal intrauterine environment increases later life CVD risk (2). Maternal nutrient reduction is a common cause of intrauterine growth restriction (IUGR) (3). IUGR is associated with increased later life risk for heart disease (4) coronary artery disease (2,5,6), hypertension, hypercholesterolemia, and stroke (5,7). There is a strong association between birthweight and mortality rate from ischemic heart disease, with smaller babies having a three-to-five-fold higher risk (8). Moreover, programming of CVD risk by adverse intrauterine conditions is transmitted across generations (9), increasing this condition’s impact.
Epidemiological studies have provided compelling evidence on etiological factors that predispose to CVD. However, epidemiological studies can only indicate an association. Demonstration of causation requires precise control of environmental confounders, randomisation, and the introduction of specific interventions. While rodent models provide useful mechanistic information, those are polytocous, and thus pregnant dams carry a much more significant nutrient burden than monotocous species. Data from monotocous species are required to identify differences between humans and the polytocous, commonly-used laboratory animal species to provide firm information for translation to humans. Studies in nonhuman primates have particular strengths concerning the production of translational data. We have developed a well-established nonhuman IUGR primate model to evaluate the effects of moderate maternal nutrient reduction (MNR) in which baboon mothers were fed 70% of the global diet eaten by controls fed ad libitum during pregnancy and lactation (10–15). At birth, MNR offspring were IUGR weighing ~89% of control offspring (16). This degree of MNR was associated with offspring dysregulation in cardiac structure with extracellular fibrosis, altered miRNA expression, and changed lipid metabolism at the cardiac structural and molecular level (13). Even when fed a regular diet after weaning, IUGR offspring of MNR mothers showed myocardial remodeling and impaired cardiac left (LV) and right ventricle (RV) physiological function in early adulthood (5.7 years, human equivalent approximately 25 years old). MNR offspring show established early adult cardiac dysfunction, impaired diastolic and systolic cardiac function (11,14), resembling changes reported in human IUGR (17,18). The dysfunction is similar to human cardiac failure with preserved ejection fraction. Notwithstanding the strong association between poor maternal nutrition and adverse fetal and life-course cardiac programming, direct and causal effects on fetal heart mitochondrial biology remain uninvestigated (19,20). Disturbance of the energy transformation process is a prime feature of heart failure in subjects with cardiomyopathy (20,21).
In this study, the innovation is to potentially explain postnatal cardiovascular health with fetal cardiac metabolism by characterizing the impact of MNR on fetal left ventricular myocardial mitochondrial function in near term fetuses, thereby providing a mechanistic framework for our findings of cardiac dysfunction and premature cardiac aging in young adult, 5.7years old MNR baboons (11,14). We hypothesized that moderate 30% global MNR during fetal development impairs fetal cardiac mitochondrial metabolism, decreasing cardiac mitochondrial reserve capacity, thereby affecting the cardiac metabolic ability to overcome postnatal life’s high demands and contribute to the compromised cardiovascular function found in older MNR offspring. To test this hypothesis, we investigated mitochondrial and metabolic markers in control and MNR fetuses at 0.9 gestation (0.9G) and determine sex-specific fetal mitochondrial remodelling resulting from MNR.
2. Materials and Methods
2.1. Detailed in vivo procedures
2.1.1. Animal care and maintenance
All animal procedures, including pain relief, were approved by the Animal Care and Use Committees of the Texas Biomedical Research Institute and the University of Texas Health Science Center at San Antonio, TX (no. 1134PC), and were conducted in Association for Assessment and Accreditation of Laboratory Animal Care-approved facilities and NIH Guide for the Care and Use of Laboratory Animals.
As previously described (22), maternal morphometric determinations were made before pregnancy to guarantee the consistency of weight and general morphometrics in animals used in the present study. Non-pregnant outbred female baboons (Papio spp.) of the similar morphometric phenotype were selected for the study. Animals were housed at the Southwest National Primate Research Center at the Texas Biomedical Research Institute (TBRI) in Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-approved facilities. Groups up to 16 female baboons were initially housed with a vasectomized male to establish a stable social group in outdoor gang cages, thereby providing full social and physical activity.
Each outdoor concrete gang cage was covered with a roof and had open sides that allowed exposure to normal lighting. Each cage holds 10–16 females and had a floor area of 21 × 37 m, being about 3.5 m high. The enrichment inside the cages included nylon bones (Nylabone, Neptune, New Jersey, USA), rubber Kong toys (Kong Company, Golden, Colorado, USA), and plastic Jolly Balls (Horseman’s Pride, Inc., Ravenna, Ohio, USA). A 0.6 m-wide platform built from expanded metal grating was placed at the height of 1.7 m and ran the entire length of the cage. A similar perch was built at the front of the cage, running the entire cage’s length. Tube perches were present at the back in the corner of each cage. Each cage had an exit into a chute 0.6 m wide by 1 m high positioned along the side of each set of cages. A fine mesh was placed on the side of the chute adjacent to the other group cages between groups of animals as they passed along the chute to the individual feeding cages. The two chutes merged and passed over a scale and into individual feeding cages, which were 0.6 by 0.9 m in floor area and 0.69 m high. All metal components were made of galvanized steel. Before pregnancy, animals were trained to be fed in individual cages. Briefly, at feeding time, all baboons passed along a chute into individual feeding cages. Once in the individual cages, they were fed with the designated amount of normal primate chow (Purina Monkey Diet 5038, Purina, St. Louis, Missouri, USA).
Each baboon’s weight was obtained while crossing an electronic scale (GSE 665; GSE Scale Systems, Milwaukee, Wisconsin, USA). A commercial software application designed to capture weight data was modified to record 50 individual measurements over 3 seconds. If the weight measurement’s standard deviation was greater than 0.01 kg of the mean weight, the weight was automatically discarded, and the weighing procedure was re-initiated.
2.1.2. Animal study groups
All female baboons were observed twice a day for well-being and three times a week for turgescence (sex skin swelling) and signs of vaginal bleeding to assess their reproductive cycle and enable timing of pregnancy (22). After a 30-day period of adaptation to the feeding system, a fertile male was introduced into each breeding cage. Pregnancy was dated initially by following the changes in the swelling of the sex skin and confirmed at 30 days of gestation by ultrasonography. On day 30 of pregnancy (term ~ 183 days gestation), twenty-four female baboons were randomly assigned to eat standard primate chow ad libitum (control diet) or to receive 70% of the average daily amount of feed eaten by the female control baboons (MNR group) on a body weight-adjusted basis at same gestational age (12 baboons/dietary group, 6 male control fetuses – C-M, 6 female control fetuses – C-F, 6 MNR male fetuses – MNR-M, and 6 MNR female fetuses – MNR-F). Animals remained in these groups until cesarean section at 165 days gestation (0.9 gestation, see Figure 1 for study timeline). Each fetus from a singleton pregnant female baboon is considered an experimental unit; in some cases, the pregnant female baboon was also assumed as the experimental unit when maternal data is presented.
Fig. 1.
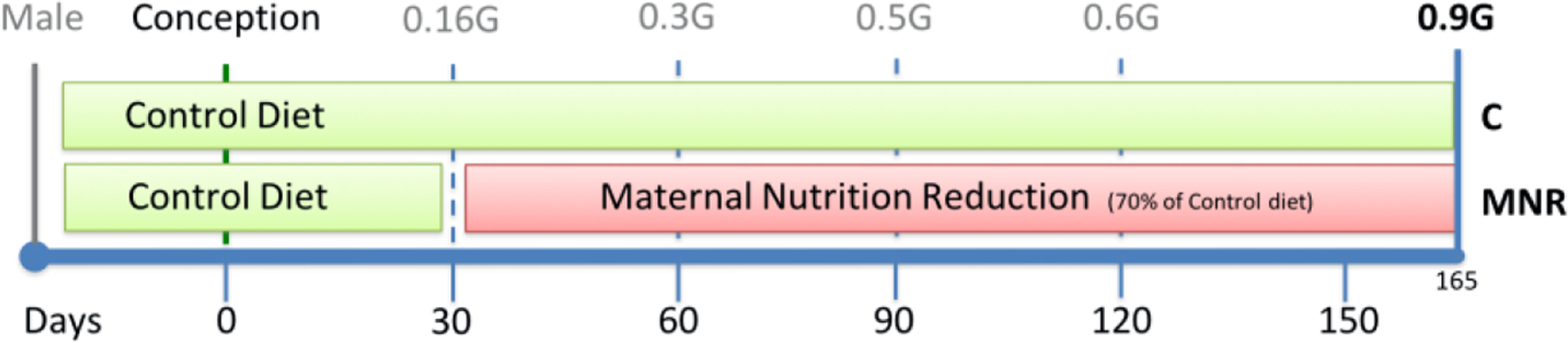
Timeline of maternal nutrition during baboon fetal development. Term baboon gestation occurs around 185 days.
Food was provided once a day as Purina Monkey Diet 5038, standard biscuits. The biscuit is described as a “complete life-cycle diet for all Old-World Primates” and contains stabilized vitamin C as well as all other required vitamins. The basic composition includes crude protein (≥ 15%), crude fat (≥ 5%), crude fiber (≤ 6%), ash (≤ 5%) and added minerals (≤ 3%) (22).
At the beginning of the feeding period, between 7–9 h or 11–13 h, each baboon received 60 biscuits in the feeding tray at the individual feeding cage. After the baboon had returned to the group cage at the end of the 2 h feeding period, the remaining biscuits in the tray and on the floor of the cage were counted and weighted. Following confirmation of pregnancy, food intake was recorded in 8 female baboons fed ad libitum and was calculated as 50.61 ± 3.61 kcal/kg of body weight per day. Before the start of the controlled diet, baboons were fed the same diet without a biscuit limit. Water was continuously available in the feeding cages via individual waterers (Lixit, Napa, California, USA) and at several locations in the group housing. Food consumption of animals, their weights, and health status were recorded daily. This feeding system allowed us to manipulate and monitor food intake in a controlled fashion while still maintaining female baboons in group housing instead of individual cages, thereby permitting regular social and physical activity. More details of housing and environmental enrichment have been previously published (22).
2.1.3. Cesarean section, fetal and maternal morphometry, and blood sampling
Animal feeding and cesarean sections were performed at specific times to avoid circadian variations between animals. Mothers were fasted from their last feeding time in the day before, until the cesarean section at 8 a.m. the next day, around 18 h (22).
Cesarean section and fetal necropsy were performed under isoflurane anesthesia (2%, 2 L/min oxygen, tracheal intubation) following tranquilization with ketamine hydrochloride (10 mg/kg intramuscularly injection) at 165 days of gestation (0.9 of gestation) using standard sterile techniques as previously described. Following hysterotomy, the umbilical cord was identified and used for fetal exsanguination with maternal and fetal baboon under general anesthesia as approved by the American Veterinary Medical Association Panel on Euthanasia (22). Fetal hearts were collected and dissected into their four chambers. Cardiac samples were taken from the free wall of the left cardiac ventricle that was cut transversely in at least four pieces. Some pieces were flash-frozen and stored at – 80°C until analyses, and one piece was fixed for histological analyses. Heparinized blood samples from the fetal umbilical vein and maternal uterine vein were obtained at cesarean section. Postoperatively mothers were placed in individual cages and watched until they were upright under their power and returned to their group cage at two weeks postoperatively (22). Maternal analgesia was administered for 3 days (buprenorphine hydrochloride intramuscularly injection; Hospira, Inc., Lake Forest, IL, USA; 0.015 mg/kg/day (23). Cesarean sections were evenly spread throughout the year and took place in the morning period, usually between 8 – 12 h. A fully certified M.D. or D.V.M. performed surgical procedures, and postsurgical care was prescribed and monitored by an accredited veterinarian.
2.1.4. Biochemical analyses
Within 1 h of collection, clotted blood was centrifuged at 10,000 g for 10 min and the serum was removed within 1 h of collection. Biochemical determinations of glucose, blood urea nitrogen (BUN), creatinine, total protein, albumin, and globulin were made in serum using a Beckman Synchron CX5CE Analyzer (Beckman Coulter, Irving, Texas, USA) by a certificated laboratory.
Heparinised plasma samples (0.1 ml) were deproteinized with 0.1 ml of 1.5 M HClO4 and neutralized with 0.05 ml of 2 M K2CO3. The solution was centrifuged at 12,000 g at 4°C for 1 min, and the supernatant was used for analyses. Amino acids were determined by HPLC involving pre-column derivatisation with ο-phthaldialdehyde, as previously described in detail (24). All amino acids were quantified using appropriate standards (Sigma-Aldrich, St. Louis, Missouri, USA) using Millenium-32 Software (Waters, Milford, Massachusetts, USA).
2.2. Analysis of mtDNA copy number by quantitative real-time PCR
Total DNA was extracted from ~20 mg of cardiac left ventricle tissue using the QIAamp DNA mini-kit (#50951304 Qiagen, Düsseldorf, Germany), following the manufacturer’s instructions and the protocol previously described (25). Briefly, the tissue was cut into small pieces and lysed with proteinase K in buffer ATL (tissue lysis buffer for use in purification of nucleic acids) provided by the kit. When the tissue was lysed entirely, buffer AL (tissue lysis buffer containing guanidine salts and detergent) and ethanol 96% were added. The mixture was applied to the QIAamp column and centrifuged at 6,000 xg for 1 min in Eppendorf 5415R benchtop centrifuge equipped with a FA-45-24-11 rotor (Eppendorf, Hamburg, Germany). After centrifugation, the column was placed in a new collection tube, and the DNA was washed sequentially with buffers AW1 (washing buffer 1) and AW2 (washing buffer 2). Ethanol was completely removed by centrifugation at full speed for 1 min. DNA was eluted with 200 μl Buffer AE and quantified using a Nanodrop 2000 device (ThermoFisher Scientific, Waltham, Massachusetts, USA).
RT-PCR was performed using the SsoFast Eva Green Supermix (Bio-Rad, Hercules, California, USA), in a CFX96 real time-PCR system (Bio-Rad), with the primers for ND1 (accession code NC_001992.1; sense sequence CCTATGAATCCGAGCAGCGT; antisense sequence GCTGGAGATTGCGATGGGTA) and for B2M (accession code NC_018158.1, sense sequence CAGGGCCCAGGACAGTTAAG; antisense sequence GGGATGGGACTCATTCAGGG) at 500 nM each. Amplification of 25 ng DNA was performed with an initial cycle of 2 min at 98°C, followed by 40 cycles of 5 sec at 98°C plus 5 sec at 60°C. At the end of each cycle, Eva Green fluorescence was recorded to enable for Ct determination. For quality control, melting temperature of the PCR products was determined after amplification by performing melting curves, and no template controls were run.
