Abstract
Endophytic bacteria represent microorganisms that live during the whole life cycle within the tissues of healthy plants without causing any obvious signs of disease. In this study, the ability of 128 endophyte bacterial isolates from some cultivated and wild grain plants (Poaceae family) in Van, Turkey, were investigated in terms of producing several extracellular hydrolytic enzymes. It was demonstrated that lipases, proteases, amylases, cellulases, pectinases, and xylanases were produced by the bacteria with relative frequencies of 74.2%, 65.6%, 55.4%, 32%, 21.8%, and 7.8%, respectively. In addition, molecular identification of a certain number of isolates selected according to their enzyme-producing capabilities was performed by 16S rRNA gene sequencing using a next-generation sequencing platform. As a result of the analysis, the isolates yielded certain strains belonging to Pseudomonas, Micrococcus, Paenibacillus, Streptococcus, Curtobacterium, Chryseobacterium, and Bacillus genera. Also, the strain G117Y1T was evaluated as a member of potential novel species based on 16S rRNA sequencing results.
Keywords: endophytic bacteria, extracellular enzymes, 16S rRNA gene, Poaceae family, Illumina MiSeq
Introduction
Although endophytes have been widely defined as microorganisms that live in the tissues of healthy plants for all or part of their life cycle, recent studies have revised this definition to include all microorganisms, including pathogens that can colonize the internal tissues of plants (Hardoim et al. 2015; Compant et al. 2021). Endophytic bacteria (EBs) have been isolated and characterized from different plant parts, including roots, stems, leaves, seeds, fruits, tubers, ovules, and nodules of various plants such as agricultural crops, meadow plants, plants grown in extreme environments, wild, and perennial plants (Afzal et al. 2019). EBs can contribute to plant health and development like Plant Growth Promoting Rhizobacteria (PGPR). In general, PGPR and EBs directly or indirectly affect the growth and development of the plant. EBs stimulate plant growth through various mechanisms such as nitrogen fixation, phytohormone production, nutrient uptake, and providing the plant with tolerance to abiotic and biotic stresses (Kandel et al. 2017). These properties make these bacteria important for various biotechnological applications in agriculture. Also, they have the potential to produce a variety of secondary metabolites like alkaloids, steroids, terpenoids, peptides, polyketones, flavonoids, quinols, and phenols with an application in agriculture, pharmaceutical and industrial biotechnology (Singh et al. 2017).
Microbial enzymes with high catalytic activities are used in many areas of the industry because they are more stable, cheaper, and can be obtained in large amounts by fermentation methods (Singh et al. 2016). Examples of industrial areas affected by discoveries of these enzymes include detergent agents, leather processing, degradation of xenobiotic compounds, food processing (bakery, meat, dairy, fruit, and vegetable products), pharmaceuticals (synthesis of pharmaceutical intermediates), biofuels (low-energy ethanol production process), and other enzyme related technologies (Singh et al. 2016). Although many bacterial isolates from various sources have been reported for the production of cellulase, protease, amylase, pectinase, lipase, asparaginase, etc., the studies involving the examination of endophytic bacteria in terms of biotechnological extracellular enzymes are relatively few (Carrim et al. 2006; Jalgaonwala and Mahajan 2011; Khan et al. 2017). Therefore, endophytic bacteria can represent a new source of enzymes with different application potentials.
In addition to entry through openings and wounds, endophytic bacteria actively colonize plant tissues using hydrolytic enzymes, such as cellulase. It was proposed that cell wall-degrading enzymes such as cellulases, xylanases, and pectinases might be responsible for plant and microbe interactions and intercellular colonization of roots (Verma et al. 2001; Kandel et al. 2017). Therefore, more knowledge on their production is also needed to understand the relationship between endophytic bacteria and plants.
The aim of this study was to examine endophyte bacteria isolated from various cultivated and wild plants of Poaceae family in Van province, Turkey, in terms of their potential to produce industrially important proteases, amylases, lipases, cellulases, xylanases, and pectinases and to perform a phylogenetic affiliation of the strains possessing relatively high enzyme activity profiles by 16S rRNA gene sequence analysis.
Experimental
Materials and Methods
Bacterial isolates. In this study, endophyte bacteria isolated from some culture and wild grain plants (Poaceae family) stored at bacteriology laboratory, Department of Plant Protection, Faculty of Agriculture, Van Yuzuncu Yil University, were used. Endophytic bacteria had been isolated according to the method described by Ozaktan et al. (2015). The trituration technique with effective surface sterilization of the plant tissues was applied in this method. The plant species and tissues from which the bacteria were isolated were shown in Table I. All strains were grown either in Nutrient Broth (NB) broth (Difco, Detroit, MI, USA) or on Nutrient Broth agar plates at 25°C.
