Abstract
Grb10 has been described as a cellular partner of several receptor tyrosine kinases, including the insulin receptor (IR) and the insulin-like growth factor I (IGF-I) receptor (IGF-IR). Its cellular role is still unclear and a positive as well as an inhibitory role in mitogenesis depending on the cell context has been implicated. We have tested other mitogenic receptor tyrosine kinases as putative Grb10 partners and have identified the activated forms of platelet-derived growth factor (PDGF) receptor β (PDGFRβ), hepatocyte growth factor receptor (Met), and fibroblast growth factor receptor as candidates. We have mapped Y771 as a PDFGRβ site that is involved in the association with Grb10 via its SH2 domain. We have further investigated the putative role of Grb10 in mitogenesis with four independent experimental strategies and found that all consistently suggested a role as a positive, stimulatory signaling adaptor in normal fibroblasts. (i) Complete Grb10 expression from cDNA with an ecdysone-regulated transient expression system stimulated PDGF-BB-, IGF-I, and insulin- but not epidermal growth factor (EGF)-induced DNA synthesis in an ecdysone dose-responsive fashion. (ii) Microinjection of the (dominant-negative) Grb10 SH2 domain interfered with PDGF-BB- and insulin-induced DNA synthesis. (iii) Alternative experiments were based on cell-permeable fusion peptides with the Drosophila antennapedia homeodomain which effectively traverse the plasma membrane of cultured cells. A cell-permeable Grb10 SH2 domain similarly interfered with PDGF-BB-, IGF-I-, and insulin-induced DNA synthesis. In contrast, a cell-permeable Grb10 Pro-rich putative SH3 domain binding region interfered with IGF-I- and insulin- but not with PDGF-BB- or EGF-induced DNA synthesis. (iv) Transient overexpression of complete Grb10 increased whereas cell-permeable Grb10 SH2 domain fusion peptides substantially decreased the cell proliferation rate (as measured by cell numbers) in normal fibroblasts. These experimental strategies independently suggest that Grb10 functions as a positive, stimulatory, mitogenic signaling adapter in PDGF-BB, IGF-I, and insulin action. This function appears to involve the Grb10 SH2 domain, a novel sequence termed BPS, and the Pro-rich putative SH3 domain binding region in IGF-I- and insulin-mediated mitogenesis. In contrast, PDGF-BB-mediated mitogenesis appears to depend on the SH2 but not on the Pro-rich region and may involve other, unidentified Grb10 domains. Distinct protein domains may help to define specific Grb10 functions in different signaling pathways.
The Grb10 family (42) is a recent addition to the group of growth factor receptor binding proteins, which represent a diverse collection of signaling mediators (25, 39). Based on structural similarities, Grb10 belongs to a superfamily of related proteins that include the adapter protein Grb7, which has been implicated in breast cancer as a partner of the epidermal growth factor (EGF) receptor (EGFR) family, in particular of HER2/erbB-2 (26, 47). Grb7 has also been shown to associate with the activated platelet-derived growth factor (PDGF) receptors α and β (PDGFRα and -β) in vitro and in vivo in a complex which may also involve the signaling mediator Shc (55). The superfamily includes Grb14, which is overexpressed in certain human cancer cell lines and for which a possible role has been suggested in PDGF signaling as a substrate of a PDGF-regulated serine kinase (6) as well as in insulin action (18). An additional member is the expression product of the Caenorhabditis elegans gene mig-10, which has been implicated in the development of the excretory canal (24).
The Grb10 superfamily members share a Pro-rich putative SH3 domain binding region at the amino terminus, a 300-amino-acid (aa) region termed GM (Grb/Mig), which contains a pleckstrin homology (PH) region at the center and a newly defined BPS (or IPS) domain between the PH and SH2 domains (11, 15). All members carry an SH2 domain at the carboxyl terminus except Mig-10, which contains a Pro-rich region instead (24). Both the SH2 and BPS domains have been implicated in the association with receptor tyrosine kinases (11, 15). Sequence variants have been reported for most superfamily members (1, 24, 49). Grb10, including a second variant, was first identified in mice, but neither variant has been identified in humans (20, 39). Additional identified human variants have been termed Grb-IR or hGrb10α, GRB10/IR-SV1 or hGRB10β, hGrb10γ, and hGrb10δ (10, 13, 21, 38). This preliminary and inconsistent nomenclature is now being replaced by an emerging consensus (5, 42).
Initially, Grb10 was discovered as a partner of the EGFR (39) and of the Ret receptor tyrosine kinase, which has been implicated in the development of the enteric nervous, endocrine, and renal systems and in papillary thyroid cancer (12, 40). Grb10 has been shown to associate with the insulin receptor (IR) (13, 14, 21) and the insulin-like growth factor I (IGF-I) receptor (IGF-IR) (6, 9, 10, 30), which carry out important metabolic and mitogenic functions, respectively. A preference was observed for the IR in a direct comparison (20). Grb10 has also been identified as a target of the Eph-related receptor tyrosine kinase ELK, which is involved in axonal guidance, neuronal bundling, and angiogenesis (48), and as a target of the growth hormone receptor (33). The interaction of Grb10 with the IR and IGF-IR was investigated in a number of independent studies (22). Different approaches have implicated different receptor sites in the association with Grb10, and the underlying mechanism is still unclear (8–11, 13, 14, 20, 30, 35, 38).
A role of Grb10 has been implicated in mitogenesis, positive or negative, depending on the specific cellular context in response to insulin or IGF-I (31, 38). In addition, it affects the transformed phenotype in IGF-IR- or Bcr-Abl-mediated malignant cell transformation (2, 31). At a more mechanistic level, Grb10 overexpression has been reported to negatively regulate tyrosine phosphorylation of GTPase-activated protein (GAP)-associated protein p60, IR substrate IRS-1, and phosphatidylinositol 3′-kinase (21). Grb10 associates with Jak2 and interferes with growth hormone-mediated gene expression independently of Stat5 (33). The Grb10 SH2 domain has been reported to associate, independently of phosphotyrosine, with Raf1 constitutively and with MEK1 in response to insulin (35). The oncogenic tyrosine kinase Bcr-Abl associates with Grb10 in a phosphotyrosine-dependent fashion (2). Basal phosphorylation on serine has been reported for Grb10 which was stimulated in response to EGF; similarly, PDGF and fibroblast growth factor (FGF) caused a mobility shift in the migration of Grb10 on sodium dodecyl sulfate (SDS) gels which was reversible by phosphatase treatment (39). Basal serine phosphorylation of one isoform was also stimulated by insulin, which was reversible by phosphatase, the MEK1 inhibitor PD98059, or the phosphatidylinositol 3′-kinase inhibitor wortmannin (10). Grb10 has been described as a direct substrate of the Tec tyrosine kinase, which represents the first evidence for a role of phosphorylation on tyrosine in Grb10 function and provides a potential link to cytokine action in the hematopoietic system (23). The formation of Grb10 tetramers has been suggested for the inactive state involving its BPS, SH2, and PH domains, which may convert to a monomeric form in the activated state (11). Combined, the available data suggest a role of Grb10 in the mitogenic signaling pathways downstream of several growth factor receptors.
To further elucidate the physiologic role of Grb10, we have tested other mitogenic receptor tyrosine kinases as putative Grb10 partners and have identified the activated forms of PDGFRβ, hepatocyte growth factor receptor (HGFR) (Met), and FGF receptor (FGFR) as candidates. We have mapped Y771 as a PDGFRβ site that is involved in the association with Grb10 via its SH2 domain. We have further investigated the putative role of Grb10 in mitogenesis with four independent experimental strategies and found that all consistently suggested a role as a positive, stimulatory signaling adapter in normal fibroblasts. (i) Complete Grb10 expression from cDNA with an ecdysone-regulated transient expression system stimulated PDGF-BB-, IGF-I-, and insulin-induced, but not EGF-induced, DNA synthesis in an ecdysone dose-responsive fashion. (ii) Microinjection of the (dominant-negative) Grb10 SH2 domain interfered with PDGF-BB- and insulin-induced DNA synthesis. (iii) Alternative experiments were based on cell-permeable fusion peptides with the Drosophila antennapedia homeodomain which effectively traverse the plasma membrane of cultured cells. A cell-permeable Grb10 SH2 domain similarly interfered with PDGF-BB-, IGF-I-, and insulin-induced DNA synthesis. In contrast, a cell-permeable Grb10 Pro-rich putative SH3 domain binding region interfered with IGF-I- and insulin- but not PDGF-BB- or EGF-induced DNA synthesis. (iv) Transient overexpression of complete Grb10 increased whereas cell-permeable Grb10 SH2 domain fusion peptides substantially decreased the cell proliferation rate (as measured by cell numbers) in normal fibroblasts.
These experimental strategies independently suggest that Grb10 functions as a positive, stimulatory, mitogenic signaling adaptor in PDGF-BB, IGF-I, and insulin action. This function appears to involve the Grb10 SH2 domain, a novel sequence termed BPS, and the Pro-rich putative SH3 domain binding region in IGF-I- and in insulin-mediated mitogenesis. In contrast, PDGF-BB-mediated mitogenesis appears to depend on the SH2 but not on the Pro-rich region and may involve other, unidentified Grb10 domains. Distinct protein domains may help to define specific Grb10 functions in different signaling pathways.
MATERIALS AND METHODS
Antibodies and peptides.
Rabbit polyclonal antibodies directed against the cytoplasmic domains of PDGFRβ, IGF-IR, or IR were obtained from Upstate Biotechnology, rabbit polyclonal Grb10 antibody directed against peptide AWRNGSTRMNILSSQSPL at the mouse Grb10 carboxyl terminus (used for Fig. 1 and 2C) was from Santa Cruz Biotechnology, monoclonal antiphosphotyrosine antibody PY20 was from Transduction Laboratories, mouse monoclonal 3′-bromo-5′-deoxyuridine (BrdU) antibody was from Amersham, and horseradish peroxidase-coupled anti-immunoglobulin G (IgG) antibody was from Kirkegaard & Perry Laboratories. Additional Grb10 antiserum (used for Fig. 7A) was produced by Hazelton Research Products Inc. (Denver, Pa.) in rabbits against a glutathione S-transferase (GST) fusion protein containing the SH2 domain of mouse Grb10 (14). Human recombinant PDGF-BB, IGF-I, and insulin were obtained from Upstate Biotechnology. Synthetic peptides were obtained from American Peptide Company, Inc. (Sunnyvale, Calif.) or as gifts from Lewis C. Cantley and his collaborators (Harvard Medical School, Boston, Mass.). Synthetic peptides representing the 16-aa cell-permeable sequence of the Drosophila antennapedia homeodomain protein (RQIKIWFQNRRMKWKK) and fusion peptides of this domain with a 16-aa fragment representing the amino-terminal Pro-rich region of Grb10 (RQIKIWFQNRRMKWKK-TASLPAIPNPFPELTG) were used to interfere with DNA synthesis. Phosphopeptides representing IR pY960 (phosphotyrosine 960) (SSNPEpYLSASD), pY1146 (DIpYETDYYRKG), pY1150 (DIYETDpYYRKG), pY1316 (KRSpYEEHIPY), pY1322 (HIPpYTHMNGG), or PDGFRβ pY716 and pY771 (SSNpYMAPYDNY) at a concentration of approximately 1 mM were mixed with the GST-Grb10 SH2 domain fusion protein to study their competition with receptor binding.
