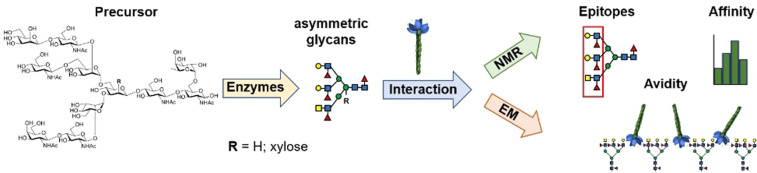
Abstract
The importance of multivalency for N‐glycan‐protein interactions has primarily been studied by attachment of minimal epitopes to artificial multivalent scaffold and not in the context of multi‐antennary glycans. N‐glycans can be modified by bisecting GlcNAc, core xylosides and fucosides, and extended N‐acetyl lactosamine moieties. The impact of such modifications on glycan recognition are also not well understood. We describe here a chemoenzymatic methodology that can provide N‐glycans expressed by the parasitic worm S. mansoni having unique epitopes at each antenna and containing core xyloside. NMR, computational and electron microscopy were employed to investigate recognition of the glycans by the human lectin DC‐SIGN. It revealed that core xyloside does not influence terminal epitope recognition. The multi‐antennary glycans bound with higher affinity to DC‐SIGN compared to mono‐valent counterparts, which was attributed to proximity‐induced effective concentration. The multi‐antennary glycans cross‐linked DC‐SIGN into a dense network, which likely is relevant for antigen uptake and intracellular routing.
Keywords: chemoenzymatic synthesis, cryo-EM, glycan, glycosyl transferase, NMR
The molecular recognition of chemoenzymatically synthesized glycans derived from the parasitic worm S. mansoni by the human lectin DC‐SIGN was investigated by a combination of NMR, computational and electron microscopy. It uncovered the importance of glycan complexity for lectin recognition including the influence of glycan valency, core xylosylation and length of oligo‐LacNAc residues.
Introduction
Glycan binding proteins play key roles in the battle between host and pathogens. Pathogens often express glyco‐epitopes that can be detected by glycan binding proteins of the host such as the mannose‐binding lectin, macrophage mannose receptor, or C‐type lectins, resulting in a diverse range of immune responses. The host also expresses glycan binding proteins such as the Siglec's and complement factor H that can detect self‐glycan signatures to maintain immune‐homeostasis.[1] A number of microbes have developed ways to achieve molecular mimicry of host glycans to avoid immune detection and establish infections.[2] Binding and structural studies have indicated that glycan binding proteins recognize relatively small oligosaccharide motifs often found at termini of complex glycans.[3] There are, however, indications that the complex architecture of glycans can modulate recognition of minimal epitopes.[4] For example, N‐glycans which have branched structures can potentially present multiple minimal epitopes that can engage with multiple glycan binding proteins resulting in increased binding avidities. It may also facilitate glycoconjugate clustering, which in turn may influence several downstream processes. Multivalency has primarily been studied by attachment of minimal epitopes to artificial multivalent scaffold,[5] and not in the context of natural multi‐antennary glycans. In addition, N‐glycans can be modified by bisecting N‐acetyl glucosamine (GlcNAc), core xylosides and fucosides, and extended N‐acetyl lactosamine (LacNAc) moieties. The impact of such modification on glycan recognition are also not well understood. These deficiencies are mainly due to inaccessibility of structurally defined complex glycans. Here, we report a synthetic methodology that can provide N‐glycans expressed by the parasitic worm S. mansoni. It includes compounds that have asymmetrical architectures and contain core xylose and terminal epitopes such as GalNAcβ1,4GlcNAc (Lac‐di‐NAc) and mono‐ and di‐Lewis X (Lex). NMR, computational and transmission electron microscopy were employed to investigate the importance of glycan complexity for recognition by the human lectin Dendritic cell‐specific ICAM‐3 grabbing nonintegrin (DC‐SIGN). It revealed that the core xyloside does not influence terminal epitope presentation and recognition. Furthermore, it was found that the multi‐antennary glycans bind with higher affinity to DC‐SIGN compared to mono‐valent minimal epitopes, which was attributed to proximity‐induced effective concentration. Finally, the studies uncovered that the multi‐antennary glycan can cross‐link DC‐SIGN into a dense network, which is likely relevant for antigen uptake and intracellular routing.
Schistosomes are parasitic helminths that cause chronic infections in humans associated with high morbidity.[6] N‐glycans of S. mansoni exhibit structural heterogeneity due variations in core modifications, the number of antennae, and their extensions into various epitopes. The expression is regulated in stage‐specific manner,[7] and for example during the egg and cercaria stage, Schistosomes abundantly decorate the core of N‐glycans by xylose.[8]
The termini of Schistosome glycans are often fucosylated to present epitopes such as Lewis X (Lex), GalNAc β1,4(Fuc α1,3)GlcNAc (LDN‐F) and di‐Lewis X (di‐Lex).[7] These epitopes can be recognized by DC‐SIGN, which is expressed on dendritic cells,[9] and interacts with conserved molecular patterns shared by a large group of microorganisms. It facilitates internalization of pathogens for processing and antigen presentation.[10] Pathogens can also exploit DC‐SIGN for infection and dissemination making it an important therapeutic target.[11]
DC‐SIGN is a homo‐tetrameric Ca2+ dependent lectin in which each monomer presents a carbohydrate recognition domain (CRD).[12] Potentially, it can engage in multivalent interactions resulting to high avidity of binding.[13] In vivo, multivalent glycans on the pathogen surface promote DC‐SIGN clustering, thus facilitating antigen uptake. There are indications that the density of surface exposed glycans influences cellular signaling, which in turn leads to enhancement or suppression of proinflammatory responses.[14] The molecular details by which these complexes are formed are poorly understood. Moreover, little is known about the preference of DC‐SIGN for specific S. mansoni derived glycans, and if there are restrictions related to glycan valency or size. The latter is due to the difficulties of synthesizing highly complex glycans that are modified by core xylose and have unusual epitopes such as Lac‐di‐NAc and LDN‐F.
