Abstract
Background
Sulfadoxine-pyrimethamine (SP) is recommended for intermittent preventive treatment in Africa against Plasmodium falciparum infection. However, increasing SP resistance (SPR) of P. falciparum affects the therapeutic efficacy of SP, and pfdhfr (encoding dihydrofolate reductase) and pfdhps (encoding dihydropteroate synthase) genes are widely used as molecular markers for SPR surveillance. In the present study, we analyzed single nucleotide polymorphisms (SNPs) of pfdhfr and pfdhps in P. falciparum isolated from infected Chinese migrant workers returning from Africa.
Methods
In total, 159 blood samples from P. falciparum-infected workers who had returned from Africa to Anhui, Shangdong, and Guangxi provinces were successfully detected and analyzed from 2017 to 2019. The SNPs in pfdhfr and pfdhps were analyzed using nested PCR. The genotypes and linkage disequilibrium (LD) were analyzed using Haploview.
Results
High frequencies of the Asn51Ile (N51I), Cys59Arg(C59R), and Ser108Asn(S108N) mutant alleles were observed, with mutation frequencies of 97.60, 87.43, and 97.01% in pfdhfr, respectively. A triple mutation (IRN) in pfdhfr was the most prevalent haplotype (86.83%). Six point mutations were detected in pfdhps DNA fragment, Ile431Val (I431V), Ser436Ala (S436A), Ala437Gly (A437G), Lys540Glu(K540E), Ala581Gly(A581G), Ala613Ser(A613S). The pfdhps K540E (27.67%) was the most predominant allele, followed by S436A (27.04%), and a single mutant haplotype (SGKAA; 62.66%) was predominant in pfdhps. In total, 5 haplotypes of the pfdhfr gene and 13 haplotypes of the pfdhps gene were identified. A total of 130 isolates with 12 unique haplotypes were found in the pfdhfr-pfdhps combined haplotypes, most of them (n = 85, 65.38%) carried quadruple allele combinations (CIRNI-SGKAA).
Conclusion
A high prevalence of point mutations in the pfdhfr and pfdhps genes of P. falciparum isolates was detected among Chinese migrant workers returning from Africa. Therefore, continuous in vitro molecular monitoring of Sulfadoxine-Pyrimethemine combined in vivo therapeutic monitoring of artemisinin combination therapy (ACT) efficacy and additional control efforts among migrant workers are urgently needed.
Keywords: Plasmodium falciparum, sulfadoxine-pyrimethamine, dihydropteroate synthase, dihydrofolate reductase, migrants from Africa
Introduction
Imported Plasmodium falciparum infections are of great concern in China during malaria elimination and post-elimination stages, because China achieved zero indigenous malaria cases in 2017 (Feng et al., 2018). Appropriate treatment for P. falciparum according to the national plan is important; however, some studies have demonstrated mutations in molecular markers related to artemisinin combination therapy (ACT) resistance among isolates from Africa, suggesting that careful surveillance of African parasite populations is still warranted (Feng et al., 2015a; Feng et al., 2019, Yan et al., 2020).
Sulfadoxine-pyrimethamine (SP) is a second-line antimalarial for uncomplicated P. falciparum malaria treatment, as recommended by the World Health Organization (WHO), which is used as an intermittent preventive treatment in pregnancy (IPTp) and as an intermittent preventive treatment in infants (IPTi) in malaria-endemic regions (Gosling et al., 2010; Konate et al., 2011). Resistance to SP is caused mainly by point mutations in pfdhfr (encoding dihydrofolate reductase) and pfdhps (encoding dihydropteroate synthase). Mutations in pfdhfr and pfdhps have been associated with decreased parasite sensitivity to the SP, because the products of these genes could incrementally increase the parasite’s tolerance to the drugs in vitro (Chulay et al. 1984). Studies have identified point mutations in codons N51I, C59R, and S108N, I164L of pfdhfr located on chromosome 4 and codons I431V, S436A, A437G, K540E, A581G, and A613S of pfdhps located on chromosome 8, all of which were associated with P. falciparum SP treatment failure (Triglia et al., 1997; Berglez et al., 2004; Pearce et al., 2009). Monitoring drug resistance and the pattern of mutations is essential for early detection and subsequent prevention of the spread of drug resistance. The present study identified the polymorphisms in pfdhfr and pfdhps in P. falciparum among returned migrant workers from Africa in 2017–2019 reported in eastern China. The results provide a deeper understanding of the disease as well as baseline information on antimalarial drug resistance among imported P. falciparum in China.
Materials and Methods
Sample Sites
Sample Collection and DNA Extraction
The study was conducted in Anhui, Shandong, and Guangxi Provinces in Eastern and Southern China, where imported P. falciparum cases were predominantly. Anhui Province covers 105 counties with 70.6 million people and experienced a malaria resurgence in 2005–2008 that was mainly caused by the accumulation of residual foci of P. vivax (Feng et al., 2015b). Shandong Province, located in eastern China, has a long coastline measuring 3,024.4 kilometers. It contains 137 counties and has a population of 97.9 million. Economic trade overseas is frequent. Another province, Guangxi, is known for its gold miners who returned from Ghana in 2013, which was mainly reported in Shanglin County (Feng et al., 2015a). The number of imported P. falciparum cases, especially those from Africa, has increased significantly in these three provinces, and 360 P. falciparum cases were reported in 2019, accounting for 18.5% of all P. falciparum cases nationwide (Zhang L et al., 2020).
