Abstract
We have mapped the 5′ and 3′ boundaries of the region of the human telomerase RNA (hTR) that is required to produce activity with the human protein catalytic subunit (hTERT) by using in vitro assembly systems derived from rabbit reticulocyte lysates and human cell extracts. The region spanning nucleotides +33 to +325 of the 451-base hTR is the minimal sequence required to produce levels of telomerase activity that are comparable with that made with full-length hTR. Our results suggest that the sequence approximately 270 bases downstream of the template is required for efficient assembly of active telomerase in vitro; this sequence encompasses a substantially larger portion of the 3′ end of hTR than previously thought necessary. In addition, we identified two fragments of hTR (nucleotides +33 to +147 and +164 to +325) that cannot produce telomerase activity when combined separately with hTERT but can function together to assemble active telomerase. These results suggest that the minimal sequence of hTR can be divided into two sections, both of which are required for de novo assembly of active telomerase in vitro.
Telomerase is a reverse transcriptase that maintains telomeres by adding a G-rich repeat (TTAGGG for vertebrates) to the 3′ single-stranded overhang at the ends of chromosomes (16, 38, 39). Human telomerase requires at least two components for the synthesis of telomeric DNA: a protein catalytic subunit (hTERT) and an integral RNA template (hTR) (4, 13, 22, 26, 35, 40, 42, 56). The limiting component for producing active telomerase in normal cells is hTERT (10, 22, 42, 56). This protein contains reverse transcriptase motifs that are essential for enzymatic activity (4, 22, 42, 56) and are conserved among diverse organisms such as Saccharomyces cerevisiae (Est2) (11), Saccharomyces pombe (Sp_Trt1p) (40), Euplotes aediculatus (Ea_p123) (31), Tetrahymena (Tt_TERT or p133) (7, 8), Oxytricha trifallax (Ot_TERT) (7), and Mus musculus (mTERT) (15, 32).
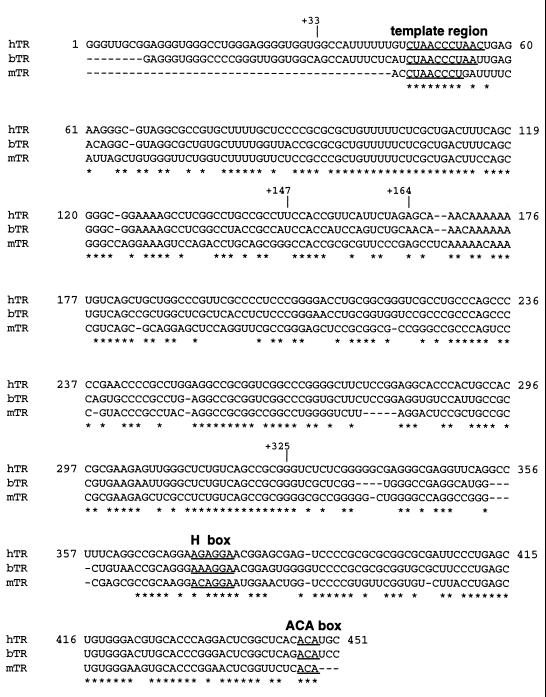
In contrast, there is little primary sequence conservation across species for the integral RNA component of telomerase. Furthermore, there is a tremendous size variation for these RNAs among divergent organisms: the ciliate RNAs are uniformly small (between 148 to 209 nucleotides [nt]) (17, 30, 33, 48, 51), while those from two yeast strains are an order of magnitude larger (1.3 kb for both S. cerevisiae [52] and Kluyveromyces lactis [34]). In the cow (55), mouse (6), and human (13), intermediate sizes (approximately 400 to 450 nt) are found (Fig. 1). While little is known about the secondary structure of telomerase RNA in higher eukaryotes, a conserved structure is present in an evolutionarily divergent group of ciliate telomerase RNAs, as deduced by biochemical probing and phylogenetic comparative analysis (2, 5, 30, 33, 48, 53). In brief, most ciliate RNAs fold to form four helices (I to IV) that are conserved not only in size but also in relative spacing. Helices I, II, and III extend from a central unpaired region that contains the template (between helices II and III); helix IV is an extension of helix I. The overall topology of the ciliate RNAs is dictated by helix I, which closes the unpaired region. One key feature of the ciliate RNAs is the presence of a conserved sequence, 5′-(CU)GUCA-3′, that is critical in establishing the 5′ template boundary (1, 2, 30, 33).
FIG. 1.
Sequence comparison of human, bovine, and mouse template RNA (hTR, bTR, and mTR). The sequence alignment was prepared by Clustal W version 1.4 (54). Nucleotides that are conserved between the three RNAs are denoted with a asterisks. The sequences that encompass the template region and that correspond to the putative H and ACA boxes in hTR are marked.
Previous studies of the human telomerase RNA component have identified regions within the RNA that are critical for telomerase activity (Fig. 1), yet details of the secondary structure await phylogenetic analysis. One feature of hTR that has already been established is the location of the template, within +46 to +56, in a region that is accessible to oligonucleotides and peptide nucleic acids and therefore likely to be single stranded (18, 43, 45, 50). Additionally, a motif similar to the small nucleolar RNA H/ACA box has been identified in the 3′ end of hTR and appears to be important in hTR accumulation and 3′ end processing (36). Previously, a reconstitution assay in which the endogenous RNA component of partially purified telomerase was removed by digestion with micrococcal nuclease (MNase) and telomerase activity was regenerated by the addition of exogenous recombinant hTR was developed and used along with site-directed mutagenesis of hTR to identify the region between nt +170 and +200 as critical for catalytic activity (3). This assay was also used to map the minimal functional region of hTR required to reconstitute detectable levels of telomerase activity. These studies indicated that this sequence lies between nt +44 and +203, with truncations containing sequences between nt +1 and +203 reconstituting levels of activity similar to those produced with full-length hTR (3). However, MNase may leave inaccessible fragments of hTR that are essential for reconstituting telomerase activity intact.
We used two strategies to map the minimal domain of hTR that is required for de novo synthesis of telomerase activity in vitro. In a previously described system, in vitro-transcribed hTR was added to hTERT that was newly synthesized in a rabbit reticulocyte lysate to produce an active enzyme (24, 56). A second novel system was developed by using only human components in which exogenous hTERT was synthesized in intact human VA13 cells which lack endogenous hTR and hTERT (57). Active telomerase was produced when recombinant hTR was combined with S100 extracts prepared from stable transformants of VA13 cells expressing hTERT. The sequence spanning nt +33 to +325 was required to generate levels of activity comparable with that produced using full-length hTR. This segment contains over 100 more nt of 3′ sequence than previously shown to be required to efficiently reconstitute activity with MNase-treated extracts (3). Our results also suggest that nt +33 to +43 contribute to efficient assembly of telomerase activity. Furthermore, the +33 to +325 fragment can be subdivided into two regions (one from nt +33 to +147, which contains the template, and the other from nt +164 to +325) that are individually inactive but together efficiently assemble active telomerase with hTERT. These pieces may represent two functional domains within the hTR molecule.
