Summary
Circulating donor‐specific antibodies (DSA) do not necessarily indicate antibody‐mediated rejection (ABMR). Here, we evaluated the diagnostic value of donor‐derived cell‐free DNA (dd‐cfDNA) as an add‐on to DSA detection. The study included two independent cohorts of DSA+ kidney allograft recipients, 45 subclinical cases identified by cross‐sectional antibody screening (cohort 1), and 30 recipients subjected to indication biopsies (cohort 2). About 50% of the DSA+ recipients had ABMR and displayed higher dd‐cfDNA levels than DSA+ABMR− recipients (cohort 1: 1.90% [median; IQR: 0.78–3.90%] vs. 0.52% [0.35–0.72%]; P < 0.001); (cohort 2: 1.20% [0.82–2.50%] vs. 0.59% [0.28–2.05%]; P = 0.086). Receiver operating characteristic (ROC) analysis revealed an area under the curve (AUC) of 0.89 and 0.69 for dd‐cfDNA, and 0.88 and 0.77 for DSA mean fluorescence intensity (MFI), respectively. In combined models, adding dd‐cfDNA to DSA‐MFI or vice versa significantly improved the diagnostic accuracy. Limited diagnostic performance of dd‐cfDNA in cohort 2 was related to the frequent finding of other types of graft injury among ABMR− recipients, like T cell‐mediated rejection or glomerulonephritis. For dd‐cfDNA in relation to injury of any cause an AUC of 0.97 was calculated. Monitoring of dd‐cfDNA in DSA+ patients may be a useful tool to detect ABMR and other types of injury.
Keywords: antibody‐mediated rejection, biomarker, donor‐derived cell‐free DNA, kidney transplantation
Analyzing two independent cohorts of kidney allograft recipients, we found that detection of donor‐derived cell‐free DNA as an add‐on to donor‐specific antibody detection may be a useful non‐invasive surveillance tool to uncover antibody‐mediated kidney allograft rejection.

Introduction
Antibody‐mediated rejection (ABMR) is a major diagnostic and therapeutic challenge [1, 2]. This type of rejection presents as a continuous process of transplant injury, often subclinical at the time of diagnosis, but associated with accelerated progression to graft failure [3]. For detection of ABMR, allograft biopsies are current gold standard, and, in the last two decades, the Banff group has continuously refined and updated its diagnostic criteria [4]. The Banff scheme includes the detection of circulating donor‐specific antibodies (DSA) as a key criterion [4]. A positive DSA result, however, has limited diagnostic accuracy, and does not inevitably indicate an active rejection process. Distinct DSA characteristics, including mean fluorescence intensity (MFI) or, in tight association with MFI, complement fixing capability [5, 6], may slightly improve the diagnostic performance of HLA single antigen testing. Nevertheless, there is still an unmet need for monitoring tools that accurately predict the presence or absence of rejection.
One interesting molecular biomarker reflecting active transplant injury may be the detection of donor‐derived cell‐free DNA (dd‐cfDNA) in peripheral blood [7, 8]. Recent studies have demonstrated associations of dd‐cfDNA release with active allograft rejection, especially with ABMR, where dd‐cfDNA fractions were shown to be particularly high [9, 10, 11, 12, 13, 14, 15, 16, 17, 18]. In a recent meta‐analysis [17], a composite weighted dd‐cfDNA median of 2.89% was reported. The diagnostic accuracy of dd‐cfDNA may be enhanced when evaluated in the context of HLA antibody testing. In a small sub‐study of a multicenter trial [9], Jordan et al. [10], found that, among 33 DSA‐positive patients with graft dysfunction, detection of dd‐cfDNA discriminated well between ABMR and no ABMR, with a receiver operator characteristic area under the curve (ROC‐AUC) of 0.86. Test performance was thereby discussed to exceed that reported for DSA MFI. Available studies, however, are small and there is scarce information on the role of dd‐cfDNA monitoring in the context of specific DSA characteristics.
The primary objective of this study was to investigate the diagnostic accuracy of dd‐cfDNA detection, in presence of a positive DSA result, in relation to the finding of ABMR versus no ABMR in corresponding transplant biopsies. We evaluated two study cohorts (i) renal allograft recipients with a silent clinical course late after transplantation, who had been subjected to cross‐sectional DSA/ABMR screening and (ii) a retrospective series of patients who underwent indication biopsies at any time post‐transplantation. For retrospective evaluation of biobanked material, we established a technique of cell‐free DNA extraction that allowed for reliable dd‐cfDNA measurement in plasma that had been collected in routine ethylenediaminetetraacetic acid (EDTA) collection tubes.
Patients and methods
Study design and patients
For this single‐center study (Medical University of Vienna), two independent cohorts of DSA+ kidney allograft recipients were analyzed retrospectively for plasma dd‐cfDNA fractions. The first cohort (cohort 1) consisted of 45 DSA+ kidney transplant recipients recruited upon ABMR screening within a prospective randomized controlled trial of bortezomib in late silent ABMR (BORTEJECT; ClinicalTrials.gov: NCT01873157) [19]. As illustrated in Fig. 1, study patients were identified by cross‐sectional HLA antibody screening of 741 prevalent adult patients in outpatient care at our unit (screening period from October 2013 through February 2015). The study protocol has earlier been detailed [19, 20]. Key inclusion criteria were as follows: age >18 years; stable allograft function ≥180 days post‐transplantation, and an estimated glomerular filtration rate (eGFR) ≥20 ml/min/m2. Exclusion criteria relevant for this study included no acute graft dysfunction or prior rejection therapy and no active viral infection including BK viremia. ABMR screening revealed 111 HLA class I and/or II DSA+ recipients, of which 86 underwent transplant biopsies, on average 23 days [median; interquartile range (IQR): 15–42 days] after DSA detection. For 45 of these subjects biobanked plasma samples collected at the day of index biopsy (shortly before biopsy) were sufficient for dd‐cfDNA measurement, allowing for their inclusion in this study (Fig. 1). The second cohort (cohort 2) consisted of kidney transplant recipients selected from our transplant database, according to the following criteria: age >18 years; a renal allograft biopsy for acute or chronic graft dysfunction, proteinuria, and/or DSA formation at any time post‐transplantation (biopsies performed after <1 month were not included to avoid an influence of reperfusion injury [21]) between January 2012 (initiation of systematic biobanking at our unit) and December 2019; presence of HLA class I and/or II DSA at time of biopsy. As shown in Fig. 1, of 879 biopsied patients 138 had a positive post‐transplant DSA result documented in our database. For 58 patients, biologic material obtained at the time of biopsy was available and the presence of DSA could be confirmed upon re‐testing of stored serum samples. Finally, 30 of them had sufficient specimens (plasma samples) for valid dd‐cfDNA measurement (Fig. 1). Twenty‐seven of these specimens were obtained shortly before biopsy. In three patients, samples were taken after the biopsy (1, 11, and 16 days, respectively). All included patients had previously consented to participate in the “Vienna Kidney Transplant Cohort Study” for prospective biobanking. Baseline characteristics, immunological and biopsy results obtained for the two cohorts are provided in Tables 1 and 2. The study was approved by the institutional ethics committee (registration numbers: 267/2011, 1515/2012, and 1887/2020) and conducted in compliance with the Good Clinical Practice Guidelines, the principles of the Declaration of Helsinki 2008, and the Declaration of Istanbul.
