Abstract
Purpose:
Membrane transport protein organic anion transporting polypeptide (OATP) 1B1 mediates hepatic uptake of many drugs (e.g. statins). The OATP1B1 c.521T>C (p. V174A) polymorphism has reduced transport activity. Conflicting in vitro results exist regarding whether V174A-OATP1B1 has reduced plasma membrane localization; no such data has been reported in physiologically relevant human liver tissue. Other potential changes, such as phosphorylation, of the V174A-OATP1B1 protein have not been explored. Current studies characterized the plasma membrane localization of V174A-OATP1B1 in genotyped human liver tissue and cell culture and compared the phosphorylation status of V174A- and wild-type (WT)-OATP1B1.
Methods:
Localization of V174A- and WT-OATP1B1 were determined in OATP1B1 c.521T>C genotyped human liver tissue (n=79) by immunohistochemistry and in transporter-overexpressing human embryonic kidney (HEK) 293 and HeLa cells by surface biotinylation and confocal microscopy. Phosphorylation and transport of OATP1B1 was determined using 32P-orthophosphate labeling and [3H]estradiol-17β-glucuronide accumulation, respectively.
Results:
All three methods demonstrated predominant plasma membrane localization of both V174A- and WT-OATP1B1 in human liver tissue and in cell culture. Compared to WT-OATP1B1, the V174A-OATP1B1 has significantly increased phosphorylation and reduced transport.
Conclusions:
We report novel findings of increased phosphorylation, but not impaired membrane localization, in association with the reduced transport function of the V174A-OATP1B1.
Keywords: Plasma membrane localization, OATP1B1, polymorphism, phosphorylation, genotype
INTRODUCTION
Organic anion transporting polypeptide (OATP) 1B1 is a membrane transport protein localized to the basolateral membrane of hepatocytes in human liver. It mediates the hepatic uptake of many important drugs (1) (e.g., lipid-lowering statins (2-4), antibiotics (5), anti-hypertensives (6-8), and anticancer drugs (9)) and endogenous compounds (3, 10). The role of OATP1B1 in hepatic drug disposition and transporter-mediated drug-drug interactions (DDIs) has been increasingly recognized. Impaired OATP1B1 transport activity due to genetic variation often causes increased systemic exposure of OATP1B1 substrates (e.g., statins), and leads to severe adverse events. In vitro, the nonsynonymous single-nucleotide polymorphism (SNP) in OATP1B1, c.521 T>C (rs4149056), which results in an amino acid substitution (Val174Ala, V174A), has significantly reduced transport activity compared with the wild-type (WT) OATP1B1 (11-14). In vivo, human subjects bearing this V174A-OATP1B1 polymorphism had increased systemic exposure of many drugs that are OATP1B1 substrates including statins (15-17) and repaglinide (18). OATP1B1 c.521 T>C is the most robust and important predictor of statin-induced myopathy (19), and is recognized as a transporter polymorphism with significant clinical relevance (20).
The initial study from Tirona et al. reported that the V174A-OATP1B1 has markedly reduced plasma membrane levels, while has similar total protein levels compared with the WT-OATP1B1, when transporters were over-expressed in HeLa cells infected with vaccinia virus (11). Although it is frequently cited (by >66 citations by literature search) that the mechanism responsible for the reduced transport function of the V174A-OATP1B1 is due to reduced transporter surface localization based on this initial finding (11), controversial results exist. Two later studies (12, 13) reported that when over-expressed in human embryonic kidney (HEK) 293 cells using a non-viral approach, V174A-OATP1B1 had significantly reduced transport function when compared with the WT-OATP1B1, however, was predominantly expressed on the plasma membrane. Currently, the mechanism underlying the reduced transport function of the V174A variant of OATP1B1 (abbreviated as V174A-OATP1B1) is obscure. It is not known whether the plasma membrane localization of V174A-OATP1B1 is cell-type dependent or if the conflicting results are due to different protocols used in different laboratories. Disregarding these apparently conflicting results, the in vivo plasma membrane localization of V174A-OATP1B1 in physiologically relevant hepatocytes of human liver tissue has not been characterized.
Altered post-translational modification of proteins has also been reported to be associated with SNPs, including transport proteins. For instance, the SNP Q141K (rs2231142) in the breast cancer resistance protein (BCRP) has altered ubiquitination-mediated degradation compared to the wild-type BCRP, leading to reduced plasma membrane levels of BCRP (21). The significantly reduced transport function of the SNP A369P in the human norepinephrine transporter (hNET) is associated with complete abolishment of the fully glycosylated form and reduced plasma membrane localization (22). Altered phosphorylation has also been associated with SNPs of many proteins leading to the gain or loss of phosphorylation at the sites of genetic variation themselves or phosphorylation at adjacent sites (23). Phosphopeptides of OATP1B1 were identified previously in a large-scale phosphoproteomic study from a non-genotyped human liver tissue (24), suggesting that OATP1B1 is also a phosphorylated protein. The phosphorylation status of OATP1B1 in an in vitro cell culture system, and particularly the potential difference between V174A- and WT-OATP1B1, have not been characterized.
The primary aims of the current study are twofold: 1) to characterize the plasma membrane localization of V174A-OATP1B1 in vivo in genotyped human liver tissue and in vitro in cell culture systems; 2) and to compare the phosphorylation status of V174A-OATP1B1 with that of the WT-OATP1B1. Immunohistochemistry (IHC), surface biotinylation, immunofluorescence staining followed by confocal microscopy were used to determine the plasma membrane localization of OATP1B1. 32P-orthophosphate labeling was used to compare the phosphorylation status between V174A- and WT-OATP1B1. Another common variant of OATP1B1, the c. 388A>G (rs2306283, N130D) variant, has unaltered transport function in most in vitro studies (11-14, 25-28), while it has increased OATP1B1 protein levels in liver tissues (29). IHC of OATP1B1 expression in the c. 388A>G genotyped human liver tissues were also evaluated for comparison purposes.
MATERIALS AND METHODS
Materials.
[3H]E217βG (specific activity 41.4 Ci/mmol) and 32P-orthophosphate (10 mCi) were purchased from PerkinElmer Life Science (Waltham, MA). Complete protease inhibitor cocktail tablets, Phosstop phosphatase inhibitor tablets were purchased from Roche Diagnostics (Indianapolis, IN). Geneticin (G418) and EZ-Link™ Sulfo-NHS-SS-Biotin were purchased from Thermo Fisher Scientific (Carlsbad, CA). Protein A/G beads were purchased from Santa Cruz Biotechnology (Dallas, TX). Unlabeled estradiol 17β-glucuronide (E217βG), lactic acid dehydrogenase (LDH) cytotoxicity kit and fetal bovine serum (FBS) were purchased from Sigma Aldrich (St. Louis, MO). Bio-Safe II liquid scintillation mixture and 1,4-dithiothreitol (DTT) were purchased from Research Products International (Mt. Prospect, IL). Other reagents, unless specified, were purchased from Sigma Aldrich (St. Louis, MO), Thermo Fisher Scientific (Carlsbad, CA), or VWR International (Radnor, PA).
Human liver tissue.
79 Formalin-fixed, paraffin-embedded (FFPE) archived human liver ( 42 from surgical resection and 37 from liver biopsy) and normal kidney tissue blocks were obtained from OUHSC Stephenson Cancer Center Biospecimen Acquisition Core and Bank from the Department of Pathology at the University of Oklahoma Health Sciences Center. All tissues were pathologically determined by certified pathologists at OUHSC, among which 8 were from surgical resections of non-cancer patients (e.g. gunshot wounds). All donor information including age, sex and diagnosis are provided in Supplemental Table S I. Use of human tissues was approved by the Institutional Review Board at the University of Oklahoma Health Sciences Center.
Genotyping of human liver samples for OATP1B1 c.521 T>C and c. 388 A>G polymorphism.
Genotyping of OATP1B1 c.521 T>C and c. 388 A>G were conducted in all 79 liver FFPE tissues. The FFPE tissue blocks were sliced into paraffin rolls (minimal thickness of 2 μm) at the OUHSC Stephenson Cancer Center Tissue Pathology Shared Resource facilities. Total genomic DNA was isolated from tissue slices using the Quick-DNA FFPE Kit (Zymo Research, Irvine, CA). Samples were genotyped for the OATP1B1 c.521 T>C polymorphism using the following primer pairs: forward, 5’-AAATTACCCAGTCTCAGGTATG-3’ and reverse, 5’-TGTCCTTCTTTAGCGAAATC-3’ to produce a 339-BP product. The same genomic DNA samples were also genotyped for the OATP1B1 c.388 A>G polymorphism using the following primer pairs: forward: 5’-GCAAATAAAGGGGAATATTTCTC-3’ and reverse, 5’-AGAGATGTAATTAAATGTATAC-3’ (11) to produce a 278-BP product. Polymerase chain reaction (PCR) was conducted using a Techne TC-312 Thermocycler (Techne, Burlington, NJ) in a 20-μl reaction system containing 50 ng of genomic DNA template, deoxyribonucleotide triphosphates (dNTP) (0.5 mM each), the primer pair (0.5 μM each), 1X Phusion buffer, and 2 units of Phusion DNA polymerase (Thermo Scientific, Carlsbad, CA). The PCR was conducted at 94°C for 5 min, then 94°C for 30 s, 54°C for 2 min, and 72°C for 1 min for 40 cycles for c.521 T>C and at 94°C for 5 min, 94°C for 30 s, 50°C for 2 min and 72°C for 1 min for 40 cycles for c.388 A>G, followed by 72°C for 10 min for the final extension. PCR products were visualized on a 2% agarose gel stained with ethidium bromide. Bands of the expected size were removed from the gel and purified using the Zymoclean Gel DNA Recovery kit (Zymo Research, Irvine, CA). The recovered PCR product was then sequenced using the reverse primers described above at the OUHSC Laboratory for Molecular Biology and Cytometry Research DNA sequencing core using a 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA).
Immunohistochemistry (IHC) and digital image analysis for OATP1B1 and OATP1B3.
The IHC staining was conducted by the OUHSC Stephenson Cancer Center Tissue Pathology Shared Resource facilities using a method similar to published previously (25, 30, 31). The immunohistochemistry protocol was based on the manufacturer’s protocol for the Leica Bond-III™ Polymer Refine Detection system (DS 98000). Briefly, FFPE tissue blocks were cut into 4-μm sections and were mounted on positively charged slides at room temperature. After deparaffinization and rehydration with an automated Multistainer (Leica ST5020) (Leica, Buffalo Grove, IL), slides were transferred to the Leica Bond-III™ automated IHC stainer (Leica, Buffalo Grove, IL). Antigen was retrieved at 100°C for 20 min in citrate buffer (pH 6). After blocking with normal goat serum (Invitrogen, Carlsbad, CA), slides were incubated for 30 min with rabbit polyclonal OATP1B1 or OATP1B3 (1:1000 dilution of each) antibody that were custom-generated in our laboratory (32, 33). Titration of the OATP1B1 and OATP1B3 antibodies for IHC was performed in pilot studies. After incubation with horseradish peroxidase (HRP)-conjugated secondary antibody, 3, 3’-diaminobenzidine (DAB) was used to detect OATP1B1 and OATP1B3 expression. The nuclei were counterstained using hematoxylin. The same tissue sections with blocking buffer lacking the primary antibody served as the negative control. Human kidney tissue was used as a negative control tissue for both OATP1B1 and OATP1B3. Hematoxylin and eosin (H&E) staining was performed for pathological examination of tissue blocks from each subject. Slides after IHC and H&E staining were scanned using the Aperio ScanScope (Leica, Buffalo Grove, IL). Images of digital slides were processed using ImageScope software (Leica, Buffalo Grove, IL).