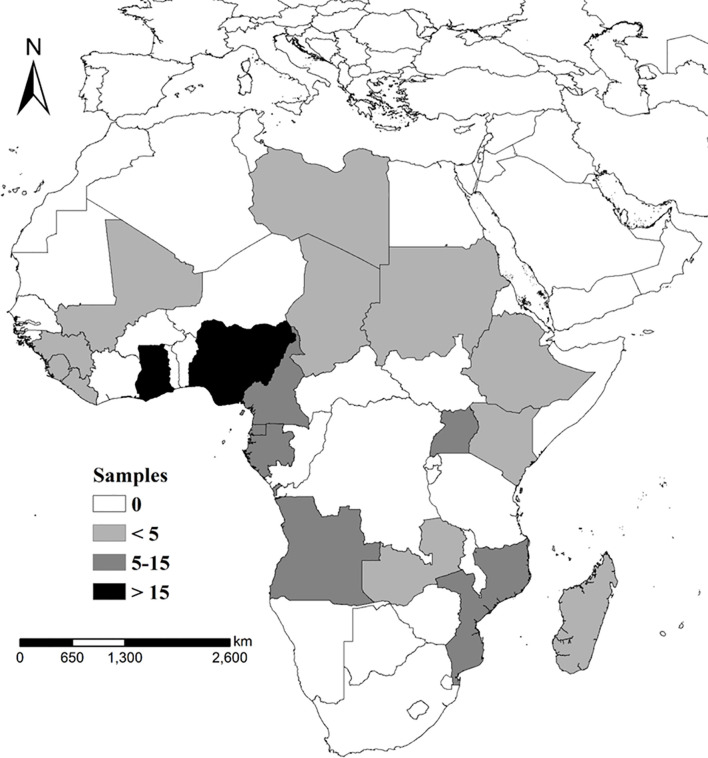
A total of 206 P. falciparum-infected blood samples were collected from the travelers returning from Africa from 2017 to 2019. The samples distribution were shown as Figure 1. Approximately 100 μl of finger-prick blood was spotted onto a piece of 3MM Whatman filter paper (GE Healthcare, Boston, MA, USA) and air dried.
Figure 1.
Sample collection and distribution, samples collection sites and sample screening and follow-up analysis of subject patterns.
The Plasmodium falciparum genomic DNA from the approximately 100 μl of collected blood sample was extracted using a QIAamp DNA blood kit (QIAGEN, Valencia, CA, USA) as described previously (Yan et al., 2020). Each of the samples was labeled with a study number and stored at −4°C until extraction. Individual epidemiological information was also collected using a web-based reporting system (China Information System for Diseases Control and Prevention) and analyzed.
Detection of pfdhfr and pfdhps Polymorphisms
Point mutations at codons 16, 51, 59, 108, and 164 of the pfdhfr gene and codons 431, 436, 437, 540, 581, and 613 of the pfdhps gene were evaluated using nested PCR amplification. The sequences of the primers used for pfdhfr and pfdhps genotyping were as described previously (Zhang et al., 2014). The primary amplification was performed using the following parameters: 1 cycle of 95°C for 3 min; 35 cycles of 95°C for 30 s, 55°C for 30 s, and 65°C for 6 for 30 s, and 1 cycle of 65°C for 60 s; and 65°C for 5 min. The second amplifcation was performed using the following parameters: 1 cycle of 95°C for 3 min; 35 cycles of 95°C for 30 s, 52°C for 30 s, and 65°C for 60 s; and 1 cycle of 65°C for 5 min. 750-bp product of pfdhps were sent for Sanger sequencing (Shanghai Bunan Biological Co., Ltd., Shanghai, China).
Data Analysis
Sequences were analyzed using the Blast program (http://blast.ncbi.nlm.nih.gov/). Multiple nucleotide sequence alignments and analysis were carried out using the MAFFT web-based tool with the Cluster Omega Sequence Alignment Editor (https://www.ebi.ac.uk/Tools/msa/clustalo/). Sequences with poor quality after three sequencing attempts or those with more than one peak at one locus were not included in the analysis. The map showing the imported of countries with number of the isolates was created by ArcGIS 10.1 (Environmental Systems Research Institute, Inc., Redlands, CA, USA). SPSS18.0 (IBM Corp., Armonk, NY, USA) was used to conduct the statistical analyses, and the Chi-squared test was employed to test the different constituent ratios of pfdhfr and pfdhps gene polymorphisms. The Fisher’s precision probability test was used as the sample size is less 50; For the sample size less than five would be discarded when analyzing the geographical distribution difference. The inter and intragenic SNP Linkage disequilibrium (LD) associations of pfdhfr and pfdhps were analyzed using Haploview 15 (Patel et al., 2017).
Ethical Considerations
This study was reviewed and approved by the ethical committee of the National Institute of Parasitic Diseases, Chinese Centre for Disease Control and Prevention (NIPD, China CDC, No. 2019008).