MATERIALS AND METHODS
Synthesis of full-length and truncated hTR.
RNAs corresponding to hTR sequences were produced with the Megascript T7 in vitro transcription system (Ambion) from templates amplified from the hTR clone pTRC3 (13). The numbering system for hTR was determined from the GenBank sequence (accession no. U86046) and is shown in Fig. 1. Truncation mutants were amplified with oligonucleotides encoding the T7 promoter and hTR sequences initiating at nt +1 (T7HTR+1), +33 (T7HTR+33), +44 (T7HTR+44), +58 (T7HTR+58), +164 (T7HTR+164), +206 (T7HTR+206), and +250 (T7HTR+250) (Table 1). These oligonucleotides were used in amplification reactions with primers that terminate at nt +451 (HTR+451), +354 (HTR+354), +325 (HTR+325), +300 (HTR+300), +280 (HTR+280), +205 (HTR+205); +163 (HTR+163), +147 (HTR+147), +104 (HTR+104), and +73 (HTR+73).
TABLE 1.
Primers for amplifying hTR sequences
| Primer | Description | Sequencea |
|---|---|---|
| T7HTR+1 | T7 promoter plus +1 to +20 | cgtaatacgactcactataGGGTTGCGGAGGGTGGGCCT |
| T7HTR+33 | T7 promoter plus +33 to +55 | cgtaatacgactcactatagGGCCATTTTTTGTCTAACCCTAA |
| T7HTR+44 | T7 promoter plus +44 to +63 | cgtaatacgactcactataggGTCTAACCCTAACTGAGAAG |
| T7HTR+58 | T7 promoter plus +58 to +74 | cgtaatacgactcactataggGAGAAGGGCGTAGGCGC |
| T7HTR+164 | T7 promoter plus +164 to +185 | cgtaatacgactcactatagggAGCAAACAAAAAATGTCAGCTG |
| T7HTR+206 | T7 promoter plus +206 to +221 | cgtaatacgactcactataGGGGACCTGCGGCGGG |
| T7HTR+250 | T7 promoter plus +250 to +265 | cgtaatacgactcactatagGGAGGCCGCGGTCGGC |
| T7HTRmt+33 | T7 promoter plus +33 to +64 | cgtaatacgactcactatagGGCCATTTTTTGTCaAACCCaAACTGAGAAGG |
| (TTTGGG template mutant) | ||
| HTR+451 | Complement of +432 to +451 | GCATGTGTGAGCCGAGTCCT |
| HTR+354 | Complement of +337 to +354 | CCTGAACCTCGCCCTCGC |
| HTR+325 | Complement of +306 to +325 | CCGCGGCTGACAGAGCCCAA |
| HTR+300 | Complement of +281 to +300 | CGCGGTGGCAGTGGGTGCCT |
| HTR+280 | Complement of +261 to +280 | CCGGAGAAGCCCCGGGCCGA |
| HTR+205 | Complement of +189 to +205 | GGGAGGGGCGAACGGGC |
| HTR+163 | Complement of +143 to +163 | CTAGAATGAACGGTGGAAGGC |
| HTR+147 | Complement of +130 to +147 | AAGGCGGCAGGCCGAGGC |
| HTR+104 | Complement of +87 to +104 | GAAAAACAGCGCGCGGGG |
| HTR+73 | Complement of +55 to +73 | CGCCTACGCCCTTCTCAGT |
Sequences corresponding to hTR are in uppercase; the T7 promoter (taatacgactcactataggg) is underlined; the first G of the triplet GGG is the first transcribed nucleotide (+1). Nucleotide mismatches in T7HTRmt+33 are denoted by lowercase italic letters.
Site-directed mutations were introduced into hTR sequences by engineering nucleotide changes into the primers used for DNA template amplification. The DNA template for synthesizing hTR(33-163/T3G3) was amplified with an oligonucleotide that contains a T7 promoter, initiates at nt +33, and directs the synthesis of TTTGGG (T7HTRmt+33) in conjunction with an oligonucleotide that terminates at nt +163 (HTR+163) (Table 1).
The amplified products were purified on 2% agarose gels and used as templates for T7 transcription reactions. RNA products for defining the minimal sequence of hTR required to assemble active telomerase with rabbit reticulocyte extracts were purified on 6% polyacrylamide–8 M urea gels, thereby eliminating the possibility of wild-type RNA contamination. Subsequent to this mapping, truncated RNAs that did not contain this minimal core sequence were tested to confirm that they could not assemble active telomerase, making gel isolation of these RNAs unnecessary. The concentration of all RNAs was quantitated by spectrophotometric analysis and confirmed by direct examination on 6 to 12% polyacrylamide–8 M urea gels.
Assembly of active telomerase.
To assemble active telomerase by using hTERT that was synthesized in a rabbit reticulocyte extract, the entire hTERT coding sequence was excised from the original vector (pGRN121; Geron Corporation, Menlo Park, Calif.) with EcoRI and subcloned into pcDNA3.1/His C (Invitrogen Corporation, Carlsbad, Calif.) to generate the clone pXhTRTE (56). This construct has an engineered consensus Kozak sequence and yields a recombinant protein containing two amino-terminal tags (a polyhistidine tag and an Xpress epitope). Full-length hTERT was synthesized by using a rabbit reticulocyte lysate transcription-translation system (Promega) as described by the manufacturer. Telomerase was assembled in a reaction containing in vitro-transcribed hTR sequences, 0.2 μl of in vitro-synthesized hTERT, and 2 μl of rabbit reticulocyte lysate in 4 μl (total volume) (24). Some reactions contained yeast tRNA (3 μg) to keep the total amount of RNA constant, as noted. To determine the effect of combining hTR(33-163) and full-length hTR on the assembly of active telomerase, hTR-wt(1-451) was added to the assembly reaction in a quantity (10 ng) sufficient to produce levels of enzymatic activity at the lower end of the linear range (data not shown). In this experiment, human U2 snRNA was included as a control for a nonspecific structural RNA that is comparable in size to hTR. This RNA was transcribed with T7 polymerase from a BstXI-linearized pGEM-U2 subclone in which the human U2 snRNA fragment was excised from pBSU2 (28) with PstI and SacI and cloned into these sites in pGEM5zf(+) (Promega). After incubation for 90 min at 30°C, the assembly reactions were diluted 10- to 25-fold in 1× CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} lysis buffer (Intergen Inc., Gaithersburg, Md.), and 1 to 2 μl (constituting 1 to 5% of the starting material) was used for the telomerase activity assay.