As hepatitis C virus (HCV)-infected liver tissue and cirrhosis liver tissues have been reported to have reduced OATP1B1 and OATP1B3 expression (34, 35), 13 HCV-positive and 6 cirrhotic liver tissues were not included in the semi-quantitative analysis of OATP1B1 and OATP1B3 staining. For all other non-HCV and non-cirrhotic liver tissue, 2-4 images of OATP1B1 and OATP1B3 IHC staining were captured (~300 x 300 μm, 200X) in nonmalignant areas of each liver tissue identified by a pathologist, and were processed for subsequent semi-quantitative analysis similarly to published previously (36, 37). In brief, for color deconvolution of IHC images, DAB and hematoxylin staining were digitally separated using ImageJ Fiji software with a color deconvolution plugin (version 1.2; WS Rasband, National Institute of Health, Bethesda, MD) (38), similar to previously published (36). Deconvoluted images with DAB staining were subjected to measurement of mean gray values, with the lower and upper thresholds set at 120 and 220 for OATP1B1 and 150 and 220 for OATP1B3, respectively. The number of nuclei in the same field were determined from the hematoxylin staining by analyzing particle numbers using ImageJ Fiji software. The mean gray values of OATP1B1 and OATP1B3 were then normalized by the number of cell nuclei in the same field using a similar strategy as published previously (37). The expression levels of OATP1B1 and OATP1B3 were determined as the average of the gray values normalized by nuclei number from 2-4 images per liver determined. When comparing OATP1B1 or OATP1B3 total protein levels among OATP1B1 c.521 TT, TC and CC genotypes, data for homozygous c.521 CC and heterozygous c.521 TC were pooled together for statistical purposes due to the low number of donors (total 2) that had the c.521 CC genotype. OATP1B1 and OATP1B3 expression in separate genotypes of c.521 TT, TC and CC are provided in Supplemental Fig. S II.
Construct and transfection.
A mammalian plasmid expression vector encoding C-terminus FLAG-tagged V174A-OATP1B1 (pCMV6-FLAG-V174A-OATP1B1) was constructed at Origene (Rockville, MD) and was verified by sequencing. The pCMV6-FLAG-WT-OATP1B1 vector has been published previously (32).
Transfection and Cell culture.
HEK293 stable cell line expressing FLAG-tagged WT-OATP1B1 was published previously (32). The HEK293-Mock cells were kindly provided by Dr. Dietrich Keppler (10, 39). The HEK293 stable cell line overexpressing FLAG-tagged V174A-OATP1B1 (HEK293-V174A-OATP1B1) was established by transfection of the pCMV6-FLAG-V174A-OATP1B1 using Lipofectamine 2000 (Thermo Fisher, Carlsbad, CA), followed by G418 selection at 600 μg/ml. HeLa cells were plated at a seeding density of 2.5 x 104 cells per well in a 24-well plate coated with Poly-L-lysine. Twenty-four hours after seeding, cells were transiently transfected with pCMV6-FLAG-WT-OATP1B1 (32) and pCMV6-FLAG-V174A-OATP1B1 using FuGENE HD transfection reagent (Promega, Madison, WI). All cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% FBS and 1% antibiotic-antimycotic solution (AA) in a humidified atmosphere (95% O2 and 5% CO2) at 37°C. HEK293 stable cell lines were supplemented with 600 μg/ml G418.
Transport assays.
HEK293-FLAG-WT-OATP1B1, -V174A-OATP1B1, and -Mock cells were seeded at a seeding density of 1.5 x 105 cells/well in 24-well plates. [3H]E217βG accumulation (1 μM, 2 min) was determined 48 h after seeding of HEK293 stable cell lines or 48 h post-transfection of HeLa cells, as published previously (32). [3H]E217βG accumulation was determined by scintillation counting and normalized by protein concentration as determined by BCA assay. To correct for differences in protein levels of WT- and V174A-OATP1B1 expressed in the HEK293 stable cell lines and in transiently transfected HeLa cells, FLAG immunoblot was conducted in separate triplicate wells from the same experiment, with immunoblot of β-actin as the loading control. [3H]E217βG accumulation was further normalized by the protein levels of FLAG-WT-OATP1B1 and FLAG-V174A-OATP1B1 that were previously normalized by β-actin, using a strategy similar to that published previously (31).
Immunoblot.
Immunoblotting was conducted similarly to those published previously (32, 33). Briefly, cells were washed once with pre-warmed 1X PBS and then lysed directly with ice-cold lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% (v/v) NP-40, 0.5% sodium-deoxycholate, and Complete™ protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Whole cell lysates (50 μg) were resolved on a 10% SDS-PAGE (BioRad, Hercules, CA) and transferred to a nitrocellulose membrane (Santa Cruz Biotechnology, Inc., Dallas, TX). The membrane was probed with the following antibodies: mouse polyclonal anti-FLAG antibody (1:4000 dilution), mouse monoclonal anti-β-actin antibody (1:5000 dilution) (Sigma Aldrich, St. Louis, MO), and mouse monoclonal anti-GAPDH antibody (1:5000 dilution) (Santa Cruz Biotechnology, Inc., Dallas, TX). The nitrocellulose membrane was then probed with an anti-mouse HRP-conjugated secondary antibody (Cell Signaling, Danvers, MA). The signal was detected using Supersignal West Dura (Pierce, Rockford, IL) with a BioRad ChemiDoc XRS imaging system (BioRad, Hercules, CA). All primary and secondary antibodies were diluted in 1X TBST. Expression of protein and loading control densitometry analysis was performed using Image Lab v4.1 software (BioRad, Hercules, CA).
Surface biotinylation.
Surface biotinylation experiments were conducted similarly to those published previously (40). HEK293-FLAG-WT-OATP1B1 and -V174A-OATP1B1 cells were seeded in 100 mm2 dishes at a seeding density of 2.5 x 106 cells per dish and were grown for 48 h. HeLa cells were seeded in 24-well plates at a seeding density of 2.5x104 cells/well and were transfected with WT- or V174A-OATP1B1 (0.5 μg DNA/well). Surface biotinylation was conducted 48 h after seeding (for HEK293-WT- and –V174A-OATP1B1 stable cell lines) or post-transfection of HeLa cells. Briefly, cells were washed three times with ice-cold phosphate-buffered saline (PBS) containing calcium and magnesium (PBS-Ca/Mg, pH 7.5; Corning, Corning, NY), followed by incubation with sulfo-NHS-SS-biotin (1 mg/ml) in PBS-Ca/Mg for 1 h at 4°C. Cells were then washed three times with ice-cold PBS-Ca/Mg containing 100 mM glycine, followed by one additional wash with ice-cold PBS with calcium and magnesium and lysed with lysis buffer containing 10 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, and Complete™ protease inhibitor cocktail. Whole cell lysates were adjusted to 1 μg/μL, 900 μg (for HEK293 stable cell lines) or 350 μg (for HeLa cells) of which was adsorbed with NeutrAvidin beads (Pierce, Rockford, IL) overnight at 4°C. After centrifugation at 15,000 rpm for 2 min, the supernatant was kept on ice and the NeutrAvidin beads were further washed five times with ice-cold lysis buffer and eluted with 2X Laemmli buffer (Biorad, Hercules, CA) containing 0.4% SDS, 50mM DTT, and 5% (v/v) β-mercaptoethanol (BME) for 1 h at room temperature. The elution was divided into three aliquots (triplicate for HEK293 cells) or two aliquots (duplicate for HeLa cells). The whole cell lysate before NeutrAvidin adsorption and supernatant after the NeutrAvidin adsorption were denatured by incubation with Laemmli buffer at room temperature for 10 min. The eluted surface biotinylated fraction (300 and 175 μg per lane for HEK293 and HeLa, respectively) and denatured whole cell lysates before (from 50 and 20 μg whole cell lysates for HEK293 and HeLa cells, respectively) and after NeutrAvidin bead adsorption (from 50 and 20 μg whole cell lysates for HEK293 and HeLa cells, respectively) were then resolved on a 10% SDS-PAGE gel (BioRad Laboratories, Inc., Hercules, CA, USA) and immunoblotted with FLAG (1:4000 dilution) and GAPDH (1:5000 dilution) antibodies. The blots were probed with a GAPDH antibody to determine whether the biotinylated protein (surface fraction) had any intracellular protein contamination, similar to previously published approaches (40). After incubation with goat anti-mouse secondary antibody, immunoblot images were captured before saturation using a BioRad Molecular XRS Imager (BioRad, Hercules, CA). Densitometry was conducted using the Image Lab v4.1 software (Biorad, Hercules, CA). Densitometry of OATP1B1 from the surface fraction was divided by the sum of that from surface fraction and supernatant (after scale up to the same amount as surface fraction (i.e. from 50 to 300 μg for HEK293 cells and 20 to 175 μg for HeLa cells). Densitometry of both the 98kd main band and minor upper band in the surface biotinylation fraction of OATP1B1 were included in the analysis. Data are expressed as fold change vs. WT-OATP1B1 control. To verify that NeutrAvidin beads have similar pull down efficiency toward a surface biotinylated protein in HEK293-WT- and HEK293-V174A-OATP1B1 cells, in a separate experiment, following surface biotinylation and NeutrAvidin adsorption, immunoblot of sodium potassium (Na/K)-ATPase, an endogenous plasma membrane protein, was conducted with a mouse anti-alpha 1 Na/K-ATPase antibody (Abcam, Cambridge, MA) (1:10,000 dilution) in the surface and supernatant fractions.
Immunocytochemistry.
Immunocytochemistry (ICC) experiments were performed similar to those published previously (32), with slight modifications. Cells were grown to confluence on coverslips coated with poly-L-lysine in 24-well plates. After fixation with ice-cold 100% methanol for 20 min at room temperature (RT), cells were permeabilized with 0.25% Triton X-100 in PBS for 5 min at RT and blocked with 5% milk in PBS overnight at 4°C. Cells were co-stained with rabbit monoclonal anti-FLAG antibody (1:100 dilution, Sigma Aldrich, St. Louis, MO) and mouse anti-alpha 1 sodium potassium (Na/K)-ATPase antibody (1:100, Abcam, Cambridge, MA) for 2 h at 37°C. After washing three times with 1X PBS, cells were incubated with Alexa Fluor 488-conjugated goat-anti-mouse IgG (1:200 dilution, Life Technologies, Grand Island, NY) and Alexa Fluor 594-conjugated goat-anti-rabbit IgG (1:200 dilution, Life Technologies, Grand Island, NY) for 1 h at 37°C protected from light. After washing, nuclei were counterstained with 300 μM diamidino-2-phenylindole (DAPI) (Life Technologies, Grand Island, NY) for 5 min at RT, and mounted on glass slides using ProLong Gold Antifade (Life Technologies, Grand Island, NY). Images were captured using the Olympus FV10i (Olympus Scientific Solutions Americas Corp., Waltham, MA) at a resolution of 1024x1024 and objective of 120X (optical section of 1.02 μm). Co-localization was quantified using the Manders method (41) in the ImageJ Fiji software with Coloc2 plugin, by which the FLAG-OATP1B1 signal (red) coincident with a signal in the green channel of Na/K-ATPase over the total intensity of FLAG-OATP1B1 was measured, similarly as published previously (42, 43).
32P-orthophosphate labelling and immunoprecipitation.
The 32P-orthophosphate labelling method was used to determine the phosphorylation status of WT-OATP1B1 and V174A-OATP1B1, similar to that published previously, with slight modifications (44). Cells were plated in 100 mm2 dishes coated with poly-L-lysine at a seeding density of 2-2.5 x 106 cells per dish and allowed to grow for 48 h. Cells were first incubated in phosphate-free DMEM containing 10% FBS for 1 h, and then were labeled in fresh phosphate-free DMEM medium containing 32P-orthophosphate (0.1mCi/ml) and 10% FBS for 5 h at 37°C. After washing, cells were lysed with ice-cold lysis buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 100 mM NaF, 1% NP40 (v/v), 0.2 mM Na3VO4, Complete™ protease inhibitor cocktail, and Phosstop tablets. Whole cell lysates were subjected to immunoprecipitation with mouse FLAG antibody, and adsorbed by protein A/G beads. After washing, the immunocomplexes were eluted in 2X Laemmli buffer at 100°C for 5 min, resolved on 10% SDS-PAGE gel and transferred to nitrocellulose membrane. The 32P-signal on the blot was determined by autoradiography and imaged with Molecular Imager (BioRad, Hercules, CA). Subsequently, FLAG immunoblot (1:4000, 1 h) was conducted to determine the protein levels of FLAG-WT-OATP1B1 or FLAG-V174A-OATP1B1. Densitometry of both the 98 kd main band and minor upper band were determined using Image Lab v4.1 software (BioRad, Hercules, CA), similar to the surface biotinylation assay. The densitometry of 32P-labeled OATP1B1 was normalized by that of OATP1B1 protein levels from the same sample on the blot.