Results
Epidemiological Information
Among the 206 P. falciparum isolates collected in this study, 202 and 177 isolates were successfully amplified and sequenced for the pfdhfr and pfdhps genes, respectively; however, only single infections and both of pfdhfr and pfdhps successful amplicons were involved in the final analysis. Therefore, a total of 159 P. falciparum isolates were successfully detected and analyzed in this study (Figure 1). Their distribution was identified as 2 from North Africa, 23 from East Africa, 54 from West Africa, and 80 from Central Africa. Among them, the Democratic Republic of the Congo (n = 28), Nigeria (n = 27), and Angola (n = 16) were considered as the top three imported source countries. The average patient age was 42 years, and 154 patients (154/159, 96.86%) were male. The numbers of cases reported in 2017, 2018, and 2019 were 28, 58, and 73, respectively.
Prevalence of pfdhfr Polymorphisms
For pfdhfr, no polymorphism was found for codons 50 or 164. Compared with mutations N51I and C59R, S108N had a higher SNP prevalence, whereas, the difference between N51I and C59R was not significant. Only three isolates were sequenced as wild-type Asparagine-Cysteine-Serine (NCS) (accounting for 1.89%), whereas, the triple-mutant genotype, Isoleucine-Arginine-Asparagine (IRN), comprised 80.50% (n = 128), the others were NCN (n = 3, 1.89%), ICN (n = 10, 6.29%), and NRN (n = 15, 9.43%), respectively (Table 1).
Table 1.
Prevalence of pfdhfr and pfdhps polymorphisms in imported African isolates.
| Gene | Position | Wild type | Mutant type | Mutant isolates(n) | Frequency(%) |
|---|---|---|---|---|---|
| pfdhfr | 51 | AAT | ATT | 138 | 86.79 |
| 59 | TGT | CGT | 143 | 89.94 | |
| 108 | AGC | AAC | 156 | 98.11 | |
| pfdhps | 431 | ATA | GTA | 12 | 7.55 |
| 436 | TCT | GCT | 43 | 27.04 | |
| 437 | GCT | GGT | 15 | 9.43 | |
| 540 | AAA | GAA | 44 | 27.67 | |
| 581 | GCG | GGG | 14 | 8.81 | |
| 613 | GCC | TCC | 3 | 1.89 |
The bold values stand for mutant nucleic acid base.
Prevalence of pfdhps Polymorphisms
The SNPs of pfdhps were relatively scattered, S436A (27.07%) and K540E (27.67%) carried a higher allele frequency, which were statistically significant than others (the average mutant frequency was 6.92%). The frequencies of the other three alleles, I431V, A437G, and A581G, were lower than those of S436A or K540E, but higher than that of S613A, which was carried by only two isolates. In all, among 159 P. falciparum isolates, 13 kinds of mutants of pfdhps were detected. Further sequencing showed that 73 single mutant isolates, including ISAEAA (the bold residue represents the mutated site; n = 41, 25.79%), IAAKAA (n = 24, 15.09%), ISGKAA (n = 6, 3.77%), and ISAKGA (n = 2, 1.26%); 13 double mutant isolates, including IAGKAA (n = 7, 4.40%), VAAKAA (n = 2, 1.26%), ISAEGA (n = 2, 1.26%), IAAKAS (n = 1, 0.63%), and IAAEAA (n = 1, 0.63%); 9 triple mutant genotypes, comprising 8 examples of VAAKGA, 1 of VSGKGA; and only 1 quadruple mutation, as VAAKGS, were identified. There were more wild-type isolates of pfdhps than pfdhfr (63 pfdhps isolates, compared with three in pfdhfr). Three genotypes, ISAKAA (39.62%, 63/159), ISAEAA (25.79%, 41/159), and IAAKAA (15.09%, 24/159), accounted for 80.50% of all pfdhps genotypes (Table 2).
Table 2.
Prevalence of pfdhfr and pfdhps haplotypes.
| Gene | Genotype | Mutations | Sample size | Frequency (%) |
|---|---|---|---|---|
| pfdhfr | NCS | 0 | 3 | 1.89% |
| NCN | 1 | 3 | 1.89% | |
| ICN | 2 | 10 | 6.29% | |
| NRN | 2 | 15 | 9.43% | |
| IRN | 3 | 128 | 80.50% | |
| pfdhps | ISAKAA | 0 | 63 | 39.62% |
| ISAEAA | 1 | 41 | 25.79% | |
| IAAKAA | 1 | 24 | 15.09% | |
| ISGKAA | 1 | 6 | 3.77% | |
| ISAKGA | 1 | 2 | 1.26% | |
| VAAKAA | 2 | 2 | 1.26% | |
| IAGKAA | 2 | 7 | 4.40% | |
| ISAEGA | 2 | 2 | 1.26% | |
| IAAKAS | 2 | 1 | 0.63% | |
| IAAEAA | 2 | 1 | 0.63% | |
| VAAKGA | 3 | 8 | 5.03% | |
| VSGKGA | 3 | 1 | 0.63% | |
| VAAKGS | 4 | 1 | 0.63% |
The bold values stand for mutant nucleic acid base.