For absolute quantification and amplification efficiency, standards at known copy numbers were produced by purifying PCR products. After optimizing the annealing temperature, products were amplified for each primer pair using the HotstarTaq Master Mix Kit (#203445 Qiagen). Briefly, 1 μl of a DNA sample was added to a PCR tube containing the HotStar Taq Master Mix and the specific primers and placed in a CFX96 real time-PCR system. The amplification protocol started with an initial activation step of 15 min at 95°C degrees, followed by 35 cycles of 1 min at 94°C (denaturation) plus 1 min at 60°C (annealing), plus 1 min at 72°C (extension), and a final extension step of 10 min at 72°C. After amplification, the products were purified using the MiniElute PCR purification kit (#280006 Qiagen) following the manufacturer’s instructions. Eluted DNA was quantified in a Nanodrop 2000 device, the copy numbers were adjusted to 5 × 109 copies/μl, and tenfold serial dilutions were prepared. mtDNA copy number was determined by the ratio between the absolute amounts of mitochondrial gene ND1 versus the absolute amount of the B2M nuclear gene in each sample, using the CFX96 Manager software (v. 3.0; Bio-Rad).
2.3. Gene expression analysis by PCR array
RNA extraction was performed following the protocol previously described by Cox et al.(26). Briefly, approximately 20 mg transversal section of frozen tissue was cut. The tissue was homogenized in 1 ml Trizol Reagent using a Power General Homogenizer (Omni International, Wilmington, Delaware, USA). Genomic DNA in the sample was sheared by passing the homogenate three times through a 22-gauge needle attached to a 1 ml syringe. The homogenized samples were incubated for 5 min at 25°C. Two hundred μl of chloroform was added to each sample, and the samples were shaken vigorously by hand for 15 s and incubated at 25°C for 3 min. Samples were then centrifuged at 12,000 xg for 15 min at 4°C. The aqueous phase containing RNA was transferred to a fresh tube, and RNA precipitated by the addition of 0.5 ml of isopropyl alcohol. Samples were incubated for 10 min at 25°C and then centrifuged at 12,000 xg for 10 min at 4°C. The RNA precipitate was washed with 1 ml of 75% ethanol and centrifuged at 7,500 xg for 5 min at 4°C. After air-drying, the RNA pellet was dissolved in diethylpyrocarbonate (DEPC)-treated ddH2O. The RNA was quantified spectrophotometrically using Thermo Scientific NanoDrop 2000 spectrophotometer (ThermoFisher Scientific) and stored at −80°C. The RNA purity and quality were checked by Ultraviolet spectrophotometry by the ratios of A260/A280 and A260/A230 and electrophoretically by visualization of the ribosomal band integrity for both the 18S and 28S ribosomal RNA. Only RNA samples that demonstrated consistent quality were used.
After RNA preparation, the samples were treated with DNase to eliminate genomic DNA, while extracted RNA was converted to cDNA using the RT2 First Strand Kit from SuperArray Bioscience Corporation (SA Biosciences, Qiagen, Valencia, California, USA) according to the manufacturer’s instructions. Briefly, 1 μg RNA was combined with 2 μL gDNA elimination buffer and brought up to a final volume of 10 μL using RNAse-free H20. This mixture was incubated at 42°C for 5 min, then chilled in ice. Ten μL of RT Cocktail was then added to this mixture and incubated at 42°C for precisely 15 min followed by 5 min at 95°C to stop the reaction. Ninety-one μL ddH20 was added to each 20 μL cDNA synthesis reaction and well mixed. The cDNA mixture was stored at −20°C until used for gene expression profiling.
The RT2 Profiler polymerase chain reaction (PCR) Array System (SuperArray Bioscience, SA Biosciences, Qiagen), was used to evaluate the different cardiac mitochondrial transcripts between control and MNR fetuses. We used the Human Mitochondrial Energy Metabolism and the Human Mitochondria PCR Pathway Arrays. Real-time PCR detection was carried out per the manufacturer’s instructions. The experimental cocktail was prepared by adding 1,350 μL of the SuperArray RT2 qPCR master mix and 1,248 μL ddH20 to 102 μL of the diluted cDNA mixture. For real-time PCR detection, 25 μL of this cocktail was added to each well in the 96-well PCR array. The array was then processed on a real-time thermal cycler (Applied Biosystems StepOnePlus, ThermoFisher Scientific, Applied Biosystems) by using the following program: 1 cycle of 10 min at 95°C followed by 40 cycles of 15 seconds at 95°C and 1 min at 60°C. A melting curve program for quality control was immediately performed after the cycling program. SYBR Green fluorescence was detected from each well during the annealing step of each cycle, and values were exported to an Excel template file for analysis. Each PCR array contained 84 transcripts of the corresponding signaling pathway, a set of five housekeeping genes as internal controls, and additional controls for efficiency of reverse transcription, PCR, and the absence of contaminating genomic DNA. Data were normalized with three endogenous controls that did not differ between groups [hypoxanthine phosphoribosyltransferase 1 (HPRT1), ribosomal protein L13a (RPL13A), and Beta-actin (ACTB) and analyzed with the ΔΔCt method (where Ct is threshold cycle) using the PCR Array Data Analysis Web Portal (SA Biosciences).
2.4. Protein analyses by Western Blotting
Protein analyses by Western Blotting was performed following standard protocols (27). A small piece of frozen tissue (≈30 mg) was used for whole tissue protein extraction. All the extraction procedures were performed on ice. Tissue was homogenized in a 20% (w/v) RIPA buffer (150 mM NaCl, 50 mM Tris pH 8.0 (HCl), 0.5% sodium deoxycholate (DOC), 1% IGEPAL (CA-630) and 0.1% sodium dodecyl sulfate (SDS)), supplemented with 5 μl/100 mg (tissue) of protease inhibitors cocktail (P8340, Sigma-Aldrich) and sodium orthovanadate, a phosphatase inhibitor, using an electric homogenizer PowerGen Model 125 (ThermoFisher Scientific, Fisher Scientific). The suspension was kept on ice for 5 min and then centrifuged at 14,000 xg for 5 min at 4°C to remove cellular debris. The pellet was discarded and protein concentration in the supernatant was determined by the Bicinchoninic acid assay (BCA) using the commercial Pierce BCA assay kit protocol (#9981, ThermoFisher Scientific, Fisher Scientific), using bovine serum albumin (BSA type V, Sigma-Aldrich) ranging from 0.25 to 2 mg/ml as standard. The amount of protein was calculated after determining the absorbance of the dye at 545 nm in a Victor X3 plate reader (PerkinElmer, Waltham, Massachusetts, USA). Standards and unknown samples were performed in triplicates. After protein determination, all the proteins were diluted for the same final concentration with RIPA and stored at −80°C until further use.
Extracted proteins were solubilized to achieve a working concentration of 1mg/ml or 2 mg/ml of protein with Laemmli buffer (62.5mM Tris pH 6.8 (HCl), 50% glycerol, 2% SDS, 0.005% bromophenol blue, supplemented with 5%β-mercaptoethanol) and boiled for 5 min in a water bath and then centrifuged at 14,000 xg for 5 min to remove cellular debris. Equivalent amounts of total protein (10 μg per lane) were loaded in a 10–20% gradient Tris-HCl polyacrylamide gel as well two different standards for molecular weight estimation and for monitoring electrophoresis progress, the Precision Plus Protein Dual Color Standards (Bio-Rad) and the SeeBlue Plus2 Pre-Stained Standard (ThermoFisher Scientific, Invitrogen). Electrophoresis was carried at room temperature in a Criterion system (Bio-Rad) using 150 V until the sample buffer (blue) reaches the bottom of the gel (≈ 90 min).
After separation by SDS-PAGE, proteins were electrophoretically transferred in a TransBlot Cell system (Bio-Rad) to a polyvinylidene difluoride (PVDF) membrane previously activated, a constant amperage (0.5 A) during 2 h at 4°C using a CAPS transfer buffer (10 mM 3-(Cyclohexylamino)-1-propanesulfonic acid pH 11 (NaOH), 10% methanol). Good electrophoretic transfer was indicated by the complete transfer of pre-stained molecular weight markers below 100 kDa and by Ponceau staining. Ponceau results were also used to confirm an equal amount of protein loading and to normalize band density. After Ponceau removal, the membranes were blocked in 5% non-fat milk/PBS overnight at 4°C with agitation. Before incubation with primary antibodies, the membrane was washed for 10 min in PBS 0.05% Tween-20 (PBS-T). Primary antibodies described in the supplemental table were prepared in 1% non-fat milk/PBS to a final volume of 5 ml and incubated overnight at 4°C. After incubation with primary antibodies, membranes were washed with PBS-T solution three times, 5 min each, and incubated with the correspondent alkaline phosphatase-conjugated secondary antibodies for 2 h at room temperature with stirring (see Suppl. Table S1 and S2).
For immunodetection, membranes were washed three times for 5 min each with PBS-T, rinsed in PBS to remove any Tween-20, which can be inhibitory to the detection method, dried and incubated with an enhanced chemifluorescence (ECF) system (#RPN5785, GE Healthcare, Little Chalfont, Buckinghamshire, UK) during a maximum of 5 min. Density analysis of bands was carried out with VisionWorks.LS Image Acquisition and Analysis Software (UVP).
Resulting images were analyzed and densities were normalized to Ponceau (28–30). The average value of the control males (C-M) group was assumed as one unit, and the values of each sample were determined proportionally.
2.5. Tissue Immunohistochemistry
Tissue immunohistochemistry was performed by a standard avidin-biotin histochemical technique, as previously described (31), and using the antibodies listed in Table S3. After optimizing the final dilution of the primary antibody that yielded the cleanest immunostaining achievable, all sections were immunostained in the same assay to assure identical conditions. Briefly, fixed tissues sections (5 μm) were deparaffinized with xylene and rehydrated to decrease ethanol grades to water (100, 95, 70, and 50%). Antigen retrieval was performed for 15 min using boiling citrate buffer (0.01 M citrate buffer, pH 6.0). After cooling for 15 min, the section was rinsed for 5 min in potassium PBS (KPBS; 0.04 M K2HPO4, 0.01 M KH2PO4, 0.154 M NaCl, pH 7.4) and washed for 10 min in a solution of 1.5% H2O2/methanol and then for 5 min in KPBS. Sections were placed in diluted (10%) normal serum for 10 min and covered with primary antibody (Table S3) overnight at 4°C and incubated for 1 h at room temperature with the appropriate biotinylated secondary antibody (1:1,000; Vector Laboratories (Burlingame, California, USA)): goat anti-rabbit (BA-1000) and horse anti-mouse (BA-2000). Negative controls were also run in the absence of the primary antibody but in normal serum. Three slides per animal were analyzed, and six pictures/slide per section were randomly taken and analyzed with ImageJ software (National Institutes of Health, New York, USA) for the fraction (area immunostained/area of the field of interest × 100%) and the density (arbitrary density units).
2.6. Enzymatic activity of mitochondrial proteins
Mitochondrial respiratory chain activities were determined in LV tissue homogenates according to previously published methods.
The activity of complex I was measured by the method described by Long (32) with some modifications. Briefly, 30 μg of tissue homogenate was resuspended in reaction buffer containing 25 mM KH2PO4 pH 7.5, 5 mM MgCl2, 300 μM KCN, 4 μM antimycin A, 3 mg/mL BSA, 60 μM coenzyme Q1, 160 μM 2,6-dichlorophenolindophenol (DCPIP). Complex I activity was determined by the measurement of DCPIP absorbance at 600 nm, 37°C, in a Victor X3 plate reader, upon the addition of 100 μM freshly-prepared NADH. Enzymatic activity was determined through the mean of slopes obtained during the linear phase of duplicates. Specific mitochondrial complex I activity was computed as the difference among basal activity in the absence or presence of 10 μM rotenone, a specific inhibitor of complex I. Normalization was performed to the protein concentration and an ε600 = 19.1 mM−1.cm−1. Complex I activity is expressed as nmol DCPIP/min/mg protein.
The activity of complex II/III was analyzed as defined by Tisdale (33), with minor modifications. Concisely, 100 μg of tissue homogenate was pre-incubated in 200 μL of phosphate buffer (166 mM KH2PO4/K2HPO4, pH 7.4) supplemented with 100 mM KCN and 500 mM sodium succinate for 5 min at 37°C. The reaction was initiated by adding 120 μL of phosphate buffer supplemented with 2 mM oxidized cytochrome c (cyt c ox) plus 15 mM EDTA-dipotassium. Complex II/III activity was determined by following the reduction of cyt c ox (increased absorbance at 550 nm), using a Victor X3 plate reader. Enzyme activity was measured through the mean of slopes obtained during the linear phase for duplicates. Mitochondrial complex II/III specific activity was calculated as the difference between basal activity in the absence or presence of 4 mM antimycin A (a specific inhibitor of complex III). Normalization was performed to the protein concentration and an ε550 = 18.5 mM−1.cm−1. Results are expressed as nmol cyt c ox/min/mg protein.
The activity of complex III was analyzed according to Luo (34), with minor adaptations. Briefly, 100 μg of tissue homogenate was suspended in reaction buffer containing 25 mM KH2PO4 pH 7.5, 4 μM rotenone, 0.025% Tween-20, 100 μM fresh decylubiquinone solution at 37°C. Enzymatic activity was measured as an increase in absorbance of cyt c ox at 550 nm, upon addition of 75 μM cyt c ox in a VICTOR X3 plate reader. Enzyme activity was determined through the mean of slopes obtained during the linear phase for duplicates. For determination of the specific complex III activity, 2.5 mM antimycin A (complex III specific inhibitor) was used, and the difference between basal activity in the absence or presence of antimycin A was determined. Normalization was performed to the protein concentration and an ε550 = 18.5 mM−1.cm−1. Results were expressed as nmol cyt cox min/mg protein.
The activity of complex IV was measured by adapting the method described by Brautigan (35). Briefly, 25 μg of tissue homogenate was suspended in reaction buffer containing 50 mM KH2PO4 pH 7.0, 4 μM antimycin A, 0.05% n-dodecyl-β-D-maltoside at 37°C. Enzymatic activity was followed in a VICTOR X3 plate reader as a decrease in absorbance of reduced cytochrome c (cyt c red) at 550 nm, upon addition of 57 μM freshly-prepared cyt c red. Enzyme activity was calculated through the mean of slopes obtained during the linear phase for duplicates. Cyanide, a complex IV-specific inhibitor, was used to determine mitochondrial complex IV specific activity, that was calculated as the difference between basal activity in the absence or presence of 10 mM of KCN. Normalization was performed to the protein concentration and an ε550 = 18.5 mM−1.cm−1. Activity is expressed as nmol cyt c red min/mg protein.
The activity of citrate synthase was analyzed by adapting the method described by Core (36). Concisely, 25 μg of tissue homogenate was suspended in a reaction buffer containing 100 mM Tris pH 8.0 plus 200 μM Acetyl-CoA, 200 μM 5,5-dithiobis-2-nitrobenzoic acid at 37°C. Enzymatic activity was measured by following the increase in absorbance (412 nm) upon adding 100 μM freshly-prepared oxaloacetate in a VICTOR X3 plate reader 37°C. Enzyme activity was calculated through the mean of slopes obtained during the linear phase for duplicates. Specific citrate synthase activity was determined by subtracting the basal activity in the presence of 0.1% Triton-X100. Normalization was performed to the protein concentration and an ε412 = 13.6 mM−1·cm−1. Enzyme activity is expressed as nmol of oxaloacetate min/mg protein.