Data analysis.
Fold changes and associated standard errors (SE) of [3H]E217βG accumulation (Fig. II B and. IV B), total (Fig. II A and IV A) and surface levels (Fig. III A and V A) of OATP1B1, and phosphorylation (Fig. VI B) in V174A-OATP1B1-expressing cells vs. WT-OATP1B1-expressing control were estimated by linear mixed effects models with a fixed group effect and a random effect (experiment date), allowing for group-specific variances, similarly as that published previously (32, 45). A two-sided p-value of < 0.05 denoted statistical significance. SAS software (version 9.4, Cary, NC) was used for data analyses.
Fig. II. Establishment of the HEK293-FLAG-V174A-OATP1B1 stable cell line and comparison of transport function between V174A- and WT-OATP1B1 in HEK293 stable cell lines.
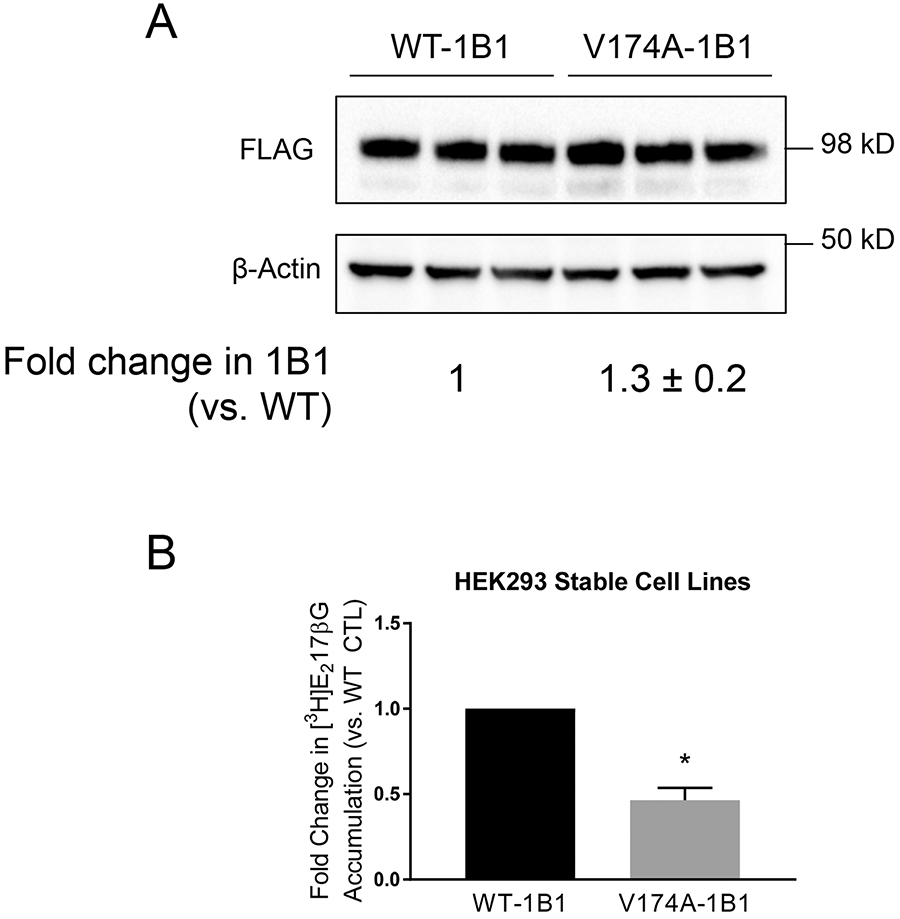
A. FLAG immunoblot in HEK293-FLAG-WT- and –V174A-OATP1B1 cells. Cells were seeded in 24-well plates at a seeding density of 1.5x105 cells/well and were grown for 48 h. Whole cell lysates were subjected to FLAG immunoblot with β-actin as the loading control. The Mixed-effect model-estimated fold change and associated SE of FLAG-V174A-OATP1B1 protein levels (vs. WT-OATP1B1 control) was expressed as mean ± SE (n=3 in triplicate). B. Mixed-effect model estimated fold change and associated SE of [3H]E217βG (1 μM, 2 min) of V174A-OATP1B1 vs. WT-OATP1B1 (*, p<0.05, n=3, in 5 replicate). In each individual experiment, [3H]E217βG accumulation mediated by WT- or V174A-OATP1B1 was normalized by relative OATP1B1 protein levels in whole cell lysates.
Fig. IV. Comparison of transport function between V174A- and WT-OATP1B1 in transiently transfected HeLa cells.
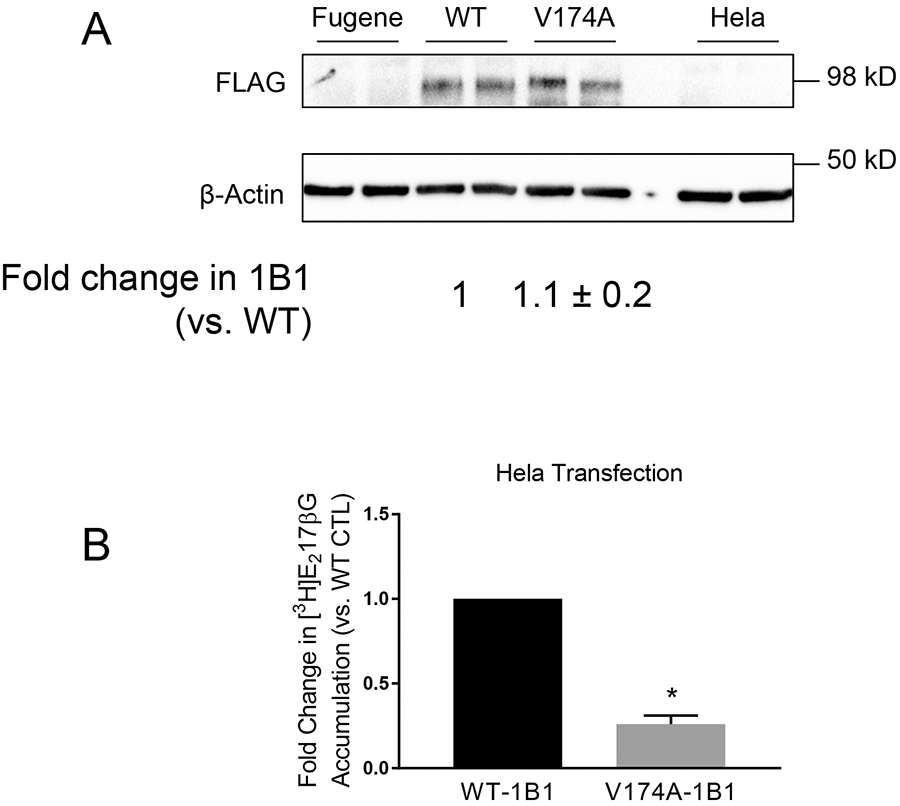
A. FLAG immunoblot in FLAG-WT- and FLAG-V174A-OATP1B1-expressing HeLa cells. HeLa cells were transfected with an expression vector encoding FLAG-WT-, FLAG-V174A-OATP1B1, or Fugene HD reagent alone. Forty-eight hours post-transfection, FLAG immunoblot was conducted with β-actin as the loading control. The mixed-effect model estimated fold change and associated SE in FLAG-OATP1B1 expression of V174A-OATP1B1 vs. WT-OATP1B1 control was expressed as mean ± SE (n=3 in duplicate). B. Mixed-effect model-estimated fold change and associated SE of [3H]E217βG (1 μM, 2 min) of V174A-OATP1B1 vs. WT-OATP1B1 (*, p<0.05, n=3, in triplicate). In each individual experiment, [3H]E217βG accumulation mediated by WT- or V174A-OATP1B1 was normalized by relative OATP1B1 protein levels in whole cell lysates.
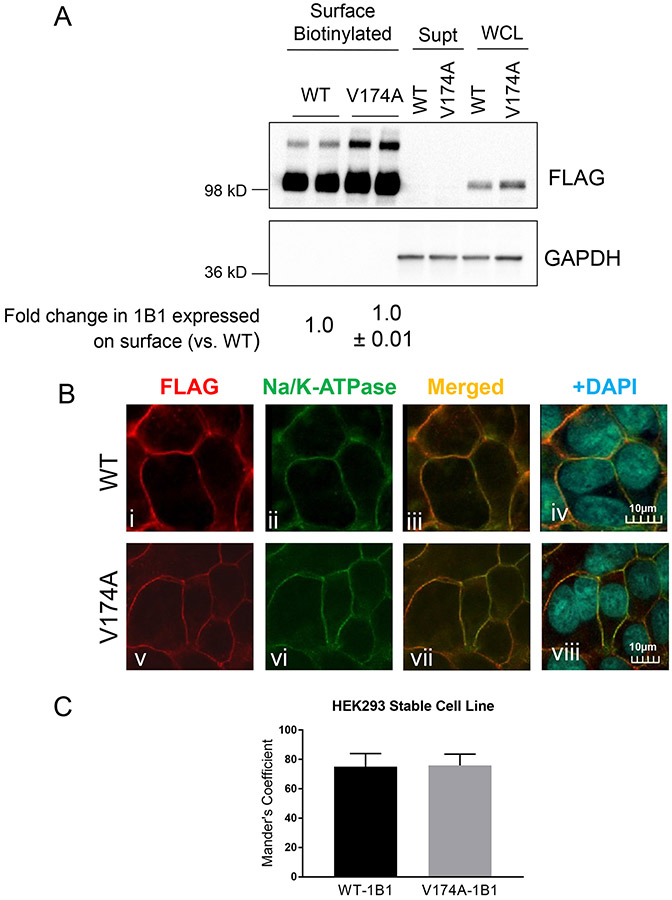
Fig. III. Determining plasma membrane localization of V174A- and WT-OATP1B1 in HEK293 stable cell lines.
A. Protein levels of FLAG-WT- and FLAG-V174A-OATP1B1 in whole cell lysates (WCL), biotinylated plasma membrane fraction, and supernatant after NeutrAvidin adsorption. GAPDH was used as a cytoplasmic protein marker. Densitometry of WT- and V174A-OATP1B1 was determined in each fraction. Densitometry of OATP1B1 in the surface fraction was divided by the sum of the densitometry in the surface fraction and supernatant after scaling up to the same amount of proteins as was used in the surface fraction. A mixed-effect model was used to compare the surface levels of OATP1B1 between V174A- and WT-OATP1B1 as described in the Materials and Methods. Model-estimated fold change and associated SE of the surface expression of V174A-OATP1B1 vs. –WT-OATP1B1 is shown (n=3 in duplicate or triplicate). B. Immunofluorescence staining of FLAG and Na/K-ATPase in HEK293 stable cell lines expressing FLAG-WT-OATP1B1 (WT) and FLAG-V174A-OATP1B1 (V174A). Co-immunofluorescence staining of FLAG-tagged WT- or V174A-OATP1B1 (red) and Na/K-ATPase (green) was performed in HEK293 stable cell lines as indicated. Nuclei were counterstained with DAPI (blue). Yellow shows co-localization of FLAG-tagged OATP1B1 and Na/K ATPase. Images were captured using an Olympus FV10i confocal microscope. Representative images are shown from three independent experiments. To quantify the percentage of FLAG-OATP1B1 co-localized with the Na/K-ATPase surface marker, the Mander’s Coefficient was calculated in (C) HEK293 stable cell lines as detailed in the Materials and Methods. Data represent mean ± SD (n=14 images in each group).
