Abstract
Stress granules (SGs) are membraneless organelles composed of mRNAs and RNA binding proteins which undergo assembly in response to stress-induced inactivation of translation initiation. In general, SG recruitment is limited to a subpopulation of a given mRNA species and RNA-seq analyses of purified SGs revealed that signal sequence-encoding (i.e., endoplasmic reticulum [ER]-targeted) transcripts are significantly underrepresented, consistent with prior reports that ER localization can protect mRNAs from SG recruitment. Using translational profiling, cell fractionation, and single molecule mRNA imaging, we examined SG biogenesis following activation of the unfolded protein response (UPR) by 1,4-dithiothreitol (DTT) and report that gene-specific subsets of cytosolic and ER-targeted mRNAs can be recruited into SGs. Furthermore, we demonstrate that SGs form in close proximity to or directly associated with the ER membrane. ER-associated SG assembly was also observed during arsenite stress, suggesting broad roles for the ER in SG biogenesis. Recruitment of a given mRNA into SGs required stress-induced translational repression, though translational inhibition was not solely predictive of an mRNA's propensity for SG recruitment. SG formation was prevented by the transcriptional inhibitors actinomycin D or triptolide, suggesting a functional link between gene transcriptional state and SG biogenesis. Collectively these data demonstrate that ER-targeted and cytosolic mRNAs can be recruited into ER-associated SGs and this recruitment is sensitive to transcriptional inhibition. We propose that newly transcribed mRNAs exported under conditions of suppressed translation initiation are primary SG substrates, with the ER serving as the central subcellular site of SG formation.
Keywords: stress granule, mRNA, endoplasmic reticulum, translational regulation, unfolded protein response, oxidative stress
INTRODUCTION
Environmental, pathogen, and nutrient stressors can disrupt proteostasis, leading to toxic protein aggregation and, in scenarios of unresolved stress, cell death (Harding et al. 2003; Sakaki et al. 2012; Wang and Kaufman 2012; Costa-Mattioli and Walter 2020). Reflecting the pathological consequences of dysregulated proteostasis, eukaryotic cells express a family of stress response eukaryotic initiation factor 2α (eIF2α) kinases whose activation results in the inhibition of protein synthesis, as a key step in the resolution of unfolded protein accumulation (Pakos-Zebrucka et al. 2016; Wek 2018). This eIF2α kinase activity is central to the unfolded protein response (UPR), which comprises a translational regulatory arm mediated by the eIF2α kinase PERK and a stress response transcriptional program which supports the restoration of proteostasis and promotes stress tolerance (Wek et al. 2006; Taniuchi et al. 2016).
In response to stress-induced inactivation of eIF2α, translationally suppressed mRNAs can undergo recruitment into stress granules (SGs), membraneless organelles comprised of mRNAs, and RNA binding proteins (RBPs) (Kedersha et al. 1999; Decker and Parker 2012; Wolozin and Ivanov 2019; Mateju et al. 2020). SG formation is driven by the structural properties of SG-resident RBPs, in particular multivalent RNA binding motifs and intrinsically disordered domains, that in the presence of RNA support granule assembly (Kato et al. 2012; Guillen-Boixet et al. 2020; Matheny et al. 2020). mRNA recruitment into SGs is not, however, a simple process; the biochemical criteria for mRNA recruitment are complex and include intrinsic and stress-regulated translation efficiencies, transcript length, AU-rich element abundance, as well as other undefined cis-encoded elements (Khong et al. 2017; Namkoong et al. 2018; Matheny et al. 2020). In addition to these global criteria, deep sequencing analyses of SG RNAs revealed that endoplasmic reticulum (ER)-targeted transcripts are substantially under-represented (Khong et al. 2017). This finding is consistent with earlier reports that ER-targeted mRNAs can be excluded from SGs (Unsworth et al. 2010). Intriguingly, the ER engages in dynamic contact-site interactions with processing bodies (PBs) and SGs that are thought to regulate granule fission (Lee et al. 2020). How the ER contributes to PB and SG biology while its associated mRNAs are largely sequestered from these regulatory organelles is unknown.
As an ER-localized physiological stress response pathway that targets eIF2α (Sidrauski et al. 2015), the UPR provides a useful biological model for examining the functional interface between the ER and SG biology. Here we report that ER-targeted mRNAs can undergo gene-selective recruitment into SGs in response to UPR activation by the reducing agent 1,4-dithiothreitol (DTT) or the oxidizing agent sodium arsenite (Braakman et al. 1992; Li et al. 2011; Oslowski and Urano 2011; Srivastava et al. 2013; Delaney et al. 2020). Combined cell fractionation and single molecule fluorescence in situ hybridization (smFISH) studies revealed that SGs formed on or near the ER membrane, identifying the ER as a subcellular site of SG formation. Of the four ER-targeted mRNAs examined, SG accumulation was observed for HSP90B1/GRP94 and CTGF/CCN2, but not HSPA5/GRP78 or B2M. Selective recruitment into ER-associated SGs was also observed for cytoplasmic mRNAs, where NCL/nucleolin was recruited into SG, but GAPDH was not. Intriguingly, mRNAs experiencing translational suppression in response to UPR activation showed differential recruitment into SGs, indicating that translational suppression alone is insufficient for SG recruitment. SG formation was, however, highly sensitive to the transcriptional inhibitors actinomycin D (ActD) and triptolide, which prevented SG formation for all mRNAs examined. We summarize these data in a working model of SG assembly on the ER, emphasizing a role for newly exported mRNAs as preferred substrates for SG recruitment.
RESULTS
Selective recruitment of ER-targeted mRNAs into stress granules
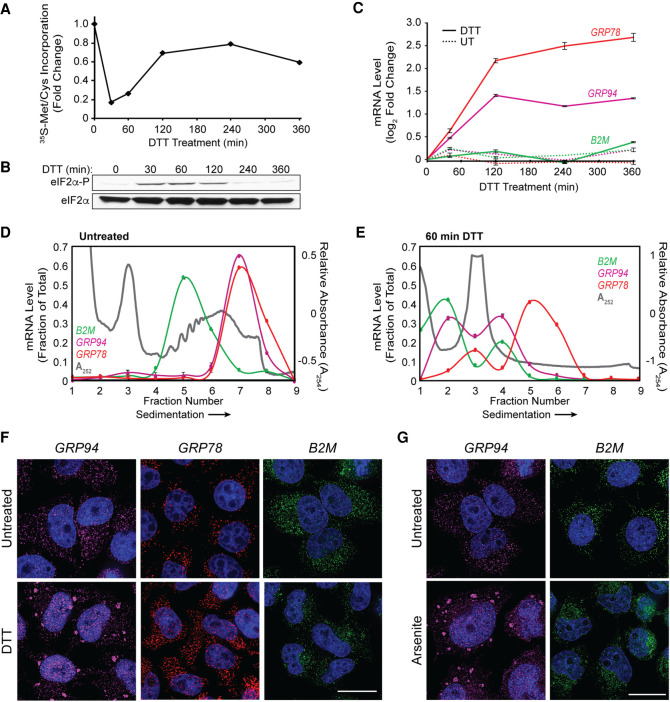
To examine the intersection between the ER, the UPR, and ribonucleoprotein (RNP) granule dynamics, we first performed polyribosome profiling and single molecule RNA imaging of three ER-targeted mRNAs which encode ER-resident stress response or secretory cargo proteins and vary in their transcriptional and translational responses to UPR activation. UPR activation was initiated by treatment of HeLa cells with the reducing agent DTT, which induces ER stress by preventing disulfide bond formation and thereby promoting the accumulation of misfolded proteins in the ER lumen (Braakman et al. 1992; Oslowski and Urano 2011). DTT addition elicited a rapid reduction in cellular translation to ∼20% of control levels at 30 min, followed by recovery to a reduced steady state, as measured by [35S]Met/Cys incorporation (Fig. 1A). This biphasic translation inhibitory response was mirrored in the kinetics of eIF2α phosphorylation (Fig. 1B), where phospho-eIF2α levels peaked at 30–60 min of DTT treatment and gradually resolved after 120 min. Activation of the UPR transcriptional arm was assayed by RT-qPCR analysis of ER chaperone GRP78 and GRP94 mRNAs and revealed the expected induction of both transcript levels by 60 min post DTT addition (Fig. 1C; Lee 1987; Lee et al. 2003). Levels of B2M mRNA were not altered in response to DTT, consistent with prior studies demonstrating that B2M is not a UPR response gene (Fig. 1C; Reid et al. 2014; Rendleman et al. 2018). Sucrose density gradient polysome profiling studies confirmed global UPR-elicited translation inhibition as evidenced by the pronounced collapse of heavy polysomes (fractions 5–8, ribosome density >4) to a predominately monosome profile (fractions 2–4, 80S monosome) following 60 min of DTT treatment (Fig. 1D,E, gray line). As with global polysome remodeling, UPR activation had a substantial impact on GRP94 and B2M translation profiles, with parallel shifts of their mRNAs from heavy to light polysome fractions, indicating reduced translation initiation frequencies for these mRNAs (Fig. 1D,E). Notably, UPR activation had comparatively modest effects on the GRP78 translation profile, with a less pronounced redistribution of GRP78 mRNAs to light polysomes (Fig. 1D,E). The blunted translational response of GRP78 mRNAs to UPR activation is consistent with previous reports that its 5′ UTR encodes an internal ribosome entry site, enabling translation initiation under conditions of elevated eIF2α phosphorylation (Starck et al. 2016).
FIGURE 1.
