Abstract
Melanomas harboring BRAF mutations can be treated with BRAF inhibitors (BRAFi), but responses are varied and tumor recurrence is inevitable. Here we used an integrative approach of experimentation and mathematical flux balance analyses in BRAF-mutated melanoma cells to discover that elevated antioxidant capacity is linked to BRAFi sensitivity in melanoma cells. High levels of antioxidant metabolites in cells with reduced BRAFi sensitivity confirmed this conclusion. By extending our analyses to other melanoma subtypes in TCGA, we predict that elevated redox capacity is a general feature of melanomas, not previously observed. We propose that redox vulnerabilities could be exploited for therapeutic benefits and identify unsuspected combination targets to enhance the effects of BRAFi in any melanoma, regardless of mutational status.
STATEMENT OF SIGNIFICANCE:
An integrative bioinformatics, flux-balance-analysis, and experimental approach identifies targetable redox vulnerabilities and shows the potential for modulation of cancer antioxidant defense to augment the benefits of existing therapies in melanoma.
Keywords: Systems biology, Redox capacity, Drug insensitivity, Combination targets, Antioxidants
INTRODUCTION:
Targeted therapy has been a major breakthrough for melanoma patients harboring BRAFV600 mutations, present in 50% of malignant melanomas (1). However, the clinical responses are highly variable and short-lived, and relapse is almost universal (1). Overcoming reduced sensitivity and acquired resistance to targeted therapy is a major goal of current melanoma research. Several mechanisms of reduced sensitivity have been proposed (2), which have led to the development of several combination regimens in melanoma either with other targeted therapies (3) or in conjunction with immunotherapies (4). While these therapies improve responses, treatment outcomes still vary and benefits remain transient and unpredictable (4).
Some resistance can be attributed to genetic mutations (5), but accumulating evidence indicates that nongenetic processes play a critical role in response of cancer cells to drug treatment (6–8). Metabolic reprogramming, recognized as a hallmark of cancer (9), has recently emerged as a potential nongenetic process that contributes to the emergence of drug-tolerant cancer cells. Otto Warburg first reported a link between tumor and metabolism in his influential observation that cancer cells convert most intracellular glucose to lactate--that is aerobic glycolysis (10). Recent studies have shown that cancer cells can utilize glycolysis, mitochondrial respiration, or both, depending on their environment (11). Furthermore, cancer cells can adapt metabolically in response to external perturbations (12). This metabolic flexibility provides cancer cells with energy, and necessary intermediates for biosynthetic processes required for survival and to maintain redox balance under changing environments (12,13). Furthermore, metabolic pathways are complex and interconnected, warranting a systems level approach to examine their relative importance, and how they change in cancer cells. Flux Balance Analysis (FBA) is the most commonly used mathematical modeling approach for genome-scale metabolic network reconstructions to estimate the role of metabolic reactions in a network (14). Such a quantitative approach can be utilized to predict global metabolic states of cancer cells under various conditions.
Melanoma cells upon BRAF-inhibition have been shown to induce an enhanced oxidative phosphorylation (7,15). Byproducts of augmented mitochondrial activity are reactive oxygen species (ROS), and this metabolic switch promotes oxidative stress in cells (16). However, the cellular response to ROS is rather complex: low levels facilitate intracellular signaling, while high levels may cause cell death (17). Therefore, cells require a robust antioxidant defense system to respond to an accumulation of ROS. By invoking antioxidant systems, cancer cells can clear excess ROS levels within a range that is not detrimental to them--a high redox homeostatic state (18). Nicotinamide adenine dinucleotide phosphate (NADPH), and glutathione (GSH) are two major antioxidants that maintain redox homeostasis in cells (19,20). NADPH is produced via the Pentose Phosphate Pathway (PPP), and other sources (21), and acts as a shared substrate for both GSH regeneration, and ROS production, thus maintaining an optimum redox balance within cancer cells. However, optimum redox balance in cancer is context-dependent, with tissue type, stroma, greatly affecting how cancer cells survive, metastasize and respond to therapies (20). Even within cancer subtypes, this redox state could be heterogeneous (22), motivating further research to examine how this balance could be altered for therapeutic benefits.
The link between oxidative stress and drug response in melanoma is beginning to be explored. BRAF-inhibitor resistant melanomas were shown to upregulate NRF2-mediated antioxidant response to maintain cell survival (16). Nonetheless, it still remains to be examined how redox potential of melanoma cells affects their drug sensitivity, and how it is maintained under BRAF-inhibition. These are important considerations, raising the possibility that redox balance can be modulated to enhance the effects of existing therapies.
Here, we show that antioxidant capacity of melanoma cells is linked to their drug sensitivity. Using an integrative approach through bioinformatics and FBA, we show that melanoma cells with reduced sensitivity to BRAFi exhibit an enhanced capacity for anti-oxidation and redox buffer, specifically through NADPH oxidizing enzymes. By directly quantifying the redox-related metabolites, we confirm that drug-insensitive melanoma cells can maintain higher levels of antioxidant metabolites during treatment. Furthermore, we report that pharmacological disruption of redox axis involving glutathione enhances the effects of targeted therapies. We also extended our analysis to other melanoma subtypes in The Cancer Genome Atlas (TCGA), and found that elevated redox capacity could be a general feature of melanoma. Our results thus provide a proof-of-principle that redox vulnerabilities could be exploited for therapeutic benefits in melanoma.
MATERIALS AND METHODS:
Cell Culture and Chemical Reagents:
All BRAF-mutated, NRAS-mutated, NF1-mutated melanoma cells were engineered to express either histone 2B-mRFP using pHIV-H2B-mRFP plasmid and/or Fluorescent Ubiquitination Cell Cycle Indicator (FUCCI) using Geminin1–110 monomeric Azami Green plasmid as previously described (23). Single-cell derived clonal populations of BRAF-mutated melanoma cells, SKMEL5 were selected by limiting dilution as previously described (8). Single-cell derived BRAF-mutated SKMEL5 subclones were derived as previously described (8). BRAF-mutated melanoma cells (SKMEL5, WM88), including the SKMEL5 subclones, NRAS-mutated melanoma cells (SKMEL2), and NF1-mutated melanoma cells (MeWO) were grown and cultured in Dulbecco’s modified Eagle’s medium and Ham’s F-12 media (DMEM:F12, 1:1, Cat. No. 11330–032). Primary Epidermal Melanocytes, Normal Human Adult (HEMa) (PCS-200–013) were obtained directly from ATCC (Manassas, VA) and cultured in Dermal Cell Basal Medium (ATCC-PCS-200–030) supplemented with Adult Melanocyte Growth Kit (ATCC-PCS-200–042). Media were obtained from Gibco (Grand Island, NY), and supplemented with 10% fetal bovine serum. All cells were cultured in humidified incubators that were CO2 and temperature (37°C) controlled. Cells were passaged 1–2 times per week and were maintained as exponentially growing cultures for a maximum of less than 20 passages. All cells were tested for mycoplasma, and tested negative. Chemical reagents used are listed in Supplementary Information (SI).
Bioinformatics Analysis Pipeline:
RNASeq of melanoma cell lines (SKMEL5 subclones) was performed as previously reported (24). Transcript count data from Gene Expression Omnibus (Accession number: GSE122041, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122041) was used for downstream analysis performed in R (https://www.r-project.org). Differential expression analyses were performed using Bioconductor (https://www.bioconductor.org) packages DESeq2 and edgeR. Transcripts counts for BRAF-mutated melanoma cell lines were obtained from CCLE database. Refer to SI for more details.
Flux Balance Analysis (FBA):
FBA is a computational approach for predicting genome-scale steady-state metabolic fluxes in samples of interest (14). Reaction fluxes throughout the metabolic network are predicted by solving the optimization problem:
where if m and r are the number of metabolites and reactions in the metabolic network, respectively, then v is a r x 1 vector of reaction fluxes to be solved for, S is the m x r stoichiometric matrix of the metabolic network, vmin,j and vmax,j are constraints imposed on each individual reaction flux, and f is an objective function that is maximized to maximize a particular metabolic phenotype of interest. Recon3D version 3.01 was used as the core metabolic network (25).
To maximize the oxidation of NADPH to NADP+ in the metabolic network, the following objective function was used in all cellular compartments:
This artificial sink on NADP+ in turn maximizes the flux through existing metabolic reactions that oxidize NADPH to NADP+. Individual reaction flux values were predicted by averaging the minimum and maximum predicted fluxes from Flux Variance Analysis (FVA), which computes the minimum and maximum obtainable flux for each reaction under the model constraints and while maximizing the objective function above.