2.7. Analysis of adenine nucleotides by HPLC
Adenine nucleotide levels were measured according to the method described by Santos et al. (37). Briefly, tissue was homogenized in 0.3 M perchloric acid (equal parts of PBS and 0.6 M perchloric acid) and kept for 5 min on ice. The acid homogenates were centrifuged at 14,000 xg for 10 min and at 4°C. Supernatants were brought to neutral pH with 3 M KOH in 1.5 mM Tris, centrifuged at 14,000 xg, for 10 min at 4°C, and stored at −80°C. Then, the supernatants were assayed for ATP, ADP, and AMP by separation in a reverse-phase high-performance liquid chromatography (HPLC), as described by Stocchi and collaborators (38). The chromatographic apparatus used was a Beckman-System Gold (Beckman Instruments, Fullerton, California, USA), consisting of a binary pump (model 126) and a variable UV detector (model 166), controlled by a computer. The detection wavelength was 254 nm, and the column used was a LiChrospher 100 RP-18 (5 μm) from Merck. An isocratic elution with 100 mmol/l phosphate buffer (KH2PO4; pH 6.5) and 1.0% methanol was performed with a flow rate of 1 ml/min. The required time for each analysis was 6 min. Peak identity was determined by the retention time compared with standards. A concentration standard curve was used to determine the concentrations of nucleotides and metabolites. Concentration of adenylates was expressed as nmol/mg of protein and adenylate energy charge (AEC) was determined according to the formula (ATP + 1/2 ADP) /(ATP + ADP + AMP).
2.8. Oxidative stress evaluation
Oxidative stress was evaluated by measuring malondialdehyde (MDA) levels and oxidized glutathione (GSSG), while the antioxidant capacity was evaluated by determination of the contents on reduced glutathione (GSH) and vitamin E. The enzymatic activity of glutathione peroxidase (Gl-Px) and glutathione reductase (Gl-Red) was also determined.
Lipid peroxidation was given by the fluorimetric measurement of MDA adducts separated by high-performance liquid chromatography (HPLC; Gilson, Lewis Center, Ohio, USA) with the ClinRep complete kit (RECIPE, Munich, Germany), at an excitation wavelength of 515 nm and an emission wavelength of 553 nm (FP-2020/2025, Jasco, Tokyo, Japan), according to the method described by Draper et al. (39). Results are expressed as mM of MDA.
Reduced and oxidized glutathione (GSH and GSSG) were evaluated according to Tsao et al. (40), with minor adaptations, by using a commercial kit (Immunodiagnostik AG, Bensheim, Germany) and an HPLC system (Gilson) with fluorimetric detection (excitation at 385 nm and emission at 515 nm; FP-2020/2025, Jasco). Results are expressed as mM of GSH and mM of GSSG.
Vitamin E present in tissue was extracted in n-hexane (Merck) and quantified by reverse-phase HPLC (Gilson), using an analytic column Spherisorb S10w (250 × 4.6 mm), eluted at 1.5 ml/min with n-hexane modified with 0.9% of methanol (Merck) and spectrophotometric detection at 287 nm. Results are expressed as mM vitamin E.
The activity of glutathione peroxidase (Gl-Px) was evaluated spectrophotometrically as described by Palia et al. (41) with minor adaptations by using tert-butylperoxide (Sigma-Aldrich) as substrate. The formation of oxidized glutathione was given by quantifying the oxidation of NADPH (Sigma-Aldrich) to NADP+ at 340 nm in a thermostatized spectrophotometer (UVIKON 933 double bean UV/Visible spectrophotometer, Kontron instruments, Milan, Italy). Results are expressed in international units of enzyme per liter (U/l).
Glutathione reductase (Gl-Red) activity was evaluated according to Goldberg et al. (42), with some modifications, by following the reduction of GSSG (Sigma) to GSH through the quantification of NADPH (Sigma) oxidation at 37°C at 340 nm, in a thermostatized spectrophotometer (UVIKON 933 double bean UV/Visible spectrophotometer). Results are expressed in international units of enzyme per liter (U/l).
2.9. Transmission Electron Microscopy
Samples were processed by the certificated Electron Microscopy Laboratory at the Department of Pathology at UTHSCSA using the Transmission Electron Microscope (TEM) JEOL 1230 (JEOL, Peabody, Massachusetts, USA). After the primary fixation at the cesarean section samples were rinsed with PBS and post-fixed for 30 min with 1% buffered osmium tetroxide, according to Malhotra (43). Samples were dehydrated in increasing grades of ethanol (70, 95 and 100%) and placed in propylene oxide for 20 min. Then, samples were infiltrated with a mixture 1:1 of propylene oxide/resin followed by 30 min in 100% resin under 25 psi vacuum. Longitudinal pieces were flat-embedding in molds and filled to the top with resin. The resin was polymerized at 80°C overnight. Tissues were sectioned in 0.5–1 μm sections and stained with T-blue for 10 seconds on a hot plate. Sections quality was checked using a light microscope. Cardiac sections were either left unstained or stained with uranyl acetate followed by Reynold’s Lead Citrate stain for 20 s. A series of 5–6 images at 3,300x magnification demonstrating areas of interest were obtained.
2.10. Data analyses and statistics
The hypothesis to be tested in this study is that MNR by 30% during gestation affects cardiac mitochondria heritage and function in the progeny. A secondary question is whether these effects are dependent on fetal sex.
In this study, each pregnant female baboon and the correspondent fetus was considered as an experimental unit. Outbred pregnant female baboons were randomly assigned to control or MNR groups. Whenever possible, we performed a blind assessment of the diet effects and blind determination of the parameters to be statistically analyzed. Data are expressed as mean ± standard error of the mean. Normality and homoscedasticity were assessed by the Kolmogorov-Smirnov normality test and Levene variance homogeneity test. Since the variables do not comply with the assumptions required for parametric tests, namely lack of normality and homoscedasticity, the equivalent non-parametric Mann-Whitney U test was employed. Statistical tests were performed considering a significance level of α=0.05. Statistical analyses were performed using SPSS version 17.0 (IBM corporation, Armonk, New York, USA) with significance set at P<0.05 by two independent investigators to minimize the influence of natural human biases and corroboration. Graphical representations were obtained using GraphPad Prism version 8.0 (GraphPad Software, San Diego, California, USA).
3. Results
3.1. Pregnancy-related morphometric and blood biochemistry changes
Non-pregnant outbred female baboons (Papio spp.) of the similar morphometric phenotype were studied in this nutritional intervention and randomly assigned to the Control (C) or MNR group, resulting in groups with similar pre-pregnancy body weight (16.3±0.7vs16.6±1.2 kg, mean+SEM, p<0.05, Table S4). At 0.9G, control mothers weighed more than MNR mothers, controls gained 12.8±2.1% of their body weight during pregnancy while MNR mothers lost weight (−3.1±3.0%). Maternal weight loss was greater in MNR mothers carrying male than female fetuses (MNR-M vs., MNR-F, −7.1±3.7vs1.8±4.2%). Placental weight was also decreased with MNR-M fetuses.
Aspartate aminotransferase (AST) increased approximately 72% in MNR mothers (Table S5), while creatine phosphokinase (CPK) increased by 27.2%. Remarkably, circulating enzyme levels were significantly higher in control mothers carrying male fetuses (Table S5).
Cortisol levels agreed with previously published data (31,44) (Figure S1A) being increased in MNR mothers and with a more pronounced effect in MNR mothers carrying male fetuses. No difference was observed in glucose levels between control and MNR fetuses when sexes were either combined or separated (Figure S1B). Only MNR-M mothers showed an increase in glucose levels compared with C-M mothers. MNR reduced fetal insulin in male fetuses, while sex differences were observed for MNR-M vs. MNR-F for maternal and fetal samples, with MNR-F presenting higher insulin levels than MNR-M (Figure S1C).
3.2. Cardiac left ventricular mitochondrial DNA copy number
To evaluate MNR effects on fetal LV cardiac mitochondrial capacity, we measured mtDNA copy number by qRT-PCR. mtDNA copy number was calculated by determining the ratio between the absolute amounts of mitochondrial gene ND1versus the absolute amount of the B2M nuclear gene in each sample (Figure 2A). The average mtDNA in control fetuses was 714.5±84.9 copies per nucleus. Increased mtDNA was measured in MNR-F (1351±160) but not in MNR-M (836±117) (Figure 2A).
Fig. 2.
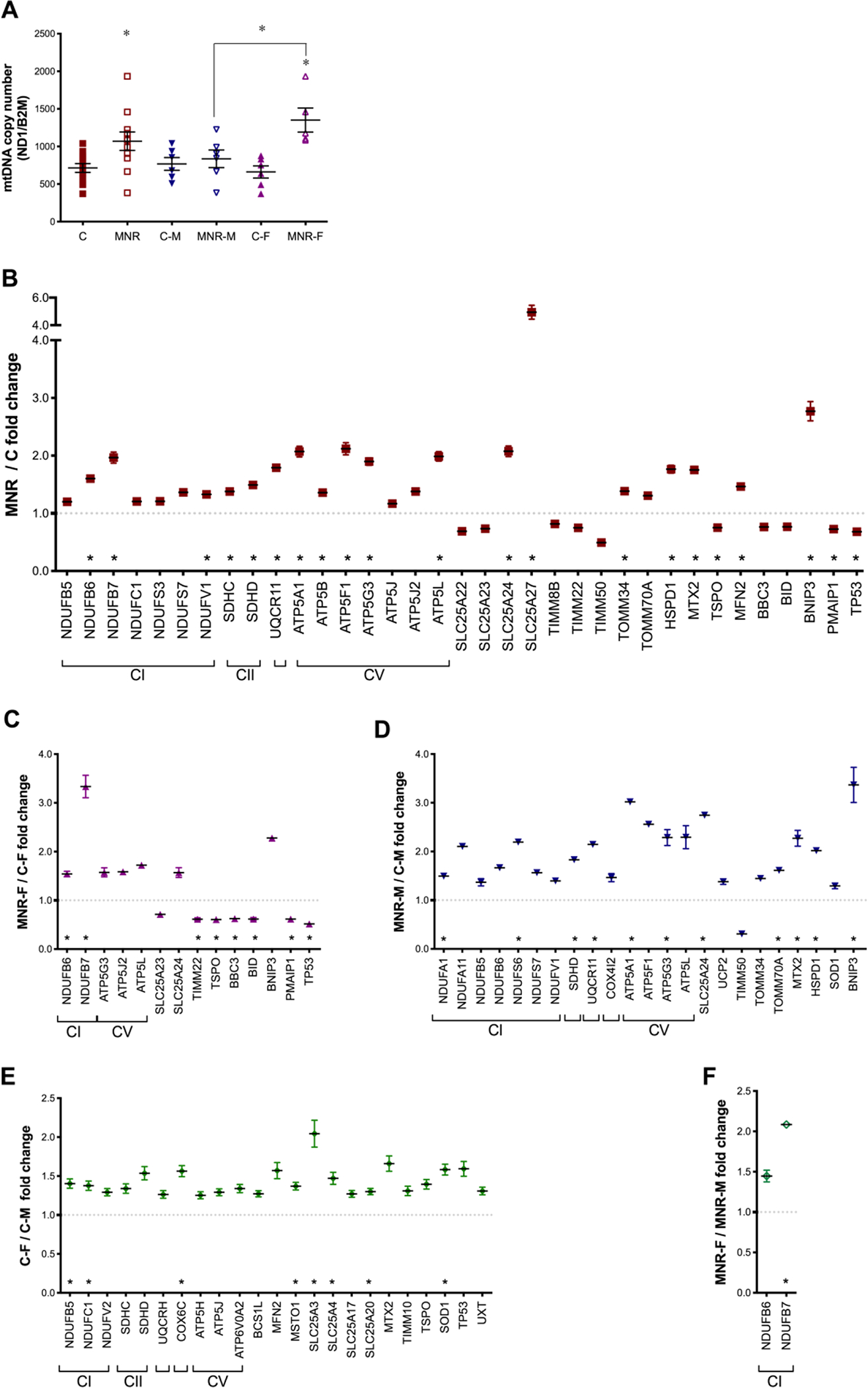
Variation of mitochondrial DNA (mtDNA) copy number and gene expression analysis of fetal cardiac left ventricle (LV) tissue from fetuses of control (C) and maternal nutrient reduction (MNR) mothers.
A: mtDNA was measured in fetal baboon cardiac LV tissue at 0.9G from control and NMR pregnancies in which mothers ate 70% of the food consumed by control mothers on a weight-adjusted basis. mtDNA copy number was calculated as the ratio between the mitochondrially encoded NADH dehydrogenase 1 (ND1) gene and the nuclear-encoded gene for beta-2-microglobulin (B2M). B-F: Gene expression analysis of control and MNR baboon fetuses at 0.9G. mRNA abundance for mitochondrial transcripts was assessed by PCR arrays in cardiac LV samples from offspring of C and MNR mothers. B: global diet-dependent effects in the mitochondrial gene expression profile; C and D comparison of transcripts expression between different maternal diets for the same sex [female fetuses (C) and male fetuses (D)]; and E and F: Sexual dimorphism in the mitochondrial profile of control (E) and MNR (F) fetuses. Transcripts related to oxidative phosphorylation system (OXPHOS), complex I (CI; NADH dehydrogenase), complex II (CII; succinate dehydrogenase), complex III (CIII; ubiquinol cytochrome c oxidoreductase), complex IV [CIV; cytochrome c oxidase (COX)], and complex V (CV; ATP synthase). Values were normalized to endogenous controls [hypoxanthine phosphoribosyltransferase 1 (HPRT1), ribosomal protein L13a (RPL13A), and Beta-actin (ACTB)] and are expressed relative to their normalized values. All transcripts presented have P<0.1 vs. respective paired group. C-M, male fetuses from control group (n=6); C-F, female fetuses from control group (n=6); MNR-M, male fetuses from MNR group (n=6); MNR-F, female fetuses from MNR group (n=6) or n=12 (sexes combined) animals/group. Means±SEM Comparison between groups was performed using a non-parametric Mann-Whitney test. P-value<0.05 was considered significant. *P<0.05 vs. respective controls or as indicated. Consult Table S6 for gene abbreviations and also Figures S2 and S3.