Selective recruitment of ER-targeted mRNAs into UPR-induced stress granules. (A) Representative time course of UPR-induced inhibition of protein synthesis assayed by [35S]Met/Cys incorporation. HeLa cell cultures were treated with DTT and protein synthesis rates assayed at the the indicated time points. (B) Immunoblot analysis of eIF2α and phospho-eIF2α levels following DTT treatment of cell cultures for the indicated times. (C) Representative time course of UPR-elicited transcriptional activation of the UPR response genes GRP94 and GRP78 and the ER-targeted gene B2M. Cell cultures were treated with DTT and total RNA was extracted for RT-qPCR analysis of transcript levels at the indicated time points. Data points are mean log2 fold-change ± SD, normalized to GAPDH levels. (D,E) Polyribosome profiling of GRP94, GRP78, and B2M translational status by sucrose density gradient velocity sedimentation. HeLa cell cultures at time zero (D) or following DTT treatment (E) were detergent extracted, and total polyribosome profiles were determined by the A254 nm absorbance traces (gray). mRNA distributions were determined by RT-qPCR analysis of GRP78 (red), GRP94 (magenta), and B2M (green) mRNAs extracted from the gradient fractions. Data are representative of three biological replicates; RT-qPCR data are mean fraction of total mRNA for the given gene across all gradient fractions ± SD. (F) Representative smFISH visualization of GRP94 (magenta), GRP78 (red), and B2M (green) mRNAs in untreated and DTT-treated (60 min) HeLa cells. (G) As in F but treatment with sodium arsenite (60 min). DAPI nuclear stain (blue) is indicated for all images. Scale bar = 20 µm.
With translation initiation inhibition being a primary trigger for SG formation (Kedersha et al. 1999), we examined the subcellular distributions of GRP94, GRP78, and B2M mRNAs by single molecule RNA fluorescence in situ hybridization (smFISH) before and after UPR activation (Fig. 1F). For the two mRNAs whose translation was strongly repressed in response to UPR activation, that is, GRP94 and B2M, divergent smFISH patterns were observed. GRP94 mRNAs transitioned from diffuse, diffraction-limited single foci at steady state to prominent perinuclear granules following UPR activation, whereas the subcellular distribution of B2M mRNAs remained as single foci in both untreated and UPR-activated states. As expected for actively translating mRNAs, GRP78 smFISH patterns were largely unaltered by the UPR (Fig. 1F). These data indicate that mRNA recruitment into UPR-elicited granules is not solely driven by mRNA translational status, and suggest that gene-specific phenomenon may contribute to granule formation. Importantly, these findings were not unique to DTT stress as treatment of HeLa cell cultures with sodium arsenite, which has been demonstrated to activate the UPR via induction of oxidative stress, also elicited GRP94, but not B2M, mRNA granule formation (Fig. 1G; Li et al. 2011; Srivastava et al. 2013; Delaney et al. 2020). This finding is consistent with prior studies which indicate that SG RNA composition can be conserved across different stressors (Khong et al. 2017).
Stress granules containing ER-targeted mRNAs can be ER-associated
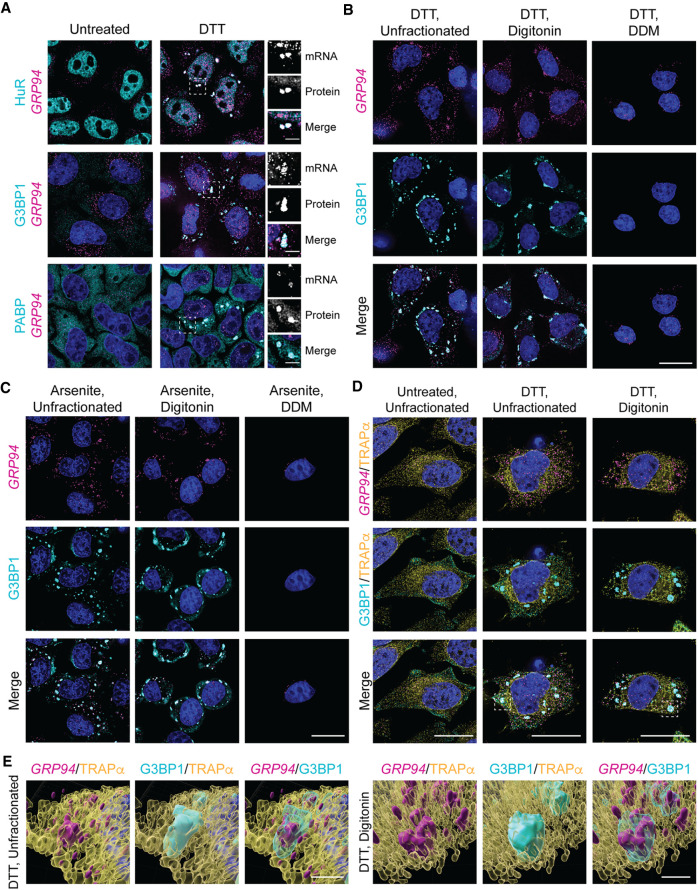
With prior studies reporting that ER-targeted mRNAs were under-represented and/or excluded from SGs (Unsworth et al. 2010; Khong et al. 2017), we considered that the UPR-elicited GRP94 mRNA granules could comprise a novel RNP granule, distinct from canonical SGs. To examine this hypothesis, immunofluorescence colocalization studies were performed for the SG marker proteins HuR, G3BP1, and PABP in parallel with GRP94 smFISH (Stoecklin and Kedersha 2013; Protter and Parker 2016; Youn et al. 2018). GRP94 smFISH profiles costained with the three SG components examined following but not prior to UPR activation, consistent with the recruitment of GRP94 mRNAs into canonical, rather than unique, SGs (Fig. 2A). In light of this finding, we hereafter refer to GRP94 mRNA granules as GRP94 SGs, or simply SGs.
FIGURE 2.
UPR activation elicits ER-associated stress granules. (A) Representative GRP94 smFISH (magenta) with immunofluorescence costaining for the stress granule protein markers HuR, G3BP1, and PABP (cyan) in untreated and DTT-treated HeLa cell cultures. Dotted boxes indicate regions of grayscale insets for mRNA and protein channels, as well as color merge, for DTT-treated cells (right). (B) Representative GRP94 smFISH (magenta) and G3BP1 immunofluorescence (cyan) costaining in DTT-treated cells. Following DTT treatment, cells were permeabilized with digitonin-supplemented buffer to release cytosolic contents (digitonin) or sequentially treated with digitonin and n-dodecyl-β-d-maltoside (DDM) buffers to solubilize organelle membranes. See also Supplemental Figure S1 for detergent permeabilization protocol validation. (C) As in B but with arsenite stress. (D) Representative micrographs of ER membrane protein TRAPα immunofluorescence (yellow) with GRP94 smFISH (magenta) and/or G3BP1 immunofluorescence (cyan) costaining in unfractionated and cytosol-depleted (digitonin-permeabilized) cells following DTT treatment or untreated control. Boxes indicate regions of interest for (E). (E) 3D renderings of representative granules from (D) in unfractionated (Supplemental Movie 1) and digitonin-permeabilized (Supplemental Movie 2) cells. DAPI staining (blue) is indicated for all images. Full cell scale bars = 20 µm, inset and 3D scale bars = 4 µm.
With GRP94 mRNAs displaying high enrichment on the ER under both homeostatic and UPR-activated conditions (Chen et al. 2011; Reid and Nicchitta 2012; Reid et al. 2014), we asked if GRP94 SGs were also ER-associated. ER-association was assayed by an established sequential detergent fractionation method where cells are incubated on ice to depolymerize the microtubule network and subsequently treated with a digitonin-supplemented buffer to permeabilize the plasma membrane, releasing the cytosolic contents of the cells while leaving the ER membrane and its associated RNAs intact (Fig. 2B; Supplemental Fig. S1; Lerner et al. 2003; Jagannathan et al. 2011). This fractionation is illustrated in Supplemental Figure S1, depicting the loss of monomeric β-Tubulin and retention of the ER-membrane protein TRAPα following digitonin treatment. With subsequent treatment of the digitonin-permeabilized cells with the nonionic detergent n-dodecyl-β-d-maltoside (DDM), the ER membrane is solubilized while leaving the nucleus largely intact (Supplemental Fig. S1).
This cell fractionation protocol was utilized to examine the subcellular distribution of GRP94 SGs in UPR-activated HeLa cell cultures via GRP94 smFISH and G3BP1 immunofluorescence costaining of intact, cytosol-extracted, and cytosol/ER-extracted cells. We observed that GRP94 SGs are intimately associated with the ER membrane, as they are retained following digitonin permeabilization and released upon treatment with DDM, following either DTT- (Fig. 2B) or arsenite-induced stress (Fig. 2C). The minor intranuclear GRP94 smFISH signal retained following DDM treatment demonstrates a selective loss of ER-associated GRP94 SGs coincident with ER membrane solubilization.
Though sequential detergent fractionation is a useful tool for examining membrane-association of cellular components, it does not allow us to examine the cytosolic contents released during digitonin permeabilization. To assess the possible compartmentalization of SGs between cytosolic and ER-associated populations, GRP94 smFISH patterns were examined in cells coimmunostained for G3BP1 and the ER-resident membrane protein TRAPα in both intact and cytosol-depleted UPR-activated cells (Fig. 2D). As in the experiments depicted in Figure 2B and Supplemental Figure S1, the parallel insensitivity of GRP94 SGs, G3BP1, and TRAPα to digitonin extraction supports the conclusion that GRP94 SG formation occurs in close physical proximity to or in direct association with the ER membrane. The close proximity of all observed GRP94 SGs to the ER membrane (TRAPα) in the UPR-activated, unfractionated experimental condition suggests that GRP94 SGs are predominately, if not exclusively, ER-associated (Fig. 2D). Surprisingly, this was also the case for all observed G3BP1 granules. 3D-reconstructions of ER-proximal SGs in unfractionated (Supplemental Movie 1) and cytosol-depleted (Supplemental Movie 2) UPR-activated cells were consistent with these findings and revealed SG complexes engaged in apparent contact sites with the ER (Fig. 2E). By the criteria of detergent sensitivity and fluorescence colocalization, these data indicate that ER-targeted mRNAs can be recruited to ER proximal/ER-associated SGs and suggest that the ER membrane may be a general site of SG formation (Khong et al. 2017; Lee et al. 2020).