Table I.
The plant species and the tissues from which the endophytic bacteria were isolated and enzymatic indexes (EIs) of hydrolytic enzymes of 16 strains selected for the 16S rRNA gene amplicon sequence analysis.
| Isolate No | Host Plant | Plant Tissue | Protease | Lipase | Amylase | Cellulase | Pectinase | Xylanase |
|---|---|---|---|---|---|---|---|---|
| G90Y2 | Aegilops sp. | Leaf | 3.46 ± 0.15efg | 9.80 ± 0.20a | 2.14 ± 0.03de | 6.10 ± 0.16cd | 1.73 ± 0.03c | – |
| G90S1 | Aegilops sp. | Stem | 2.94 ± 0.08gh1 | 6.79 ± 2.01bc | 3.23 ± 0.09bc | 5.02 ± 0.27de | – | – |
| G88K1 | Triticum aestivum L. | Root | 3.78 ± 0.06def | 1.90 ± 0.11e | – | – | – | 2.88 ± 0.38ns |
| G83S3 | Triticum aestivum L. | Stem | 2.85 ± 0.05h1 | 3.67 ± 0.15de | 3.91 ± 0.37ab | 4.40 ± 0.10e | 2.05 ± 0.05bc | – |
| G105Y1 | Dactylis glomerata L. | Leaf | 7.29 ± 0.71a | 1.87 ± 0.34e | 3.03 ± 0.29bcd | 12.75 ± 1.38a | 3.81 ± 0.38a | – |
| G105S1 | Dactylis spp. | Stem | – | – | – | – | – | – |
| G100Y1 | Festuca spp. | Leaf | 3.40 ± 0.12fgh | 6.96 ± 0.54b | 2.18 ± 0.08de | 7.02 ± 0.46c | – | – |
| G80K3 | Secale cereale L. | Root | 4.03 ± 0.17de | – | 3.05 ± 0.13bcd | – | 4.44 ± 0.90a | – |
| G70K2 | Secale cereale L. | Root | 2.73 ± 0.341 | 7.24 ± 0.78b | 2.69 ± 0.04cd | 4.07 ± 0.13ef | 2.34 ± 0.18bc | – |
| G42K2 | Cultivated Poaceae spp. | Root | 3.57 ± 0.20ef | 4.89 ± 0.22bcd | 2.68 ± 0.09cd | 2.66 ± 0.04f | 1.76 ± 0.14c | – |
| G119Y1T | Eremopoa sp. | Leaf | – | 4.37 ± 0.15cd | 1.29 ± 0.04e | 7.50 ± 0.00c | – | – |
| G118S2T | Eremopoa songarica L. | Stem | 4.22 ± 0.16cd | 4.46 ± 0.22cd | 1.69 ± 0.08de | 3.46 ± 0.19ef | – | 2.65 ± 0.41ns |
| G117Y1T | Eremopoa sp. | Leaf | 3.22 ± 0.13fgh1 | 6.32 ± 1.78bc | 2.81 ± 0.01cd | 2.46 ± 0.12f | – | 1.90 ± 0.27ns |
| G116K1T | Eremopoa songarica L. | Root | 4.68 ± 0.25bc | 1.91 ± 0.18e | – | – | – | – |
| G113Y3 | Triticum aestivum L. | Leaf | 5.12 ± 0.07b | 3.15 ± 0.13de | 4.70 ± 0.17a | 9.77 ± 0.42b | 3.48 ± 0.29ab | 1.75 ± 0.25ns |
| G107Y2 | Triticum aestivum L. | Leaf | 3.26 ± 0.09fgh1 | 7.33 ± 0.67b | 2.68 ± 0.27cd | 4.95 ± 0.30de | – | – |
Means of four replicates (Mean ± Std. Errors). Values within a column followed by different lowercase letters are significantly different (p < 0.05).
ns – not significant
Determination of enzyme activities. The presence of the following enzymes were analyzed: amylases, lipases, cellulases, proteases, pectinases, and xylanases. The pure cultures of the isolates were inoculated onto solid diagnostic media by four isolated droplets. Enzyme Index (EI) is a practical tool that compares the enzymatic production of different isolates (Carrim et al. 2006; Jena and Chandi 2013). The EI for each enzyme was calculated at the end of a specific incubation time. EIs were calculated as a mean ratio of opaque zone diameter to colony diameter.
