Abstract
We examined the reproducibility of a second-generation branched-DNA (bDNA) assay (Quantiplex HIV RNA 2.0) for quantification of human immunodeficiency virus type 1 (HIV-1) RNA in plasma by retesting 325 specimens on separate runs and on different lots. The performance of the bDNA test was also assessed by data analysis obtained during routine testing of 15,365 specimens. Upon retesting, 96 and 86% of specimens displaying RNA levels above 5,000 and between 500 and 5,000 copies/ml, respectively, showed less than a 0.3 log10 (twofold) difference with their initial values. Assay variability was found to increase as viral load decreased. Overall, the bDNA version 2.0 assay was found to be a reproducible and efficient test for routine quantification of HIV-1 RNA in plasma.
Human immunodeficiency virus (HIV) RNA levels and CD4+ T-lymphocyte counts are important biological markers for the management of HIV-infected patients (1). HIV RNA levels in plasma have been shown to be a strong determinant of disease progression and serve as a guideline to determine when antiviral therapy should be initiated and changed (5, 6, 8). The clinical significance of HIV RNA levels will depend on the biological variability within individuals and on the variability of the test employed. In clinically stable patients, HIV RNA levels measured by branched-DNA (bDNA) technology have been found to vary by 0.4 log10 over a 48-h period (2). Several limited studies have shown that the reproducibility of the currently available viral load assays can vary by as much as 0.5 log10 (4, 9, 10). However, the actual variation and the performance of these assays in routine testing have not been substantially documented.
We performed HIV viral load measurements on a routine basis for the follow-up of HIV-infected patients in the Canadian province of Quebec. EDTA-plasma samples were obtained from hospitals and private clinics. Specimens were frozen at −70°C within 6 h of collection and transported on dry ice to the Laboratoire de Santé Publique du Québec, where they were stored at −70°C until testing. In most cases, two aliquots of at least 2.0 ml were obtained for each specimen. Viral RNA levels were measured by a second-generation bDNA assay (Quantiplex HIV RNA 2.0; Chiron Corporation, Emeryville, Calif.) according to the manufacturer’s instructions (3). Briefly, in this assay clinical specimens, positive and negative controls, and DNA standards are each processed in duplicate. The positive control consists of human plasma containing inactivated HIV type 1 (HIV-1). Following a series of nucleic acid hybridization steps, alkaline phosphatase-labeled probes are incubated with a chemiluminescent substrate. The amount of light emitted is measured in a luminometer and reported as relative luminescent units (RLU). The quantity of HIV-1 RNA is determined from a standard curve defined by light emission from four DNA standards. The DNA standards are calibrated against an HIV RNA transcript standard as follows (in RNA copies/milliliter): A, 800,000; B, 54,000; C, 6,000; D, 250. The result reported as RNA copies/milliliter is derived from the mean of the two readings. The extent of the divergence between duplicate readings is provided by the coefficient of variation (CV). The quantification limit of the test is set at 500 RNA copies/ml. Specimens were retested, if sufficient volume was available, when yielding >500 RNA copies/ml with a CV above 35% or when yielding <500 RNA copies/ml with a CV above 35% if the RLU of one of the readings was above the RLU of one of the D standards, as recommended by Chiron personnel. Retesting of specimens was performed within 5 working days on a fresh aliquot.
Between April 1, 1997, and March 31, 1998, an average of 301 specimens were received weekly. This volume of testing required 2.5 operators per week working a 7-hour day, 5 days a week. The mean turnaround time from reception of specimens to result printout was 6 days (including weekend delays). Operator tasks included unpacking of specimens on dry ice, specimen labeling, revision of patient data, testing, entering of results, and data collection. Tests were performed by a pool of four trained operators.