Figure 1.
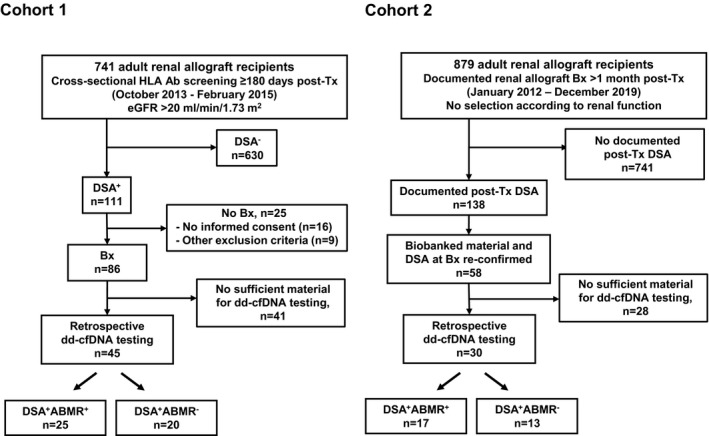
Study flow chart for cohorts 1 and 2. ABMR, antibody‐mediated rejection; Bx, biopsy; dd‐cfDNA, donor‐derived cell‐free DNA; DSA, donor‐specific antibody; eGFR, estimated glomerular filtration rate.
Table 1.
Baseline characteristics.
| Variables | Cohort 1 (n = 45) | Data available (n) | Cohort 2 (n = 30) | Data available (n) |
|---|---|---|---|---|
| Variables recorded at the time of transplantation | ||||
| Female sex, n (%) | 20 (44.4) | 45 | 16 (53.3) | 30 |
| Recipient age (years), median (IQR) | 52.4 (44.1–55.6) | 45 | 54.7 (42.6–62.2) | 30 |
| Deceased donor, n (%) | 42 (93.3) | 45 | 24 (80.0) | 30 |
| Living donor, n (%) | 3 (6.7) | 45 | 6 (20.0) | 30 |
| Prior kidney transplant, n (%) | 14 (31.1) | 45 | 10 (33.3) | 30 |
| Current CDC panel reactivity ≥10%, n (%) | 7 (20.0) | 35 | 4 (19.0) | 21 |
| Preformed anti‐HLA DSA, n (%)* | 17 (63.0) | 27 | 8 (30.8) | 26 |
| Donor age (years), median (IQR) | 46.0 (31.8–59.0) | 44 | 55 (42–59) | 30 |
| HLA mismatch (A, B, DR), median (IQR) | 3 (2–4) | 44 | 3 (3–4) | 30 |
| Cold ischemia time (hours), median (IQR) | 11.9 (9–17) | 43 | 12.9 (9.1–17.1) | 30 |
| Initial immunosuppression | ||||
| Induction with antithymocyte globulin, n (%) | 17 (37.8) | 45 | 9 (30.0) | 30 |
| Induction with IL‐2 receptor antibody induction, n (%) | 15 (33.3) | 45 | 18 (60.0) | 30 |
| Tacrolimus‐based immunosuppression, n (%) | 31 (68.9) | 45 | 26 (86.7) | 30 |
| Cyclosporine A‐based immunosuppression, n (%) | 13 (28.9) | 45 | 4 (13.3) | 30 |
| mTOR inhibitor‐based immunosuppression, n (%) | 1 (2.2) | 45 | 0 (0) | 30 |
| Peri‐transplant immunoadsorption, n (%) | 19 (42.2) | 45 | 8 (26.7) | 30 |
| Variables recorded at the time of index biopsy | ||||
| Age of study patients (years), median (IQR) | 56.4 (50.4–63.4) | 45 | 58.5 (45.0–65.3) | |
| Years after transplantation, median (IQR) | 4.0 (1.3–10.9) | 45 | 2.8 (0.3–5.3) | 30 |
| Renal parameters | ||||
| Serum creatinine (mg/dl), median (IQR) | 1.45 (1.19–1.95) | 45 | 1.50 (1.18–2.59) | 30 |
| Urinary protein/creatinine ratio (mg/g), median (IQR) | 216 (101–411) | 45 | 432 (235–2258) | 29 |
| Maintenance immunosuppression | ||||
| Triple immunosuppression | 36 (80.0) | 45 | 30 (100) | 30 |
| Dual immunosuppression | 9 (20.0) | 45 | 0 (0) | 30 |
| Tacrolimus, n (%) | 28 (62.2) | 45 | 26 (86.7) | 30 |
| Cyclosporine A ression, n (%) | 14 (31.1) | 45 | 3 (10) | 30 |
| mTOR inhibitor, n (%) | 2 (4.4) | 45 | 1 (1) | 30 |
| Belatacept, n (%) | 1 (2.2) | 45 | 0 (0) | 30 |
| MPA, n (%) | 39 (86.7) | 45 | 28 (93.0) | 30 |
| Steroids, n (%) | 41 (91.1) | 45 | 100 (100) | 30 |
DSA, donor‐specific antibody; CDC, complement‐dependent cytotoxicity; IQR, interquartile range; MPA, mycophenolic acid; mTOR, mammalian target of rapamycin.
*For recipients transplanted before 2009, solid‐phase HLA antibody screening on the wait list was not available [34].
Table 2.
Renal, immunologic and biopsy results.
| Variables | Cohort 1 (N = 45) | P value | Cohort 2 (N = 30) | P value | ||
|---|---|---|---|---|---|---|
| ABMR+ (n = 25) | ABMR− (n = 20) | ABMR+ (n = 17) | ABMR‐ (n = 13) | |||
| Renal parameters | ||||||
| Serum creatinine (mg/dl), median (IQR) | 1.71 (1.23–2.41) | 1.34 (1.19–1.81) | 0.10 | 1.41 (1.15–2.51) | 1.77 (1.17–2.81) | 0.77 |
| Protein/creatinine ratio (mg/g), median (IQR) | 343 (85–863) | 167 (115–224) | 0.041 | 1003 (305–2985) | 318 (221–411) | 0.073 |
| DSA characteristics | ||||||
| HLA class I DSA only, n (%) | 4 (16.0) | 9 (45.0) | 0.03 | 3 (17.6) | 5 (38.5) | 0.20 |
| HLA class II DSA only, n (%) | 14 (56.0) | 7 (35.0) | 0.14 | 11 (64.7) | 5 (38.5) | 0.15 |
| HLA class I and II DSA, n (%) | 7 (28.0) | 4 (20.0) | 0.40 | 3 (17.6) | 3 (23.1) | 0.53 |
| Anti‐DQ DSA, n (%) | 15 (60.0) | 10 (50.0) | 0.36 | 10 (58.8) | 5 (38.5) | 0.23 |
| Number of DSA, median (IQR) | 1 (1–2) | 1 (1–2) | 0.32 | 1 (1–2) | 1 (1–2) | 0.71 |
| DSA‐MFI*, median (IQR) | 4407 (2819–9886) | 1390 (1108–1781) | <0.001 | 8843 (3254–18 195) | 2120 (1461–5345) | 0.007 |
| Biopsy results | ||||||
| ABMR, n (%) | ||||||
| Active ABMR, i (%) | 8 (32.0) | – | – | 3 (17.6) | – | – |
| Chronic active ABMR, n (%) | 16 (64.0) | – | – | 13 (76.5) | – | – |
| Chronic ABMR (inactive), n (%) | 1 (4.0) | – | – | 1 (5.9) | – | – |
| C4d‐positive ABMR, n (%) | 11 (44.0) | – | – | 7 (41.2) | – | ‐ |
| Molecular ABMR score, median (IQR) | 0.65 (0.45–0.87) | 0.08 (0.04–0.13) | <0.001 | – | – | – |
| Banff borderline lesion, n (%) | 3 (12.0) | 2 (10.0) | 0.61 | 1 (5.9) | 0 (0) | 0.57 |
| TCMR, n (%) | 0 (0) | 0 (0) | – | 1 (5.9) | 2 (15.4) | 0.40 |
| BK virus nephropathy, n (%) | 0 (0) | 0 (0) | – | 0 (0) | 1 (7.7) | 0.43 |
| Glomerulonephritis, n (%) | 1 (4.0) | 2 (10.0) | 0.42 | 0 (0) | 3 (23.1) | 0.07 |
ABMR, antibody‐mediated rejection; DSA, donor‐specific antibody; IQR, interquartile range; MFI, mean fluorescence intensity.