Geographical Genetic Analysis
The mutant frequency in all targeted pfdhfr gene fragments were all above 86.79%, and there was no significant difference (P > 0.05) among isolates from West, East, and Central Africa. For the pfdhps gene, the mutant genotypes carried a relatively high number of polymorphisms. The two P. falciparum samples from North Africa were wild-types (Table 3). Two of six loci were detected as site mutations in samples from East Africa, with K540E representing 69.57% (16/23) of the mutations, which was a markedly higher frequency than that for S436A (4.34%, 1/23). Each locus carried a sense mutation associated with West Africa, the mutant frequencies were 14.81% (8/54, I413V), 51.85% (28/54, S436V), 9.26% (5/54, A437G), 11.11% (6/54, K540E), 12.96% (7/54, A581G), and 5.56% (3/54, A613S). The site mutations in central African region were all different for the S436V (17.5%, 14/80, P < 0.001) and K540E (27.5%, 22/80, P = 0.017) loci, which was significantly different compared with the Western African Region isolates (P < 0.05). Moreover, the mutant frequency of K540E varied between Western and Eastern African Region isolates (P <0.01). A613S only occurred in Ghana (n = 2) and Nigeria (n = 1) in West Africa.
Table 3.
Geographical genetic analysis of pfdhfr/pfdhps genotypes.
| Region | No. samples | Country | No. samples | No. of mutations | pfdhfr/pfdhps genotypes | No. of genotypes |
|---|---|---|---|---|---|---|
| North Africa | 2 | Libya | 1 | 3 | IRN/ISAKAA | 1 |
| Sudan | 1 | 3 | IRN/ISAKAA | 1 | ||
| East Africa | 23 | Kenya | 1 | 3 | IRN/ISAKAA | 1 |
| Madagascar | 2 | 2 | NRN/ISAKAA | 2 | ||
| Ethiopia | 3 | 3 | IRN/ISAKAA | 1 | ||
| 4 | IRN/IAAKAA | 1 | ||||
| 4 | IRN/ISAEAA | 1 | ||||
| Zambia | 4 | 3 | NRN/ISAEAA | 1 | ||
| 4 | IRN/ISAEAA | 3 | ||||
| Uganda | 6 | 4 | IRN/ISAEAA | 6 | ||
| Mozambique | 7 | 2 | ICN/ISAKAA | 1 | ||
| 4 | IRN/ISAEAA | 6 | ||||
| West Africa | 54 | Mali | 1 | 3 | IRN/ISAKAA | 1 |
| Sierra Leone | 2 | 2 | NCN/IAAKAA | 1 | ||
| 3 | IRN/ISAKAA | 1 | ||||
| Guinea | 2 | 3 | IRN/ISAKAA | 1 | ||
| 4 | IRN/ISGKAA | 1 | ||||
| Liberia | 3 | 1 | NCN/ISAKAA | 1 | ||
| 4 | IRN/IAAKAA | 2 | ||||
| Cote d'lvoire | 8 | 0 | NCS/ISAKAA | 1 | ||
| 2 | NRN/ISAKAA | 1 | ||||
| 3 | IRN/ISAKAA | 2 | ||||
| 4 | IRN/IAAKAA | 3 | ||||
| 4 | IRN/ISAEAA | 1 | ||||
| Ghana | 16 | 1 | NCS/IAAKAA | 1 | ||
| 2 | NRN/ISAKAA | 1 | ||||
| 3 | IRN/ISAKAA | 3 | ||||
| 3 | ICN/ISAEAA | 1 | ||||
| 3 | NRN/IAAKAA | 1 | ||||
| 4 | IRN/IAAKAA | 3 | ||||
| 4 | IRN/ISAEAA | 2 | ||||
| 4 | ICN/IAGKAA | 1 | ||||
| 4 | NRN/IAAKAS | 1 | ||||
| 5 | IRN/IAAEAA | 1 | ||||
| 5 | IRN/IAGKAA | 1 | ||||
| Nigeria | 22 | 2 | NRN/ISAKAA | 1 | ||
| 3 | IRN/ISAKAA | 6 | ||||
| 4 | IRN/IAAKAA | 5 | ||||
| 4 | IRN/ISAEAA | 1 | ||||
| 5 | IRN/IAGKAA | 1 | ||||
| 5 | IRN/VAAKAA | 1 | ||||
| 5 | NRN/VAAKGS | 1 | ||||
| 6 | IRN/VAAKGA | 6 | ||||
| Central Africa | 80 | Chad | 1 | 6 | IRN/VSGKGA | 1 |
| Central Africa | 3 | 2 | NRN/ISAKAA | 2 | ||
| 3 | IRN/ISAKAA | 1 | ||||
| Congo | 4 | 2 | NRN/ISAKAA | 2 | ||
| 3 | IRN/ISAKAA | 2 | ||||
| Equatorial Guinea | 6 | 3 | IRN/ISAKAA | 3 | ||
| 4 | IRN/ISAKGA | 1 | ||||
| 5 | IRN/ISAEGA | 1 | ||||
| 6 | IRN/VAAKGA | 1 | ||||
| Gabon | 10 | 3 | IRN/ISAKAA | 7 | ||
| 4 | IRN/ISAEAA | 3 | ||||
| Angola | 14 | 2 | ICN/ISAKAA | 2 | ||
| 2 | NCN/ISGKAA | 1 | ||||
| 3 | IRN/ISAKAA | 4 | ||||
| 4 | IRN/IAAKAA | 2 | ||||
| 4 | IRN/ISAKGA | 1 | ||||
| 4 | IRN/ISAEAA | 3 | ||||
| 4 | IRN/ISGKAA | 1 | ||||
| Cameroon | 14 | 3 | IRN/ISAKAA | 5 | ||
| 4 | IRN/IAAKAA | 4 | ||||
| 4 | IRN/ISAEAA | 1 | ||||
| 5 | IRN/IAGKAA | 2 | ||||
| 5 | IRN/VAAKAA | 1 | ||||
| 6 | IRN/VAAKGA | 1 | ||||
| DR Congo | 28 | 2 | ICN/ISAKAA | 1 | ||