Drug Combination Assay and Drug Response:
The dose-response curves are generated using a 2-fold dilution of single drugs at concentrated indicated (highest) to zero (solvent the drug was dissolved in, either DMSO or H2O), all concentrations contain equal percentage of the solvent used. For combination with PLX4720, 8 μM PLX4720 was used as diluent, and serially diluted for the second drug from highest concentration to lowest. Direct measurements of cell counts were made using Cellavista software and ImageJ macros as previously described (8,23,26). Primary melanocytes were seeded into 96-well plates (~3,000 cells/well) and drug treatments applied the following day, including DMSO or PBS control (all concentrations contained equal percentage of DMSO or PBS solvent). A 96-well plate was fixed at time 0, and was used as control for the treatment plates. Drug combination assay was performed as above and cells were treated for 24 h., 48 h., and 72 h. At each time point, cells were fixed with 3% Paraformaldehyde (PFA) for 20 minutes at room temperature, washed with PBS three times, and stored in a dark room until all plates were processed. When all plates were fixed, cells were stained with NucBlue Hoechst 33342 dye (Invitrogen, Ref# R37605, Lot# 2139329) at a concentration of 1 drop/mL of PBS as per the manufacturer’s instructions. Cells were then imaged on the Evos M700 (Invitrogen) microscope using the DAPI and trans-luminescence channels for nuclear and whole cell imaging respectively. The center 25% area of the well was imaged for all the plates. Hoechst stained nuclei were segmented with a custom macro in FIJI (ImageJ), and cell nuclei counts for all treatments were then transformed on a log2-scale and normalized to log2 cell count at time 0.
siRNA Delivery and Drug Combination:
For gene silencing experiments, ON-TARGETplus Human NOX5 siRNA SMARTPool (Cat# L-010195–00) was used. ON-TARGETplus Non-Targeting pool (Cat# D-001810–10-05) was used as scrambled control. NOX5 siRNA was used at the final concentration of 25 nM. Both were obtained from Dharmacon (Lafayette, CO) and stored at −20°C until use. Transfection was carried out according to manufacturer’s instructions using DharmaFECT1 Transfection Reagent (Cat# T-2001–02). For combination with PLX4720, either scrambled mix or siRNA mix was used as diluent and the dose-response curves are generated using 2-fold dilution of PLX4720 at concentrated indicated (highest, 32 μM) to zero (either scrambled or siRNA alone), all concentrations contained equal percentage of the solvent used, and equal concentrations of either scrambled mix or siRNA mix. Melanoma cells were treated with siRNA or scrambled at day 0, and replaced with fresh reagents at day 3. Direct measurements of cell counts were made using Cellavista software and ImageJ macros as previously described (8,23,26). RT-qPCR Quantification of NOX5 expression was done as previously described (24). For all drug responses including combination assay, primary melanocytes, and siRNA-drug combination, the drug-induced proliferation (DIP) rates are calculated using the slope of the log2-normalized population curves after 24 h.
Cellular ROS Staining Assay and Quantification:
CellRox™ DeepRed Reagent was used according to the manufacturer’s instructions. Refer to SI for more details.
Antioxidant Rescue Assay:
For drug-combinations indicated, the dose-response curves are generated as described above. For the indicated concentrations, two known antioxidants--reduced glutathione (GSH, 5 mM), and ferrostatin-1 (FER, 5 μM) are added to the combinations at day 0, and replenished at 72 h. along with growth medium, inhibitors combination for the duration of the assay upto 6 days.
Metabolomics Analysis:
Global untargeted-targeted metabolomic analysis on BRAF-mutated melanoma cells (WM88, SKMEL5-SC10) treated with either DMSO or 8 μM PLX4720 for 24 h. were done as described in Supplementary Information (SI).
DATA AND CODE AVAILABILITY:
All data, calculated DIP rates, DEGs between subclones, SCGs for CCLE melanoma panel, expression data, codes used to generate the figures are available in the GitHub repo: GitHub Repo. Metabolomics data is available at the NIH Common Fund’s National Metabolomics Data Repository (NMDR) Web site, the Metabolomics Workbench, https://www.metabolomicsworkbench.org where it has been assigned study ID (ST001339). RNASeq of melanoma cell lines (SKMEL5 subclones) is available at the Gene Expression Omnibus (Accession number: GSE122041).
SUPPLEMENTAL INFORMATION:
Supplemental information including Supplementary Figures, Tables, Methods can be found at: Supplementary Information (SI).
RESULTS:
Gene expressions and Flux Balance Analyses reveal enhanced capacity of redox balance in cells with reduced sensitivity to BRAF-inhibition
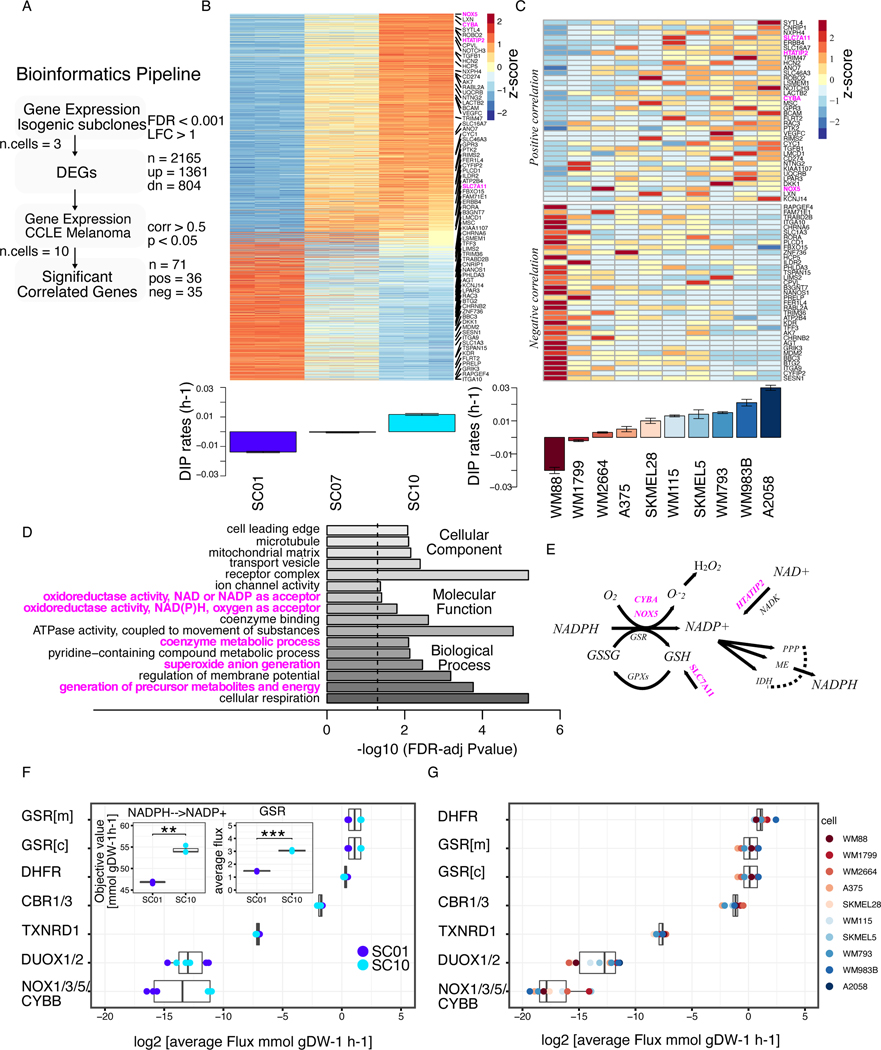
We recently reported that BRAF-mutated melanoma cell lines, including isogenic single-cell derived subclones, exhibit varying drug sensitivities to a small molecule BRAF kinase inhibitor (BRAFi) (8,26). Using Drug-Induced Proliferation (DIP) rates (Supplementary Table S1) (23), as a measure of drug effects, we examined the molecular correlates of BRAFi sensitivity, and reported that top 200 differentially expressed genes (DEGs) in drug-insensitive cells were enriched in processes and functions related to redox metabolism (24). Here, we extended our analysis to examine all significant DEGs among isogenic subclones (Supplementary Table S2) using their RNASeq profiles. Among 2,165 DEGs, 1361 (⅔) were up-regulated, while 804 (⅓) were down-regulated (Fig. 1A, B). Next, we projected all DEGs onto ten BRAF-mutated melanoma cells from CCLE datasets (27), and calculated their correlation to DIP rates. We selected significantly correlated genes using a criterion as outlined in Fig. 1A, and identified both positively and negatively correlated genes (Fig. 1C & Supplementary Table S3). Gene Ontology (GO) analysis on positively correlated genes in both subclones (Fig. 1B), and CCLE melanoma panel (Fig. 1C) showed enrichment of molecular functions related to redox balance, coenzyme metabolic process, and oxidoreductase activity (Fig. 1D & Supplementary Table S4). Specifically, we observed enrichment of gene signatures (CYBA, NADPH Oxidase 5 (NOX5), HTATIP2, SLC7A11, highlighted in magenta in Fig. 1B & C) related to redox balance and modulation, oxidoreductase activity, and reactions that utilize or consume antioxidant NADPH as a substrate (Fig. 1E). To quantify the contribution of redox enzymes toward NADPH oxidation, we next predicted steady-state fluxes through several major NADPH-oxidizing reactions using a previously published framework for integrating transcriptomic, kinetic, and thermodynamic data into Flux Balance Analysis (FBA) models of individual cancer cell lines (14,21). The drug-insensitive SC10 model showed significantly greater total conversion of NADPH to NADP+ compared to the drug-sensitive SC01 model. While most reactions did not display significant differences in predicted flux distributions, the SC10 model had significantly higher fluxes through Glutathione Disulfide Reductase (GSR), dihydrofolate reductase (DHFR), and NADPH oxidase (NOX1/3/5/CYBB) (Fig. 1F & Supplementary Table S5A). Consistent with the results in isogenic subclones, the subset of CCLE melanoma cell lines which are insensitive to MAPK-pathway inhibition also had higher predicted fluxes through GSR, DUOX1/2 reactions (Fig. 1G & Supplementary Table S5B–C). Taken together, these results suggest that melanoma cells with reduced drug sensitivity, as measured by DIP rates, exhibit enhanced anti-oxidation and redox balance. Specifically, we found that NADPH oxidation reactions coupled to glutathione exhibited higher gene expression and predicted fluxes in drug-insensitive melanoma cells.
Figure 1: BRAF-mutated melanoma cells with reduced sensitivity to BRAF-inhibition show elevated redox capacity.