Results and Discussion
Chemoenzymatic Synthesis
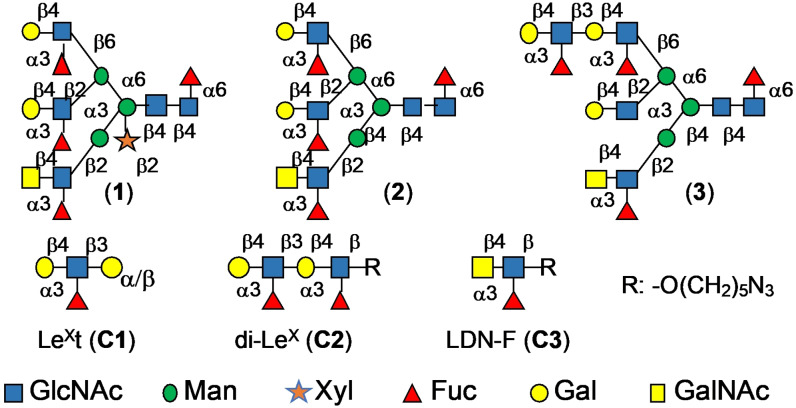
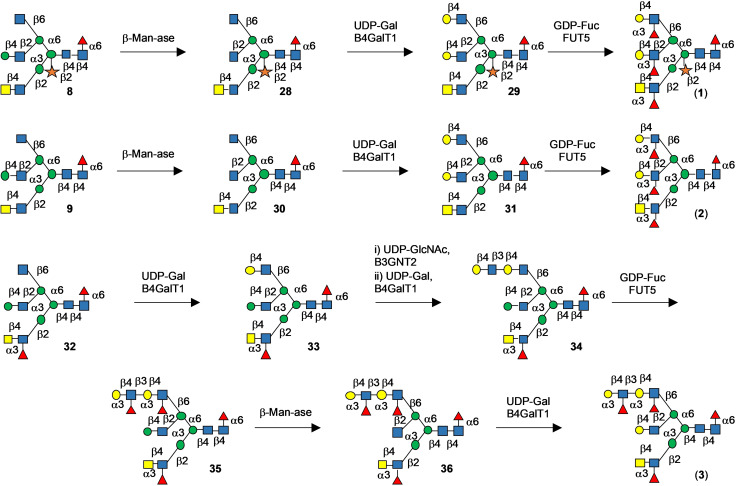
We set out to develop a chemoenzymatic methodology that can provide asymmetric complex glycans expressed by S. mansoni such as compounds 1, 2 and 3 (Figure 1). The compounds are modified by core xyloside, a Lac‐di‐NAc moiety and various patterns of fucosylation.
Figure 1.
Schematic representation of the different glycans (1–3) and corresponding fragments (C1–C3).
In parallel, reference compounds LeXt (C1, Galβ1,4‐(Fucα1,3)GlcNAcβ1,3Galα‐O‐aminopropyl; LeX tetraose, Elicityl), di‐LeX (C2, Galβ1,4‐(Fucα1,3)GlcNAcβ1,3Galβ1,4‐(Fucα1,3)GlcNAcβ‐O‐azidopentyl) and LDN‐F (C3, GalNAcβ1,4‐(Fucα1,3)GlcNAcβ‐O‐azidopentyl) were prepared or purchased for NMR studies described below. Challenging aspects of the preparation of compounds such as 1 are the installation of a core xyloside and the decoration of each antennae by unique appendages. The β‐1,2‐xylosyltransferase from Arabidopsis thaliana (XYLT), which potentially can install a core xyloside, operates early in the biosynthesis of N‐glycans and has narrow substrate specify. In particular, it cannot transfer xylose when an antenna is modified by a galactoside, which greatly complicates the construction of asymmetrical glycans.[15] Thus, we opted for a synthetic strategy in which the xyloside was introduced by chemical glycosylation to give precursors that can be elaborated by glycosyl transferases into asymmetrical multi‐antennary glycans.
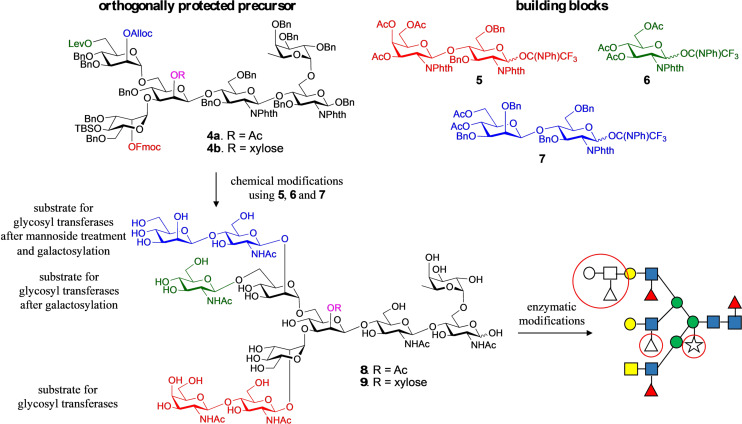
Thus, heptasaccharide 4 a was prepared, which is modified by a core xyloside, and at potential branching points is modified by the orthogonal protecting groups levulinoyl (Lev), fluorenylmethyloxycarbonyl (Fmoc), allyloxycarbonyl (Alloc), and t‐butyl‐dimethylsilyl (TBS).[16] Sequential removal of these protecting groups will give acceptors that can be extended by glycosyl donors such as 5, 6 and 7 to provide, after deprotecting, a compound such as 8 and 9 (Figure 2). It was anticipated that the Lac‐di‐NAc moiety of 8 and 9 can selectively be modified by glycosyl transferases. In the next stage of synthesis, the terminal Gal can be converted into LacNAc, and then enzymatically extended into a complex structure.[16a] It was expected that compound 4 b would give entry into complex glycans lacking core xylose. The synthesis of the donors 5, 6 and 7 is presented in the SI (Scheme S4).
Figure 2.
Overview of the synthetic strategy for the preparation of asymmetrical glycans with and without core xyloside. The coupling of the common precursors, 4 a and 4 b, which display orthogonal protecting groups at key branching points, with glycosyl donors 5–7 followed by global deprotection afforded asymmetric tri‐antennary glycans, 8 and 9, respectively. The latter intermediates were used as substrates for enzymatic extensions of each antenna to yield the asymmetrical glycans 1, 2 and 3, which differ for in presence or absence of core‐xylose, fucosylation of the arms and for the presence of a di‐LeX moiety. Antenna selective arm extension was possible because the unnatural β‐mannoside at the C‐2′ arm temporarily blocks it from enzymatic modification. It can, however, be unmasked by a β‐mannosidase and after enzymatic galactosylation, a precursor is obtained that can be elaborated by many glycosyl transferases into a complex structure. The strategy also exploits that many glycosyl transferases can modify LacNAc but not GlcNAc. The latter can, however, be converted into LacNAc by galactosylation with GalT1. Key structural features are highlighted by red circles.
