As of May 3, 2021, more than 154 million individuals around the world have been infected with the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), and 3.22 million have succumbed to coronavirus disease (COVID-19) (1). One major contributor to its morbidity and mortality is venous thromboembolism (VTE), which affects ∼17% of hospitalized patients with COVID-19 and ∼28% of those who require ICU support (2). Promisingly, a recent interim analysis of a multiplatform randomized controlled trial (N = 2,895) consisting of participants in the ATTACC (Antithrombotic Therapy to Ameliorate Complications of COVID-19), ACTIV-4 (Anti-thrombotics for Adults Hospitalized with COVID-19) and REMAP-CAP (Randomized, Embedded, Multifactorial Adaptive Platform Trial for Community-Acquired Pneumonia) studies indicates that full-dose anticoagulation therapy results in significantly improved outcomes, including reduced mortality and prevention of organ failure, compared with low-dose (prophylactic) anticoagulation therapy in hospitalized patients with COVID-19 (3). Although preliminary, these data provide a compelling rationale to explore several important mechanistic questions regarding the relation between COVID-19 and coagulopathy. These include 1) What are the molecular pathways by which SARS-CoV-2 infection induce coagulopathy in COVID-19? and 2) Is the coagulopathy unique to SARS-CoV-2 infections or is it generic to all severe viral respiratory tract infections?
In this issue of the Journal, FitzGerald and colleagues (pp. 687–697) provide data that fill in some crucial gaps in knowledge (4). Although COVID-19 affects multiple organs, the most severe form occurs when SARS-CoV-2 infects the lower respiratory tract, leading to pneumonia and, in certain cases, venous thromboembolic disease, including pulmonary embolism (5). To understand this mechanism, FitzGerald and colleagues reanalyzed data from several publicly available transcriptomic databases. Using bulk sequencing data of human bronchial epithelial cells infected with SARS-CoV-2 (6), they found that 10 genes (EDN1 [endothelin 1], SERPINB2 [serpin family B member 2], PLAU [plasminogen activator, urokinase], F3 [coagulation factor III, tissue factor], LYN [LYN proto-oncogene, Src family tyrosine kinase], C1QTNF1 [C1q and TNF related 1], PLAUR [plasminogen activator urokinase receptor], PDPN [podoplanin], CEACAM1 [CEA cell adhesion molecule 1], and NFE2L2 [nuclear factor, erythroid 2 like 2]) within the regulation of blood coagulation biologic process (7) were differentially expressed in human bronchial epithelial cells infected by SARS-CoV-2 (adjusted P < 0.05). Importantly, the SARS-CoV-2 infection significantly increased the protein expression of TF (tissue factor), which is the master regulator of the extrinsic pathway of blood coagulation. In contrast, SARS-CoV-2 infection had no significant effect on the expression of TFP1, a TF inhibitor, which led to a significant imbalance in the ratio between TF and TFP1 in favor of thrombosis. Another notable gene dysregulated by SARS-CoV-2 infection was PROS1, which encodes protein S. Protein S is a cofactor for protein C; together, they inactivate factors Va and VIIIa, leading to the inhibition of the coagulation cascade. By downregulating PROS1, SARS-CoV-2 infection creates a prothrombotic milieu in COVID-19 lungs. Interestingly, there are approved drugs (e.g., menadione and warfarin), which can modulate PROS1 (8). This may present an opportunity to repurpose these compounds for treating or preventing venous thromboembolic complications in patients with severe COVID-19.
These in vitro findings were largely recapitulated in human BAL samples from subjects with and without COVID-19. Using bulk and single-cell sequencing data, FitzGerald and colleagues identified bronchial epithelial cells as the predominant source of F3 (the gene that encodes TF), and macrophages were the main source for PLAUR (which encodes plasminogen urokinase-localizing protein) and SERPINB2 (which encodes PAI-1 [plasminogen activator inhibitor-2]), which are both negative regulators of plasmin that inhibit cross-linking of the fibrin clot. In the circulating immune cells of patients with COVID-19, however, none of the genes in the coagulation pathway were significantly dysregulated (compared with those of control subjects). Finally, FitzGerald and colleagues showed that this prothrombotic response may be unique to SARS-CoV-2 and not shared by other respiratory viruses such as influenza A, which did not significantly alter the expression levels of genes involved in the coagulation pathway. Consistent with this notion, a recent study has shown a significant relationship of genes related to coagulation (e.g., F3, PROS1, ITGB3, and TFPI2) with SARS-CoV-2 but not to other human coronaviruses (e.g., Middle East Respiratory Syndrome coronavirus [MERS-CoV] and severe acute respiratory syndrome coronavirus [SARS-CoV]) (9). Together, these data suggest that SARS-CoV-2 coaxes a unique response in the epithelial cells and alveolar macrophages that leads to a prothrombotic milieu in the lungs, which can result in thrombosis in situ or pulmonary embolism in susceptible individuals (Figure 1).