FIG. 1.
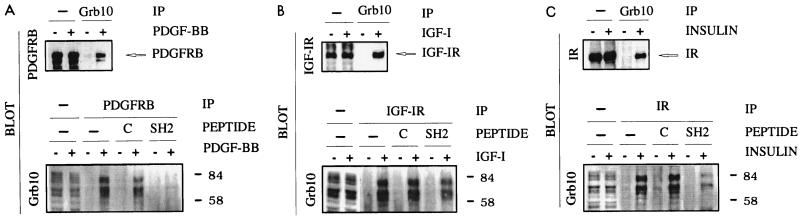
Association of cellular Grb10 with PDGFRβ (A), IGF-IR (B), and IR (C). Normal NIH 3T3 fibroblasts (A), IGF-IR-overexpressing mouse fibroblasts (R+) (B), or IR-overexpressing NIH 3T3 fibroblasts (C) were starved and stimulated (+) with the indicated ligand (PDGF-BB, IGF-I, or insulin) or were left untreated (-). Detergent cell lysates were immediately separated by SDS-PAGE (—) or were first incubated (PEPTIDE) with purified cell-permeable Grb10 SH2 domain fusion peptide (SH2) or with control eluate (C) or were left untreated (—) and immunoprecipitated (IP) with Grb10-specific (Grb10) or receptor-specific (PDGFRB, IGF-IR, or IR) antibodies prior to SDS-PAGE. Proteins were transferred to nitrocellulose and identified by immunoblotting (BLOT) with specific antibodies directed against Grb10, PDGFRβ, IGF-IR, or IR. Positions of molecular markers in kilodaltons and positions of PDGFRβ (PDGFRB), IGF-IR, and IR are indicated on the right.
FIG. 2.
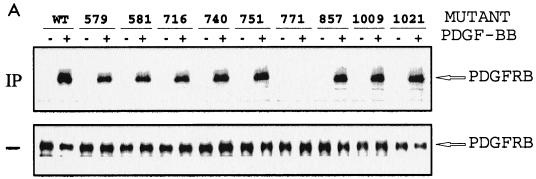
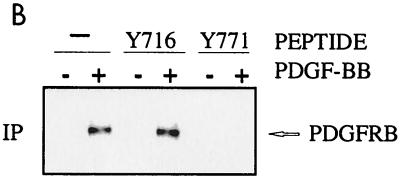
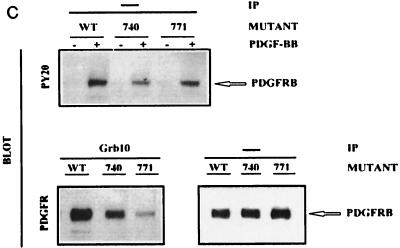
(A and B) Mapping of Grb10 SH2 domain binding to PDGFRβ phosphotyrosine 771. TRMP canine kidney epithelial cells overexpressing wild-type (WT) PDGFRβ or mutations Y579F, Y581F, Y716F, Y740F, Y751F, Y771F, Y857F, Y1009F, or Y1021F (A) or wild-type PDGFRβ (19) (B) were stimulated with 25 ng of PDGF-BB per ml (+) or were left untreated (-) after starvation. (A) Detergent cell lysates were immediately separated by SDS PAGE (—) or were first incubated (IP) with GST-Grb10 SH2 domain fusion protein alone or with phosphopeptides representing major autophosphorylation motif (Y716 or Y771) of PDGFRβ (B and C). Complexes were precipitated with glutathione-Sepharose prior to SDS-PAGE. Proteins were transferred to nitrocellulose and identified by immunoblotting with PDGFRβ antibody. The position of the receptor (PDGFRB) is indicated on the right. (C) Mutation Y771F interferes with the association between PDGFRβ and complete cellular Grb10. TRMP canine kidney epithelial cells overexpressing wild-type (WT) PDGFRβ or mutation Y740F or Y771F were starved and stimulated with 50 ng of PDGF-BB per ml (+ or unlabeled) or left untreated (-). Detergent cell lysates were immediately separated by SDS-PAGE (—) or were first immunoprecipitated (IP) with Grb10-specific antibody (Grb10) prior to SDS-PAGE. Proteins were transferred to nitrocellulose and identified by immunoblotting (BLOT) with specific antibodies directed against phosphotyrosine (PY20) or PDGFRβ (PDGFR). The position of the receptor (PDGFRB) is indicated on the right.
FIG. 7.
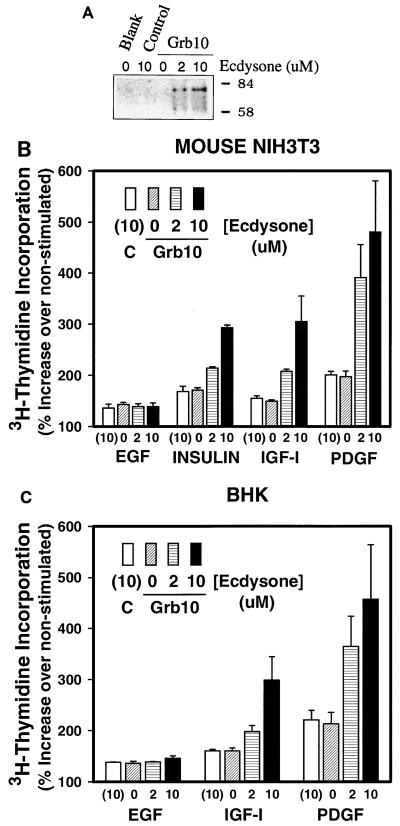
Ecdysone-regulated Grb10 expression results in ecdysone dose-responsive stimulation of PDGF-BB-, IGF-, or insulin-mediated DNA synthesis. Mouse NIH 3T3 fibroblasts or BHK fibroblasts were cotransfected with 1 μg of each plasmid of the Grb10 expression system per ml or with a control expression plasmid (Control) for 18 h in the presence of Lipofectamine or were left untreated (Blank). After a 5-h recovery in complete medium, cells were incubated with the indicated concentrations of ecdysone analog ponasterone A for 39 h. (A) NIH 3T3 detergent cell lysates were analyzed by SDS-PAGE. Proteins were transferred to nitrocellulose and identified by immunoblotting with a specific rabbit antiserum raised against the Grb10 SH2 domain. Positions of migration of size markers are indicated in kilodaltons on the right. (B and C) Cells were starved in low serum for the last 24 h of the induction period and were subsequently incubated with 25 ng of PDGF-BB, 100 ng of IGF-I, 100 ng of insulin, or 100 ng of EGF per ml or were left untreated. Cells were labeled 18 h later with 0.5 μCi of [methyl-3H]thymidine at 3 Tbq/mmol for 5 h, and acid (10% TCA)-precipitated radioactivity was quantified by liquid scintillation spectroscopy. Error bars indicate the variation between duplicate measurements. DNA synthesis in the absence of growth factor has been defined as zero.
GST fusion protein construct and protein purification.
A Grb10 SH2 domain-encoding cDNA fragment of the carboxyl-terminal 108 aa was ligated to the BamHI and EcoRI sites of plasmid pGEX-11T (Pharmacia), using PCR primers 5′-CCGGGATCCATTCACAGGACTCAGCATTG-3′ and 5′-GCCGAATTCTTCTATCTATCTAGCG-3′, and the final construct was confirmed by DNA sequence analysis. The GST-Grb10 SH2 domain fusion and control GST protein were expressed in Escherichia coli DH5α, purified on a glutathione-agarose column (Pharmacia), eluted in 10 mM reduced glutathione in 50 mM Tris-HCl (pH 8.0), and stored after addition of 10 mM dithiothreitol and 1 mM EDTA as described by the manufacturer (14).
Construction and expression of cell-permeable Grb10 SH2 domain fusion peptides.
Optimal E. coli codons were used in the design of oligonucleotides which contained a 5′-trityl group for column purification (Fisher Scientific). Oligonucleotides including the 16-aa cell membrane transfer sequence (RQIKIWFQNRRMKWKK) of the antennapedia homeodomain (5′ GGC GGC AGC CAT ATG CGT CAG ATC AAA ATG TGG TTC CAG AAC CGT CGT ATG AAA TGG AAA AAA GGA TCC 3′ [membrane transfer sequence with 5′ NdeI and 3′ BamHI sites] and 5′ CCC CAA GCT TCA CTT AAT TAA GAG CTC TTA CTG CCA AGA CGG CGG CGG CGG CGG CAG CGC CGG CGG AGA CGG GAA GGA TCC TTT TTT 3′ [Pro-rich sequence with 5′ HindIII and 3′ BamHI sites]) were hybridized via a 12-nucleotide overlap and extended by mutually primed DNA synthesis. NdeI and HindIII restriction sites had been introduced at the 5′ and 3′ ends, respectively. A BamHI restriction site had been introduced at the 3′ end of the membrane transfer sequence. The complete sequence was inserted into the NdeI and HindIII sites of plasmid pET 28a(+) (Novagen, Madison, Wis.). This expression plasmid is based on the strong T7 transcriptional promoter, confers kanamycin resistance, and expresses an amino-terminal 6-aa His-tag peptide for purification of the recombinant protein. The Grb10 SH2 domain was amplified between aa 521 and 621, using PCR primers (5′ TAT TTT GGA TCC TGG TTC CAT GGA CAT ATC TCC CGC 3′ and 5′ TAT TTT AAG CTT TTA CAC GCG GAT GCA GTG GTG TTT CAG 3′) which introduced a 5′ BamHI site and a 3′ HindIII site into the final PCR product. The PCR product was digested with BamHI and HindIII, isolated by electrophoresis followed by electroelution, and inserted into the BamHI and HindIII sites of the pET 28a(+) construct. All recombinant plasmids were transformed, isolated, and characterized by restriction and DNA sequence analyses. The plasmid containing the membrane transfer sequence fused to the Grb10 SH2 domain was introduced into expression host E. coli BL21(DE3), which carries the lacUV5 promoter for induction by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were grown in Luria-Bertani medium containing kanamycin (30 μg/ml) at 37°C to an optical density of 0.2 to 0.3 at 600 nm. IPTG induction was carried out for 5 h before cells were harvested and resuspended in 0.5 M Tris-HCl (pH 6.8). Cells were disrupted by French press (SLM Instruments, Inc.) lysis and centrifuged at 15,000 × g at 4°C for 20 min. The cleared lysate was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) on a 15% gel. The cell-permeable Grb10 fusion peptide was purified by affinity chromatography on a nickel column (Novagen) and eluted with 0.4 M imidazole in the presence of 6 M urea. The peptide was dialyzed by using a molecular mass cutoff of 3.5 kDa (Pierce, Ill.). Precipitation was observed after dialysis, and the peptide was resolubilized by addition of 1% dimethyl sulfoxide to a final concentration of 0.1% in 50 mM Tris-HCl buffer (pH 6.8). The peptide was stored for several months at 4°C, and the protein concentration was determined by Bradford analysis.
Ecdysone-inducible Grb10 cDNA expression.