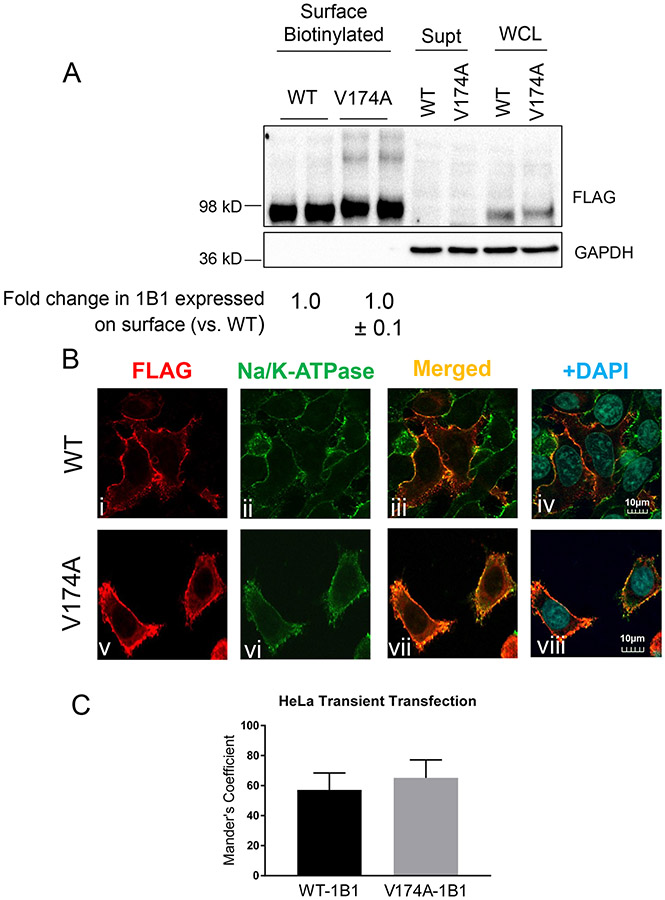
Fig. V. Determining plasma membrane localization of V174A- and WT-OATP1B1 in HeLa cell lines.
A. Protein levels of FLAG-WT- and FLAG-V174A-OATP1B1 in whole cell lysates (WCL), biotinylated plasma membrane fraction, and supernatant after NeutrAvidin adsorption. GAPDH was used as a cytoplasmic protein marker. Densitometry of WT- and V174A-OATP1B1 was determined in each fraction. Densitometry of OATP1B1 in the surface fraction was divided by the sum of the densitometry in the surface fraction and supernatant after scaling up to the same amount of proteins as was used in the surface fraction. A mixed-effect model was used to compare the surface levels of OATP1B1 between V174A- and WT-OATP1B1 as described in the Materials and Methods. Model-estimated fold change and associated SE of the surface expression of V174A-OATP1B1 vs. –WT-OATP1B1 is shown (n=3 in duplicate). B. Immunofluorescence staining of FLAG and Na/K-ATPase in HeLa cells transfected with FLAG-WT-OATP1B1 (WT) or FLAG-V174A-OATP1B1 (V174A). Co-immunofluorescence staining of FLAG-tagged WT- or V174A-OATP1B1 (red) and Na/K-ATPase (green) was performed in transfected HeLa cells as indicated. Nuclei were counterstained with DAPI (blue). Yellow shows co-localization of FLAG-tagged OATP1B1 and Na/K-ATPase. Images were taken using an Olympus FV10i confocal microscope at a resolution of 1024x1024 and 120X objective. Representative images from the same experiment are shown (n=3 independent experiments). To quantify the percentage of FLAG-OATP1B1 co-localized with the Na/K-ATPase surface marker, the Mander’s Coefficient was calculated in (C) transiently transfected HeLa cells as detailed in the Methods section. Data represent mean ± SD (n≥8 images in each group).
Fig. VI. Increased phosphorylation status of OATP1B1 with V174A variant in the HEK293 stable cell line.
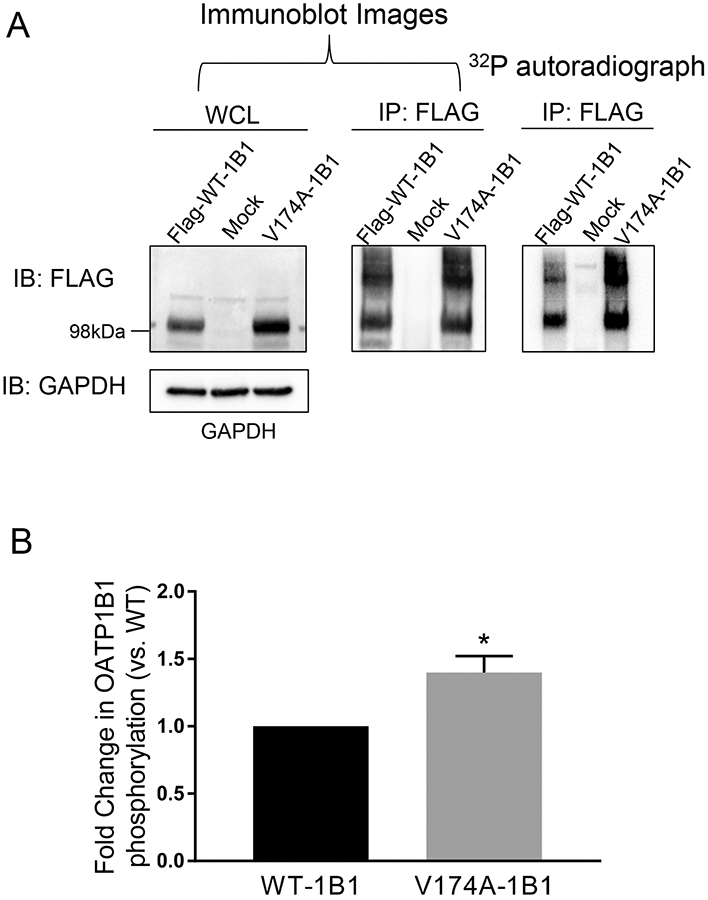
A. Phosphorylation of WT- and V174A-OATP1B1. HEK293-FLAG-WT- and –V174A-OATP1B1 cells were seeded at a density of 2-2.5x106 cells/100mm2 dish. Forty-eight hours after seeding, cells were metabolically labelled with 32P-orthophosphate for 5 h at 37°C. After labelling, cells were lysed and whole cell lysates (500 μg) were immunoprecipitated (IP) with FLAG antibody and subjected to autoradiography and subsequent immunoblot (IB) with FLAG and GAPDH antibodies. Representative images from four independent experiments are shown. B. Densitometry of 32P-labelled WT-OATP1B1 and V174A-OATP1B1 was normalized by its respective protein amount, as detected by FLAG immunoblot. A mixed-effect model was used to compare the phosphorylation of OATP1B1 between V174A- and WT-OATP1B1 as described in the Materials and Methods. Model-estimated fold change and associated SE of phosphorylation of V174A-OATP1B1 vs. WT-OATP1B1 is shown (* p<0.05, n=4).
RESULTS
Immunohistochemistry of OATP1B1 and OATP1B3 in human liver tissues.
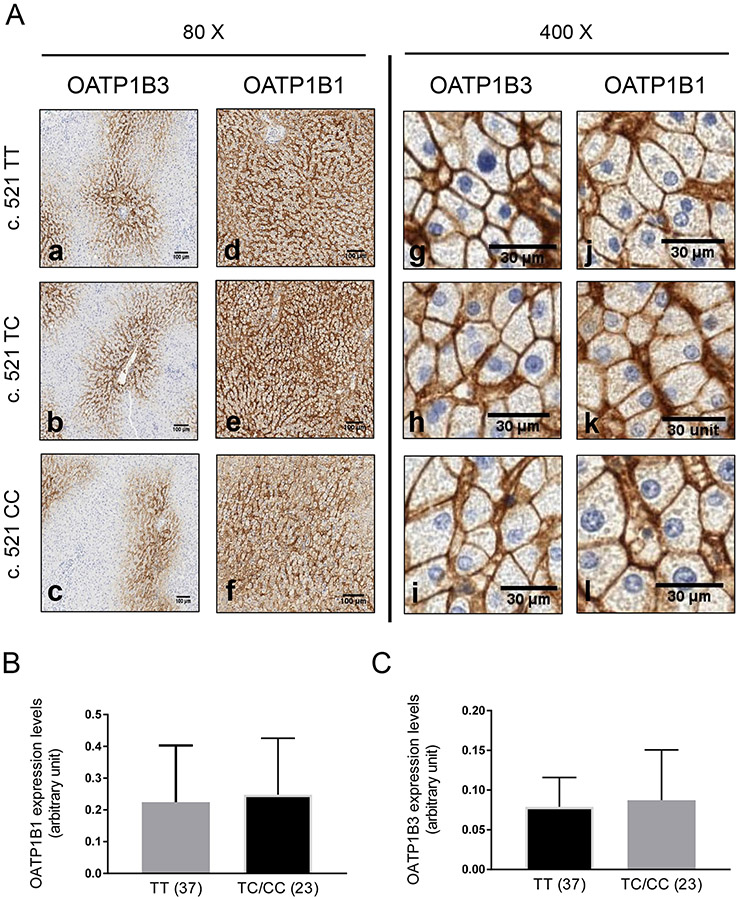
Among the 79 human liver tissues, 54 (68.4%), 23 (29.1%), and 2 (2.5%) tissues were genotyped as c.521 TT homozygous, c.521 TC heterozygous, and c.521 CC homozygous, respectively (Table I). Among the same 79 human liver tissue, 25 (31.6%), 35 (44.3%), and 19 (24.1%) tissues were genotyped as c. 388 AA homozygous, c.388 AG heterozygous, and c.388 GG homozygous (Table I). Human kidney tissue has been reported to lack OATP1B1 and OATP1B3 expression (10, 39). Thus, human kidney tissue was used as the negative control to determine the specificity of the OATP1B1 and OATP1B3 antibodies for IHC. IHC of OATP1B1 and OATP1B3 detected specific staining in human liver tissues (Fig. I A brown color), but not in the negative control (Supplemental Fig. S I). Similar to what was published previously (25), OATP1B3 was highly expressed around the central vein area (Fig. I a-c), while OATP1B1 had a more diffused expression pattern compared to OATP1B3 (Fig. I d-f). In the two OATP1B1 c.521 CC liver tissues (Fig. I A, Supplemental Fig. S X livers 41 and 58), both tissues are heterozygous for c.388 AG (Supplemental Table S I). OATP1B1 (Fig. I A j-l) and OATP1B3 (Fig. I A g-i) are primarily localized on the plasma membrane in both tissues. OATP1B1 c.388A>G and c.521T>C form four haplotypes, namely *1A (C.388A-C.521T), *1B (c.388G-c.521T), *5 (c.388A-c.521C), and *15 (c.388G-c.521C) (1). These haplotypes of OATP1B1 determined from the current studies are included in supplemental Table S1. Plasma membrane localization of OATP1B1 and OATP1B3 can be observed in all tissues with varied IHC staining intensity among tissues (Supplemental Fig. S X). A semi-quantitative analysis was conducted to compare OATP1B1 and OATP1B3 total expression levels between the c.521 TT and c.521 TC/CC genotypes (Fig. I B and C and Supplemental Fig. S II). There was no significant difference in either OATP1B1 or OATP1B3 expression levels between the c.521 TT and c.521 TC/CC genotypes. OATP1B1 expression levels in the c. 388 GG genotype were 1.5 ± 1.1 fold of those in the c. 388 AA genotype (Supplemental Fig. S II C). OATP1B1 expression levels in *1B/1B were 1.9 ±1.6 fold of those of *1A/1A) (Supplemental Fig. S II D). There is an increased trend of OATP1B1 protein levels in the c. 388GG variant vs. the c. 388AA variant (p=0.1) and *1B/1B vs. *1A/1A (p=0.08). However, this increase was not statistically significant.
Table I.