Transcriptional inhibitors block GRP94 stress granule formation
The data in Figure 1F demonstrate that mRNA recruitment into SGs was selective for one of the three ER-targeted mRNAs examined, GRP94. mRNAs can, however, display temporal variations in their translational sensitivity to eIF2α phosphorylation state and so we examined GRP94 and B2M mRNA organization over a time course of elevated eIF2α phosphorylation (Fig. 1A,B; Andreev et al. 2015; Sidrauski et al. 2015; Young and Wek 2016). In these experiments, GRP94 granules were observed as early as 30 min post-DTT addition whereas B2M mRNAs remained as diffuse single foci throughout the 2 h time course (Fig. 3A) despite the similar and sustained inhibition of translation for the two mRNAs (Fig. 3B). The smFISH data depicted in Figure 3A also highlights the divergent transcriptional responses of the two genes to UPR activation. For GRP94, UPR-elicited transcriptional induction was detected by smFISH as intranuclear transcriptional foci at the 30 min time point, with prominent intranuclear mRNA staining at the 120 min time point (Fig. 3A). In contrast, UPR activation did not alter B2M intranuclear smFISH patterns, consistent with the data presented in Figure 1C. These smFISH data provide orthogonal validation that GRP94, but not B2M, is transcriptionally up-regulated in response to UPR activation and indicate that gene transcriptional state may be a criterion for mRNA recruitment into SGs.
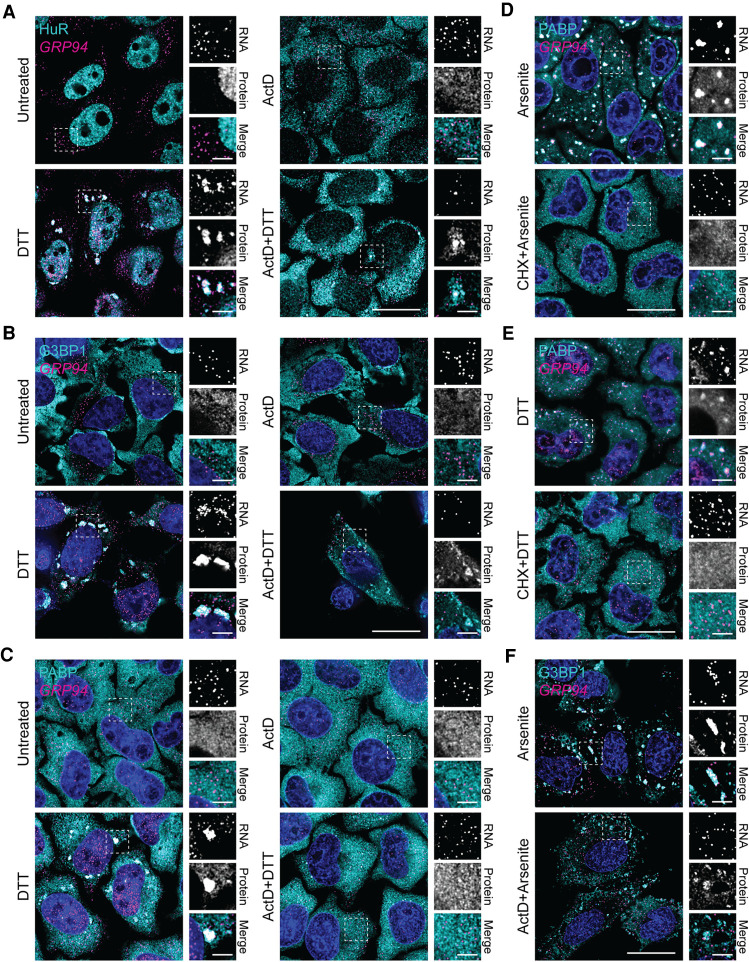
FIGURE 3.
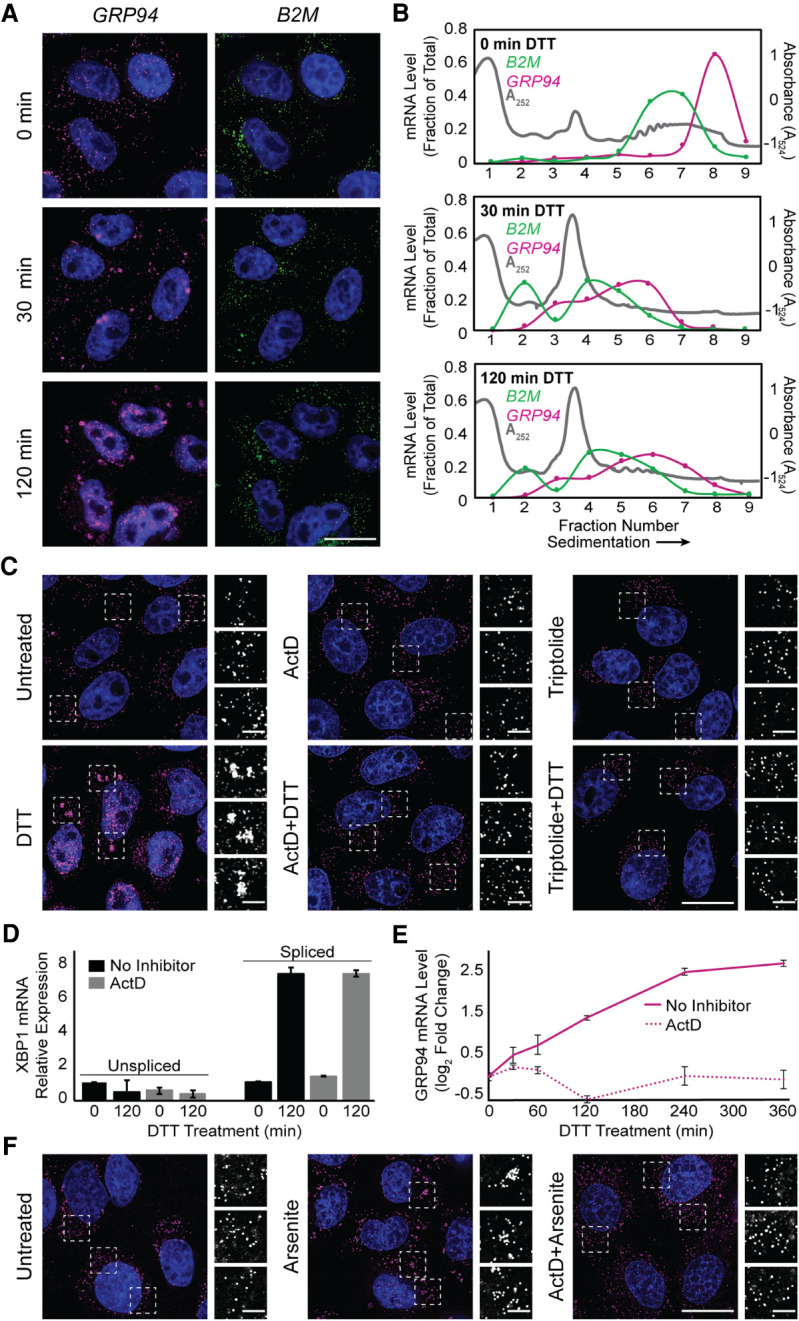
Transcriptional inhibitors actinomycin D and triptolide prevent RNA recruitment into stress granules. (A) Representative time course analysis of GRP94 (magenta) and B2M (green) smFISH staining patterns over the course of maximal inhibition of eIF2α activity (see Fig. 1A). (B) Sucrose density velocity sedimentation gradients and RT-qPCR analysis as in Figure 1D at the indicated time points following DTT addition. Data are representative of three biological replicates. (C) Representative GRP94 smFISH in control (untreated, ActD, triptolide) and stressed (treatment with DTT, with and without treatment with indicated transcriptional inhibitor, ActD or triptolide) conditions. Boxes indicate regions of grayscale insets of mRNA distributions (right). (D) RT-qPCR analysis of spliced and unspliced XBP1 mRNA in control (untreated or ActD) and stressed (treatment with DTT, with and without ActD) conditions. Data are expression level relative to 0 min without ActD after normalization to GAPDH ± SEM from three biological replicates. (E) RT-qPCR analysis of GRP94 mRNA levels over a time course of DTT treatment with or without ActD addition. Data are mean log2 fold change of expression relative to GAPDH ± SEM from three biological replicates. (F) Representative GRP94 smFISH in untreated and sodium arsenite-treated cells with or without ActD addition. Dotted boxes indicate regions of grayscale insets of mRNA distribution (right). DAPI staining (blue) is included for all images. Full cell scale bar = 20 µm, inset scale bar = 4 µm.
To explore potential links between gene transcription and mRNA recruitment into SGs, we examined SG formation in cells treated with the transcriptional inhibitors actinomycin D (ActD), a DNA intercalating agent, or triptolide, an RNA polymerase II inhibitor (Titov et al. 2011). As shown in Figure 3C, GRP94 SG formation was efficiently inhibited in the presence of either ActD or triptolide. To exclude possible off-target effects of transcriptional inhibition on UPR signaling, we assayed for IRE1-mediated splicing of transcription factor XBP1 mRNA, a signature event in UPR activation, in the absence or presence of ActD (Fig. 3D; Cox and Walter 1996; Cox et al. 1997; Walter and Ron 2011). As expected, ActD addition resulted in decreased levels of unspliced XBP1 mRNA at the 2 h time point, a consequence of mRNA decay in the absence of ongoing transcription. ActD did not, however, alter IRE1 activation, as demonstrated by similar levels of spliced XBP1 mRNA in both control and ActD-treated cells following DTT addition (Fig. 3D). Furthermore, ActD treatment has previously been shown not to inhibit eIF2α phosphorylation (Bounedjah et al. 2014). These data demonstrate that transcriptional inhibition did not disrupt UPR signaling; ActD treatment did, however, block transcriptional up-regulation of GRP94 (Fig. 3E). ActD treatment also prevented arsenite-elicited GRP94 SG formation (Fig. 3F), demonstrating this inhibition was not a stressor-specific effect.
Given the established translocation of nuclear HuR protein into the cytosol following ActD treatment, and the corresponding inhibition of cytosolic RBP aggregation in combination with arsenite stress (Bounedjah et al. 2014), we investigated the effects of ActD treatment on HuR, G3BP1, and PABP granule formation following DTT-induced UPR activation (Fig. 4). HuR accumulation in the cytosol was evident following treatment with ActD alone or in combination with DTT (Fig. 4A). Furthermore, ActD treatment markedly inhibited, but did not entirely block, formation of HuR-positive granules following DTT addition (Fig. 4A). In the instances of observed HuR granule formation, there was minimal colocalization of GRP94 mRNA (Fig. 4A), consistent with the finding presented in Figure 3.