3.3. Mitochondrial transcript changes in the fetal cardiac left ventricular myocardium
Human Mitochondrial Energy Metabolism and the Human Mitochondria Pathway Arrays were used for fetal LV RNA expression profiling. Summarized data are shown in Figure2 (see also Table S6). The heat maps in Figure S2 and S3 provide a graphical summary representation of RNA expression fold regulation in response to the maternal diet in the fetal LV (Figure S2) and sex-dependent differences in control fetuses (Figure S3). Our results support the general conclusion that MNR increased fetal cardiac mitochondrial transcripts, more noticeable in MNR-F fetuses. When sexes were combined (Figure 2B) twenty-one transcripts were differentially expressed between C and MNR fetuses, with 85% of significant differences being upregulation in MNR. Most upregulated transcripts encoded subunits of the mitochondrial oxidative phosphorylation system (OXPHOS), three complex I subunits: NDUFB6, NDUFB7, and NDUFV1; two complex II subunits: SDHC and SDHD; one complex III subunits: UQCR11; and five ATP synthase subunits: ATP5A1, ATP5B, ATP5F1, ATP5G3 and ATP5L. Besides, transcripts for regulators and mediators of mitochondrial molecular transport, namely small-molecule transporters (SLC25A24 and SLC25A27); one member of the inner membrane translocation system (TOMM34); mitochondrial outer membrane import complex protein 2 (MTX2); a mediator of mitochondrial fusion (MFN2); heat shock protein 1 (HSPD1) and pro-apoptotic factor and autophagy mediator (BNIP3) were also upregulated in MNR fetuses. Finally, two members related to the apoptosis pathway (PMAIP1 and TP53) and one related to cholesterol transporter (TSPO) were downregulated.
Maternal diet-induced transcript differences were also observed within the same sex (Figures 2C, 2D). Multiple components of the mitochondrial respiratory chain were increased in MNR-M fetuses, including two complex I subunits (NDUFA1, NDUFS6), one complex II subunit (SDHD), one complex III subunit (UQCR11) and two ATP synthase subunits (ATP5A1, ATP5G3). Increased transcripts also included SLC25A24, MTX2, HSPD1, BNIP3, and one member of the outer membrane translocase complex (TOMM70A) (Figure 2D).
Comparing MNR-F and C-F fetuses, increased abundance of two mitochondrial respiratory chain transcripts, NDUFB6 and NDUFB7 and decreased transcripts related to cell death pathways, e.g. pro-apoptotic Bcl-2-binding component 3 (BBC3), BID, a mediator of mitochondrial damage induced by caspase-8, PMAIP1, which is related to the activation of caspases and apoptosis, TP53, TIMM22, an inner mitochondrial membrane protein translocase and TSPO, involved in cholesterol homeostasis, were observed (Figure 2C).
Significant sexual dimorphism was present in the cardiac mitochondrial-related gene expression profile in control fetuses (C-F vs C-M, Figure 2E). Female fetuses presented a higher content of transcripts for NDUFB5 and NDUFC1, complex I subunits; COX6C, a cytochrome c oxidase subunit; MSTO1, a regulator of mitochondrial morphology and distribution; SLC25A3, SLC25A4, SLC25A20, regulators and mediators of mitochondrial molecular transport, and SOD1, encoding for cytosolic superoxide dismutase. However, sex differences were attenuated by MNR, with only one transcript being different between sexes for this group, complex I subunit NDUFB7 (MNR-F vs MNR-M, Figure 2F).
3.4. Left ventricle cardiac mitochondrial protein content
To assess whether transcriptional changes were translated into altered protein production, we performed Western blot (WB) and immunohistochemical analyses. In agreement with the observed increase in transcripts related to OXPHOS, WB analysis (Figure 3) revealed an increase in cardiac mitochondrial respiratory chain proteins in MNR fetuses, including complex I subunit NDUFB8, complex II subunit UQCRC1, and cytochrome c (CYT C), a major factor in cell death regulation. Protein for the outer membrane channel VDAC1 (a structural protein), and cyclophilin D (CYC D), a modulator of the mitochondrial permeability transition pore, increased in MNR fetuses. However, the most significant increase was observed for the mitochondrial fission 1 protein (Fis1, 0.89±0.08vs1.33±0.16 A.U.).
Fig. 3.
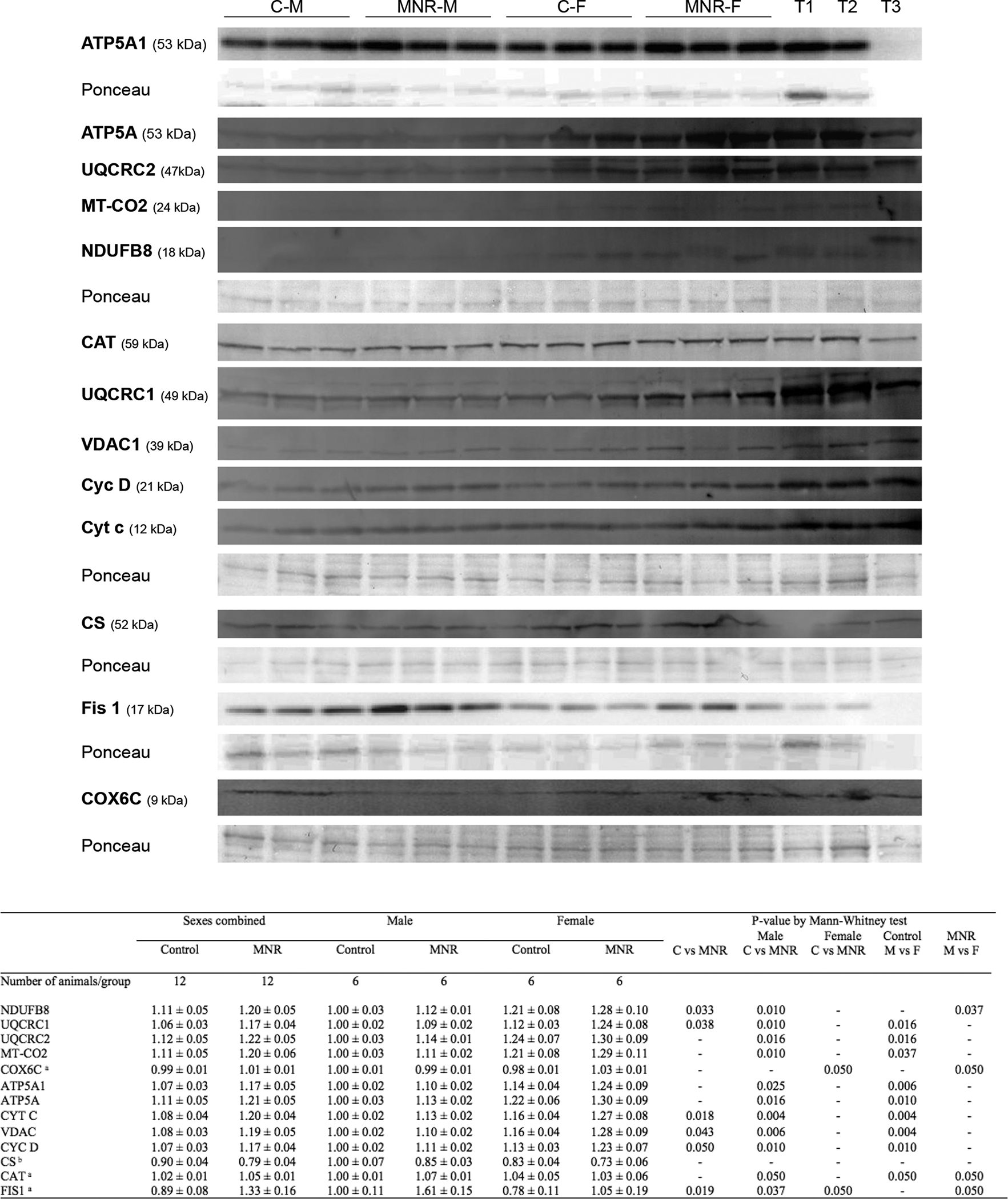
Protein content by immunoblot detection were determined in fetal cardiac LV tissue of control and MNR pregnancies, the latter characterized as 70% of the food consumed by control mothers on a weight-adjusted basis of baboons at 0.9G. C-M, male fetuses from the control group; C-F, female fetuses from the control group; MNR-M, male fetuses from MNR group; MNR-F, female fetuses from MNR group; T1, LV cardiac sample from an adult male baboon; T2, LV cardiac sample from an adult female baboon; T3, human cardiac sample, not used in all membranes. Ponceau staining for the respective membrane was used for normalization and as a loading control. Data are presented as arbitrary units and represent densitometry analysis of membranes by immunoblot detection after image acquisition. Data are means ± SEM; n=6 (when separated by sex) or n=12 (sexes combined) animals/group. Comparison between groups was performed using a non-parametric Mann-Whitney test. P-value less than 0.05 was considered significant. See also Table S1–S2.
a for these proteins, the sample size is different from the one previously indicated, being n=3 (when separated by sex) or n=6 (sexes combined) animals/group
b for the CS protein, the sample size is different from the one previously indicated, being n=4 (when separated by sex) or n=8 (sexes combined) animals/group.
Maternal diet-induced cardiac protein expression differences were also observed within the same sex. In summary, from the 14 mitochondrial proteins tested, 11 increased in MNR-M vs C-M fetuses and only two in MNR-F vs C-F fetuses (COX6C and Fis1), denoting a more significant effect of MNR on male fetuses at the protein level. Pronounced sexual dimorphism was present in control fetuses, with female fetuses displaying a higher content in nine mitochondrial proteins (C-M vs C-F, UQCRC1, UQCRC2, MT-CO2, ATP5A1, ATP5A, CYT C, VDAC, CYC D, and CAT). Interestingly, MNR attenuated this sex-related difference decreasing to four the number of mitochondrial proteins whose expression differed between sexes in MNR fetuses (MNR-M vs MNR-F, NDUFB8, COX6C, CAT, and FIS1).
Tissue content of mitochondrial proteins (COX6C, CYC1, MFN2, and TIMM9A) was further measured by immunohistochemistry (Figure 4). In agreement with the overall mitochondrial proteins pattern, mitofusin 2 (MFN2), which participates in mitochondrial fusion, was increased in the LV of MNR female fetuses (C-F vs. MNR-F, Figure 4A, F). Although under control diet conditions, female fetuses had decreased content of MFN2 compared to male fetuses (C-M vs. C-F, % fraction stained and density (AU)), Figure 4A, E), there were no other differences between diets or sexes in the quantitative immunohistochemistry of CYC1, COX6C or TIMM9A in LV cardiac tissue (Figure 4B–D).
Fig. 4.
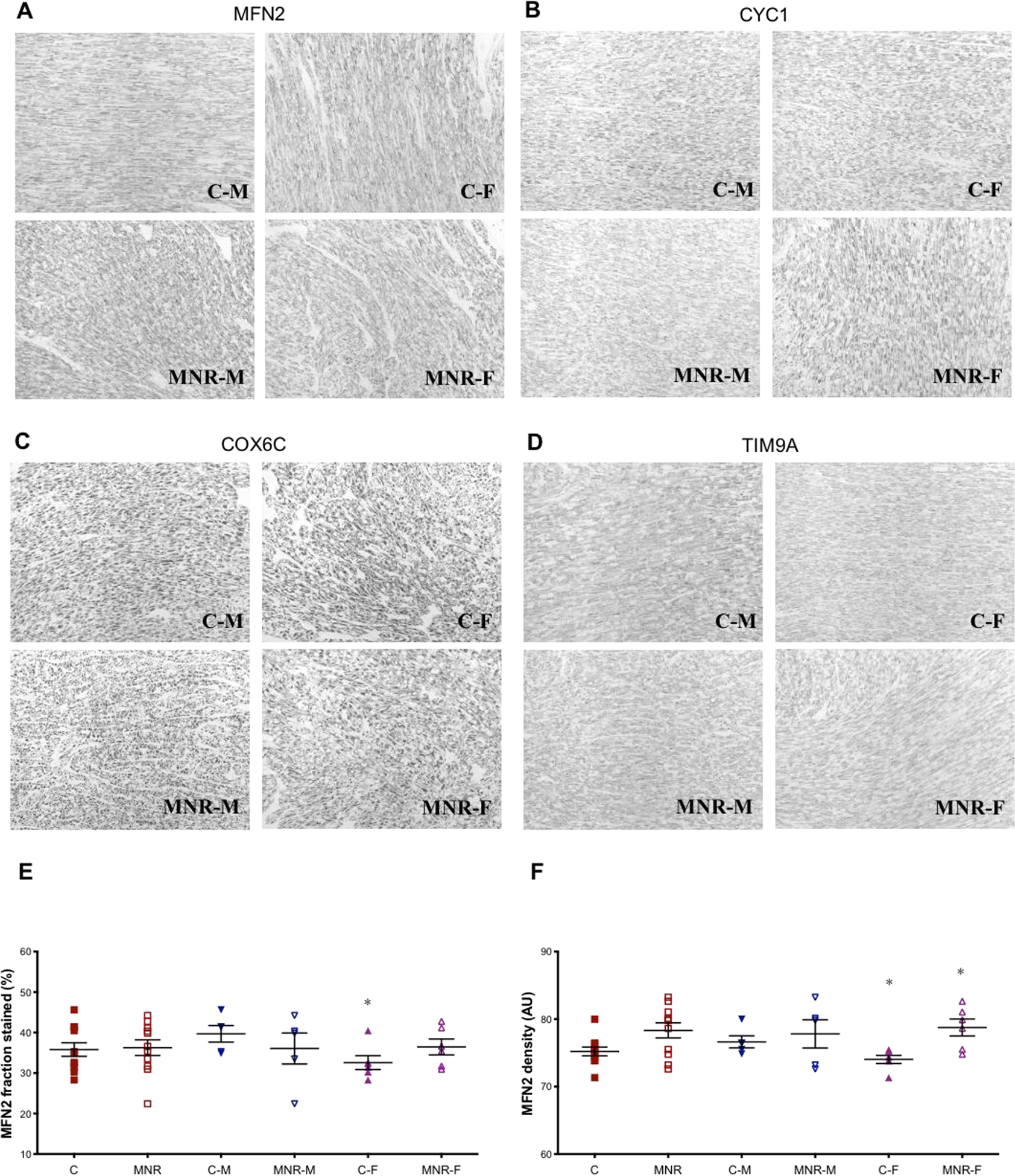
Quantitative immunohistochemistry of mitochondrial proteins in fetal cardiac LV tissue of control and MNR pregnancies, the latter characterized as 70% of the food consumed by control mothers on a weight-adjusted basis of baboons at 0.9G. A-D: The mitochondrial subunits MFN2 (A), CYC1 (B), COX6C (C) and TIMM9A (D) were analyzed in cardiac left ventricle tissue of fetal baboons from mothers that were fed ad libitum (control group) or fed with 70% of the control (MNR group). A-D: representative micrographs (magnification: x20) from cardiac LV sections of C-M, male fetuses from control group; C-F, female fetuses from control group; MNR-M, male fetuses from MNR group; MNR-F, female fetuses from MNR group. Immunoreactivity of MNF2 was expressed as fraction stained (in %; E) and density [F; in arbitrary units (AU)]. Data are expressed as mean + SEM; n=5 (when separated by sex) or n=10 (sexes combined) animals/group. Comparison between groups was performed using a non-parametric Mann-Whitney test. P-value less than 0.05 was considered significant. *P<0.05 vs. respective controls.