Demographic information of human liver tissues genotyped for c. 521T>C and c. 388 A>G
| c.521 T>C Genotype (n, %) |
c.388 A<G Genotype (n, %) |
||||||
|---|---|---|---|---|---|---|---|
| Total (n) | c.521 TT | c.521 TC | c.521 CC | c.388 AA | c.388 AG | c.388 GG | |
| Age | |||||||
| <55 | 42 | 28 (66.7%) | 14 (33.3%) | 0 (0.0%) | 14 (33.3%) | 18 (42.9%) | 10 (23.8%) |
| 55-63 | 25 | 20 (80.0%) | 3 (12.0%) | 2 (8.0%) | 8 (32.0% | 11 (44.0%) | 6 (24.0%) |
| ≥64 | 12 | 6 (50.0%) | 6 (50.0%) | 0 (0.0%) | 3 (25.0%) | 6 (50.0% | 3 (25.0%) |
| Sex | |||||||
| Female | 42 | 29 (69.0%) | 12 (28.6%) | 1 (2.4%) | 12 (28.6%) | 21 (50.0%) | 9 (21.4%) |
| Male | 37 | 25 (67.6%) | 11 (29.7%) | 1 (2.7%) | 13 (35.2%) | 14 (37.8%) | 10 (27.0%) |
| Total | |||||||
| n | 79 | 54 | 23 | 2 | 25 | 35 | 19 |
| % | 100% | 68.4% | 29.1% | 2.5% | 31.6% | 44.3% | 24.1% |
Percentages indicate the number of each genotype out of the total in each group.
Fig. I. OATP1B1 and OATP1B3 expression in OATP1B1 c. 521 T>C genotyped human liver tissues.
A. Paraffin-embedded liver sections (4 μm) stained (brown signal) with OATP1B3 (a-c and g-i) and OATP1B1 (d-f and j-l) antibody, detected by DAB-HRP complex. Representative images from n=79 genotyped human liver tissue are shown. Semi-quantification of OATP1B1 (B) and OATP1B3 (C) expression in non-HCV and non-cirrhotic human livers genotyped for c.521 TT (n=37) and TC/CC (n=23) as detailed in Materials and Methods. Statistical analysis was conducted by student t-test.
Establishment of the V174A-OATP1B1 stable cell line in HEK293 cells.
A stable cell line expressing FLAG-V174A-OATP1B1 was first developed in HEK293 cells. Protein levels of FLAG-V174A-OATP1B1 were determined by FLAG immunoblot and were compared with that of FLAG-WT-OATP1B1. The FLAG antibody specifically detected both the FLAG-tagged-V174A- and -WT-OATP1B1, but not the negative control (Supplemental Fig. S III A). [3H]E217βG accumulation of FLAG-V174A-OATP1B1 had significantly higher (~70 fold) accumulation when compared to HEK293-Mock cells (Supplemental Fig. S III B).
We next compared the [3H]E217βG transport mediated by V174A- and WT-OATP1B1 in the HEK293-FLAG-V174A- and -FLAG-WT-OATP1B1 stable cell lines, respectively. To take into consideration expression levels of OATP1B1 protein in the two stable cell lines (Fig. II A), [3H]E217βG accumulation (pmol/mg protein) was further normalized by the relative protein levels of V174A-OATP1B1 vs. WT-OATP1B1 as determined by immunoblot, using a strategy similar to that published previously (31). As shown in Fig. II B, [3H]E217βG accumulation (1 μM, 2 min) in the HEK293-FLAG-V174A-OATP1B1 cells was significantly reduced to 0.46 ± 0.07 (mean ± SE, n=3) fold of that in the HEK293-FLAG-WT-OATP1B1 cells, suggesting that the V174A-OATP1B1 has reduced transport activity compared with that of the WT-OATP1B1.
Determining the plasma membrane localization of V174A-OATP1B1 and WT-OATP1B1 in HEK293-stable cell lines by surface biotinylation and immunofluorescence staining.
The percentage of WT- and V174A-OATP1B1 proteins expressed on the plasma membrane vs. their respective total proteins was estimated by surface biotinylation assay, as detailed in the Materials and Methods. FLAG-V174A- and FLAG-WT-OATP1B1 protein levels were determined by FLAG immunoblot of the biotinylated surface fraction and whole cell lysates before and after Neutravidin adsorption (supernatant). There was no significant difference in the fraction of OATP1B1 expressed on the plasma membrane between V174A- and WT-OATP1B1-expressing HEK293 stable cell lines (p>0.05, n=3), with a 1.0 ± 0.01 fold change in V174A-OATP1B1 expression on the surface when compared to WT-OATP1B1 (Fig. III A). GAPDH, a cytosolic protein marker (46), was not detected in the surface biotinylation fraction, suggesting that there was no contamination of intracellular protein in the surface biotinylated fraction (Fig. III A, full blots provided in Supplemental Fig. S IV A). Both the minor upper band (> 98 kD) and the major band (~98 kD) in the surface fraction are FLAG-OATP1B1 as these bands were not detected in the in the surface fraction of a negative control HEK293-Mock cells (Fig. S IV B). As shown in Supplemental Fig. S V, surface levels of Na/K-ATPase in HEK293-V174A-OATP1B1 cells was similar to that in the HEK293-WT-OATP1B1 cells (~1.17-fold vs. WT control), suggesting that there was similar NeutrAvidin adsorption/elution efficiency toward an endogenous control plasma membrane protein Na/K-ATPase in HEK293-WT- and -V174A-OATP1B1 cell lines.
Cellular distribution of FLAG-V174A- and FLAG-WT-OATP1B1 in HEK293 stable cell lines was further determined by immunofluorescence staining, followed by confocal microscopy. The FLAG antibody specifically detected the WT- and V174A-OATP1B1 in HEK293 stable cell lines expressing their respective transport proteins (Fig. III B i and v), but not in the negative control HEK293-Mock cells (Supplemental Fig. S VI A). Both FLAG-WT-OATP1B1 (Fig. III B i, red) and FLAG-V174A-OATP1B1 (Fig. III B v, red) proteins were primarily expressed on the plasma membrane in transporter-expressing HEK293 stable cell lines, as determined by FLAG immunofluorescence staining. Na/K-ATPase has been reported to be abundantly expressed on the plasma membrane (47), and was used as a plasma membrane protein control for immunofluorescence staining in the current studies. The results indicate that the Na/K-ATPase was primarily localized on the plasma membrane (Fig. III B ii and vi, green), and were co-localized with the FLAG-WT- (Fig. III B iii and iv, yellow) and FLAG-V174A-OATP1B1 (Fig. IV B vii and viii, yellow) in HEK293-FLAG-WT- and HEK293-FLAG-V174A-OATP1B1 stable cell lines, respectively. The percentage of WT- and V174A-FLAG-OATP1B1 (red) that are co-localized with Na/K-ATPase (green) out of their respective total FLAG-OATP1B1 staining were 75.0 ± 8.9 and 75.9 ± 7.7% (mean ± SD from 3 separate experiments; n=3-4 fields each experiment) in WT- and V174A-expressing HEK293 stable cell lines, respectively (Fig. III C); there is no significant difference in percentage of OATP1B1 co-localized with Na/K-ATPase between WT- and V174A-OATP1B1 (p>0.05 by student t-test).
Determining transport function and plasma membrane localization of WT- and V174A-OATP1B1 transiently transfected in HeLa cells.
The initial report by Tirona et al. regarding the transport function and surface levels of V174A-OATP1B1 determined by surface biotinylation was conducted in transiently transfected HeLa cells after vaccinia viral infection (11). The current studies used a commercially available, non-viral reagent, FuGENE HD, for transient transfection. In transiently transfected HeLa cells, FLAG immunoblot specifically detected FLAG-WT- and -V174A-OATP1B1 expression in HeLa cells transfected with plasmid vectors encoding these two proteins, respectively, but not in the reagent alone or non-transfected HeLa cells (Fig. IV A). Protein levels of FLAG-V174A-OATP1B1 were 1.1 ± 0.2 fold (mean ± SE) of that of the FLAG-WT-OATP1B1 (Fig. IV A). To compare the transport function of FLAG-V174A-OATP1B1 with the FLAG-WT-OATP1B1, a similar strategy was used as in the HEK293 stable cell lines described above, i.e., normalizing the [3H]E217βG accumulation (pmol/mg protein) by the protein levels of FLAG-WT- and -V174A-OATP1B1, in order to take into account differences in protein levels. As shown in Fig. IV B, [3H]E217βG accumulation (1 μM, 2 min) in FLAG-V174A-OATP1B1-expressing HeLa cells was significantly reduced to 0.26 ± 0.05 (mean ± SE, p<0.05) of that in the FLAG-WT-OATP1B1-expressing HeLa cells, suggesting that the V174A-OATP1B1 has reduced transport activity compared with the WT-OATP1B1.
As shown in Fig. V A, there was no significant difference in the levels of OATP1B1 expressed on the plasma membrane between V174A- and WT-OATP1B1-expressing HeLa cells (p>0.05, n=3), with a 1.0 ± 0.1 (mean ± SE, n=3) fold change in V174A-OATP1B1 surface levels when compared to WT-OATP1B1. No GAPDH, a cytosolic protein, was detected in the surface biotinylated fraction.
The FLAG antibody specifically detected the WT- and V174A-OATP1B1 transiently expressed in HeLa cells (Fig. V B i and v), but not in the cells transfected with vehicle control pCMV-6 (Supplemental Fig. S VI B). FLAG-WT- (Fig. V B i, red) and FLAG-V174A-OATP1B1 (Fig. V B v, red) were primarily localized on the plasma membrane in HeLa cells. Similar to what was observed in the HEK293 stable cell lines (Fig. III B ii and vi), Na/K-ATPase was primarily localized on the plasma membrane, as indicated by fluorescence staining (Fig. V B ii and vi, green) in HeLa cells. Co-localization of Na/K-ATPase with the FLAG-WT- (Fig. V B iii and iv, yellow) and FLAG-V174A-OATP1B1 (Fig. V B vii and viii, yellow) on the plasma membrane were observed in FLAG-WT- and FLAG-V174A-OATP1B1-expressing HeLa cells. The percentage of WT- and V174A-FLAG-OATP1B1 (red) that are co-localized with Na/K-ATPase (green) out of respective total FLAG-OATP1B1 staining were 57.1±11.2 and 65.1±12% (mean ± SD, from 3 separate experiments; 2-3 fields each experiment) in WT- and V174A-expressing HeLa cells, respectively (Fig. V C); there is no significant difference in percentage of OATP1B1 co-localized with Na/K-ATPase between WT- and V174A-OATP1B1 (p>0.05 by student t-test).
Comparison of phosphorylation status of WT- and V174A-OATP1B1 in HEK293-stable cell lines.
The assay to determine phosphorylation of FLAG-OATP1B1 through a 32P-orthophosphate labelling approach was first established in a pilot study using HEK293-Mock cells as the negative control. As shown in Supplemental Fig. S VIII, after 32P-orthophosphate labeling for 5 h, phosphorylated OATP1B1 detected by autoradiography and the immunoprecipitated (IP) total OATP1B1 proteins detected by FLAG immunoblotting were superimposable. The 32P-labelling followed by immunoprecipitation with FLAG specifically detected FLAG-WT-OATP1B1 in the HEK293-FLAG-WT-OATP1B1 cell line, but not in the HEK293-Mock cells (Supplemental Fig. S VIII). The phosphorylation status of WT- and V174A-OATP1B1 was subsequently compared in HEK293-WT-OATP1B1 and –V174A-OATP1B1 stable cell lines under the same experimental condition as in Supplemental Fig. S VIII. As shown in Figs. VI A and B, phosphorylation of V174A-OATP1B1 was significantly increased to 1.4 ± 0.1 fold of the WT-OATP1B1 (Fig. VI B, mean ± SE, p < 0.05, a full representative blot is shown in Supplemental Fig. S IV C).