| 2 | NRN/ISAKAA | 1 | ||||
| 3 | IRN/ISAKAA | 6 | ||||
| 3 | NRN/ISAEAA | 1 | ||||
| 3 | ICN/ISAEAA | 3 | ||||
| 4 | IRN/IAAKAA | 1 | ||||
| 4 | IRN/ISGKAA | 3 | ||||
| 4 | ICN/ISAEGA | 1 | ||||
| 4 | IRN/ISAEAA | 9 | ||||
| 5 | IRN/IAGKAA | 2 |
Linkage Disequilibrium (LD) Analysis
In total, 25 genotypes of pfdhfr/pfdhps were detected among the 159 P. falciparum isolates. For pfdhfr, no SNP was found for codons 16 or 164. Ultimately, 5 and 13 genotypes of pfdhfr (involving codons 51, 59, and 108) and pfdhps (involving codons 431, 436, 437, 540, 581, and 613) were detected and analyzed. To evaluate the SNP LD associations of pfdhfr and pfdhps, several statistically significant associations were found among the SNPs located in both the pfdhfr and pfdhps genes (Figure 2). For the pfdhfr gene, N51I was significantly associated with the SNPs (t175c, C59R; and g323a, S108N) with a D’ value of 0.84 (P < 0.05) and 1.0 (P < 0.05), respectively. Similarly, t175c was significantly associated with the g323a (0.71, P < 0.05). For the pfdhps gene, t1482g, c1486g, a1794g, and g2013t formed an LD block. The sole SNP (t1482g, S436A) was significantly associated with the SNPs (c1486g, A437G; a1794g, K540E; and g2013t, A613S) with D’ values of 0.75, 1.0, and 0.73, respectively. The SNP (t152a) of the pfdhfr gene encoding N51I was significantly associated with c1486g, with a D’ value of 0.68. No associations were detected for the other SNPs of either the pfdhfr or pfdhps genes.
Figure 2.
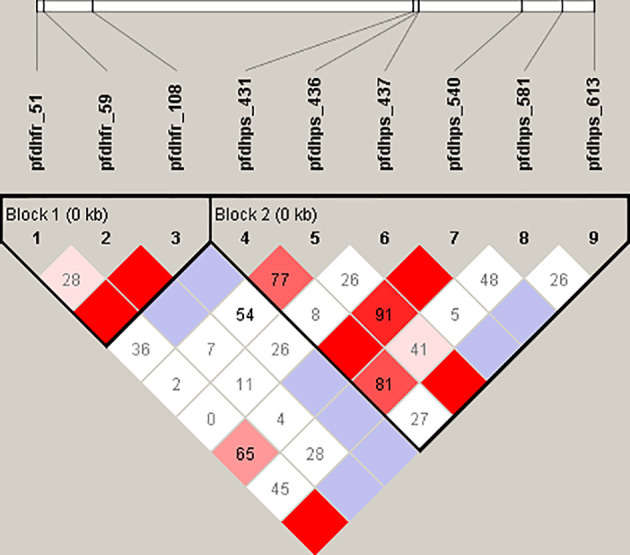
Linkage disequilibrium (LD) analysis of pfdhfr and pfdhps SNPs. For the pfdhfr gene, the single amino acid mutations were N51I, C59R, and S108N, respectively. Likewise, pfdhps gene are related to the mutations S436A, A437G, K540E, and A613S, respectively. The dark and light red squares indicate a linkage that was statistically significant (P < 0.05). Cambridge blue squares indicate a linkage that is present but is not statistically significant (P > 0.05). A white square with indicates that no linkage is present.
Discussion
The emergence and spread of Sulfadoxine-Pyrimethamine (SP) resistance has narrowed its usage to IPTp and IPTi in Africa (Rieckmann and Cheng, 2002). In 2019, the WHO covered 33 African countries for IPTp, in which at least nearly 62% of pregnant women received a first dose of IPTp (of four doses of ITPp)) (World Health Organization, 2020). The presents study aimed to determine the prevalence of SP resistance-associated pfdhfr and pfdhps genes in P. falciparum isolated from returned migrant workers from Africa reported in China. The study showed the pfdhfr triple haplotype mutation (IRN, including N51I, C59R, S108N) was highly prevalent, which was similar to other publications concerning African isolates (Jiang et al., 2019; Gikunju et al., 2020; Quan et al., 2020). This high prevalence was also found in the migrant workers returning to Guangxi from Ghana, suggesting that SP resistant genotype was widespread in Central and West Africa (Zhao et al., 2020). It was noted pfdhfr I164L, which was also associated with high resistance to cycloguanil, was detected in Ghanaian isolates, but was not found in our study (Zhao et al., 2020). It is worth noting that I164L is common in East Africa and Asia (Basuki et al., 2018,; Lynch et al., 2017), but rarely seen in Central and West Africa. One of the explanations was that this mutation site carries a high fitness cost to the parasite and therefore it is unable to survive the immune response of hosts in West Africa (Nzila et al., 2002). Further studies are needed to assess the effect of this mutation on the phenotype of parasites carrying this haplotype.
