Abstract
Human pre-mRNA splicing is primarily catalyzed by the major spliceosome, comprising five small nuclear ribonucleoprotein complexes, U1, U2, U4, U5, and U6 snRNPs, each of which contains the corresponding U-rich snRNA. These snRNAs are encoded by large gene families exhibiting significant sequence variation, but it remains unknown if most human snRNA genes are untranscribed pseudogenes or produce variant snRNAs with the potential to differentially influence splicing. Since gene duplication and variation are powerful mechanisms of evolutionary adaptation, we sought to address this knowledge gap by systematically profiling human U1, U2, U4, and U5 snRNA variant gene transcripts. We identified 55 transcripts that are detectably expressed in human cells, 38 of which incorporate into snRNPs and spliceosomes in 293T cells. All U1 snRNA variants are more than 1000-fold less abundant in spliceosomes than the canonical U1, whereas at least 1% of spliceosomes contain a variant of U2 or U4. In contrast, eight U5 snRNA sequence variants occupy spliceosomes at levels of 1% to 46%. Furthermore, snRNA variants display distinct expression patterns across five human cell lines and adult and fetal tissues. Different RNA degradation rates contribute to the diverse steady state levels of snRNA variants. Our findings suggest that variant spliceosomes containing noncanonical snRNAs may contribute to different tissue- and cell-type–specific alternative splicing patterns.
Keywords: snRNA, snRNA biogenesis, snRNA variants, spliceosomes, pre-mRNA splicing
INTRODUCTION
Intron removal is a key step in the maturation of eukaryotic messenger RNAs (mRNAs), and the generation of multiple transcript isoforms through alternative splicing enables an expanded proteome (Nilsen and Graveley 2010; Zheng and Black 2013; Ule and Blencowe 2019). Human pre-mRNA splicing is primarily catalyzed by the major spliceosome, a complex macromolecular assemblage that is constructed de novo in a step-wise manner on each intron (Fig. 1A; Kastner et al. 2019; Wilkinson et al. 2020). The core of the spliceosome consists of five small nuclear ribonucleoprotein complexes (snRNPs), U1, U2, U4, U5, and U6, each of which contains the corresponding U-rich snRNA as well as common and snRNP-specific proteins. Many additional proteins associate with the pre-mRNA and snRNPs to execute intron removal. U1 and U2 snRNAs initially base-pair with the 5′ splice site (5SS) and the branch site (BS) upstream of the 3′ splice site (3SS), respectively. The U4/U6.U5 tri-snRNP then joins U1 and U2, whereupon rearrangements lead to a catalytically active spliceosome and removal of the intron (Fig. 1A). In a subset of eukaryotes, including humans, a much less abundant “minor” spliceosome recognizes <1% of pre-mRNA introns. Minor introns are characterized by noncanonical splice sites that are recognized by divergent sequence paralogs of the major snRNAs, termed U11, U12, U4atac, and U6atac (Patel and Steitz 2003; Padgett 2012; Turunen et al. 2013). U5 is the only snRNA that is shared between the major and minor spliceosomes. In both major and minor spliceosomes, conserved snRNA sequence motifs direct RNA:RNA and RNA:protein interactions required for splicing.
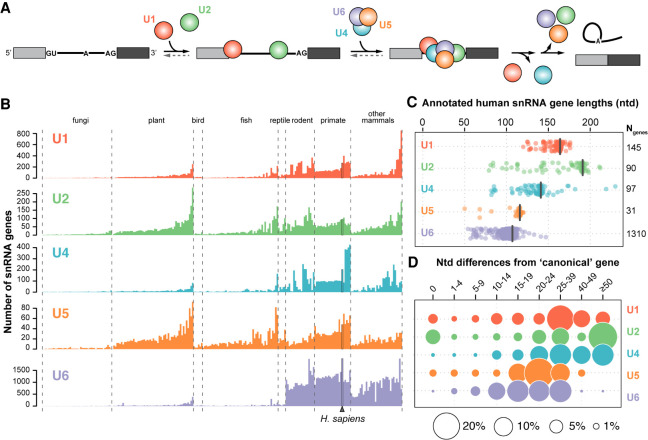
FIGURE 1.