3.5. Activity of mitochondrial proteins and tissue adenine nucleotide content in the fetal cardiac left ventricle
Citrate synthase (CS), a mitochondrial matrix enzyme commonly used as a mitochondrial marker, was decreased in MNR (C 1798.7±145.9vsMNR 1011.4±189.9 nmol/min/mg protein, Table 1), with female fetuses demonstrating a more significant effect, reaching a difference of 51% between C-F and MNR-F (1527±187vs741±98 nmol/min/mg protein). CS activity was also sex-dependent in control fetuses, being 26% lower in C-F than C-M. Considering that the decrease in CS activity may reflect inherent mitochondrial dysfunction caused by MNR, we calculated the activities of mitochondrial respiratory chain complexes before and after normalization with CS (Table 1).
Table 1.
Effects of maternal diet on enzymatic activity of mitochondrial respiratory chain complex and changes in the fetal cardiac LV tissue adenine nucleotides and energy charge at 0.9G in control and MNR pregnancies, the latter characterized as a 70% reduction of the food eaten by the control mothers on a weight-adjusted basis.
| Sexes combined | Male | Female | P-value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | MNR | Control | MNR | Control | MNR | C vs MNR | Male C vsMNR | Female C vsMNR | Control M vs F | MNR M vs F | |
| Number of animals/group | 10 | 10 | 5 | 5 | 5 | 5 | |||||
| Citrate Synthase (nmol/min/mg) | 1799±146 | 1011±190 | 2070±154 | 1281±341 | 1527±187 | 741±98 | 0.008 | - | 0.016 | 0.047 | - |
| Complex I (nmol/min/mg) | 1814±84 | 1339±131 | 1955±124 | 1441±142 | 1673±79 | 1237±227 | 0.013 | 0.028 | - | - | - |
| Complex II / III (nmol/min/mg) | 839±143 | 170±66 | 1157±78 | 260±123 | 521±187 | 81±21 | 0.001 | 0.009 | 0.009 | 0.047 | - |
| Complex III (nmol/min/mg) | 1265±153 | 1111±278 | 929±108 | 834±291 | 1601±193 | 1388±475 | - | - | - | 0.028 | - |
| Complex IV (nmol/min/mg) | 154±55 | 845±51 | 219±89 | 761±69 | 90±60 | 928±59 | <0.001 | 0.016 | 0.009 | - | - |
| CompIex I / Citrate Synthase | 1.1±0.1 | 1.6±0.2 | 1.0±0.1 | 1.4±0.4 | 1.2±0.3 | 1.7±0.3 | - | - | - | - | - |
| (Complex II / III) / Citrate Synthase | 0.47±0.07 | 0.19±0.06 | 0.58±0.08 | 0.3±0.1 | 0.4±0.1 | 0.14±0.04 | 0.013 | - | - | - | - |
| Compex III / Citrate Synthase | 0.82±0. | 1.7±0.5 | 0.45±0.04 | 1.0±0.4 | 1.2±0.3 | 2.3±0.9 | - | - | - | 0.009 | - |
| Complex IV / Citrate Synthase | 0.08±0.03 | 1.1±0.2 | 0.11±0.04 | 0.8±0.2 | 0.063±0.04 | 1.4±0.2 | <0.001 | 0.009 | 0.009 | - | - |
| Number of animals/group | 12 | 12 | 6 | 6 | 6 | 6 | |||||
| ATP (nmol/mg) | 3±2 | 0.70±0.05 | 5±4 | 0.76±0.08 | 0.82±0.08 | 0.63±0.05 | 0.028 | - | - | - | - |
| ADP (nmol/mg) | 11±2 | 7.9±0.7 | 14±5 | 8.7±0.8 | 8.0±1 | 7±1 | - | - | - | - | - |
| AMP (nmol/mg) | 75±8 | 77±7 | 70±13 | 84±8 | 81±8 | 69±11 | - | - | - | - | - |
| TAN (nmol/mg) | 89±8 | 86±7 | 89±14 | 94±7 | 90±8 | 77±12 | - | - | - | - | - |
| AEC | 0.09±0.03 | 0.06±0.006 | 0.13±0.06 | 0.05±0.01 | 0.05±0.005 | 0.06±0.002 | - | 0.05 | - | 0.033 | - |
Abbreviations: ATP-adenosine triphosphate; ADP-adenosine diphosphate; AMP-adenosine diphosphate; TAN-total adenine nucleotide pool; AEC-adenylate energy charge.
Data are means±SEM; n=5 or 6 (when separated by sex) or =10 or 12 (sexes combined) animals/group. Comparison between groups was performed using a non-parametric Mann-Whitney test. P-value<0.05 was considered significant and presented.
Mitochondrial complex I, complex II/III, and complex IV activities were decreased in MNR fetuses, with complex II/III being the most severely affected, decreasing 80% in activity (Table 1). In MNR-M fetuses, complex I and complex II/III activities were decreased, with complex II/III activity decreasing 77%. Conversely, complex IV activity increased 3.5 fold in male MNR fetuses. We found a similar pattern in female fetuses with MNR inducing an 84% decrease in complex II/III activity and a 10.4-fold increase in complex IV activity. Sexual dimorphism was present in control fetuses, with female fetuses presenting higher complex III and lower II/III activity. MNR abolished these sex-related differences (Table 1).
After normalization for CS activity, an effect of MNR on respiratory chain activities persisted with a significant decrease in complex II/III and an increase in complex IV activity in both sexes. However, only complex III activity remained different between sexes in control fetuses (C-M vs. C-F, 0.45±0.04vs1.18±0.32).
We assessed possible differences in adenine nucleotide levels between control and MNR fetuses (Table 1). Although mitochondrial ADP and AMP levels were similar in C and MNR fetuses, ATP decreased by 73% in MNR fetuses. Notably, this difference mainly resulted from MNR-M fetuses. Diet-induced reduction of adenylate energy charge was also observed for male fetuses. Once again, sex dimorphism was noted in control fetuses, with males exhibiting 5.2-fold higher ATP content and an increase of 2.2-fold in adenylate energy charge than females.
3.6. Cardiac left ventricle redox state
We evaluated cardiac LV oxidative stress with the lipid peroxidation marker malondialdehyde (MDA) and measured antioxidant enzymes and molecules, including activity of glutathione peroxidase (Gl-Px), glutathione reductase (Gl-Red), and quantification of reduced and oxidized glutathione (GSH and GSSG), and vitamin E (Table 2). There was a 40% increase in MDA in MNR fetuses, which was significantly higher in males (60% increase). GSH quantification also showed sex-related differences in MNR. MNR-F fetuses showed a 2.4-fold greater GSH concentration than MNR-M.
Table 2.
Effects of maternal diet on indicators of antioxidant capacity and oxidative stress in fetal cardiac left ventricle from control ad libitum-fed pregnancies and in the presence of maternal nutrient reduction (MNR), based on a 70% reduction of the food eaten by the control mothers on a weight-adjusted basis at 0.9 gestation.
| Sexes combined | Male | Female | P-value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | MNR | Control | MNR | Control | MNR | C vs MNR | Male C vs MNR | Female C vs MNR | Control M vs F | MNR M vs F | |
| Number of animals/group | 10 | 10 | 5 | 5 | 5 | 5 | |||||
| GSH (mM) | 12±2 | 14±3 | 10±2 | 8±2 | 14±5 | 19±5 | - | - | - | - | 0.047 |
| GSSG (mM) | 4±2 | 3.1±0.9 | 3±1 | 1.6±0.5 | 5±3 | 5±2 | - | - | - | - | - |
| GSH/GSSG | 2.9±0.6 | 7±2 | 3.4±0.8 | 6±1 | 2.4±0.7 | 8±3 | 0.021 | ||||
| Gl-Px (U/l) | 28.5±2.5 | 23±2 | 27±3 | 23±2 | 30±4 | 23±3 | - | - | - | - | - |
| Gl-Red (U/l) | 79±7 | 83±5 | 79±6 | 87±6 | 78±14 | 80 ±9 | - | - | - | - | - |
| Vit E (mM) | 80±12 | 50±3 | 110±16 | 55±1 | 51±2 | 45±4 | - | - | - | - | - |
| MDA (mM) | 1.1±0.1 | 1.5±0.2 | 0.9±0.1 | 1.4±0.1 | 1.2±0.2 | 1.6±0.4 | 0.041 | 0.028 | - | - | - |
Data are means ± SEM; n=5 (when separated by sex) or =10 (sexes combined) animals/group. Comparison between groups was performed using a non-parametric Mann-Whitney test. P-value less than 0.05 was considered significant and presented.
3.7. Cardiac left ventricle mitochondrial morphology
We used transmission electron microscopy (TEM) to analyze changes in mitochondrial morphology and overall organization (Figure 5). MNR markedly altered mitochondrial ultrastructure, especially the number and shape of the cristae. Abundant cristae were found in mitochondria from control fetal cardiac LV of both sexes, whereas in MNR fetal LV samples, both sexes displayed mitochondria with sparse, disarranged, and distorted cristae, partial or total cristolysis, and electron-lucent matrix. Maternal diet-induced mitochondrial morphological alterations were more prominent in MNR-M fetuses, presenting defective mitochondria with few cristae, some resembling an onion-like structure, characterized by multi-layered inner membranes suggesting concentric spherical rings (arrow in Figure 5). Multiple panels in the NMR group of images show several double-membrane structures, resembling autophagic vacuoles.
Fig.5.
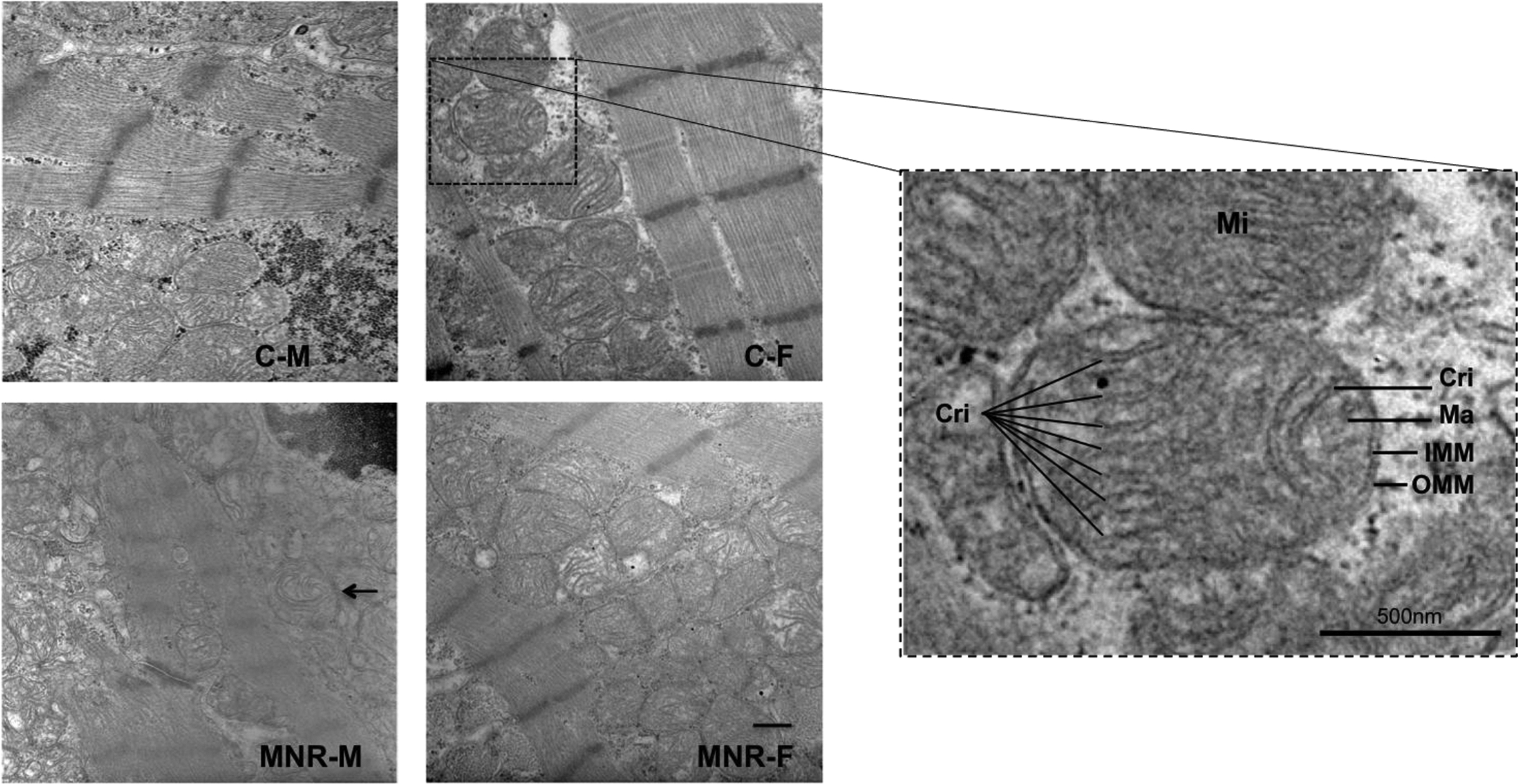
Representative transmission electron microscopy of cardiac LV tissue of fetal baboon from mothers that were fed ad libitum (control group) or 70% of the control (MNR group). C-M, male fetuses from the control group; C-F, female fetuses from the control group; MNR-M, male fetuses from the MNR group; MNR-F, female fetuses from the MNR group. Arrow indicates one mitochondrion with multiple concentric sheets of the inner membrane. Method of staining: uranyl acetate/lead citrate. The rightmost panel is the image magnification of a sample from a female fetus of a control mother. Mitochondria are delimited by double membranes, the inner mitochondrial membrane (IMM) and the outer mitochondrial membrane (OMM), enclosing the matrix (Ma), the section that contains the mitochondrial DNA. The topology of the IMM is dynamically controlled, allowing a greater variation in the morphology of the cristae (Cri). The OMM is more uniform and establishes the organelle border. Method of staining: uranyl acetate/lead citrate. Micrographs were taken with a magnification of 3,300x. Scale bar = 500 nm.
4. Discussion
Suitable healthy human fetal tissues, uncomplicated by confounding pathology, are very rarely, if ever, available to investigate prenatal mitochondrial bioenergetics. Human fetal cardiac samples are obtained at autopsy in pathological situations after a variable period without a functioning circulation. A further limitation of human studies is the lack of homogenous subjects and ensure that the challenges introduced are very carefully controlled. To overcome these limitations, we have developed and characterized a baboon, nonhuman primate model of IUGR that shows an overall offspring cardiac phenotype similar to that described in human IUGR (11,14,45) in order to investigate the nature of the specific in utero nutritional-induced alterations in fetal cardiac mitochondrial function that could explain the greater susceptibility of IUGR offspring to CVD later in life.