*MFI of the immunodominant DSA.
Measurement of dd‐cfDNA
Plasma samples were obtained from peripheral blood collected in BD Vacutainer® K3‐EDTA collection tubes (Becton Dickinson, Franklin Lakes, NJ, USA), separated within <2 h (which precluded relevant white blood cell lysis and release of recipient cfDNA [22]), transferred to barcoded polypropylene tubes and stored at a mean temperature of equal or below −70 °C, following a uniform predefined protocol (Biobank of the Medical University of Vienna [23]). Cell‐free DNA was extracted from ≥0.5 ml plasma using Qiagen’s QIAamp Circulating Nucleic Acid Kit, followed by a double AMPure clean‐up step to remove contaminating DNA. Following an earlier described protocol [9], cell‐free DNA was analyzed by the AlloSeq cfDNA assay (CareDx, Fremantle, WA, Australia), a targeted next‐generation sequencing assay that employs allelic single nucleotide polymorphisms to quantify dd‐cfDNA without a need for separate recipient or donor genotyping. Negative control samples obtained from a group of 10 DSA‐negative patients (nine samples were taken shortly before biopsy, one 6 days after biopsy collection) who showed no rejection or other causes of graft injury in matched biopsies had median levels of 0.21% (IQR: <0.15–0.34%; range: <0.15–0.42%). For comparative analysis of results obtained with plasma collected in BD Vacutainer® tubes versus standard Cell‐Free DNA BCT blood collection tubes (Streck, La Vista, NE, USA), we prospectively collected material from 10 adult kidney transplant recipients who underwent allograft biopsies between June 2020 and August 2020, >1 month after transplantation. Plasma samples were obtained shortly before biopsy collection. As shown in Figure S1, parallel measurement of dd‐cfDNA revealed a tight correlation between the results obtained with the two different sample types (Spearman rho = 0.99, P < 0.0001).
HLA antibody detection
As earlier detailed [6], we used LABscreen Single Antigen assays (One Lambda, Thermo Fisher Scientific, Canoga Park, CA, USA) to characterize HLA reactivity patterns. For cohort 1, HLA antibody tests were performed prospectively upon cross‐sectional screening, and for cohort 2, testing was done retrospectively on biobanked sera obtained at the time of biopsy (to confirm and characterize post‐transplant DSA documented in our database). Serum samples were heat‐inactivated (30 min at 56 °C; cohort 1) or treated with EDTA (10 mm; cohort 2) to counteract complement interference. DSA (MFI threshold >1000) were defined in the context of serological and/or low‐ or high‐resolution donor/recipient HLA typing (HLA‐A, ‐B, ‐Cw, ‐DR, ‐DQ and/or DP). Test results were documented as the MFI of the immunodominant DSA (DSA‐MFI).
Biopsies
Histomorphology and C4d staining were evaluated on formalin‐fixed paraffin‐embedded sections. For microarray‐based molecular analysis of biopsy samples (cohort 1), a fraction of one biopsy core was used for gene expression analysis using the Molecular Microscope Diagnostic (MMDx) system as previously described [24]. A classifier related to ABMR (molecular ABMR score) was generated using a reference set of 1208 biopsy specimens [25]. ABMR was defined according to the Banff classification, based on histomorphologic (glomerulitis [g], peritubular capillaritis [ptc], transplant glomerulopathy [cg], C4d), ultrastructural (multilayering of basement membranes of peritubular capillaries), serological (DSA detection), and for cohort 1 (BORTEJECT trial) also molecular results (molecular ABMR score ≥ 0.2), respectively. For the present analysis, documented single lesions of ABMR (types and scores) were re‐interpreted following the rules of the Banff 2017 update [26].
Statistics
Continuous data are presented as median and IQR, and categorical variables as absolute and relative frequencies. For inter‐group comparisons we applied nonparametric testing (Mann–Whitney U test). Bivariate correlations were calculated using the Spearman coefficient. Receiver‐operating characteristic (ROC) analyses were performed to display the area under the curve (AUC) and sensitivity/specificity of significant biomarkers and to determine the respective thresholds with the highest accuracy (highest sum of true‐positive and true‐negative predictions). Random forest analysis was performed using R package randomForestSRC to calculate the relative variable importance (RVI), using the permutation method. Logistic regression, likelihood ratio tests, and bootstrapped AUC validation were done using the rms package. A two‐sided P < 0.05 was considered statistically significant. All analyses were performed using IBM SPSS Statistics Version 24 (IBM, Armonk, NY, USA) or R version 3.6.1 (https://www.r‐project.org, Vienna, Austria).
Results
Patient characteristics and disposition
The study included two separate cohorts of DSA+ kidney allograft recipients who all underwent allograft biopsies and had paired dd‐cfDNA results (Fig. 1). Cohort 1 included 45 DSA+ recipients with a silent clinical course late after transplantation. Cohort 2 consisted of 30 DSA+ recipients subjected to indication biopsies early or late after transplantation. Baseline data are provided in Tables 1 and 2. In cohort 1, biopsies were performed after a median of 4.0 years, and in cohort 2 after 2.8 years after transplantation. Median serum creatinine and protein/creatinine ratio at the time of index biopsy were 1.45 mg/dl and 216 mg/g in cohort 1, and 1.50 mg/dl and 432 mg/g in cohort 2, respectively. DSA‐MFI were 3007 (median; IQR: 1390–5137) and 5110 (1826–12 206), respectively. About 50% had anti‐HLA‐DQ DSA. In cohort 1, 25 recipients (55.6%) were diagnosed with ABMR, the majority showing chronic active ABMR (n = 16). In cohort 2, 17 of the 30 patients (56.7%) had ABMR (chronic active rejection: n = 13; Table 2).