The human genome contains more than a thousand snRNA gene variants. (A) Simplified splicing schematic. Shown are two exons (filled rectangles) flanking an intron (black line). The U1 snRNP (red) binds the 5′ splice site (GU) and the U2 snRNP (green) binds the branch site (A), upstream of the 3′ splice site (AG). The U4/U6.U5 tri-snRNP (blue, purple, orange) then joins and the U1 and U4 snRNPs are released upon catalytic activation. The intron is excised as a branched “lariat” concomitant with exon ligation and the snRNPs are recycled. For simplicity, non-snRNP splicing factors are not shown. (B) Number of snRNA genes per haploid genome identified based on a computational genome annotation and comparison of sequence homology with the main snRNA genes. Only genes annotated in both the Ensembl (Zerbino et al. 2018) and Rfam (Kalvari et al. 2018) databases were included. Genomes within each clade are sorted according to their average number of snRNA genes, excluding U6. The number of genomes analyzed in each clade is: fungi (46), plant (54), bird (6), fish (50), reptile (5), rodent (19), primate (24), and other mammals (34). (C) Distribution of annotated human snRNA gene lengths. The length in nucleotides of the canonical snRNA of each type is indicated by a black bar and corresponds to: U1 (164), U2 (191), U4 (141), U5 (116), U6 (107). The total number of genes per snRNA family are noted on the right. (D) Number of nucleotide mismatches or indels compared to the canonical snRNA. Differences were calculated according to a Needleman–Wunsch alignment (Needleman and Wunsch 1970). A nucleotide difference of zero indicates snRNA genes with the canonical sequence. Circle area is proportional to the percentage of genes for a given type of snRNA.
U1, U2, U4, and U5 snRNA genes and their minor spliceosome paralogs are transcribed by RNA polymerase II (RNAP II), directed by a set of snRNA gene-specific transcription factors (Hernandez 2001; Guiro and Murphy 2017). During synthesis, pre-snRNAs are 5′-capped, 3′-trimmed, and bound by factors that promote efficient nuclear export (Matera and Wang 2014; Gruss et al. 2017). Delivery to the cytoplasm sets in motion several steps of snRNP biogenesis, including the addition of a ring formed of seven different Sm proteins by the SMN complex, 7-methylguanosine cap hypermethylation to a 2,2,7-trimethylguanosine (TMG) cap, and further trimming of the 3′ end (Matera and Wang 2014; Gruss et al. 2017). Together, the Sm ring and TMG cap function to direct the pre-snRNP back into the nucleus for final 3′-trimming (Fischer et al. 2011). RNAP III-made U6 snRNAs receive different 5′ and 3′ modifications and were not thought to be actively exported from the nucleus (Didychuk et al. 2018), although a recent study suggests otherwise (Becker et al. 2019). U6 snRNAs are loaded with a paralogous Lsm (“like Sm”) ring instead of an Sm ring. Despite these distinct biogenesis pathways, final maturation steps for all snRNAs occur in the nuclear Cajal bodies, where remaining snRNP-specific proteins are loaded, base and ribose modifications occur, and the U4/U6 di-snRNP and U4/U6.U5 tri-snRNP complexes are formed prior to spliceosome assembly (Matera and Wang 2014).
Prior studies showed that the abundances of protein components of the splicing machinery are highly variable across human tissues (Grosso et al. 2008), and that changing the expression level of individual proteins can affect splicing in an intron-specific manner (Tejedor et al. 2015; Han et al. 2017). Similarly, recent work demonstrated that depleting individual snRNAs in vivo leads to altered splice-site selection or reduced splicing efficiency for specific introns, rather than a global decrease in splicing transcriptome-wide (Dvinge et al. 2019). The resulting splicing profiles were specific to each snRNA, suggesting that splice sites are not uniformly sensitive to the cellular abundance of the individual snRNPs. This finding directly implicates snRNA abundance as an additional level of alternative splicing regulation.