FIGURE 4.
Actinomycin D inhibits RNA binding protein granulation during the UPR. (A) Representative GRP94 smFISH (magenta) with immunofluorescence costaining for HuR (cyan) in control (untreated or ActD) and stressed (treatment with DTT, with and without ActD) HeLa cell cultures. Note redistribution of HuR from the nucleoplasm to the cytoplasm in response to ActD treatment. Boxes indicate regions of grayscale insets for mRNA and protein channels, as well as color merge, for each condition (right). (B) As in A but immunofluorescence staining for G3BP1 (cyan). (C) As in A but immunofluorescence staining for PABP (cyan). (D) Representative GRP94 smFISH (magenta) with immunofluorescence costaining for PABP (cyan) in cells treated with arsenite with and without cycloheximide (CHX). Boxes indicate regions of grayscale insets for mRNA and protein channels, as well as color merge, for each condition (right). (E) As in D but with DTT stress. (F) Representative GRP94 smFISH (magenta) with immunofluorescence costaining for G3BP1 (cyan) in cells treated with arsenite with and without ActD. Boxes indicate regions of grayscale insets for mRNA and protein channels, as well as color merge, for each condition (right). DAPI nuclear stain (blue) is included in all images. Full cell scale bar = 20 µm, inset scale bar = 4 µm.
To examine the specificity of the ActD-induced RNA binding protein granule blockade, we also investigated the effect of ActD treatment on G3BP1 and PABP granule formation (Fig. 4B,C). G3BP1 staining mirrored the aggregation patterns of HuR, with decreased protein granule formation and little to no GRP94 mRNA recruitment when treated with ActD + DTT compared to DTT alone (Fig. 4B). PABP staining, however, which can serve as a proxy for mRNA detection, did reveal a complete block of both PABP and GRP94 mRNA granule formation following combined ActD + DTT treatment (Fig. 4C). The degree of PABP and GRP94 SG formation inhibition by ActD was similar to the established effect of cycloheximide (CHX) treatment during arsenite stress (Fig. 4D; Kedersha et al. 2000). The inhibition of SG formation by CHX treatment was also confirmed with DTT stress (Fig. 4E). Together these data suggest that DTT-induced SG protein aggregation is suppressed but not blocked by ActD treatment, whereas RNA recruitment into SGs is effectively prevented by ActD, similarly to CHX, despite total extra-nuclear RNA concentrations being similar with and without short term ActD exposure (Bounedjah et al. 2014). These data indicate that SG proteins, such as G3BP1, initiate granule nucleation during combined ActD + DTT treatment similar to stress-induced conditions without transcriptional inhibitors (Yang et al. 2020). In contrast, preexisting mRNAs are not recruited into these nucleation sites, as demonstrated by +ActD conditions (Fig. 4). The lack of RNA recruitment to the nucleation sites would prevent further growth of the SGs suggesting a role for de novo RNA transcription in SG biogenesis. In support of this interpretation, a similar inhibition of G3BP1 granule formation was seen when cells were treated with both ActD and arsenite, while GRP94 mRNA remained dispersed compared to arsenite treatment alone (Fig. 4F).
Transcriptional inhibition suppresses stress granule recruitment of cytoplasmic and ER-targeted mRNAs
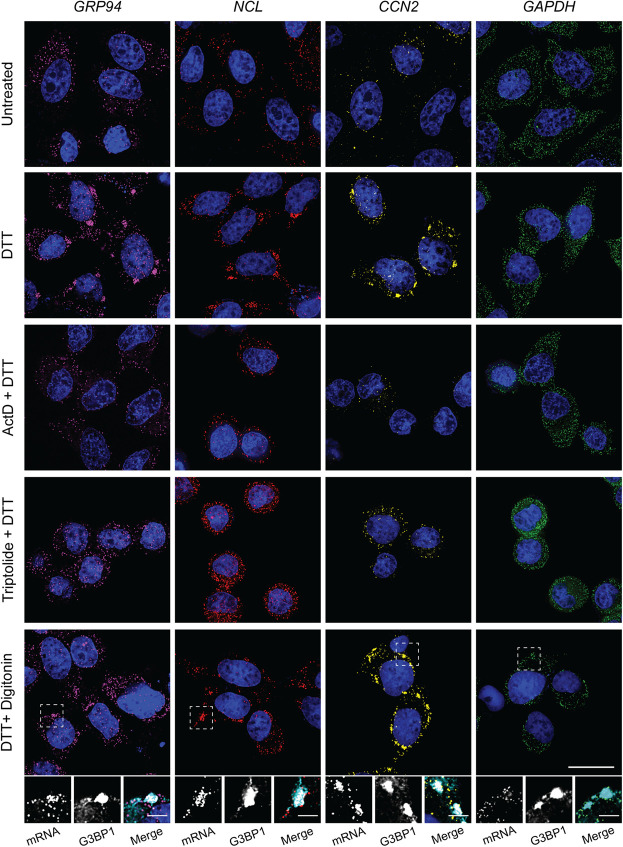
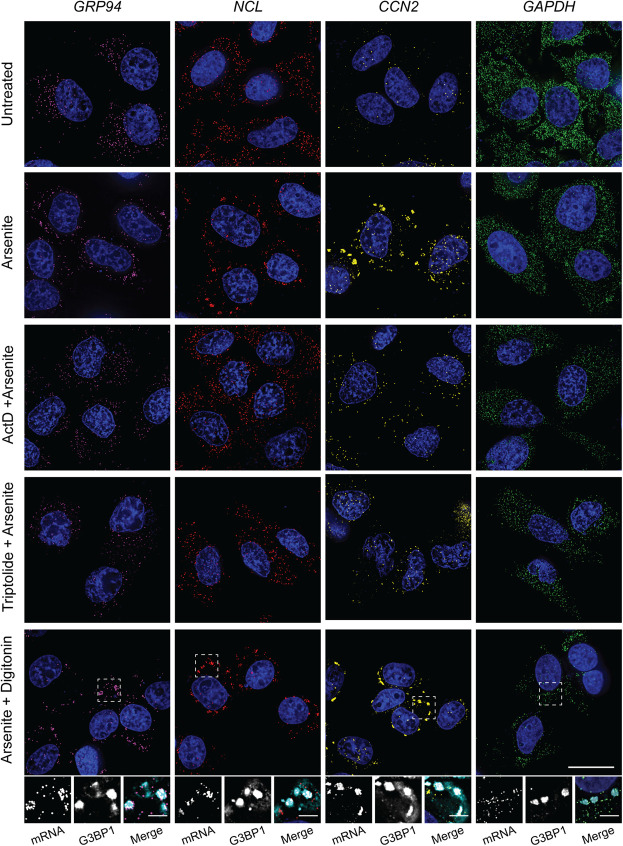
The data presented in Figures 3 and 4 indicate that pharmacological inhibition of transcription disrupts SG formation by preventing RNA recruitment into granules. We also considered that this phenomenon may be restricted to ER-targeted mRNAs and/or ER-associated SG biogenesis. To further explore the association between ER-targeting, SG recruitment, and sensitivity to transcriptional inhibition, we selected additional genes for smFISH studies of SG biogenesis. With SG recruitment efficiencies positively correlating with CDS length (Khong et al. 2017), transcript length was considered during gene selection. Three genes were chosen based on the presence or absence of a signal sequence, known translational repression following UPR-activation (Reid et al. 2014; Rendleman et al. 2018), high basal or UPR-induced transcriptional status, and relative CDS length: nucleolin (NCL), connective tissue growth factor (CTGF/CCN2), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). NCL (CDS = 2133 nt) encodes a transcript of similar length to GRP94 (CDS = 2412 nt) and lacks an encoded signal sequence, therefore identifying it as a cytoplasmic mRNA, allowing for assessment of the requirement for ER-targeting in recruitment into ER-associated SG. CCN2 (CDS = 1050 nt) was previously identified as an ER-targeted UPR-responsive gene (Rendleman et al. 2018), but encodes a substantially shorter transcript than GRP94, providing a useful test of the contribution of gene transcriptional status versus transcript length to SG recruitment among ER-targeted genes (Khong et al. 2017). GAPDH (CDS = 1008 nt) was selected as a length-matched comparator for CCN2 that, like NCL, lacks an encoded signal sequence and whose transcription is not UPR-responsive (Rendleman et al. 2018).
Paired smFISH of GRP94, NCL, CCN2, and GAPDH mRNAs were performed at steady state and following UPR activation by DTT (Fig. 5) or arsenite (Fig. 6) treatment, with and without transcriptional inhibition. As depicted in Figures 5 and 6, GRP94, NCL, and CCN2 mRNAs were recruited to perinuclear SGs following UPR activation by either stressor, whereas GAPDH mRNAs displayed a diffuse subcellular distribution pattern similar to the untreated control. Consistent with data reported in Figures 3 and 4, treatment of cell cultures with either ActD or triptolide blocked UPR-induced SG formation for GRP94, NCL, and CCN2 alike, further suggesting a functional link between newly transcribed/exported mRNAs and SG biogenesis. To determine if UPR-induced NCL and CCN2 SGs were ER-associated, as was observed for GRP94 SGs (Fig. 2), cells were treated with either DTT or arsenite and mRNA distributions examined following digitonin extraction of the cytosol (DTT + digitonin) (Figs. 5, 6). These data demonstrate that NCL and CCN2 granules were also retained in perinuclear locales in digitonin-permeabilized cells, consistent with an ER association. To confirm that these ER-localized NCL and CCN2 granules were in fact canonical SGs, as was demonstrated for GRP94 (Fig. 2), costaining for G3BP1 was included following digitonin extraction and confirmed that these granules contain the canonical SG marker (Figs. 5, 6 insets).
FIGURE 5.