We developed a novel assembly system for telomerase that uses only human components. Exogenous hTERT was synthesized in the VA13 cell line, which contains no detectable levels of hTERT mRNA, hTR, or telomerase activity (57) (this study and data not shown). VA13 cells were infected with a retrovirus expressing hTERT (pBabepuro-hTERT) (44), and the infected population (VA13-hTERT) was selected with puromycin (750 ng/ml). VA13-hTERT cells were harvested and resuspended in water at a concentration of 100,000 cells/μl. The sample was sonicated with two pulses at 50 J/W · s and then centrifuged at 100,000 × g for 1 h. Glycerol was added to the S100 supernatant to a final concentration of 20%, and the extract was stored at −80°C. To assemble active telomerase, 2 μl of the S100 extract from VA13-hTERT cells was combined with 200 ng of in vitro-transcribed hTR in a 5-μl final volume with 1 mM (final concentration) ATP. The need for ATP in the assembly reaction is consistent with the requirement for ATP turnover in the functioning of the foldasome, a complex of proteins required for the assembly of catalytically active telomerase (24). After incubation for 90 min at 30°C, 5% of the assembly reaction was assayed for telomerase activity by using a PCR-based telomerase assay (TRAP assay; Intergen).
Northern analysis.
The integrity of hTR and its truncation mutants were verified by Northern analysis following the assembly reaction using a 1.5% agarose–2.2 M formaldehyde gel. The RNA was transferred to a Hybond-N+ membrane (Amersham) and probed with 32P-labeled oligonucleotide complementary to hTR nt +143 to +163 (hTR+163 (Table 1) in Rapid-Hyb buffer (Amersham) according to the manufacturer’s protocol. The blot was washed twice for 15 min each time at room temperature with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate and exposed to a PhosphorImager screen (Molecular Dynamics).
RT-PCR.
The absence of detectable levels of hTR and hTERT mRNA in VA13 cells was confirmed by reverse transcription-PCR (RT-PCR) using previously described procedures (13, 40). RNA was extracted from VA13 and telomerase-positive HL-60 cells by using Trizol Reagent (Gibco BRL) and treated with DNase I to avoid genomic DNA contamination. Equivalent amounts of total RNA (1 μg) were reverse transcribed with a Retroscript kit (Ambion) and amplified by using primers for hTR (13) or hTERT mRNA (40). The products were loaded onto a 5% polyacrylamide gel, stained with ethidium bromide, and detected with UV light.
Telomerase assay.
For most samples, the TRAP assay was performed as originally described (27), with minor modifications (25, 58). The TRAP-eze telomerase detection kit (Intergen), which includes a 36-bp internal standard for semiquantitative measurements, was used as recommended by the manufacturer. After telomerase extension at room temperature for 30 min, samples were subjected to a 94°C hot start, followed by a two-step PCR (94°C for 30 s, 60°C for 30 s) for 27 or 28 cycles. Telomerase products were electrophoresed on 10% polyacrylamide gels for 2 h at 300 V and exposed to a PhosphorImager screen. Quantitative estimates of telomerase activity were calculated by determining the ratio of the band intensities of the 36-bp internal standard to that of the characteristic 6-bp telomerase-specific ladder. This method of quantitation is accurate over an approximately 300-fold range of activities (Intergen).
To test whether hTR(33-163/T3G3) can serve as a template in the presence of full-length hTR, the assembly reaction was incubated at 30°C for 90 min and then diluted 12.5-fold in 1× CHAPS lysis buffer (Intergen). A 0.2-μl aliquot of this diluted sample was added to a 5-μl extension reaction that contained 50 μM (final concentration) dGTP and dTTP along with reaction buffer and substrate (TS primer) provided by the TRAP-eze telomerase detection kit (Intergen). After telomerase extension for 30 min at room temperature, the reaction mixture was heated to 94°C for 5 min to inactivate telomerase. The products were then amplified in a 50-μl reaction in the presence of all four deoxynucleoside triphosphates with reagents from Intergen except the primer mix, which contains the template and amplification primer for synthesizing the internal standard and the comeback primer for amplifying telomerase extension products. In its place, we substituted a primer mix containing the primer T/G-ACXII (GCGCGGCAAACCCAAACCCAAACCCAAACC) for efficient amplification of TTTGGG extension products.
RESULTS
Minimal contiguous sequence of hTR necessary for assembly of active telomerase in vitro.
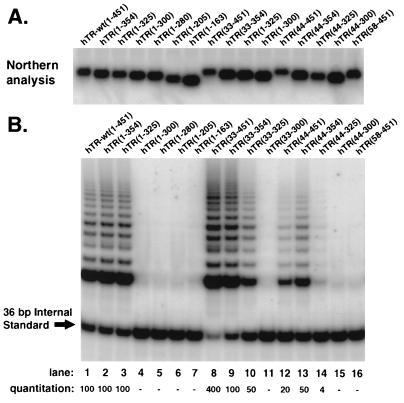
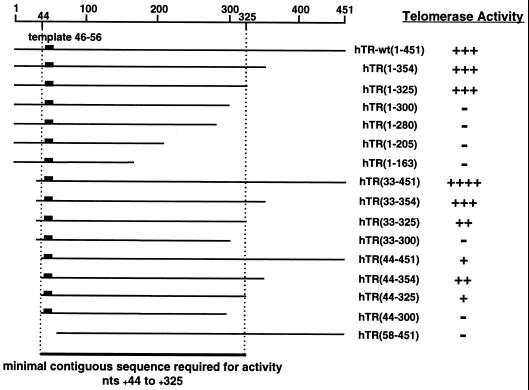
Each of the hTR truncation mutants examined in this study was transcribed from a PCR template that was generated with a primer that contained sequences for the T7 promoter as well as sequences complementary to the 5′ region of hTR and a primer for the appropriate truncation at the 3′ end of hTR (Table 1). The truncations are denoted by the nucleotide position of the 5′ and 3′ ends (GenBank accession no. U86046). These PCR products were used to produce the corresponding RNAs with an in vitro transcription system (see Materials and Methods). Each truncated RNA was incubated with newly synthesized hTERT, and the assembly reaction was analyzed for both RNA integrity (Fig. 2A) and telomerase activity (Fig. 2B). Northern blot analysis revealed that each of the mutants was of the appropriate size and concentration (30 to 100% of input) (Fig. 2A), indicating that under these conditions most of the synthesized RNAs were intact. Reactions containing the truncated RNA hTR(1-325) produced levels of telomerase activity that were similar to those produced with hTR-wt(1-451) (Fig. 2B, lane 3). However, hTR(1-300), hTR(1-280), hTR(1-205), and hTR(1-163) were unable to yield detectable levels of active telomerase under the assay conditions used (Fig. 2B, lanes 4 to 7). It has been reported that in a reticulocyte lysate assembly system in which hTERT is synthesized in the presence of a pretranscribed RNA, the truncation spanning nt +10 to +159 of hTR acts as a template with reduced efficiency compared to wild-type hTR (4). Therefore, we assayed more of the assembly reaction to see if we could detect similar activity with our truncations. Using five times more extract, corresponding to the maximum amount of extract that does not inhibit PCR amplification, we were able to detect a faint ladder with hTR(1-300), but this activity was exceptionally weak (less than 0.2%) compared to full-length hTR (data not shown). The same cutoff was observed when hTERT was synthesized in the presence of hTR (data not shown). Additional truncations at the 5′ end of the RNA demonstrated that hTR(44-325) could assemble with hTERT to produce 3 to 5% of maximal activity (Fig. 2B, lane 14). Interestingly, when the first 32 nt of hTR were deleted without disturbing the 3′ end, a four- to fivefold increase in telomerase activity was consistently observed (Fig. 2B, lane 8). The smallest hTR fragment that yielded levels of activity close to that obtained with full-length RNA was hTR(33-325) (Fig. 2B, lane 10). As expected, when the template region for hTR (nt +46 to +56) was deleted to produce hTR(58-451), telomerase activity was abolished (Fig. 2B, lane 16). A schematic representation of each hTR truncation (Fig. 3) shows that the minimal region of hTR for assembling at least 3 to 5% of wild-type activity includes the template region and approximately 270 nt 3′ to the template.