Donor‐derived cell‐free DNA and biopsy results
Cohort 1
As shown in Fig. 2, DSA+ABMR+ patients had significantly higher levels than DSA+ABMR‐ patients (dd‐cfDNA: 1.90% [median; IQR: 0.78–3.90%] versus 0.52% [0.35–0.72%]; P < 0.001). Levels of dd‐cfDNA were increased both in active (1.20%; 0.78–4.80%) and chronic active ABMR (2.05%; 1.65–4.50%), while the single patient with chronic (inactive) ABMR had a level of 0.60%. Among DSA+ABMR‐ patients, two patients showed borderline rejection and another two patients IgA nephropathy. These patients had dd‐cfDNA levels below 1% (Fig. 2). As shown in Table 2 and Figure S2 median levels of DSA‐MFI (4407 vs. 1390; P < 0.001) and urinary protein/creatinine ratio (343 vs. 167 mg/g; P = 0.041) were significantly higher among DSA+ABMR+ versus DSA+ABMR− patients. Inter‐group differences in DSA number and serum creatinine were not different (Table 2). Using an earlier described 1% cut‐off for dd‐cfDNA [9], we found profound differences between dd‐cfDNA levels >1% vs. <1% with respect to ABMR diagnosis (n = 18/20 [90%] vs. n = 7/25 [28%]; P < 0.001), microcirculation injury (g+ptc sum score: 3 [median: 1–5] vs. 0 [IQR: 0–2]; P < 0.001) and molecular ABMR scores (0.67 [0.45–0.87] vs. 0.09 [0.04–0.20]; P < 0.001) (data not shown).
Figure 2.
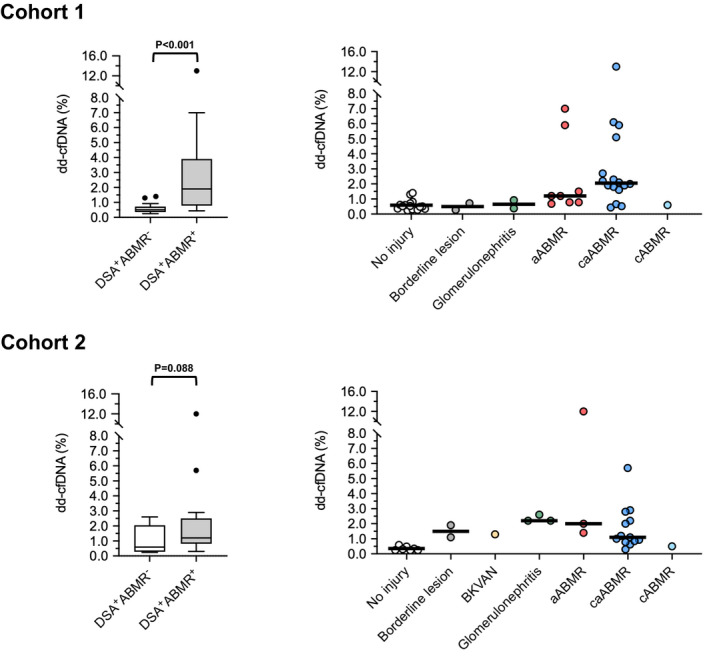
Levels of dd‐cfDNA in cohorts 1 and 2, in relation to biopsy results. Box plots indicate median, IQR, and range. We used the unpaired Mann‐Whitney U test for group comparisons. ABMR, antibody‐mediated rejection; aABMR, active ABMR; BKVAN, BK virus nephropathy; BL, borderline lesion; caABMR, chronic active ABMR; cABMR, chronic ABMR (inactive).
Cohort 2
There was a trend towards higher dd‐cfDNA levels in DSA+ABMR+ compared to DSA+ABMR− patients (1.20% [median; IQR: 0.82–2.50%] vs. 0.59% [0.28–2.05%]; P = 0.086; Fig. 2). Sub‐phenotyping of ABMR revealed the highest levels among patients with active ABMR (1.40% [2.0–12.0%]), followed by chronic active ABMR (1.10% [0.82–2.50%]), and a level of 0.50% in a patient with chronic (inactive) ABMR. Six DSA+ABMR− patients (46%) showed other types of graft injury: chronic TCMR (n = 2), glomerulonephritis (IgA nephropathy: n = 2, FSGS: n = 1), and BK virus nephropathy (BKVAN) plus C4d positivity without morphologic evidence of rejection (n = 1). These subjects had dd‐cfDNA levels above 1%, while those who did not show any features of active injury had levels in the range of DSA− nonrejecting control subjects (Fig. 2). As detailed in Table 2 and Figure S2, DSA+ABMR+ patients had higher DSA‐MFI than DSA+ABMR− patients (median: 8843 vs. 2120; P = 0.007) and there was a trend towards higher urinary protein/creatinine ratio among DSA+ABMR+ patients (1003 vs. 318; P = 0.073). Serum creatinine and DSA number were not significantly different (Table 2). Numerical differences between patients with dd‐cfDNA fractions >1% versus <1% with respect to ABMR diagnosis (ABMR versus no ABMR: n = 11/17 [64.7%] vs. n = 6/13 [46.2%]; P = 0.26) or microcirculation inflammation (g+ptc sum score: 4 [1–5] vs. 0 [0–4]; P = 0.13) did not achieve statistical significance (not shown).
Diagnostic performance of dd‐cfDNA in relation to ABMR
Cohort 1
As shown in Fig. 3 and Table 3, ROC analysis of dd‐cfDNA in relation to ABMR diagnosis revealed an AUC of 0.89 (P < 0.001). MFI of the immunodominant DSA showed an AUC of 0.88 (P < 0.001), and protein/creatinine ratio an AUC of 0.75 (P = 0.03). Serum creatinine and DSA number failed to discriminate ABMR from no ABMR. Evaluating characteristics of predictive power in relation to continuous threshold values, dd‐cfDNA and DSA‐MFI were found to have the highest accuracy at thresholds of 0.78% (0.80; specificity and sensitivity: 0.80) and 1986 (0.87; sensitivity: 0.92; specificity: 0.80), respectively (Table 3). We performed a random forest analysis to demonstrate the RVI contributing to ABMR prediction (Fig. 4). A first model included all five parameters: dd‐cfDNA, DSA‐MFI, DSA number, serum creatinine and protein/creatinine ratio. The most important variable in this analysis were DSA‐MFI, followed by dd‐cfDNA and protein/creatinine ratio. Serum creatinine and DSA number had no importance here. In a second model reduced to dd‐cfDNA and DSA‐MFI, DSA‐MFI remained the most important predictor (Fig. 4).
Figure 3.