Transcript length and transcriptional state influence mRNA recruitment into ER-associated stress granules following DTT stress. Representative smFISH of long transcript size-paired genes GRP94 (magenta, 2412 nt) and NCL (red, 2133 nt), and short transcript size-paired genes CCN2 (yellow, 1050 nt) and GAPDH (green, 1008 nt) mRNAs in untreated and DTT-treated HeLa cell cultures. Where indicated, cell cultures were treated with ActD or triptolide in addition to DTT. To evaluate the ER-association and presence of the SG protein marker G3BP1 in observed RNA granules, cells were permeabilized in digitonin-supplemented buffers to extract cytosolic contents while leaving the ER membrane intact, and costained for G3BP1 by immunofluorescence. Dotted boxes indicate regions of grayscale insets for mRNA and G3BP1 protein channels, as well as color merge, for digitonin permeabilized cells (below). DAPI nuclear stain (blue) included in all images. Full cell scale bars = 20 µm, inset scale bars = 4 µm.
FIGURE 6.
Transcript length and transcriptional state influence mRNA recruitment into ER-associated stress granules following arsenite stress. Representative smFISH of long transcript size-paired genes GRP94 (magenta, 2412 nt) and NCL (red, 2133 nt), and short transcript size-paired genes CCN2 (yellow, 1050 nt) and GAPDH (green, 1008 nt) mRNAs in untreated and arsenite-treated HeLa cell cultures. Where indicated, cell cultures were treated with ActD or triptolide in addition to arsenite. To evaluate the ER-association and presence of the SG protein marker G3BP1 in observed RNA granules, cells were permeabilized in digitonin-supplemented buffers to extract cytosolic contents while leaving the ER membrane intact, and costained for G3BP1 by immunofluorescence. Dotted boxes indicate regions of grayscale insets for mRNA and G3BP1 protein channels, as well as color merge, for digitonin permeabilized cells (below). DAPI nuclear stain (blue) included in all images. Full cell scale bars = 20 µm, inset scale bars = 4 µm.
Combined, the data presented in Figures 5 and 6 support a model where SG biogenesis can occur on or in immediate physical proximity to the ER regardless of an encoded signal sequence, and is disabled following transcriptional inhibition. These data are also consistent with the prior finding that transcript length is positively correlated with SG recruitment, where longer transcripts (e.g., GRP94 and NCL) were recruited to SGs and shorter transcripts (e.g., GAPDH, B2M) were refractory to SG recruitment (Khong et al. 2017). CCN2, as a short transcript that did assemble into SGs, provides an interesting case and suggests that transcriptional activation may outweigh the transcript length predictions for mRNA recruitment into SGs. As discussed below, these data also confirm prior studies reporting a broad representation of the mRNA transcriptome on the ER (Diehn et al. 2000; Lerner et al. 2003; Lerner and Nicchitta 2006; Chen et al. 2011; Reid and Nicchitta 2012, 2015a; Jan et al. 2014; Chartron et al. 2016).
DISCUSSION
Activation of the UPR, an ER-centric stress response signaling pathway, causes dramatic remodeling of translation on the ER (Stephens and Nicchitta 2008; Reid et al. 2014; Reid and Nicchitta 2015a). Via the UPR sensor/eIF2α kinase, PERK, translation initiation is reduced following UPR activation, and stress granule formation is enabled (Namkoong et al. 2018; Reich et al. 2020). Notably, however, ER-targeted mRNAs are underrepresented in SG transcriptome analyses and there are examples of ER-targeted mRNAs which are excluded from SGs (Unsworth et al. 2010; Khong et al. 2017). Here we report examples of ER-targeted mRNAs that are efficiently recruited into SGs in response to UPR activation by either DTT or arsenite stress. With the constraint of a limited number of mRNAs examined, this phenomenon was found to be gene-specific, where the signal sequence-encoding mRNAs GRP94/HSP90B1 and CCN2/CTGF were recruited into SGs whereas GRP78/BiP/HSPA5 and B2M were refractory to SG recruitment. GRP78 mRNA exclusion from SGs was expected based on its 5′ UTR internal ribosome entry site (Starck et al. 2016); the exclusion of B2M mRNA revealed interesting characteristics of SG formation, as discussed below. Unexpectedly, GRP94 and CCN2 SGs were found in close apposition to, or in direct association with, the ER membrane, suggesting the ER may serve as an important subcellular locale for SG biogenesis. Although little is currently known regarding the subcellular organization of SG dynamics, the recent finding that ER contact sites regulate PB and SG fission supports a role for the ER in SG biology (Lee et al. 2020). In this study, we also observed that transcriptional inhibition blocked DTT- and arsenite-elicited SG formation for both ER-targeted and cytoplasmic mRNAs. The sensitivity of SG biogenesis to ongoing transcription suggests that gene transcriptional state may be a relevant criterion in the process of mRNA recruitment into SGs.
The finding that ER-targeted mRNAs vary in their SG recruitment patterns indicates that, like translational status, an encoded signal sequence is not itself prognostic of mRNA recruitment into SGs. The conclusion that select ER-targeted mRNAs can assemble into SGs was somewhat unexpected, given that ER-targeted mRNAs are under-represented in SG transcriptomes and that ER markers are nearly absent in purified SGs (Khong et al. 2017; Namkoong et al. 2018). Given the limited number of RNAs assessed in the present study, further data on purified ER-associated SGs are needed to assess the representation of ER-targeted mRNAs in UPR-elicited SGs. Indeed, there are a multitude of reasons by which SGs containing ER-targeted mRNAs may have been missed in previous studies. In the case of SG transcriptome analyses, the biochemical purification methods used for SG isolation may not yield a fully representative SG population, as noted by these authors (Khong et al. 2017). Another potentially important consideration is that, at the mRNA level, ER-targeted genes, which largely encode secreted or integral membrane proteins, are expressed at relatively low levels and are more cell-type variable, as compared to housekeeping genes (Reid and Nicchitta 2012, 2015a,b). These factors may preclude detection of ER-targeted genes from purified subsets of RNA populations in a given cell type. Transcript abundance itself, however, is unlikely to be a primary driver for SG recruitment as is evident by experiments with GAPDH mRNAs (Fig. 5), which, although highly abundant, did not undergo SG assembly in the stress conditions examined here.
Bioinformatic analyses of the SG transcriptome have identified a positive correlation between SG recruitment efficiency and transcript length. This positive correlation was supported by the mRNAs examined in the current study; longer transcripts such as GRP94 and NCL mRNAs were recruited into SGs whereas shorter transcripts such as B2M and GAPDH were refractory to SG recruitment (Khong et al. 2017; Namkoong et al. 2018). Exceptions to this length-based correlation are also apparent in that MDR1 encodes long transcripts (CDS: 3843 nt), is ER-associated, and yet escapes SG recruitment (Unsworth et al. 2010), and CCN2 transcripts reported within are relatively short, ER-associated, and are efficiently recruited into SGs. One element that could explain these exceptions is UPR-induced transcriptional up-regulation; GRP94 and CCN2 mRNAs are UPR responsive, whereas MDR1 is not (Rendleman et al. 2018). This hypothesis is supported by the current finding that transcriptional inhibition prevents SG formation. Additionally, NCL is not a UPR responsive gene, but maintains high levels of transcription both at homeostasis and during stress, further supporting the hypothesis that relative transcription/export rate correlates with SG recruitment.
Although substantial progress has been made in defining the protein and RNA compositions of SGs, as well as their exchange dynamics, little is known regarding their subcellular localization. The discovery of ER contact sites that regulate RNP granule formation and fission is consistent with the view that the ER may contribute to SG biology (Lee et al. 2020). To this point, the mRNA transcriptome and translatome is broadly represented on the ER membrane, supporting the observations within of both ER-targeted and cytoplasmic mRNAs being recruited into ER-associated SGs (Diehn et al. 2000; Chen et al. 2011; Reid and Nicchitta 2012, 2015a; Jan et al. 2014; Chartron et al. 2016; Voigt et al. 2017). Furthermore, mRNAs can undergo direct (i.e., ribosome-independent) anchoring to the ER, and comprehensive proteomic screens for RNA-interacting proteins have identified a number of candidate ER integral membrane RBPs that, by virtue of their ability to localize mRNAs to the cytoplasmic surface of the ER, may assist in ER-associated SG formation (Cui et al. 2012; Jagannathan et al. 2014; Castello et al. 2016; Hsu et al. 2018; Queiroz et al. 2019; Trendel et al. 2019). Notably, one of these proteins (AEG1/MTDH) interacts with both ER-targeted and cytoplasmic-encoding mRNAs and, although lacking known RNA binding motifs, contains a large intrinsically disordered domain, a frequently identified motif believed to function in SG nucleation (Hsu et al. 2018). Other, ER-resident proteins may also contain intrinsically disordered domains or other binding motifs which could contribute to SG nucleation on the ER membrane. These observations suggest that insights into the subcellular locale(s) of SGs, their dynamic interactions with intracellular membranes, and the biochemical mechanism of membrane tethering may represent important avenues of future study.
In consideration of the criteria which may predict mRNA recruitment into SGs, the identification of two ER-targeted mRNAs whose transcription is up-regulated in response to UPR activation and which undergo recruitment into SGs prompted us to ask if de novo transcription may contribute to SG biogenesis. Using two inhibitors of transcription with different modes of action, as well as two separate stress conditions, we observed transcriptional inhibition efficiently prevented SG formation. Transcriptional inhibition did not disrupt induction of the UPR as indicated by activation of the stress sensor/endonuclease IRE1 and production of the UPR transcription factor XBP1s, indicating that the lack of SG formation under these conditions was either a direct or indirect consequence of the transcriptional block. Mechanistic links between gene transcriptional status and SG recruitment have not been widely investigated. However, a prior study examining SG formation in the context of poliovirus infection reported that ActD treatment blocked poliovirus infection-dependent SG formation (Piotrowska et al. 2010). These authors also observed that ActD treatment reduced SG formation, assayed via oligo(dT) FISH, in response to arsenite and heat shock stress, and suggested that newly synthesized and preexisting mRNAs may undergo separate and selective modes of recruitment into SGs. Bridging from this report, and noting that transcriptional up-regulation is an integral element of the UPR, the finding that transcriptional inhibition blocks mRNA recruitment into ER-associated SGs led us to favor the view that newly exported mRNAs are a preferred substrate for SG assembly. The link between SG formation and UPR activation also suggests that transcriptional status may be relevant to SG recruitment, where genes undergoing relatively high transcriptional rates, either via basal transcription or through UPR-activated transcription, represent high probability candidates for SG recruitment. Alternatively, Bounedjah et al. (2014), who also reported that ActD treatment suppresses SG formation during arsenite stress, proposed that the ActD-sensitive accumulation of HuR and TIA-1, another SG protein, in the cytoplasm blocks SG formation via promoting their disassembly. Although neither HuR nor TIA-1 are essential for SG formation (Bley et al. 2015), a role for nucleoplasmic protein translocation in SG dynamics remains an open question.