Mutations in pfdhps haplotypes at S436A and K540E, which are associated with decreased parasite sensitivity to Sulfadoxine drugs (Berglez et al., 2004), carried a higher allele frequency, which was also found in Uganda and Tanzania (Alifrangis et al., 2009; Mbonye et al., 2015). The WHO recommended that IPTp should not implemented in the regions when K540E exceed 50%; in our study, this mutation was present at 27.67%, which might favor the continued efficacy of IPTp treatment in these countries. Another mutation, A581G, is considered to have an important modulatory role in SP resistance. IPTp and SP could not protect pregnant women from delivering low birth weight infants when the frequency of this mutation is above 10% (Chico et al., 2015). In our study, the frequency of this mutation was 8.81% and half of these mutated isolates were found in Nigeria. For the mutation A437G, which was associated with resistance to Sulfadoxine in endemic regions because of the drug pressure selection, showed a frequency of only 9.43% in our study, which was lower than that reported in Equatorial Guinea, Pakistan, and Iran (Rouhani et al., 2015; Yaqoob et al., 2018; Jiang et al., 2019). The I431V mutation was detected in Nigeria, Cameroon, Equatorial Guinea, and Chad. The most frequent haplotype of I431V was VAAKGA, similar to that found in isolates from Cameroon and Nigeria (Chauvin et al., 2015; Oguike et al., 2016). These haplotypes occurring in Central and West Africa suggested that SP conferred a selective advantage, and ongoing drug pressure is relative strong because SP was used as IPTp in these regions. It is noted A437G was widely spread in central and western African countries, which indicating in vivo SP drug resistance in these regions (Pearce et al., 2009). Some other studies combined with the clinical study indicated the pfdhfr triple mutant genotype was associated with SP treatment resistance (Basco et al, 2000; Naidoo et al., 2013). However, in this study, the A437G was not widely spread in central and western African region, even less than S436A, which was also different with our previous study in China-Myanmar border (Zhang et al., 2014), which may partially ascribe as the sample size limitation, the loss of drug pressure, and time passage may also the potential reason.
In our study, the frequency of the quadruple mutation IRN/ISAEAA was significantly higher in East Africa (100% in Uganda, 85.7% in Mozambique) than western and central African countries (P < 0.01), suggesting that the clinical implications of such haplotypes require further combined genotype and phenotype analysis. SP was limited for malaria control among the general population in many countries in Africa because of the high frequency of drug resistance developed by P. falciparum; therefore, we expected to obtain SP sensitive strains. Indeed, in our study, 39.62% of the isolates harbored the non-mutated pfdhps gene. This may be similar to the process for chloroquine, which tends to recover its effectiveness against the parasite after a long period during which its use in malaria control activities is halted (Lu et al., 2017).
Conclusion
This study showed a high frequency of SP-resistance associated SNPs in the pfdhfr and pfdhps genes of P. falciparum isolated since 2017 in returned migrant workers from Africa. The high resistance may be linked to the unsuccessful withdrawal of the SP treatment, and thus might affect the efficacy of IPTp for pregnant women and IPTi for infants. Mutations such as K540E, and the pfdhfr-pfdhps haplotype IRN/ISAEAA, which occurred at moderate frequencies in East Africa, such as in Uganda and Mozambique, the other regions showed as high frequency triple mutations of pfdhfr, but relatively scatter site mutations in pfdhps gene. The present data could provide the evidence for molecular surveillance in the post-elimination stage in China, focusing on the risk population among returning migrant workers from Africa, and could be used to determine the treatment policy for imported malaria in China.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the ethical committee of the National Institute of Parasitic Diseases (NIPD), Chinese Centre for Disease Control and Prevention, Chinese Center for Tropical Diseases Research. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
HY conceived the study. J-hY, FH, X-yF, and JF performed the study and analyzed the data. JF assisted with editing the manuscript. J-pC provided strategic advice and revised the manuscript. All authors assisted with draft revisions. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by the Key Techniques in Collaborative Prevention and Control of Major Infectious Diseases in the Belt and Road (Grant No. 2018ZX10101002-004) and the National Natural Science Foundation of China (Grant No. 81602904). The study had also been supported by “the Fifth Round of Three-Year Public Health Action Plan of Shanghai (No. GWV-10.1-XK13)”.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Alifrangis M., Lusingu J. P., Mmbando B., Dalgaard M. B., Vestergaard L. S., Ishengoma D., et al. (2009). Five-Year Surveillance of Molecular Markers of Plasmodium Falciparum Antimalarial Drug Resistance in Korogwe District, Tanzania: Accumulation of the 581G Mutation in the P. Falciparum Dihydropteroate Synthase Gene. Am. J. Trop. Med. Hyg. 80, 523–527. 10.4269/ajtmh.2009.80.523 [DOI] [PubMed] [Google Scholar]