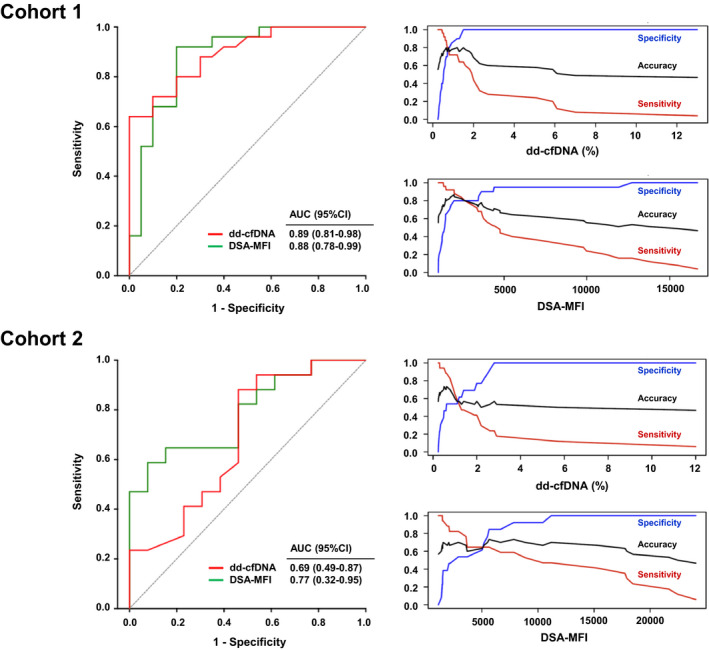
Diagnostic accuracy of dd‐cfDNA and DSA‐MFI in relation to ABMR diagnosis. ROC analysis and characteristics of prediction in relation to continuous threshold values (accuracy, sensitivity and specificity) are shown for cohorts 1 and 2. AUC, area under the curve; CI, confidence interval.
Table 3.
ROC analysis of biomarkers and clinical variables predicting ABMR.
| Variables | AUC (95%CI) | P value | Threshold with maximum accuracy | Threshold value | ||
|---|---|---|---|---|---|---|
| Max. accuracy | Sensitivity | Specificity | ||||
| Cohort 1 | ||||||
| dd‐cfDNA | 0.89 (0.81–0.98) | <0.001 | 0.80 | 0.80 | 0.80 | 0.78 |
| DSA‐MFI | 0.88 (0.78–0.99) | <0.001 | 0.87 | 0.92 | 0.80 | 1986 |
| DSA number | 0.58 (0.40–0.75) | 0.39 | 0.58 | 0.48 | 0.70 | 2 |
| Serum creatinine, mg/dl | 0.64 (0.48–0.81) | 0.08 | 0.64 | 0.56 | 0.75 | 1.65 |
| Protein/creatinine ratio, mg/g | 0.75 (0.52–0.84) | 0.03 | 0.71 | 0.64 | 0.80 | 231 |
| Cohort 2 | ||||||
| dd‐cfDNA | 0.69 (0.49–0.87) | 0.12 | 0.73 | 0.88 | 0.54 | 0.62 |
| DSA‐MFI | 0.77 (0.32–0.95) | <0.001 | 0.73 | 0.65 | 0.87 | 5636 |
| DSA number | 0.54 (0.33–0.75) | 0.70 | 0.50 | 0.35 | 0.69 | 2 |
| Serum creatinine, mg/dl | 0.47 (0.25–0.69) | 0.78 | 0.57 | 0.71 | 0.39 | 1.31 |
| Protein/creatinine ratio, mg/g | 0.70 (0.49–0.91) | 0.06 | 0.79 | 0.77 | 0.83 | 432 |
ABMR, antibody‐mediated rejection; AUC, area under the curve; CI, confidence interval; dd‐cfDNA, donor‐derived cell‐free DNA; ROC, receiver‐operator curve.
Figure 4.
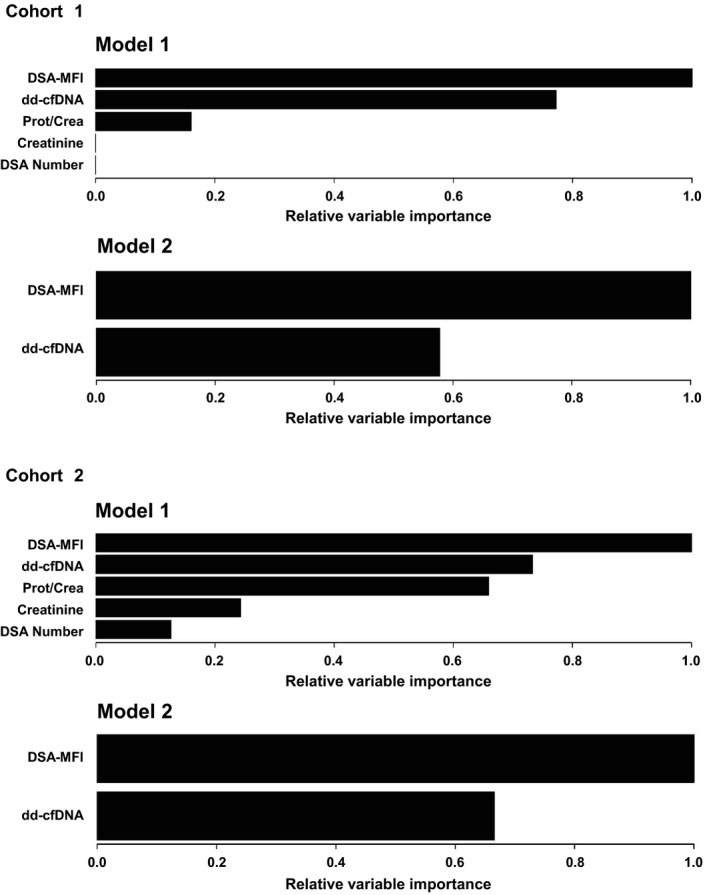
RVI in two different models of random forest analysis. Results are shown for cohorts 1 and 2. The RVI of a certain variable is determined by randomly shuffling the values of this particular variable in the out‐of‐bag‐sample while keeping all other variables the same. The decrease in diagnostic accuracy is a measure of the importance of this variable. In model 1, the following laboratory biomarkers were included: dd‐cfDNA, DSA‐MFI, DSA number, serum creatinine and protein/creatinine ratio. In model 2, two variables, dd‐cfDNA and DSA‐MFI, were included.
Cohort 2
The AUC calculated for dd‐cfDNA in relation to ABMR was 0.69 (P = 0.12; Fig. 3, Table 3). Among other variables, AUC levels were highest for DSA‐MFI (0.77; P < 0.001), followed by protein/creatinine ratio (0.70; P = 0.06). DSA number and protein/creatinine ratio were not discriminative (Table 3). A random forest analysis including dd‐cfDNA, DSA‐MFI, DSA number, serum creatinine, and protein/creatinine ratio revealed the highest RVI for DSA‐MFI, followed by dd‐cfDNA and protein/creatinine ratio. Serum creatinine and DSA number had by far less importance. Reducing the model to dd‐cfDNA and DSA‐MFI, DSA‐MFI was found to have highest importance (Fig. 4).
Combined biomarkers and ABMR in DSA+ patients
We used likelihood ratio tests to compare logistic regression models for diagnosing ABMR (Table 4). Comparisons were between single variable models (cfDNA or MFI alone) and models combining cfDNA or MFI with each of the other variables in turn. As shown in Table 4, adding DSA‐MFI to dd‐cfDNA (or vice versa) resulted in significantly better models, with an increase in AUC from 0.90 to 0.92 (cohort 1) and 0.68 to 0.84 (cohort 2), respectively.
Table 4.