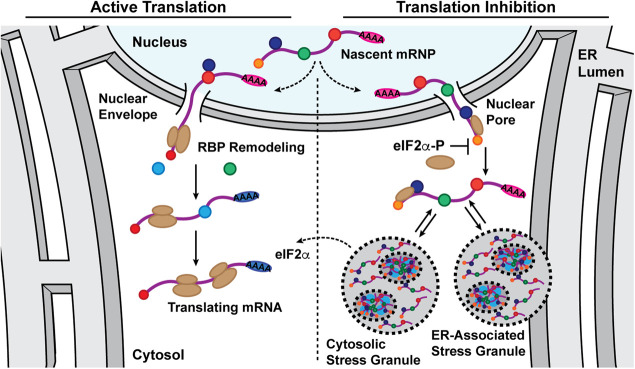
We present a working model for SG formation that emphasizes functional links between transcription, nuclear export, translation-driven mRNP remodeling, and the subcellular trafficking fate of exported mRNAs (Fig. 7; Moore 2005; Palazzo et al. 2007; Martin and Ephrussi 2009; Maquat et al. 2010; Blower 2013). As illustrated, in the homeostatic state, newly processed mRNPs undergo nuclear export-coupled translational remodeling of their RBP composition, followed by recruitment into the ribosome-engaged mRNA pool (Lejeune et al. 2002; Maquat et al. 2010; Trcek et al. 2013; Halstead et al. 2015; He and Jacobson 2015). In response to stress-elicited inhibition of translation initiation, in contrast, newly exported mRNAs are inefficiently translated and thus retain their nuclear RBP signature (Dostie and Dreyfuss 2002; Maquat et al. 2010; Matheny et al. 2020). Reflecting the relative propensity of nuclear RBPs for higher order assembly, this cohort of newly exported, translationally repressed mRNAs could then be efficiently recruited into SGs. As previously reported, CDS and overall transcript length positively contribute to this process, perhaps through proportionately higher RBP binding densities per transcript and enhanced sterically driven RNA–RNA interactions (Matherly et al. 1989; Namkoong et al. 2018; Hofmann et al. 2021). Following recovery from stress, translation-driven RBP remodeling is predicted to then confer resistance of an mRNA to subsequent SG recruitment (Piotrowska et al. 2010). By this mechanism, SGs may serve as storage depots for newly exported transcripts during stress, in particular for mRNAs comprising stress response gene programs, and subsequently as sites for translation-driven mobilization of these transcripts into the ribosome-engaged mRNA pool during recovery to cellular homeostasis. With the ER being physically continuous with the outer nuclear envelope, physical proximity may favor recruitment of newly exported mRNAs into ER-associated SGs, a view supported by recent work implicating the ER in SG and PB biogenesis (Lee et al. 2020).
FIGURE 7.
Model depicting partitioning of newly exported mRNAs between translation-engaged and stress granule-associated states in response to UPR activation. A working model depicting the functional states of newly exported mRNAs in homeostatic (active translation, low phospo-eIF2α) or stress-activated (reduced translation, elevated phospo-eIF2α) cellular states. This model highlights a role for translation-dependent RNA binding protein remodeling of newly exported mRNAs in determining mRNA recruitment into polyribosomes or SGs. As illustrated, under conditions of stress-induced translational inhibition, the nuclear RNA binding protein signature of newly exported mRNAs would be relatively long-lived and thus serve as a signal for ribonucleoprotein recruitment into stress granules, which could include both cytoplasmic and ER-associated forms. Once eIF2α phosphorylation is resolved, stress granule-associated mRNAs would resume translation, undergo RNA binding protein remodeling, and enter the polyribosome-associated pool. In this way, stress granules could serve as a triage station for newly exported mRNAs and undergo rapid mobilization following stress adaptation.
MATERIALS AND METHODS
Cell culture and treatments
HeLa cells were cultured in DMEM plus 10% FBS at 37°C in 5% CO2 and experiments performed with cultures at 70%–80% confluence. For experiments involving optical imaging, cells were seeded onto 12 mm round #1 glass coverslips. Polylysine (Sigma, Cat. No. P8920) coated glass coverslips were used for all optical imaging experiments where cells were subjected to sequential detergent fractionation prior to paraformaldehyde fixation. Where indicated, cell stress was elicited by treating cell cultures with 1–5 mM DTT or 1 mM arsenite for the noted time period (0–360 min). All DTT treatment times and concentrations were titrated for each batch of cells and DTT stock to ensure phenotypic consistency. For RNA transcription inhibitor experiments, cell cultures were treated with 10 µM actinomycin D or 30 nM triptolide for 30 min prior to stress induction and inhibitors were included in all media through the experimental time course. For translation inhibitor experiments, cell cultures were treated with 100 µg/mL cycloheximide for 10 min prior to stress induction, and cycloheximide was included in all media through the experimental time course.
[35S]-Met/Cys biosynthetic labeling
For determination of protein synthesis rates under conditions of UPR activation, cell cultures were treated with DTT for 0–6 h and pulse-labeled for 10 min at the indicated time points with 75 µCi/mL [35S]-Met/Cys in methionine- and cysteine-free culture media. At the termination of the pulse labeling period, cell cultures were washed with ice-cold PBS containing 100 µg/mL cycloheximide to block further translation and subsequently lysed on ice for 5 min in 1% Triton X-100/PBS containing 100 µg/mL cycloheximide and protease inhibitor cocktail. Detergent lysates were adjusted to 10% (vol:vol) trichloroacetic acid, samples were incubated on ice, and precipitated proteins were recovered by centrifugation (15 min, 15,000g, 4°C). Protein pellets were washed with 100% acetone to remove residual TCA, allowed to air dry, and solubilized by addition of 0.5 M Tris, pH 11, 5% SDS. [35S]-Met/Cys levels were determined by liquid scintillation counting of the resuspended protein fraction, and corresponding protein concentrations were determined by BCA assay (Pierce). Data are reported as scintillation counts per minute (CPM) per milligram (mg) of protein.
Immunoblotting
For immunoblot analysis, HeLa cell cultures were placed on ice and lysed in ice-cold 1% CHAPSO, 150 mM NaCl, 20 mM K-HEPES, pH 7.5, 10% glycerol, 1 mM EDTA, 100 mM NaF, 17.5 mM B-glycerophosphate, and 1× protease inhibitor cocktail for 5 min. Lysates were TCA precipitated as noted above, washed with 100% acetone, resuspended in 0.5 M Tris, pH 11, 5% SDS, and protein concentrations determined via BCA assay (Pierce). Equivalent protein mass per sample was separated by SDS-PAGE, transferred to nitrocellulose membranes, blocked in 10% nonfat dry milk/Tris-buffered saline and processed per antibody supplier's recommendations. Antibodies used include eIF2α (Santa Cruz, Cat. No. sc11386; 1:500 dilution) and phospho-eIF2α (Cell Signaling Technologies, Cat. No. 3398; 1:1500 dilution).
Polysome gradients, RNA extraction, and RT-qPCR analyses
Polyribosomes from cells lysed in dodecylmaltoside (DDM) lysis buffer (200 mM KCl, 25 mM K-HEPES, pH 7.2, 10 mM MgOAc2, 2 mM DTT, 50 µg/mL cycloheximide, 1× protease inhibitor cocktail, 40 U/mL RNaseOUT, and 2% DDM) were resolved on 15%–50% sucrose gradients and fractionated using a Teledyne/Isco gradient fractionator as described in Stephens and Nicchitta (Stephens and Nicchitta 2007, 2008). Total RNA was extracted from collected gradient fractions or total cell lysates by guanidinium thiocyanate-phenol-chloroform extraction and RNA concentrations determined by UV spectrometry (Chomczynski and Sacchi 2006). An equivalent mass of RNA was used for cDNA synthesis, conducted with Moloney murine leukemia virus reverse transcriptase (Promega) and random hexamers (Roche Applied Science) or an iScript cDNA Synthesis Kit (Bio-Rad). cDNA was diluted fivefold, and quantitative PCR (RT-qPCR) was performed using iQ SYBR Green Supermix (Bio-Rad) per manufacturer's protocol on a 7900HT Sequence Detection System (Applied Biosystems) or with Luna Universal qPCR Master Mix (New England Biolabs) on a CFX Connect Real-Time PCR Detection System (Bio-Rad). RT-qPCR data from polysome fractions was reported as fraction of total mRNA for the indicated gene across the entire gradient (2−Ct for a given fraction divided by the sum of 2−Ct for all fractions). RT-qPCR data from total cell RNA samples was presented as the indicated gene mRNA level relative to GAPDH mRNA level (2−ΔCt). Primers used include GRP94, CTGGAAATGAGGAACTAACAGTCA (forward) and TCTTCTCTGGTCATTCCTACACC (reverse); GRP78, CAACCAACTGTTACAATCAAGGTC (forward) and CAAAGGTGACTTCAATCTGTGG (reverse); B2M, TTCTGGCCTGGAGGCTATC (forward) and TCAGGAAATTTGACTTTCCATTC (reverse); GAPDH, TCATCAGCAATGCCTCCTGC (forward) and GATGGCATGGACTGTGGTCA (reverse); XBP1, CAGCACTCAGACTACGTGCA (forward) and ATCCATGGGGAGATGTTCTGG (reverse); XBP1-spliced, CTGAGTCCGAATCAGGTGCAG (forward) and ATCCATGGGGAGATGTTCTGG (reverse).