Amylase activity. The strains were inoculated onto nutrient agar supplemented with 1% (w/v) starch. After incubation for two days at 25°C, agar plate surfaces were treated with iodine solution, which allowed to observe unstained zone around active amylase colonies (Hankin and Anagnostakis 1975).
Lipase activity. Lipase activity was determined according to the method described by Hankin and Anagnostakis (1975) with minor modifications. The strains were inoculated onto the medium containing (g/l): Nutrient Broth 8 g, CaCl2 H2O 0.1 g, agar 15 g, pH 6.0, and 20 ml Tween 20. Tween 20 was separately added into the medium after sterilization. Cultures were incubated at 25°C for two or three days and the plates were kept at +4°C for 30 min. Variants showing opaque zone around colonies were evaluated as lipase positive.
Cellulase activity. Cellulase activity was determined by the method reported by Amore et al. (2015) with some modifications. The isolates were inoculated onto the medium containing (g/l): NaNO3 1 g, K2HPO4 1 g, KCl 1 g, MgSO4 0.5 g, yeast extract 0.5 g, glucose 1 g, carboxymethylcellulose (CMC) 5 g, and agar 15 g. The plates were incubated at 25°C for 5–8 days. At the end of the incubation, 0.2% (w/v) Congo Red solution was added to Petri dishes and kept at ambient temperature for 20 min. Then the Petri dishes were washed by adding 5 M NaCl solution to remove excess dye and kept at room temperature for another 30 min. Colonies with a light-yellow zone around the colony on a red background were evaluated as cellulase positive.
Protease activity. Protease activity was studied with modified method of Carrim et al. (2006). Nutrient Agar containing 1% (g/l) skimmed milk powder was used to prepare a protease substrate. Milk powder (10 g/100 ml) was sterilized at 110°C for 5 minutes, cooled to 45°C, and added to a basal medium in aseptic conditions. Strains inoculated onto the above medium were kept for two or three days at 25°C. A transparent zone formation around the colonies indicated a protease activity.
Pectinase activity. Pectinase activity was determined according to the method of Kobayashi et al. (1999). The isolates were inoculated onto the medium containing (g/l): yeast extract 2 g, ammonium sulfate 2 g, Na2HPO4 6 g, KH2PO4 3 g, pectin 5 g, and agar 15 g. The plates were incubated at 25°C for three days. At the end of incubation, after adding 1% (w/v) cetyltrimethylammonium bromide (CTAB) solution, the Petri dishes were kept at room temperature for 10 min. Transparent zone formation around the colony indicated a pectinase activity.
Xylanase activity. Xylanase activity was studied with a modified method of Amore et al. (2015). The isolates were inoculated onto the medium containing (g/l): NaNO3 1 g, K2HPO4 1 g, KCl 1 g, MgSO4 0.5 g, yeast extract 0.5 g, glucose 1 g, agar 15 g, and xylan 5 g. After the isolates were inoculated onto the medium, they were incubated at 25°C for two or four days. At the end of the incubation, 0.1% (w/v) Congo Red solution was poured into the Petri dish and staining was performed for 20 min. To remove the excessive dye, 5 M NaCl solution was added to the Petri dishes and kept at room temperature for 30 min. A light-colored zone on a red background indicated a xylanase activity.
Genotypic characterization of the selected isolates. Based on enzyme activities determined using solid selective media, 16 isolates were selected for diagnosis processes, giving successful and different EI values. The selected strains were identified by the 16S rRNA gene amplicon sequencing. DNA isolation was performed by the modified method of Govindarajan et al. (2007), and the 16S rRNA gene was amplified by polymerase chain reaction (PCR) using the genomic DNA as a template and universal bacterial primers, 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-TACGGTTACCTTGTTACGACTT-3’) (Frank et al. 2008). A 50 µl reaction mixture contained 2.5 U Taq polymerase (Thermo Fisher Scientific, Waltham, MA, USA), 0.3 mM dNTPs, 25 mM MgCl2, 20 pmol of each primer, 5 µl of 10 x reaction buffer (Thermo Fisher Scientific), and 20 ng of template DNA. The step-up PCR procedure included denaturation at 95°C for 5 min, followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min, with a final extension at 72°C for 10 min. Amplification products were electrophoresed on a 1.5% agarose gel in 1 × TBE buffer.