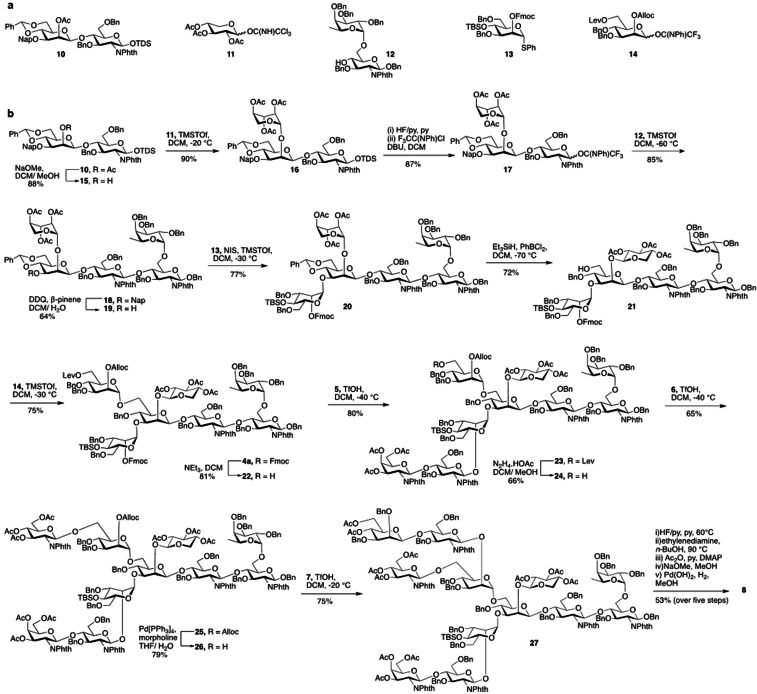
Compound 10 (Scheme 1) is an appropriate starting material for the synthesis of core xylosylated N‐glycan 4 a. Its mannosyl moiety is protected at C‐2 by an acetyl ester, which can selectively be cleaved to generate an acceptor that can be xylosylated with donor 11 to provide glycan 16. The anomeric thexyl dimethylsilyl (TDS) can be cleaved by HF pyridine to give a lactol that can be converted into donor 17 for glycosylation with acceptor 12 to give pentasaccharide 18. The naphthylmethyl (NAP) ether and the benzylidene acetal of the resulting compound can selectively be cleaved or reductively opened, respectively to give acceptors that can be glycosylated with thiophenyl mannoside donor 13 and N‐phenyl trifluoroacetimidate donor 14 to provide fully protected heptasaccharide 4 a, which is an appropriate precursor for antennae selective extension. Alternatively, the acetyl ester of compound 10 can be left intact to give access to non‐xylosylated N‐glycan precursor 4 b (see SI, Scheme S2 and S3). Thus, disaccharide 10 was treated with sodium methoxide in methanol to give acceptor 15, which was coupled with trichloroacetimidate 11 to provide xyloside bearing trisaccharide 16 as only the β‐anomer. The xylosyl moiety adopted a 1C4 conformation as evident from the coupling constant between H‐1 and H‐2 (J 1,2=3.7 Hz) indicating a di‐equatorial orientation. Others have observed that xylosides protected by acetyl esters in apolar solvent can adopt a 1C4 conformation.[17] It has been suggested that the resulting anomeric effect overcomes unfavorable steric interactions of axial substituents. It appears that protecting groups at neighboring glycosyl moieties can influence the conformation of the xyloside, indicating that steric hindrance of these entities exert a conformational control. Compound 16 was treated with HF in pyridine to remove the anomeric dimethylthexylsilyl group (TDS), and the resulting anomeric lactol was reacted with 2,2,2‐trifluoro‐N‐phenylacetimidoyl chloride in presence of DBU to form N‐phenyl trifluroacetimidate[18] donor 17 in a yield of 87 % over two steps. The later compound was glycosylated with disaccharide acceptor 12 [19] in presence of TMSOTf as the catalyst to furnish 18 (85 %). Next, the Nap ether of 18 was oxidatively removed by DDQ in the presence of β‐pinene as acid scavenger[20] to afford acceptor 19 (64 %). A NIS/TMSOTf mediated glycosylation of thio‐manosyl donor 13, modified by the orthogonal protecting groups Fmoc and TBS at C‐2 at C‐4, respectively (see SI, Scheme S5) with acceptor 19 gave hexasaccharide 20 in a yield of 77 % as only the α‐anomer. The benzylidene acetal of 20 was regioselectively opened by treatment with triethylsilane (Et3SiH) and dichlorophenylborane (PhBCl2) in DCM at −78 °C to furnish acceptor 21 having a hydroxyl at the C‐6 position of the central mannoside. As previously observed,[17a, 21] upon removal of a benzylidene acetal, the xyloside moiety adopted a 4C1 conformation, which was reflected by an increase in the 3 J 1,2 coupling constant from a di‐equatorial (3.7 Hz) to a di‐axial orientation (7 Hz). Finally, a TMSOTf‐mediated glycosylation of mannosyl trichoroacetimidate 14, modified by the orthogonal protecting groups Alloc and Lev (see SI, Scheme S5), with acceptor 21 gave heptasaccharide 4 a in a yield of 75 %. The corresponding non‐xylosylated hexasaccharide core 4 b was synthesized following a similar glycosylation strategy (See SI, Scheme S2).
Scheme 1.
Synthesis of N‐glycan precursor 8 starting from key building blocks 10–14.