We show here for the first time that MNR adversely impacts fetal cardiac LV mitochondria in a sex-dependent fashion. MNR increased 50% of the fetal LV mtDNA content, which was more pronounced in female fetuses, in which a two-fold increase in mtDNA was observed. Transcription of key mitochondrial genes involved in mitochondrial dynamics and oxidative phosphorylation was up-regulated in NMR fetuses, resulting in increased content of several mitochondrial proteins, namely components of the mitochondrial respiratory chain (NDUFB8, UQCRC1, and cytochrome c) and ATP synthase. However, the activity of OXPHOS enzymes was significantly decreased in MNR fetuses (complex I and complex II/III activities), possibly contributing to the 73% decreased ATP content and increased oxidative stress in the cardiac left ventricle tissues, as seen by increased lipid peroxidation marker, MDA. Microscopy of the fetal cardiac left ventricles reflected the mitochondrial dysmorphology induced by MNR, revealing mitochondria with sparse and disarranged cristae.
In our baboon model, we observed a fetal brain-to-liver weight ratio of 3.31 for control fetuses and 4.18 for MNR fetuses was determined, supporting the view that fetal IUGR was present in the MNR fetuses. Our previous study demonstrated that 5.7 years old offspring of MNR mothers exhibited myocardial remodeling with reduced systolic and diastolic function, as detected by magnetic resonance imaging. Of importance, when compared with aged baboons (mean age 15.9 years – normal baboon life span 23 years), levels of dysfunction similar to a premature ageing cardiac phenotype were observed (14). Both LV and RV dysfunction, with reduced wall thickness, reduced filling rates, prolonged diastolic filling times, reduced ejection fraction, reduced 3D sphericity indices, and decreased cardiac output with lower stroke volume, was determined (14). Similar changes have been shown in IUGR human fetuses and children (46–52). In humans, IUGR is significantly associated with preterm birth. Human cardiac magnetic resonance imaging studies reported altered cardiac postnatal growth after preterm birth. Preterm born babies had increased left ventricular mass at 3 months postnatal age, and exhibited at 20 to 39 years increase in LV free wall mass, abnormal LV wall geometry, and impaired LV systolic/diastolic function relative to term-born subjects (53). Bensley et al showed an inverse relationship between the percentage of proliferating cardiac cells and gestational age, with a reduction in the proliferation of cardiomyocytes in the hearts of the preterm infants. In contrast, cardiomyocyte proliferation was still ongoing in age-matched control fetuses. This reduced cardiomyocyte proliferation in preterm infants may adversely impact the final number of cardiomyocytes which may reduce cardiac functional reserve and impair the reparative capacity of the myocardium (54). Analogous results were described in animal models such as rodents and sheep (52,55–60), suggesting cross-species similarity for IUGR cardiac effects.
Here, we sought to identify cardiac LV mitochondrial phenotype changes in MNR-induced IUGR fetuses of both sexes.
Disruption of mitochondrial function will have more significant effects on organs with higher metabolic demands and/or limited regenerative capacity, such as the heart. Impairment of mitochondrial oxidative phosphorylation and fatty acid oxidation have been measured in dilated cardiomyopathies in young infants (61,62). Effects on cardiac function of MNR programming are likely exacerbated during the transition from pre-natal to postnatal life, a critical period in which mitochondrial ATP synthesis is determinant (63).
We observed a significant increase in mtDNA copy number in MNR fetuses, possibly a compensatory response to decreased glucose influx into MNR fetal cardiac tissue due to the decreased insulin signaling. Increased circulating mtDNA levels was also found in diabetic patients and fetal blood of IUGR and premature pregnancies (64–66). We demonstrate that fetal undernutrition up-regulates several relevant mitochondrial transcripts and proteins in the fetal baboon cardiac LV, suggesting an increased mitochondrial capacity. However, the measured activity of mitochondrial proteins is significantly decreased in MNR fetuses, accompanied by 73% decreased ATP content and increased oxidative stress. Thus, reduced mitochondrial enzyme activity and ATP production is likely not due to a reduction in mitochondrial content or the expression of the respiratory chain complexes. Instead, observed differences may be due to functionally damaged mitochondria that produce less ATP and produce more reactive oxygen species (ROS), since MNR induced a 40% increase in MDA, reflecting the pro-oxidative environment in the MNR fetal cardiac LV tissue.
ROS play a physiological role at low concentrations (67) but are an important cause of cellular dysfunction and CVD at high concentrations (68,69). Mitochondria are also a key target of oxidative stress, resulting from ROS generation by the respiratory chain under stress conditions (70). The fact that oxidized mtDNA is not repaired as efficiently as nuclear DNA, contributes to a loss of mtDNA fitness because of the higher persistence of damaged mitochondrial genomes (71). Damaged mitochondria and increased ROS production may also lead to cell death, features equally described in CVD and cardiac aging (72), and in line with the increased mRNA levels for the mitochondrial-specific protein BNIP3 found in the hearts of MNR fetuses. Recent studies demonstrated that cardiomyocytes BNIP3 overexpression is associated with aberrant mitochondrial function, loss of mitochondrial membrane potential, induction of mitochondrial permeability transition pore, development of cardiac hypertrophy, activation of aberrant mitophagy, and induction of cardiac cell death (73,74). Growing evidence also attests significant relationship between BNIP3 and mitochondrial morphology (75).
TEM demonstrated that LV MNR fetal cardiac tissues display mitochondrial cristae abnormalities. Mitochondrial cristolysis is related to mitochondrial inner membrane potential decrease and severe respiratory chain defects (76). The inner mitochondrial membrane hosts key oxidative phosphorylation enzymes, so there is a close relationship between the number of cristae and the surface area of that membrane and cellular metabolic activity capacity (77). Abnormalities in mitochondrial morphology, reflected as disorganized cristae, have been described in cardiac and skeletal muscle biopsies from children aged 0.5 to 12 years with non-compaction cardiomyopathy (78). The degree of cristolysis observed in MNR baboon fetuses suggests that the capacity of the fetal LV to generate energy by mitochondrial OXPHOS is critically compromised, priming cardiomyocytes to a low energy bioenergetic state, in agreement with the observed low adenine nucleotides and energy charge.
In agreement with the mitochondrial abnormalities observed, we found alterations of mitochondrial fission/fusion involved proteins in the cardiac LV of MNR offspring. Imbalance in mitochondrial fission/fusion process leads to mitochondrial deformities associated with numerous human diseases (79,80). We found elevated MFN2 transcripts in the MNR fetal LV, accompanied by increased immunoreactive protein in MNR-F fetuses. Outer mitochondrial membrane MFN2 induces the fusion of this membrane with the membranes of neighboring organelles, being also a mitochondrial assembly regulatory factor (79,80). Changes in mitochondrial fission/fusion may contribute to the mitochondrial degeneration in the cardiac LV of MNR near-term offspring observed by TEM.
The present work clearly indicates fetal sex-dependent outcomes. Male MNR fetuses seem more affected by in utero nutritional deprivation. These findings agree with the considerable body of data documenting sex dissimilarities in the frequency and severity of coronary artery disease, cardiac hypertrophy, heart failure and sudden cardiac death with mitochondrial dysfunction playing a role in these diseases (81–83).
Sex-based differences in human disease are caused by the levels of endogenous sex steroid hormones that now we know regulate mitochondrial metabolism (83,84).
We have demonstrated increased myocardial fibrosis and autophagy in male MNR fetuses by term, indicating impaired stiffness and predisposition to diastolic dysfunction (13). This increased stiffness in the fetal hearts is demonstrated in the postnatal dynamic cardiac MRI data (14,85). Sex-specific hypertensive effects have been reported in MNR-M offspring (86). Sex differences can be in part explained by intrinsic sex-specific mitochondrial differences (87,88). It has been described that estrogens and androgens protect mitochondria against aging-related degenerative effects in a tissue-specific manner by activating their respective receptors (89). While the mechanisms and targets by which estrogens act directly and indirectly to regulate mitochondrial function are not entirely clarified, it is clear that estradiol regulates mitochondrial metabolism and morphology via nuclear and mitochondrial-mediated events, including stimulation of nuclear respiratory factor-1 (NRF-1) transcription, one of NRF-1 target is TFAM that binds mtDNA to regulate its transcription (reviewed in (84)). However, mechanisms for sex differences include not only estrogens and androgens but also sex chromosome-encoded genes.
Since overall effects were more severe in MNR-M fetuses, energy production, cardiac function, and the ability to withstand “second hits” later in life would likely be more compromised in males who were IUGR at birth, decreasing metabolic resilience.
MNR-M fetuses exhibited higher MDA levels than females indicating a more elevated prooxidative environment in male cardiac LV tissue, a critical factor in the pathogenesis development of CVD (69). In a rat model of MNR (86), plasma from 21-day-old male offspring showed increased carbonyls content, decreased GSH, and decreased superoxide anion scavenging activity while MNR female plasma did not show a prooxidative status. Women show slower progress of atherosclerosis, lesser incidence of heart failure (90,91), and usually acquire heart disease advanced in life than men (92). Also, abnormal cardiac morphology and function in healthy humans and animal models are plausible risk factors for sex-associated CVD development (93). Additionally, hearts from females show greater contractility (93) and better calcium handling (94), functions in which mitochondria play important roles. Our data strengthen the view that pregnancy responses to metabolic stressors can have differential LV mitochondrial effects in male and female fetal hearts that reflect sex differences in production and handling of oxidative stress.
To our knowledge, the present study is the first demonstrating the sensitivity of fetal LV cardiac mitochondrial function to moderate MNR. Our model consistently shows important human parallels regarding responses to prenatal stressors that lead to later life disease vulnerabilities. In this model, postnatal dysfunction has also been demonstrated in offspring carbohydrate metabolism by the early development of peripheral insulin resistance (95). We demonstrate here several fetal alterations that potentially predispose to the early adult impaired cardiac ventricular dysfunction previously reported (11,14,85). Numerous studies establish that mitochondrial alterations produced by gestational exposure persist into adulthood or across generations (20).
This study offers a unique view of the sensitivity of cardiac mitochondria during fetal development in an animal model that has consistently shown important parallels with the human in terms of responses to stressors in the womb that lead to later life disease vulnerabilities. This model enables qualitative and quantitative assessment of biological processes, exposing new mechanisms that regulate cardiac tissue metabolism and function. Potential therapeutic interventions that address the decreased mitochondrial function have been investigated in rodents. There is a need for similar studies in nonhuman primates, addressing interventions to prevent the programmed mitochondrial dysfunction in utero as well as to reverse the postnatal effects due to lifestyle habits. Among the future studies, we propose dietary and/or exercise interventions to improve cardiac mitochondrial function and increase reserve cardiovascular response in MNR/IUGR offspring and measurement of cardiac function and mitochondrial capacity throughout in longitudinal studies in this baboon model.
Although this is a relatively short-term study, perform a longer-term follow-up will better understand the sex-specific outcomes, identify sex-specific biomarkers for cardiac dysfunction, and evaluate the effect of developing programming in premature cardiac aging and mortality. Afterward, the longstanding implications on well-being and illness risk is accumulative throughout time, so any alteration (relative insufficiencies) in cardiac biology that arise earlier in the life continuum tend to produce larger magnitudes than those (with equivalent magnitude) that happen later in life. Mitochondria are the heart of cellular bioenergetics, so we expect that gestational dysfunction of cardiac mitochondria will have pronounced consequences later in life. Significant knowledge is missing, such as the extent and duration of the long-term consequences of developmental conditions on initial mitochondrial function, the clinical implication of these effects on pathophysiology susceptibility over the life span, the molecular mechanism(s) perpetuating long-term effects and their plasticity, the establishment of non-invasive biomarkers of mitochondrial function across tissues, and the influence of sex differences on long-term mitochondrial function and outputs. Both sexes were studied and revealed fetal sex-dependent outcomes. Male MNR fetuses were more severely affected by in utero nutritional deprivation. Understanding the sex-specific underlying pathogenesis of IUGR and the sex-specific cellular mechanisms responsible for in utero programmed predisposition to cardiac disease will allow the development of sex-specific biomarkers for early diagnosis in both sexes provide an opportunity for more timely and sex-target interventions to improve life course cardiovascular health.
5. Conclusion
Alterations in cardiac mitochondrial structure/function are implicated in cardiac developmental programming and likely influence long-term cardiac health (Figure 6). A better understanding of the underlying pathogenesis of IUGR and the cellular mechanisms responsible for in utero programmed predisposition to cardiac disease will allow the development of biomarkers for early diagnosis that provides an opportunity for more timely interventions to improve life course cardiovascular health.
Fig. 6.
In utero cardiac mitochondrial alterations due to IUGR in a non-human primate model may explain the previously described sex-dependent later life cardiac pathologies.
6. Study Limitations
The sample size used in this study (n=6 by sex and treatment, total n=12 with sexes pooled) may appear small compared with rodent studies; however, the power of our analysis is very high compared with other primate studies. We published a paper surveying different publications with primates as research models (96), which averaged 6 with sexes pooled, lower than our study.
Although we have shown early left and right ventricular heart failure with preserved ejection fraction in MNR offspring in the same model (11,14,85,97), we did not perform cardiac mitochondrial functional analysis in the fetal or adult cardiac muscle under basal or stress conditions. However, we reported a significant reduction in maximum respiration rate of adult skin-derived fibroblasts from MNR compared to control (98). MNR-induced mitochondrial dysfunction may be defined as priming pathological deviance from the physiologic program of a healthy cell. The effects of the MNR-induced state may range from mild to severe alteration of metabolic and signaling pathways leading to increased or accelerated cell death, depending on the cell type, its energy requirements, its underlying expression program, and its accumulation of second-hit stresses. We do not anticipate that MNR-programmed effects to be exclusively cardiac, but instead affect the whole organism leading to tissue-specific MNR-implications. This raises the pertinence to study other tissues.
Further studies are warranted to establish how early cardiac implications of IUGR can be detected. It is primordial to establish cardiac-impairment IUGR biomarkers to use in a clinical setting. More research in this area is clearly needed. Hence, it may be prudent for clinicians to consider periodic echocardiographic examinations of IUGR offspring to monitor cardiac performance and potentially intervene early, as this group may be at greater risk of cardiovascular disease at an earlier age.
Supplementary Material
7. Clinical Perspectives.
There is a lack of knowledge on the role of mitochondria during fetal development on later-life cardiac dysfunction caused by maternal nutrient reduction (MNR).
Our nonhuman primate baboon model of moderate MNR revealed a sex-dependent relationship between MNR, fetal cardiac mitochondrial remodeling and bioenergetic imbalance that may ultimately predispose the offspring to cardiometabolic disorders later in life.
Uncovering the precise mechanisms underlying the IUGR pathogenesis and the in utero programmed predisposition to cardiac disease will allow the development of biomarkers for an early diagnosis, and to establish timely and efficient interventions to improve a life course cardiovascular health.
Acknowledgments
The authors dedicate this paper to the late Dr. Thomas J. McDonald from University of Texas Health Science Center at San Antonio. The authors acknowledge the contributions by Paulina Quezada and Greg Langone, Michelle Zavala, Ana Maria Silva, Leslie Myatt, Nagarjun Kasaraneni, Chunming Guo, Balasubashini Muralimanoharan, James Mele, Karen Moore and Susan Jenkins.