Logistic regression models of single and combined parameter analysis predicting ABMR in DSA+ recipients.
| Comparison* | Likelihood ratio | P value | AUC (combined variables) | AUC (single variable) |
|---|---|---|---|---|
| Cohort 1 | ||||
| Model 1 | ||||
| dd‐cfDNA vs. [dd‐cfDNA + DSA‐MFI] | 5.50 | 0.008 | 0.92 | 0.90 (dd‐cfDNA) |
| dd‐cfDNA vs. [dd‐cfDNA + serum creatinine] | 0.72 | 0.38 | 0.89 | |
| dd‐cfDNA vs. [dd‐cfDNA + protein/creatinine ratio] | 3.59 | 0.19 | 0.88 | |
| Model 2 | ||||
| DSA‐MFI vs. [DSA‐MFI + dd‐cfDNA] | 19.1 | <0.0001 | 0.93 | 0.88 (DSA‐MFI) |
| DSA‐MFI vs. [DSA‐MFI + serum creatinine] | 3.10 | 0.78 | 0.85 | |
| DSA‐MFI vs. [DSA‐MFI + protein/creatinine ratio] | 11.2 | <0.001 | 0.89 | |
| Cohort 2 | ||||
| Model 1 | ||||
| dd‐cfDNA vs. [dd‐cfDNA + DSA‐MFI] | 9.96 | 0.001 | 0.84 | 0.68 (dd‐cfDNA) |
| dd‐cfDNA vs. [dd‐cfDNA + serum creatinine] | 0.10 | 0.72 | 0.63 | |
| dd‐cfDNA vs. [dd‐cfDNA + protein/creatinine ratio] | 0.23 | 0.13 | 0.74 | |
| Model 2 | ||||
| DSA‐MFI vs. [DSA‐MFI + dd‐cfDNA] | 4.83 | 0.028 | 0.83 | 0.79 (DSA‐MFI) |
| DSA‐MFI vs. [DSA‐MFI + serum creatinine] | 3.09 | 0.79 | 0.80 | |
| DSA‐MFI vs. [DSA‐MFI + protein/creatinine ratio] | 0.28 | 0.60 | 0.75 | |
ABMR, antibody‐mediated rejection; dd‐cfDNA, donor‐derived cell‐free DNA; DSA, donor‐specific antibody.
*For logistic regression analysis, dd‐cfDNA levels and protein/creatinine ratios were log transformed.
Donor‐derived cell‐free DNA and active allograft injury of any cause
Finally, in a separate analysis, we evaluated the diagnostic performance of biomarkers and clinical variables in relation to any features of transplant injury versus normal histology (cohort 1: n = 29 vs. n = 16 patients; cohort 2: n = 23 vs. n = 7 patients). Inter‐group comparisons regarding dd‐cfDNA levels, DSA characteristics and renal parameters are depicted in Figure S3. In both cohorts, transplant injury was associated with increased fractions of dd‐cfDNA levels as compared to patients with normal histology (cohort 1: 1.60% [median; IQR: 0.70–2.50%] vs. 0.52% [0.35–0.69%]; P < 0.001; cohort 2: 1.4 [0.93–2.20] vs. 0.31 [0.25–0.48]; P < 0.001). For both cohorts, also DSA‐MFI and protein/creatinine ratio were significantly different. Differences with respect to serum creatinine reached statistical significance only in cohort 1 (Figure S3). As shown in Fig. 5, AUC levels turned out to be highest for dd‐cfDNA, showing a marked increase in the diagnostic performance in cohort 2 (as compared to analysis in relation to ABMR; cohort 1: 0.85 [0.73–0.96] P < 0.001; cohort 2: 0.97 [0.92–1.0]; P < 0.001).
Figure 5.
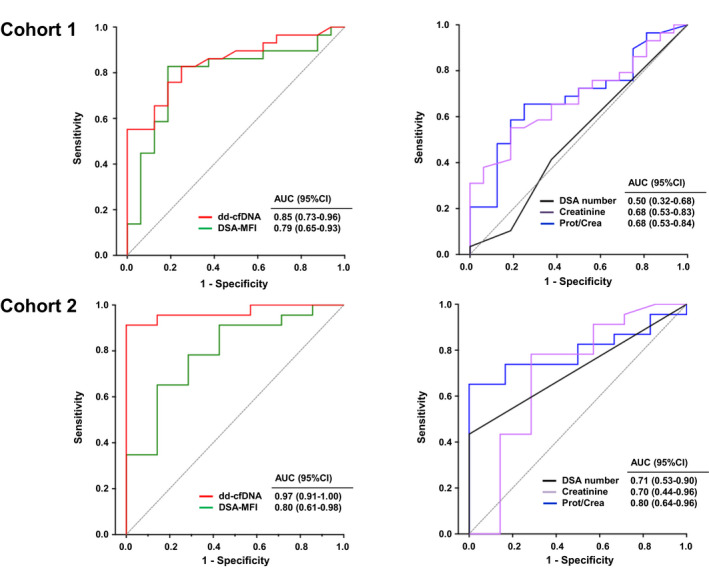
Prediction of transplant injury of any cause by ROC analysis. ROC analysis is shown for DSA characteristics and renal parameters. AUC, area under the curve; CI, confidence interval; dd‐cfDNA, donor‐derived cell‐free DNA, DSA, donor‐specific antibody; MFI, mean fluorescence intensity.
Discussion
The primary objective of our study was to determine the diagnostic accuracy of dd‐cfDNA as a noninvasive marker identifying biopsy‐diagnosed ABMR in the context of a positive post‐transplant DSA result. To address this question, we evaluated two separate cohorts of DSA+ recipients, the first one including patients with a silent clinical course late after transplantation, and the second one including patients that have been subjected to early or late biopsies for cause. For these two cohorts, we calculated a maximum accuracy of 0.80 and 0.73 at threshold levels of 0.78% and 0.62%, respectively, with a RVI below that calculated for DSA‐MFI. Applying combined models, the addition of dd‐cfDNA to DSA‐MFI results (or vice versa) was found to significantly improve the diagnostic accuracy.