A 2.5-kb NruI-HindIII restriction fragment containing the complete protein-coding mouse Grb10 cDNA was removed from plasmid pRK5-Grb10 (39) and inserted with adapter sequences into the BamHI and EcoRV sites of plasmid pIND (Invitrogen). The proper ligation product was confirmed by restriction and DNA sequence analyses. This plasmid carries a heat shock transcriptional promoter and ecdysone response elements to allow ecdysone dose-dependent regulation of Grb10 expression. Ponasterone A (Invitrogen), an ecdysone analog, was used as an inducer in all experiments. The expression system employs a second plasmid, pVgRXR, for the expression of the ecdysone receptor from a cytomegalovirus (CMV) promoter which was cotransfected at the same concentration. Subconfluent NIH 3T3 fibroblasts were washed with serum- and antibiotic-free medium and incubated for 5 h at 37°C in 1 ml to which a transfection mix containing 1 μg of each plasmid and 10 μl of Lipofectamine had been added according the instructions of the manufacturer (Life Technologies, Gaithersburg, Md.). Then 1 ml of complete culture medium was added, and cells were incubated for 14 h and subsequently for 5 h in fresh complete medium. Ponasterone A was added at various concentrations for 24 h. For protein analysis by SDS-PAGE and immunoblotting, cells were cultured in fresh complete medium for an additional 24 h in the presence of ponasterone A before being lysed. Alternatively, for subsequent mitogenic analysis, cells were treated as described below.
Thymidine incorporation and interference by cell-permeable peptides.
To subsequently assay for DNA synthesis, cells were starved for 20 h in the presence of ponasterone A. PDGF-BB at 25 ng/ml or IGF-I, insulin, or EGF at 100 ng/ml was added for 18 h, and finally 0.5 μCi of [methyl-3H]thymidine at 3 Tbq/mmol was added for 5 h. Cells were rinsed three times in ice-cold phosphate-buffered saline and incubated in 10% trichloroacetic acid (TCA) for 1 h at 4°C. Cells were rinsed, and 0.5 ml of 0.2 N NaOH–0.1% SDS was added at 37°C for 1 h. The pH was neutralized by addition of 0.5 ml of 2 M Tris (pH 6.8), and 2 ml of cocktail (ScintiSafe Econo-1; Fisher) was added before the incorporated radioactivity was quantified by liquid scintillation spectroscopy.
To test the effect of cell-permeable peptides on DNA synthesis, NIH 3T3 fibroblasts were starved for 20 h. Growth factors were added at the concentrations indicated above in addition to cell-permeable peptides at various concentrations for 18 h; 0.5 μCi of [methyl-3H]thymidine was added for 5 h, and the incorporated radioactivity was determined as described above. Normal fibroblasts were used for all growth factors except for insulin, which was tested in IR-overexpressing fibroblasts (3).
Microneedle injection and BrdU incorporation.
NIH 3T3 fibroblasts overexpressing IR (3) were grown on 12-mm-diameter coverslips and starved for 24 h in serum-free medium; 150 cells were microinjected for each condition with about 10−14 liters of GST fusion protein at 3 μg/μl by using glass microcapillaries (Eppendorf) in 5 mM NaH2PO4–100 mM KCl (pH 7.4) (45). Two hours later, cells were stimulated with PDGF-BB (25 ng/ml), insulin (100 ng/ml), or 10% fetal bovine serum for 16 h in the presence of BrdU. Cells were fixed in 90% ethanol–5% acetic acid for 20 min at 22°C and incubated with BrdU mouse monoclonal antibody for 1 h at 22°C. Cells were stained with fluorescein-labeled donkey anti-mouse IgG antibody and mounted, and indirect fluorescence was analyzed with an Axiovert fluorescence microscope (Zeiss).
Cell culture, immunoprecipitation, and immunoblotting.
Normal fibroblasts or cell lines overexpressing various wild-type or mutant receptors were used as indicated in the figure legends. Cells were incubated in serum-free medium for 16 h and stimulated with PDGF-BB (25 ng/ml), IGF-I (100 ng/ml), or insulin (100 ng/ml) for 15 min. Cells were rinsed twice with PBS and harvested in ice-cold lysis buffer containing 50 mM HEPES (pH 7.4), 1% Triton X-100, 10% glycerol, 137 mM NaCl, 2 mM EDTA, 10 mM NaF, 100 mM Na3VO4, 10 mM sodium pyrophosphate, 10 μg of leupeptin per ml, 10 μg of aprotinin per ml, and 1 mM phenylmethylsulfonyl fluoride. Proteins (typically 25 μg) were directly subjected to SDS-PAGE or (400 μg) were first mixed with cell-permeable Grb10 SH2 domain fusion protein (10 μg), control eluate, or phosphopeptides along with antibodies directed against Grb10, PDGFRβ, IGF-IR, or IR and coprecipitated with glutathione-Sepharose or protein A-Sepharose beads, respectively. Precipitates were washed with lysis buffer, separated by SDS-PAGE (8% gel), and analyzed by immunoblotting with the indicated specific antibodies, using the Amersham ECL (enhanced chemiluminescence) detection system (14).
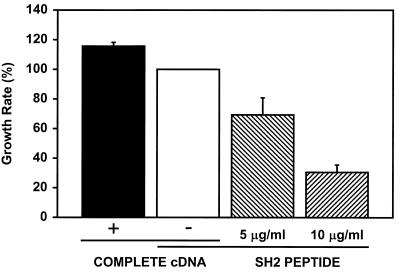
Constitutive Grb10 expression and cell proliferation.
A 2.5-kb NruI-HindIII restriction fragment containing the complete protein-coding mouse Grb10 cDNA was removed from plasmid pRK5-Grb10 (39), and after end filling, the fragment was inserted into the SmaI site of plasmid pHook2 (Invitrogen). The proper ligation product was confirmed by restriction and DNA sequence analyses. This plasmid carries a CMV transcriptional promoter for constitutive expression of mouse Grb10. Subconfluent mouse fibroblasts (R− or NIH 3T3) were rinsed with antibiotic-free medium and incubated for 24 h at 37°C on 60-mm-diameter culture plates in 3 ml of medium to which transfection mix containing 2 μg of Grb10 expression plasmid or the corresponding control plasmid, 12 μl of Lipofectamine, and 8 μl of Plus reagent had been added according to the instructions of the manufacturer (Life Technologies). The transfection medium was subsequently replaced with complete culture medium supplemented with 10% newborn calf serum. Then 70,000 cells of transfected cultures were split into 35-mm-diameter wells (approximately 10% confluent). To test the effect of Grb10 SH2 domain fusion peptide at 5 or 10 μg/ml (or of control eluate) on the cell proliferation rate, equal numbers (∼2,000) of cells were grown in 96-well plates in the presence of purified cell-permeable SH2 domain fusion peptide or an equal volume of control eluate in Dulbecco modified Eagle medium–F-12 medium supplemented with 10% newborn calf serum for 6 days. Cells were either trypsinized and manually counted with a hemocytometer or quantified biochemically by measuring the colorimetic change of 10% 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) into blue-colored formazan by the mitochondrial enzyme succinate dehydrogenase (7, 32).
RESULTS
Association of cellular Grb10 with the activated PDGFRβ, IGF-IR, or IR.
Based on earlier reports which implicated a role of Grb10 in mitogenesis, we first tested the association of Grb10 with PDGFRβ, a major, mitogenic growth factor receptor, in coprecipitation experiments followed by immunoblotting. After starvation, normal NIH 3T3 fibroblasts were stimulated with PDGF-BB before detergent cell lysates were precipitated with a rabbit polyclonal antibody directed against the mouse Grb10 carboxyl terminus (Santa Cruz Biotechnology). Precipitates were washed and separated by SDS-PAGE, and proteins were identified by immunoblotting with a PDGFRβ-specific rabbit polyclonal antibody. One major protein band of 190 kDa and one smaller, minor band were identified (Fig. 1A), compatible with the predicted migration of PDGFRβ (19). The signal was only found in response to PDGF-BB stimulation indicating that cellular Grb10 associates with activated PDGFRβ in a PDGF-BB-dependent fashion (Fig. 1A). Complementary experiments were carried out by immunoprecipitation with PDGFRβ-specific antibody followed by immunoblotting with rabbit polyclonal antibody directed against the mouse Grb10 carboxyl terminus. Grb10 antibodies frequently detect at least three major protein bands that may represent differently processed forms of Grb10 and have repeatedly been reported to range from 65 to 85 kDa (20, 39). Several bands were also observed in our detergent cell lysates with the Grb10 antibody described above (Fig. 1) or with our own raised Grb10 antiserum but not with preimmune serum (not shown). At least three major bands were found to associate with the activated PDGFRβ in a strictly PDGF-BB-dependent fashion (Fig. 1A). Consequently, both complementary experimental strategies show an association of cellular Grb10 with PDGFRβ in response to PDGF-BB stimulation. When we compared the interaction of Grb10 with IGF-IR and IR, which had been demonstrated before (8–10, 13, 20, 21, 30, 38) by using the same experimental strategy, at least three major forms of Grb10 were similarly found to associate with the activated receptors dependent on IGF-I and insulin stimulation in fibroblasts (Fig. 1B and C).
Mapping of Grb10 SH2 domain association to PDGFRβ Y771.
To address whether the Grb10 SH2 domain was sufficient to associate with PDGFRβ, a GST fusion protein representing the carboxyl-terminal 108 aa of Grb10 (14) was prepared and mixed with detergent cell lysates of TRMP canine kidney cells overexpressing PDGFRβ. Complexes were precipitated with glutathione-Sepharose, separated by SDS-PAGE, and identified by immunoblotting with PDGFRβ-specific antibody. The Grb10 SH2 domain was found to effectively associate and coprecipitate with activated PDGFRβ, strictly in response to PDGF-BB stimulation (Fig. 2A).
This approach was extended to define the PDGFRβ motif which is involved in the association with the Grb10 SH2 domain. A series of PDGFRβ point mutants lacking any one of the major tyrosine autophosphorylation sites were compared in detergent cell lysates of overexpressing TRMP canine kidney cells which lack endogenous PDGFRβ. Each lysate was mixed with GST-Grb10 SH2 domain fusion protein, and glutathione-Sepharose precipitates were analyzed by immunoblotting with PDGFRβ-specific antibody. All mutants were found to coprecipitate with the Grb10 fusion protein except for PDGFRβ mutation Y771F (Fig. 2A). These data implicate phosphotyrosine 771 in the association with the Grb10 SH2 domain. Whether this PDGFR motif actually associates with the Grb10 SH2 domain was determined in peptide competition experiments. Phosphopeptides representing Y771 and Y716 as a control were mixed with GST-Grb10 SH2 domain fusion protein in detergent cell extracts to study their impact on PDGFRβ association. Immunoblotting of glutathione-Sepharose precipitates with PDGFRβ-specific antibody demonstrated that the phosphopeptide representing phosphotyrosine 771 competes with PDGFRβ for Grb10 SH2 domain association (Fig. 2B) and confirms a role of the sequences around Y771 in Grb10 binding.
Mutation Y771F interferes with PDGFRβ association with complete cellular Grb10.