The 16S rRNA gene amplicon sequencing was performed by the Sentebiolab Biotechnology Company (Turkey) using the Miseq (Illumina) next-generation sequencing platform. The sequences obtained were analyzed using the database on the website (https://www.ezbiocloud.net), and then the sequences were logged in to the GenBank site and accessed “GenBank accession” numbers (Table II). The phylogenetic tree was created by the GGDC web server at http://ggdc.dsmz.de using the phylogenomic data line DSMZ (German Collection of Microorganisms and Cell Cultures GmbH) adapted to single genes (Meier-Kolthoff et al. 2013). Multiple sequence alignment was done with the “MUSCLE” (Edgar 2004), and the phylogenetic tree was created using the Maximum Likelihood method (Stamatakis 2014).
Table II.
Identification of strains according to the results of sequence analysis using the EzBioCloud database and GenBank accession numbers.
| Code of the isolates | Top-hit reference species | Top-hit reference strain | Similarity (%) |
Coverage (%) |
GenBank Accession Numbers |
|---|---|---|---|---|---|
| G119Y1T | Bacillus toyonensis | BCT-7112 | 100.00 | 70.10 | MW752891 |
| G118S2T | Pseudomonas congelans | DSM 14939 | 100.00 | 100.0 | MW752990 |
| G117Y1T | Streptococcus thermophilus | ATCC 19258 | 94.58 | 89.30 | MW774413 |
| G116K1T | Micrococcus luteus | NCTC 2665 | 99.58 | 100.00 | MW755305 |
| G113Y3 | Bacillus halotolerans | ATCC 25096 | 99.93 | 100.00 | MW753050 |
| G107Y2 | Curtobacterium flaccumfaciens | LMG 3645 | 100.00 | 100.00 | MW753051 |
| G105Y1 | Bacillus subtilis subsp. inaquosorum | KCTC 13429 | 99.92 | 84.50 | MW753052 |
| G105S1 | Bacillus idriensis | SMC 4352-2 | 99.58 | 100.00 | MW753132 |
| G100Y1 | Paenibacillus nuruki | TI45-13ar | 99.25 | 100.00 | MW753131 |
| G90Y2 | Paenibacillus tundrae | A10b | 99.84 | 83.30 | MW753134 |
| G90S1 | Curtobacterium flaccumfaciens | LMG 3645 | 100.00 | 100.00 | MW757038 |
| G88K1 | Pseudomonas orientalis | CFML 96-170 | 99.62 | 89.20 | MW753212 |
| G83S3 | Paenibacillus seodonensis | DCT19 | 99.23 | 88.00 | MW753225 |
| G80K3 | Paenibacillus xylanexedens | B22a | 99.80 | 100.00 | MW753226 |
| G70K2 | Paenibacillus xylanexedens | B22a | 99.80 | 100.00 | MW753223 |
| G42K2 | Chryseobacterium luteum | DSM 18605 | 99.44 | 100.00 | MW753224 |
Statistical analysis. All enzyme measurement experiments were performed in four replicates, and each measurement on Petri dishes was repeated twice. The Statistical Analysis System (SAS version 9.4 SAS, Cary, NC) was used to analyze the data. General linear model (GLM) analysis was used to determine differences between the averages of the groups, and Duncan’s multiple comparison test was used to determine differences between the groups. P values < 0.05 were considered statistically different.
Results and Discussion
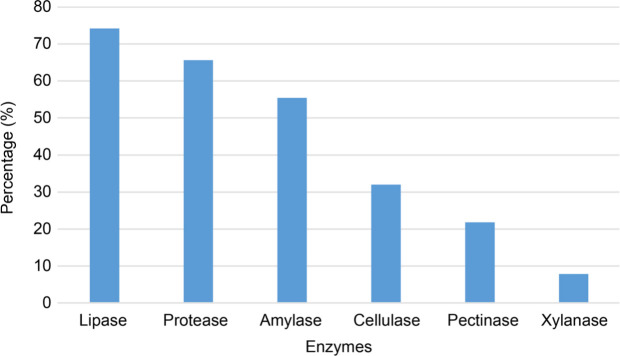
In this study, a total of 128 endophyte bacteria isolated from various cultivated and wild grain plants (Poaceae family) were used. For all the isolates, the EI of each enzyme activity is given in Table SI. Since endophytic bacteria offer a relatively new source of genes, enzymes, and secondary metabolites, we aimed to investigate several biotechnologically important extracellular enzymes of endophytic origin. By this purpose, endophytic bacteria isolated from Van province, Turkey, were evaluated for the presence of hydrolytic enzymes, including cellulases, xylanases, pectinases, amylases, proteases, and lipases (Fig. 1). They successfully demonstrated a variety of enzyme activities. It was revealed that lipases, proteases, amylases, cellulases, pectinases, and xylanases were produced with relative frequencies of 74.2%, 65.6% and 55.4%, 32%, 21.8%, and 7.8%, respectively (Fig. 2).
