Abstract
This study investigates the timeframe that optimizes saliva sensitivity for SARS-CoV-2 detection using reverse transcriptase–polymerase chain reaction (RT-PCR) testing.
While real-time reverse transcriptase–polymerase chain reaction (RT-PCR) on nasopharyngeal swabs is the current standard for SARS-CoV-2 detection, saliva is an attractive alternative for diagnosis and screening due to ease of collection and minimal supply requirements.1,2 Studies on the sensitivity of saliva-based SARS-CoV-2 molecular testing have shown considerable variability.3 We conducted a prospective, longitudinal study to investigate the testing time frame that optimizes saliva sensitivity for SARS-CoV-2 detection.
Methods
Between June 17, 2020, and February 15, 2021, a convenience sample of individuals exposed to a household member with RT-PCR–confirmed SARS-CoV-2 within 2 weeks were recruited from Children’s Hospital Los Angeles and nearby community testing sites into the Household Exposure and Respiratory Virus Transmission and Immunity Study (HEARTS). Paired nasopharyngeal and saliva samples were collected every 3 to 7 days for up to 4 weeks or until 2 negative nasopharyngeal test results. RT-PCR for SARS-CoV-2 N1 and N2 genes was performed; cycle threshold less than 40 defined a positive result. A nasopharyngeal N1 cycle threshold of 34 or less was defined as high viral load.4 Detailed specimen collection and RT-PCR methods are reported in the eMethods in the Supplement.
Saliva sensitivity was calculated using nasopharyngeal-positive RT-PCR as the reference standard. COVID-19 onset was defined as the earlier date between first symptom (collected by questionnaire daily) or first RT-PCR positivity. Pre- and postsymptomatic were defined as asymptomatic time points before and after a symptomatic interval, respectively. Saliva sensitivity by week of collection and between symptomatic and asymptomatic individuals were compared using the χ2 test or the Fisher exact test. Generalized estimating equations were used to determine clinical characteristics (Table) associated with saliva sensitivity in nasopharyngeal-positive pairs while accounting for repeated samples from the same individuals. Analyses were performed using SPSS version 27.0 (IBM Corp) with a 2-sided P < .05 considered significant. Written informed consent was obtained from participants. The study was approved by the institutional review board of Children’s Hospital Los Angeles.
Table. Characteristics Predicting Higher Odds of Saliva RT-PCR Positivity at COVID-19–Positive Time Pointsa.
| Participant characteristics (n = 256) | No. (%) | Odds ratio (95% CI) | P value |
|---|---|---|---|
| Sex | |||
| Female | 148 (57.8) | 1 [Reference] | .02 |
| Male | 108 (42.2) | 1.78 (1.09-2.91) | |
| Age | |||
| Adult (≥18 y) | 181 (70.7) | 1 [Reference] | .60 |
| Child (<18 y) | 75 (29.3) | 1.16 (0.66-2.04) | |
| Ethnicityb | |||
| Hispanic/Latinx | 239 (93.4) | 0.19 (0.03-1.48) | .11 |
| Non-Hispanic/Latinx | 17 (6.6) | 1 [Reference] | |
| Raceb | |||
| Asian | 9 (3.5) | 0.33 (0.04-3.09) | .33 |
| Black | 0 | ||
| White | 245 (95.7) | 1 [Reference] | |
| Multiple | 2 (0.8) | 0.10 (0.02-0.62) | .01 |
| Comorbiditiesc | |||
| No | 210 (82.0) | 1 [Reference] | .26 |
| Yes | 46 (18.0) | 1.49 (0.75-2.94) | |
| Smokerd | |||
| No | 243 (94.9) | 1 [Reference] | .005 |
| Yes | 13 (5.1) | 3.18 (1.43-7.09) | |
| Characteristic at time of sample collection (n = 524) | |||
| COVID-19–associated symptom presentatione | |||
| No | 402 (76.7) | 1 [Reference] | <.001 |
| Yes | 122 (23.3) | 2.84 (1.58-5.11) | |
| Nasopharyngeal swab viral loadf | |||
| Low | 131 (25.0) | 1 [Reference] | <.001 |
| High | 393 (75.0) | 5.16 (2.87-9.28) | |
| Sample collection timing | |||
| Days since COVID-19 onset at time of specimen collection | 524 (100) | 0.94 (0.91-0.96)g | <.001 |
Abbreviation: RT-PCR, reverse transcriptase–polymerase chain reaction.
From all nasopharyngeal-positive paired samples (n = 524), generalized estimating equations analysis (goodness-of-fit quasilikelihood information criterion, 570.9) were used to determine different likelihoods of saliva SARS-CoV-2 PCR positivity. The odds ratio of having a positive RT-PCR result in saliva while holding all other variables constant is shown.
Race and ethnicity were self-reported by the participants with the groups provided. Participants who identified with more than 1 race are reported in the “multiple” category.