(A) Bioinformatics pipeline to identify genes correlated with drug sensitivity, extension of an approach in previously published report (24). Briefly, Differentially Expressed Genes (DEGs) were identified from gene expression of isogenic subclones. Expressions of the identified genes were then correlated with DIP rates in CCLE melanoma panel. Statistical criteria used shown in the pipeline. (B) Heatmap of DEGs among subclones (top); Barplot of DIP rates of three subclones, SC01, SC07, and SC10 in 8 μM PLX4720 (bottom); (C) Heatmap of 71 genes from melanoma cell lines in CCLE panel that show a significant correlation between their expression and drug sensitivity. Genes in the upper panel show positive correlation, while genes in the lower panel show negative correlation to DIP rates. Highlighted in magenta are the redox-related genes involved in NADPH oxidation (top); Barplot of quantified DIP rates for the indicated melanoma cells in 8 μM PLX4720 (bottom). (D) Enriched Gene Ontology (GO) terms of the significant, and positively correlated genes from Figs. 1B, and C, and their FDR-adjusted p-values on barplot. (E) Schematic of the redox axis of NADPH oxidation, genes highlighted in magenta are the genes that are positively correlated in Fig. 1C. (F) Average FBA predicted metabolic fluxes through NADPH oxidation reactions between two single-cell-derived SKMEL5 subclones models, SC01, and SC10; three replicates of the model were considered. Inset (left) shows the objective value for net conversion of NADPH to NADP+ for two subclones (p-value = 3.79e-3); inset (right) average fluxes through one of NADPH oxidizing reactions, GSR (p-value = 2.72e-5)) (G) Average FBA predicted metabolic fluxes through NADPH oxidation reactions in CCLE melanoma cell line models. * p <= 0.05, ** p <= 0.01, *** p <= 0.001, **** p <= 0.0001.
Redox metabolites with antioxidant properties are maintained at higher levels after BRAF-inhibition in melanoma cells with reduced sensitivity to BRAF-inhibition
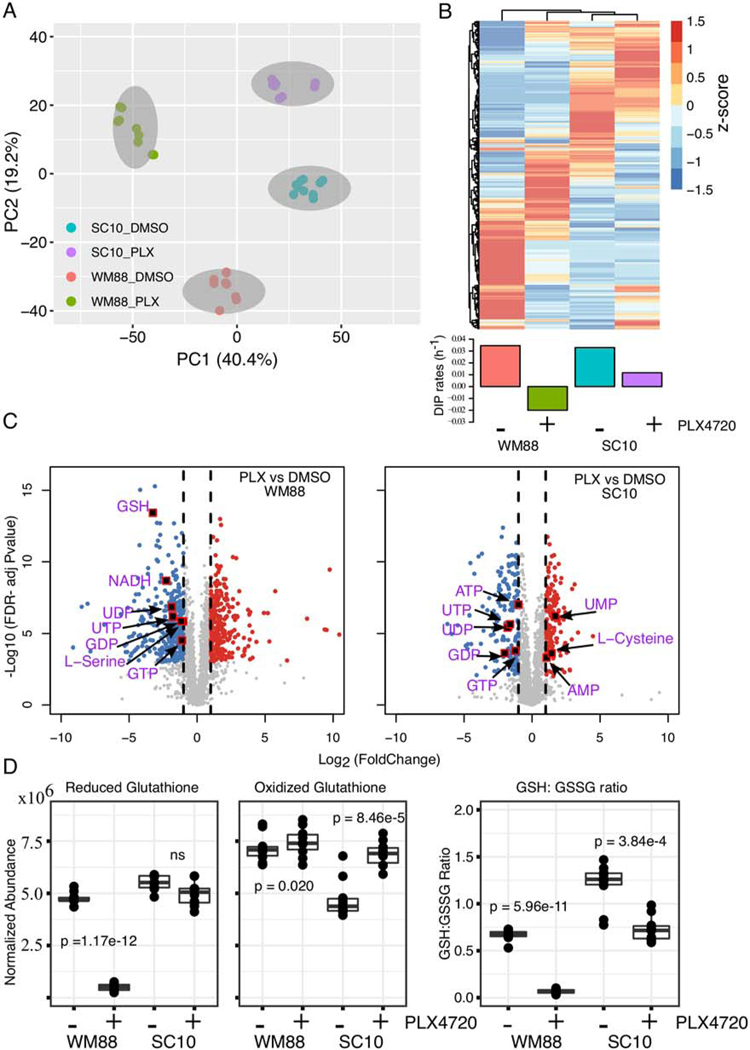
To directly assess the metabolite abundance, we performed a combined global untargeted-targeted liquid chromatography-mass spectrometry (LC-MS/MS) metabolomics analysis in BRAF-mutated melanoma cells under BRAF-inhibition. In particular, we selected one drug-sensitive (WM88) and one drug-insensitive (SC10) melanoma cell line, and compared their metabolite profiles both in the absence and presence of BRAF-inhibitors (Supplementary Table S6-7). In total, 2687 features or metabolites (unique retention time and m/z pairs) were detected, quantified and analyzed in these studies. Unsupervised Principal Component Analysis across all detected metabolites clustered sensitive melanoma cells away from insensitive cells at baseline along principal component 1 (PC1), while the drug-treated samples are separated along principal component 2 (PC2) (Fig. 2A). In general, the cells exhibit distinct metabolite profiles, with BRAF-inhibition inducing a marked metabolite profile shift (Fig. 2A, B). In these studies, 18 validated (Level 1) redox metabolites were quantified (Supplementary Table S8). These metabolites were validated through accurate mass measurements (<5 ppm error), similarities in isotope distribution (>90%), matched retention times, and matched fragmentation spectra based on in-house databases (28). In sum, we observed 2687 metabolites and using the combined global metabolomic profiles were able to mine the data for high confidence metabolite identification for molecules of interest (redox, GSH synthesis and energy metabolites).
Figure 2: Global metabolomic analysis reveals distinct metabolite profiles in BRAF-mutated melanoma cells with differing drug sensitivities.
(A) Principal component analysis (PCA) of metabolite profiles for two BRAF-mutated melanoma cells, WM88, and SC10 (SKMEL5-SC10), treated either in DMSO or 8 μM PLX4720 for 24 h. The two cell lines cluster and separate along PC1, while the drug-treated samples separate along PC2. (b) Heatmap analysis of identified metabolites in two melanoma cells (WM88 and SC10) either in DMSO or PLX4720 treatment (top) provides a global comparison of the relative abundances of individual metabolite compounds across the different groups; Barplot of DIP rates in WM88, and SC10 in DMSO or in 8 μM PLX4720 (bottom). (C) Volcano plots showing differentially expressed metabolites after treatment of two melanoma cells (WM88 or SC10) with 8 μM PLX4720 for 24 h. The negative log10 transformed Bonferroni corrected P-values (y-axis) are plotted against the average log2 fold changes in metabolite abundance (x-axis). Grey represents metabolites not differentially expressed, red are upregulated metabolites, and blue are downregulated metabolites. Vertical dotted lines represent log2 fold change of 1 or −1. Statistical cutoff of FDR-adjusted p-values < 0.001, and log-fold change greater or equal to 1 was used. (D) Box plots showing normalized abundances of reduced glutathione (GSH), oxidized glutathione (GSSG), and the ratio of GSH and GSSG in indicated melanoma cells treated with either DMSO or 8μM PLX4720 for 24 h: the solid line is the median, the box spans the first, and third quartiles, the whiskers extend to 1.5x the interquartile range--total of 10 replicates shown. Statistical significance calculated by paired t test, p-values as indicated in the figure.
To probe the treatment-dependent changes in metabolite abundances, we also performed pairwise comparisons between BRAFi treated and control samples within each cell line. In particular, we focused on the redox-related metabolites based on our earlier results on anti-oxidation and redox balance. Differentially expressed metabolites were determined based on a statistical cutoff of FDR-adjusted p-values < 0.001, and log-fold change greater or equal to 1. PLX4720 treatment significantly depleted the levels of energy metabolites such as GTP, UTP, ATP, GDP in both cell lines (Fig. 2C, Supplementary Fig. S1A). Among the redox metabolites, there was an increase in monophosphate nucleotides such as UMP, AMP in SC10 (Fig. 2C). Interestingly, the GSH level was significantly reduced in WM88 but not in SC10 upon BRAFi treatment (Fig. 2C). GSH levels, however, were comparable between two cells at baseline (Fig. 2D, Supplementary Fig. S1B). Because glutathione can exist in both reduced (GSH) and oxidized form (GSSG), we also quantified the levels of GSSG in both WM88 and SC10 cells. GSSG levels were lower in SC10 compared to WM88 at baseline, but similar in PLX4720 treatment (Fig. 2D). The ratio of reduced and oxidized glutathione, often used as an indicator of oxidative stress in cells, were also calculated. These data show that SC10 cells had a significantly higher GSH:GSSG ratio at baseline compared to drug-sensitive WM88 cells, rendering them a robust antioxidant capacity (Fig. 2D). Upon treatment, SC10 showed a slight decrease in GSH:GSSG ratio (μ_DMSO = 1.20, μ_PLX = 0.732, decrease of 1.6 fold). In contrast, WM88 showed an approximately 10-fold decrease in its ratio upon BRAF-inhibition (μ_DMSO = 0.67, μ_PLX = 0.067) (Fig. 2D).