Next, attention was focused on the installation of the various branching points to give tri‐antennary precursor glycan 8 by subsequent removal of the orthogonal protecting groups, glycosylations with donors 5, 6 and 7 followed by deprotection. Treatment of heptasaccharide 4 a with Et3N resulted in the selective removal of the Fmoc protecting group without affecting other functionalities to give glycosyl acceptor 22 in a good yield of 81 %. The latter acceptor was glycosylated with N‐phenyl trifluoroacetimidate donor 5 in the presence of trifluoromethanesulfonic acid (TfOH) at −40 °C to afford 23 (80 %). The Lev ester of 23 was selectively removed by treatment with hydrazine acetate to provide acceptor 24 (66 %), which was coupled with N‐phenyl trifluoroacetimidate glycosyl donor 6 using TfOH as promoter at −40 °C to afford 25 in a yield of 65 %. Next, the Alloc protecting group was removed by treatment with tetrakis(triphenylphosphine)‐palladium‐0 (Pd[PPh3]4) and morpholine to afford acceptor 26, which was then glycosylated with donor 7 using the standard procedure to afford tri‐antennary glycan 27 in 75 % yield. The latter compound was subjected to global deprotection over five steps entailing treatment with HF/pyridine to remove TBS ether, followed by heating under reflux with ethylenediamine in n‐butanol to cleave the phthalimide‐protecting groups. The exposed free amines and hydroxyls were acetylated by acetic anhydride in pyridine followed by cleavage of the esters by sodium methoxide. Finally, the benzyl ethers were removed by catalytic hydrogenation in the presence of palladium hydroxide (Pd[OH]2) which afforded the required tri‐antennary glycans 8 containing core xylose. The non‐xylosylated hexasaccharide core 4 b was extended by a similar glycosylation sequence to generate glycan 9 (See SI, Scheme S3).
Compounds 8 and 9 were treated with Helix pomatia β‐mannosidase and the inhibitor 1‐deoxyfuconojirimycin,[22] to avoid the cleavage of core α‐fucoside, to yield 28 and 30, respectively (Scheme 2). The two GlcNAc termini of the α6‐arm were simultaneously galactosylated by B4GalT1 and UDP‐galactose to give derivatives 29 and 31 having terminal LacNAc moieties. The latter glycans were fucosylated at all three arms by using FUT5 which modified the LacNAc and Lac‐di‐NAc substrates, affording compounds 1 and 2. The synthesis of asymmetric glycan 3 started by treatment of 9 with FUT5 to transform the LDN moiety into LDN‐F epitope providing 32. A key point in our chemoenzymatic strategy was the capping of the α6β2‐arm by a mannosyl moiety, and thus exposure of 32 to B4GalT1 resulted in the selective galactosylation of the α6β6‐arm. Exposure of the resulting compounds to B3GNT2 and then B4GalT1 resulted in the installation of a di‐LacNAc moiety to give compound 34. The di‐LacNAc moiety was transformed into LexLex epitope by treatment with FUT5 to give 35. The GlcNAc residue of the α6β2‐arm of 35 was unmasked by treatment with β‐mannosidase to give 36 which was galactosylated by B4GalT1 to provide target compound 3.
Scheme 2.
Enzymatic extension of precursor glycans 8 and 9 to give asymmetric complex glycan 3.
NMR Studies
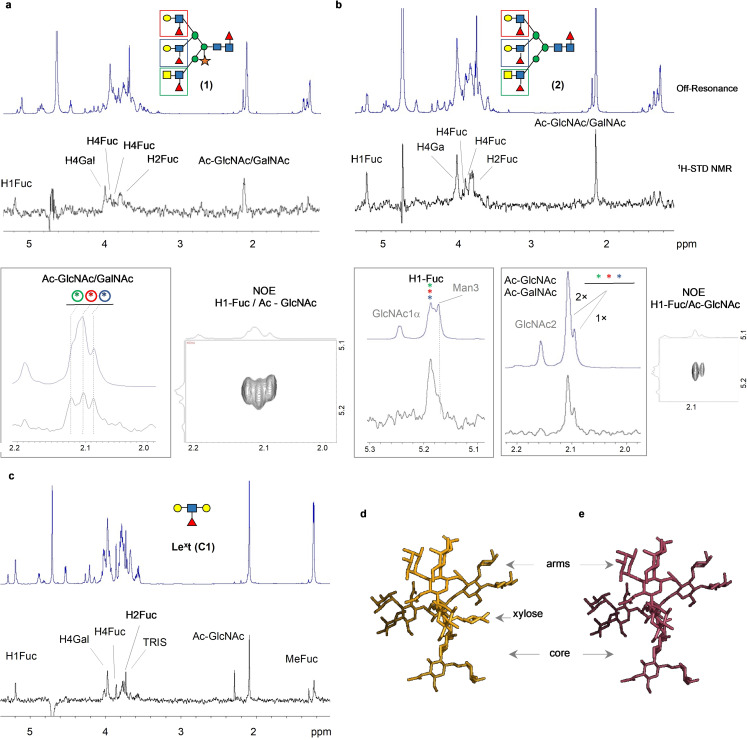
The interaction of DC‐SIGN with the xylosylated (1) and non‐xylosylated (2) glycans was examined by NMR. Saturation Transference Difference (1H‐STD‐NMR) experiments were performed using recombinant extracellular domain (ECD) of DC‐SIGN as receptor, which is organized as a tetramer in solution. The resulting 1H‐STD NMR profiles for 1 and 2 were very similar (Figure 3 a,b). No signals originating from the xyloside were observed in the 1H‐STD spectrum of compound 1, indicating that this residue does not participate in lectin binding. Comparing the 1H‐STD profiles of the complex glycans 1 and 2 with those of Lex (C1) showed that the contact between the lectin and glycans 1 and 2 takes place exclusively at the terminal epitopes (Figure 3 c).
Figure 3.
1H STD‐NMR spectra obtained for the complexes of DC‐SIGN (ECD) with (a) glycan (1), (b) glycan (2) and (c) the minimal epitope: the Lex trisaccharide (C1). The specific and unique 1H‐STD NMR signals are highlighted, along with the NOE cross peaks employed for their assignment. In particular the 2D sections correspond to the NOE correlations between the anomeric protons of the fucose residues and the methyl protons of the corresponding acetamide moieties for each of the three arms. The 1H‐STD NMR profiles are derived from the double difference between the STD spectrum in the presence and in absence of the protein. Irradiation frequency was set at 0.4 ppm. Protein saturation was achieved with a Gaussian‐shaped pulse of 49 ms (Gauss 1.1000, with a power of 1 e−05 W. Water suppression was applied by using the excitation sculpting. (d) and (e). Representative 3D models for glycan (1) and (2) respectively, obtained from MD simulations analysis. The models support the accessibility of the three arms for providing interactions with the lectin. Alternative views are provided in the SI Figure S8.