Comorbid conditions included preexisting lung, heart, kidney, liver, or neurologic disease; diabetes; cancer; or other immunosuppression.
Smoking status refers to self-reports of current use of tobacco, marijuana, or vaping products.
Participants were considered symptomatic for COVID-19 if they reported at least 1 of the following: fever, chills, headache, fatigue, muscle aches, runny nose, congestion, cough, sore throat, shortness of breath, wheeze, altered smell, altered taste, vomiting, diarrhea, or abdominal pain.
A high nasopharyngeal swab viral load was defined as cycle threshold ≤34 and a low viral load as cycle threshold >34 in the SARS-CoV-2 N1 gene.4
For each day after COVID-19 onset, the odds of saliva RT-PCR positivity decreased by a factor of 0.94.
Results
We tested 889 paired nasopharyngeal swab-saliva samples from 404 participants, of which SARS-CoV-2 was detected in 524 nasopharyngeal (58.9%) and 318 saliva (35.7%) specimens. SARS-CoV-2 was detected in both specimens in 258 pairs (29.0%). Of the 256 nasopharyngeal SARS-CoV-2–positive participants (63.4%), the mean age was 28.2 years (range, 3.0-84.5 years); 108 (42.2%) were male. Participants returned for a median of 3 visits (interquartile range, 2-4). Ninety-three participants (36.3%) were asymptomatic throughout their infection; 126 (77.3%) of 163 symptomatic individuals reported mild severity.
Saliva sensitivity was highest in samples collected during the first week of infection at 71.2% (95% CI, 62.6%-78.8%) but decreased each subsequent week (Figure, A). Participants who presented with COVID-19–associated symptoms on the specimen collection day during week 1 of infection had significantly higher saliva sensitivity compared with asymptomatic participants (88.2% [95% CI, 77.6%-95.1%] vs 58.2% [95% CI, 46.3%-69.5%]; P < .001). Saliva sensitivity remained significantly higher in symptomatic participants in week 2 (83.0% [95% CI, 70.6%-91.8%] vs 52.6% [95% CI, 42.6%-62.5%]; P < .001). No difference was observed more than 2 weeks after COVID-19 onset (Figure, B). Sensitivities did not significantly differ for never-symptomatic (34.7% [95% CI, 27.3%-42.7%]), presymptomatic (57.1% [95% CI, 31.7%-80.2%]), and postsymptomatic (42.9% [95% CI, 36.8%-49.1%]) time points (P = .26).
Figure. Saliva Sensitivity by Collection Timing After COVID-19 Onset Overall and in Symptomatic and Asymptomatic Individuals.
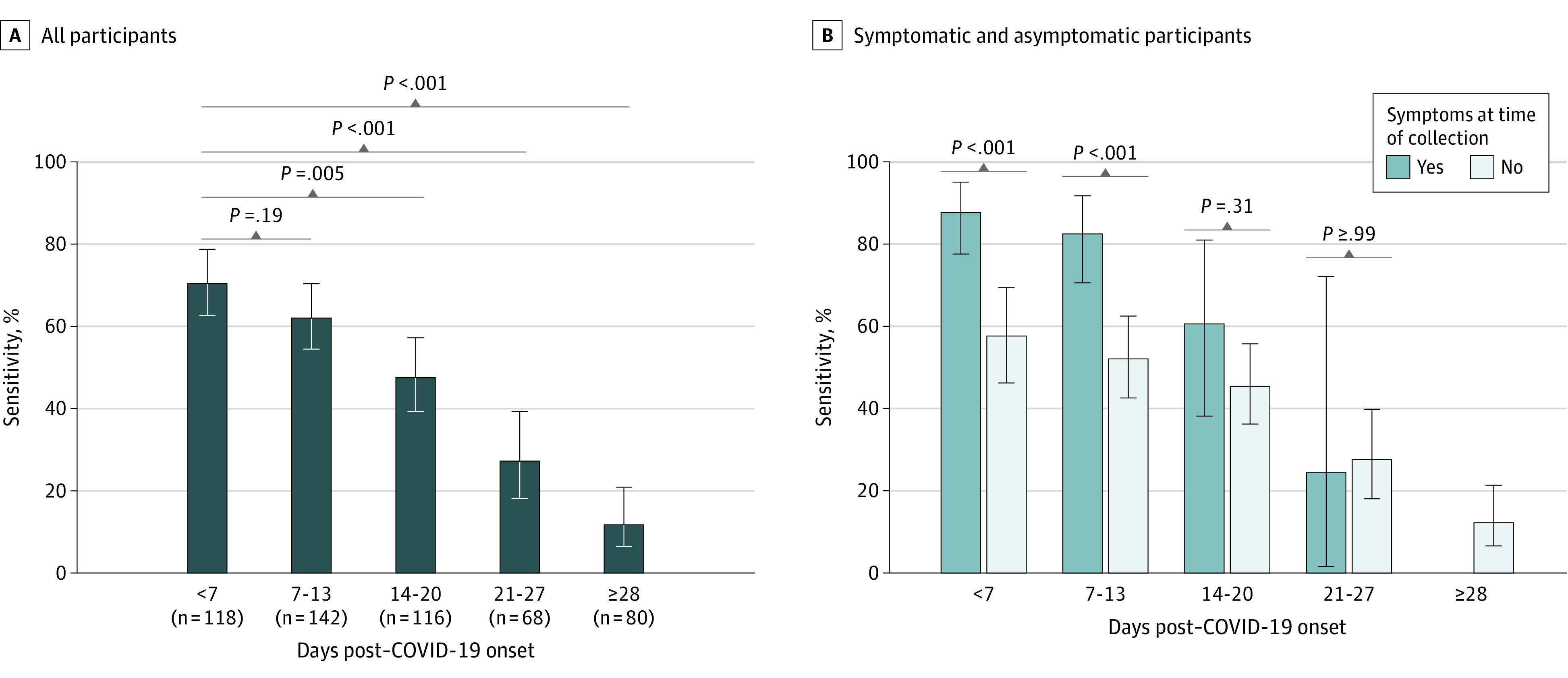
Saliva sensitivity in all 524 nasopharyngeal-positive paired samples from 256 participants (A) and participants who were symptomatic vs asymptomatic at time of specimen collection (B) grouped by collection timing after COVID-19 onset, defined as the earliest of either first symptom or first reverse transcriptase–polymerase chain reaction positivity. Error bars indicate 95% CIs.