In the last years, several groups have evaluated the value of dd‐cfDNA as a rejection marker, mostly in small studies evaluating different patient cohorts [7, 8]. Most of these studies have revealed favorable diagnostic accuracy of the assay, and a particularly high negative predictive value—in some studies >90%—may help avoid unnecessary biopsies [27]. AUC for discriminating ABMR versus no ABMR were reported to be in a range between 0.82 and 0.91 [9, 11, 12, 15], and in a recent meta‐analysis including five studies, pooled AUC levels of 0.89 were calculated [18]. To date, there is only scarce information on the diagnostic performance of dd‐cfDNA testing as an add‐on to DSA detection. Our results are in line with an earlier small study performed in DSA+ patients [10]. In this sub‐study of a multicentric trial [9], Jordan et al. [10] evaluated 87 patients with paired dd‐cfDNA results for whom DSA results had been reported. Among those, 33 had current/prior DSA with a 48% prevalence of active or chronic active ABMR. As in our study, dd‐cfDNA were elevated in patients diagnosed with ABMR (median 2.9%), resulting in an AUC of 0.89. Superior test performance as compared to our study may be explained by a high frequency of mixed rejections reported for ABMR cases (n = 5/16) [10]. A limited discriminative potential in our second cohort of patients subjected to indication biopsies—comparable to that observed in a study by Huang et al. [11]—seemed to result from a high prevalence of different features of injury other than ABMR, that are, TCMR, glomerulonephritis and a case of BKVAN associated with isolated C4d staining. Each of these five patients had dd‐cfDNA levels beyond 1%, reinforcing that dd‐cfDNA release is not limited to rejection but may also include other disease states. In contrast, patients without such lesions had dd‐cfDNA levels comparable to negative control subjects with normal histology and absence of DSA. Accordingly, when morphologic transplant injury of any cause was analyzed in relation to normal histology, dd‐cfDNA had a superior diagnostic accuracy with an AUC of 0.96. In the same analysis, DSA‐MFI lost its superior diagnostic value. Our results may argue against induction of significant endothelial injury and dd‐cfDNA release triggered by DSA binding in absence of visible microcirculation inflammation. Interestingly, in some contrast, in our subclinical cohort, tubulo‐interstitial infiltrates (classified as borderline) or glomerulonephritis did not trigger meaningful increases in dd‐cfDNA fractions, which may reflect the silent clinical presentation. Some [9, 13, 14] but not all [11, 12, 28] previous studies have shown moderate elevations of dd‐cfDNA fractions in patients with borderline lesions and/or TCMR, commonly below those found in ABMR. In a multicenter study by Stites et al. [14], among 79 patients with lower grade TCMR/borderline lesion, only 42 patients had elevated dd‐cfDNA (>0.5%), and these patients showed adverse graft function, more frequent de novo DSA formation and future/persistent rejection. In contrast, in a recent meta‐analysis of four studies evaluating dd‐cfDNA in relation to TCMR (35 samples), dd‐cfDNA levels were not elevated [17]. The elevated fraction of dd‐cfDNA in the single patient with BKVAN may be in line with an earlier report of 10 patients with BK viremia, where dd‐cfDNA levels correlated tightly with viral load and biopsy‐diagnosed BKVAN [29]. To our knowledge, there is no published data on dd‐cfDNA in glomerulonephritis, but our data suggest that at least in some instances recurrent or de novo glomerulonephritis may associate with allograft injury leading to an increase in its levels.
To the best of our knowledge, this is the first study evaluating dd‐cfDNA as an adjunct to distinct DSA characteristics, such as DSA‐MFI or the number of detected DSA. A major finding was thereby, that applying likelihood ratio tests to compare logistic regression models, DSA‐MFI (but not the number of DSA) added significantly to the diagnostic performance of dd‐cfDNA measurement (and vice versa). Nevertheless, using DSA‐MFI as a variable, we are aware that even after serum treatment to prevent complement interference, HLA single antigen testing to characterize DSA is prone to in vitro artifacts and MFI scores will remain an inaccurate measure of antibody concentration and binding properties.
Recent studies have suggested a role of ABMR in absence of detectable DSA [30, 31]. Molecular gene expression signatures were thereby reported to be very similar between DSA‐positive and DSA‐negative ABMR [31]. Our study did not address the diagnostic performance of dd‐cfDNA in this specific context, but very recently, a small study has suggested that this marker is discriminative irrespective of whether DSA are positive or negative [15].
We are aware of the limitations of our study, in particular, as in previous studies, a limited sample size. A strength of this monocentric study was a standardized evaluation of dd‐cfDNA and HLA single antigen testing in a single lab (for cohort 2, samples were tested retrospectively in a single batch to preclude day‐to‐day variations in test results). Moreover, for the first cohort, which was recruited in the context of a prospective randomized trial, biopsies were evaluated by two dedicated pathologists and, in parallel, subjected to molecular gene expression analysis using the MMDx platform (molecular criteria were included as a criterion of ABMR) to add precision and reduce the influence of sampling error [32, 33]. In this context, we want to mention a recent study by Gupta et al. [16] demonstrating better prediction of rejection with MMDx versus histomorphology. For the second cohort, however, molecular data were not systematically available, and ABMR diagnosis was based solely on morphologic criteria as the diagnostic gold‐standard, which may have led to less accuracy. Moreover, for this cohort, biopsies were evaluated by different members of our routine nephropathology team, which may have led to a higher degree of variability regarding biopsy interpretation due to inter‐observer variability. Finally, in our study, dd‐cfDNA fractions were analyzed in relation to total cfDNA, and we did not assess absolute concentrations. One may argue that this approach may not account for variations in the amount of recipient cfDNA. Such variations may for example be due to exercise, infections or changes in leukocyte counts [27]. Recent studies have suggested that determination of absolute concentrations of dd‐cfDNA fragments could be a strategy to improve diagnostic test performance [13, 27].
In conclusion, our results reinforce a discriminative potential of dd‐cfDNA in relation to ABMR diagnosis. Combined measurement of dd‐cfDNA and DSA‐MFI may thereby, significantly enhance test performance.
Authorship
KAM, KD, SK, RR‐S, and GAB: participated in the research design, performance of the research, data analysis, interpretation of results, and writing of the manuscript. ME, FE, MW and HR: participated in data analysis and writing of the manuscript. AT, TV and SH: participated in performance of the research. SC: participated in writing of the manuscript.
Funding
The study was funded by an investigator‐initiated unrestricted grant from CareDx Inc., Brisbane, South San Francisco, CA, USA (to G.A. Böhmig).
Conflict of interest
T. Viard, A. Tillgren and S. Casas were employed by CareDx Inc., Brisbane, San Francisco, CA, USA. All remaining authors have nothing to disclose. The results presented in this article have not been published previously in whole or part, except in abstract format.
Supporting information
Figure S1. Correlation between dd‐cfDNA results obtained with plasma collected in BD Vacutainer® tubes versus Streck Cell‐Free DNA blood collection tubes.
Figure S2. Biomarkers and renal parameters in relation to ABMR.
Figure S3. Biomarkers and renal parameters in relation to transplant injury of any cause.
Acknowledgements
The study was funded by an investigator‐initiated unrestricted grant from CareDx Inc., Brisbane, South San Francisco, CA, USA (to G.A. Böhmig). The authors wish to thank Dr. Jeff Reeve for his valuable scientific advice.
Contributor Information
Katharina A. Mayer, Email: katharina.mayer@meduniwien.ac.at.
Georg A. Böhmig, Email: georg.boehmig@meduniwien.ac.at.