Due to severe 1H NMR signal overlap, untangling a possible preference of DC‐SIGN for the two Lex moieties and the LDN‐F epitope (Figure 1) at the different branches of the glycans was challenging. However, for compound 1, the NMR signals of the acetyl moieties at the GlcNAc residues of the three arms appear at different NMR chemical shifts, allowing unequivocal assignment by NOE spectroscopy (Figure 3 a). The results from the 1H‐STD NMR experiment showed that all GlcNAc residues contribute equally to the binding. A model of the two glycans in solution further supported that the three branches are equally accessible for lectin binding (Figure 3 d,e).
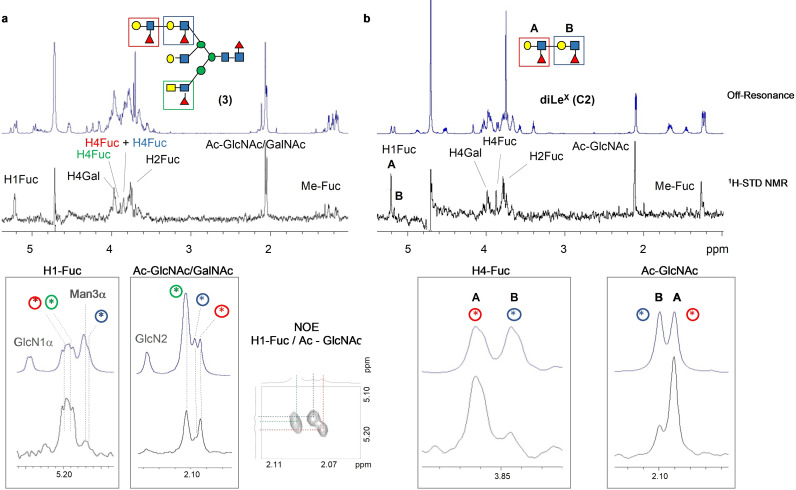
S. mansoni often presents extended N‐acetyl lactosamine (Galβ1,4‐GlcNAcβ1,3‐Galβ1,4‐GlcNAc; poly‐LN) chains which can be modified by fucosylation to form poly‐Lewisx moieties (poly‐Lex).[23] The asymmetric glycan 3 presents an extended arm with two repeating Lex motifs, an LDN‐F epitope and a non‐fucosylated LacNAc epitope (Figure 1). The 1H‐STD NMR analysis of 3 with DC‐SIGN (ECD) (Figure 4 a) showed a similar profile as observed for glycans 1 and 2 indicating that the terminal Lex and LDN‐F epitopes are equally well recognized. In contrast, the STD signals from the internal Lex motif were barely visible, indicating this moiety is less available for lectin recognition. This notion was further assessed by analyzing the interaction of the hexasaccharide C2 (di‐Lex, Figure 1) to DC‐SIGN (Figure 4 b). The simpler NMR spectra for this compound allowed a clear differentiation of the contributions of the internal and terminal Lex moieties. In particular, the 1H‐STD intensities from the internal Lex moiety only accounted for ≈15 % of those from terminal Lex.
Figure 4.
1H STD‐NMR spectra recorded for the interaction of DC‐SIGN (ECD) with (a) glycan (3) and (b) diLex hexasaccharide (C2). The specific and unique 1H‐STD NMR signals are highlighted, along with the NOE cross peaks employed for their assignments. Irradiation frequency was set at 0.4 ppm. Protein saturation was achieved with a Gaussian‐shaped pulse of 49 ms (Gauss 1.1000, with a power of 1 e−05 W. Water suppression was applied by using the excitation sculpting.
Next, the interactions of DC‐SIGN with the complex glycans 1, 2 and 3 were examined from the protein perspective using receptor‐based NMR methods. To this end, the chemical shift perturbation (CSP) analysis of the 15N labeled DC‐SIGN carbohydrate recognition domain (CRD) was performed (SI section 5.5). The tri‐antennary glycans 1, 2 and 3 showed similar CSPs, which did not extend beyond the canonical carbohydrate binding site, supporting that despite the complex structures, there are no additional contacts between the protein and glycans. However, the induced CSPs were significantly stronger than those observed for LDN‐F (C3)[24] and Lex (C1)[25] for the same number of equivalents.
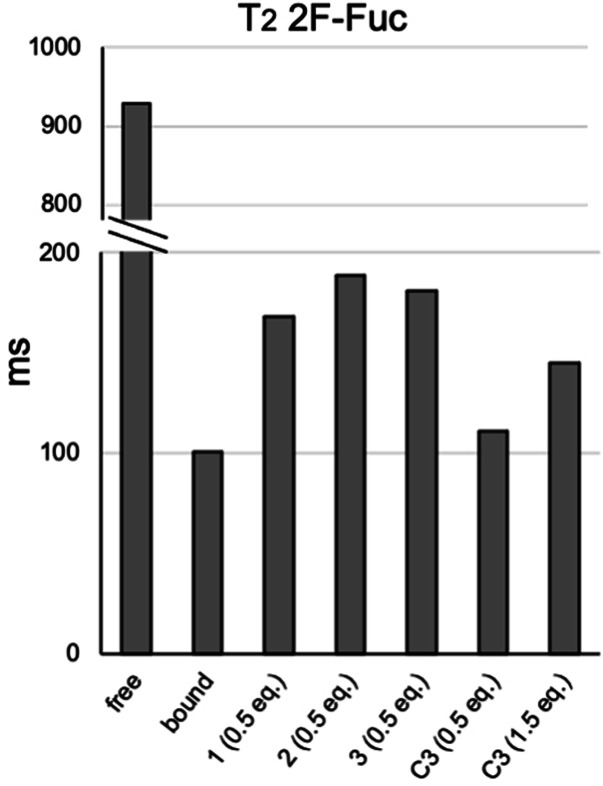
To confirm that the multi‐antennary glycans bind stronger than their monovalent counterparts, we used 2‐fluoro‐fucose (2‐F‐Fuc) as probe in competition NMR experiments to determine relative binding affinities. The addition of the lectin to the fluorinated probe causes a dramatic decrease in 19F transverse relaxation time (T2) with respect to the free form, which is indicative of 2‐F‐Fuc binding. Next 0.5 equivalent with respect to the probe of compounds 1, 2 and 3 were added. The addition of a competitor molecule causes a recovery in T2 for the fluorinated probe, which is proportional to the relative affinities of the tested compounds (Figure 5). Although subtle differences were observed, the results indicate that the three glycans bind DC‐SIGN with a similar affinity.