FIG. 2.
Mapping the 5′ and 3′ boundaries of hTR that define the minimal sequence for assembling active telomerase with hTERT. The templates for hTR truncations were made by PCR using appropriate 5′ and 3′ primers (see Materials and Methods), gel isolated, and subjected to in vitro transcription. Transcribed RNAs were gel isolated to ensure that wild-type hTR was not carried over into the assembly reaction. Mutant nomenclature refers to the 5′ and 3′ nucleotides within hTR. Each mutant (0.2 μg) was added for telomerase assembly under optimized conditions (0.2 μl of hTERT, 200 mM KCl, and 50% rabbit reticulocyte lysate in 4 μl at 30°C for 90 to 120 min). The assembly reaction was analyzed by Northern blot analysis (A) and the TRAP assay (B). The Northern analysis demonstrates that equal amounts of the hTR mutants were still present after the assembly reaction. Relative quantitation is shown for this representative TRAP assay and reflects the ratio between the abundance of extended products versus the 36-bp internal standard.
FIG. 3.
Schematic representation of hTR truncations and their effects on telomerase activity. The 451-nt hTR sequence is represented schematically with respect to the template region (nt +46 to +56). Truncations are denoted by the transcribed nucleotides within hTR. Relative activity was determined by defining the level of activity with full-length hTR as 100%. The amount of activity for all truncations represents a minimum of three independent experiments and is shown symbolically as a relative range of average activity: ++++, greater than or equal to 400%; +++, between 100 and 399%; ++, between 25 and 99%; +, between 1 and 24%; −, undetectable.
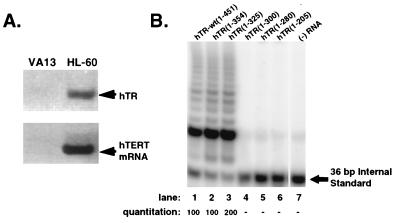
To confirm the 3′ boundary of functional hTR in a human cellular milieu, we set up an assembly system for telomerase in which hTERT is synthesized in intact human cells. VA13 cells are human fibroblasts that have been immortalized with simian virus 40 large T antigen, lack detectable hTERT mRNA and hTR by RT-PCR analysis (Fig. 4A), and maintain their telomeres through an alternative pathway (57). We tested whether VA13 cellular extracts were capable of assembling telomerase activity. S100 extracts from VA13 cells that express exogenous hTERT were prepared as described in Materials and Methods. These extracts tested negative for telomerase activity but could assemble active enzyme when combined with hTR (Fig. 4B). As found previously for hTERT that was synthesized in a rabbit reticulocyte extract, the levels of activity produced in reactions with hTR-wt(1-451), hTR(1-354), and hTR(1-325) were similar. However, no detectable activity was produced in reactions containing hTR(1-300), hTR(1-280), or hTR(1-205) as the template RNA (Fig. 4B). We conclude that the 3′ boundary for the minimal functional region in hTR necessary to generate levels of activity comparable those obtained with full-length hTR lies between nt +300 to +325.
FIG. 4.
Mapping the 3′ boundary of hTR required to produce active telomerase with hTERT that was synthesized in intact human cells. Truncation mutants of hTR were analyzed in assembly reactions in which hTERT was synthesized in intact VA13 cells that lack endogenous hTR and hTERT. (A) RT-PCR was performed to demonstrate that the VA13 human fibroblast cell line immortalized with simian virus 40 large T antigen lacks both hTR and mRNA for hTERT. The telomerase-positive strain HL-60 served as a positive control for amplification. (B) Truncation mutants of hTR were incubated with S100 extracts of VA13 cells that synthesize exogenous hTERT. A fraction of the assembly reaction was analyzed by the TRAP assay. Relative quantitation is shown for this representative TRAP assay and reflects the ratio between the abundance of extended products versus the 36-bp internal standard.
The hTR truncation hTR(33-163) serves as a template in the presence of full-length hTR.
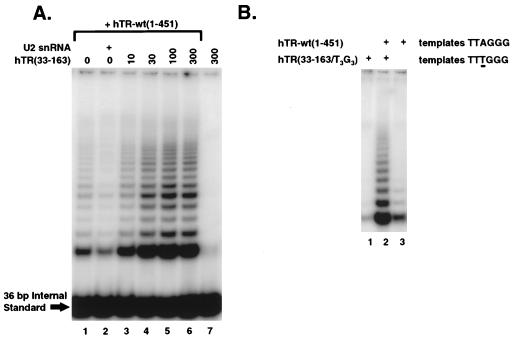
Although hTR(33-163) alone does not generate active telomerase with hTERT (Fig. 5A, lane 7), the addition of 10- to 300-fold molar excess of hTR(33-163) into an assembly reaction in combination with full-length hTR resulted in an increase in telomerase activity over full-length hTR alone (Fig. 5A; compare lane 1 to lanes 3 to 6). In striking contrast, when human U2 snRNA was added to the reaction, using a mass equivalent to the highest concentration of hTR(33-163) examined, telomerase activity was reduced, perhaps through nonspecific protein-RNA contacts that inhibit telomerase assembly or catalytic activity (Fig. 5A, lane 2).
FIG. 5.
hTR(33-163) enhances telomerase activity in presence of full-length hTR. (A) Full-length hTR and hTR(33-163) were transcribed separately and then combined in an assembly reaction with hTERT. Lanes 1 to 6 contain 10 ng of hTR-wt(1-451); lanes 3 to 6 also contain hTR(33-163) at 10-, 30-, 100-, and 300-fold molar excess over hTR-wt(1-451) (corresponding to 29, 86, 290, 860 ng, respectively); lane 2 contains 860 ng of human U2 snRNA; lane 7 contains 860 ng of hTR(33-163) alone. A portion of the assembly reaction was analyzed by the TRAP assay. (B) Assembly reactions were prepared from 300 ng of hTR(33-163/T3G3) (lanes 1 and 2) and/or 10 ng of hTR-wt(1-451) (lanes 2 and 3). To measure activity from the mutant template [in hTR(33-163/T3G3)] but not the wild-type template [in hTR-wt(1-451)], the telomerase extension reaction was performed in the presence of only dTTP and dGTP. After extension, the enzyme was heat inactivated and the products were amplified by PCR. Because a unique comeback primer was required for efficient amplification (see Materials and Methods), the internal standard provided with the Intergen TRAP-eze kit could not be used in this experiment.