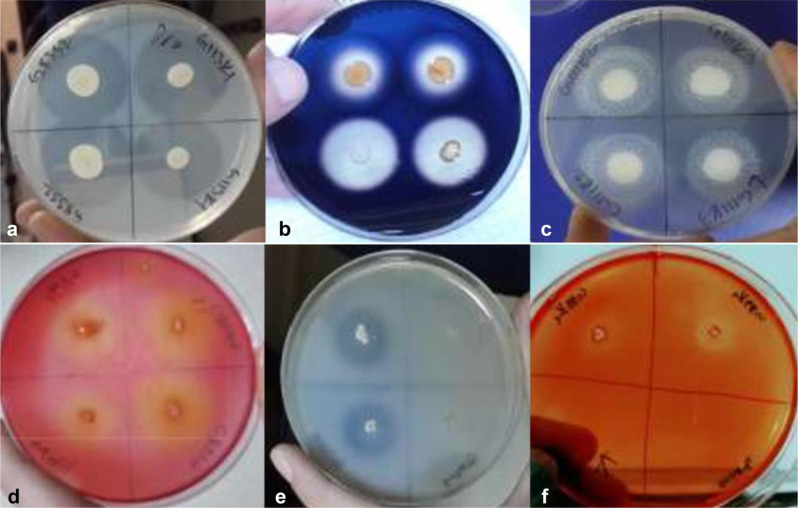
Fig. 1.
Petri dishes with colonies surrounded by zones of extracellular enzymatic activity; a) protease, b) amylase, c) lipase, d) cellulase, e) pectinase, f) xylanase.
Fig. 2.
Relative frequency (%) of strains (from a total of 128) producing individual hydrolytic enzymes.
After the enzyme activity measurements were completed, 16 isolates revealing relatively high EI value for at least one enzyme tested were selected to perform a phylogenetic affiliation based on the 16S rRNA gene amplicon sequencing analysis. Also, among these selected strains, one producing none of the enzymes was selected for the identification (Table I).
The 16S rRNA gene amplicon sequencing of 16 isolates was successfully achieved. The ~ 1,500 bp 16S rRNA gene contains nine variable regions (V1–V9) in a highly conserved order. Since next-generation sequencing platforms provide an appropriate read of full-length the 16S rRNA gene intragenomic variants, they provide a better taxonomic resolution at species or strain level (Johnson et al. 2019). Illumina MiSeq method yielded full-length reading of the 16S rRNA gene amplicons for almost all strains. The lowest 16S rRNA gene reading length belongs to the strain G119Y1T with 70.1%, which nevertheless covers the V1‒V5 regions (Johnson et al. 2019) (Table II). As a result of pairwise comparisons of the 16S rRNA gene sequences on EzBioCloud server, five Paenibacillus sp. (G100Y1, G90Y2, G83S3, G80K3, G70K2), four Bacillus sp. (G119Y1T, G105S1, G113Y3, G105Y1), two Pseudomonas sp. (G88K1, G118S2T), two Curtobacterium sp. (G107Y2, G90S1), one Micrococcus sp. (G116K1T), one Streptococcus sp. (G117Y1T), one Chryseobacterium sp. (G42K2) were identified (Table II).
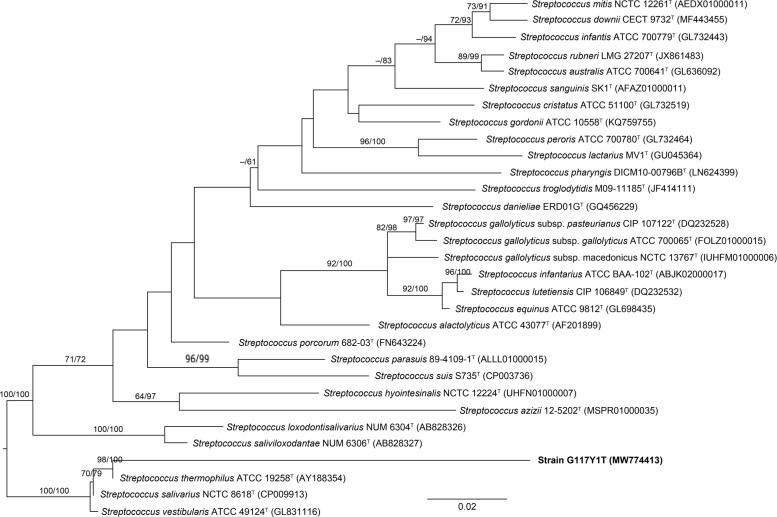
Except for strain G117Y1T, the 16S rRNA gene amplicon sequencing results of all strains yielded 99‒100% similarity (Table II). The 16S rRNA gene sequences alone may not be sufficient to identify a new species, but it can indicate that a new species is isolated (Tindall et al. 2010). The 94.58% similarity with G117Y1T is far below the threshold necessary to identify a new species (Stackebrandt and Goebel 1994; Stackebrandt and Ebers 2006), and, thus, this strain may represent a new species or even genus (Fig. 3). Noteworthy, strain G117Y1T gave positive results in terms of all enzymes except pectinase (Table I).