To address whether PDGFRβ association with complete cellular Grb10 would be similarly affected by mutation Y771F, we stimulated TRMP cells expressing wild-type and mutant PDGFRβ with ligand and precipitated detergent cell lysates with Grb10 antibody. Proteins were separated by SDS-PAGE, and coprecipitated PDGFRβ was evaluated in immunoblots with PDGFRβ-specific antibody. Compared to wild-type PDGFRβ, mutant Y740F showed reduced but significant association with cellular Grb10 (Fig. 2C), which had also been observed with the Grb10 SH2 domain, where it was similar in reduction to several other PDGFRβ mutants when compared to wild-type PDGFRβ (Fig. 2A). In contrast, mutant Y771F showed only marginally detectable association with cellular Grb10 (Fig. 2C) which had been below the level of detection in experiments with the Grb10 SH2 domain (Fig. 2A). Consequently, Y771F association experiments with cellular Grb10 as well as with the Grb10 SH2 domain indicate an important role of the region around PDGFRβ tyrosine 771 in Grb10 binding.
Differential interference of cell-permeable Grb10 SH2 domain fusion peptides with the association between cellular Grb10 and PDGFRβ, IGF-IR, or IR.
To evaluate the importance of the Grb10 SH2 domain in PDGF binding in a complementary experimental strategy, we tested the effect of competing cell-permeable SH2 domain fusion peptides (described below) on the association between PDGFRβ and cellular Grb10. NIH 3T3 fibroblasts were starved and stimulated with PDGF-BB, and coimmunoprecipitation of cellular Grb10 was assayed in the presence or absence of cell-permeable SH2 domain fusion peptides with PDGFRβ-specific antibody. Immunoblots with Grb10-specific antibody demonstrated that the SH2 domain peptide virtually abolished the association of any form of cellular Grb10 with PDGFRβ (Fig. 1A). In contrast, we observed only a minimal effect of the Grb10 SH2 domain on the association with cellular Grb10 in analogous experiments with activated IGF-IR (Fig. 1B) but a significant reduction for activated IR (Fig. 1C). These findings are fully consistent with earlier studies (15) which reported that the IR associates with the SH2 and BPS domains of Grb10, resulting in high-affinity binding, which explains the significant but partial interference of the SH2 domain (Fig. 1C), whereas the IGF-IR primarily associates with the BPS domain, which explains the observed minimal interference by the SH2 domain (Fig. 1B). In the absence of published studies on the association with PDGFRβ, our results demonstrate the importance of the Grb10 SH2 domain, which essentially abolished binding of cellular Grb10 (Fig. 1A), in line with results of the Grb10 SH2 domain association studies shown in Fig. 2.
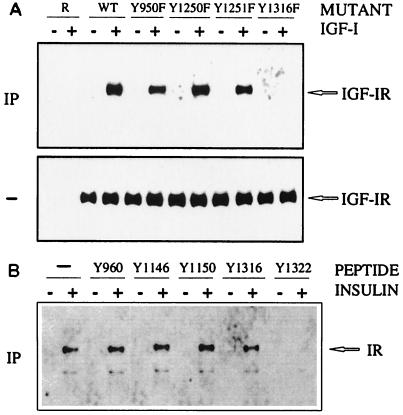
Mapping of Grb10 SH2 domain association to IGF-IR Y1316/IR Y1322.
In a complementary approach, sequences of the IGF-IR which are involved in the association with the Grb10 SH2 domain were characterized by receptor mutant analysis. Tyrosine mutation Y1316F at the carboxyl terminus abolished the association with the Grb10 SH2 domain, whereas mutations in the juxtamembrane and activation loop regions of the IGF-IR did not interfere (Fig. 3A). This finding is compatible with an analysis of the homologous IR motif Y1322, which had been implicated in the association with Grb10 in our earlier studies by receptor mutation and phosphopeptide binding analysis (14). Phosphopeptides representing major IGF-IR autophosphorylation motifs were not available for our studies. Instead phosphopeptides representing various major IR autophosphorylation sites were mixed with GST-Grb10 SH2 domain fusion protein in detergent cell extracts to study their impact on IR association. Of the tested motifs, only Y1322 (which is homologous to IGF-IR Y1316 [50]) interfered with IR binding of the Grb10 SH2 domain (Fig. 3B). Other tested phosphotyrosine motifs in the carboxyl-terminal, juxtamembrane, or activation loop region were not found to interfere with Grb10 SH2 domain binding. These data implicate homologous sites at the carboxyl termini of the IGF-I and insulin receptors in the association with Grb10. Our results demonstrate that the Grb10 SH2 domain binds to both IR and IGF-IR, while this may only represent a minor aspect of the Grb10 association with IGF-IR (Fig. 1B) as reported previously (15).
FIG. 3.
Mapping of Grb10 SH2 domain association to IGF-IR Y1316/IR Y1322. R− mouse embryo fibroblasts carrying an IGF-IR gene disruption and overexpressing wild-type (WT) IGF-IR or IGF-IR mutations Y950F, Y1250F, Y1251F, and Y1316F (27, 28) (A) or CHO cells overexpressing wild-type IR (34) (B) were stimulated (+) with the indicated ligand (IGF-I or insulin) or were left untreated (-). Detergent cell extracts were immediately separated by SDS-PAGE (-) (A) or were incubated for coimmunoprecipitation (IP) with GST-Grb10 SH2 domain fusion protein alone or with phosphopeptides representing major autophosphorylation motifs of IR (as indicated by Y positions) (B). Complexes were precipitated with glutathione-Sepharose prior to SDS-PAGE. Proteins were transferred to nitrocellulose and identified by immunoblotting with antibodies directed against IGF-IR (A) or IR (B). Positions of IGF-IR and IR are marked on the right.
Association of other activated receptor tyrosine kinases with the Grb10 SH2 domain.
Many signaling adapters are shared among a number of receptor tyrosine kinases, which encouraged us to test whether Grb10 associates with other growth factor receptors. We evaluated one member of most receptor tyrosine kinase subfamilies (52) and compared activated and nonactivated receptors in most cases. Of those examples tested, a clear association with the Grb10 SH2 domain was observed for PDGFRβ, HGFR, and FGFR (Fig. 4) and as described earlier for IR and IGF-IR (8–10, 13, 14, 20, 21, 30, 38). In contrast, an association could not be demonstrated for nerve growth factor (NGF) receptor (TrkA) or EGFR (Fig. 4). The presence of all receptors in cell lysates and their activation had been demonstrated with receptor-specific antibodies and phosphotyrosine-specific antibodies, respectively. These results suggest that the assay used can specifically discriminate between different receptor tyrosine kinase interactions and are compatible with a potential role of Grb10 as a general mitogenic signaling adapter.
FIG. 4.
Association of activated receptor tyrosine kinases with the Grb10 SH2 domain. Cell lines overexpressing the various indicated receptor tyrosine kinases were incubated (+) with specific ligand (Activated) or were left untreated (-). For FGFR, only constitutively activated samples (+) were available. Detergent extracts were immediately separated by SDS-PAGE (Lysate) or were first mixed with control GST (—) or GST-Grb10 SH2 domain fusion protein, and complexes were precipitated with glutathione-Sepharose prior to SDS-PAGE. Proteins were transferred to nitrocellulose and identified by immunoblotting with phosphotyrosine antibodies (pTyr) or with specific antibodies directed against the respective receptor tyrosine kinase (Receptor-specific). Protein bands are identified as the respective receptor types shown on the left. IGF-IR was overexpressed in IGF-IR gene-disrupted R− mouse embryo fibroblasts (27, 28) that had been stimulated with IGF-I. IR was overexpressed in CHO cells that had been stimulated with insulin (34). PDGFRβ was overexpressed in TRMP canine epithelial kidney cells that had been stimulated with PDGF-BB (19). HGFR/c-Met experiments were performed with constitutively activated v-Met (+) and a kinase-inactivated mutant of v-Met (43) (-). Constitutively activated (+) Xenopus (type 1, two-Ig-loop) FGFR had been expressed with the baculovirus system in Sf9 insect cells; nonstimulated forms were not available (indicated for one lane [na]). EGFR was overexpressed in A431 human epidermoid carcinoma cells that had been stimulated with EGF. TrkA-overexpressing NIH 3T3 cells had been stimulated with 7S NGF. The receptor tyrosine kinases (52) were gifts from various colleagues as indicated in the text.
Microinjection of dominant-negative GST-Grb10 SH2 domain interferes with growth factor-mediated DNA synthesis.
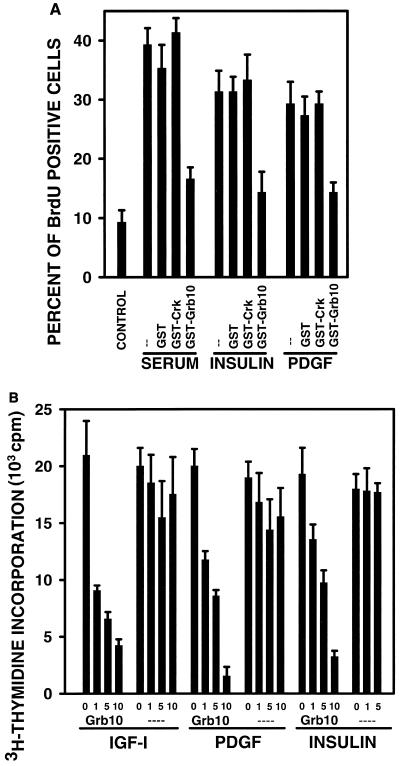
To demonstrate a role of Grb10 in mitogenesis specifically, we used four independent experimental strategies to interfere with the function of cellular Grb10 or to enhance the function of Grb10 by overexpression. First, we used the GST-Grb10 SH2 domain fusion protein in a microinjection approach (45). It is expected that the SH2 domain will act like a dominant-negative form of Grb10 when introduced into cells at high concentration (37). NIH 3T3 fibroblasts overexpressing human IR were starved and stimulated with various growth factors, and GST-Grb10 SH2 domain fusion protein was microinjected. The effect on DNA synthesis was evaluated at the single-cell level by fluorescence microscopy analysis of incorporated BrdU. Fetal bovine serum, insulin, and PDGF-BB all stimulated DNA synthesis, as judged by the increased number of fluorescence-positive cells (Fig. 5A). This remained unaffected by microinjection of GST protein alone or a GST fusion protein with the Crk SH2 domain, which served as a control for the specificity of the assay (3). Microinjection of the GST-Grb10 SH2 domain fusion protein typically reduced DNA synthesis to less than 50% of growth factor-stimulated levels (Fig. 5A). The significant mitogenic response to insulin, which exceeds the response to PDGF-BB in this experiment, is explained by the overexpressed human IR levels, whereas normal NIH 3T3 fibroblasts did not significantly respond to insulin (not shown).
FIG. 5.