Cell fractionation
Sequential detergent cell fractionation was performed as described in (Jagannathan et al. 2011). Briefly, cell cultures were washed with ice-cold PBS, incubated on ice for 20 min, and permeabilized with a cytosol extraction buffer containing 110 mM KCl, 25 mM K-HEPES, pH 7.2, 2.5 mM MgOAc2, 1 mM EGTA, 1 mM DTT, 50 µg/mL cycloheximide, 1× protease inhibitor cocktail, 40 U/mL RNaseOUT, and 0.015% digitonin for 10 min on ice. Digitonin-treated cells were then washed in an identical buffer with the digitonin concentration reduced to 0.004%. The ER membrane was subsequently solubilized by addition of a detergent extraction buffer containing 200 mM KCl, 25 mM K-HEPES, pH 7.2, 10 mM MgOAc2, 2 mM DTT, 50 µg/mL cycloheximide, 1× protease inhibitor cocktail, 40 U/mL RNaseOUT, and 2% DDM on ice for 10 min. Samples were then fixed and processed for smFISH and/or immunofluorescence as described below.
Single molecule RNA fluorescence in situ hybridization and immunofluorescence
Single molecule mRNA fluorescence in situ hybridization (smFISH) was performed with Stellaris FISH probes (LCG Biosearch Technologies) as per the manufacturer's recommendations. All reagents were prepared in DEPC-treated deionized water. Cell cultures were fixed in 3.7% paraformaldehyde in PBS for 15 min at room temperature, washed twice in PBS, and permeabilized in ice-cold 70% ethanol at 4°C. Following fixation, cells were washed in 2× saline-sodium citrate (SSC) buffer containing 10% deionized formamide (VWR) and hybridized with fluorescently labeled oligonucleotide probe mixture for 4–16 h at 37°C in hybridization buffer (2× SSC, 10% deionized formamide, 10% wt/vol dextran sulfate). At the end of the probe hybridization period, cells were washed twice for 30 min in 2× SSC plus 10% deionized formamide at 37°C with DAPI included in the second wash. Following equilibration in 2× SSC, processed coverslips were mounted onto slides using VectaShield Antifade Mounting Medium (Vector Laboratories).
Combined immunofluorescence and smFISH costaining studies were performed using the protocol recommended by LCG Biosearch Technologies for sequential Stellaris FISH and immunofluorescence for adherent cell cultures. Briefly, cell cultures were fixed as above and permeabilized in 0.1% Triton X-100/PBS for 5 min on ice, followed by two sequential washes in ice-cold PBS. For cells that had undergone sequential detergent fractionation, the Triton X-100 permeabilization step was omitted. Detergent-permeabilized cells were blocked in 1% RNAse-free UltraPure BSA (Ambion) for 1 h at room temperature, stained for 1 h in primary antibody diluted in 1% BSA/PBS, washed with PBS, stained for 30 min in secondary antibody diluted in 1% BSA/PBS, washed with PBS, and post-fixed in 100% cold methanol for 5 min on ice. Cells were then washed with 2× SSC plus 10% deionized formamide, hybridized with fluorescent smFISH probes for 4–16 h at 37°C, washed, equilibrated, and mounted as described above. Antibodies used include HuR (a gift from Dr. Jack Keene, Duke University School of Medicine), G3BP1 (Santa Cruz, Cat. No. sc-365338), PABP (a gift from Dr. Jack Keene, Duke University School of Medicine), TRAPα (Migliaccio et al. 1992), β-tubulin (Developmental Studies Hybridoma Bank, Cat. No. E7), Alexa Fluor 488 goat anti-mouse (Thermo Fisher, Cat. No. A11001), Alexa Fluor 555 goat anti-mouse (Thermo Fisher, Cat. No. A21422), Alexa Fluor 488 goat anti-rabbit (Thermo Fisher, Cat. No. A11008), Alexa Fluor 555 goat anti-rabbit (Thermo Fisher, Cat. No. A21428), and Alexa Fluor 647 goat anti-mouse (Thermo Fisher, Cat. No. A 21235).
Fluorescence imaging and image processing
All imaging experiments were performed on a DeltaVision Elite deconvolution microscope (Applied Precision) equipped with 100× NA 1.4 oil immersion objective (UPlanSApo 100XO; Olympus) and a high-resolution CCD camera (CoolSNAP HQ2; Photometrics) and conducted at the Duke Light Microscopy Core Facility. Images were acquired as Z-stacks (0.2 µm intervals) at identical exposure conditions across samples for a given protein or smFISH probe. The data were deconvolved using the SoftWoRx program (v6.1 with system-level queuing) (Applied Precision) and processed on ImageJ/FIJI software (Schindelin et al. 2012) to merge channels and pseudocolor images or Imaris 9.6 software for 3D renderings. Only linear changes were made to the brightness/contrast values of the images as required for visualization of patterns and distributions. For smFISH data, brightness/contrast settings were adjusted to ensure optimal visualization of the RNA molecules, while not altering the number of foci in a given sample.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
COMPETING INTEREST STATEMENT
David Reid is an employee and shareholder of Moderna, Inc.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to Molly Hannigan and JohnCarlo Kristofich for critical comments and feedback on the manuscript and to Alyson Hoffman for scientific contributions to the early phases of the project. This work was supported by grants from the National Institutes of Health (NIH; GM101533, GM118630, C.V.N.) and by shared instrumentation grant NIH 1S10RR027867 (DeltaVisionElite).
Author contributions: J.R.C.: conceptualization, data curation, formal analysis, investigation, methodology, visualization, writing—original draft, review, and editing. Q.C.: investigation, data curation. D.W.R.: data curation, formal analysis, software, visualization, writing—review and editing. S.J.: conceptualization, formal analysis, investigation, methodology, writing—review and editing. C.V.N.: conceptualization, formal analysis, funding acquisition, methodology, supervision, writing—original draft, review, and editing.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078858.121.
REFERENCES
- Andreev DE, O'Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, Baranov PV. 2015. Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. Elife 4: e03971. 10.7554/eLife.03971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bley N, Lederer M, Pfalz B, Reinke C, Fuchs T, Glass M, Moller B, Huttelmaier S. 2015. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res 43: e26. 10.1093/nar/gku1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD. 2013. Molecular insights into intracellular RNA localization. Int Rev Cell Mol Biol 302: 1–39. 10.1016/B978-0-12-407699-0.00001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounedjah O, Desforges B, Wu TD, Pioche-Durieu C, Marco S, Hamon L, Curmi PA, Guerquin-Kern JL, Pietrement O, Pastre D. 2014. Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res 42: 8678–8691. 10.1093/nar/gku582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Helenius J, Helenius A. 1992. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J 11: 1717–1722. 10.1002/j.1460-2075.1992.tb05223.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Horos R, Strein C, Fischer B, Eichelbaum K, Steinmetz LM, Krijgsveld J, Hentze MW. 2016. Comprehensive identification of RNA-binding proteins by RNA interactome capture. Methods Mol Biol 1358: 131–139. 10.1007/978-1-4939-3067-8_8 [DOI] [PubMed] [Google Scholar]
- Chartron JW, Hunt KC, Frydman J. 2016. Cotranslational signal-independent SRP preloading during membrane targeting. Nature 536: 224–228. 10.1038/nature19309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Jagannathan S, Reid DW, Zheng T, Nicchitta CV. 2011. Hierarchical regulation of mRNA partitioning between the cytoplasm and the endoplasmic reticulum of mammalian cells. Mol Biol Cell 22: 2646–2658. 10.1091/mbc.e11-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. 2006. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc 1: 581–585. 10.1038/nprot.2006.83 [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Walter P. 2020. The integrated stress response: from mechanism to disease. Science 368: eaat5314. 10.1126/science.aat5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87: 391–404. 10.1016/S0092-8674(00)81360-4 [DOI] [PubMed] [Google Scholar]
- Cox JS, Chapman RE, Walter P. 1997. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell 8: 1805–1814. 10.1091/mbc.8.9.1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XA, Zhang H, Palazzo AF. 2012. p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PLoS Biol 10: e1001336. 10.1371/journal.pbio.1001336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. 2012. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 4: a012286. 10.1101/cshperspect.a012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney P, Ramdas Nair A, Palmer C, Khan N, Sadler KC. 2020. Arsenic induced redox imbalance triggers the unfolded protein response in the liver of zebrafish. Toxicol Appl Pharmacol 409: 115307. 10.1016/j.taap.2020.115307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M, Eisen MB, Botstein D, Brown PO. 2000. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nat Genet 25: 58–62. 10.1038/75603 [DOI] [PubMed] [Google Scholar]
- Dostie J, Dreyfuss G. 2002. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol 12: 1060–1067. 10.1016/S0960-9822(02)00902-8 [DOI] [PubMed] [Google Scholar]
- Guillen-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlussler R, Kim K, Trussina I, Wang J, Mateju D, et al. 2020. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181: 346–361.e317. 10.1016/j.cell.2020.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead JM, Lionnet T, Wilbertz JH, Wippich F, Ephrussi A, Singer RH, Chao JA. 2015. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science 347: 1367–1671. 10.1126/science.aaa3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633. 10.1016/S1097-2765(03)00105-9 [DOI] [PubMed] [Google Scholar]
- He F, Jacobson A. 2015. Nonsense-mediated mRNA decay: degradation of defective transcripts is only part of the story. Annu Rev Genet 49: 339–366. 10.1146/annurev-genet-112414-054639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Kedersha N, Anderson P, Ivanov P. 2021. Molecular mechanisms of stress granule assembly and disassembly. Biochim Biophys Acta Mol Cell Res 1868: 118876. 10.1016/j.bbamcr.2020.118876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JC, Reid DW, Hoffman AM, Sarkar D, Nicchitta CV. 2018. Oncoprotein AEG-1 is an endoplasmic reticulum RNA-binding protein whose interactome is enriched in organelle resident protein-encoding mRNAs. RNA 24: 688–703. 10.1261/rna.063313.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan S, Nwosu C, Nicchitta CV. 2011. Analyzing mRNA localization to the endoplasmic reticulum via cell fractionation. Methods Mol Biol 714: 301–321. 10.1007/978-1-61779-005-8_19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan S, Hsu JC, Reid DW, Chen Q, Thompson WJ, Moseley AM, Nicchitta CV. 2014. Multifunctional roles for the protein translocation machinery in RNA anchoring to the endoplasmic reticulum. J Biol Chem 289: 25907–25924. 10.1074/jbc.M114.580688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Williams CC, Weissman JS. 2014. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346: 1257521. 10.1126/science.1257521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149: 753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J Cell Biol 147: 1431–1442. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151: 1257–1268. 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R. 2017. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell 68: 808–820 e805. 10.1016/j.molcel.2017.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS. 1987. Coordinated regulation of a set of genes by glucose and calcium ionophores in mammalian cells. Trends Biochem Sci 12: 20–23. 10.1016/0968-0004(87)90011-9 [DOI] [Google Scholar]