Figure 5.
19F‐NMR relaxation filter experiments performed for the fluorine‐containing monosaccharide 2‐fluoro‐fucose (2F‐Fuc) in absence and in presence of the tetrameric lectin DC‐SIGN. The addition of competitor molecules, 1–3 and C3 causes a recovery in the transverse relaxation time (T2) for the fluorinated probe, which is proportional to the relative affinities of the tested compounds.
Next, we compared the relative affinity of the synthetized multi‐antennary N‐glycans with respect to the monovalent counterpart. Thus, in a new sample tube containing 2‐F‐Fuc and DC‐SIGN (ECD), we added 0.5 equivalents of LDN‐F (C3), again with respect to the fluorinated probe. In this case, the competition with the monovalent compound caused only a marginal recovery in transverse relaxation time (T2), which indicates a much lower affinity of LDN‐F (C3) for the lectin. Because the multivalent molecules contain three putative epitopes for lectin binding, we further added the compound LDN‐F (C3) to reach 1.5 equivalents with respect to the fluorinated probe, equalizing in this way the concentration of binder epitopes with respect to the multivalent N‐glycans. Nevertheless, the recovery in T2 transverse relaxation time of the fluorinated probe is lower than that observed using the multivalent ligands, which demonstrate that a multivalent presentation enhances glycan‐lectin binding.
Multivalency is often attributed to a favorable spatial organization of a multivalent ligand that that can make multiple interactions with a multivalent protein. DC‐SIGN is a tetramer with their CRD binding sites separated by ≈40 Å.[26] The spatial distance between two terminal epitopes at the same glycan was estimated as approx. 20 Å (Figure 3 d,e). Thus, the possibility that one glycan can simultaneously bind to two different CRDs within the same tetramer through a chelating effect is unlikely. It has been proposed that “rebinding” can also contribute to cooperativity,[27] and in this model, as soon as a single ligand‐receptor complex dissociates, the presence of another ligand will increase the probability of another binding event. It is likely that the trimeric glycans exhibit higher affinities due to rebinding.
Multivalent Presentation and Lattice Formation
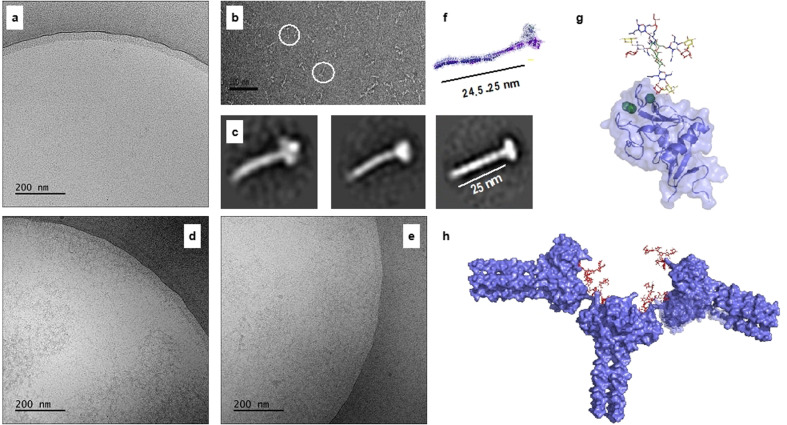
It is possible that the antenna of compounds 1–3 engage with more than one DC‐SIGN tetramer to form a higher ordered complexes.[28] To examine such a mode of binding, transmission electron microscopy (TEM) was employed to provide support for the existence of glycan‐lectin complexes. The apo DC‐SIGN (ECD) was used as control. In this format, lectin alone gave a soluble monodisperse organization (Figure 6 a–c). Conversely, incubation of the multivalent glycans 1 and 2 with DC‐SIGN (ECD) generated a tangled network (Figure 6 d,e). It is likely, the aggregates arise from the cross‐linking of two or more DC‐SIGN CRDs at different ECD tetramers by the multivalent glycans. In fact, we observed protein aggregation in the sample containing glycans 1, 2 and 3 but not in those containing the monovalent ligands LeX (C1) and LDN‐F (C3), (Figure S15). These observations further support our hypothesis that multi‐antennary glycans may be able to cross‐link DC‐SIGN (EDC) tetramers. Molecular modelling studies support that the glycans have appropriate geometries to engage with two DC‐SIGN tetramers (Figure 6 g,h). The 3D model of DC‐SIGN ECD assembled into tetramers was generated as previously described[13] (see SI, Section 6.3).
Figure 6.
Electron microscopy of DC‐SIGN interacting with multivalent glycans. Cryo‐TEM images at 50,000× magnification of apo DC‐SIGN (a) and in the presence of compound 1 (d) and 2 (e). (b) apo DC‐SIGN visualised by negative‐stain; the white circles mark the individual molecules. Scale bar is 100 nm. (c) three class‐averages resulting from 2D classification of extracted particles from negative‐stain 2D images and indicating the tetrameric oligomerization of DC‐SIGN (see SI). (f) 3D model of DC‐SIGN ECD assembled into tetramers. (g) 3D model of DC‐SIGN CRD showing the interaction with compound 1 as a single molecule. (h) 3D models of DC‐SIGN ECD (assembled into tetramers) showing the putative interaction with compound 1 and forming an inter‐connected network.