(A) Microinjection of dominant-negative GST-Grb10 SH2 domain interferes with growth factor-mediated DNA synthesis. IR-overexpressing mouse NIH 3T3 fibroblasts were starved in 0.1% fetal bovine serum for 16 h, control GST (GST), GST-Crk SH2, or GST-Grb10 SH2 domain fusion proteins were microinjected into 150 cells per sample, and cells were stimulated with 10% serum, insulin, or PDGF-BB for 16 h (--, noninjected controls). BrdU incorporation was visualized by indirect immunofluorescence. Fluorescence-positive cells were counted, and the mitogenic response is presented as the fraction of positive cells in response to growth factor stimulation. Basal BrdU incorporation in the absence of growth factors is shown on the left (CONTROL). The means of three independent experiments are shown; variations are indicated by error bars. (B) Cell-permeable fusion peptides of the Grb10 SH2 domain interfere with PDGF-BB-, IGF-I, and insulin-mediated DNA synthesis. Mouse NIH 3T3 fibroblasts were starved in 0.1% serum-containing medium for 20 h. Cells were subsequently incubated either with cell-permeable Grb10 SH2 domain fusion peptide purified from E. coli (Grb10) at concentrations of 0, 1, 5, and 10 μg/ml or with a purified control sample (----) lacking fusion peptide (see Materials and Methods). In parallel, cells were stimulated with either IGF-I (100 ng/ml), PDGF-BB (25 ng/ml), or insulin (100 ng/ml) for 18 h; 0.5 μCi of [methyl-3H]thymidine at 3 Tbq/mmol was then added for 5 h, and acid (10% TCA)-precipitated radioactivity was quantified by scintillation spectroscopy. Error bars indicate the variation between triplicate measurements.
Cell-permeable fusion peptides of the Grb10 SH2 domain interfere with PDGF-BB, IGF-I-, and insulin-mediated DNA synthesis.
In a second experimental strategy, we introduced the Grb10 SH2 domain into NIH 3T3 fibroblasts with an alternative technique and measured DNA synthesis with a different assay (Fig. 5B). We used cell-permeable peptides including a 16-aa motif of the Drosophila antennapedia homeodomain protein, which effectively transfers fusion peptides of up to 100 aa in length into any cell type tested (41) and allowed us to interfere with mitogenesis in large numbers of cultured cells. A cDNA encoding the Grb10 SH2 domain was isolated by PCR, fused to a sequence encoding a cell-permeable 16-aa peptide, and introduced into an E. coli expression plasmid under T7 transcriptional promoter control [pET 28(a+); Novagen]. Protein expression was induced, the fusion peptide was purified by nickel column affinity chromatography, dialyzed, and resolubilized. NIH 3T3 fibroblasts were starved, stimulated with PDGF-BB, IGF-I, or insulin, and purified cell-permeable Grb10 SH2 domain fusion peptide was added. DNA synthesis was evaluated by the quantification of incorporated [3H]thymidine. For all growth factors tested, we observed a substantial reduction in DNA synthesis in a fusion peptide dose-responsive fashion (Fig. 5B). Control isolates lacking fusion peptides did not alter DNA synthesis significantly (Fig. 5B). These data fully support the results obtained with GST-Grb10 SH2 domain fusion protein by the microinjection approach (Fig. 5A).
Cell-permeable fusion peptides of the Grb10 Pro-rich region interfere with IGF-I- and insulin- but not PDGF-BB-mediated DNA synthesis.
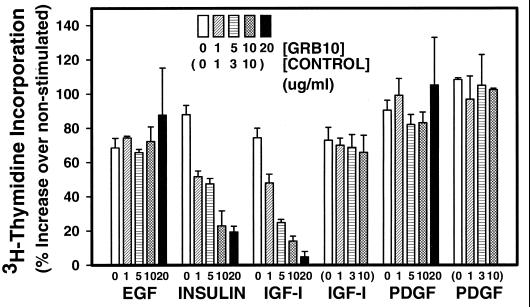
If Grb10 functions as a signaling adapter, we reasoned that other Grb10 domains should participate in the mitogenic response. We tested the Pro-rich amino-terminal putative SH3 domain binding region of Grb10 in a similar experimental strategy involving synthetic cell-permeable fusion peptides. Efforts to express this short domain in E. coli as described above had not been successful. NIH 3T3 fibroblasts were starved and stimulated with various growth factors, and synthetic cell-permeable fusion peptides with a 16-aa fragment of the Grb10 amino-terminal Pro-rich region were added. DNA synthesis was evaluated by the quantification of incorporated [3H]thymidine. Most growth factors stimulated DNA synthesis up to twofold. For control cell-permeable peptides lacking any Grb10 domain, DNA synthesis was not significantly altered at any tested concentration of peptide (Fig. 6). Similarly, EGF-stimulated DNA synthesis was not significantly reduced by increasing doses of fusion peptide, compatible with the lack of evidence for a role of Grb10 in EGF mitogenic action (Fig. 4), which remains to be resolved (39). Increasing doses of fusion peptide resulted in a dose-dependent decrease in DNA synthesis in NIH 3T3 fibroblasts in response to insulin (IR overexpressing) and IGF-I (normal) down to as little as 10% of stimulated levels (Fig. 6). These data suggest that the Grb10 Pro-rich region participates in IGF-I and insulin action, possibly by interacting with the SH3 domain of an unidentified signaling mediator. All experimental strategies used, microinjection or introduction of cell-permeable peptides of the Grb10 SH2 domain and introduction of a cell-permeable Pro-rich region, are expected to result in dominant-negative forms of Grb10 (37). All strategies were shown to interfere with DNA synthesis, which suggests that complete cellular Grb10 functions as a positive, stimulatory, mitogenic signaling adapter.
FIG. 6.
Cell-permeable fusion peptides of the Grb10 Pro-rich region interfere with IGF-I- and insulin-mediated, but not PDGF-BB-mediated, DNA synthesis. Mouse NIH 3T3 fibroblasts were starved in 0.5% serum-containing medium for 24 h, incubated with 0, 1, 5, 10, or 20 μg of the Grb10 fusion peptide (RQIKIWFQNRRMKWKK-TASLPAIPNPFPELTG) per ml or with 0, 1, 3, or 10 μg of a control peptide (RQIKIWFQNRRMKWKK) per ml (in parentheses), and stimulated with 25 ng of PDGF-BB, 100 ng of IGF-I, 100 ng of insulin, or 100 ng of EGF per ml or left untreated for 18 h; 0.5 μCi of [methyl-3H]thymidine was then added for 5 h, and acid (10% TCA)-precipitated radioactivity was quantified by scintillation spectroscopy. The data presented are based on three independent experiments. Error bars indicate the variation between duplicate measurements of the representative experiment shown. DNA synthesis in the absence of growth factor has been defined as zero.
For PDGF-BB-mediated DNA synthesis, no significant reduction was observed with increasing doses of fusion peptide (Fig. 6). This result suggests that in contrast to the SH2 domain of Grb10 (Fig. 5), the Pro-rich region may not participate in PDGF-BB-mediated mitogenesis.
Ecdysone-regulated Grb10 expression results in ecdysone dose-responsive stimulation of PDGF-BB-, IGF-I-, or insulin-mediated DNA synthesis.
If Grb10 represents a positive mitogenic signaling mediator, increased expression should stimulate DNA synthesis. To regulate Grb10 expression, we used an ecdysone-controlled cDNA expression system which was introduced into fibroblasts by lipofection. In response to increasing doses of ecdysone, increased expression of several protein bands was reproducibly observed in immunoblots with Grb10 rabbit polyclonal antiserum compared to basal levels in the absence of ecdysone or in cells which had been transfected with a control plasmid or remained untransfected (Fig. 7A). Background protein levels are not visible due to the limited cell numbers used in the transfection assay. Overall, the expressed proteins migrated similarly to those observed in untransfected cells at higher concentrations of cell lysate (Fig. 1); however, differences likely exist.
To address the impact of increased Grb10 expression levels on mitogenesis, NIH 3T3 or baby hamster kidney (BHK) fibroblasts were transfected with the expression system, starved, and stimulated with various growth factors, and Grb10 expression was induced by varying doses of ecdysone. DNA synthesis was quantified through the incorporation of [3H]thymidine. All growth factors were found to stimulate DNA synthesis between 2.5- and 3-fold (Fig. 7B). Increasing doses of ecdysone led to a dose-dependent increase in DNA synthesis for all growth factors except EGF, confirming results which failed to provide evidence for a role of Grb10 in EGF mitogenic action (Fig. 4 and 6). Maximum DNA synthesis levels were elevated more than twofold by the highest dose of ecdysone compared to noninduced levels for PDGF-BB, which also typically showed the highest mitogenic response (Fig. 7B). The significant insulin stimulation of DNA synthesis is explained by the highly elevated levels of human IR in the NIH 3T3 fibroblasts used for this peptide hormone (3). In the absence of ecdysone or after ecdysone induction of control plasmids, even at the highest dose, no change in DNA synthesis was observed. These experiments directly show a role of Grb10 as a positive mitogenic signaling adapter (Fig. 7B), a finding supported by the interference with DNA synthesis demonstrated by various dominant-negative experimental strategies (Fig. 5 and 6).
Overexpression of complete Grb10 or interference (by cell-permeable SH2 domain fusion peptides) with cellular Grb10 proportionally increases or decreases the cell proliferation rate, respectively.
To expand these studies, we carried out additional experiments to formally address the effect of Grb10 on cell proliferation. For this purpose, mouse fibroblasts (NIH 3T3 or R−) were transfected with a constitutive CMV promoter-controlled Grb10 expression plasmid and replated at low density. Cell proliferation was monitored by determining cell numbers daily by cell counts or by measuring metabolic activity, which correlates with cell numbers, colorimetrically (MTT assay). Alternatively, cell-permeable peptides with the Grb10 SH2 domain were tested for their effects on the cell proliferation rate. As shown in Fig. 8, Grb10 overexpression increased whereas introduction of the Grb10 SH2 domain substantially decreased the cell proliferation rate of the tested fibroblasts in a peptide dose-responsive fashion. At the highest peptide dose, basal levels decreased by up to 70%, while the increase measured as a result of cDNA overexpression did not exceed 20% of basal levels. Cell-permeable fusion peptides have been reported to enter most cells efficiently and have been detected for as long as 8 days after treatment (41), whereas transient cDNA expression experiments typically reach a smaller fraction of the treated cells (less than 30% in our experiments based on green fluorescent protein expression [data not shown]), and expression is typically seen for a time period significantly shorter than a week for up to 3 days (data not shown). The measured increase in the cell proliferation rate of the transfected culture by transient Grb10 cDNA expression will consequently represent a significant underestimate of the real effect per cell when increased levels of Grb10 are actually present. In this context, the observed opposite effects of Grb10 overexpression and the dominant-negative SH2 domain peptides are complementary, which confirms and expands the observed changes in DNA synthesis shown in Fig. 5 to 7. All data are compatible with a positive, stimulatory role of Grb10 in mitogenesis. Further analysis of the effect of Grb10 on cell proliferation will focus on stably expressing cell lines.
FIG. 8.
Transient overexpression of complete Grb10 or interference (by cell-permeable SH2 domain fusion peptides) with cellular Grb10 proportionally increases or decreases the cell proliferation rate, respectively. Mouse NIH 3T3 fibroblasts were incubated with 5 or 10 μg of cell-permeable SH2 domain fusion peptide (cross-hatched columns) or control eluate (open column; resulting basal proliferation rate has been defined as 100%) per ml. Alternatively, fibroblasts were transfected with a CMV promoter-controlled expression plasmid (pHook2; Invitrogen) containing either a complete protein-coding Grb10 cDNA (black column) or no cDNA insert (open column; resulting basal proliferation rate has been defined as 100%). Cell proliferation was quantified for up to 1 week by either measuring cell numbers daily or by a colorimetric (MTT) assay. Changes in the cell proliferation rate are shown as percentage of the basal rate (open column).