Funding
This work was supported by FEDER funds through the Operational Programme Competitiveness Factors-COMPETE and national funds by FCT-Foundation for Science and Technology, Portugal under fellowships [SFRH/BD/64694/2009 and SFRH/BPD/116061/2016 to S.P.P.; Post-Doctoral Researcher contract DL57/2016 #SFRH/BPD/84473/2012 to A.I.D]; [PTDC/DTP-DES/1082/2014 and POCI-01-0145-FEDER-016657 to P.J.O.]; [UID/NEU/04539/2020 to CNC]; Program Project Grant (P01) from the National Institutes of Health, USA [PO1-HD21350 to P.W.N., C.L. and M.J.N.] and R24 RR021367. Resources and facilities were also supported by the Southwest National Primate Research Center, San Antonio,Texas, USA grant [P51-RR013986] from the National Center for Research Resources, NIH, currently supported by the Office of Research Infrastructure Programs through [P51-OD011133] and [C06-RR013556]. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of this document.
Footnotes
Tweet: Maternal nutrition reduction causes fetal sex-dependent cardiac mitochondrial remodelling, bioenergetic imbalance and later-life cardiac disease.
Conflicts of interest/Competing interests
The authors declare that they have no conflict of interest. relationship with industry.
Ethics approval
All animal procedures were approved by the Animal Care and Use Committees of the Texas Biomedical Research Institute and the University of Texas Health Science Center at San Antonio, TX (no. 1134PC), and were conducted in Association for Assessment and Accreditation of Laboratory Animal Care-approved facilities and NIH Guide for the Care and Use of Laboratory Animals as detailed in the supplemental material.
Consent for publication
All authors agreed with the content and gave explicit consent to submit and that they obtained consent from the responsible authorities at the institute/organization where the work has been carried out, before the work was submitted.
8. Availability of data and material
All data and materials are available upon request.
9 References
- 1.WHO. Noncommunicable diseases [Internet]. 2018. [cited 2020 May 13]. Available from: https://www.who.int/en/news-room/fact-sheets/detail/noncommunicable-diseases
- 2.Thornburg KL. The programming of cardiovascular disease. J Dev Orig Health Dis [Internet]. 2015. October 15 [cited 2018 Jan 7];6(05):366–76. Available from: http://www.journals.cambridge.org/abstract_S2040174415001300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masoumy EP, Sawyer AA, Sharma S, Patel JA, Gordon PMK, Regnault TRH, et al. The lifelong impact of fetal growth restriction on cardiac development. Pediatr Res [Internet]. 2018;84(4):537–44. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29967522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rich-Edwards JW, Stampfer MJ, Manson JE, Rosner B, Hankinson SE, Colditz G a, et al. Birth weight and risk of cardiovascular disease in a cohort of women followed up since 1976. BMJ. 1997;315(7105):396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker DJ, Osmond C, Golding J, Kuh D, Wadsworth ME. Growth in utero, blood pressure in childhood and adult life, and mortality from cardiovascular disease. BMJ. 1989;298(6673):564–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rich-Edwards J, Kleinman K, Michels K, Stampfer M, Manson J, Rexrode K, et al. Longitudinal study of birth weight and adult body mass index in predicting risk of coronary heart disease and stroke in women. BMJ. 2005;330(7500):1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker DJ, Martyn CN, Osmond C, Hales CN, Fall CH. Growth in utero and serum cholesterol concentrations in adult life. BMJ. 1993;307(6918):1524–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Syddall H, Aihie Sayer A, Dennison E, Martin H, Barker D, Cooper C. Cohort Profile: The Hertfordshire Cohort Study. Int J Epidemiol. 2005December1;34(6):1234–42. Available from: http://academic.oup.com/ije/article/34/6/1234/707357/Cohort-Profile-The-Hertfordshire-Cohort-Study [DOI] [PubMed] [Google Scholar]
- 9.Cheong JN, Wlodek ME, Moritz KM, Cuffe JSM. Programming of maternal and offspring disease: impact of growth restriction, fetal sex and transmission across generations. J Physiol. 2016September1;594(17):4727–40. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26970222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop AC, Libardoni M, Choudary A, Misra B, Lange K, Bernal J, et al. Nonhuman primate breath volatile organic compounds associate with developmental programming and cardio-metabolic status. J Breath Res. 2018May14;12(3):036016. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29593130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuo AH, Li C, Huber HF, Schwab M, Nathanielsz PW, Clarke GD. Maternal nutrient restriction during pregnancy and lactation leads to impaired right ventricular function in young adult baboons. J Physiol. 2017;13(13):4245–60. Available from: http://doi.wiley.com/10.1113/JP273928%0Ahttp://www.ncbi.nlm.nih.gov/pubmed/28439937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo AH, Li C, Huber HF, Clarke GD, Nathanielsz PW. Intrauterine growth restriction results in persistent vascular mismatch in adulthood. J Physiol. 2017November3; Available from: http://doi.wiley.com/10.1113/JP275139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muralimanoharan S, Li C, Nakayasu ES, Casey CP, Metz TO, Nathanielsz PW, et al. Sexual dimorphism in the fetal cardiac response to maternal nutrient restriction. J Mol Cell Cardiol. 2017July;108:181–93. Available from: 10.1016/j.yjmcc.2017.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo AH, Li C, Li J, Huber HF, Nathanielsz PW, Clarke GD. Cardiac remodelling in a baboon model of intrauterine growth restriction mimics accelerated ageing. J Physiol [Internet]. 2017;595(4):1093–110. Available from: http://doi.wiley.com/10.1113/JP272908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, Jenkins S, Mattern V, Comuzzie AG, Cox LA, Huber HF, et al. Effect of moderate, 30 percent global maternal nutrient reduction on fetal and postnatal baboon phenotype. J Med Primatol. 2017;46(6):293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li C, McDonald TTJ, Wu G, Nijland MMJ, Nathanielsz PWP. Intrauterine growth restriction alters term fetal baboon hypothalamic appetitive peptide balance. J Endocrinol. 2013June;217(3):275–82. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23482706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mäkikallio K, Räsänen J, Mäkikallio T, Vuolteenaho O, Huhta JC. Human fetal cardiovascular profile score and neonatal outcome in intrauterine growth restriction. Ultrasound Obstet Gynecol. 2008January1;31(1):48–54. Available from: http://doi.wiley.com/10.1002/uog.5210 [DOI] [PubMed] [Google Scholar]
- 18.Severi FM, Rizzo G, Bocchi C, D’Antona D, Verzuri MS, Arduini D. Intrauterine growth retardation and fetal cardiac function. Fetal Diagn Ther. 2000;15(1):8–19. Available from: http://www.ncbi.nlm.nih.gov/pubmed/10705209 [DOI] [PubMed] [Google Scholar]
- 19.Lock MC, Tellam RL, Botting KJ, Wang KCW, Selvanayagam JB, Brooks DA, et al. The role of miRNA regulation in fetal cardiomyocytes, cardiac maturation and the risk of heart disease in adults. J Physiol. 2018;596(23):5625–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gyllenhammer LE, Entringer S, Buss C, Wadhwa PD. Developmental programming of mitochondrial biology: a conceptual framework and review. Proc R Soc B Biol Sci [Internet]. 2020. May 13;287(1926):20192713. Available from: https://royalsocietypublishing.org/doi/10.1098/rspb.2019.2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheuermann-Freestone M, Madsen PL, Manners D, Blamire AM, Buckingham RE, Styles P, et al. Abnormal cardiac and skeletal muscle energy metabolism in patients with type 2 diabetes. Circulation. 2003;107(24):3040–6. [DOI] [PubMed] [Google Scholar]
- 22.Schlabritz-Loutsevitch NE, Howell K, Rice K, Glover EJ, Nevill CH, Jenkins SL, et al. Development of a system for individual feeding of baboons maintained in an outdoor group social environment. J Med Primatol. 2004June;33(3):117–26. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15102068 [DOI] [PubMed] [Google Scholar]
- 23.Schlabritz-Loutsevitch NE, Hubbard GB, Dammann MJ, Jenkins SL, Frost P a, McDonald TJ, et al. Normal concentrations of essential and toxic elements in pregnant baboons and fetuses (Papio species). J Med Primatol. 2004June;33(3):152–62. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15102072 [DOI] [PubMed] [Google Scholar]
- 24.Wu A, Ying Z, Gomez-Pinilla F. Dietary omega-3 fatty acids normalize BDNF levels, reduce oxidative damage, and counteract learning disability after traumatic brain injury in rats. J Neurotrauma. 2004October;21(10):1457–67. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15672635 [DOI] [PubMed] [Google Scholar]
- 25.Pereira SP, Deus CM, Serafim TL, Cunha-Oliveira T, Oliveira PJ. Metabolic and Phenotypic Characterization of Human Skin Fibroblasts After Forcing Oxidative Capacity. Toxicol Sci. 2018July1;164(1):191–204. Available from: https://academic.oup.com/toxsci/article/164/1/191/4947784 [DOI] [PubMed] [Google Scholar]
- 26.Cox L, Nijland M, Gilbert J, Schlabritz-Loutsevitch N, Hubbard G, McDonald T, et al. Effect of 30 per cent maternal nutrient restriction from 0.16 to 0.5 gestation on fetal baboon kidney gene expression. J Physiol. 2006April;572(Pt 1):67–85. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1779656&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machado NG, Baldeiras I, Pereira GC, Pereira SP, Oliveira PJ. Sub-chronic administration of doxorubicin to Wistar rats results in oxidative stress and unaltered apoptotic signaling in the lung. Chem Biol Interact. 2010December;188(3):478–86. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20932959 [DOI] [PubMed] [Google Scholar]
- 28.Romero-Calvo I, Ocón B, Martínez-Moya P, Suárez MD, Zarzuelo A, Martínez-Augustin O, et al. Reversible Ponceau staining as a loading control alternative to actin in Western blots. Anal Biochem. 2010June15;401(2):318–20. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20206115 [DOI] [PubMed] [Google Scholar]
- 29.Dittmer A, Dittmer J. β-Actin is not a reliable loading control in Western blot analysis. Electrophoresis. 2006;27(14):2844–5. Available from: http://doi.wiley.com/10.1002/elps.200500785 [DOI] [PubMed] [Google Scholar]
- 30.Aldridge GM, Podrebarac DM, Greenough WT, Weiler IJ. The use of total protein stains as loading controls: An alternative to high-abundance single-protein controls in semi-quantitative immunoblotting. J Neurosci Methods. 2008;172(2):250–4. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2567873&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pereira SP, Oliveira PJ, Tavares LC, Moreno AJ, Cox L a., Nathanielsz PW, et al. Effects of moderate global maternal nutrient reduction on fetal baboon renal mitochondrial gene expression at 0.9 gestation. Am J Physiol - Ren Physiol. 2015June1;308(11):F1217–28. Available from: http://ajprenal.physiology.org/lookup/doi/10.1152/ajprenal.00419.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long J, Ma J, Luo C, Mo X, Sun L, Zang W, et al. Comparison of two methods for assaying complex I activity in mitochondria isolated from rat liver, brain and heart. Life Sci. 2009August12;85(7–8):276–80. Available from: 10.1016/j.lfs.2009.05.019 [DOI] [PubMed] [Google Scholar]
- 33.Tisdale HD. Preparation and properties of succinic-cytochrome c reductase (complex II-III). Methods Enzymol. 1967;10(C):213–5. [Google Scholar]
- 34.Luo C, Long J, Liu J. An improved spectrophotometric method for a more specific and accurate assay of mitochondrial complex III activity. Clin Chim Acta. 2008September;395(1–2):38–41. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18502205 [DOI] [PubMed] [Google Scholar]
- 35.Brautigan DL, Ferguson-Miller S, Margoliash E. Mitochondrial cytochrome c: Preparation and activity of native and chemically modified cytochromes c. Methods Enzymol. 1978;53(C):128–64. [DOI] [PubMed] [Google Scholar]
- 36.Coore HG, Denton RM, Martin BR, Randle PJ. Regulation of adipose tissue pyruvate dehydrogenase by insulin and other hormones. Biochem J. 1971;125(1):115–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos MS, Moreno AJ, Carvalho AP. Relationships between ATP depletion, membrane potential, and the release of neurotransmitters in rat nerve terminals. An in vitro study under conditions that mimic anoxia, hypoglycemia, and ischemia. Stroke. 1996;27(5):941–50. [DOI] [PubMed] [Google Scholar]
- 38.Stocchi V, Cucchiarini L, Magnani M, Chiarantini L, Palma P, Crescentini G. Simultaneous extraction and reverse-phase high-performance liquid chromatographic determination of adenine and pyridine nucleotides in human red blood cells. Anal Biochem. 1985;146(1):118–24. [DOI] [PubMed] [Google Scholar]
- 39.Draper HH, Hadley M. Malondialdehyde determination as index of lipid peroxidation. Methods Enzymol. 1990;186:421–31. [DOI] [PubMed] [Google Scholar]
- 40.Tsao S-MM, Yin M-CC, Liu W-HH. Oxidant stress and B vitamins status in patients with non-small cell lung cancer. Nutr Cancer. 2007;59(1):8–13. [DOI] [PubMed] [Google Scholar]
- 41.Paglia DE, Valentine WN. Studies on the quantitative and qualitative characterization of erythrocyte glutathione peroxidase. J Lab Clin Med. 1967;70(1):158–69. [PubMed] [Google Scholar]
- 42.Goldberg D, Spooner R. Glutathione reductase. In: Bergmerye, editor. Methods in Enzymatic Analysis. New York.: Verlag Chemie Weinheim Academic Pres; 1983. p. 258–65. [Google Scholar]
- 43.Malhotra BSK. Experiments on fixation for electron microscopy. Small. 1962;103(March):5–15. [Google Scholar]
- 44.Li C, Ramahi E, Nijland MJ, Choi J, Myers D a., Nathanielsz PW, et al. Up-regulation of the fetal baboon hypothalamo-pituitary-adrenal axis in intrauterine growth restriction: Coincidence with hypothalamic glucocorticoid receptor insensitivity and leptin receptor down-regulation. Endocrinology. 2013;154(7):2365–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuo AH, Li J, Li C, Huber HF, Nathanielsz PW, Clarke GD. Poor perinatal growth impairs baboon aortic windkessel function. J Dev Orig Health Dis. 2017October11;5038:1–6. Available from: https://www.cambridge.org/core/product/identifier/S2040174417000770/type/journal_article [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crispi F, Hernandez-Andrade E, Pelsers MMAL, Plasencia W, Benavides-Serralde JA, Eixarch E, et al. Cardiac dysfunction and cell damage across clinical stages of severity in growth-restricted fetuses. Am J Obstet Gynecol. 2008;199(3). Available from: https://ac.els-cdn.com/S000293780800687X/1-s2.0-S000293780800687X-main.pdf?_tid=ef09df74-f411-11e7-98b1-00000aab0f6b&acdnat=1515374532_c2b22053d76d7dcb2792d04a3b4fbd87 [DOI] [PubMed] [Google Scholar]