Additionally, we also quantified the abundance of cysteine, cystine—precursors of de novo GSH synthesis. Transport of cystine into cells to form cysteine depends on the activity of cystine/glutamate antiporter, SLC7A11, one of the genes positively correlated with DIP rates. Both WM88 and SC10 have relatively similar low abundance values for the cystine molecule. Since cystine is at/near the limit of detection and below the limit of quantification of this particular LC-MS/MS analytical method, we did not detect cystine, therefore, we quantified the ratio of cystine and cysteine in drug-treated conditions. The ratio of cystine and cysteine under treatment was significantly higher in WM88 than in SC10, indicating a rapid accumulation of extracellular cystine (Supplementary Fig. S1C). Additionally, BRAFi treatment significantly increased the levels of cysteine in SC10, and not in WM88 (Fig. 2C). Additionally, serine and glycine can be used for a substrate for GSH synthesis (29). Levels of serine were substantially higher in SC10 compared to WM88 at baseline, and upon BRAFi treatment were maintained in SC10, and significantly decreased in WM88 (Fig. 2C, Supplementary Fig. S1A, Supplementary Fig. S1D). Furthermore, we examined the levels of cAMP at baseline and drug-treated conditions in these cells, and found that in both sensitive (WM88) and drug-insensitive (SC10), BRAFi treatment significantly decreased the levels of cAMP (Supplementary Fig. S1E). Interestingly, the levels of cAMP are much higher in SC10 compared to WM88 at baseline (Supplementary Fig. S1E). Among other energy metabolites, glycine increased in abundance in both cells upon BRAFi treatment, whereas glutamate decreased in both (Supplementary Fig. S1F-G). Collectively, these results indicate that drug-insensitive cells have a robust anti-oxidation and redox system that can be maintained under BRAF-inhibition, perhaps through both an efficient regeneration and de novo synthesis of GSH.
Pharmacological inhibition of redox enzymes synergizes with MAPK-inhibition in BRAF-mutated melanoma cells
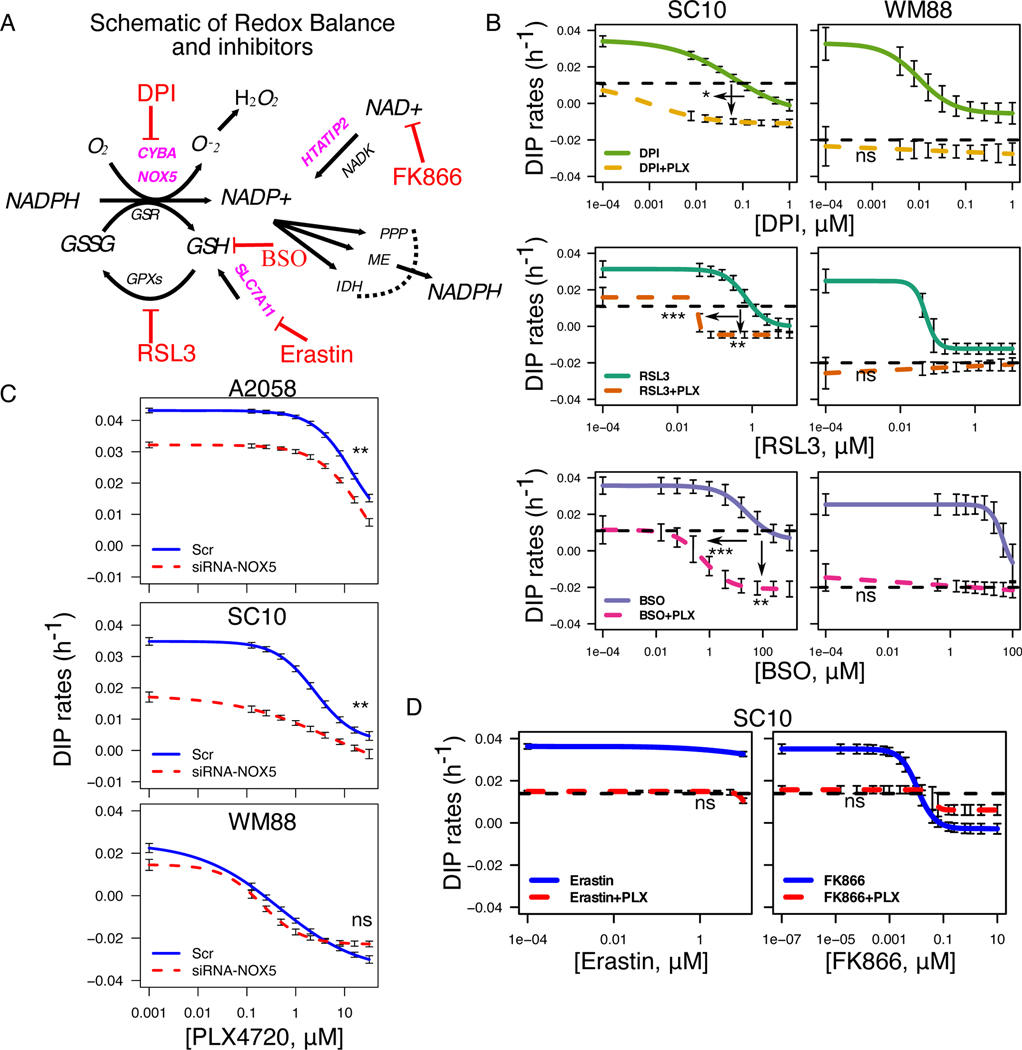
Based on our results involving antioxidants NADPH (Fig. 1), and GSH (Fig. 2), we carefully constructed a schematic of reactions and identified the known inhibitors of the redox components (Fig. 3A). Because our data suggest a positive correlation between enhanced anti-oxidation balance in melanoma cells and reduced drug sensitivity, we examined whether we could alter sensitivity of melanoma cells to BRAFi by using redox inhibitors. To evaluate the role of different redox components, we tested BRAF-inhibitors in combination with known redox inhibitors such as DPI, RSL3, Buthionine sulfoximine (BSO), Erastin, and FK866. We subjected two cell lines, one sensitive (WM88), and one insensitive (SC10) to increasing concentrations of either DPI, RSL3, or BSO alone or in combination with 8 μM PLX4720 (Fig. 3B). While both cells exhibited concentration-dependent anti-proliferative effects to single redox inhibitors (DPI, RSL3, BSO), enhanced effects of combination were observed only in the insensitive, and not in cells already sensitive to PLX4720 (Fig. 3B). Similar effects were seen in A2058, considered as largely insensitive BRAF-mutated melanoma cells (Supplementary Fig. S2A). Depending on the drugs, the combination either increased efficacy (maximum effect), or potency (more effect with less drug) or both in drug-insensitive SC10. Because of the known off-target effects of the pharmacological agents, we used small interfering RNA (siRNA)-mediated knockdown of the identified targets to validate the effects of the combinations. We selected two drug-insensitive cell lines, A2058, and SC10, based on their NOX5 expression at baseline (Supplementary Fig. S2B). Consistent with the pharmacological inhibition, siRNA-knockdown of NOX5 enhanced the effects of BRAFi in NOX5hi BRAF-mutated melanoma cells (Fig. 3C & Supplementary Fig. S2C). Interestingly, the combination increased both potency and efficacy in drug-insensitive cells (SC10, A2058) and was similar for most concentrations in drug-sensitive cells (WM88) (Fig. 3C & Supplementary Fig. S2C). However, not all redox inhibitors enhanced the effects of PLX4720. Erastin (SLC7A11 inhibitor) alone did not exhibit any effects on SC10, and its combination with PLX4720 was similar to the effects of PLX4720 alone (Fig. 3D, black dotted line). Similarly, the combination of FK866 (which inhibits the enzyme NAMPT, and thus depletes NAD+) with PLX4720 was not advantageous compared to single agents alone. At lower concentrations, the effect of combination was similar to PLX4720 alone, and at higher concentrations, similar to FK866 alone, indicating that the combination did not enhance the effects of single agents (Fig. 3D). This is an interesting result, as it suggests that inhibition of only certain redox components may enhance the effects of BRAF-inhibitors in BRAF-mutated melanoma cells.
Figure 3: Pharmacological inhibition of redox enzymes enhances the effects of BRAFi in BRAF-mutated melanoma cells.
(A) Schematic of reactions identified in Figure 1 with pharmacological inhibitors targeting each node as shown. (B) Drug-dose response curves of SC10 (left), and WM88 (right) either in single agents or in combination with 8μM PLX4720. Arrows show the enhanced effects of combinations, indicating that combination enhances the effects of single agents in drug-insensitive (SC10), and not in WM88 cells. (C) Dose-response curves of A2058, SC10 and WM88 cells either in scrambled control or in NOX5-siRNA at increasing concentrations of PLX4720 (32uM and 2-fold decrease), solid-blue and dotted-red lines represent DIP rates for scrambled control+PLX4720, and siRNA+PLX4720 respectively. (D) Drug-dose response curves of SC10 either in single agents or in combination with 8μM PLX4720. In both (B), and (D), black dotted lines represent the DIP rates in 8μM PLX4720; colored solid lines denote the DIP rates in increasing concentrations of single agents (DPI: NADPH Oxidase (NOX) inhibitor, RSL3: GPX4 inhibitor, BSO: GCLC inhibitor, potent depletion of GSH, Erastin: SLC7A11 inhibitor, FK866: NAD+ depletion, NAMPT inhibitor); while the colored dotted lines denote the drug responses of combination of 8μM PLX4720 and specific single agents corresponding to the colored solid curve (3+ biological replicates). For SC10: DPI-PLX (IC50_single=0.062μM, IC50_double=0.0084μM, p1=0.04), RSL3-PLX (IC50_single=1.22μM, IC50_double=0.029μM, p1=6.73e-7, p2=1.15e-3), BSO-PLX (IC50_single=13.8μM, IC50_double=0.209μM, p1=2.66e-4, p2=1.13e-3); for WM88: DPI-PLX (p=ns), RSL3-PLX (p=ns), BSO-PLX (p=ns); * p <= 0.05, ** p <= 0.01, *** p <= 0.001, p >= 0.05 (ns); p1 and p2 represent p-value for difference in IC50 (potency) and difference in maximum effect (efficacy) respectively. Significance tests for siRNA-knockdown are presented in Fig. S2C.