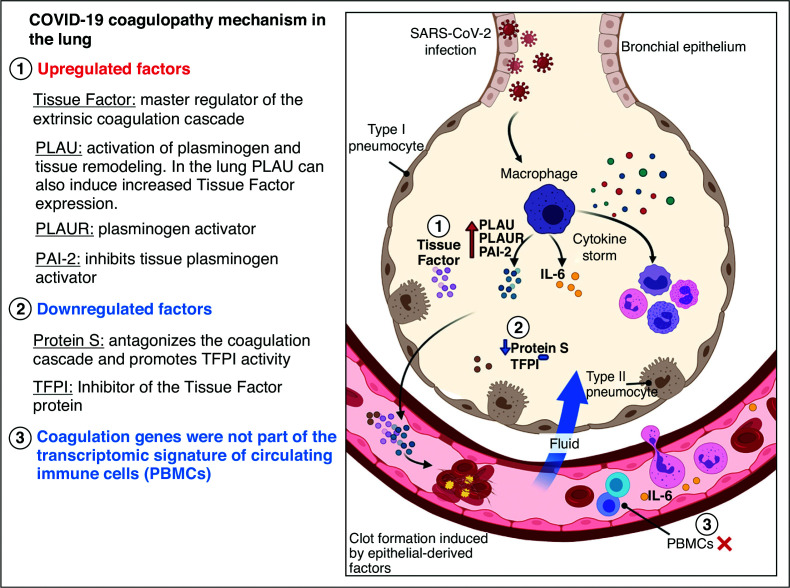
Figure 1.
Proposed mechanism of coagulopathy in lungs of patients with coronavirus disease (COVID-19). Figure was created with BioRender.com. PAI-2 = plasminogen activator inhibitor-2; PBMCs = peripheral blood mononuclear cells; PLAU = plasminogen activator urokinase; PLAUR = plasminogen activator urokinase receptor; SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2; TFPI = tissue factor pathway inhibitor.
There are some limitations to FitzGerald and colleagues’ work. First, their results were derived from experiments that contained small sample sizes. Second, statistical significance of the results was based on nominal, rather than adjusted, P values. The latter is preferred, as transcriptomics data may contain many false positives because of multiple comparisons and multidimensionality of the datasets. Third, the authors weaved together the analysis on the basis of results from several original experiments, which were conducted by different investigators using different cohorts. This could have introduced measured and unmeasured errors and thereby confounded their analyses. Fourth, the authors did not consider systemic factors such as proinflammatory cytokines, which can increase the thrombotic tendencies of patients, as this was beyond the purview of the study. IL-6 is a notable cytokine in this pathway, as it may be responsible for COVID-19–related cytokine storm and affect production of many proteins in the coagulation cascade, such as fibrinogen and various clotting factors by the liver (10).
Notwithstanding these limitations, the work of FitzGerald and colleagues is scientifically sound and clinically relevant. Thromboembolic disease is common among hospitalized patients with COVID-19, affecting 1 in 5 patients. Their work highlights an important mechanism by which this occurs in COVID-19 and, importantly, proffers novel targets for therapeutic discoveries and approved drugs, which can be potentially repurposed. New or repurposed therapies are desperately needed in the global fight against the COVID-19 pandemic.
The diagram provides an overview of potential mechanism of COVID-19 coagulopathy based on the work by FitzGerald and colleagues (4). SARS-CoV-2 infects the lung epithelial cells causing COVID-19 pneumonia. The immune cells (e.g., alveolar macrophages) sense the virus and produce cytokines (e.g., IL-6), which attract other immune cells (e.g., granulocytes) and recruit additional cytokines, thus creating a vicious cycle of inflammation that can damage the lung tissue. Blood vessels become leaky and allow exudative fluid to rush into the interstitium and alveolar units. IL-6 can transmigrate from the alveolar space into the systemic circulation through the pulmonary capillaries, where it can impact other tissues (e.g., liver) and promote the production of clotting factors and fibrinogen by hepatocytes. IL-6 can also orchestrate a COVID-19–related cytokine storm, causing macrophages to overproduce proteins that promote thrombosis, including PLAUR and the PAI-2. PLAU protein upregulates the production of TF in type II pneumocytes. The TF protein is a master regulator of the extrinsic coagulation cascade. Protein S also decreases in COVID-19. The imbalance between the TF, its inhibitor (TFPI), and protein S favors thrombosis. Genes implicated in the coagulation pathway are not dysregulated in peripheral blood mononuclear cells (PBMCs [i.e., T cells]) during SARS-CoV-2 infection.
Footnotes
Originally Published in Press as DOI: 10.1165/rcmb.2021-0134ED on March 30, 2021
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Worldometer https://www.worldometers.info/coronavirus/.
- 2. Jiménez D, García-Sanchez A, Rali P, Muriel A, Bikdeli B, Ruiz-Artacho P, et al. Incidence of VTE and bleeding among hospitalized patients with coronavirus disease 2019: a systematic review and meta-analysis. Chest. 2021;159:1182–1196. doi: 10.1016/j.chest.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.REMAP-CAP Trial Melbourne, Australia: REMAP-CAP; https://www.remapcap.org/media. [Google Scholar]
- 4. FitzGerald ES, Chen Y, Fitzgerald KA, Jamieson AM. Lung epithelial cell transcriptional regulation as a factor in COVID-19–associated coagulopathies. Am J Respir Cell Mol Biol. 2021;64:687–697. doi: 10.1165/rcmb.2020-0453OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blanco-Melo D, Nilsson-Payant BE, Liu W-C, Uhl S, Hoagland D, Møller R, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045, e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cotto KC, Wagner AH, Feng YY, Kiwala S, Coffman AC, Spies G, et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46:D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jha PK, Vijay A, Halu A, Uchida S, Aikawa M. Gene expression profiling reveals the shared and distinct transcriptional signatures in human lung epithelial cells infected with SARS-CoV-2, MERS-CoV, or SARS-CoV: potential implications in cardiovascular complications of COVID-19. Front Cardiovasc Med. 2021;7:623012. doi: 10.3389/fcvm.2020.623012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen LYC, Hoiland RL, Stukas S, Wellington CL, Sekhon MS. Confronting the controversy: interleukin-6 and the COVID-19 cytokine storm syndrome. Eur Respir J. 2020;56:2003006. doi: 10.1183/13993003.03006-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]