The major spliceosomal snRNAs are encoded by multigene families, comprising expressed snRNA sequence variants and putatively untranscribed retroposon-like pseudogenes (Denison et al. 1981; Berstein et al. 1985; Dahlberg and Lund 1988). Studies in mice, frogs, and humans found expression changes of the canonical U1 and multiple U1 variant genes during cellular differentiation and development (Lund et al. 1985, 1987; O'Reilly et al. 2013). Furthermore, these changes have recently been implicated in regulating developmental gene expression (Vazquez-Arango et al. 2016). Likewise, it has been found that multiple U5 variants are expressed in human cells (Krol et al. 1981; Sontheimer and Steitz 1992) and can be assembled into ribonucleoprotein complexes (Sontheimer and Steitz 1992). In other organisms, U5 snRNA variants have also been found to change in expression level during cellular differentiation and development in a cell-type–specific manner (Morales et al. 1997; Chen et al. 2005; Lu and Matera 2015). Additional evidence has emerged suggesting that snRNAs have cell-type–specific functions (Jia et al. 2012; Ishihara et al. 2013), and that alterations in snRNA stoichiometry can cause mRNA splicing defects (Zhang et al. 2008). It has been suggested that low abundance human snRNA sequence variants can form functional “variant snRNPs” (Vazquez-Arango and O'Reilly 2017).
Despite these findings, it remains unknown if the majority of human snRNA genes are simply inactive pseudogenes or whether they have the potential to influence alternative splicing. Splicing may be impacted either by gain or loss of function imposed by snRNA variants. The latter could include variants that are incorporated into snRNPs but are unable to assemble into spliceosomes, thus sequestering splicing factors, or that cannot undergo all of the conformational changes and interactions required during splicing, thereby arresting the splicing cycle (Dvinge 2018). Even low abundance variant snRNAs may influence cellular splicing programs by being preferentially recruited to specific introns, analogous to the minor spliceosomal snRNAs.
In this study, we systematically profiled human U1, U2, U4, and U5 snRNA variants, including expression level, RNA stability, subcellular localization, and incorporation into snRNPs and spliceosomes. We used available RNA-seq data on small noncoding RNAs from numerous human cell lines to identify 55 expressed variant snRNA genes. Although most expressed snRNA variants are detected in the cytosol, a subset either remains in the nucleus or is rapidly degraded in the cytosol. All snRNA variants that are detected in the cytoplasm associate with Sm proteins; however, some are less efficiently incorporated into mature snRNPs. The majority of expressed variants assemble into mature snRNPs and spliceosomes, although often at very low levels reflective of their low abundance. In contrast, we observed more comparable abundances of eight U5 gene variants in snRNPs and spliceosomes. Furthermore, we demonstrated that snRNA variant genes are variably expressed in human cell lines and display distinct expression in pooled adult and fetal human tissues. Our findings suggest that variant snRNAs may contribute to differential tissue- and cell-type–specific splicing patterns, which may have implications for human development and disease.
RESULTS
snRNA gene number, length, and sequence vary greatly among eukaryotes
The number of snRNA genes identified across eukaryotes varies by orders of magnitude (Fig. 1B). Many fungi have only a single locus for each snRNA, but large snRNA gene families are found in plants and animals (Egeland et al. 1989; Mount et al. 2007; Marz et al. 2008). This expansion of snRNA genes is particularly evident in mammals, especially for U6 genes (Hayashi 1981). Interestingly, a subclade of New World monkeys (Supplemental Fig. 1A) has about fourfold more U4 genes than most other primates and twofold more than humans, without a concerted increase in the number of U6 genes (Fig. 1B). The mechanism of this expansion of U4 genes is unknown.
Most snRNA genes have been identified using computational approaches and annotated as being pseudogenes due to sequence differences. However, only a small number have been manually evaluated or studied experimentally (Denison et al. 1981; O'Reilly et al. 2013; Vazquez-Arango et al. 2016; Kosmyna et al. 2020). Here, we refer to the most abundantly expressed and biochemically studied snRNA within each family as the “canonical” snRNA, and the remaining snRNAs as “variants.” Since U5A and U5B are expressed at similar levels (Krol et al. 1981), we arbitrarily specify U5A as canonical.
The human genome contains 1674 individual snRNA genes annotated as either U1, U2, U4, U5, or U6 (Supplemental Table S1), which are spread throughout the chromosomes as either tandem arrays or single genes (Supplemental Fig. 1B). U6 snRNA genes are the most abundant in vertebrate genomes, although many are 3′ truncated and may originate from chimeric fusions with the LINE-1 transposable element or other transcripts (Buzdin et al. 2002; Gilbert et al. 2005; Doucet et al. 2015; Moldovan et al. 2019). The length of the predicted snRNA coding regions varies substantially relative to each canonical snRNA (Fig 1C), suggesting that many of the variant genes are truncated and/or incorporated into the genome as fusion genes with other transcripts. The divergence from the canonical sequence ranges from a single nucleotide to more than 50 nt difference across genes of a given snRNA (Fig. 1D). In the case of U2 snRNA variant genes, the high rate of nucleotide divergence is due primarily to the presence of numerous truncated variant U2 genes (Fig. 1C,D).