A total of 15,365 specimens from 5,339 patients were tested: 58.4% of specimens showed RNA levels above 500 copies/ml, while 41.6% displayed RNA levels below this value. A CV of >35% was observed in 3.3 and 9.8% of specimens yielding RNA levels above and below 500 copies/ml, respectively. The positive control displayed a CV above 35% in 12.4% of runs. Accordingly, it varied more than the clinical specimens with >500 RNA copies/ml (P < 0.001). A total of 415 runs were performed, and 13 (3.1%) were rejected. The kit negative control yielded a quantifiable result in 6 (1.4%) runs, and the positive control was outside the prescribed range in 7 (1.7%) runs.
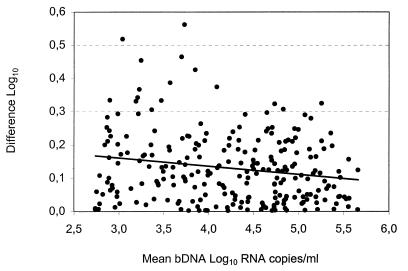
To evaluate the in-house reproducibility of the bDNA assay, a second aliquot of a previously tested specimen showing a CV below 35% was included in subsequent assay runs. Each specimen was retested on a separate run, on a different lot, and in different positions of the microwell plate. Overall, 325 specimens were retested: 255 had initial RNA levels above 500 copies/ml, and 70 had levels below this value. The former were selected as to cover the dynamic range of the standard curve and, upon repeat testing, 243 of 255 yielded results above 500 RNA copies/ml. Figure 1 illustrates the difference obtained between duplicate tests. Variance analysis indicates that this difference increased as the viral load decreased (P < 0.005). For clinical and comparative reasons, the data was stratified according to RNA levels (Table 1). The mean difference between duplicate tests was within 0.3 log10 (twofold). Nonetheless, several specimens displayed a difference of >0.3 log10, indicating that results can occasionally vary by more than twofold. The 12 specimens which initially showed >500 RNA copies/ml but whose RNA levels became undetectable upon repeat testing were excluded from this analysis as they would have minimized the actual variation. Only one showed an initial viral load above 1,000 RNA copies/ml. Finally, upon repeat testing of the 70 specimens displaying <500 RNA copies/ml and a CV below 35%, 8 (11.4%) showed viral loads above this value. Of these 8 specimens, 4 displayed RNA values between 500 and 999 copies/ml and 4 showed values between 1,000 and 1,999 copies/ml. This observation is consistent with variation being greater towards the lower limit of the assay.
FIG. 1.
Log10 difference between duplicate tests against their means for 243 clinical specimens. The regression line is included in the figure.
TABLE 1.
Interassay variability of bDNA version 2.0 assay according to RNA level
| RNA levels (copies/ml) | No. of specimensa | Log10 difference (mean ± SD) | No. (%) showing a difference of:
|
||
|---|---|---|---|---|---|
| <0.3 log10 | ≥0.3–<0.5 log10 | ≥0.5 log10 | |||
| 500–4,999 | 69 | 0.16 ± 0.12 | 59 (85.5) | 9 (13.0) | 1 (1.4) |
| 5,000–49,999 | 86 | 0.13 ± 0.10 | 82 (95.3) | 3 (3.5) | 1 (1.2) |
| 50,000–499,999 | 88 | 0.11 ± 0.09 | 85 (96.6) | 3 (3.4) | 0 |
Specimens classified according to mean of initial and repeat values.
The results obtained upon repeat testing of specimens that showed more than 500 RNA copies/ml and a CV above 35% were compared to their initial values. Table 2 shows the data for the 141 of 203 specimens that maintained RNA levels above 500 copies/ml after retesting. Specimens displaying a CV between 35 and 50%, of which 40% had RNA levels below 5,000 copies/ml, showed a mean difference of 0.13 log10, with a standard deviation (SD) of 0.11 (Table 2). This value is similar to that observed for specimens showing a CV below 35% (Table 1). An increase in the proportion of specimens displaying more than a 0.3-log10 difference upon repeat testing was observed for those showing a CV above 50%. Of the 62 (30.5%) specimens whose RNA levels became undetectable upon repeat testing, 53 (85.5%), 7 (11.3%), and 2 (3.2%) displayed initial values between 500 and 1,499, 1,500 and 2,499, and 2,500 and 3,499 RNA copies/ml, respectively.