For each day after COVID-19 onset, the odds ratio for saliva detection was 0.94 (95% CI, 0.91-0.96) compared with the previous day (P < .001) (Table). Participants presenting with COVID-19–associated symptoms at the time of specimen collection or with high nasopharyngeal viral loads had 2.8 (95% CI, 1.6-5.1; P < .001) and 5.2 (95% CI, 2.9-9.3; P < .001) higher odds of having a saliva-positive RT-PCR result compared with those with asymptomatic presentation or low nasopharyngeal viral loads, respectively.
Discussion
Saliva was sensitive for detecting SARS-CoV-2 in symptomatic individuals during initial weeks of infection, but sensitivity in asymptomatic SARS-CoV-2 carriers was less than 60% at all time points. As COVID-19 testing strategies in workplaces, schools, and other shared spaces are optimized, low saliva sensitivity in asymptomatic infections must be considered.5 This study suggests saliva-based RT-PCR should not be used for asymptomatic COVID-19 screening.
This study has limitations. Samples were collected following household exposure; therefore, pretest probability was high. Nasopharyngeal swab testing was the reference standard, but this is not a perfect test for SARS-CoV-2 infection, and a positive RT-PCR result from any sample past 10 days of infection may not be predictive of viral replication or infectivity.6
Section Editors: Jody W. Zylke, MD, Deputy Editor; Kristin Walter, MD, Associate Editor.
eMethods
References
- 1.Centers for Disease Control and Prevention . Interim guidelines for collecting and handling of clinical specimens for COVID-19 testing. Accessed February 23, 2021. https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html
- 2.Moreno-Contreras J, Espinoza MA, Sandoval-Jaime C, et al. Saliva sampling and its direct lysis, an excellent option to increase the number of SARS-CoV-2 diagnostic tests in settings with supply shortages. J Clin Microbiol. 2020;58(10):e01659-e20. doi: 10.1128/JCM.01659-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riccò M, Ranzieri S, Peruzzi S, et al. RT-qPCR assays based on saliva rather than on nasopharyngeal swabs are possible but should be interpreted with caution: results from a systematic review and meta-analysis. Acta Biomed. 2020;91(3):e2020025. doi: 10.23750/abm.v91i3.10020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaafar R, Aherfi S, Wurtz N, et al. Correlation between 3790 quantitative polymerase chain reaction-positives samples and positive cell cultures including 1941 severe acute respiratory syndrome coronavirus 2 isolates. Clin Infect Dis. 2021;72(11):e921. doi: 10.1093/cid/ciaa1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nacher M, Mergeay-Fabre M, Blanchet D, et al. Prospective comparison of saliva and nasopharyngeal swab sampling for mass screening for COVID-19. Front Med (Lausanne). 2021;8:621160. doi: 10.3389/fmed.2021.621160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owusu D, Pomeroy MA, Lewis NM, et al. ; Household Transmission Study Team . Persistent SARS-CoV-2 RNA shedding without evidence of infectiousness: a cohort study of individuals with COVID-19. J Infect Dis. Published online February 27, 2021. doi: 10.1093/infdis/jiab107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods


