Abstract
Various RNA‐targeting approaches have been engineered to modify specific sites on endogenous transcripts, breaking new ground for a variety of basic research tools and promising clinical applications in the future. Here, we combine site‐directed adenosine‐to‐inosine RNA editing with chemically induced dimerization. Specifically, we achieve tight and dose‐dependent control of the editing reaction with gibberellic acid, and obtain editing yields up to 20 % and 44 % in the endogenous STAT1 and GAPDH transcript in cell culture. Furthermore, the disease‐relevant MECP2 R106Q mutation was repaired with editing yields up to 42 %. The introduced principle will enable new applications where temporal or spatiotemporal control of an RNA‐targeting mechanism is desired.
Keywords: ADAR, chemically induced dimerization, gibberellic acid, RNA targeting, site-directed RNA editing
RNA editing: Site‐directed adenosine‐to‐inosine RNA editing was engineered to be under control of the plant hormone gibberellic acid, applying the mechanism of chemically induced dimerization. Tight control and editing yields up to 44 % where achieved on endogenous targets in human cell culture.
RNA base editing enables the rewriting of genetic information with high efficiency and without the risk of permanent off‐target effects and thus has high prospects for clinical application.[1] Furthermore, the reversibility of an editing event on the transient (m)RNA copy allows to tune the yield of base exchange and might be used to introduce otherwise lethal mutations suddenly and/or temporally restricted.[2] The SNAP‐ADAR approach was engineered for site‐directed adenosine‐to‐inosine (A‐to‐I) RNA editing.[3] For this, the dsRNA binding domains responsible for substrate recognition in wildtype adenosine deaminase acting on RNA (ADAR)[4] are replaced by the self‐labeling SNAP‐tag. The SNAP‐tag binds covalently to guideRNAs carrying its substrate, O 6‐benzylguanine[5] (BG, snap‐guideRNAs), which then allows for recruitment of the fused ADAR deaminase domain to a specific target via Watson‐Crick base pairing. The approach has been shown to be rationally programmable, to achieve high editing yields in cell culture and in vivo,[6] to be very precise,[2] and to be efficient enough to enable concurrent editing.[6] The extension of the approach by further layers of control is desirable. Recently, we achieved photo‐control in developing embryos by application of guideRNAs carrying a nitropiperonyloxymethyl‐protected BG moiety.[7] Here, we now include control of the editing reaction by chemically induced dimerization[8] with a small molecule. This opens many new opportunities to run editing under temporal, spatial or dose control.
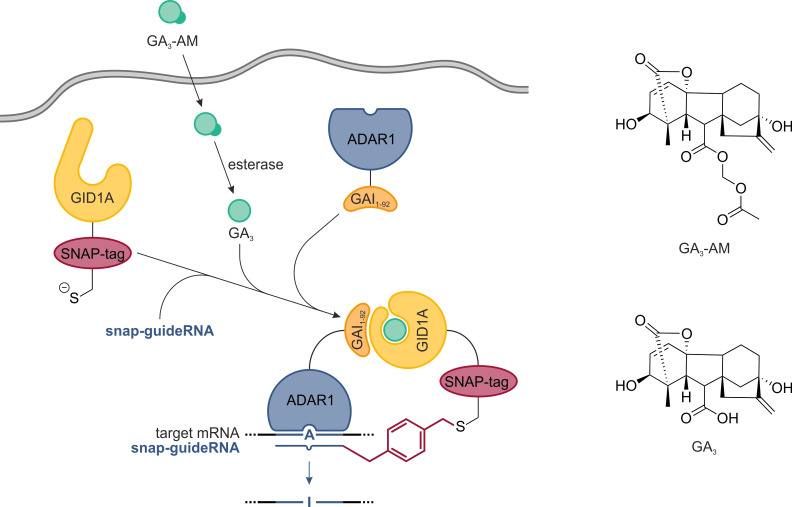
Specifically, we decided to use gibberellic acid (GA3) for chemically induced dimerization. GA3 is a plant hormone that can be delivered as a cell‐permeable prodrug (GA3−AM), that has been shown to induce the heterodimerization of the two plant proteins GAI (gibberellic acid insensitive) and GID1A (gibberellin insensitive dwarf 1A, Figure 1) on a timescale of seconds to minutes inside live cells.[9] Binding of GA3 to GID1A induces a conformational change that leads to recruitment of GAI. In order to control the SNAP‐ADAR‐based editing reaction by GA3‐induced dimerization, the SNAP‐tag and the ADAR deaminase domain needed engineering into two separate fusion proteins with GAI and GID1A, respectively. We decided to use a GAI1–92−ADAR1 fusion, applying a 92 amino acid fragment of GAI sufficient for dimerization,[9] and a SNAP‐GID1A fusion. In our design, we kept the SNAP‐tag and ADAR deaminase domain at their respective N‐ and C‐terminal position. We combined the ADAR deaminase domain with the GAI fragment to also place the latter in accordance with its native N‐terminal position. The GID1A protein has recently been applied in fusion with an N‐terminal eGFP‐tag.[9, 10] We expected that the exchange of the eGFP‐tag with a SNAP‐tag will not interfere with the function of the plant protein. Finally, both transgenes have the same size (59 kDa). We engineered four plasmid constructs (I–IV) that contain both transgenes in one expression cassette (Figure 2a). This design was chosen to obtain a balanced expression of both transgenes after stable genetic integration of the respective single plasmid into a cell line. Furthermore, under transient expression conditions such constructs would help to reduce the transfection bias and to improve the balance in the expression of both transgenes. The two transgenes were either put consecutively, each with its own CMV promotor and bGH terminator, or they were expressed as a single P2A[11] construct from one promotor using a translational skipping mechanism to create two separate proteins from one transcript in a nearly 1 : 1 stoichiometry. For the editase fusion, we either chose the catalytic deaminase domain of wildtype human ADAR1 (GAI1–92−ADAR1) or of a hyperactive mutant (GAI1–92−ADAR1Q), carrying a well‐known E>Q single point mutation.[12] To create duo cell lines expressing both transgenes stably under doxycycline induction, we applied the 293 Flp‐In T‐REx system. For each construct, I–IV, we generated a separate duo cell line, 1–4, by a plasmid transfection and antibiotic selection procedure, as described before.[6] The doxycycline‐dependent expression of both transgenes was confirmed by Western blot with antibodies against ADAR1 deaminase (Figure 2b) and SNAP‐tag (Figure 2c). Notably, the expression levels were comparably low in relation to the stable expression of SNAP‐ADAR1Q after integration into the 293 Flp‐In T‐REx cell line (Figure 2b,c).[6]
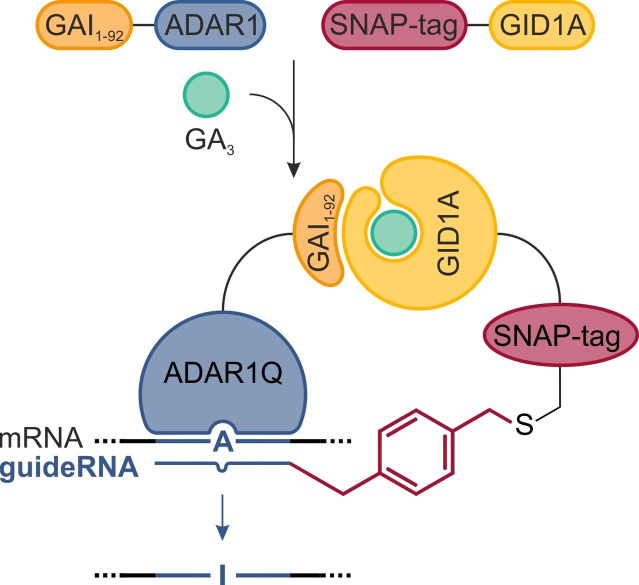
Figure 1.
Principle of gibberellic acid‐induced site‐directed RNA editing with the SNAP‐ADAR platform. Covalent conjugation of an O 6‐benzylguanosine (BG)‐modified guideRNA (snap‐guideRNA) with the SNAP‐tagged deaminase ADAR enables the steering of A‐to‐I deaminase activity to any arbitrary mRNA to achieve programmable, RNA‐guided site‐specific RNA editing. To place the process under control of gibberellic acid, the SNAP‐ADAR protein is split into a GAI1–92−ADAR and a SNAP‐GID1A fusion, separating the editing activity from the RNA‐targeting mechanism. Gibberellic acid, delivered in the form of a cell‐permeable acetoxymethyl ester (GA3−AM), enforces heterodimerization of GAI1–92 and GID1A by binding to the latter, and thereby recruits the ADAR deaminase to the guideRNA/mRNA substrate duplex.
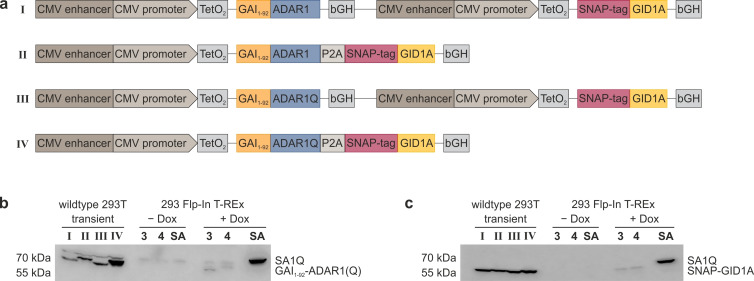
Figure 2.
Expression constructs for gibberellic acid‐induced RNA editing with SNAP‐ADAR and analysis of transgene expression. a) Constructs I–IV were designed to create transgenic 293 Flp‐In T‐REx cell lines 1–4, stably co‐expressing GAI1–92−ADAR1(Q) and SNAP‐GID1A from one cassette under doxycycline control. TetO2: tet operator, leads to repression of expression in the absence of a tetracycline;[13] bGH: bovine growth hormone terminator; P2A: porcine teschovirus‐1 self‐cleaving 2A peptide.[11] The protocol for the generation of stable cell lines 1–4 from constructs I–IV and details on the constructs can be found in the Supporting Information. b) Characterization of GAI1–92−ADAR1(Q) expression via Western Blot (α‐ADAR1 deaminase domain). Wildtype 293T cells were transiently transfected with constructs I–IV and stable cell lines 3 and 4 were examined without (−Dox) and with 24 h (+Dox) doxycycline induction. Previously established SA1Q 293 Flp‐In T‐REx cell line (SA) shown for comparison. c) Same as (b) but for expression analysis of SNAP‐GID1A (α‐SNAP‐tag).
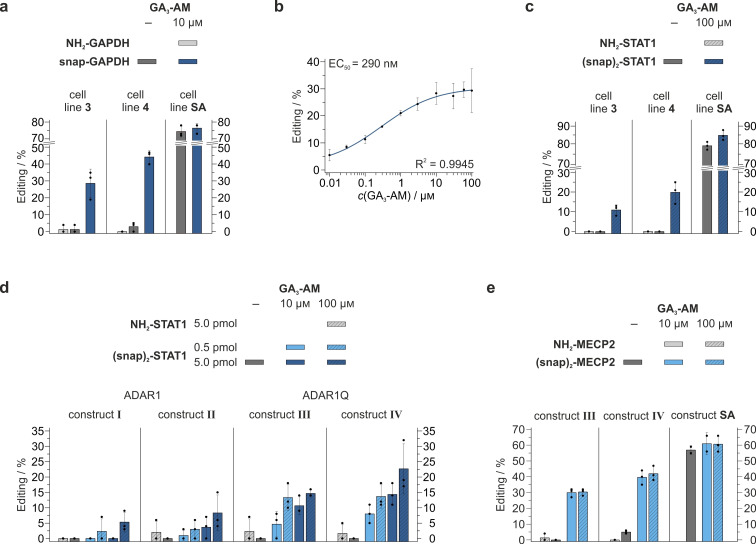
First, we tested the editing reaction in cell lines 3 and 4 expressing the hyperactive ADAR1Q fusion. Specifically, we targeted a 5′‐UAG codon in the 3′ untranslated region (UTR) of the endogenous GAPDH transcript (Figure 3a). Beside the snap‐GAPDH guideRNA, carrying the BG moiety required for covalent reaction with the SNAP‐tag, we also applied an NH2−GAPDH guideRNA as control, comprising of the same sequence and modification pattern, but lacking the BG moiety, thus incompetent of forming a conjugate. The control guideRNA (NH2−GAPDH) gave no detectable editing, highlighting the requirement for covalent guideRNA attachment to recruit ADAR deaminase activity. This clean negative control is a hallmark of RNA‐targeting with the SNAP‐ADAR approach.[2, 6] Notably, in the absence of the inducer GA3−AM, no GAPDH editing above the threshold for accurate detection (5 %) was detected with the snap‐GAPDH guideRNA. However, in presence of 10 μm GA3−AM in the medium, editing levels of 29±9 % and 44±4 % were achieved in cell line 3 and 4, respectively. We therefore estimate the dynamic change of the editing yield by GA3−AM induction to be >10 fold. Nevertheless, the editing efficiency stayed clearly below the one obtained with the analogous SNAP‐ADAR1Q cell line,[6] which, as expected, yielded high editing independent of GA3−AM (74±3 % versus 76±3 %, Figure 3a). This loss of efficiency might be either due to the low expression of the GAI and GID1A fusion proteins compared to the SNAP‐ADAR1Q fusion (Figure 2b,c), or it could be a drawback resulting from the necessity to bring not only one, but two proteins, a guideRNA, and an mRNA together for editing. To make sure that the applied GA3−AM amount was sufficient to induce maximum editing, we determined the dose‐response of the editing yield in cell line 4 over a concentration range from 10 nm to 100 μm GA3−AM (Figure 3b). We determined the EC50 to approximately 290 nm, indicating that the editing yield was already close to saturation at 10 μm GA3−AM. The determined EC50 value fits to earlier reports from different applications in literature.[9]
Figure 3.
Controlling site‐directed RNA editing with gibberellic acid. a) A snap‐GAPDH guideRNA (5.0 pmol) targeting a 5′‐UAG codon in the 3′‐UTR of endogenous GAPDH was transfected into cell lines 3 and 4, as indicated. An NH2‐guideRNA lacking the BG moiety required for covalent conjugation to the SNAP‐GID1A fusion served as negative control. Simultaneously, GA3−AM (10 μm) was added to the medium, as indicated. RNA editing yields were determined 24 h after transfection by RT‐PCR and Sanger sequencing, as described in the Supporting Information. Editing clearly depended on GA3−AM. b) Determination of the dose‐response to GA3−AM effecting the RNA editing yield in cell line 4. Editing was performed as described in panel a) on endogenous GAPDH, but with GA3−AM concentrations ranging from 10 nm to 100 μm. The EC50 was determined to 290 nm by applying a logistic fit. c) Analogous to panel a), but with a (snap)2‐STAT1 guideRNA (5.0 pmol) targeting the phosphorylation site Tyr(Y)701 (5′‐UAU codon) in the endogenous STAT1 transcript in cell lines 3 and 4, induced with 100 μm GA3−AM, as indicated. d) Editing of Y701 in endogenous STAT1 in wildtype 293T cells and under transient plasmid transfection of the expression cassettes I–IV (300 ng/well). Cells were treated with GA3−AM and guideRNAs in indicated concentrations and amounts. e) Repair of transiently plasmid transfected MECP2 R106Q in wildtype 293T cells under transient plasmid transfection of expression cassette III or IV with a (snap)2‐MECP2 guideRNA (1.0 pmol) targeting the disease relevant R106Q mutation, induced with 10 μM or 100 μM GA3−AM as indicated. In panel (a)–(e), the data is shown as the mean±s.d. of N=3 independent experiments.
In the past, we found A‐to‐I RNA editing in the open reading frame (ORF) considerably more challenging compared to editing in the 3′‐UTR.[6] Thus, we tested editing in the ORF of the endogenous STAT1 transcript (Figure 3c). Specifically, we designed a guideRNA targeting the phosphorylation site Tyr(Y)701 (5′‐UAU codon), which is important for activation of said transcription factor upon interferon signaling.[14] Again, we found no detectable editing with an NH2‐guideRNA lacking the self‐labeling moiety. However, we obtained reasonable editing levels when applying the (snap)2–STAT1 guideRNA, able to recruit two SNAP‐GID1A proteins per guideRNA. In presence of 100 μm GA3−AM, editing levels of 11±3 % and 20±6 % were achieved in cell line 3 and 4, respectively. Again, cell line 4 outperformed cell line 3 by means of editing yields. Due to the lack of detectable editing in absence of GA3−AM, the dynamic range for the induction with GA3−AM was estimated to be very high again. However, the overall editing yields were moderate compared to levels obtained in the analogous SNAP‐ADAR1Q 293 Flp‐In T‐REx cell line (79±2 % and 84±3 %, without versus with GA3−AM, Figure 3c).[6] We wondered if this was due to the low expression levels of the GAI and GID1A fusion proteins, and if editing could be fostered by stronger expression of the fusion proteins and further optimization of conditions. We thus tested the editing of all four constructs under transient transfection into wildtype 293T cells, and varied the amount of (snap)2–STAT1 guideRNA (0.5 pmol versus 5.0 pmol) and of the inducer (10 μm versus 100 μm, Figure 3d). We made several observations. First, we found wildtype constructs I and II to give considerably less editing than the hyperactive constructs III and IV. This is in accordance with literature for the analogous SNAP‐ADAR1 293 Flp‐In T‐REx cells.[6] Second, editing worked better with higher amounts of guideRNA. Third, higher amounts of inducer also fostered editing. Similar to what we had seen for cell line 4 versus cell line 3, construct IV gave better editing yields than construct III under most conditions. Taken together, the data suggests that editing yield is boosted by every component that assists in the formation of the tertiary complex (guideRNA+mRNA+two proteins), for example, more guideRNA, more inducer, and higher protein expression. The latter was supported by Western blot (Figure 2b), showing that more GAI1–92−ADAR1Q was expressed by construct IV than by construct III. Notably, the editing yields at endogenous STAT1 under transient transgene expression did hardly exceed the levels obtained in the stable cell lines (Figure 3c,d). Obviously, the balanced transgene expression in the stable cell lines, even at very low expression levels, is more powerful for targeting endogenous transcripts than the strong, but uneven transgene expression upon plasmid transfection.
Finally, we aimed to repair the R106Q mutation in the transcription factor Methyl CpG Binding Protein 2 (MECP2), which is known to cause Rett syndrome. The underlying G‐to‐A mutation is located in the DNA binding domain of MECP2 and leads to reduced protein stability and therefore decreased expression levels, as well as reduced binding to heterochromatin.[15] Since healthy expression levels of MECP2 vary between different neural cell types[16] and duplication of MECP2 causes MECP2 duplication syndrome,[17] repair of R106Q under tight, precisely doseable control at the transcript level is highly desirable. We thus transfected wildtype 293T cells with MECP2 R106Q and either construct III or IV and tested a guideRNA targeting the R106Q site. Upon induction with 10 μm or 100 μm GA3−AM, we achieved good editing yields for both construct III (30±3 % and 30±2 %, respectively) and construct IV (40±5 % and 42±5 %, respectively, Figure 3e). Contrary to the editing in the STAT1 transcript (Figure 3d), the editing yields for MECP2 in presence of 10 μM and 100 μM GA3−AM were equal, indicating saturation at 10 μM inducer for this target, which fits well to the dose‐dependence curve shown for the GAPDH target (Figure 3b). Once again, construct IV performed better than construct III. Notably, the MECP2 editing levels of our constructs came close to the editing levels with the transfected SNAP‐ADAR1Q construct (57±3 %, 61±7 % and 60±5 % without, with 10 μm and with 100 μm GA3−AM, respectively). Importantly, the editing yields obtained with construct IV are in the range of editing yields reported to suffice for significant enrichment of heterochromatic MECP2 (37–52 %) in vivo in murine neural cells[18] with the λN‐ADAR2Q system[19] and therefore might lead to significant diminution of the Rett syndrome phenotype.
In summary, we achieved tight control of site‐directed RNA editing by chemically induced dimerization with a small molecule plant hormone. The dimerization occurs promptly (seconds to minutes) after addition of the inducer[9] and elicits a tunable, dose‐dependent response. This temporal and dose‐dependent control of the RNA editing reaction may break new ground for attractive applications, for example, to trigger targeted editing during embryogenesis after microinjection of all components,[7] to trigger editing to measure RNA lifetimes with RNA timestamp approaches more accurately,[20] or to modulate the pharmacological (adverse) effect of targeted editing.[21] Importantly, we demonstrated that our engineered system, based on the SNAP‐ADAR approach in combination with gibberellic acid‐induced GID1A‐GAI heterodimerization, works not only via transient overexpression, but also under stable genetic integration of the components, which, as we had shown before, reduces artifacts[22] and global off‐target editing.[2, 6] Furthermore, the editing reaction was strongly dependent on the small molecule – virtually lacking any reaction in the absence of gibberellic acid. Even though the splitting of the SNAP‐ADAR editing enzyme into two separate fusion proteins was required to engineer small molecule control, editing of lowly expressed endogenous transcripts was possible in reasonable yields, as demonstrated for the editing of the functionally important phosphorylation site Y701 in STAT1. This is even more remarkable given the comparably low expression levels of the engineered components. We assume that the SNAP‐ADAR RNA‐targeting approach is particularly suited for the engineering of this kind of small molecule‐control as the covalent conjugation of guideRNA and SNAP‐tag pre‐organizes two components permanently, thus reducing the number of components which need to encounter for editing. Additionally, the disease‐relevant R106Q mutation in MECP2 could be repaired to an extent that has been reported[18] to significantly enhance MECP2 function. We expect that the approach can be further improved, for example the editing yields may be amplified by optimizing the expression levels of the fusion proteins. Furthermore, the approach could be extended by one‐ or two photon‐decaging of gibberellic acid[10] to enable spatiotemporal control in the future.[8, 23] Finally, the design principle could be included into further tools that apply RNA‐guided proteins to manipulate the (epi)transcriptome[24] or could be integrated into existing SNAP‐tag‐based sensors[25] to include a further layer of control.
Experimental Section
Detailed experimental procedures for Western Blotting (including full blots) and editing experiments, as well as further details on constructs I–IV, the generation of stable cell lines and guideRNAs along with their sequences can be found in the Supporting Information.
Conflict of interest
T.S. holds patents on site‐directed RNA editing.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
We gratefully acknowledge funding by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – projects 430214260, STA1053/7‐1, STA1053/11‐1 for TS. The work was supported by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement no. 647328 to TS). We thank Richard Wombacher and Gail Mandel for kindly providing GAI1–92 and GID1A coding sequences and mMECP2 R106Q in pEGFP−N3, respectively. Open access funding enabled and organized by Projekt DEAL.