Conclusion
S. Mansoni expresses fucosylated glycans that can be recognized by DC‐SIGN on the cell surface of DCs. The structures of these antigenic glycans and their density on pathogen surface probably represents a first barrier for host detection. The molecular basis of complex formation between DC‐SIGN and glycans is not well understood. Moreover, little is known about a possible preference of DC‐SIGN for S. mansoni derived glycans, and it is unclear whether glycan complexity can modulate binding. The latter is due to difficulties of synthesizing highly complex glycans expressed by S. Mansoni required for structural and functional studies. Current approaches are limited to relatively simple epitopes or symmetrical structures.[23, 24, 28] To address these limitations, we have developed a chemoenzymatic approach that can provide N‐glycans derived from S. mansoni including compounds having core xylose and antenna with unusual structures such as Lac‐di‐NAc and Lex‐Lex. The compounds made it possible to examine the influence of glycan complexity on DC‐SIGN recognition. It revealed that the core xyloside does not influence terminal epitope presentation and lectin binding. Recently, the interaction of DC‐SIGN with symmetric bi‐antennary glycans with and without core‐xylose was examined by microarray technology.[29] It showed core xylose abolished lectin binding for the inner‐core mannosyl residues. Together with the NMR data presented here, it supports a model in which the xyloside masks or distort the conformation of the glycan‐core but does not impact the presentation of the Lex epitopes. Xylosylated glycans are highly expressed from the egg to cercaria stage of development, but substantially decreases during the schistosomula and adult worm stages.[8] The stage‐specific expression of core xylosylated glycans implies a role in snail‐schistosome interactions but may be less relevant during intra‐mammalian stages of development. The NMR studies also uncovered that the multi‐antennary glycans bind with higher affinity to DC‐SIGN compared to mono‐valent minimal epitopes. The STD experiments indicated that only the terminal epitopes of compounds 1–3 engage with DC‐SIGN and no further interactions were observed between protein and glycan. Modeling studies showed that the distance between the terminal epitopes is too short to engage with two different CRDs within the same tetramer. Thus, it is unlikely that avidity enhancement occurs through a classical chelate effect.[30] Although enhancement of affinity through proximity‐induced effective concentrations has received relatively little attention, it is likely that the higher affinities of 1–3 can be accounted to such an effect.
Multiple biological properties of DC‐SIGN, such as antigen uptake and processing, can be attributed to receptor clustering. Recently, it was shown that DC‐SIGN mediated uptake of glycoconjugates by immature moDCs does not directly correlate with their affinity,[14] and instead it appeared that the size of the cluster is critical for antigen uptake and routing.[31] Based on this study, we propose that multi‐antennary glycans may be able to cross‐link DC‐SIGN (EDC) tetramers into a dense network, where the nodes are tetramers of DC‐SIGN connected through the multivalent glycans. Physiologically, DC‐SIGN is embedded in the cellular membrane and, recently, the relevance of proper protein presentation was demonstrated.[28] Although our in‐solution studies do not represent N‐glycan presentation on the pathogen surface, it is conceivable that lectins and glycans presented at a surface can form similar latices and that the branched nature of N‐glycans contribute to cluster size, which in turn will impact update and intracellular transport. Future studies will focus on investigating DC‐SIGN clustering and internalization using nano‐ or micro‐structures, decorated with the types of complex glycans presented herein. The complex glycans will also be important to unravel structural elements required for triggering different signaling pathways during S. mansoni infection, as well as for the further developing of glycan‐based vaccines.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
This research was supported by the Netherlands Organization for Scientific Research (NWO; TOP‐PUNT grant 718.015.003 to G.J.B.), the Human Frontier Science Program Organization (HFSP; grant LT000747/2018‐C to L.U.), Ramón y Cajal (RYC) Contract (to A.A), the Agencia Estatal Investigacion of Spain (AEI; grants RTI2018‐095700‐B‐I00 (to NGAA) and RTI2018‐094751‐B‐C21) and the Severo Ochoa Excellence Accreditation (SEV‐2016‐0644). The staff of the Electron Microscopy Platform at the CIC bioGUNE and David Gil‐Carton are acknowledged for valuable technical assistance. Dr. Sean Connell is acknowledged for the support in the 2D classification of DC‐SGN ECD tetramers (EM). Dr. Prof. Jesús Jiménez‐Barbero is acknowledged for insightful discussions, suggestions and comments.
A. D. Srivastava, L. Unione, M. Bunyatov, I. A. Gagarinov, S. Delgado, N. G. A. Abrescia, A. Ardá, G.-J. Boons, Angew. Chem. Int. Ed. 2021, 60, 19287.
References
- 1.
- 1a.Geijtenbeek T. B. H., Gringhuis S. I., Nat. Rev. Immunol. 2009, 9, 465–479; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1b.Crocker P. R., Paulson J. C., Varki A., Nat. Rev. Immunol. 2007, 7, 255–266. [DOI] [PubMed] [Google Scholar]
- 2.Varki A., Gagneux P., in Essentials of Glycobiology, Cold Spring Harbor (NY), 2015, pp. 77–88. [PubMed] [Google Scholar]
- 3.Rillahan C. D., Paulson J. C., Annu. Rev. Biochem. 2011, 80, 797–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H., Xu C.-F., Blais S., Wan Q., Zhang H.-T., Landry S. J., Hioe C. E., J. Immunol. 2009, 182, 6369–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.
- 5a.Penades S., Davis B. G., Seeberger P. H., in Essentials of Glycobiology, Cold Spring Harbor (NY), 2015, pp. 743–753; [Google Scholar]
- 5b.Kiessling L. L., Gestwicki J. E., Strong L. E., Angew. Chem. Int. Ed. 2006, 45, 2348–2368; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2006, 118, 2408–2429. [Google Scholar]
- 6.Payet B., Chaumentin G., Boyer M., Amaranto P., Lemonon-Meric C., Lucht F., Scand. J. Infect. Dis. 2006, 38, 572–575. [DOI] [PubMed] [Google Scholar]
- 7.
- 7a.Hokke C. H., Deelder A. M., Hoffmann K. F., Wuhrer M., Exp. Parasitol. 2007, 117, 275–283; [DOI] [PubMed] [Google Scholar]
- 7b.Mickum M. L., Prasanphanich N. S., Heimburg-Molinaro J., Leon K. E., Cummings R. D., Front. Genet. 2014, 5, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smit C. H., van Diepen A., Nguyen D. L., Wuhrer M., Hoffmann K. F., Deelder A. M., Hokke C. H., Mol. Cell. Proteomics 2015, 14, 1750–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.
- 9a.Kapsenberg M. L., Nat. Rev. Immunol. 2003, 3, 984–993; [DOI] [PubMed] [Google Scholar]
- 9b.Mosser D. M., Edwards J. P., Nat. Rev. Immunol. 2008, 8, 958–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geijtenbeek T. B., Gringhuis S. I., Nat. Rev. Immunol. 2016, 16, 433–448. [DOI] [PubMed] [Google Scholar]
- 11.van Kooyk Y., Geijtenbeek T. B., Nat. Rev. Immunol. 2003, 3, 697–709. [DOI] [PubMed] [Google Scholar]
- 12.van Kooyk Y., Geijtenbeek T. B., Immunol. Rev. 2002, 186, 47–56. [DOI] [PubMed] [Google Scholar]
- 13.