- 47.Crispi F, Bijnens B, Figueras F, Bartrons J, Eixarch E, Le Noble F, et al. Fetal growth restriction results in remodeled and less efficient hearts in children. Circulation. 2010June8;121(22):2427–36. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20497977 [DOI] [PubMed] [Google Scholar]
- 48.Leipälä JA, Boldt T, Turpeinen U, Vuolteenaho O, Fellman V. Cardiac hypertrophy and altered hemodynamic adaptation in growth-restricted preterm infants. Pediatr Res. 2003;53(6):989–93. Available from: https://www.nature.com/articles/pr2003440.pdf [DOI] [PubMed] [Google Scholar]
- 49.Fouzas S, Karatza AA, Davlouros PA, Chrysis D, Alexopoulos D, Mantagos S, et al. Neonatal cardiac dysfunction in intrauterine growth restriction. Pediatr Res. 2014May12;75(5):651–7. Available from: http://www.nature.com/articles/pr201422 [DOI] [PubMed] [Google Scholar]
- 50.Comas M, Crispi F, Cruz-Martinez R, Figueras F, Gratacos E. Tissue Doppler echocardiographic markers of cardiac dysfunction in small-for-gestational age fetuses. YMOB. 2011;205:57.e1–57.e6. Available from: https://ac.els-cdn.com/S000293781100319X/1-s2.0-S000293781100319X-main.pdf?_tid=460bcaa6-f40f-11e7-bc81-00000aab0f01&acdnat=1515373390_0b26ed4eb9e7ae04d32fedda05434e98 [DOI] [PubMed] [Google Scholar]
- 51.Verburg BO, Jaddoe VWV, Wladimiroff JW, Hofman A, Witteman JCM, Steegers EAP. Fetal Hemodynamic Adaptive Changes Related to Intrauterine Growth: The Generation R Study. Circulation. 2008February5;117(5):649–59. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7805194 [DOI] [PubMed] [Google Scholar]
- 52.Darby JRT, Varcoe TJ, Orgeig S, Morrison JL. Cardiorespiratory consequences of intrauterine growth restriction: Influence of timing, severity and duration of hypoxaemia. Theriogenology. 2020;150:84–95. Available from: 10.1016/j.theriogenology.2020.01.080 [DOI] [PubMed] [Google Scholar]
- 53.Lewandowski AJ, Augustine D, Lamata P, Davis EF, Lazdam M, Francis J, et al. Preterm heart in adult life: Cardiovascular magnetic resonance reveals distinct differences in left ventricular mass, geometry, and function. Circulation. 2013;127(2):197–206. [DOI] [PubMed] [Google Scholar]
- 54.Bensley JG, Moore L, De Matteo R, Harding R, Black MJ. Impact of preterm birth on the developing myocardium of the neonate. Pediatr Res. 2018;83(4):880–8. Available from: 10.1038/pr.2017.324 [DOI] [PubMed] [Google Scholar]
- 55.Wang KCW, Zhang L, Mcmillen IC, Botting KJ, Duffield JA, Zhang S, et al. Fetal growth restriction and the programming of heart growth and cardiac insulin-like growth factor 2 expression in the lamb. J Physiol. 2011;589(19):4709–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrison JL, Botting KJ, Dyer JL, Williams SJ, Thornburg KL, McMillen IC. Restriction of placental function alters heart development in the sheep fetus. Am J Physiol - Regul Integr Comp Physiol. 2007;293(1). [DOI] [PubMed] [Google Scholar]
- 57.Bubb KJ, Cock ML, Black MJ, Dodic M, Boon WM, Parkington HC, et al. Intrauterine growth restriction delays cardiomyocyte maturation and alters coronary artery function in the fetal sheep. J Physiol. 2007;578(3):871–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Master JS, Zimanyi MA, Yin KV., Moritz KM, Gallo LA, Tran M, et al. Transgenerational left ventricular hypertrophy and hypertension in offspring after uteroplacental insufficiency in male rats. Clin Exp Pharmacol Physiol. 2014;41(11):884–90. [DOI] [PubMed] [Google Scholar]
- 59.Dong F, Ford SP, Fang CX, Nijland MJ, Nathanielsz PW, Ren J. Maternal nutrient restriction during early to mid gestation up-regulates cardiac insulin-like growth factor (IGF) receptors associated with enlarged ventricular size in fetal sheep. Growth Horm IGF Res. 2005;15(4):291–9. [DOI] [PubMed] [Google Scholar]
- 60.Ge W, Hu N, George LA, Ford SP, Nathanielsz PW, Wang XM, et al. Maternal nutrient restriction predisposes ventricular remodeling in adult sheep offspring. J Nutr Biochem. 2013;24(7):1258–65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Lipshultz SE, Sleeper LA, Towbin JA, Lowe AM, Orav EJ, Cox GF, et al. The Incidence of Pediatric Cardiomyopathy in Two Regions of the United States. N Engl J Med. 2003April24;348(17):1647–55. Available from: http://www.nejm.org/doi/abs/10.1056/NEJMoa021715 [DOI] [PubMed] [Google Scholar]
- 62.Kelly DP, Strauss AW, Strauss AW. Inherited cardiomyopathies. N Engl J Med. 1994March31;330(13):913–9. Available from: http://www.nejm.org/doi/abs/10.1056/NEJM199403313301308 [DOI] [PubMed] [Google Scholar]
- 63.Porter GA, Hom JR, Hoffman DL, Quintanilla RA, de Mesy Bentley K, Sheu S-SS, et al. Bioenergetics, mitochondria, and cardiac myocyte differentiation. Prog Pediatr Cardiol. 2011May;31(2):75–81. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3096664&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Novielli C, Mandò C, Tabano S, Anelli GM, Fontana L, Antonazzo P, et al. Mitochondrial DNA content and methylation in fetal cord blood of pregnancies with placental insufficiency. Placenta. 2017;55:63–70. [DOI] [PubMed] [Google Scholar]
- 65.Czajka A, Ajaz S, Gnudi L, Parsade CK, Jones P, Reid F, et al. Altered Mitochondrial Function, Mitochondrial DNA and Reduced Metabolic Flexibility in Patients With Diabetic Nephropathy. EBioMedicine. 2015;2(6):499–512. Available from: 10.1016/j.ebiom.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou M, Zhu L, Cui X, Feng L, Zhao X, He S, et al. Reduced peripheral blood mtDNA content is associated with impaired glucose‐stimulated islet β cell function in a Chinese population with different degrees of glucose tolerance. Diabetes Metab Res Rev. 2016October30;32(7):768–74. Available from: http://libweb.anglia.ac.uk/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brand MD. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic Biol Med. 2016;100:14–31. Available from: 10.1016/j.freeradbiomed.2016.04.001 [DOI] [PubMed] [Google Scholar]
- 68.Cadenas S ROS and redox signaling in myocardial ischemia-reperfusion injury and cardioprotection. Free Radic Biol Med. 2018March;117:76–89. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0891584918300340 [DOI] [PubMed] [Google Scholar]
- 69.Ballinger SW. Mitochondrial dysfunction in cardiovascular disease. Free Radic Biol Med. 2005;38(10):1278–95. [DOI] [PubMed] [Google Scholar]
- 70.Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial Reactive Oxygen Species (ROS) and ROS-Induced ROS Release. Physiol Rev. 2014;94(3):909–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94(2):514–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9012815%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC19544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, Li Y, He C, Gou B, Song M. Mitochondrial regulation of cardiac aging. Biochim Biophys Acta - Mol Basis Dis. 2019;(December):0–1. Available from: 10.1016/j.bbadis.2018.12.008 [DOI] [PubMed] [Google Scholar]
- 73.Regula KM, Ens K, Kirshenbaum LA. Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ Res. 2002;91(3):226–31. [DOI] [PubMed] [Google Scholar]
- 74.Dhingra A, Jayas R, Afshar P, Guberman M, Maddaford G, Gerstein J, et al. Ellagic acid antagonizes Bnip3-mediated mitochondrial injury and necrotic cell death of cardiac myocytes. Free Radic Biol Med. 2017;112(August):411–22. Available from: 10.1016/j.freeradbiomed.2017.08.010 [DOI] [PubMed] [Google Scholar]
- 75.Gao A, Jiang J, Xie F, Chen L. Bnip3 in mitophagy: Novel insights and potential therapeutic target for diseases of secondary mitochondrial dysfunction. Clin Chim Acta [Internet]. 2020;506(December 2019):72–83. Available from: 10.1016/j.cca.2020.02.024 [DOI] [PubMed] [Google Scholar]
- 76.Arismendi-Morillo G Electron microscopy morphology of the mitochondrial network in gliomas and their vascular microenvironment. Biochim Biophys Acta [Internet]. 2010. November 9 [cited 2011 Oct 11];1807(6):602–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21070743 [DOI] [PubMed] [Google Scholar]
- 77.Modica-Napolitano JS, Singh KK. Mitochondria as targets for detection and treatment of cancer. Expert Rev Mol Med. 2002;4(9):1–19. [DOI] [PubMed] [Google Scholar]
- 78.Pignatelli RH, McMahon CJ, Dreyer WJ, Denfield SW, Price J, Belmont JW, et al. Clinical Characterization of Left Ventricular Noncompaction in Children: A Relatively Common Form of Cardiomyopathy. Circulation. 2003;108(21):2672–8. [DOI] [PubMed] [Google Scholar]
- 79.Filadi R, Pendin D, Pizzo P. Mitofusin 2: from functions to disease. Cell Death Dis [Internet]. 2018. March 28 [cited 2018 Aug 14];9(3):330. Available from: https://www.nature.com/articles/s41419-017-0023-6.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bach D, Pich S, Soriano FX, Vega N, Baumgartner B, Oriola J, et al. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism: A novel regulatory mechanism altered in obesity. J Biol Chem. 2003May9;278(19):17190–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12598526 [DOI] [PubMed] [Google Scholar]
- 81.Gilbert JS, Nijland MJ. Sex differences in the developmental origins of hypertension and cardiorenal disease. Am J Physiol Regul Integr Comp Physiol. 2008December;295(6):R1941–52. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2685301&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khalifa ARM, Abdel-Rahman EA, Mahmoud AM, Ali MH, Noureldin M, Saber SH, et al. Sex-specific differences in mitochondria biogenesis, morphology, respiratory function, and ROS homeostasis in young mouse heart and brain. Physiol Rep. 2017;5(6):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Silaidos C, Pilatus U, Grewal R, Matura S, Lienerth B, Pantel J, et al. Sex-associated differences in mitochondrial function in human peripheral blood mononuclear cells (PBMCs) and brain. Biol Sex Differ. 2018;9(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klinge CM. Estrogenic control of mitochondrial function. Redox Biol [Internet]. 2020;31(August 2019):101435. Available from: 10.1016/j.redox.2020.101435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuo AH, Li C, Huber HF, Nathanielsz PW, Clarke GD. Ageing changes in biventricular cardiac function in male and female baboons (Papio spp.). J Physiol. 2018;596(21):5083–98. Available from: http://doi.wiley.com/10.1113/JP276338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rodríguez-Rodríguez P, de Pablo ALL, Condezo-Hoyos L, Martín-Cabrejas MA, Aguilera Y, Ruiz-Hurtado G, et al. Fetal undernutrition is associated with perinatal sex-dependent alterations in oxidative status. J Nutr Biochem. 2015;26(12):1650–9. Available from: 10.1016/j.jnutbio.2015.08.004 [DOI] [PubMed] [Google Scholar]
- 87.John C, Grune J, Ott C, Nowotny K, Deubel S, Kühne A, et al. Sex Differences in Cardiac Mitochondria in the New Zealand Obese Mouse. Front Endocrinol (Lausanne) [Internet]. 2018. December 4;9(December):1–9. Available from: https://www.frontiersin.org/article/10.3389/fendo.2018.00732/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cardinale DA, Larsen FJ, Schiffer TA, Morales-Alamo D, Ekblom B, Calbet JAL, et al. Superior intrinsic mitochondrial respiration in women than in men. Front Physiol. 2018;9(AUG):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.López-Lluch G Mitochondrial activity and dynamics changes regarding metabolism in ageing and obesity. Mech Ageing Dev. 2017;162:108–21. [DOI] [PubMed] [Google Scholar]
- 90.Vassalle C, Simoncini T, Chedraui P, Pérez-López FR. Why sex matters: the biological mechanisms of cardiovascular disease. Vol. 28, Gynecological Endocrinology. 2012. p. 746–51. [DOI] [PubMed] [Google Scholar]
- 91.Czubryt MP, Espira L, Lamoureux L, Abrenica B. The role of sex in cardiac function and disease. Can J Physiol Pharmacol. 2006;84(1):93–109. [DOI] [PubMed] [Google Scholar]
- 92.Mosca L, Manson JE, Sutherland SE, Langer RD, Manolio T, Barrett-Connor E. Cardiovascular Disease in Women : A Statement for Healthcare Professionals From the American Heart Association. Circulation. 1997;96(7):2468–82. [DOI] [PubMed] [Google Scholar]
- 93.Merz CN, Moriel M, Rozanski A, Klein J, Berman DS. Gender-related differences in exercise ventricular function among healthy subjects and patients. Am Heart J. 1996;131(4):704–9. [DOI] [PubMed] [Google Scholar]
- 94.Regitz-Zagrosek V, Oertelt-Prigione S, Seeland U, Hetzer R. Sex and gender differences in myocardial hypertrophy and heart failure. Circ J. 2010;74(7):1265–73. [DOI] [PubMed] [Google Scholar]
- 95.Choi J, Li C, McDonald TJ, Comuzzie A, Mattern V, Nathanielsz PW. Emergence of insulin resistance in juvenile baboon offspring of mothers exposed to moderate maternal nutrient reduction. Am J Physiol Regul Integr Comp Physiol. 2011September; 301(3):R757–62. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3174762&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pantham P, Rosario FJ, Nijland M, Cheung A, Nathanielsz PW, Powell TL, et al. Reduced placental amino acid transport in response to maternal nutrient restriction in the baboon. Am J Physiol Integr Comp Physiol. 2015;309(7):R740–6. Available from: http://ajpregu.physiology.org/lookup/doi/10.1152/ajpregu.00161.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kuo AH, Li C, Nathanielsz PW. Accelerated aging of cardiac function in offspring of undernourished pregnant baboons. Reprod Sci. 2015;22(March):97A–98A. Available from: http://ovidsp.ovid.com/ovidweb.cgi?T=JS&PAGE=reference&D=emed13&NEWS=N&AN=71847347 [Google Scholar]
- 98.Salmon AB, Dorigatti J, Huber HF, Li C, Nathanielsz PW. Maternal nutrient restriction in baboon programs later-life cellular growth and respiration of cultured skin fibroblasts: a potential model for the study of aging-programming interactions. GeroScience. 2018;40(3):269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and materials are available upon request.