To determine if hTR(33-163) can synthesize telomeric repeats from its own template in the presence of full-length hTR, the template region of hTR(33-163) was mutated to direct the synthesis of TTTGGG, creating the RNA hTR(33-163/T3G3). To confirm that the modified template region is able to direct the synthesis of TTTGGG, we first engineered this mutation within the hTR truncation that spans nt +33 to +451. Telomerase extension products made from an assembly reaction containing this mutant template RNA consist of a ladder of TTTGGG repeats, as demonstrated by sequencing cloned DNA prepared from the resulting extension products (data not shown). Synthesis of products from this mutant template requires only dTTP and dGTP, whereas synthesis from the wild-type template has an additional dATP requirement. If the extension reaction is conducted in the presence of only dTTP and dGTP, a ladder should be seen only if telomerase is using the mutant template. For efficient product amplification in the TRAP assay, we found it necessary to design a new reverse primer that anneals perfectly to the TTTGGG repeat sequence. When added to the assembly reaction alone, hTR(33-163/T3G3) was not able to direct the synthesis of TTTGGG repeats (Fig. 5B, lane 1). Similarly, under these extension conditions, hTR-wt(1-451) did not produce a typical product ladder, although a weak pattern consisting of small bands was observed which may reflect misincorporation with the wild-type template or contamination of the nucleotides with small amounts of dATP (Fig. 5B, lane 3). In marked contrast, when both of these RNAs were included in the assembly reaction, a strong ladder was produced (Fig. 5B, lane 2). These results demonstrated that the truncated RNA hTR(33-163/T3G3) promotes the synthesis of TTTGGG in the presence of full-length hTR.
Efficient assembly of active telomerase from hTERT and two functionally inactive hTR truncations.
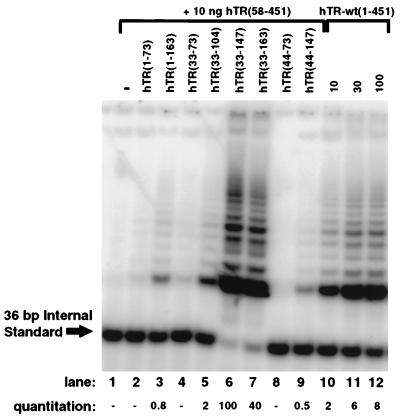
In S. cerevisiae, telomerase can contain two RNA components that functionally interact such that wild-type telomerase RNA rescues an inactive mutant RNA to enable both RNAs to act as templates (46, 47). We tested whether the template region of hTR-wt(1-451) was essential to provide the complementation that we observed with our inactive RNAs. Using hTR(58-451), which lacks the template but which may provide other required structural components of hTR, we examined short RNA truncations containing the template for the ability to generate active telomerase (Fig. 6, lanes 1 to 9). None of these RNAs assembled active telomerase when tested individually with hTERT (data not shown). In the presence of hTR(58-451), hTR(33-163) and hTR(33-147) synthesized the telomeric repeat with similar efficiencies (Fig. 6, lanes 6 and 7). This combination of RNAs produced levels of activity that were substantially greater than the maximal amount of activity obtained with full-length hTR [corresponding to 100 ng of hTR-wt(1-451) (Fig. 6, lane 12, and data not shown)]. In contrast, equivalent molar concentrations of the template truncations hTR(1-163), hTR(33-104), and hTR(44-147) yielded less than 5% of the levels of activity obtained with hTR(33-163) and hTR(33-147) (Fig. 6, lanes 3, 5, and 9), while hTR(1-73), hTR(33-73), and hTR(44-73) generated only very weak or no discernible ladders (Fig. 6, lanes 2, 4, and 8). The RNA hTR(1-163) is much less efficient at assembling active telomerase than the shorter truncation hTR(33-163). Although this effect is more dramatic for these template truncations, it is consistent with our previous observation that removal of the first 32 nt of full-length hTR increases the amount of activity produced (Fig. 2B). Furthermore, the differences in activity for truncations hTR(33-147) and hTR(44-147) reiterates the previous observation that deleting the sequence between nt +33 and +43 in full-length hTR reduces the amount of activity (Fig. 2B). This result suggests that nt +33 and +147 constitute the boundaries of the minimal template-containing region that can effectively produce active telomerase with hTR(58-451).
FIG. 6.
Complementation of hTR(58-451) with template-containing hTR truncations to assemble active telomerase. Assembly reactions for lanes 1 to 9 contained 10 ng of hTR(58-451) and either no additional RNA (lane 1) or a template-containing RNA present as the molar equivalent to 1 μg of full-length hTR, as indicated (lanes 2 to 9). Reactions for lanes 10 to 12 contain increasing quantities of hTR-wt(1-451): 10 ng (lane 10), 30 ng (lane 11) and 100 ng (lane 12). To compensate for differences in RNA quantities, 3.3 μg of yeast tRNA was included in each sample. A fraction (1/20) of the assembly reaction product was examined by the TRAP assay. Relative quantitation is shown for this representative TRAP assay and reflects the ratio between the abundance of extended products versus the 36-bp internal standard.