Enhanced effects of combination are due to increased oxidative stress
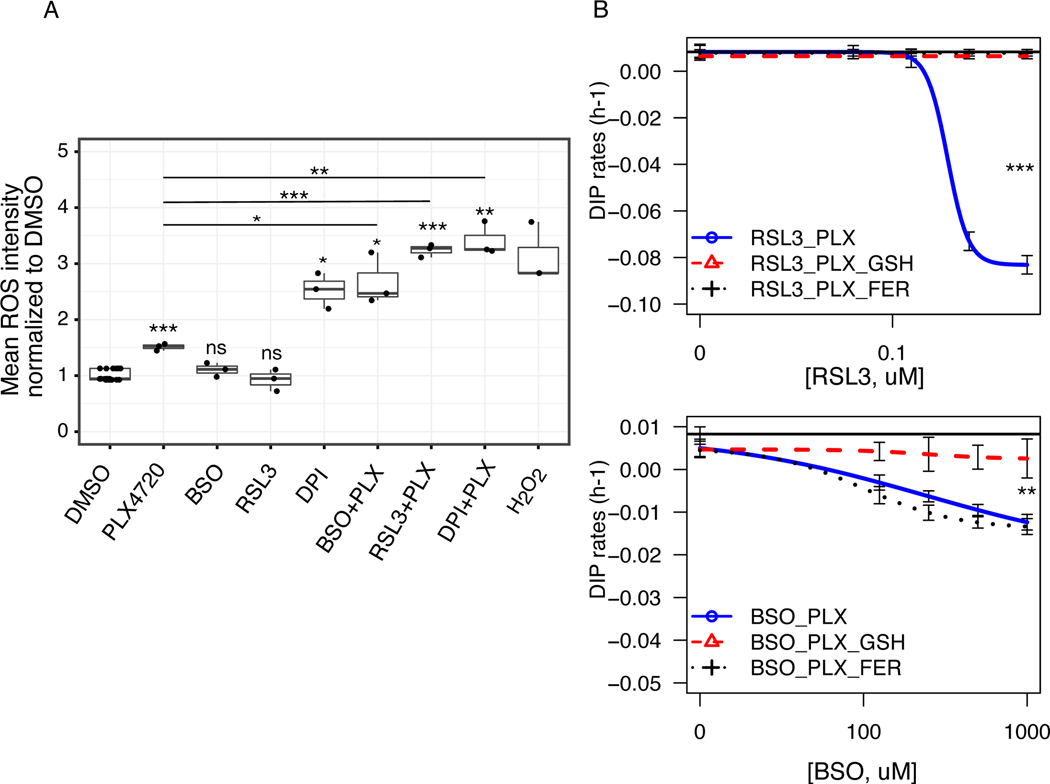
Our drug combinations suggest that co-administration of certain redox inhibitors with PLX4720 enhance the benefits of targeted therapies in melanoma (Fig. 3). To investigate how co-administrations affect redox balance in melanoma cells, we stained BRAF-mutated melanoma cells, SKMEL5 treated with either single agents or combinations with CellROX® Deep Red Reagent. Specifically, we tested three drug combinations that showed increased drug effects with BRAF-inhibitors: DPI, RSL3, and BSO. Unlike in BSO and RSL3, the total cellular reactive oxygen species (ROS) levels were increased upon PLX4720 treatment (p < 0.001) (Fig. 4A & Supplementary Fig. S3A), consistent with the results in earlier reports (19). Although counter-intuitive, DPI treatment as a single agent induced a significant increase in ROS intensity compared to DMSO control (Fig. 4A & Supplementary Fig. S3A). This result is consistent with studies that suggest DPI is a non-specific NOX-inhibitor, is shown to inhibit xanthine oxidase, eNOS and causes an enhanced oxidative stress in cells (30). Similarly, all three drug combinations (BSO+PLX, RSL3+PLX, & DPI+PLX) resulted in a significant increase in total cellular ROS compared to DMSO control, PLX4720 alone or the respective single agents (Fig. 4A & Supplementary Fig. S3A). To examine whether ROS induction in drug combination differs by cell lines, we compared total ROS intensity in four different melanoma cells: two drug-insensitive cells (SC10, A2058) and two drug-sensitive cells (WM88, SC01). Our analyses suggest that the ROS induction due to combination varies by cell lines. In the concentrations tested, we saw a general trend towards an increase in ROS intensity in combinations in all cell lines. While only PLX4720+DPI induced significant ROS compared to DMSO in all three cells (A2058, SC10, SC01), other treatment groups exhibited significant increase only in SC01 (Supplementary Fig. S3B). Due to substantial cell death in WM88, we didn’t have enough cells to examine total ROS. Our results reveal that co-administration of redox inhibitors with PLX4720 disrupts redox balance in cells, and causes an increase in oxidative stress that varies among cell lines based on their inherent antioxidant capacity. Therefore, we sought to test whether the lethal effects of these drug combinations could be mitigated by exogenous antioxidants. We excluded the combination of DPI and PLX4720, because of the broad off-target effects of DPI. For the other two combinations, we co-treated melanoma cells with two known antioxidants: reduced Glutathione (GSH), and lipophilic antioxidant Ferrostatin-1 (FER). The effects of PLX4720 alone could not be rescued by either GSH, and FER (Supplementary Fig. S4), indicating that the antiproliferative effects of PLX4720 is not due to oxidative stress alone. Interestingly, supplementation of the culture medium with GSH substantially rescued both RSL3, and BSO-induced cell death in combination (Fig. 4B), while Ferrostatin-1 rescue was specific to RSL3 only, confirming the glutathione levels as a mechanistic link for reduced drug sensitivity in BRAF-mutated melanoma cells.
Figure 4: Co-treatment of redox inhibitors and BRAFi induces oxidative stress.
(A) Box plots showing total cellular ROS mean intensity per cell for either single agents or in combinations normalized to intensity in DMSO control for melanoma cells (SKMEL5 fluorescently tagged with GFP-H2B) stained with CellROX® Deep Red Reagent. Cells were treated with DMSO control, 8 μM PLX4720, and H2O2 (1mM incubated for an hour); middle panel has BSO (500 μM), RSL3 (78.125 nM), and DPI (1.25 μM); in box plot, the solid line is the median, the box spans the first, and third quartiles, the whiskers extend to 1.5x the interquartile range. (B) Fitted drug-dose response curves of quantified DIP rates of SKMEL5 melanoma cells in drug combinations (RSL3+PLX, left), and combinations (BSO+PLX, right). Two known antioxidants--reduced glutathione (GSH, 5mM), and ferrostatin-1 (FER, 5μM) were added to the combinations at day 0, and replenished at 72 h along with growth medium, and inhibitor. Solid black lines denote the DIP rates in 8 μM PLX4720. The effects can be rescued by GSH in both, while Ferrostatin-1 rescues only in RSL3 combination (3+ biological replicates). For CellRox staining in (A), p-values of treatment groups compared to DMSO control are: PLX4720 (6.16e-4), BSO (ns), RSL3 (ns), DPI (0.013), BSO+PLX (0.023), RSL3+PLX (3.60e-4), DPI+PLX (4.60e-3); p-values of combination groups compared to PLX4720 alone are: BSO+PLX (0.046), RSL3+PLX (1.58e-4), DPI+PLX (6.36e-3); * p <= 0.05, ** p <= 0.01, *** p <= 0.001, p >= 0.05 (ns).