- Basuki S., F., Risamasu P. M., Kasmijati A. P., Riyanto S., Hidayat A., et al. (2018). Origins and Spread of Novel Genetic Variants of Sulfadoxine-Pyrimethamine Resistance in Plasmodium Falciparum Isolates In Indonesia. Malaria J. 17, 475. 10.1186/s12936-018-2597-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basco L. K., Tahar R., Keundjian A., Ringwald P. (2000). Sequence Variations in the Genes Encoding Dihydropteroate Synthase and Dihydrofolate Reductase and Clinical Response to Sulfadoxine-Pyrimethamine in Patients With Acute Uncomplicated Falciparum Malaria. J. Infect. Dis. 182, 624–628. 10.1086/315731 [DOI] [PubMed] [Google Scholar]
- Berglez J., Iliades P., Sirawaraporn W., Coloe P., Macreadie I. (2004). Analysis in Escherichia Coli of Plasmodium Falciparum Dihydropteroate Synthase (DHPS) Alleles Implicated in Resistance to Sulfadoxine. Int. J. Parasitol. 34, 95–100. 10.1016/j.ijpara.2003.09.009 [DOI] [PubMed] [Google Scholar]
- Chauvin P., Menard S., Iriart X., Nsango S. E., Tchioffo M. T., Abate L., et al. (2015). Prevalence of Plasmodium Falciparum Parasites Resistant to Sulfadoxine/Pyrimethamine in Pregnant Women in Yaounde, Cameroon: Emergence of Highly Resistant Pfdhfr/Pfdhps Alleles. J. Antimicrob. Chemother. 70, 2566–2571. 10.1093/jac/dkv160 [DOI] [PubMed] [Google Scholar]
- Chico R. M., Cano J., Ariti C., Collier T. J., Chandramohan D., Roper C., et al. (2015). Influence of Malaria Transmission Intensity and the 581G Mutation on the Efficacy of Intermittent Preventive Treatment in Pregnancy: Systematic Review and Meta-Analysis. Trop. Med. Int. Health TM IH 20, 1621–1633. 10.1111/tmi.12595 [DOI] [PubMed] [Google Scholar]
- Chulay J. D., Watkins W. M., Sixsmith D. G. (1984). Synergistic Antimalarial Activity of Pyrimethamine and Sulfadoxine Against Plasmodium Falciparum in Vitro. Am. J. Trop. Med. Hyg. 33, 325–330. 10.4269/ajtmh.1984.33.325 [DOI] [PubMed] [Google Scholar]
- Feng J., Kong X., Xu D., Yan H., Zhou H., Tu H., et al. (2019). Investigation and Evaluation of Genetic Diversity of Plasmodium Falciparum Kelch 13 Polymorphisms Imported From Southeast Asia and Africa in Southern China. Front. Public Health 7, 95. 10.3389/fpubh.2019.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Li J., Yan H., Feng X., Xia Z. (2015. a). Evaluation of Antimalarial Resistance Marker Polymorphism in Returned Migrant Workers in China. Antimicrob Agents Chemother 59, 326–330. 10.1128/AAC.04144-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Xiao H., Xia Z., Zhang L., Xiao N. (2015. b). Analysis of Malaria Epidemiological Characteristics in the People’s Republic of China 2004-2013. Am. J. Trop. Med. Hyg. 93, 293–299. 10.4269/ajtmh.14-0733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Zhang L., Huang F., Yin J. H., Tu H., Xia Z. G., et al. (2018). Ready for Malaria Elimination: Zero Indigenous Case Reported in the People’s Republic of China. Malaria J. 17, 315. 10.1186/s12936-018-2444-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gikunju S. W., Agola E. L., Ondondo R. O., Kinyua J., Kimani F., LaBeaud A. D., et al. (2020). Prevalence of Pfdhfr and Pfdhps Mutations in Plasmodium Falciparum Associated With Drug Resistance Among Pregnant Women Receiving IPTp-SP at Msambweni County Referral Hospital, Kwale County, Kenya. Malaria J. 19, 190. 10.1186/s12936-020-03263-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosling R. D., Cairns M. E., Chico R. M., Chandramohan D. (2010). Intermittent Preventive Treatment Against Malaria: An Update. Expert Rev. Anti Infect. Ther. 8, 589–606. 10.1586/eri.10.36 [DOI] [PubMed] [Google Scholar]
- Jiang T., Chen J., Fu H., Wu K., Yao Y., Eyi J. U. M., et al. (2019). High Prevalence of Pfdhfr-Pfdhps Quadruple Mutations Associated With Sulfadoxine-Pyrimethamine Resistance in Plasmodium Falciparum Isolates From Bioko Island, Equatorial Guinea. Malaria J. 18, 101. 10.1186/s12936-019-2734-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konate A. T., Yaro J. B., Ouedraogo A. Z., Diarra A., Gansane A., Soulama I., et al. (2011). Intermittent Preventive Treatment of Malaria Provides Substantial Protection Against Malaria in Children Already Protected by an Insecticide-Treated Bednet in Burkina Faso: A Randomised, Double-Blind, Placebo-Controlled Trial. PloS Med. 8, e1000408. 10.1371/journal.pmed.1000408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F., Zhang M., Culleton R. L., Xu S., Tang J., Zhou H., et al. (2017). Return of Chloroquine Sensitivity to Africa? Surveillance of African Plasmodium Falciparum Chloroquine Resistance Through Malaria Imported to China. Parasites Vectors 10, 355. 