DISCUSSION
In this study, we have demonstrated Grb10 association with activated PDGFRβ in response to PDGF-BB stimulation (Fig. 1A). The Grb10 SH2 domain was found to be sufficient for this interaction (Fig. 2A) and, when used as a competitor, to essentially block the interaction with cellular Grb10 (Fig. 1A). Of the major tyrosine autophosphorylation sites of PDGFRβ, only one, Y771 when mutated, resulted in the lack of association with the Grb10 SH2 domain (Fig. 2A). Synthetic phosphopeptides representing Y771 interfered with PDGFRβ binding of the Grb10 SH2 domain (Fig. 2B) and indicated that the motif around phosphotyrosine 771 actually binds to Grb10. PDGFRβ mutation Y771F similarly interfered with the association of complete cellular Grb10 (Fig. 2C). This binding site is shared by Ras GAP, which has been shown to be involved in the chemotactic response to PDGF (51). Since transgenic mice lacking GAP demonstrate normal PDGF-BB-mediated mitogenesis, GAP does not appear to participate in that response unless it plays an unidentified putative role by down-modulating Grb10 action, possibly by competing for the same PDGFRβ binding motif (51). It is conceivable that Grb10 may modulate GAP action or otherwise participate in the chemotactic signal, but this issue is outside the scope of this study.
Grb10 association with the IGF-IR had been demonstrated earlier (8, 30) and was confirmed in this study (Fig. 1B); however, a preference of Grb10 for the IR had been observed in a direct comparison (20). This was investigated by others (15), who reported that IR associates with the SH2 and BPS domains of Grb10, resulting in high-affinity binding, which explains the significant but partial interference by the SH2 domain (Fig. 1C), whereas IGF-IR primarily associates with the BPS domain, which explains the minimal interference observed by the SH2 domain (Fig. 1B). Of a number of point mutations at major IGF-IR autophosphorylation sites, only mutation Y1316F at the carboxyl terminus interfered with the association of the Grb10 SH2 domain (Fig. 3A). This finding is supported by our earlier studies which implicated the homologous carboxyl-terminal autophosphorylation motif Y1322 of IR in Grb10 SH2 domain binding, based on analysis of receptor mutants and the binding of GST-Grb10 SH2 domain fusion proteins to immobilized IR phosphopeptides (14). Phosphopeptides representing the major IGF-IR autophosphorylation sites were not available for our studies; however, new experiments with IR phosphopeptides confirmed that only a peptide representing Y1322 (homologous to IGF-IR Y1316 [50]) specifically interfered with IR binding to the Grb10 SH2 domain and showed that this sequence actually interacts with Grb10 (Fig. 3B). However, the SH2 domain association likely represents only a minor aspect of the interaction between Grb10 and IGF-IR, which appears to be largely carried out by the Grb10 BPS domain (15). The consensus receptor phosphotyrosine sequence motif which is recognized by the Grb10 SH2 domain has not yet been established to our knowledge, and our comparison of the implicated PDGFRβ, IR, and IGF-IR sites of association has not been able to define it (not shown).
In independent studies of several research teams, the exact sites of interaction with Grb10 are controversial for both IGF-IR and IR. Using a yeast two-hybrid interaction approach, an 800-bp fragment of Grb10 was independently found to associate with the IGF-IR carboxyl terminus, but at a site reported to lie between aa 1229 and 1245 (30). In contrast to that study, the activation loop had been implicated in the association with IR and IGF-IR by using a yeast two-hybrid approach combined with receptor mutants (9). Based on IR mutants and phosphopeptides, the kinase activation loop and the juxtamembrane region have been implicated in the interaction with Grb10 in another study (13). However, the juxtamembrane region and the carboxyl terminus were not found to be essential in a further study based on yeast two-hybrid mapping and IR mutant analysis of Grb10 association (38). Part of the differences may be explained by the observation that in addition to the SH2 domain, other Grb10 sequences have been implicated in the interaction with IR and IGF-IR (13). A new domain, termed BPS (or IPS) to reflect its location between the PH and SH2 domains at aa 358 to 434, has been implicated in Grb10 binding to the insulin, IGF-I, and EGF receptors (11, 15). Whereas our studies are exclusively based on the association of Grb10 SH2 domain fusion peptides, other studies have used complete Grb10 or larger fragments of Grb10 and included the interactions of the BPS domain. A recent study has compared various IR mutants for their interaction with the Grb10 SH2, the BPS, and a combination of both domains (15). Only mutation Y1150/1151F in the activation loop was found to essentially abolish the association of IR with Grb10, surprisingly with any isolated domain either SH2 or BPS, or both in combination. This finding suggests either that both Grb10 domains bind to the same IR site, which is not supported by the model presented in this study (15), or that the mutation in the activation loop abolishes other putative IR binding sites of Grb10, which would impair the specificity of the analysis. Since phosphorylation of the activation loop is one of the first steps in IR activation and a requirement for subsequent steps such as phosphorylation of carboxyl-terminal sites (53) and since carboxyl-terminal mutations have not been tested, the reported finding does not address the specific binding of Grb10 to other receptor sites.
Other factors which may help to explain some of the differences observed by different research teams are the distinct Grb10 sequence variants which have been studied and may be explained by differential splicing; more than one Grb10 protein band has typically been identified in many experiments, which may also be due in part to variable translation starts and/or modification (9, 14, 20, 21, 24, 31, 39, 48). In addition to the original mouse Grb10 (39), a related human protein termed Grb-IR or hGrb10α had been identified as an IR partner with a truncated PH domain (21). Several human sequence variants at the amino terminus termed Grb10/IR-SV1 or hGrb10β, hGrb10γ, hGrb10δ (10, 13, 38) and an additional variant in the mouse which lacks a sequence upstream of the amino-terminal end of the GM region (20) have been reported (42). Most available polyclonal antibodies are expected to cross-react with several of the known variants; however, only specific forms will be expressed in specific cells and tissues. The set of three major protein bands which are frequently identified in fibroblasts unlikely represent three distinct splice variants since mouse Grb10 (39), when overexpressed, results in overall similar patterns of intensified protein bands which must all result from the same cDNA (Fig. 7A). Many cell types including NIH 3T3 fibroblasts may express only few Grb10 variants which may be represented by several distinct protein bands due to the use of alternative translation start signals and differences in posttranslational modification (20, 39).
To learn more about the role of Grb10 as a mitogenic mediator, we investigated its interaction with other receptor tyrosine kinases. In addition to the observed association with PDGF, insulin, and IGF-I receptors, we found the SH2 domain to associate with activated HGFR (Met) and FGFR but not with EGFR and NGF receptors (TrkA) (Fig. 4). Since our experiments were based on GST-Grb10 SH2 domain fusion proteins, it is possible that association with TrkA involves additional Grb10 sequences such as the BPS domain. This probably does not apply to the putative association with the EGFR (15). When combined with the reported Grb10 association with other receptor tyrosine kinases such as Ret (12, 40) and the Eph-related receptor tyrosine kinase ELK (48), our data suggest a role for Grb10 downstream of most receptor tyrosine kinase subfamilies (52) except for the EGF and NGF subfamilies. This inference is based on at least one member of each subfamily (52) which was tested for Grb10 association (Fig. 4) and is compatible with a putative role of Grb10 as a general mitogenic mediator, in addition to other putative functions of Grb10 downstream of the ELK receptor in axonal guidance, neuronal bundling, or angiogenesis (48).
The role of Grb10 in EGF action is still controversial. Grb10 was originally cloned with an activated carboxyl-terminal EGFR fragment as a probe but was found to interact only weakly with the activated receptor (13, 15, 39). However, serine phosphorylation of Grb10 has been reported in response to EGF stimulation (39). Interaction with the EGFR has not been observed in our study (Fig. 4) despite the fact that the Grb10 SH2 domain had been implicated in this association (15), and changes in Grb10 function were not found to affect EGF-stimulated DNA synthesis (Fig. 6 and 7B).
To test the putative mitogenic role of Grb10 directly, we used four independent experimental strategies to interfere with cellular Grb10 function or increase cellular Grb10 levels. Initially, we microinjected GST-Grb10 SH2 domain fusion protein to test its impact on PDGF-BB- and insulin-mediated mitogenesis in NIH 3T3 fibroblasts. The high fusion protein concentration in the injected cells is expected to exert a dominant-negative effect on cellular Grb10 function (37). The observed reduction in DNA synthesis suggests a positive, stimulatory role of cellular Grb10 in that pathway (Fig. 5A). Microinjection of GST-Grb10/IR-SV1 or hGrb10β had been shown earlier to interfere with IGF-I- and insulin-mediated, but not with EGF-mediated, DNA synthesis in Rat-1 fibroblasts (38). The observed reduction in DNA synthesis (Fig. 5A) was specific for the Grb10 SH2 domain since a control Crk SH2 domain did not exert a significant effect.
In an alternative, dominant-negative strategy, the Grb10 SH2 domain was introduced into NIH 3T3 fibroblasts as a cell-permeable fusion peptide with a 16-aa fragment of the Drosophila antennapedia homeodomain protein, which mediates the effective transfer of fusion peptides across the cell membrane (41). A tyrosine-phosphorylated, cell-permeable Grb2 binding peptide based on EGFR sequences was shown to specifically block the mitogenic response to PDGF and EGF but not to FGF, demonstrating that selective pathways are inhibited by this approach (54). Activation of the mitogen-activated protein kinase cascade was inhibited in response to up to 10-ng/ml but not to higher 50-ng/ml) EGF concentrations, suggesting that the peptide does not exert an excessive inhibitory function (54). In our experiments, we observed a substantial decrease in PDGF-BB-, IGF-I-, and insulin-mediated DNA synthesis, as measured by the incorporation of [3H]thymidine into DNA, which corresponded to the dose of the cell-permeable Grb10 SH2 domain fusion peptide (Fig. 5B). These results directly confirmed those shown in Fig. 5A from microinjection studies using an alternative approach.
In addition, these studies indicate a role of the Grb10 SH2 domain in IGF-I-mediated mitogenesis (Fig. 5B) but not through binding to the IGF-IR (Fig. 1B), since this is largely carried out by the Grb10 BPS domain (15). The SH2 domain is consequently implicated in the association with another unknown signaling mediator, probably at an activated phosphotyrosine which may be represented by another (receptor) tyrosine kinase or by an alternative signaling mediator. Such a mechanism could assemble different receptors into a joint signaling complex and regulate cross talk between distinct receptor pathways. Grb10 may represent a shortcut in the Raf-mediated signaling cascade by directly interacting with Raf1 and MEK1 via its SH2 domain and linking growth factor receptors to these mediators (35).