Pan-melanoma metabolic models exhibit altered redox metabolism and enhanced antioxidative capacity
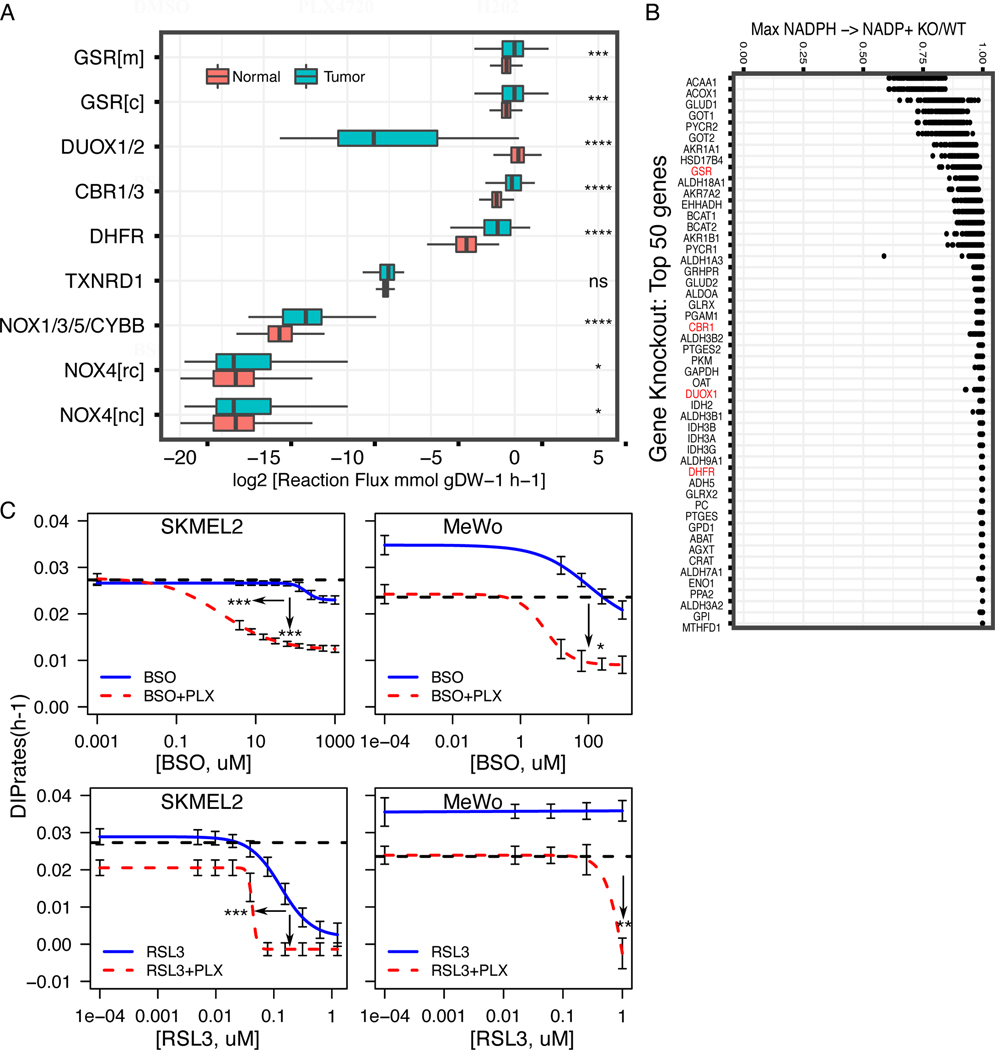
While melanomas with reduced sensitivity to BRAF-inhibition show enhanced antioxidative capacity potentiating redox-targeting therapies, it remains unclear whether other melanoma subtypes display similar or varying redox phenotypes. To investigate whether alterations in redox metabolism are common attributes across melanomas, FBA models of 103 melanoma tumors from The Cancer Genome Atlas (TCGA) and 860 normal skin tissues from the Genotype-Tissue Expression (GTEx) project were developed using their respective gene expression profiles (21,31,32). Melanoma tumor models displayed greater steady-state fluxes through major NADPH-oxidizing reactions including GSR and DHFR compared to normal tissue models (Fig. 5A). Interestingly, normal tissue models predicted significantly greater flux through DUOX1/2, an NADPH-oxidizing enzyme involved in cellular ROS generation; thus, increased activity of antioxidant-generating GSR and decreased activity of ROS-generating DUOX1/2 likely contribute to the enhanced antioxidative capacity in melanoma tumors compared to normal tissues. These findings are similar to the comparison in predicted metabolic fluxes between drug-insensitive SC10 cells and drug-sensitive SC01 cells (Fig. 1F), suggesting a possible progression in redox capacity from normal skin tissues (low capacity) to drug-sensitive melanomas (medium capacity) to drug-insensitive melanomas (high capacity).
Figure 5: Average FBA predicted metabolic flux distributions in TCGA melanoma tumors compared to normal skin tissues identify key redox vulnerabilities in pan-melanoma tumors.
(A) Box plots showing estimated average reaction fluxes through different enzymes in NADPH oxidation pathways in melanoma tumors (red box-plot) compared to normal skin (grey-green box-plot). Each dot represents a distinct tumor (expression from TCGA melanoma patient) or normal tissue (expression from Genotype-Tissue Expression (GTEx). (B) Effect of in-silico gene knockdown on predicted net conversion of NADPH to NADP+ in TCGA melanoma tumors, expressed as a ratio of net conversion after (KO) versus before knockdown (WT) (KO/WT). Shown are top 50 genes with greatest effects on NADPH net conversion, enzymes highlighted in red are genes in Fig. 5A. (C) Drug-dose response curves of NRAS-mutated melanoma cells (SKMEL2, left), and NF1-mutated melanoma cells (MeWo, right) in single agents or in combination with 8μM PLX4720. In both (left, and right), black dotted lines represent the DIP rates in 8μM PLX4720; colored solid lines denote the DIP rates in increasing concentrations of single agents (RSL3: GPX4 inhibitor, BSO: GCLC inhibitor, potent depletion of GSH); while the colored dotted lines denote the drug responses of combination of 8μM PLX4720 and specific single agents corresponding to the colored solid curve (3+ biological replicates). * p <= 0.05, ** p <= 0.01, *** p <= 0.001, **** p <= 0.0001. For SKMEL2: BSO-PLX (IC50_single=188 μM, IC50_double=1.39 μM, p1=1.37e-4, p2=5.64e-20), RSL3-PLX (IC50_single=0.131 μM, IC50_double=0.0429 μM, p1=9.40e-4, p2=ns). For MeWO; BSO-PLX (p2 = 0.01), RSL3-PLX (p2 = 6.6e-3); p1 and p2 represent p-value for difference in IC50 (potency) and difference in maximum effect (efficacy) respectively.
To measure redox fluxes under MAPKi treatment, we integrated MAPK-pathway inhibition in our FBA models. Based on recent studies (33), we assumed that BRAFi inhibits glycolysis in implementing our models. Parmenter et al. demonstrated that BRAFi inhibits glycolysis by suppressing expression of glucose transporters (GLUT1/3) as well as hexokinase (HK2). To simulate this effect in FBA models, we examined the effect of both eliminating cellular glucose uptake and hexokinase activity on the ability of melanoma TCGA samples to oxidize NADPH to NADP+. We observed a large stratification when comparing predicted BRAFi sensitivity (maximal NADPH oxidation before versus after BRAFi implementation) between melanoma tumor models (Supplementary Fig. S5A). We then performed Flux Variance analysis to identify individual reaction fluxes that correlate with predicted BRAFi sensitivity. Fluxes through DHFR, NOX1/3/5/CYBB, and NOX4 were significantly greater in predicted BRAFi-resistant tumors (Supplementary Fig. S5B). Additionally, using our FBA models of TCGA tumors, a simulated metabolome-wide gene knockout screen was performed, where for each of the 3,268 genes in the Recon3D metabolic reconstruction, the effect of gene knockout on total NADPH oxidation in each melanoma model was determined (Supplementary Fig. 5B). Many of the gene knockouts with greatest effects on NADPH oxidation across TCGA melanoma models included major genes involved in the reduction of NADP+ to NADPH (e.g. GLUD1, GLUD2, IDH2, MTHFD1), as well as genes utilizing reduced NADPH to increase cellular antioxidant stores or decrease levels of reactive oxygen species (e.g. GSR, CBR1) (21). To determine whether simulated gene knockdown results are biased towards genes proximal to NADPH reduction in the metabolic network, next we calculated the distance between each metabolic reaction to the closest NADPH-reducing reaction. Gene distance has a mean value of 1.31 with a maximum value of 3. Although all of the top 50 genes from the simulated knockdown screen have distance values of 0 or 1, we observed small distance values for all genes suggesting a high degree of connectivity to NADPH metabolism for all genes across the human metabolic network (Supplementary Fig. S5C). Together, these analyses suggest that our results are not biased by network-distance and it is relevant that genes encoding enzymes of Fig. 5A, are featured in the top 50 genes shown in Fig. 5B.
Next, we sought to test whether our identified redox-targeting therapies work in other subtypes of melanoma. We applied the drug combinations (BSO+PLX, and RSL3+PLX) in NRAS-mutated (SKMEL2), and NF1-mutated (MeWo) melanoma cells. The combinations increased efficacy of BRAF-inhibitors in both SKMEL2, and MeWo cells, consistent with results above (Fig. 5C). Although PLX4720 has some off-target effects at moderate to high concentrations in non-BRAF-mutated melanoma cells (34), we observed these effects are minimal in terms of growth rates (DIP rates) in PLX4720 alone in both SKMEL2 and MeWo cells and only enhanced when it is combined with redox inhibitors (Fig. 5C). Furthermore, to investigate the effects of these agents on normal cells, we treated Primary Human Epidermal Melanocytes, PCS-200–013 with increasing concentrations of BSO, RSL3, and combined them with 8μM PLX4720. The effects of drugs, both single, and double agents are statistically insignificant compared to DMSO control, suggesting that the primary cells are largely unaffected by these agents (Supplementary Fig. S6). Collectively, these findings suggest that altered redox metabolism and enhanced antioxidant production are common features among melanomas, and that inhibition of redox enzymes may be a viable treatment strategy for melanomas in general.
DISCUSSION:
Overall, we report here that higher antioxidant ability of cells correlates to lower drug sensitivity to targeted therapies in melanoma. Our analyses reveal an association between an elevated redox capacity, involving two reducing cofactors NADPH and GSH, and reduced drug sensitivity in melanoma. Furthermore, we show that drug-insensitive melanoma cells not only possess a robust redox system, but also maintain key antioxidant metabolites during treatment. Indeed, we show that the disruption of this redox buffer improves the outcomes of targeted therapies. In summary, our findings suggest that modulation of cancer antioxidant defense could be exploited to augment the benefits of existing therapies in melanoma (16). This strategy may extend beyond melanoma, as antioxidant pathways have been implicated in tumor progression, and drug resistance (22,35,36).