- 13a.Gao A., Shrinivas K., Lepeudry P., Suzuki H. I., Sharp P. A., Chakraborty A. K., Proc. Natl. Acad. Sci. USA 2018, 115, E11053–E11060; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13b.Fasting C., Schalley C. A., Weber M., Seitz O., Hecht S., Koksch B., Dernedde J., Graf C., Knapp E.-W., Haag R., Angew. Chem. Int. Ed. 2012, 51, 10472–10498; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2012, 124, 10622–10650; [Google Scholar]
- 13c.Mammen M., Choi S.-K., Whitesides G. M., Angew. Chem. Int. Ed. 1998, 37, 2754–2794; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 1998, 110, 2908–2953. [Google Scholar]
- 14.Jarvis C. M., Zwick D. B., Grim J. C., Alam M. M., Prost L. R., Gardiner J. C., Park S., Zimdars L. L., Sherer N. M., Kiessling L. L., Proc. Natl. Acad. Sci. USA 2019, 116, 14862–14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kajiura H., Okamoto T., Misaki R., Matsuura Y., Fujiyama K., J. Biosci. Bioeng. 2012, 113, 48–54. [DOI] [PubMed] [Google Scholar]
- 16.
- 16a.Wang Z., Chinoy Z. S., Ambre S. G., Peng W., McBride R., de Vries R. P., Glushka J., Paulson J. C., Boons G. J., Science 2013, 341, 379–383; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16b.Li T., Huang M., Liu L., Wang S., Moremen K. W., Boons G. J., Chem. Eur. J. 2016, 22, 18742–18746; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16c.Gagarinov I. A., Li T., Torano J. S., Caval T., Srivastava A. D., Kruijtzer J. A., Heck A. J., Boons G. J., J. Am. Chem. Soc. 2017, 139, 1011–1018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16d.Liu L., Prudden A. R., Capicciotti C. J., Bosman G. P., Yang J. Y., Chapla D. G., Moremen K. W., Boons G. J., Nat. Chem. 2019, 11, 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.
- 17a.Brzezicka K., Echeverria B., Serna S., van Diepen A., Hokke C. H., Reichardt N. C., ACS Chem. Biol. 2015, 10, 1290–1302; [DOI] [PubMed] [Google Scholar]
- 17b.Laurière M., Laurière C., Chrispeels M. J., Johnson K. D., Sturm A., Plant Physiol. 1989, 90, 1182–1188; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17c.Kurosaka A., Yano A., Itoh N., Kuroda Y., Nakagawa T., Kawasaki T., J. Biol. Chem. 1991, 266, 4168–4172; [PubMed] [Google Scholar]
- 17d.Prenner C., Mach L., Glossl J., Marz L., Biochem. J. 1992, 284, 377–380; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17e.Wilson I. B., Harthill J. E., Mullin N. P., Ashford D. A., Altmann F., Glycobiology 1998, 8, 651–661. [DOI] [PubMed] [Google Scholar]
- 18.
- 18a.Yu B., Tao H., Tetrahedron Lett. 2001, 42, 2405–2407; [Google Scholar]
- 18b.Yu B., Tao H., J. Org. Chem. 2002, 67, 9099–9102. [DOI] [PubMed] [Google Scholar]
- 19.Sun B., Srinivasan B., Huang X., Chem. Eur. J. 2008, 14, 7072–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloyd D., Bylsma M., Bright D. K., Chen X., Bennett C. S., J. Org. Chem. 2017, 82, 3926–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.
- 21a.Kerékgyártó J., van der Ven J. G. M., Kamerling J. P., Lipták A., Vliegenthart J. F. G., Carbohydr. Res. 1993, 238, 135–145; [DOI] [PubMed] [Google Scholar]
- 21b.Crich D., Dai Z., Tetrahedron 1999, 55, 1569–1580. [Google Scholar]
- 22.Winchester B., Barker C., Baines S., Jacob G. S., Namgoong S. K., Fleet G., Biochem. J. 1990, 265, 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srivatsan J., Smith D. F., Cummings R. D., J. Biol. Chem. 1992, 267, 20196–20203. [PubMed] [Google Scholar]
- 24.Srivastava A. D., Unione L., Wolfert M. A., Valverde P., Ardá A., Jiménez-Barbero J., Boons G.-J., Chem. Eur. J. 2020, 26, 15605–15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pederson K., Mitchell D. A., Prestegard J. H., Biochemistry 2014, 53, 5700–5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.
- 26a.Mitchell D. A., Fadden A. J., Drickamer K., J. Biol. Chem. 2001, 276, 28939–28945; [DOI] [PubMed] [Google Scholar]
- 26b.Garber K. C. A., Wangkanont K., Carlson E. E., Kiessling L. L., Chem. Commun. 2010, 46, 6747–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber M., Bujotzek A., Haag R., J. Chem. Phys. 2012, 137, 054111. [DOI] [PubMed] [Google Scholar]
- 28.Porkolab V., Pifferi C., Sutkeviciute I., Ordanini S., Taouai M., Thépaut M., Vivès C., Benazza M., Bernardi A., Renaudet O., Fieschi F., Org. Biomol. Chem. 2020, 18, 4763–4772. [DOI] [PubMed] [Google Scholar]
- 29.Brzezicka K., Vogel U., Serna S., Johannssen T., Lepenies B., Reichardt N.-C., ACS Chem. Biol. 2016, 11, 2347–2356. [DOI] [PubMed] [Google Scholar]
- 30.Cecioni S., Imberty A., Vidal S., Chem. Rev. 2015, 115, 525–561. [DOI] [PubMed] [Google Scholar]
- 31.Unger W. W., van Beelen A. J., Bruijns S. C., Joshi M., Fehres C. M., van Bloois L., Verstege M. I., Ambrosini M., Kalay H., Nazmi K., Bolscher J. G., Hooijberg E., de Gruijl T. D., Storm G., van Kooyk Y., J. Controlled Release 2012, 160, 88–95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information