Fig. 3.
The phylogenetic tree constructed by the maximum likelihood method and rooted by midpoint-rooting. Branches are scaled in terms of the expected number of nucleotide substitutions per site. Numbers at nodes indicate the bootstrap support values larger than 60% from maximum likelihood (left) and maximum parsimony (right) bootstrapping. GenBank accession numbers are given in brackets.
Different studies in the literature show that our identified strains belonging to seven different genera were endophytes (Verma et al. 2001; Rashid et al. 2012; Khan et al. 2017; Afzal et al. 2019). The different species of these genera produce high-potential bioactive compounds such as antimicrobials and enzymes to be used in the fields such as medicine and bioremediation, especially in agriculture (Doddamani and Ninnekar 2001; Schallmey et al. 2004; Lacava et al. 2007; Grady et al. 2016; Roy et al. 2018). Although the number of strains that we identified molecularly comprise a small cluster within all 128 isolates, they could reveal the diversity and support the literature data.
Carrim et al. (2006) presented the enzymatic activity of endophytic bacteria ranking as follows: protease (60%), amylase (60%), and lipase (40%). They did not detect cellulase and pectinase activities. Jalgaonwala and Mahajan (2011) detected 50% cellulase-positive endophytic bacteria in their study. On the other hand, our results revealed a high number of bacterial isolates with cellulase, lipase, and protease activities. Also, we have found a significant number of pectinase-positive isolates (Fig. 2). Despite the relatively limited number of studies, the percentage of endophyte bacteria with the positive scores for each of these enzymes varied due to the high species diversity.
Among the identified strains, Bacillus spp. (B. toyonensis, B. halotolerans, B. subtilis subsp. inaquosorum) except B. idriensis showed especially high cellulase activity among six tested enzymes (Tables I and II). The strain G105S, which, in contrast to other strains, did not produce the above enzymes, was closely related to B. idriensis (99.58%) (Table II). However, B. idriensis that possessed protease, cellulase, and pectinase activities, was isolated as an endophyte in the study conducted by Afzal et al. (2017). Pseudomonas spp. (P. congelans, P. orientalis) were the main xylanase producers among identified strains. In general, the number of strains demonstrating xylanase activity was relatively low. For this reason, these strains belonging to the genus Pseudomonas are valuable as xylanase enzyme producers. Xylanases produced by bacteria (Bacillus spp., Pseudomonas spp., Streptomyces spp.) are efficient in a broad pH and temperature range. Therefore, they are very useful in different industries reciprocally (Burlacu et al. 2016). Among the isolates we described, Paenibacillus spp. was observed as the most productive group of lipases and cellulases. Paenbacillus species are known to produce different hydrolytic enzymes (Sakiyama et al. 2001; El-Deeb et al. 2013). Cho et al. (2008) isolated two cellulose hydrolase genes (cel5A and cel5B) from endophytic Paenibacillus polymyxa. The strain belonging to Streptococcus, Micrococcus, Curtobacterium, and Chryseobacterium showed high activity of proteases, lipases, and xylanases. Generally, in this study, Gram-positive bacteria displayed broader hydrolytic enzyme potential than Gram-negative bacteria. Published data revealed that endophyte diversity varies according to different territories, plants, and even different plant tissues (Akinsanya et al. 2015).
Although this study was carried out in line with the biotechnological perspective, extracellular enzymes should also be evaluated and discussed in terms of the relationship between endophyte bacteria and the plant hosts. For example, different levels of cellulases and pectinases were reported to be important in endophytic diazotrophic bacteria during plant cells colonization (Verma et al. 2001). Considering that the plant pathogen bacteria also synthesize the enzymes that break down the cell wall, more information about the expression and regulation of these enzymes in both groups could be crucial to understand and distinguish between these two groups of bacteria.