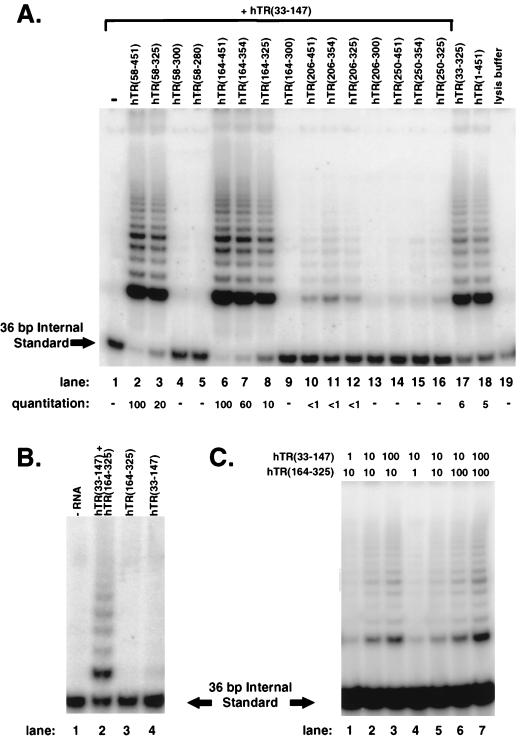
To map the boundaries of the minimal structural component of hTR that facilitates assembly of human telomerase activity in conjunction with hTERT and hTR(33-147), we created an extensive panel of hTR truncations containing sequences downstream of the template region (Fig. 7A). As before, assembly reactions with hTERT and hTR(33-147) did not produce detectable activity (Fig. 7A, lane 1); similarly, none of the RNAs without a template region could assemble active telomerase with hTERT (data not shown). However, the combination of hTR(33-147) and the truncations which span nt +164 to +325 of hTR, including hTR(58-451), hTR(58-325), hTR(164-451), hTR(164-354), and hTR(164-325), efficiently assembled active telomerase with hTERT (Fig. 7A, lanes 2, 3, and 6 to 8, respectively), with RNAs terminating between +326 to +451 functioning more efficiently than hTR(164-325). A low level of activity, which corresponds to less than 1% of that produced in reactions with hTR(33-147) and hTR(58-451), was detected when the 5′ boundary for the template-lacking RNA was positioned at nt +206 in hTR(206-451), hTR(206-354), and hTR(206-325) (Fig. 7A, lanes 10 to 12). Very weak or no activity was observed when this boundary was moved to nt +250 in hTR(250-451), hTR(250-354), and hTR(250-325) (Fig. 7A, lanes 14 to 16). Complementation was seen for truncations terminating at nt +325 but not at +300 or +280 (Fig. 7A; compare lane 3 with lanes 4 and 5; compare also lanes 8 and 9 with lanes 12 and 13). This 3′ boundary is identical to the minimal contiguous sequence of hTR required to assemble active telomerase (Fig. 2). These results indicate that active telomerase can be assembled when two independently inactive RNA fragments, hTR(33-147) and hTR(164-325), are combined with hTERT. We confirmed that these two RNAs also function synergistically in assembly reactions containing hTERT that was synthesized in intact human cells (Fig. 7B). In this analysis, the addition of 200 ng of either hTR(164-325) or hTR(33-147) into the assembly reaction did not yield detectable levels of telomerase activity (Fig. 7B, lanes 3 and 4). However, inclusion of both RNAs in the assembly reaction produced telomerase activity (Fig. 7B, lane 2). The boundaries of these RNAs further indicate that the sequence between nt +148 and +163 of hTR is not essential for the assembly of telomerase activity in vitro.
FIG. 7.
Complementation of hTR(33-147) with hTR truncations that lack a template to assemble active telomerase. Assembly reactions were performed with hTERT that was synthesized in rabbit reticulocyte lysate (A and C) or in VA13 cells (B), and 1/20 of each assembly reaction product was examined by the TRAP assay. (A) Assembly reactions for lanes 1 to 16 contained 250 ng of hTR(33-147) (7 pmol, the molar equivalent to 1 μg of full-length hTR) and either no additional RNA (lane 1) or a 1/10 molar equivalent amount (0.7 pmol) of the nontemplating RNAs, as indicated (lanes 2 to 16). The assembly reaction for lane 17 contained 70 ng (0.7 pmol) of gel-isolated hTR(33-325); lane 18 contained 100 ng (0.7 pmol) of hTR(1-451). Yeast tRNA (3.3 μg) was included in each sample. Lane 19 is a lysis buffer control to demonstrate the specific assembly of active telomerase. Relative quantitation is shown for this representative TRAP assay and reflects the ratio between the abundance of extended products versus the 36-bp internal standard. (B) S100 extracts prepared from VA13 cells that express exogenous hTERT did not yield telomerase activity when assayed alone (lane 1) or when mixed with either hTR(164-325) (lane 3) or hTR(33-147) (lane 4). However, combining hTR(33-147) and hTR(164-325) with VA13-hTERT extracts yielded telomerase activity in vitro (lane 2). (C) Titration experiments were performed with hTR(33-147) and hTR(164-325). Relative amounts of the RNAs included in the assembly reactions are shown, with 1 U representing 0.07 pmol, which is the molar equivalent of 10 ng of hTR(1-451). When 7 pmol of hTR(33-147) or hTR(164-325) was tested individually in the assembly reactions, no telomerase products were detected (data not shown).
The relative amounts of hTR(33-147) and hTR(164-325) were varied in order to determine whether they were interacting stoichiometrically or whether one fragment might be acting catalytically to promote the correct folding of either the other fragment or the hTERT protein. Figure 7C demonstrates that roughly equal amounts of the two RNA components were required per unit of activity produced.
DISCUSSION
The minimal region of the human telomerase RNA component that is required for activity.
Previous efforts to map the minimal functional region of the human telomerase RNA employed a reconstitution system where MNase was used to remove endogenous hTR from human telomerase that was partially purified from 293 cells (3). The nuclease was then inactivated, and hTR was added to reconstitute activity. In those studies, nt +1 to +203 defined the functional region of hTR necessary to reconstitute levels of activity similar to those produced with full-length hTR. For our assembly reactions where hTERT and hTR were synthesized separately and then coassembled to generate active telomerase, hTR residues beyond nt +203 and extending to nt +325 are required for efficient de novo assembly. Activity resulting from nt +1 to +300 of hTR was less than 0.2% of that produced with full-length hTR, while shorter 3′ truncations (containing nt +1 to +280, +1 to +203, or +1 to +163) did not produce detectable levels of activity in our assay. It is possible that for endogenous telomerase, sequences in the hTR molecule are inaccessible to MNase cleavage. These buried fragments may contain sequences between nt +203 (the 3′ boundary identified by Autexier et al. [3]) to +325 (the 3′ boundary identified in this study) that enable short RNAs to serve as a template and efficiently produce active enzyme. Alternatively, nt +203 to +325 may be critical for the correct folding of hTERT (see below), but once the telomerase ribonucleoprotein is assembled, the protein conformation may remain stable even after removal of these sequences. A variant of this possibility is that auxiliary factors associated with the telomerase holoenzyme stabilize the complex after MNase degradation, thereby allowing short hTR pieces to reconstitute activity. All of these possibilities are consistent with the observation that efficient de novo formation of active telomerase requires sequences that were previously thought to be dispensable. We find it intriguing that there are also discrepancies between mapping studies of the Tetrahymena telomerase template RNA using the MNase and de novo assembly protocols (29). Some phylogenetically conserved portions of the Tetrahymena telomerase template RNA that are dispensable for reconstituting activity in the MNase protocol (2) were required for production of active enzyme when the catalytic protein was synthesized in a rabbit reticulocyte lysate system (29).
Although the sequence between nt +33 to +325 produced almost full activity when combined with hTERT in vitro, additional hTR sequences are essential for the production of telomerase activity in vivo (36). Deletion of only 23 nt from the 3′ end of hTR prevented any detectable accumulation of the RNA in cells, presumably because it destroys a domain in hTR that is similar to the H/ACA box of small nucleolar RNAs (36). In cells, this motif is involved in both hTR accumulation and 3′ end processing and is therefore critical for the production of active telomerase (36). Additional elements in hTR could play other essential roles in the production of the active holoenzyme. Such elements could mediate contacts with proteins such as TP1, the mammalian homolog of Tetrahymena p80 (9, 21, 41).