A. S. Stroppel, R. Lappalainen, T. Stafforst, Chem. Eur. J. 2021, 27, 12300.
References
- 1.Rees H. A., Liu D. R., Nat. Rev. Genet. 2018, 19, 770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel P., Stafforst T., Curr. Opin. Biotechnol. 2019, 55, 74–80. [DOI] [PubMed] [Google Scholar]
- 3.Stafforst T., Schneider M. F., Angew. Chem. Int. Ed. 2012, 51, 11166–11169; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 11329–11332. [Google Scholar]
- 4.Nishikura K., Annu. Rev. Biochem. 2010, 79, 321–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keppler A., Gendreizig S., Gronemeyer T., Pick H., Vogel H., Johnsson K., Nat. Biotechnol. 2003, 21, 86–89. [DOI] [PubMed] [Google Scholar]
- 6.Vogel P., Moschref M., Li Q., Merkle T., Selvasaravanan K. D., Li J. B., Stafforst T., Nat. Methods 2018, 15, 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanswillemenke A., Kuzdere T., Vogel P., Jékely G., Stafforst T., J. Am. Chem. Soc. 2015, 137, 15875–15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voß S., Klewer L., Wu Y.-W., Curr. Opin. Chem. Biol. 2015, 28, 194–201. [DOI] [PubMed] [Google Scholar]
- 9.Miyamoto T., DeRose R., Suarez A., Ueno T., Chen M., Sun T.-p., Wolfgang M. J., Mukherjee C., Meyers D. J., Inoue T., Nat. Chem. Biol. 2012, 8, 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schelkle K. M., Griesbaum T., Ollech D., Becht S., Buckup T., Hamburger M., Wombacher R., Angew. Chem. Int. Ed. 2015, 54, 2825–2829; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2015, 127, 2867–2871. [Google Scholar]
- 11.Kim J. H., Lee S.-R., Li L.-H., Park H.-J., Park J.-H., Lee K. Y., Kim M.-K., Shin B. A., Choi S.-Y., PLoS One 2011, 6, e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuttan A., Bass B. L., Proc. Natl. Acad. Sci. USA 2012, 109, E3295–E3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao F., Svensjö T., Winkler T., Lu M., Eriksson C., Eriksson E., Hum. Gene Ther. 1998, 9, 1939–1950. [DOI] [PubMed] [Google Scholar]
- 14.Darnell J. E., Kerr I. M., Stark G. R., Science 1994, 264, 1415–1421. [DOI] [PubMed] [Google Scholar]
- 15.Sinnamon J. R., Kim S. Y., Corson G. M., Song Z., Nakai H., Adelman J. P., Mandel G., Proc. Natl. Acad. Sci. USA 2017, 114, E9395–E9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shahbazian M. D., Antalffy B., Armstrong D. L., Zoghbi H. Y., Hum. Mol. Genet. 2002, 11, 115–124. [DOI] [PubMed] [Google Scholar]
- 17.Van Esch H., Bauters M., Ignatius J., Jansen M., Raynaud M., Hollanders K., Lugtenberg D., Bienvenu T., Jensen L. R., Gécz J., Moraine C., Marynen P., Fryns J.-P., Froyen G., Am. J. Hum. Genet. 2005, 77, 442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinnamon J. R., Kim S. Y., Fisk J. R., Song Z., Nakai H., Jeng S., McWeeney S. K., Mandel G., Cell Rep. 2020, 32, 107878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montiel-Gonzalez M. F., Vallecillo-Viejo I., Yudowski G. A., Rosenthal J. J. C., Proc. Natl. Acad. Sci. USA 2013, 110, 18285–18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriques S. G., Chen L. M., Liu S., Zhong E. D., Scherrer J. R., Boyden E. S., Chen F., Nat. Biotechnol. 2020, 39, 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivera V. M., Clackson T., Natesan S., Pollock R., Amara J. F., Keenan T., Magari S. R., Phillips T., Courage N. L., F. C. Jr, Holt D. A., Gilman M., Nat. Med. 1996, 2, 1028–1032. [DOI] [PubMed] [Google Scholar]
- 22.Vogel P., Hanswillemenke A., Stafforst T., ACS Synth. Biol. 2017, 6, 1642–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ankenbruck N., Courtney T., Naro Y., Deiters A., Angew. Chem. Int. Ed. 2018, 57, 2768–2798; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 2816–2848. [Google Scholar]
- 24.
- 24a.Liu X.-M., Zhou J., Mao Y., Ji Q., Qian S.-B., Nat. Chem. Biol. 2019, 15, 865–871; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24b.Rau K., Rösner L., Rentmeister A., RNA 2019, 25, 1311–1323; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24c.Zhao B. S., Roundtree I. A., He C., Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.
- 25a.Yu Q., Xue L., Hiblot J., Griss R., Fabritz S., Roux C., Binz P.-A., Haas D., Okun J. G., Johnsson K., Science 2018, 361, 1122–1126; [DOI] [PubMed] [Google Scholar]
- 25b.Podewin T., Ast J., Broichhagen J., Fine N. H. F., Nasteska D., Leippe P., Gailer M., Buenaventura T., Kanda N., Jones B. J., M'Kadmi C., Baneres J.-L., Marie J., Tomas A., Trauner D., Hoffmann-Röder A., Hodson D. J., ACS Cent. Sci. 2018, 4, 166–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information