In this study, a potentiality of endophytic bacteria isolated from several grain plants (Poaceae family) in Van province, Turkey, to produce biotechnologically important enzymes, was revealed for the first time. Endophyte bacteria are rich sources of enzymes and new secondary metabolites for many industries due to their high species diversity and adaptation to different environments. Therefore, investigation of these isolates not only in terms of extracellular enzymes but also in terms of specific and industrially important secondary metabolites should be among the future.
Supplementary Material
Acknowledgments
This work was supported by the Scientific Research Project Units of Van Yuzuncu Yil University, Turkey (Project number: FYL-2018-7557). Also we sincerely thank Assist. Prof. Dr. Ahmet Akkopru, Van Yuzuncu Yil University, Turkey for his support.
Footnotes
Conflict of interest
The authors do not report any financial or personal connections with other persons or organizations, which might negatively affect the contents of this publication and/or claim authorship rights to this publication.
Literature
- Afzal I, Iqrar I, Shinwari ZK, Yasmin A. Plant growth-promoting potential of endophytic bacteria isolated from roots of wild Dodonaea viscosa L. Plant Growth Regul. 2017;81(3):399–408. 10.1007/s10725-016-0216-5 [DOI] [Google Scholar]
- Afzal I, Shinwari ZK, Sikandar S, Shahzad S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol Res. 2019April;221:36–49. 10.1016/j.micres.2019.02.001 [DOI] [PubMed] [Google Scholar]
- Akinsanya MA, Goh JK, Lim SP, Ting AS. Diversity, antimicrobial and antioxidant activities of culturable bacterial endophyte communities in Aloe vera. FEMS Microbiol Lett. 2015December;362(23):fnv184. 10.1093/femsle/fnv184 [DOI] [PubMed] [Google Scholar]
- Amore A, Parameswaran B, Kumar R, Birolo L, Vinciguerra R, Marcolongo L, Ionata E, La Cara F, Pandey A, Faraco V. Application of a new xylanase activity from Bacillus amyloliquefaciens XR44A in brewer’s spent grain saccharification. J Chem Technol Biotechnol. 2015March;90(3):573–581. 10.1002/jctb.4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlacu A, Cornea CP, Israel-Roming F. Screening of xylanase producing microorganisms. Res J Agric Sci. 2016;48(2):8–15. [Google Scholar]
- Carrim AJI, Barbosa E, Vieira JDG. Enzymatic activity of endophytic bacterial isolates of Jacaranda decurrens Cham (Carobinhado-campo). Braz Arch Biol Technol. 2006May;49:353–359. 10.1590/S1516-89132006000400001 [DOI] [Google Scholar]
- Cho KM, Hong SJ, Math RK, Islam SM, Kim JO, Lee YH, Kim H, Yun HD. Cloning of two cellulase genes from endophytic Paenibacillus polymyxa GS01 and comparison with cel44C-man 26A. J Basic Microbiol. 2008December;48(6):464–472. 10.1002/jobm.200700281 [DOI] [PubMed] [Google Scholar]
- Compant S, Cambon MC, Vacher C, Mitter B, Samad A, Sessitsch A. The plant endosphere world – bacterial life within plants. Environ Microbiol. 2021April;23(4):1812–1829. 10.1111/1462-2920.15240 [DOI] [PubMed] [Google Scholar]
- Doddamani HP, Ninnekar HZ. Biodegradation of carbaryl by a Micrococcus species. Curr Microbiol. 2001July;43(1):69–73. 10.1007/s002840010262 [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004March19;32(5): 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Deeb B, Fayez K, Gherbawy Y. Isolation and characterization of endophytic bacteria from Plectranthus tenuiflorus medicinal plant in Saudi Arabia desert and their antimicrobial activities. J Plant Interact. 2013;8(1):56‒64. 10.1080/17429145.2012.680077 [DOI] [Google Scholar]
- Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008April;74(8):2461–2470. 10.1128/AEM.02272-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan M, Kwon SW, Weon HY. Isolation, molecular characterization and growth-promoting activities of endophytic sugarcane diazotroph Klebsiella sp. GR9. World J Microbiol Biotechnol. 2007July;23:997–1006. 10.1007/s11274-006-9326-y [DOI] [Google Scholar]
- Grady EN, MacDonald J, Liu L, Richman A, Yuan ZC. Current knowledge and perspectives of Paenibacillus: a review. Microb Cell Fact. 2016December1;15(1):203. 10.1186/s12934-016-0603-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankin L, Anagnostakis SL. The use of solid media for detection of enzyme production by fungi. Mycologia. 1975;67(3):597–607. 10.2307/3758395 [DOI] [Google Scholar]
- Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015September;79(3):293–320. 10.1128/MMBR.00050-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalgaonwala RE, Mahajan RT. Evaluation of hydrolytic enzyme activities of endophytes from some indigenous medicinal plants. Int J Agr Sci Tech. 2011;7(6):1733–1741. [Google Scholar]
- Jena SK, Chandi CR. Optimization of culture conditions of phosphate solubilizing activity of bacterial sp. isolated from Similipal biosphere reserve in solid-state cultivation by response surface methodology. Int J Curr Microbiol App Sci. 2013;2(5):47–59. [Google Scholar]
- Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta HO, Gerstein M, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019November6;10:5029. 10.1038/s41467-019-13036-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel SL, Joubert PM, Doty SL. Bacterial endophyte colonization and distribution within plants. Microorganisms. 2017November25; 5(4):77. 10.3390/microorganisms5040077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan LA, Shahzad R, Al-Harrasi A, Lee JI. Endophytic microbes: A resource for producing extracellular enzymes. In: Maheshwari D, Annapurna K, editors. Endophytes: Crop productivity and protection. Sustainable Development and Biodiversity, vol. 16. Cham (Switzerland): Springer International Publishing; 2017. p. 95–110. 10.1007/978-3-319-66544-3_5 [DOI] [Google Scholar]
- Kobayashi T, Koike K, Yoshimatsu T, Higaki N, Suzumatsu A, Ozawa T, Hatada Y, Ito S. Purification and properties of a low-molecular-weight, high-alkaline pectate lyase from an alkaliphilic strain of Bacillus. Biosci Biotechnol Biochem. 1999January;63(1):65–72. 10.1271/bbb.63.65 [DOI] [PubMed] [Google Scholar]
- Lacava PT, Li W, Araújo WL, Azevedo JL, Hartung JS. The endophyte Curtobacterium flaccumfaciens reduces symptoms caused by Xylella fastidiosa in Catharanthus roseus. J Microbiol. 2007October; 45(5):388–393. [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013February21;14:60. 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaktan H, Çakır B, Gül A, Yolageldi L, Akköprü A, Fakhraei D, Akbaba M. Isolation and evaluation of endophytic bacteria against Fusarium oxysporum f. sp. cucumerinum infecting cucumber plants. Austin J Plant Biol. 2015;1(1):1003. [Google Scholar]
- Rashid S, Charles TC, Glick BR. Isolation and characterization of new plant growth promoting bacterial endophytes. Appl Soil Ecol. 2012October;61:217–224. 10.1016/j.apsoil.2011.09.011 [DOI] [Google Scholar]
- Roy K, Dey S, Uddin MK, Barua R, Hossain MT. Extracellular pectinase from a novel bacterium Chryseobacterium indologenes strain SD and its application in fruit juice clarification. Enzyme Res. 2018March21;2018:3859752. 10.1155/2018/3859752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakiyama CC, Paula EM, Pereira PC, Borges AC, Silva DO. Characterization of pectin lyase produced by an endophytic strain isolated from coffee cherries. Lett Appl Microbiol. 2001August;33(2): 17–1121. 10.1046/j.1472-765x.2001.00961.x [DOI] [PubMed] [Google Scholar]
- Schallmey M, Singh A, Ward OP. Developments in the use of Bacillus species for industrial production. Can J Microbiol. 2004January; 50(1):1–17. 10.1139/w03-076 [DOI] [PubMed] [Google Scholar]
- Singh M, Kumar A, Singh R, Pandey KD. Endophytic bacteria: a new source of bioactive compounds. 3 Biotech. 2017October;7(5):315. 10.1007/s13205-017-0942-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kumar M, Mittal A, Mehta PK. Microbial enzymes: industrial progress in 21st century. 3 Biotech. 2016December;6(2):174. 10.1007/s13205-016-0485-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Micr. 1994October; 44(4):846–849. 10.1099/00207713-44-4-846 [DOI] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014May1;30(9):1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall BJ, Rosselló-Móra R, Busse HJ, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010January;60(Pt 1):249–266. 10.1099/ijs.0.016949-0 [DOI] [PubMed] [Google Scholar]
- Verma SC, Ladha JK, Tripathi AK. Evaluation of plant growth promoting and colonization ability of endophytic diazotrophs from deep water rice. J Biotechnol. 2001October4;91(2–3):127–141. 10.1016/S0168-1656(01)00333-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.