How ROS affects cancer cells has been a contentious issue for years. ROS likely elicits a broad spectrum of cellular responses (13,17). Although still debated, its effects on cancer cells can best be described as hormesis (17): at low levels, it can mediate sustained network signaling leading to enhanced proliferation (17), while at high levels, it can induce cellular damage and cell death (37). The level of ROS that a cell can handle, thus, likely depends on how functional its antioxidant machinery is. In general, tumors can sustain a much higher level of ROS compared to normal tissues by invoking their antioxidant systems, thereby creating a high redox homeostatic state (18). Melanoma tumors could possibly exist in such a high redox homeostatic state, as we see an increased activity of antioxidant-generating GSR, and decreased activity of ROS-generating DUOX1/2 in TCGA melanoma tumors compared to normal skin. However, there exists a significant variability even within tumors, captured well in melanoma cell lines, suggesting redox capacity perhaps could be on a continuum: low capacity (normal skin), moderate capacity (drug-sensitive melanomas), and high capacity (drug-insensitive melanomas).
Based on modeling by Flux Balance Analysis (FBA), the high redox capacity in drug-insensitive melanomas is predicted to stem from elevated fluxes through metabolic reactions which generate important antioxidant molecules including glutathione. This computational approach towards predicting genome-scale metabolic phenotypes has many advantages. By integrating the most comprehensive model of human metabolism to date with transcriptomic data from cell lines or tumors of interest, quantitative predictions on the metabolic status of these samples can be obtained, which would not be available from gene expression analysis alone (21,25). Additionally, although FBA models can only make steady-state flux predictions, the interconnections between 13,000+ metabolic reactions in the human metabolic network often yield predictions with greatly improved accuracy and utility compared to smaller dynamic metabolic models which are missing these genome-scale interactions (14). Because of these advantages, FBA has led to many successful predictions in cancer metabolism, providing powerful insights into both cancer pathophysiology and treatment strategies (38–40). Additionally, we used FBA to predict metabolic fluxes for reactions in central carbon metabolism (glycolysis and TCA cycle) in TCGA melanoma tumors, and compared them with GTEx normal skin tissues (Supplementary Fig. S7A). Our analysis suggests that many reactions had a statistically significant difference between melanoma tumors and normal skin, with most glycolytic and some TCA cycle fluxes significantly increased in melanoma tumors (Supplementary Fig. S7A). Interestingly, similar analyses between BRAF-mutated TCGA melanoma tumors compared with TCGA non BRAF-mutated melanomas show no difference in predicted metabolic fluxes through central carbon metabolism (Supplementary Fig. S7B). By coupling FBA, gene expression, and metabolomics screen, we show that the drug-insensitive melanomas have an increased dependence on antioxidant systems. This increased dependence could present a therapeutic opportunity in melanomas. Indeed, our own results and recent reports inform that redox modulation could be a viable strategy to improve the treatment outcomes (41).
Glutathione (GSH) is the most abundant antioxidant in the cell. It can be synthesized either de novo or regenerated using NADPH as the substrate (42). De novo GSH biosynthesis requires cysteine, which is imported into the cells by cystine/glutamate antiporter, SLC7A11 (35,36). Our data suggest that drug-insensitive melanoma cells have both upregulated SLC7A11 expression, and higher predicted fluxes through GSH regenerating GSR, suggesting an efficient maintenance of glutathione. Intriguingly, drug-insensitive melanoma cells can also maintain higher levels of GSH, and maximally utilize cysteine during treatment. In addition, the level of serine is higher in drug-insensitive cells, and increased upon treatment. Since serine is a precursor of cysteine, it may help cells maintain necessary levels of glutathione (29). Therefore, we next questioned whether the modulation of GSH level affects drug sensitivity in melanoma. Inhibition of SLC7A11, however, did not improve the effects of BRAFi in melanoma cells, indicating that melanoma cells could maintain GSH levels through NADPH regeneration. In contrast, GSH depletion by BSO enhanced the effects of combination. Thus, GSH modulation appears as an exciting therapeutic target in melanoma. Efficient regeneration of GSH requires sufficient NADPH, another important cofactor in cells. Our work focused on NADPH oxidation (consumption), and examined the contributions of several enzymes that convert NADPH to NADP+. Sixteen out of the top 50 enzymes that had the most effect on the predicted fluxes through NADPH are known to be involved in NADPH production pathways. Examining the importance of NADPH production in melanoma drug sensitivity is a possible area of future investigation. We speculate that drug-insensitive melanoma cells may also possess a robust NADPH production pathway. It is worth noting that our current work refers to the early response of melanoma to BRAFi, and examines the relationship between inherent redox potential in a cell and its initial drug sensitivity. Our drug-insensitive cells are not drug-resistant, instead they respond to MAPKi albeit at varying levels, such that the drug responses are in a continuum. When chronically made resistant to BRAFi, melanoma cells will go through certain adaptations, including the emergence of drug resistant phenotypes. When melanoma cells adapt long-term to perturbations and oxidative stress, they exhibit a strong activation of NRF2-mediated oxidative response, and regeneration of reduced glutathione (16). Melanoma cells undergo metabolic modifications including the changes in their mitochondria, redox-alterations and metabolic plasticity to survive prolonged exposure to MAPKi (43), and ultimately develop resistance.
Consistent with earlier reports (44,45), we observed that BRAFi alone also induced ROS in melanoma. However, BRAFi induced effects on melanoma cells could not be rescued by known antioxidants, suggesting that effects of BRAFi on drug-insensitive melanoma cells are not solely due to ROS. Another possibility is that MAPKi-insensitive melanoma cells perhaps utilize their robust antioxidant system to tolerate the induced ROS. It is implicated that MAPKi inhibits glycolysis in melanoma cells (15,33,43) and leads to an increase in ROS production (44). We postulate that inhibition of glycolysis might make melanoma cells over-reliant on antioxidants. We believe that elevated antioxidant usage may be a strategy for melanoma cells to thrive initially under MAPKi. However, this very strategy makes them vulnerable to redox inhibition when redox pathways are co-targeted with MAPKi, consistent with recent studies that show metabolic vulnerability in SLC7A11hi cancer cells (36). Interestingly, this vulnerability could be a general feature of melanoma tumors regardless of the mutational status, as indicated by FBA analysis on TCGA samples, and enhanced effects of PLX4720 when combined with redox inhibitors in SKMEL2 (NRAS-mutated) and MeWo (NF1-mutated) melanoma cells. Interestingly, the effects of these drugs and their combinations are statistically insignificant in primary melanocytes. Therefore, an increase in oxidative stress due to the combinations identified here, possibly disrupts that redox balance. One such agent for combination, RSL3, is known to induce ferroptosis in cancer cells (46). Our results are, thus, consistent with recent reports that suggest ferroptosis-inducing drugs may enhance the current treatment options for melanoma (47). Ferroptosis has recently been exploited in different cancer types as an alternative way to reduce the fraction of drug-tolerant persister cells (48). Recently, redox metabolite cystine and its antiporter SLC7A11 have been linked to ferroptosis in pancreatic cancer (46). Yet, one important consideration is that ROS is not one, but a family of chemical species. Therefore, it remains to be determined whether different species of ROS have different effects on melanoma cells. Interestingly, inhibition of NOX5 by genetic knockdown (siRNA) enhanced the effects of PLX4720 in NOX5hi cells, suggesting the NOX5-generated ROS might exhibit pro-proliferation signaling. This is consistent with recent studies that indicate NOX5-mediated ROS-induced downstream signaling may contribute to the growth of melanoma cells and other human tumors (49). It is tempting to speculate the role of NADPH Oxidases in maintaining cellular bioenergetics as recently described (50). Although additional experiments are necessary, redox signaling activated in part due to altered phosphatase function has been implicated in NOX5-mediated cell proliferation (49). Further studies are required to ascertain the species of ROS that have detrimental effects on melanoma cells. Taken together, these results combined with recent reports support the hormetic nature of ROS-mediated signaling. Molecular basis for hormesis would be an exciting area of future investigation. One could imagine this could be cell-line or tumor specific, as the gene regulatory network (GRN) has been linked with the ability of cells to undergo metabolic plasticity (13). Future work seems necessary to also investigate the compensatory antioxidant pathways, and how they help maintain redox homeostasis in melanoma cells.
In summary, our integrative approach demonstrates that enhanced redox balance capacity provides an early survival advantage in melanomas treated with MAPKi. This finding extends beyond BRAF-mutated melanomas (Fig. 5) and, we speculate, other cancer types in which an overactive MAPK pathways is the oncogenic driver. Our analyses indicate that this finding is actionable, and identify new vulnerabilities in melanoma cells: Co-administration of redox inhibitors would significantly increase the effect of MAPKi in insensitive tumors. In virtually all cases, we observed both enhanced potency and efficacy (24) in such drug combinations compared to single agents. Intriguingly, no increased effects were observed in sensitive cells. These results suggest strategies for personalized interventions. Thus, biomarkers for redox potential in melanomas could be used to stratify patients, in order to identify the ones that would benefit from a triple drug combination (MAPKi plus redox inhibitors). Naturally, additional studies in large datasets are necessary to conclusively firm up correlations between BRAFi responsiveness and gene expression or fluxes involving redox pathways. We envision such studies as eminently feasible, integrating data from transcriptomics, epigenomics, and epigenetics, and possibly involving machine learning algorithms to substantiate the goodness of predictive biomarker sets.
Supplementary Material
ACKNOWLEDGMENTS:
We are grateful to Meenhard Herlyn (Wistar Institute), Kim Dahlman and Ann Richmond (Vanderbilt University) for kindly providing BRAF-, NRAS-, and NF1-mutated melanoma cell lines; to Joshua P. Fessel for insightful discussions on NADPH oxidases; to Darren R. Tyson, Christian T. Meyer, David J. Wooten, Leonard A. Harris for useful discussions; to Jing Hao and Kevin Contreras for support in reagent procurement, and experimental preparation. We would also like to thank Prof. Kevin A. Janes (University of Virginia) for critical review of this manuscript.