Sequences upstream of the template of hTR are not essential for producing active telomerase in vitro, although they do contribute to the overall level of activity (Fig. 2 and reference 3). Removal of the first 43 nt of hTR, but not the first 32 nt, reduces the level of active telomerase, suggesting that nt +33 to +43 may aid in assembly or in catalysis, perhaps by anchoring the template region of hTR on hTERT and facilitating translocation during processive elongation. Interestingly, the bovine RNA component (55) shows striking sequence similarity to the human RNA in the region 5′ to the template (Fig. 1). The 5′ start site of the mouse telomerase RNA, which has been mapped to only 2 nt from the template region (23) (Fig. 1), provides support for the observation that the sequences 5′ of the template of the human RNA are not critical for producing an active enzyme.
While some aspects of the secondary structure of the ciliate RNA components have been studied in depth (2, 5, 30, 33, 48, 53), it is unclear how many of these structural features are common to the RNA components of other organisms. Sequences 5′ to the template that are conserved in ciliate RNAs include the boundary element and the sequences within helix I that define the overall topology of the ciliate RNAs (1, 2, 30, 33, 48). These features are not present in the mouse RNA (23) and are not necessary to produce active telomerase with the human RNA (3) (Fig. 1 and 2). There are also considerable differences in the overall size of the integral RNA component. Although the ciliate RNAs are less than half the size of hTR (148 to 209 nt versus 451 nt), our data indicate that the minimal sequence for hTR needed for generating 3 to 5% of the activity assembled with full-length hTR (nt +44 to +325 = 282 nt) is slightly larger than the ciliate RNAs. However, the RNA component from two species of yeast is very large (about 1,300 nt for S. cerevisiae [52] and K. lactis [34]), even considering that approximately half of the K. lactis RNA is not required for catalysis (49).
Assembly of active telomerase from hTERT and two independently inactive segments of hTR.
The core region of hTR (between nt +33 to +325) that produces levels of telomerase activity with hTERT that are similar to those obtained with full-length hTR sequence can be separated into two distinct segments, hTR(33-147) and hTR(164-325), that function synergistically with hTERT to yield active enzyme. The sequence between hTR nt +33 to +147 contains at least three features that are required for efficient assembly of active enzyme with hTERT: the template region (between nt +46 and +56), sequence between nt +33 and +43 upstream of the template (Fig. 2), and approximately 90 nt of sequence immediately downstream of the template (Fig. 6). We have mapped the 5′ boundary of the other essential region in hTR to the sequence between nt +164 and +206 (Fig. 7), which encompasses nt +170 to +200, a region that was previously shown to be required for activity (3). Our results also identify a new region of hTR, the sequence between nt +148 and +163, that is dispensable for catalysis. This finding is consistent with the lack of telomerase inhibition observed when a peptide nucleic acid targeted to this region, but not to surrounding sequences, is included in the ribonucleoprotein assembly reaction (19).
The region between nt +164 and +325 of hTR may be essential for catalytic activity because it participates directly in synthesizing the telomeric repeat. Alternatively, this fragment might serve a structural role in the enzyme, perhaps by helping the protein to form an anchor site (20) or by positioning key residues in the catalytic site. A structural role for this RNA fragment is conceivable because the assembly of ribonucleoproteins generally occurs through an induced-fit process, such that both protein and RNA conformation are modulated through their interactions (14). The RNA component could induce gross conformational changes in the structure of disordered or partially disordered hTERT, as has been observed for bacteriophage λ N protein and bacterial Ffh proteins (14, 37, 59). This hypothesis predicts that hTERT that is synthesized in the rabbit reticulocyte lysate and in VA13 cells lacking hTR may be largely misfolded, which is consistent with our observation that efficient assembly of active complexes requires chaperone proteins (24). It is possible that the sequence from nt +203 to +325 of hTR is needed to induce the correct folding of hTERT but that once folded correctly, hTERT can maintain at least a partially folded conformation in the absence of this RNA sequence. This might explain the finding that nt +203 to +325 are not needed to reconstitute endogenous telomerase in which the RNA has been removed (3).
The catalytic core of a group I self-splicing intron can be reconstituted from its two distinct structural domains and substrate RNA (12). These pieces of RNA self-assemble, presumably through high-affinity tertiary contacts which fold the molecule into a catalytically active configuration. It is possible that hTR(33-147) and hTR(164-325) also represent topologically distinct subdomains that interact to form a structure similar in topology to the core region of full-length hTR. This RNA duplex could then assemble with the hTERT protein catalytic subunit to generate an active enzyme. An alternative possibility for how these molecules assemble is that hTERT provides the scaffold that binds these RNAs, with all three components being essential for producing an active enzyme. Our findings provide tools to investigate the process of ribonucleoprotein assembly and may facilitate the identification of the structural features within hTR.
ACKNOWLEDGMENTS
We gratefully acknowledge Joe Baur for his contributions to the development of a human extract-based assembly system for telomerase.
This work was supported by research grants from the National Institute on Aging (AG07992) and Geron Corporation. Postdoctoral support was from National Institutes of Health oncology training grant T32-CA66187 to L.P.F., National Institute on Aging grant AG05747 to S.E.H., and National Institutes of Health oncology training grant T32-CA66187 and an American Cancer Society postdoctoral fellowship to V.M.T. J.W.S. is an Ellison Medical Foundation Senior Scholar.