TABLE 2.
Comparison of initial and repeat tests for specimens displaying initial values of >500 RNA copies/ml and a CV of >35%
| %CV | No. of specimens | Log10 difference (mean ± SD) | No. (%) showing a difference of:
|
||
|---|---|---|---|---|---|
| <0.3 log10 | ≥0.3–0.5 log10 | ≥0.5 log10 | |||
| 35–49 | 93 | 0.13 ± 0.11 | 87 (93.5) | 5 (5.4) | 1 (1.1) |
| 50–69 | 37 | 0.22 ± 0.13 | 26 (70.3) | 11 (29.7) | 0 |
| 70–99 | 8 | 0.25 ± 0.15 | 5 (62.5) | 3 (37.5) | 0 |
| 100–125 | 3 | 1.03 ± 0.29 | 0 | 0 | 3 (100) |
Table 3 shows the results obtained upon repeat testing of 280 specimens displaying initial RNA levels below 500 copies/ml and a CV of >35% when the RLU of one of the readings was above the RLU of one of the D standards. The frequency of specimens yielding >500 RNA copies/ml was lower than that observed for specimens with a CV below 35% (6.1 versus 11.4%). A possible explanation is that a CV above 35% is likely to be observed more frequently for specimens with extremely low HIV RNA levels than for specimens with RNA levels just below the 500-copies/ml assay detection limit. Upon repeat testing, the latter specimens are more likely than the former to yield results above 500 copies/ml due to assay characteristics.
TABLE 3.
Analysis of specimens displaying initial values of <500 RNA copies/ml
| RLU values of initial test | No. (%) of samples according to RNA level (copies/ml) after repeat testing
|
|||||
|---|---|---|---|---|---|---|
| CV <35%
|
CV >35%
|
|||||
| <500 | >500 | Total | <500 | >500 | Total | |
| Both above one D Stda RLU | 5 (45.5) | 6 (54.5)b | 11 (15.7) | 17 (60.7) | 11 (39.3)b | 28 (10) |
| Only one above one D Std RLU | 246 (97.6) | 6 (2.4) | 252 (90) | |||
| One or both below one D Std RLU | 57 (96.6) | 2 (3.4) | 59 (84.3) | |||
| Total | 62 (88.6) | 8 (11.4) | 70 (100) | 263 (93.9) | 17 (6.1) | 280 (100) |
Std, standard.
The proportion of specimens showing >500 RNA copies/ml between RLU groupings was significantly different (P < 0.001).
In the bDNA version 2.0 assay, specimens are tested in duplicate. As a consequence, the divergence (%CV) between the two readings could be used to determine when a test should be repeated. However, no information is provided in the manufacturer’s instructions as to what degree of discordance between duplicate readings is acceptable. The data in Table 2 support retesting of specimens displaying >500 RNA copies/ml only when the CV is above 50%. However, the data excludes specimens whose RNA levels became undetectable after repeat testing, most of which (60 of 62) displayed initial RNA levels between 500 and 2,499 copies/ml. Of specimens displaying initial RNA levels between 500 and 2,499 copies/ml and a CV between 35 and 50%, 34.7% showed levels that became undetectable after repeat testing. Therefore, we recommend retesting of specimens with RNA levels between 500 and 2,500 copies/ml when the CV is above 35% and retesting of specimens with >2,500 copies/ml when the CV is >50%. Table 3 shows the results obtained upon retesting of specimens whose levels were initially below the detection limit according to initial RLU values. For these specimens the %CV is not useful in determining if a specimen should be retested. The data indicates that specimens had a tendency to yield >500 RNA copies/ml upon repeat testing only when both of their RLU values were above at least one of the D standard RLU (P < 0.001), irrespective of whether the CV was above or below 35%.