We have to consider possible cross talk of the Grb10 SH2 domain and Pro-rich fusion peptides with mechanisms involving the related mediators Grb7 and Grb14. However, functional selectivity has been reported for SH2 domains in the Grb7 family (17) and for Pro-rich SH3 domain ligands (46), which was also observed in control experiments with other Pro-rich regions (not shown). The responses shown in Fig. 5 to 8 point to a positive stimulatory role of Grb10 in the mitogenic actions of PDGF-BB, IGF-I, and insulin but have not been observed for Grb7 or Grb14 (5), suggesting that these mediators do not affect our results.
Based on the putative role of Grb10 as a signaling adapter, we tested an additional domain, the Grb10 amino-terminal Pro-rich region, as a synthetic cell-permeable fusion peptide with a 16-aa fragment of the Drosophila antennapedia homeodomain protein. With this approach, the IGF-I and insulin-stimulated mitogenic response was almost eliminated with increasing doses of peptide in a dose-responsive fashion, whereas the EGF and PDGF-BB responses were not significantly affected (Fig. 6). Since the high concentration of the peptide is expected to represent a dominant-negative form, this result is consistent with a stimulatory role of the Grb10 Pro-rich region in IGF-I and insulin action. This is compatible with a role of Grb10 as a mitogenic signaling adapter which may involve its SH2 or BPS and Pro-rich domains to form a signaling complex between an activated receptor tyrosine kinase and an SH3 domain mediator such as Abl (13). In contrast to the Grb10 SH2 domain, the Pro-rich region appears not to participate in PDGF-BB action (Fig. 6). A role as a putative signaling adapter in this pathway may consequently involve an alternative Grb10 domain, such as the PH region, which remains to be identified. The functional differences observed for these two growth factor pathways at the level of the involved Grb10 domain structure may reflect a specific role of Grb10 in either pathway which remains to be elucidated.
A stimulatory role for Grb10 in PDGF-BB-, IGF-I-, and insulin- but not EGF-mediated mitogenesis was directly shown by cDNA expression from an ecdysone-regulated Grb10 expression plasmid. Stimulation of DNA synthesis was ecdysone dose responsive, with the highest level observed for PDGF-BB (sixfold over background levels [Fig. 7B]). The Grb10-mediated mitogenesis correlated with increased Grb10 expression levels which had been reproducibly observed on immunoblots (Fig. 7A). A wide range of ecdysone-adjustable expression levels has been found in cultured cells or in whole experimental mice, which in combination with the reported lack of basal expression surpassed tetracycline-based expression systems in a direct comparison (36). Few if any side effects were described for ecdysone, including lack of toxicity at functional doses in vivo, even in whole mice (36).
The effect of Grb10 on mitogenesis is controversial. Interference of the Grb10 SH2 domain with insulin-stimulated DNA synthesis upon microinjection into fibroblasts suggested a positive role for Grb10 (38), which is also supported by the implicated stimulatory role of Grb10 in Bcr-Abl-mediated oncogenicity (2) and by its implicated protective function in apoptosis (35). On the other hand, the possible implication of Grb10 as a candidate for the Silver-Russel syndrome gene would point to a negative role in mitogenesis (29). In addition, an inhibitory effect on IGF-I-mediated cell growth was reported for increased expression levels of mouse Grb10α (39) in fibroblasts which were transformed by IGF-IR overexpression (31). These cells express very high IGF-IR levels (500,000 per cell) and grow independently of growth factors except for IGF-I. Increased levels of stably expressed Grb10 have been reported to interfere with the transformed phenotype as well as with cell growth by delaying the S and G2 phases of the cell cycle (31). Control experiments in the same cell line have not been performed; however, overexpression of Grb10 in different, normal, IGF-IR-disrupted fibroblasts did not interfere with cell growth. A Myc-tagged cDNA of Grb10 was expressed in these experiments, and it remains possible that the tag affects Grb10 function so as to result in a dominant-negative form. Alternatively, the transformed phenotype of this experimental system may have an impact on the observed function of Grb10 when compared to normal fibroblasts.
We addressed this question in our own study by monitoring the effect of increased Grb10 levels or of interference with cellular Grb10 on the cell doubling time in normal fibroblasts. We observed that Grb10 overexpression consistently increased the cell proliferation rate, whereas cell-permeable SH2 fusion peptides resulted in a dose-dependent substantial decrease (Fig. 8). The observed opposite effects of Grb10 overexpression and of the dominant-negative SH2 domain peptides are complementary, confirming and expanding the observed changes in DNA synthesis shown in Fig. 5 to 7. Consequently, all findings presented in this study support a positive, stimulatory role of Grb10 as a cellular partner of the PDGFβ, IGF-I, and insulin receptors, based on several independent dominant-negative as well as positive experimental strategies. Such a role as a general mitogenic signaling adapter is suggested in many growth factor signaling pathways by the observed association between Grb10 and other receptor tyrosine kinases (Fig. 4). It is possible that Grb10 acts positively or negatively in mitogenesis dependent on the specific cellular context. Such a variable role has been described for other signaling mediators, including Myc, one important example, and its role in apoptosis (16, 44).
ACKNOWLEDGMENTS
We are grateful to Benjamin R. Braun, Rukmani Krishnamoorthy, Yuyan Zhao, and Youhou Kang for expert technical assistance, Renato Baserga, Jonathan Cooper, Yousuke Ebina, Robert E. Friesel, Barbara L. Hempstead, Takashi Kadowaki, Masato Kasuga, Derek LeRoith, Jack Lilien, Jerrold M. Olefsky, Moraq Park, Tony Pawson, and Morris F. White for cell lines, plasmids, or expressed proteins, Gert Wolf and Zhou Songyang for support, and Nora Riedel for ideas and the critical discussion of the manuscript. We thank Jeffrey Vang, Christina Roffi, Veronica Kemerko, Isam Abbarrah, and Wissam Malouf for technical help.
Part of this work was supported by funds to H.R. from the National Science Foundation (grants MCB-9696090 and MCB-9808795) and the Juvenile Diabetes Foundation International (grant 195088).
REFERENCES
- 1.Angrist M, Bolk S, Bently K, Nallasamy S, Halushka M K, Chakravarti A. Genomic structure of the gene for the SH2 and pleckstrin homology domain-containing protein GRB10 and evaluation of its role in Hirschsprung disease. Oncogene. 1998;17:3065–3070. doi: 10.1038/sj.onc.1202226. [DOI] [PubMed] [Google Scholar]
- 2.Bai R Y, Jahn T, Schrem S, Munzert G, Weidner K M, Wang J Y J, Duyster J. The SH2-containing adapter protein Grb10 interacts with BCR-ABL. Oncogene. 1998;17:941–948. doi: 10.1038/sj.onc.1202024. [DOI] [PubMed] [Google Scholar]
- 3.Blakesley V A, Scrimgeour A, Esposito D, LeRoith D. Signaling via the insulin-like growth factor-I receptor: does it differ from insulin receptor signaling? Cytokine Growth Factor Rev. 1996;7:153–159. doi: 10.1016/1359-6101(96)00015-9. [DOI] [PubMed] [Google Scholar]
- 4.Brown S, Friesel R. Production of recombinant Xenopus fibroblast growth factor receptor-1 using a baculovirus expression system. Biochem Biophys Res Commun. 1993;193:1116–1122. doi: 10.1006/bbrc.1993.1741. [DOI] [PubMed] [Google Scholar]
- 5.Daly R J. The Grb7 family of signalling proteins. Cell Signal. 1998;10:613–618. doi: 10.1016/s0898-6568(98)00022-9. [DOI] [PubMed] [Google Scholar]
- 6.Daly R J, Sanderson G M, Janes P W, Sutherland R L. Cloning and characterization of Grb14, a novel member of the Grb7 gene family. J Biol Chem. 1996;271:12502–12510. doi: 10.1074/jbc.271.21.12502. [DOI] [PubMed] [Google Scholar]
- 7.Denizot F, Lang R. Rapid colorimetric assay for cell growth and survival. Modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J Immunol Methods. 1986;89:271–277. doi: 10.1016/0022-1759(86)90368-6. [DOI] [PubMed] [Google Scholar]
- 8.Dey B R, Frick K, Lopaczynski W, Nissley S P, Furlanetto R W. Evidence for the direct interaction of the insulin-like growth factor I receptor with IRS-1, Shc, and Grb10. Mol Endocrinol. 1996;10:631–641. doi: 10.1210/mend.10.6.8776723. [DOI] [PubMed] [Google Scholar]
- 9.Dong L Q, Farris S, Christal J, Liu F. Site-directed mutagenesis and yeast two-hybrid studies of the insulin and insulin-like growth factor-1 receptors: the Src homology-2 domain-containing protein hGrb10 binds to the autophosphorylated tyrosine residues in the kinase domain of the insulin receptor. Mol Endocrinol. 1997;11:1757–1765. doi: 10.1210/mend.11.12.0014. [DOI] [PubMed] [Google Scholar]
- 10.Dong L Q, Du H, Porter S G, Kolakowski L F, Lee A V, Mandarino J, Fan J, Yee D, Liu F. Cloning, chromosome localization, expression, and characterization of a Src homology 2 and pleckstrin homology domain-containing insulin receptor binding protein hGrb10γ. J Biol Chem. 1997;272:29104–29112. doi: 10.1074/jbc.272.46.29104. [DOI] [PubMed] [Google Scholar]
- 11.Dong L Q, Porter S, Hu D, Liu F. Inhibition of hGrb10 binding to the insulin receptor by functional domain-mediated oligomerization. J Biol Chem. 1998;273:17720–17725. doi: 10.1074/jbc.273.28.17720. [DOI] [PubMed] [Google Scholar]
- 12.Durick K, Wu R-Y, Gill G N, Taylor S S. Mitogenic signaling by Ret/ptc2 requires association with enigma via a LIM domain. J Biol Chem. 1996;271:12691–12694. doi: 10.1074/jbc.271.22.12691. [DOI] [PubMed] [Google Scholar]
- 13.Frantz J D, Giorgetti-Peraldi S, Ottinger E A, Shoelson S E. Human GRB-IR beta/GRB10: splice variants of an insulin and growth factor receptor-binding protein with PH and SH2 domains. J Biol Chem. 1997;272:2659–2667. doi: 10.1074/jbc.272.5.2659. [DOI] [PubMed] [Google Scholar]
- 14.Hansen H, Svensson U, Zhu J, Laviola L, Giorgino F, Wolf G, Smith R J, Riedel H. Interaction between the Grb10 SH2 domain and the insulin receptor carboxyl terminus. J Biol Chem. 1996;271:8882–8886. doi: 10.1074/jbc.271.15.8882. [DOI] [PubMed] [Google Scholar]