DISCUSSION
Plasma membrane localization is critical for membrane transport proteins to exert their transport function. As human liver tissue is the physiologically relevant model with which to study hepatic OATP1B1, the current study first examined the plasma membrane localization of V174A-OATP1B1 in genotyped human liver tissue. IHC of OATP1B1 has been reported previously by several laboratories (10, 25), however, these studies were conducted in non-genotyped liver tissues. The current study is the first to characterize the plasma membrane localization of OATP1B1 using the c.521 T>C and c. 388A>G OATP1B1 genotyped liver tissues and provide direct evidence showing that the V174A-OATP1B1 has predominant plasma membrane localization on the basolateral membrane of hepatocytes, similar as the WT-OATP1B1 (Fig. I A j-I). A quantitative proteomics approach has been used previously to determine the membrane levels of OATP1B1 in c.521 T>C genotyped human liver tissues (29). No significant difference in the absolute amount of total membrane-bound OATP1B1 proteins, which contains both OATP1B1 bound to the plasma membrane and to the intracellular membrane vesicles, was found between the two genotypes, c.521 TT and c.521 CC/TC (29). Similar to this previous report (29), we observed no significant difference in total expression levels of OATP1B1 or OATP1B3 between c. 521 TT and TC/CC genotypes using semi-quantitative analysis of the IHC staining results (Fig. I B and C and Supplemental Fig. S II A and B). Noticing that due to the relative low allelic frequency of the c. 521 CC, which is found at an allelic frequency of 14%, 11-16%, and 2% in European-American, Japanese, and African-American populations, respectively (11, 12), only one and two c. 521 CC homozygous liver tissue samples were identified in the previous (29) and current studies, respectively. Quantitative comparison of expression of OATP1B1 between the WT (c.521 TT) and V174A-homozygous (c. 521 CC) genotypes in human liver tissue warrants further studies by increasing the sample size using a quantitative proteomics approach (29) or by immunoblot in genotyped liver tissues (25).
The c. 388 A>G (N130D) polymorphism of OATP1B1 has a relatively high frequency of 30% in the European-American population (11). However, in vitro N130D-OATP1B1 either does not affect OATP1B1 transport function (11-14, 25-28), or has moderate effects (less than 50%) with increased (14) or decreased (48) transport function. The c. 521 T>C OATP1B1 variant has demonstrated impaired transport function both in vitro (11-14) and in vivo (15-17); therefore, it is the main focus of current study. The current in vitro studies characterizing the phosphorylation status and membrane localization focused on V174A-OATP1B1 and were not extended to N130D-OATP1B1. In vivo, decreased exposure of pravastatin (6) and repaglinide (49) was observed in subjects bearing the OATP1B1 *1B/1B compared with the *1A/1A haplotype, suggesting that N130D-OATP1B1 may have increased transport function in vivo. Absolute protein quantification showed that OATP1B1 protein levels were significantly higher in the c.388 GG genotype vs. the AA genotype (29), which may explain the apparent increased transport function of N130D-OATP1B1 in vivo. We also observed an increasing trend of OATP1B1 protein levels in the c. 388 GG vs. AA genotypes and the *1B/1B vs. *1A/1A haplotypes, although these trends were not statistically significant (29). Nevertheless, the c. 388 A>G variation does not seem to affect membrane localization of OATP1B,1 as distinct membrane localization of OATP1B1 can be observed in both the c. 388 AA and GG variants (Supplemental Table 1 and figure S X). Similar to the OATP1B1 *5/*5 (c. 521CC), OATP1B1*15/*15 has reduced transport function in vitro (50) and is associated with increased plasma concentration of statins, such as pravastatin (51), pitavastatin (51), and rosuvastatin (52, 53), in vivo. In vitro, OATP1B1 *15 is primarily expressed on the plasma membrane (50). No homozygous * 15/* 15 was identified in the current study. There is no apparent difference in plasma membrane localization of OATP1B1 in donors bearing *1A/*1A vs. *1A/*15 or *1B/*1B vs. * 1B/* 15 (Supplemental Table 1 and figure S X), most likely because the *15 does not affect in vivo OATP1B1 plasma membrane localization, similar to in vitro reports (50).
Approaches including surface biotinylation, absolute quantification of OATP1B1 in the plasma membrane fraction, and fluorescence staining followed by confocal microscopy have been used by several laboratories and in different cell lines to study the plasma membrane localization of V174A-OATP1B1 (11-14). Notably, among these approaches used, the confocal microscopy results in these studies were all reported at a qualitative level, while the absolute quantification and surface biotinylation assays provided a quantitative and semi-quantitative measurement of membrane expression, respectively. In HEK293 cells, using both surface biotinylation and confocal microscopy methods, Iwai reported that V174A-OATP1B1 had similar expression levels on the plasma membrane to those of the WT-OATP1B1 (12). Using confocal microscopy, Nozawa et al., observed a similar predominant plasma membrane localization of V174A-OATP1B1 (13). Our results in HEK293 and HeLa cells using surface biotinylation and confocal microscopy (Fig. III and V) indicate that WT- and V174A-OATP1B1 have similar plasma membrane localization pattern, consistent with reports by Iwai and Nozawa (12, 13). We also normalized the WT- and V174A-OATP1B1-mediated [3H]E217βG transport by relative surface levels of OATP1B1. [3H]E217βG transport mediated by V174A-OATP1B1 was 0.55 ± 0.05 and 0.38 ± 0.03 fold of WT-OATP1B1 after normalization by surface levels of OATP1B1 in transporter-expressing HEK293 and HeLa cells, respectively (p<0.05). The results were similar to that normalized by total protein levels (Fig. III and V).
However, reports by Tirona (11) and Kameyama (14) apparently contradict the above mentioned studies. In HeLa cells, Tirona (2001) reported that V174A-OATP1B1 has markedly reduced plasma membrane levels compared with the WT-OATP1B1, as determined by surface biotinylation assay (11). However, the intracellular fraction of OATP1B1 was not shown in the study for either wild-type or V174A. This study was conducted in HeLa cells 16.5 h post-transduction with a vaccinia virus at a multiplicity of infection (MOI) of 10 (11). The current study employed a widely used, commercially available and non-viral reagent, FuGENE HD, for transfection. We found that V174A-OATP1B1 primarily localized on the plasma membrane when transiently expressed in HeLa cells (Fig. V), similar to what we observed in the V174A-OATP1B1-expressing HEK293 stable cell line (Fig. III). The vaccinia virus has been reported to cause lytic infection (54) and to induce apoptosis or necrosis in multiple cell lines, such as Chinese hamster ovary (CHO) (55) and HeLa G cells (54). In the current study, a LDH toxicity assay in transiently transfected HeLa cells showed negligible cytotoxicity using FuGENE (Supplemental Fig. S IX). As the vaccinia virus is not readily available, we were not able to repeat Tirona’s previous study condition to determine if the contradicting results between the current and previous reports are due to different gene expression systems. Nevertheless, our in vitro finding of predominant plasma membrane localization of V174A-OATP1B1 in both HeLa and HEK293 cells is consistent with our findings in genotyped human liver tissues.
Kameyama et al. determined the plasma membrane localization of OATP1B1 using only an immunofluorescence staining/confocal microscopy approach, and concluded that V174A-OATP1B1 has less membrane localization than does the WT-OATP1B1, based on the qualitative description that the V174A-OATP1B1 was observed both on the plasma membrane and in the cytosol, while the WT-OATP1B1 was observed mainly on the membrane (14). Membrane proteins constantly undergo internalization and recycling (56) and it is anticipated that OATP1B1 would be observed both on the plasma membrane and in the cytosol. We have observed cytosolic staining of WT-OATP1B1 in our previous work (32), and as well as in the current study (Supplemental Fig. S VII A and B). In fact, both cytosolic and membrane staining of WT-OATP1B1 was observed in a later publication from the same laboratory (57). Therefore, in our opinion, the qualitative immunocytochemistry staining results alone are not sufficient to draw the conclusion that V174A-OATP1B1 has reduced plasma membrane expression compared to the WT-OATP1B1. In summary, when determining the plasma membrane expression levels of transport proteins, it is crucial to include a quantitative or semi-quantitative method (e.g. biotinylation) to evaluate its membrane expression, in addition to visual verification by confocal microscopy.
OATP1B1’s transport function has previously been reported to be downregulated by protein kinase C (PKC) activation, which has been attributed to an increase in the internalization rate and reduction of the recycling of OATP1B1 back to the surface (58). PKC activation also rapidly down-regulates OATP1B3-mediated transport (33). Such reduction in OATP1B3 transport function is associated with increased phosphorylation but not altered plasma membrane levels during the treatment time determined (33). Members of the OATP family have been reported to have putative phosphorylation sites using computer-based prediction (33, 59). In the current study, we showed for the first time that OATP1B1 is a phosphorylated protein in in vitro culture conditions (Fig. VI and Supplemental Fig. S VIII) and that V174A-OATP1B1 has a modest but significant increase in phosphorylation status compared with that of the WT-OATP1B1 (Fig. VI). Altered phosphorylation has been reported to be associated with increased or decreased activity of membrane proteins, including membrane transport proteins. For example, increased phosphorylation has been reported to be associated with increased activity of the epithelial sodium (Na) channel (60). Increased phosphorylation of OATP2B1 (59), the brain glutamate/aspartate transporter GLAST-1 (61), OATP1B3 (33) and dopamine (62) and serotonin transporters (63) following PKC activation is associated with decreased transporter function. Our study has a similar trend as the latter where increased phosphorylation of V174A-OATP1B1 is associated with decreased transport function.
In previous in vitro studies describing reduced transport activity of OATP1B1 c. 521 T>C (11, 14), V174A-OATP1B1 has been reported to have unchanged Km while has significantly decreased Vmax values (11, 14), suggesting that the V174A variation is likely to affect the capacity of OATP1B1-mediated transport but not the substrate affinity. However, it is important to notice that the reduced Vmax of a transporter is not always associated with reduced plasma membrane levels. For example, decreased Vmax and unaltered surface levels has been reported for OATP1B3 following bortezomib pretreatment (40) and rabbit intestinal Na+-glucose transporter (SGLT1) following PKC activation (64). V174A-OATP1B1 may have decreased transport capacity due to decreased translocation of substrate, similarly as speculated for SGLT1 (64). Altered phosphorylation of polymorphism variants of proteins have also been reported previously (23). Polymorphism variations occurring at amino acids that can be phosphorylated (i.e. Ser, Thr or Tyr) can lead to the addition or abolishment of phosphorylation at these sites. For example, the K897T SNP in the human ether-a-go-go-related gene 1 (hERG1) gene of the Kv11.1 channel protein creates a new phosphorylation site for AKT kinase, leading to an increase in phosphorylation of the protein (65). A polymorphic variation can also alter phosphorylation at a site near or adjacent to the SNP (23). In the case of the most common polymorphism in the human insulin receptor substrate-1 (IRS-1), variation at the amino acid that is not subjected to phosphorylation in the G972R polymorphism of IRS-1 is associated with decreased phosphorylation of two phosphorylation sites (Y941 and Y989) adjacent to the polymorphism (66). The G972R-IRS-1 variant has altered association with the responsible insulin receptor kinase, which may contribute to the altered phosphorylation of IRS-1 (66). As neither V or A in the V174A variation is a site of phosphorylation, the increased phosphorylation in the V174A-OATP1B1 protein is likely to occur at some phosphorylation sites adjacent to this variation. Scansite 2.0 (67) predicts that there are multiple phosphorylation sites adjacent to V174A, such as S168 and Y169, which are putative phosphorylation sites by protein kinase C and insulin receptor tyrosine kinase. Currently, the membrane topology of OATP1B1 from crystallography has not been developed. The V174, S168, and Y169 of OATP1B1 are predicted to be located in the 4th transmembrane (TM) domain (68) by TMPred (69). It is paradoxical, at first glance, that membranous amino acids are proposed as phosphorylation sites. However, the 4th transmembrane domain is predicted with a relative low probability or score, which measures the prediction confidence level, compared with other TM domains as predicted by Phobius (1) and TMPred (69), respectively (Supplemental Table S II). Results from quantitative phosphoproteomics identifying the kinase responsible for such altered phosphorylation of OATP1B1, as well as information about the membrane topology of OATP1B1 from crystallography data, would provide insight into how the V174A polymorphism enhances phosphorylation of OATP1B1.
Liver disease state has been reported to be associated with altered localization of transport proteins and phosphorylation status of phosphoproteins. For example, Hardwick et al. reported previously that in end-stage nonalcoholic steatohepatitis (NASH) (not fatty), ABCC2 appears to internalize into intracellular vesicles away from the biliary membrane (71). Urasaki et al reported that the amount of phosphor-Akt isoforms were significantly higher in NASH versus control livers (72). Potential effects of liver disease state on OATP1B1 membrane localization and phosphorylation are worth being characterized in further studies.