REFERENCES
- 1.Autexier C, Greider C W. Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 1995;9:2227–2239. doi: 10.1101/gad.9.18.2227. [DOI] [PubMed] [Google Scholar]
- 2.Autexier C, Greider C W. Mutational analysis of the Tetrahymena telomerase RNA: identification of residues affecting telomerase activity in vitro. Nucleic Acids Res. 1998;26:787–795. doi: 10.1093/nar/26.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Autexier C, Pruzan R, Funk W D, Greider C W. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 4.Beattie T L, Zhou W, Robinson M O, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharyya A, Blackburn E H. Architecture of telomerase RNA. EMBO J. 1994;13:5721–5723. doi: 10.1002/j.1460-2075.1994.tb06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasco M A, Funk W, Villeponteau B, Greider C W. Functional characterization and developmental regulation of mouse telomerase RNA. Science. 1995;269:1267–1270. doi: 10.1126/science.7544492. [DOI] [PubMed] [Google Scholar]
- 7.Bryan T M, Sperger J M, Chapman K B, Cech T R. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc Natl Acad Sci USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins K, Kobayashi R, Greider C W. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 10.Counter C M, Meyerson M, Eaton E N, Ellisen L W, Caddle S D, Haber D A, Weinberg R A. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene. 1998;16:1217–1222. doi: 10.1038/sj.onc.1201882. [DOI] [PubMed] [Google Scholar]
- 11.Counter C M, Meyerson M, Eaton E N, Weinberg R A. The catalytic subunit of yeast telomerase. Proc Natl Acad Sci USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doudna J A, Cech T R. Self-assembly of a group I intron active site from its component tertiary structural domains. RNA. 1995;1:36–45. [PMC free article] [PubMed] [Google Scholar]
- 13.Feng J, Funk W D, Wang S-S, Weinrich S L, Avilion A A, Chiu C-P, Adams R R, Chang E, Allsop R C, Yu J, Le S, West M D, Harley C B, Andrews W H, Greider C W, Villeponteau B. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 14.Frankel A D, Smith C A. Induced folding in RNA-protein recognition: more than a simple molecular handshake. Cell. 1998;92:149–151. doi: 10.1016/s0092-8674(00)80908-3. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg R A, Allsopp R C, Chin L, Morin G B, DePinho R A. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 16.Greider C W, Blackburn E H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 17.Greider C W, Blackburn E H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton S E, Pitts A E, Katipally R R, Jia X, Rutter J P, Davies B A, Shay J W, Wright W E, Corey D R. Identification of determinants for inhibitor binding within the RNA active site of human telomerase using PNA scanning. Biochemistry. 1997;36:11873–11880. doi: 10.1021/bi970438k. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton S E, Simmons C G, Kathiriya I S, Corey D R. Cellular delivery of peptide nucleic acids and inhibition of human telomerase. Chemistry Biol. 1999;6:343. doi: 10.1016/S1074-5521(99)80046-5. [DOI] [PubMed] [Google Scholar]
- 20.Hammond P W, Lively T N, Cech T R. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol Cell Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass M B, Arruda I, Robinson M O. A mammalian telomerase-associated protein. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 22.Harrington L, Zhou W, McPhail T, Oulton R, Yeung D S K, Mar V, Bass M B, Robinson M O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinkley C S, Blasco M A, Funk W D, Feng J, Villeponteau B, Greider C W, Herr W. The mouse telomerase RNA 5"-end lies just upstream of the telomerase template sequence. Nucleic Acids Res. 1998;26:532–536. doi: 10.1093/nar/26.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holt S E, Aisner D L, Baur J, Tesmer V M, Dy M, Ouellette M, Trager J B, Morin G B, Toft D O, Shay J W, Wright W E, White M A. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holt S E, Norton J C, Wright W E, Shay J W. Comparison of the telomeric repeat amplification protocol (TRAP) to the new TRAP-eze telomerase detection kit. Methods Cell Sci. 1996;18:237–248. [Google Scholar]
- 26.Kilian A, Bowtell D D, Abud H E, Hime G R, Venter D J, Keese P K, Duncan E L, Reddel R R, Jefferson R A. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 27.Kim N W, Piatyszek M A, Prowse K R, Harley C B, West M D, Ho P L C, Coviello G M, Wright W E, Weinrich S L, Shay J W. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 28.Konarska M M, Sharp P A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 29.Licht J D, Collins K. Telomerase RNA function in recombinant tetrahymena telomerase. Genes Dev. 1999;13:1116–1125. doi: 10.1101/gad.13.9.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lingner J, Hendrick L L, Cech T R. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- 31.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 32.Martin-Rivera L, Herrera E, Albar J P, Blasco M A. Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc Natl Acad Sci USA. 1998;95:10471–10476. doi: 10.1073/pnas.95.18.10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCormick-Graham M, Romero D P. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995;23:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McEachern M J, Blackburn E H. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 35.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoff M J, Liu Q, Bacchetti S, Haber D A, Weinberg R A. hEST2, the putative human telomerase catalytic subunit gene, is up- regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell J R, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mogridge J, Legault P, Li J, Van Oene M D, Kay L E, Greenblatt J. Independent ligand-induced folding of the RNA-binding domain and two functionally distinct antitermination regions in the phage lambda N protein. Mol Cell. 1998;1:265–275. doi: 10.1016/s1097-2765(00)80027-1. [DOI] [PubMed] [Google Scholar]
- 38.Morin G B. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 39.Moyzis R K, Buckingham J M, Cram L S, Dani M, Deaven L L, Johens M D, Meyne J, Ratliff R L, Wu J R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 41.Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- 42.Nakayama J, Tahara H, Tahara E, Saito M, Ito K, Nakamura H, Nakanishi T, Ide T, Ishikawa F. Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat Genet. 1998;18:65–68. doi: 10.1038/ng0198-65. [DOI] [PubMed] [Google Scholar]
- 43.Norton J C, Piatyszek M A, Wright W E, Shay J W, Corey D R. Inhibition of human telomerase activity by peptide nucleic acids. Nat Biotechnol. 1996;14:615–619. doi: 10.1038/nbt0596-615. [DOI] [PubMed] [Google Scholar]
- 44.Ouellette M M, Aisner D L, Savre-Train I, Wright W E, Shay J W. Telomerase activity does not always imply telomere maintenance. Biochem Biophys Res Commun. 1999;254:795–803. doi: 10.1006/bbrc.1998.0114. [DOI] [PubMed] [Google Scholar]
- 45.Pitts A E, Corey D R. Inhibition of human telomerase by 2′-O-methyl-RNA. Proc Natl Acad Sci USA. 1998;95:11549–11154. doi: 10.1073/pnas.95.20.11549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prescott J, Blackburn E H. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–8000. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prescott J, Blackburn E H. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 1997;11:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- 48.Romero D P, Blackburn E H. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 49.Roy J, Fulton T B, Blackburn E H. Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes Dev. 1998;12:3286–3300. doi: 10.1101/gad.12.20.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schnapp G, Rodi H P, Rettig W J, Schnapp A, Damm K. One-step affinity purification protocol for human telomerase. Nucleic Acids Res. 1998;26:3311–3313. doi: 10.1093/nar/26.13.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shippen-Lentz D, Blackburn E H. Functional evidence for an RNA template in telomerase. Science. 1990;247:546–552. doi: 10.1126/science.1689074. [DOI] [PubMed] [Google Scholar]
- 52.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 53.ten Dam E, van Belkum A, Pleij K. A conserved pseudoknot in telomerase RNA. Nucleic Acids Res. 1991;19:6951. doi: 10.1093/nar/19.24.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsao D A, Wu C W, Lin Y S. Molecular cloning of bovine telomerase RNA. Gene. 1998;221:51–58. doi: 10.1016/s0378-1119(98)00432-6. [DOI] [PubMed] [Google Scholar]
- 56.Weinrich S L, Pruzan R, Ma L, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichtsteiner S, Kim N W, Trager J B, Taylor R D, Carlos R, Andrews W H, Wright W E, Shay J W, Harley C B, Morin G B. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 57.Wen J, Cong Y S, Bacchetti S. Reconstitution of wild-type or mutant telomerase activity in telomerase-negative immortal human cells. Hum Mol Genet. 1998;7:1137–1141. doi: 10.1093/hmg/7.7.1137. [DOI] [PubMed] [Google Scholar]
- 58.Wright W E, Shay J W, Piatyszek M A. Modifications of a telomeric repeat amplification protocol (TRAP) result in increased reliability, linearity and sensitivity. Nucleic Acids Res. 1995;23:3794–3795. doi: 10.1093/nar/23.18.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng N, Gierasch L M. Domain interactions in E. coli SRP: stabilization of M domain by RNA is required for effective signal sequence modulation of NG domain. Mol Cell. 1997;1:79–87. doi: 10.1016/s1097-2765(00)80009-x. [DOI] [PubMed] [Google Scholar]