- 15.He W, Rose D W, Olefsky J M, Gustafson T A. Grb10 interacts differentially with the insulin receptor, insulin-like growth factor-I receptor, and epidermal growth factor receptor via the Grb10 Src homology 2 (SH2) domain and a second novel domain located between the pleckstrin homology and SH2 domains. J Biol Chem. 1998;273:6860–6867. doi: 10.1074/jbc.273.12.6860. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman B, Liebermann D A. The proto-oncogene c-myc and apoptosis. Oncogene. 1998;17:3351–3357. doi: 10.1038/sj.onc.1202592. [DOI] [PubMed] [Google Scholar]
- 17.Janes P W, Lackmann M, Church W B, Sanderson G M, Sutherland R L, Daly R J. Structural determinants of the interaction between the erbB2 receptor and the Src homology 2 domain of Grb7. J Biol Chem. 1997;272:8490–8497. doi: 10.1074/jbc.272.13.8490. [DOI] [PubMed] [Google Scholar]
- 18.Kasus-Jacobi A, Perdereau D, Auzan C, Clauser E, Van Obberghen E, Mauvais-Jarvis F, Girard J, Burnol A F. Identification of the rat adapter Grb14 as inhibitor of insulin actions. J Biol Chem. 1998;273:26026–26035. doi: 10.1074/jbc.273.40.26026. [DOI] [PubMed] [Google Scholar]
- 19.Kazlauskas A, Kashishian A, Cooper J A, Valius M. GTPase-activating protein and phosphatidylinositol 3-kinase bind to distinct regions of the platelet-derived growth factor receptor beta subunit. Mol Cell Biol. 1992;12:2534–2544. doi: 10.1128/mcb.12.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laviola L, Giorgino F, Chow J C, Baquero J A, Hansen H, Ooi J, Zhu J, Riedel H, Smith R J. The adapter protein Grb10 associates preferentially with the insulin receptor as compared to the IGF-1 receptor in mouse fibroblasts. J Clin Investig. 1997;99:830–837. doi: 10.1172/JCI119246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu F, Roth R A. Grb-IR: a SH2 domain-containing protein that binds to the insulin receptor and inhibits its function. Proc Natl Acad Sci USA. 1995;92:10287–10291. doi: 10.1073/pnas.92.22.10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu F, Roth R A. Binding of SH2 containing proteins to the insulin receptor: a new way for modulating insulin signalling. Mol Cell Biochem. 1998;182:73–78. [PubMed] [Google Scholar]
- 23.Mano H, Ohya K, Miyazato A, Yamashita Y, Ogawa W, Inazawa J, Ikeda U, Shimada K, Hatake K, Kasunga M, Ozawa K, Kajigaya S. Grb10/GrbIR as an in vivo substrate of Tec tyrosine kinase. Genes Cells. 1998;3:431–441. doi: 10.1046/j.1365-2443.1998.00201.x. [DOI] [PubMed] [Google Scholar]
- 24.Manser J, Roonprapunt C, Margolis B. C. elegans cell migration gene mig-10 shares similarities with a family of SH2 domain proteins and acts cell nonautonomously in excretory canal development. Dev Biol. 1997;184:150–164. doi: 10.1006/dbio.1997.8516. [DOI] [PubMed] [Google Scholar]
- 25.Margolis B. The GRB family of SH2 domain proteins. Prog Biophys Mol Biol. 1994;62:223–244. doi: 10.1016/0079-6107(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 26.Margolis B, Silvennoinen O, Comoglio F, Roonprapunt C, Skolnik E, Ullrich A, Schlessinger J. High-efficiency expression cloning of epidermal growth factor-receptor-binding proteins with Src homology 2 domains. Proc Natl Acad Sci USA. 1992;89:8894–8898. doi: 10.1073/pnas.89.19.8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miura M, Li S, Baserga R. Effect of a mutation at tyrosine 950 of the insulin-like growth factor I receptor on the growth and transformation of cells. Cancer Res. 1995;55:663–667. [PubMed] [Google Scholar]
- 28.Miura M, Surmacz E, Burgaud J L, Baserga R. Different effects on mitogenesis and transformation of a mutation at tyrosine 1251 of the insulin-like growth factor I receptor. J Biol Chem. 1995;270:22639–22644. doi: 10.1074/jbc.270.38.22639. [DOI] [PubMed] [Google Scholar]
- 29.Miyoshi N, Kuroiwa Y, Kohda T, Shitara H, Yonekawa H, Kawabe T, Hasegawa H, Barton S C, Surani M A, Kaneko-Ishino T, Ishino F. Identification of the Meg1/Grb10 imprinted gene on mouse proximal chromosome 11, a candidate for the Silver-Russel syndrome gene. Proc Natl Acad Sci USA. 1998;95:1102–1107. doi: 10.1073/pnas.95.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrione A, Valentinis B, Li S, Ooi J Y T, Margolis B, Baserga R. Grb10: a new substrate of the insulin-like growth factor I receptor. Cancer Res. 1996;56:3165–3167. [PubMed] [Google Scholar]
- 31.Morrione A, Valentinis B, Resnicoff M, Xu S-Q, Baserga R. The role of mGrb10a in insulin-like growth factor I-mediated growth. J Biol Chem. 1997;272:26382–26387. doi: 10.1074/jbc.272.42.26382. [DOI] [PubMed] [Google Scholar]
- 32.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 33.Moutoussamy S, Renaudie F, Lago F, Kelly P A, Finidori J. Grb10 identified as a potential regulator of growth hormone (GH) signaling by cloning of GH receptor target proteins. J Biol Chem. 1998;273:15906–15912. doi: 10.1074/jbc.273.26.15906. [DOI] [PubMed] [Google Scholar]
- 34.Myers M J, Backer J M, Siddle K, White M F. The insulin receptor functions normally in Chinese hamster ovary cells after truncation of the C-terminus. J Biol Chem. 1991;266:10616–10623. [PubMed] [Google Scholar]
- 35.Nantel A, Mohammad-Ali K, Sherk J, Posner B L, Thomas D Y. Interaction of the Grb10 adapter protein with the Raf1 and MEK1 kinases. J Biol Chem. 1998;273:10475–10484. doi: 10.1074/jbc.273.17.10475. [DOI] [PubMed] [Google Scholar]
- 36.No D, Yao T P, Evans R M. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Northrop J P, Pustelnik M J, Lu A T, Grove J R. Characterization of the roles of SH2 domain-containing proteins in T-lymphocyte activation by using dominant negative SH2 domains. Mol Cell Biol. 1996;16:2255–2263. doi: 10.1128/mcb.16.5.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Neill T J, Rose D W, Pillay T S, Hotta K, Olefsky J M, Gustafson T A. Interaction of a GRB-IR splice variant (a human GRB10 homolog) with the insulin and insulin-like growth factor I receptors. J Biol Chem. 1996;271:22506–22513. doi: 10.1074/jbc.271.37.22506. [DOI] [PubMed] [Google Scholar]
- 39.Ooi J, Yajnik V, Immanuel D, Gordon M, Moskow J J, Buchberg A M, Margolis B. The cloning of Grb10 reveals a new family of SH2 domain proteins. Oncogene. 1995;10:1621–1630. [PubMed] [Google Scholar]
- 40.Pandey A, Duan H, Di Fiore P P, Dixit V M. The Ret receptor protein tyrosine kinase associates with the SH2-containing adapter protein Grb10. J Biol Chem. 1995;270:21461–21463. doi: 10.1074/jbc.270.37.21461. [DOI] [PubMed] [Google Scholar]
- 41.Prochiantz A. Getting hydrophilic compounds into cells: lessons from homeopeptides. Curr Opin Neurobiol. 1996;6:629–634. doi: 10.1016/s0959-4388(96)80095-x. [DOI] [PubMed] [Google Scholar]
- 42.Riedel, H., and B. R. Braun. Grb10 in insulin signaling. In G. Grunberger and Y. Zick (ed.), Insulin signaling: from cultured cells to animal models, in press. Harwood Academic Press, Langhorne, Pa.
- 43.Rodrigues G A, Park M. Autophosphorylation modulates the kinase activity and oncogenic potential of Met receptor tyrosine kinase. Oncogene. 1994;9:2019–2027. [PubMed] [Google Scholar]
- 44.Rohn J L, Hueber A O, McCarthy N J, Lyon D, Navarro P, Burgering B M, Evan G I. The opposing roles of the Akt and c-Myc signalling pathways in survival from CD95-mediated apoptosis. Oncogene. 1998;17:2811–2818. doi: 10.1038/sj.onc.1202393. [DOI] [PubMed] [Google Scholar]
- 45.Sasaoka T, Rose D W, Jhun B H, Saltiel A R, Draznin B, Olefsky J M. Evidence for a functional role of Shc proteins in mitogenic signaling induced by insulin, insulin-like growth factor-I, and epidermal growth factor. J Biol Chem. 1994;269:13689–13694. [PubMed] [Google Scholar]
- 46.Sparks A B, Rider J E, Hoffman N G, Fowlkes D M, Quilliam L A, Kay B K. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, cortactin, p53bp2, PLCgamma, Crk, and Grb2. Proc Natl Acad Sci USA. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stein D, Wu J, Fuqua S A W, Roonprapunt C, Yajnik V, D’Eustachio P, Moskow J J, Buchberg A M, Osborne C K, Margolis B. The SH2 domain protein GRB-7 is coamplified, overexpressed and in a tight complex with HER2 in breast cancer. EMBO J. 1994;13:1331–1340. doi: 10.1002/j.1460-2075.1994.tb06386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stein E, Cerretti D P, Daniel T O. Ligand activation of ELK receptor tyrosine kinase promotes its association with Grb10 and Grb2 in vascular endothelial cells. J Biol Chem. 1996;271:23588–23593. doi: 10.1074/jbc.271.38.23588. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka S, Mori M, Akiyoshi T, Tanaka Y, Mafune K, Wands J R, Sugimachi K. A novel variant of human Grb7 is associated with invasive esophageal carcinoma. J Clin Investig. 1998;102:821–827. doi: 10.1172/JCI2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ullrich A, Gray A, Tam A W, Yang-Feng T, Tsubokawa M, Collins C, Henzel W, Le Bon T, Kathuria S, Chen E, Jacobs S, Francke U, Ramachandran J, Fujita-Yamaguchi Y. Insulin-like growth factor I receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity. EMBO J. 1986;5:2503–2512. doi: 10.1002/j.1460-2075.1986.tb04528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Geer P, Henkemeier M, Jacks T, Pawson T. Aberrant Ras regulation and reduced p190 tyrosine phosphorylation in cells lacking p120-Gap. Mol Cell Biol. 1997;17:1840–1847. doi: 10.1128/mcb.17.4.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Geer P, Hunter T, Lindberg R A. Receptor tyrosine kinases and their signal transduction pathways. Annu Rev Cell Biol. 1994;10:251–337. doi: 10.1146/annurev.cb.10.110194.001343. [DOI] [PubMed] [Google Scholar]
- 53.White M F, Kahn C R. The insulin signaling system. J Biol Chem. 1994;269:1–4. [PubMed] [Google Scholar]
- 54.Williams E J, Dunican D J, Green P J, Howell F V, Derossi D, Walsh F S, Doherty P. Selective inhibition of growth factor-stimulated mitogenesis by a cell-permeable Grb2-binding peptide. J Biol Chem. 1997;272:22349–22354. doi: 10.1074/jbc.272.35.22349. [DOI] [PubMed] [Google Scholar]
- 55.Yokote K, Margolis B, Heldin C-H, Claesson-Welsh L. Grb7 is a downstream signaling component of platelet-derived growth factor alpha- and beta-receptors. J Biol Chem. 1996;271:30942–30949. doi: 10.1074/jbc.271.48.30942. [DOI] [PubMed] [Google Scholar]