One of the limitations of the current study is that the causal relationship of altered phosphorylation and reduced transport of V174A-OATP1B1 has not yet been established. One cannot exclude the possibility that the increased phosphorylation status of V174A-OATP1B1 may be an epiphenomenon. Characterization of the phosphorylation site(s) of OATP1B1, identification of the phosphorylation sites of which are increased in the V174A-OATP1B1 (by using an approach such as quantitative phosphoproteomics (73, 74),) and testing the function of these sites by site-directed mutagenesis will ultimately provide the direct evidence of the role of altered phosphorylation on the reduced transport function of the V174A-OATP1B1. Such studies are ongoing in our laboratory and are beyond the scope of the current report.
CONCLUSION
In summary, the current study for the first time reported the predominant plasma membrane localization of V174A-OATP1B1 in human liver tissues. The lack of association of reduced transport of V174A-OATP1B1 with reduced plasma membrane levels in vitro suggests the existence of other, yet unidentified mechanism(s). The V174A-OATP1B1 has increased phosphorylation compared to the WT-OATP1B1 in vitro, while the potential role of phosphorylation in OATP1B1 transport function needs to be further evaluated in future studies.
Supplementary Material
ACKNOWLEDGEMENT
We would like to thank Dr. Dietrich Keppler for providing the HEK293-Mock stable cell line and Dr. Allison Gillaspy for designing the PCR primers.
This research was supported by NIH R01 GM094268 [W. Y]. The Olympus FV10i confocal microscope is supported by equipment grants from the NIH to Dr. Wei Yue (R01GM094268-06S1) and from the Presbyterian Health Foundation to Dr. Kelly Standifer. Research reported in this publication was supported in part by the National Cancer Institute Cancer Center Support Grant P30CA225520 awarded to the University of Oklahoma Stephenson Cancer Center and the Molecular Biology Shared Resource. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Alexandra Crowe is an American Foundation of Pharmaceutical Education pre-doctoral Fellow.
ABBREVIATIONS
- OATP
Organic anion transporting polypeptide
- SLCO
solute carrier organic anion
- HEK 293
human embryonic kidney
- DPBS
Dulbecco’s phosphate-buffered saline
- BSA
bovine serum albumin
- DMEM
Dulbecco’s modified Eagle medium
- DDI
drug-drug interactions
- HBSS
Hanks balanced salt solution
- E217βG
estradiol 17β-glucuronide
References
- 1.Niemi M, Pasanen MK, Neuvonen PJ. Organic anion transporting polypeptide 1B1: a genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol Rev. 2011;63(1):157–181. [DOI] [PubMed] [Google Scholar]
- 2.Hirano M, Maeda K, Shitara Y, Sugiyama Y. Contribution of OATP2 (OATP1B1) and OATP8 (OATP1B3) to the hepatic uptake of pitavastatin in humans. J Pharmacol Exp Ther. 2004;311(1):139–146. [DOI] [PubMed] [Google Scholar]
- 3.Hsiang B, Zhu Y, Wang Z, Wu Y, Sasseville V, Yang WP, Kirchgessner TG. A novel human hepatic organic anion transporting polypeptide (OATP2). Journal of Biological Chemistry. 1999;274(52):37161–37168. [DOI] [PubMed] [Google Scholar]
- 4.Simonson SG, Raza A, Martin PD, Mitchell PD, Jarcho JA, Brown CD, Windass AS, Schneck DW. Rosuvastatin pharmacokinetics in heart transplant recipients administered an antirejection regimen including cyclosporine. Clin Pharmacol Ther. 2004;76(2):167–177. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi H, Takeuchi T, Okada M, Kobayashi M, Unno M, Abe T, Goto J, Hishinuma T, Shimada M, Mano N. Screening of antibiotics that interact with organic anion-transporting polypeptides 1B1 and 1B3 using fluorescent probes. Biol Pharm Bull. 2011;34(3):389–395. [DOI] [PubMed] [Google Scholar]
- 6.Maeda K, Ieiri I, Yasuda K, Fujino A, Fujiwara H, Otsubo K, Hirano M, Watanabe T, Kitamura Y, Kusuhara H, Sugiyama Y. Effects of organic anion transporting polypeptide 1B1 haplotype on pharmacokinetics of pravastatin, valsartan, and temocapril. Clin Pharmacol Ther. 2006;79(5):427–439. [DOI] [PubMed] [Google Scholar]
- 7.Treiber A, Schneiter R, Hausler S, Stieger B. Bosentan is a substrate of human OATP1B1 and OATP1B3: inhibition of hepatic uptake as the common mechanism of its interactions with cyclosporin A, rifampicin, and sildenafil. Drug metabolism and disposition: the biological fate of chemicals. 2007;35(8):1400–1407. [DOI] [PubMed] [Google Scholar]
- 8.Nakagomi-Hagihara R, Nakai D, Kawai K, Yoshigae Y, Tokui T, Abe T, Ikeda T. OATP1B1, OATP1B3, and mrp2 are involved in hepatobiliary transport of olmesartan, a novel angiotensin II blocker. Drug metabolism and disposition: the biological fate of chemicals. 2006;34(5):862–869. [DOI] [PubMed] [Google Scholar]
- 9.Feng B, Xu JJ, Bi YA, Mireles R, Davidson R, Duignan DB, Campbell S, Kostrubsky VE, Dunn MC, Smith AR, Wang HF. Role of hepatic transporters in the disposition and hepatotoxicity of a HER2 tyrosine kinase inhibitor CP-724,714. Toxicological sciences : an official journal of the Society of Toxicology. 2009;108(2):492–500. [DOI] [PubMed] [Google Scholar]
- 10.Konig J, Cui Y, Nies AT, Keppler D. A novel human organic anion transporting polypeptide localized to the basolateral hepatocyte membrane. Am J Physiol Gastrointest Liver Physiol. 2000;278(1):G156–164. [DOI] [PubMed] [Google Scholar]
- 11.Tirona RG, Leake BF, Merino G, Kim RB. Polymorphisms in OATP-C. Identification of multiple allelic variants associated with altered transport activity among European and African-Americans. J Biol Chem. 2001;276(38):35669–35675. [DOI] [PubMed] [Google Scholar]
- 12.Iwai M, Suzuki H, Ieiri I, Otsubo K, Sugiyama Y. Functional analysis of single nucleotide polymorphisms of hepatic organic anion transporter OATP1B1 (OATP-C). Pharmacogenetics. 2004;14(11):749–757. [DOI] [PubMed] [Google Scholar]
- 13.Nozawa T, Nakajima M, Tamai I, Noda K, Nezu J, Sai Y, Tsuji A, Yokoi T. Genetic polymorphisms of human organic anion transporters OATP-C (SLC21A6) and OATP-B (SLC21A9): allele frequencies in the Japanese population and functional analysis. J Pharmacol Exp Ther. 2002;302(2):804–813. [DOI] [PubMed] [Google Scholar]
- 14.Kameyama Y, Yamashita K, Kobayashi K, Hosokawa M, Chiba K. Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics. 2005;15(7):513–522. [DOI] [PubMed] [Google Scholar]
- 15.Pasanen MK, Fredrikson H, Neuvonen PJ, Niemi M. Different effects of SLCO1B1 polymorphism on the pharmacokinetics of atorvastatin and rosuvastatin. Clin Pharmacol Ther. 2007;82(6):726–733. [DOI] [PubMed] [Google Scholar]
- 16.Chung JY, Cho JY, Yu KS, Kim JR, Oh DS, Jung HR, Lim KS, Moon KH, Shin SG, Jang IJ. Effect of OATP1B1 (SLCO1B1) variant alleles on the pharmacokinetics of pitavastatin in healthy volunteers. Clin Pharmacol Ther. 2005;78(4):342–350. [DOI] [PubMed] [Google Scholar]
- 17.Niemi M, Schaeffeler E, Lang T, Fromm MF, Neuvonen M, Kyrklund C, Backman JT, Kerb R, Schwab M, Neuvonen PJ, Eichelbaum M, Kivisto KT. High plasma pravastatin concentrations are associated with single nucleotide polymorphisms and haplotypes of organic anion transporting polypeptide-C (OATP-C, SLCO1B1). Pharmacogenetics. 2004;14(7):429–440. [DOI] [PubMed] [Google Scholar]
- 18.Niemi M, Backman JT, Kajosaari LI, Leathart JB, Neuvonen M, Daly AK, Eichelbaum M, Kivisto KT, Neuvonen PJ. Polymorphic organic anion transporting polypeptide 1B1 is a major determinant of repaglinide pharmacokinetics. Clin Pharmacol Ther. 2005;77(6):468–478. [DOI] [PubMed] [Google Scholar]
- 19.Group SC, Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359(8):789–799. [DOI] [PubMed] [Google Scholar]
- 20.Yee SW, Brackman DJ, Ennis EA, Sugiyama Y, Kamdem LK, Blanchard R, Galetin A, Zhang L, Giacomini KM. Influence of Transporter Polymorphisms on Drug Disposition and Response: A Perspective From the International Transporter Consortium. Clin Pharmacol Ther. 2018;104(5):803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furukawa T, Wakabayashi K, Tamura A, Nakagawa H, Morishima Y, Osawa Y, Ishikawa T. Major SNP (Q141K) variant of human ABC transporter ABCG2 undergoes lysosomal and proteasomal degradations. Pharm Res. 2009;26(2):469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn MK, Mazei-Robison MS, Blakely RD. Single nucleotide polymorphisms in the human norepinephrine transporter gene affect expression, trafficking, antidepressant interaction, and protein kinase C regulation. Mol Pharmacol. 2005;68(2):457–466. [DOI] [PubMed] [Google Scholar]
- 23.Ren J, Jiang C, Gao X, Liu Z, Yuan Z, Jin C, Wen L, Zhang Z, Xue Y, Yao X. PhosSNP for systematic analysis of genetic polymorphisms that influence protein phosphorylation. Mol Cell Proteomics. 2010;9(4):623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bian Y, Song C, Cheng K, Dong M, Wang F, Huang J, Sun D, Wang L, Ye M, Zou H. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J Proteomics. 2014;96:253–262. [DOI] [PubMed] [Google Scholar]
- 25.Ho RH, Tirona RG, Leake BF, Glaeser H, Lee W, Lemke CJ, Wang Y, Kim RB. Drug and bile acid transporters in rosuvastatin hepatic uptake: function, expression, and pharmacogenetics. Gastroenterology. 2006;130(6):1793–1806. [DOI] [PubMed] [Google Scholar]
- 26.Nozawa T, Minami H, Sugiura S, Tsuji A, Tamai I. Role of organic anion transporter OATP1B1 (OATP-C) in hepatic uptake of irinotecan and its active metabolite, 7-ethyl-10-hydroxycamptothecin: in vitro evidence and effect of single nucleotide polymorphisms. Drug Metab Dispos. 2005;33(3):434–439. [DOI] [PubMed] [Google Scholar]
- 27.Katz DA, Carr R, Grimm DR, Xiong H, Holley-Shanks R, Mueller T, Leake B, Wang Q, Han L, Wang PG, Edeki T, Sahelijo L, Doan T, Allen A, Spear BB, Kim RB. Organic anion transporting polypeptide 1B1 activity classified by SLCO1B1 genotype influences atrasentan pharmacokinetics. Clin Pharmacol Ther. 2006;79(3):186–196. [DOI] [PubMed] [Google Scholar]
- 28.Oswald S, Konig J, Lutjohann D, Giessmann T, Kroemer HK, Rimmbach C, Rosskopf D, Fromm MF, Siegmund W. Disposition of ezetimibe is influenced by polymorphisms of the hepatic uptake carrier OATP1B1. Pharmacogenet Genomics. 2008;18(7):559–568. [DOI] [PubMed] [Google Scholar]
- 29.Prasad B, Evers R, Gupta A, Hop CE, Salphati L, Shukla S, Ambudkar SV, Unadkat JD. Interindividual variability in hepatic organic anion-transporting polypeptides and P-glycoprotein (ABCB1) protein expression: quantification by liquid chromatography tandem mass spectroscopy and influence of genotype, age, and sex. Drug Metab Dispos. 2014;42(1):78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mondalek FG, Fung KM, Yang Q, Wu W, Lu W, Palmer BW, Frimberger DC, Greenwood-Van Meerveld B, Hurst RE, Kropp BP, Lin HK. Temporal expression of hyaluronic acid and hyaluronic acid receptors in a porcine small intestinal submucosa-augmented rat bladder regeneration model. World J Urol. 2015;33(8):1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thakkar N, Kim K, Jang ER, Han S, Kim K, Kim D, Merchant N, Lockhart AC, Lee W. A cancer-specific variant of the SLCO1B3 gene encodes a novel human organic anion transporting polypeptide 1B3 (OATP1B3) localized mainly in the cytoplasm of colon and pancreatic cancer cells. Mol Pharm. 2013;10(1):406–416. [DOI] [PubMed] [Google Scholar]