- Lee AH, Iwakoshi NN, Glimcher LH. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23: 7448–7459. 10.1128/MCB.23.21.7448-7459.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Cathey PI, Wu H, Parker R, Voeltz GK. 2020. Endoplasmic reticulum contact sites regulate the dynamics of membraneless organelles. Science 367: eaay7108. 10.1126/science.aay7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Ishigaki Y, Li X, Maquat LE. 2002. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling. EMBO J 21: 3536–3545. 10.1093/emboj/cdf345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner RS, Nicchitta CV. 2006. mRNA translation is compartmentalized to the endoplasmic reticulum following physiological inhibition of cap-dependent translation. RNA 12: 775–789. 10.1261/rna.2318906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, Nicchitta CV. 2003. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA 9: 1123–1137. 10.1261/rna.5610403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xu J, Li F, Chaudhary SC, Weng Z, Wen J, Elmets CA, Ahsan H, Athar M. 2011. Unfolded protein response signaling and MAP kinase pathways underlie pathogenesis of arsenic-induced cutaneous inflammation. Cancer Prev Res 4: 2101–2109. 10.1158/1940-6207.CAPR-11-0343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE, Tarn WY, Isken O. 2010. The pioneer round of translation: features and functions. Cell 142: 368–374. 10.1016/j.cell.2010.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. 2009. mRNA localization: gene expression in the spatial dimension. Cell 136: 719–730. 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateju D, Eichenberger B, Voigt F, Eglinger J, Roth G, Chao JA. 2020. Single-molecule imaging reveals translation of mRNAs localized to stress granules. Cell 183: 1801–1812.e1813. 10.1016/j.cell.2020.11.010 [DOI] [PubMed] [Google Scholar]
- Matheny T, Van Treeck B, Huynh TN, Parker R. 2020. RNA partitioning into stress granules is based on the summation of multiple interactions. RNA 27: 174–189. 10.1261/rna.078204.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matherly LH, Schuetz JD, Westin E, Goldman ID. 1989. A method for the synchronization of cultured cells with aphidicolin: application to the large-scale synchronization of L1210 cells and the study of the cell cycle regulation of thymidylate synthase and dihydrofolate reductase. Anal Biochem 182: 338–345. 10.1016/0003-2697(89)90605-2 [DOI] [PubMed] [Google Scholar]
- Migliaccio G, Nicchitta CV, Blobel G. 1992. The signal sequence receptor, unlike the signal recognition particle receptor, is not essential for protein translocation. J Cell Biol 117: 15–25. 10.1083/jcb.117.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. 2005. From birth to death: the complex lives of eukaryotic mRNAs. Science 309: 1514–1518. 10.1126/science.1111443 [DOI] [PubMed] [Google Scholar]
- Namkoong S, Ho A, Woo YM, Kwak H, Lee JH. 2018. Systematic characterization of stress-induced RNA granulation. Mol Cell 70: 175–187.e178. 10.1016/j.molcel.2018.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oslowski CM, Urano F. 2011. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol 490: 71–92. 10.1016/B978-0-12-385114-7.00004-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. 2016. The integrated stress response. EMBO Rep 17: 1374–1395. 10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Springer M, Shibata Y, Lee CS, Dias AP, Rapoport TA. 2007. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol 5: e322. 10.1371/journal.pbio.0050322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska J, Hansen SJ, Park N, Jamka K, Sarnow P, Gustin KE. 2010. Stable formation of compositionally unique stress granules in virus-infected cells. J Virol 84: 3654–3665. 10.1128/JVI.01320-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, Parker R. 2016. Principles and properties of stress granules. Trends Cell Biol 26: 668–679. 10.1016/j.tcb.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiroz RML, Smith T, Villanueva E, Marti-Solano M, Monti M, Pizzinga M, Mirea DM, Ramakrishna M, Harvey RF, Dezi V, et al. 2019. Comprehensive identification of RNA-protein interactions in any organism using orthogonal organic phase separation (OOPS). Nat Biotechnol 37: 169–178. 10.1038/s41587-018-0001-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich S, Nguyen CDL, Has C, Steltgens S, Soni H, Coman C, Freyberg M, Bichler A, Seifert N, Conrad D, et al. 2020. A multi-omics analysis reveals the unfolded protein response regulon and stress-induced resistance to folate-based antimetabolites. Nat Commun 11: 2936. 10.1038/s41467-020-16747-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Nicchitta CV. 2012. Primary role for endoplasmic reticulum-bound ribosomes in cellular translation identified by ribosome profiling. J Biol Chem 287: 5518–5527. 10.1074/jbc.M111.312280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Nicchitta CV. 2015a. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat Rev Mol Cell Biol 16: 221–231. 10.1038/nrm3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Nicchitta CV. 2015b. Comment on “Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling”. Science 348: 1217. 10.1126/science.aaa7257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Chen Q, Tay AS, Shenolikar S, Nicchitta CV. 2014. The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell 158: 1362–1374. 10.1016/j.cell.2014.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendleman J, Cheng Z, Maity S, Kastelic N, Munschauer M, Allgoewer K, Teo G, Zhang YBM, Lei A, Parker B, et al. 2018. New insights into the cellular temporal response to proteostatic stress. Elife 7: e39054. 10.7554/eLife.39054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaki K, Yoshina S, Shen X, Han J, DeSantis MR, Xiong M, Mitani S, Kaufman RJ. 2012. RNA surveillance is required for endoplasmic reticulum homeostasis. Proc Natl Acad Sci 109: 8079–8084. 10.1073/pnas.1110589109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, McGeachy AM, Ingolia NT, Walter P. 2015. The small molecule ISRIB reverses the effects of eIF2α phosphorylation on translation and stress granule assembly. Elife 4: e05033. 10.7554/eLife.05033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava RK, Li C, Chaudhary SC, Ballestas ME, Elmets CA, Robbins DJ, Matalon S, Deshane JS, Afaq F, Bickers DR, et al. 2013. Unfolded protein response (UPR) signaling regulates arsenic trioxide-mediated macrophage innate immune function disruption. Toxicol Appl Pharmacol 272: 879–887. 10.1016/j.taap.2013.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P. 2016. Translation from the 5′ untranslated region shapes the integrated stress response. Science 351: aad3867. 10.1126/science.aad3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens SB, Nicchitta CV. 2007. In vitro and tissue culture methods for analysis of translation initiation on the endoplasmic reticulum. Methods Enzymol 431: 47–60. 10.1016/S0076-6879(07)31004-5 [DOI] [PubMed] [Google Scholar]
- Stephens SB, Nicchitta CV. 2008. Divergent regulation of protein synthesis in the cytosol and endoplasmic reticulum compartments of mammalian cells. Mol Biol Cell 19: 623–632. 10.1091/mbc.e07-07-0677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Kedersha N. 2013. Relationship of GW/P-bodies with stress granules. Adv Exp Med Biol 768: 197–211. 10.1007/978-1-4614-5107-5_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi S, Miyake M, Tsugawa K, Oyadomari M, Oyadomari S. 2016. Integrated stress response of vertebrates is regulated by four eIF2α kinases. Sci Rep 6: 32886. 10.1038/srep32886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titov DV, Gilman B, He QL, Bhat S, Low WK, Dang Y, Smeaton M, Demain AL, Miller PS, Kugel JF, et al. 2011. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat Chem Biol 7: 182–188. 10.1038/nchembio.522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Sato H, Singer RH, Maquat LE. 2013. Temporal and spatial characterization of nonsense-mediated mRNA decay. Genes Dev 27: 541–551. 10.1101/gad.209635.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trendel J, Schwarzl T, Horos R, Prakash A, Bateman A, Hentze MW, Krijgsveld J. 2019. The human RNA-binding proteome and its dynamics during translational arrest. Cell 176: 391–403.e319. 10.1016/j.cell.2018.11.004 [DOI] [PubMed] [Google Scholar]
- Unsworth H, Raguz S, Edwards HJ, Higgins CF, Yague E. 2010. mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J 24: 3370–3380. 10.1096/fj.09-151142 [DOI] [PubMed] [Google Scholar]
- Voigt F, Zhang H, Cui XA, Triebold D, Liu AX, Eglinger J, Lee ES, Chao JA, Palazzo AF. 2017. Single-molecule quantification of translation-dependent association of mRNAs with the endoplasmic reticulum. Cell Rep 21: 3740–3753. 10.1016/j.celrep.2017.12.008 [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334: 1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Wang S, Kaufman RJ. 2012. The impact of the unfolded protein response on human disease. J Cell Biol 197: 857–867. 10.1083/jcb.201110131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC. 2018. Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb Perspect Biol 10: a032870. 10.1101/cshperspect.a032870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. 2006. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans 34: 7–11. 10.1042/BST0340007 [DOI] [PubMed] [Google Scholar]
- Wolozin B, Ivanov P. 2019. Stress granules and neurodegeneration. Nat Rev Neurosci 20: 649–666. 10.1038/s41583-019-0222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, et al. 2020. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 181: 325–345.e328. 10.1016/j.cell.2020.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JY, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, et al. 2018. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell 69: 517–532.e511. 10.1016/j.molcel.2017.12.020 [DOI] [PubMed] [Google Scholar]
- Young SK, Wek RC. 2016. Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J Biol Chem 291: 16927–16935. 10.1074/jbc.R116.733899 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.