10.1186/s13071-017-2298-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch C. A., Pearce R., Pota H., Egwang C., Egwang T., Bhasin A., et al. (2017). Travel and the Emergence of High-Level Drug Resistance in Plasmodium Falciparum in Southwest Uganda: Results From a Population-Based Study. Malaria J. 16, 150. 10.1186/s12936-017-1812-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbonye A. K., Birungi J., Yanow S. K., Shokoples S., Malamba S., Alifrangis M., et al. (2015). Prevalence of Plasmodium Falciparum Resistance Markers to Sulfadoxine-Pyrimethamine Among Pregnant Women Receiving Intermittent Preventive Treatment for Malaria in Uganda. Antimicrob Agents Chemother. 59, 5475–5482. 10.1128/AAC.00507-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo I., Roper C. (1984). Mapping 'Partially Resistant', 'Fully Resistant', and 'Super Resistant' Malaria. Trends Parasitol. 29, 505–515. 10.1016/j.pt.2013.08.002 [DOI] [PubMed] [Google Scholar]
- Nzila A. M., Mberu E. K., Nduati E., Ross A., Watkins W. M., Sibley C. H. (2002). Genetic Diversity of Plasmodium Falciparum Parasites From Kenya is Not Affected by Antifolate Drug Selection. Int. J. Parasitol. 32, 1469–1476. 10.1016/S0020-7519(02)00164-9 [DOI] [PubMed] [Google Scholar]
- Oguike M. C., Falade C. O., Shu E., Enato I. G., Watila I., Baba E. S., et al. (2016). Molecular Determinants of Sulfadoxine-Pyrimethamine Resistance in Plasmodium Falciparum in Nigeria and the Regional Emergence of Dhps 431V. Int. J. Parasitol Drugs Drug Resist. 6, 220–229. 10.1016/j.ijpddr.2016.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P., Bharti P. K., Bansal D., Ali N. A., Raman R. K., Mohapatra P. K., et al. (2017). Prevalence of Mutations Linked to Antimalarial Resistance in Plasmodium Falciparum From Chhattisgarh, Central India: A Malaria Elimination Point of View. Sci. Rep. 7, 16690. 10.1038/s41598-017-16866-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce R. J., Pota H., Evehe M. S., Ba el H., Mombo-Ngoma G., Malisa A. L., et al. (2009). Multiple Origins and Regional Dispersal of Resistant Dhps in African Plasmodium Falciparum Malaria. PloS Med. 6, e1000055. 10.1371/journal.pmed.1000055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan H., Igbasi U., Oyibo W., Omilabu S., Chen S. B., Shen H. M., et al. (2020). High Multiple Mutations of Plasmodium Falciparum-Resistant Genotypes to Sulphadoxine-Pyrimethamine in Lagos, Nigeria. Infect. Dis. Poverty 9, 91. 10.1186/s40249-020-00712-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckmann K., Cheng Q. (2002). Pyrimethamine-Sulfadoxine Resistance in Plasmodium Falciparum Must be Delayed in Africa. Trends Parasitol 18, 293–294. 10.1016/S1471-4922(02)02287-0 [DOI] [PubMed] [Google Scholar]
- Rouhani M., Zakeri S., Pirahmadi S., Raeisi A., Djadid N. D. (2015). High Prevalence of Pfdhfr-Pfdhps Triple Mutations Associated With Anti-Malarial Drugs Resistance in Plasmodium Falciparum Isolates Seven Years After the Adoption of Sulfadoxine-Pyrimethamine in Combination With Artesunate as First-Line Treatment in Iran. Infect. Genet. Evol. 31, 183–189. 10.1016/j.meegid.2015.01.020 [DOI] [PubMed] [Google Scholar]
- Triglia T., Menting J. G., Wilson C., Cowman A. F. (1997). Mutations in Dihydropteroate Synthase are Responsible for Sulfone and Sulfonamide Resistance in Plasmodium Falciparum. Proc. Natl. Acad. Sci. U. S. A. 94, 13944–13949. 10.1073/pnas.94.25.13944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2020). World Malaria Report 2020 (Geneva: World Health Organization; ). [Google Scholar]
- Yan H., Kong X., Zhang T., Xiao H., Feng X., Tu H., et al. (2020). Prevalence of Plasmodium Falciparum Kelch 13 (PfK13) and Ubiquitin-Specific Protease 1 (Pfubp1) Gene Polymorphisms in Returning Travelers From Africa Reported in Eastern China. Antimicrob. Agents Chemother. 20, 64. 10.1128/AAC.00981-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaqoob A., Khattak A. A., Nadeem M. F., Fatima H., Mbambo G., Ouattara A., et al. (2018). Prevalence of Molecular Markers of Sulfadoxine-Pyrimethamine and Artemisinin Resistance in Plasmodium Falciparum From Pakistan. Malar J. 17, 471. 10.1186/s12936-018-2620-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Feng J., Xia Z. G., Zhou S. S. (2020). Epidemiological Characteristics of Malaria and Progress on Its Elimination in China in 2019. Chin. J. Parasitol Parasit Dis. 38, 133–138. 10.12140/j.issn.1000-7423.2020.02.001 [DOI] [Google Scholar]
- Zhang Y., Yan H., Wei G., Han S., Huang Y., Zhang Q., et al. (2014). Distinctive Origin and Spread Route of Pyrimethamine-Resistant Plasmodium Falciparum in Southern China. Antimicrob. Agents Chemother. 58, 237–246. 10.1128/AAC.00972-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Pi L., Qin Y., Lu Y., Zeng W., Xiang Z., et al. (2020). Widespread Resistance Mutations to Sulfadoxine-Pyrimethamine in Malaria Parasites Imported to China From Central and Western Africa. Int. J. Parasitol. Drugs Drug Resist. 12, 1–6. 10.1016/j.ijpddr.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.