- 32.Alam K, Pahwa S, Wang X, Zhang P, Ding K, Abuznait AH, Li L, Yue W. Downregulation of Organic Anion Transporting Polypeptide (OATP) 1B1 Transport Function by Lysosomotropic Drug Chloroquine: Implication in OATP-Mediated Drug-Drug Interactions. Molecular pharmaceutics. 2016;13(3):839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powell J, Farasyn T, Kock K, Meng X, Pahwa S, Brouwer KL, Yue W. Novel mechanism of impaired function of organic anion-transporting polypeptide 1B3 in human hepatocytes: post-translational regulation of OATP1B3 by protein kinase C activation. Drug metabolism and disposition: the biological fate of chemicals. 2014;42(11):1964–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Billington S, Ray AS, Salphati L, Xiao G, Chu X, Humphreys WG, Liao M, Lee CA, Mathias A, Hop C, Rowbottom C, Evers R, Lai Y, Kelly EJ, Prasad B, Unadkat JD. Transporter Expression in Noncancerous and Cancerous Liver Tissue from Donors with Hepatocellular Carcinoma and Chronic Hepatitis C Infection Quantified by LC-MS/MS Proteomics. Drug Metab Dispos. 2018;46(2):189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogasawara K, Terada T, Katsura T, Hatano E, Ikai I, Yamaoka Y, Inui K. Hepatitis C virus-related cirrhosis is a major determinant of the expression levels of hepatic drug transporters. Drug Metab Pharmacokinet. 2010;25(2):190–199. [DOI] [PubMed] [Google Scholar]
- 36.Sysel AM, Valli VE, Nagle RB, Bauer JA. Immunohistochemical quantification of the vitamin B12 transport protein (TCII), cell surface receptor (TCII-R) and Ki-67 in human tumor xenografts. Anticancer Res. 2013;33(10):4203–4212. [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen K, Krusenstjerna-Hafstrom R, Lohse J, Petersen KH, Derand H. A novel quantitative immunohistochemistry method for precise protein measurements directly in formalin-fixed, paraffin-embedded specimens: analytical performance measuring HER2. Mod Pathol. 2017;30(2):180–193. [DOI] [PubMed] [Google Scholar]
- 38.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23(4):291–299. [PubMed] [Google Scholar]
- 39.Konig J, Cui Y, Nies AT, Keppler D. Localization and genomic organization of a new hepatocellular organic anion transporting polypeptide. J Biol Chem. 2000;275(30):23161–23168. [DOI] [PubMed] [Google Scholar]
- 40.Alam K, Farasyn T, Crowe A, Ding K, Yue W. Treatment with proteasome inhibitor bortezomib decreases organic anion transporting polypeptide (OATP) 1B3-mediated transport in a substrate-dependent manner. PloS one. 2017;12(11):e0186924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manders EMM, Verbeek FJ, Aten JA. Measurement of co - localization of objects in dual - colour confocal images. Journal of Microscopy. 1993;169(3):375–382. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Spagnol G, Naslavsky N, Caplan S, Sorgen PL. TC-PTP directly interacts with connexin43 to regulate gap junction intercellular communication. J Cell Sci. 2014;127(Pt 15):3269–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224(Pt 3):213–232. [DOI] [PubMed] [Google Scholar]
- 44.Sefton BM. Labeling cultured cells with 32Pi and preparing cell lysates for immunoprecipitation. Curr Protoc Cell Biol. 2001;Chapter 14:Unit 14 14. [DOI] [PubMed] [Google Scholar]
- 45.Pahwa S, Alam K, Crowe A, Farasyn T, Neuhoff S, Hatley O, Ding K, Yue W. Pretreatment With Rifampicin and Tyrosine Kinase Inhibitor Dasatinib Potentiates the Inhibitory Effects Toward OATP1B1- and OATP1B3-Mediated Transport. J Pharm Sci. 2017;106(8):2123–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soundararajan R, Melters D, Shih IC, Wang J, Pearce D. Epithelial sodium channel regulated by differential composition of a signaling complex. Proc Natl Acad Sci U S A. 2009;106(19):7804–7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajasekaran SA, Huynh TP, Wolle DG, Espineda CE, Inge LJ, Skay A, Lassman C, Nicholas SB, Harper JF, Reeves AE, Ahmed MM, Leatherman JM, Mullin JM, Rajasekaran AK. Na,K-ATPase subunits as markers for epithelial-mesenchymal transition in cancer and fibrosis. Mol Cancer Ther. 2010;9(6):1515–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tirona RG, Leake BF, Wolkoff AW, Kim RB. Human organic anion transporting polypeptide-C (SLC21A6) is a major determinant of rifampin-mediated pregnane X receptor activation. J Pharmacol Exp Ther. 2003;304(1):223–228. [DOI] [PubMed] [Google Scholar]
- 49.Kalliokoski A, Backman JT, Neuvonen PJ, Niemi M. Effects of the SLCO1B1*1B haplotype on the pharmacokinetics and pharmacodynamics of repaglinide and nateglinide. Pharmacogenet Genomics. 2008;18(11):937–942. [DOI] [PubMed] [Google Scholar]
- 50.van de Steeg E, Greupink R, Schreurs M, Nooijen IH, Verhoeckx KC, Hanemaaijer R, Ripken D, Monshouwer M, Vlaming ML, DeGroot J, Verwei M, Russel FG, Huisman MT, Wortelboer HM. Drug-drug interactions between rosuvastatin and oral antidiabetic drugs occurring at the level of OATP1B1. Drug Metab Dispos. 2013;41(3):592–601. [DOI] [PubMed] [Google Scholar]
- 51.Deng JW, Song IS, Shin HJ, Yeo CW, Cho DY, Shon JH, Shin JG. The effect of SLCO1B1*15 on the disposition of pravastatin and pitavastatin is substrate dependent: the contribution of transporting activity changes by SLCO1B1*15. Pharmacogenet Genomics. 2008;18(5):424–433. [DOI] [PubMed] [Google Scholar]
- 52.Choi JH, Lee MG, Cho JY, Lee JE, Kim KH, Park K. Influence of OATP1B1 genotype on the pharmacokinetics of rosuvastatin in Koreans. Clin Pharmacol Ther. 2008;83(2):251–257. [DOI] [PubMed] [Google Scholar]
- 53.Lee E, Ryan S, Birmingham B, Zalikowski J, March R, Ambrose H, Moore R, Lee C, Chen Y, Schneck D. Rosuvastatin pharmacokinetics and pharmacogenetics in white and Asian subjects residing in the same environment. Clin Pharmacol Ther. 2005;78(4):330–341. [DOI] [PubMed] [Google Scholar]
- 54.Kalbacova M, Spisakova M, Liskova J, Melkova Z. Lytic infection with vaccinia virus activates caspases in a Bcl-2-inhibitable manner. Virus Res. 2008;135(1):53–63. [DOI] [PubMed] [Google Scholar]
- 55.Ramsey-Ewing A, Moss B. Apoptosis induced by a postbinding step of vaccinia virus entry into Chinese hamster ovary cells. Virology. 1998;242(1):138–149. [DOI] [PubMed] [Google Scholar]
- 56.Hao M, Maxfield FR. Characterization of rapid membrane internalization and recycling. J Biol Chem. 2000;275(20):15279–15286. [DOI] [PubMed] [Google Scholar]
- 57.Furihata T, Matsumoto S, Fu Z, Tsubota A, Sun Y, Matsumoto S, Kobayashi K, Chiba K. Different interaction profiles of direct-acting anti-hepatitis C virus agents with human organic anion transporting polypeptides. Antimicrob Agents Chemother. 2014;58(8):4555–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hong M, Hong W, Ni C, Huang J, Zhou C. Protein kinase C affects the internalization and recycling of organic anion transporting polypeptide 1B1. Biochimica et biophysica acta. 2015;1848(10 Pt A):2022–2030. [DOI] [PubMed] [Google Scholar]
- 59.Kock K, Koenen A, Giese B, Fraunholz M, May K, Siegmund W, Hammer E, Volker U, Jedlitschky G, Kroemer HK, Grube M. Rapid modulation of the organic anion transporting polypeptide 2B1 (OATP2B1, SLCO2B1) function by protein kinase C-mediated internalization. The Journal of biological chemistry. 2010;285(15):11336–11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oh Y, Smith PR, Bradford AL, Keeton D, Benos DJ. Regulation by phosphorylation of purified epithelial Na+ channels in planar lipid bilayers. Am J Physiol. 1993;265(1 Pt 1):C85–91. [DOI] [PubMed] [Google Scholar]
- 61.Conradt M, Stoffel W. Inhibition of the high-affinity brain glutamate transporter GLAST-1 via direct phosphorylation. J Neurochem. 1997;68(3):1244–1251. [DOI] [PubMed] [Google Scholar]
- 62.Huff RA, Vaughan RA, Kuhar MJ. Phorbol esters increase dopamine transporter phosphorylation and decrease transport Vmax. J Neurochem. 1997;68(1):225–232. [DOI] [PubMed] [Google Scholar]
- 63.Jayanthi LD, Samuvel DJ, Blakely RD, Ramamoorthy S. Evidence for biphasic effects of protein kinase C on serotonin transporter function, endocytosis, and phosphorylation. Mol Pharmacol. 2005;67(6):2077–2087. [DOI] [PubMed] [Google Scholar]
- 64.Vayro S, Silverman M. PKC regulates turnover rate of rabbit intestinal Na+-glucose transporter expressed in COS-7 cells. Am J Physiol. 1999;276(5):C1053–1060. [DOI] [PubMed] [Google Scholar]
- 65.Gentile S, Martin N, Scappini E, Williams J, Erxleben C, Armstrong DL. The human ERG1 channel polymorphism, K897T, creates a phosphorylation site that inhibits channel activity. Proc Natl Acad Sci U S A. 2008;105(38):14704–14708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McGettrick AJ, Feener EP, Kahn CR. Human insulin receptor substrate-1 (IRS-1) polymorphism G972R causes IRS-1 to associate with the insulin receptor and inhibit receptor autophosphorylation. The Journal of biological chemistry. 2005;280(8):6441–6446. [DOI] [PubMed] [Google Scholar]
- 67.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31(13):3635–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niemi M Role of OATP transporters in the disposition of drugs. Pharmacogenomics. 2007;8(7):787–802. [DOI] [PubMed] [Google Scholar]
- 69.Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- 70.Claros MG, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10(6):685–686. [DOI] [PubMed] [Google Scholar]
- 71.Hardwick RN, Fisher CD, Canet MJ, Scheffer GL, Cherrington NJ. Variations in ATP-binding cassette transporter regulation during the progression of human nonalcoholic fatty liver disease. Drug metabolism and disposition: the biological fate of chemicals. 2011;39(12):2395–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Urasaki Y, Zhang C, Cheng JX, Le TT. Quantitative Assessment of Liver Steatosis and Affected Pathways with Molecular Imaging and Proteomic Profiling. Sci Rep. 2018;8(1):3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Juvvadi PR, Cole DC, Falloon K, Waitt G, Soderblom EJ, Moseley MA, Steinbach WJ. Kin1 kinase localizes at the hyphal septum and is dephosphorylated by calcineurin but is dispensable for septation and virulence in the human pathogen Aspergillus fumigatus. Biochem Biophys Res Commun. 2018;505(3):740–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zennadi R, Whalen EJ, Soderblom EJ, Alexander SC, Thompson JW, Dubois LG, Moseley MA, Telen MJ. Erythrocyte plasma membrane-bound ERK1/2 activation promotes ICAM-4-mediated sickle red cell adhesion to endothelium. Blood. 2012;119(5):1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.