This work was supported by the US National Institutes of Health Grants U54 CA217450, U01 CA215845, R01 CA186193, and U01 CA174706 (to V.Q.); U01 CA215848 (to M.L.K); Vanderbilt Institute for Clinical and Translational Research (VICTR) grants 16721, and 16721.1 (to B.B.P). J.E.L is supported by NIH/NCI F30 CA224968. C.E.H. is supported by NIH/NCI F31 CA221147. P.E.S. is supported by 2T32DK007563-31. K.N.H. is now at Houston Methodist Research Institute, Houston, TX. C.J.R. is now at Yale University School of Medicine MD-PhD Program, New Haven, CT.
Footnotes
DECLARATION OF CONFLICT OF INTERESTS:
V. Quaranta is Academic Co-Founder at Parthenon Therapeutics, Inc. B.B. Paudel and V. Quaranta have pending patent applications on drug combination targets in BRAF-mutated melanoma. No potential conflicts of interest were declared by other authors.
REFERENCES:
- 1.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. Massachusetts Medical Society; 2012;366:707–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizos H, Menzies AM, Pupo GM, Carlino MS, Fung C, Hyman J, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin Cancer Res. AACR; 2014;20:1965–77. [DOI] [PubMed] [Google Scholar]
- 3.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol. 2017;14:463–82. [DOI] [PubMed] [Google Scholar]
- 5.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017;546:431–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith MP, Brunton H, Rowling EJ, Ferguson J, Arozarena I, Miskolczi Z, et al. Inhibiting Drivers of Non-mutational Drug Tolerance Is a Salvage Strategy for Targeted Melanoma Therapy. Cancer Cell. 2016;29:270–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paudel BB, Harris LA, Hardeman KN, Abugable AA, Hayford CE, Tyson DR, et al. A Nonquiescent “Idling” Population State in Drug-Treated, BRAF-Mutated Melanoma. Biophys J. 2018;114:1499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 10.Warburg O. The Metabolism of Carcinoma Cells. J Cancer Res. American Association for Cancer Research Journals; 1925;9:148–63. [Google Scholar]
- 11.Dang CV. Links between metabolism and cancer. Genes Dev. 2012;26:877–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. [DOI] [PubMed] [Google Scholar]
- 13.Paudel BB, Quaranta V. Metabolic plasticity meets gene regulation. Proc. Natl. Acad. Sci. U. S. A. 2019. page 3370–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28:245–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23:302–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khamari R, Trinh A, Gabert PE, Corazao-Rozas P, Riveros-Cruz S, Balayssac S, et al. Glucose metabolism and NRF2 coordinate the antioxidant response in melanoma resistant to MAPK inhibitors. Cell Death Dis. 2018;9:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:R453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer D, Werth F, Nguyen HA, Kiecker F, Eberle J. Critical role of reactive oxygen species (ROS) for synergistic enhancement of apoptosis by vemurafenib and the potassium channel inhibitor TRAM-34 in melanoma cells [Internet]. Cell Death & Disease. 2017. page e2594–e2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris IS, DeNicola GM. The Complex Interplay between Antioxidants and ROS in Cancer. Trends Cell Biol. Elsevier; 2020;30:440–51. [DOI] [PubMed] [Google Scholar]
- 21.Lewis JE, Costantini F, Mims J, Chen X, Furdui CM, Boothman DA, et al. Genome-Scale Modeling of NADPH-Driven β-Lapachone Sensitization in Head and Neck Squamous Cell Carcinoma. Antioxid Redox Signal. 2018;29:937–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarmiento-Salinas FL, Delgado-Magallón A, Montes-Alvarado JB, Ramírez-Ramírez D, Flores-Alonso JC, Cortés-Hernández P, et al. Breast Cancer Subtypes Present a Differential Production of Reactive Oxygen Species (ROS) and Susceptibility to Antioxidant Treatment. Front Oncol. 2019;9:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris LA, Frick PL, Garbett SP, Hardeman KN, Paudel BB, Lopez CF, et al. An unbiased metric of antiproliferative drug effect in vitro. Nat Methods. 2016;13:497–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer CT, Wooten DJ, Paudel BB, Bauer J, Hardeman KN, Westover D, et al. Quantifying Drug Combination Synergy along Potency and Efficacy Axes. Cell Syst. 2019;8:97–108.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunk E, Sahoo S, Zielinski DC, Altunkaya A, Dräger A, Mih N, et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat Biotechnol. 2018;36:272–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardeman KN, Peng C, Paudel BB, Meyer CT, Luong T, Tyson DR, et al. Dependence On Glycolysis Sensitizes BRAF-mutated Melanomas For Increased Response To Targeted BRAF Inhibition. Sci Rep. 2017;7:42604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, McLean JA. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J Am Soc Mass Spectrom. 2016;27:1897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez AE, Ducker GS, Billingham LK, Martinez CA, Mainolfi N, Suri V, et al. Serine Metabolism Supports Macrophage IL-1β Production. Cell Metab. 2019;29:1003–11.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riganti C, Gazzano E, Polimeni M, Costamagna C, Bosia A, Ghigo D. Diphenyleneiodonium inhibits the cell redox metabolism and induces oxidative stress. J Biol Chem. 2004;279:47726–31. [DOI] [PubMed] [Google Scholar]
- 31.Weinstein JN, The Cancer Genome Atlas Research Network, Collisson EA, Mills GB, Mills Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project [Internet]. Nature Genetics. 2013. page 1113–20. Available from: 10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carithers LJ, Moore HM. The Genotype-Tissue Expression (GTEx) Project [Internet]. Biopreservation and Biobanking. 2015. page 307–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parmenter TJ, Kleinschmidt M, Kinross KM, Bond ST, Li J, Kaadige MR, et al. Response of BRAF-mutant melanoma to BRAF inhibition is mediated by a network of transcriptional regulators of glycolysis. Cancer Discov. 2014;4:423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105:3041–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji X, Qian J, Rahman SMJ, Siska PJ, Zou Y, Harris BK, et al. xCT (SLC7A11)-mediated metabolic reprogramming promotes non-small cell lung cancer progression. Oncogene. 2018;37:5007–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Olszewski K, Zhang Y, Lim EW, Shi J, Zhang X, et al. Cystine transporter regulation of pentose phosphate pathway dependency and disulfide stress exposes a targetable metabolic vulnerability in cancer. Nat Cell Biol. Nature Publishing Group; 2020;1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panieri E, Santoro MM. ROS homeostasis and metabolism: a dangerous liason in cancer cells. Cell Death Dis. 2016;7:e2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis NE, Abdel-Haleem AM. The evolution of genome-scale models of cancer metabolism. Front Physiol. 2013;4:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gatto F, Miess H, Schulze A, Nielsen J. Flux balance analysis predicts essential genes in clear cell renal cell carcinoma metabolism. Sci Rep. 2015;5:10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Folger O, Jerby L, Frezza C, Gottlieb E, Ruppin E, Shlomi T. Predicting selective drug targets in cancer through metabolic networks. Mol Syst Biol. 2011;7:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12:931–47. [DOI] [PubMed] [Google Scholar]
- 42.Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, Rabinowitz JD. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delgado-Goni T, Miniotis MF, Wantuch S, Parkes HG, Marais R, Workman P, et al. The BRAF Inhibitor Vemurafenib Activates Mitochondrial Metabolism and Inhibits Hyperpolarized Pyruvate-Lactate Exchange in BRAF-Mutant Human Melanoma Cells. Mol Cancer Ther. 2016;15:2987–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cesi G, Walbrecq G, Zimmer A, Kreis S, Haan C. ROS production induced by BRAF inhibitor treatment rewires metabolic processes affecting cell growth of melanoma cells. Mol Cancer. 2017;16:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ravindran Menon D, Das S, Krepler C, Vultur A, Rinner B, Schauer S, et al. A stress-induced early innate response causes multidrug tolerance in melanoma. Oncogene. 2015;34:4448–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Badgley MA, Kremer DM, Maurer HC, DelGiorno KE, Lee H-J, Purohit V, et al. Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science. science.sciencemag.org; 2020;368:85–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsoi J, Robert L, Paraiso K, Galvan C, Sheu KM, Lay J, et al. Multi-stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress. Cancer Cell. 2018;33:890–904.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Viswanathan VS, Ryan MJ, Dhruv HD, Gill S, Eichhoff OM, Seashore-Ludlow B, et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Antony S, Jiang G, Wu Y, Meitzler JL, Makhlouf HR, Haines DC, et al. NADPH oxidase 5 (NOX5)-induced reactive oxygen signaling modulates normoxic HIF-1α and p27Kip1 expression in malignant melanoma and other human tumors. Mol Carcinog. 2017;56:2643–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu W, Hu Y, Chen G, Chen Z, Zhang H, Wang F, et al. Novel role of NOX in supporting aerobic glycolysis in cancer cells with mitochondrial dysfunction and as a potential target for cancer therapy. PLoS Biol. 2012;10:e1001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data, calculated DIP rates, DEGs between subclones, SCGs for CCLE melanoma panel, expression data, codes used to generate the figures are available in the GitHub repo: GitHub Repo. Metabolomics data is available at the NIH Common Fund’s National Metabolomics Data Repository (NMDR) Web site, the Metabolomics Workbench, https://www.metabolomicsworkbench.org where it has been assigned study ID (ST001339). RNASeq of melanoma cell lines (SKMEL5 subclones) is available at the Gene Expression Omnibus (Accession number: GSE122041).