The HIV RNA level is an indicator of disease progression and is monitored for the management of infected patients. The variability and performance of the viral load assays can have an impact on therapeutic decisions. Thus, it is important that clinicians be informed on the reproducibility and performance of the viral load assays. In this study, the reproducibility of the bDNA version 2.0 assay was evaluated at a single site by retesting specimens on different assay runs and lots and in the majority of cases by different operators. This represents the variability likely to be encountered in the day-to-day practice, as most patients are likely to be monitored by the same clinician and to have their viral loads determined in the same laboratory. It remains to be determined if testing at different sites would further affect the quality of results (10). The data presented in this report should guide clinicians on the use of viral load measures obtained by the bDNA version 2.0 assay in patient management.
In conclusion, the bDNA version 2.0 assay was found to be a reproducible and efficient test for routine quantification of HIV-1 RNA in plasma. It is noteworthy that 41.6% of specimens tested in this study showed RNA levels below 500 copies/ml. An assay with a lower detection limit would thus provide more informative data for these specimens.
Acknowledgments
We thank Claire Sauvé, Danielle Sasseville, Jasmine Chamberland, and Micheline Lortie for performing the viral load tests and data collection.
REFERENCES
- 1.Carpenter C C J, Fischl M A, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S G, Richman D D, Saag M S, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A for the International AIDS Society-USA. Antiretroviral therapy for HIV infection in 1996. JAMA. 1996;276:146–154. [PubMed] [Google Scholar]
- 2.Deeks S G, Coleman R L, White R, Pachl C, Schambelan M, Chernoff D N, Feinberg M B. Variance of plasma human immunodeficiency virus type 1 RNA levels measured by branched DNA within and between days. J Infect Dis. 1997;176:514–517. doi: 10.1086/517278. [DOI] [PubMed] [Google Scholar]
- 3.Kern D, Collins M, Fultz T, Detmer J, Hamren S, Peterkin J J, Sheridan P, Urdea M, White R, Yeghiazarian T, Todd J. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3196–3202. doi: 10.1128/jcm.34.12.3196-3202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin H J, Pedneault L, Hollinger F B. Intra-assay performance characteristics of five assays for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1998;36:835–839. doi: 10.1128/jcm.36.3.835-839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien W A, Hartigan P M, Martin D, Esinhart J, Hill A, Benoit S, Rubin M, Simberkoff M S, Hamilton J D the Veterans Affairs Cooperative Study Group on AIDS. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. N Engl J Med. 1996;334:426–431. doi: 10.1056/NEJM199602153340703. [DOI] [PubMed] [Google Scholar]
- 7.Revets H, Marissens D, De Wit S, Lacor P, Clumeck N, Lauwers S, Zissis G. Comparative evaluation of NASBA HIV-1 RNA QT, AMPLICOR-HIV Monitor, and QUANTIPLEX HIV RNA assay, three methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:1058–1064. doi: 10.1128/jcm.34.5.1058-1064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saag M S, Holodniy M, Kuritzkes D R, O’Brien W A, Coombs R, Poscher M E, Jacobsen D M, Shaw G M, Richman D D, Volberding P A. HIV viral load markers in clinical practice. Nat Med. 1996;2:625–629. doi: 10.1038/nm0696-625. [DOI] [PubMed] [Google Scholar]
- 9.Schuurman R, Descamps D, Weverling G J, Kaye S, Tijnagel J, Williams I, van Leeuwen R, Tedder R, Boucher C A B, Brun-Vezinet F, Loveday C. Multicenter comparison of three commercial methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3016–3022. doi: 10.1128/jcm.34.12.3016-3022.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yen-Lieberman B, Brambilla D, Jackson B, Bremer J, Coombs R, Cronin M, Herman S, Katzenstein D, Leung S, Lin H J, Palumbo P, Rasheed S, Todd J, Vahey M, Reichelderfer P. Evaluation of a quality assurance program for quantitation of human immunodeficiency virus type 1 RNA in plasma by the AIDS Clinical Trials Group virology laboratories. J Clin Microbiol. 1996;34:2695–2701. doi: 10.1128/jcm.34.11.2695-2701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]