References
- 1.Loupy A, Lefaucheur C. Antibody‐mediated rejection of solid‐organ allografts. N Engl J Med 2018; 379: 1150. [DOI] [PubMed] [Google Scholar]
- 2.Böhmig GA, Eskandary F, Doberer K, Halloran PF. The therapeutic challenge of late antibody‐mediated kidney allograft rejection. Transpl Int 2019; 32: 775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irish W, Nickerson P, Astor BC, et al. Change in estimated GFR and risk of allograft failure in patients diagnosed with late active antibody‐mediated rejection following kidney transplantation. Transplantation 2021; 105: 648. [DOI] [PubMed] [Google Scholar]
- 4.Loupy A, Haas M, Roufosse C, et al. The Banff 2019 Kidney Meeting Report (I): updates on and clarification of criteria for T cell– and antibody‐mediated rejection. Am J Transplant 2020; 20: 2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loupy A, Lefaucheur C, Vernerey D, et al. Complement‐binding anti‐HLA antibodies and kidney‐allograft survival. N Engl J Med 2013; 369: 1215. [DOI] [PubMed] [Google Scholar]
- 6.Eskandary F, Bond G, Kozakowski N, et al. Diagnostic contribution of donor‐specific antibody characteristics to uncover late silent antibody‐mediated rejection‐results of a cross‐sectional screening study. Transplantation 2017; 101: 631. [DOI] [PubMed] [Google Scholar]
- 7.Paul RS, Almokayad I, Collins A, Raj D, Jagadeesan M. Donor‐derived cell‐free DNA: advancing a novel assay to new heights in renal transplantation. Transplant Direct 2021; 7: e664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filippone EJ, Farber JL. The monitoring of donor‐derived cell‐free DNA in kidney transplantation. Transplantation 2021; 105: 509. [DOI] [PubMed] [Google Scholar]
- 9.Bloom RD, Bromberg JS, Poggio ED, et al. Cell‐free DNA and active rejection in kidney allografts. J Am Soc Nephrol 2017; 28: 2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan SC, Bunnapradist S, Bromberg JS, et al. Donor‐derived cell‐free DNA identifies antibody‐mediated rejection in donor specific antibody positive kidney transplant recipients. Transplant Direct 2018; 4: e379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang E, Sethi S, Peng A, et al. Early clinical experience using donor‐derived cell‐free DNA to detect rejection in kidney transplant recipients. Am J Transplant 2019; 19: 1663. [DOI] [PubMed] [Google Scholar]
- 12.Whitlam JB, Ling L, Skene A, et al. Diagnostic application of kidney allograft‐derived absolute cell‐free DNA levels during transplant dysfunction. Am J Transplant 2019; 19: 1037. [DOI] [PubMed] [Google Scholar]
- 13.Oellerich M, Shipkova M, Asendorf T, et al. Absolute quantification of donor‐derived cell‐free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study. Am J Transplant 2019; 19: 3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stites E, Kumar D, Olaitan O, et al. High levels of dd‐cfDNA identify patients with TCMR 1A and borderline allograft rejection at elevated risk of graft injury. Am J Transplant 2020; 20: 2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, Zheng C, Li X, et al. Diagnostic performance of donor‐derived plasma cell‐free DNA fraction for antibody‐mediated rejection in post renal transplant recipients: a prospective observational study. Front Immunol 2020; 11: 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta G, Moinuddin I, Kamal L, et al. Correlation of donor‐derived cell‐free DNA with histology and molecular diagnoses of kidney transplant biopsies. Transplantation 2021. 10.1097/TP.0000000000003838 [DOI] [PubMed] [Google Scholar]
- 17.Wijtvliet V, Plaeke P, Abrams S, et al. Donor‐derived cell‐free DNA as a biomarker for rejection after kidney transplantation: a systematic review and meta‐analysis. Transpl Int 2020; 33: 1626. [DOI] [PubMed] [Google Scholar]
- 18.Xiao H, Gao F, Pang Q, et al. Diagnostic accuracy of donor‐derived cell‐free DNA in renal‐allograft rejection: a meta‐analysis. Transplantation 2021; 105: 1303. [DOI] [PubMed] [Google Scholar]
- 19.Eskandary F, Regele H, Baumann L, et al. A randomized trial of bortezomib in late antibody‐mediated rejection (BORTEJECT). J Am Soc Nephrol 2018; 29: 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eskandary F, Bond G, Schwaiger E, et al. Bortezomib in late antibody‐mediated kidney transplant rejection (BORTEJECT Study): study protocol for a randomized controlled trial. Trials 2014; 15: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen J, Zhou Y, Chen Y, et al. Dynamics of early post‐operative plasma ddcfDNA levels in kidney transplantation: a single‐center pilot study. Transpl Int 2019; 32: 184. [DOI] [PubMed] [Google Scholar]
- 22.Danesi R, Lo YMD, Oellerich M, et al. What do we need to obtain high quality circulating tumor DNA (ctDNA) for routine diagnostic test in oncology? ‐ Considerations on pre‐analytical aspects by the IFCC workgroup cfDNA. Clin Chim Acta 2021; 520: 168. [DOI] [PubMed] [Google Scholar]
- 23.Haslacher H, Gerner M, Hofer P, et al. Usage data and scientific impact of the prospectively established fluid bioresources at the hospital‐based MedUni Wien Biobank. Biopreserv Biobank 2018; 16: 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halloran PF, Reeve J, Akalin E, et al. Real time central assessment of kidney transplant indication biopsies by microarrays: the INTERCOMEX study. Am J Transplant 2017; 17: 2851. [DOI] [PubMed] [Google Scholar]
- 25.Reeve J, Böhmig GA, Eskandary F, et al. Precision molecular phenotyping of kidney transplant biopsies using archetypal analysis. JCI Insight 2017; 2: e94197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas M, Loupy A, Lefaucheur C, et al. The Banff 2017 Kidney Meeting Report: revised diagnostic criteria for chronic active T cell‐mediated rejection, antibody‐mediated rejection, and prospects for integrative endpoints for next‐generation clinical trials. Am J Transplant 2018; 18: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oellerich M, Sherwood K, Keown P, et al. Liquid biopsies: donor‐derived cell‐free DNA for the detection of kidney allograft injury. Nat Rev Nephrol, 2021. 10.1038/s41581-021-00428-0 [DOI] [PubMed] [Google Scholar]
- 28.Gielis EM, Ledeganck KJ, Dendooven A, et al. The use of plasma donor‐derived, cell‐free DNA to monitor acute rejection after kidney transplantation. Nephrol Dial Transplant 2020; 35: 714. [DOI] [PubMed] [Google Scholar]
- 29.Kant S, Bromberg J, Haas M, Brennan D. Donor‐derived cell‐free DNA and the prediction of BK virus‐associated nephropathy. Transplant Direct 2020; 6: e622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senev A, Coemans M, Lerut E, et al. Histological picture of antibody‐mediated rejection without donor‐specific anti‐HLA antibodies: Clinical presentation and implications for outcome. Am J Transplant 2019; 19: 763. [DOI] [PubMed] [Google Scholar]
- 31.Callemeyn J, Lerut E, de Loor H, et al. Transcriptional Changes in Kidney Allografts with Histology of Antibody‐Mediated Rejection without Anti‐HLA Donor‐Specific Antibodies. J Am Soc Nephrol 2020; 31: 2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madill‐Thomsen KS, Wiggins RC, Eskandary F, Böhmig GA, Halloran PF. The effect of cortex/medulla proportions on molecular diagnoses in kidney transplant biopsies: rejection and injury can be assessed in medulla. Am J Transplant 2017; 17: 2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madill‐Thomsen K, Perkowska‐Ptasinska A, Böhmig GA, et al. Discrepancy analysis comparing molecular and histology diagnoses in kidney transplant biopsies. Am J Transplant 2020; 20: 1341. [DOI] [PubMed] [Google Scholar]
- 34.Schwaiger E, Eskandary F, Kozakowski N, et al. Deceased donor kidney transplantation across donor‐specific antibody barriers: predictors of antibody‐mediated rejection. Nephrol Dial Transplant 2016; 31: 1342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Correlation between dd‐cfDNA results obtained with plasma collected in BD Vacutainer® tubes versus Streck Cell‐Free DNA blood collection tubes.
Figure S2. Biomarkers and renal parameters in relation to ABMR.
Figure S3. Biomarkers and renal parameters in relation to transplant injury of any cause.