Published RNA-seq data suggest many variant snRNA genes are expressed
Owing to their short length and lack of polyA tails, snRNAs are not detected by many RNA-seq protocols. To identify expressed snRNA variants in existing data sets, we focused on studies that specifically targeted short noncoding RNAs, which assayed 59 cell lines from the NCI-60 panel (Marshall et al. 2017) and 34 cell lines from the ENCODE consortium (Djebali et al. 2012). The U6 genes were not considered for our analysis, owing to their large number and high degree of sequence similarity. Two stringent criteria were used for RNA-seq snRNA variant mapping. First, all reads that aligned to more than one variant were discarded, since their source could not be determined. Second, only reads that aligned without mismatches were considered, to provide the highest confidence in snRNA variant detection. The alignments were combined across samples for each genomic locus containing an snRNA gene and all results were manually assessed for correct read alignment (Supplemental Fig. 2).
The uniformity and coverage of RNA-seq reads across a given gene locus depend on its expression level and the degree of sequence diversity. Many U5 snRNA variants have nucleotide variations that span most of their 120-nt length. Therefore, uniquely mapped reads could often be identified throughout the gene body, as exemplified by RNU5D-1 (Supplemental Fig. 2D, asterisk). In contrast, the transcribed U1 genes vary at fewer nucleotide positions. Thus, the uniquely mapped reads were often located within a single region within the gene, as seen in RNU1-11P (Supplemental Fig. 2A, asterisk). Variants that displayed limited unique sequence mapping and/or very low RPMs were considered unreliable and were excluded from further study (e.g., U1-14P, U2-7P, and U4-4P, Supplemental Fig. 2). Since the RNA-seq data varied considerably both in terms of number of samples per cell line and sequencing depth for each sample, we do not anticipate that the mapped reads are a precise indicator of the relative abundance of individual variants, nor a definitive answer to whether some snRNA variants are expressed. In total, 92 expressed snRNA variants were provisionally identified by RNA-seq.
RT-qPCR confirms expression of a diverse collection of variant snRNAs
In order to validate and more accurately quantify the expression of variant snRNAs, we performed reverse transcription and polymerase chain reaction (RT-PCR) on pooled total RNA from thirteen human cell lines (see Materials and Methods) using primers specific for most of the snRNA variants detected by RNA-seq (Supplemental Table S2). Due to high sequence similarity and the potential for cross-hybridization, all PCR amplicons were validated by Sanger sequencing (Supplemental Fig. 3). For several snRNA variants that had RNA-seq reads it was not possible to design specific primers that could be validated by sequencing, as the only sequence variation would be within the primer. In total, we identified 55 unique sequence variants expressed in human cell lines, while 24 variants detected by RNA-seq showed no discernable expression (7) or amplified the wrong sequence (17) by RT-PCR (Supplemental Table S2).
Since expression of snRNA variants may differ in cell lines compared to in vivo, we measured the relative expression of the five canonical major snRNAs, four minor snRNAs, 15 U1 variants, 16 U2 variants, five U4 variants, and 19 U5 variants across a pool of healthy adult or fetal human tissues by quantitative RT-PCR (RT-qPCR; Fig. 2A). We found that all snRNA variants identified in cell lines were expressed in the sampled human tissues, with the exception of U5F-6P and U5F-8P in the adult pool. Relative to control noncoding RNA genes (5S rRNA, 7SK, and 7SL RNA), significant differences in the expression of variant snRNAs are observed between fetal and adult tissues (Fig. 2B). For example, U2-70P and U5A-8P are expressed greater than or equal to fivefold higher in adult tissues, while U2-3P, U5F-6P, and U5F-8P are expressed greater than fivefold higher in fetal tissues. This finding suggests developmental regulation of snRNA variants in humans, as has been previously suggested in other organisms (Lund et al. 1985, 1987; Chen et al. 2005; Lu and Matera 2015) and for a handful of human U1 variants (O'Reilly et al. 2013; Vazquez-Arango et al. 2016). We conclude that snRNA variants are expressed in human tissues and a subset of variants display differential fetal and adult expression profiles, suggestive of potential developmental- and/or tissue-specific function.
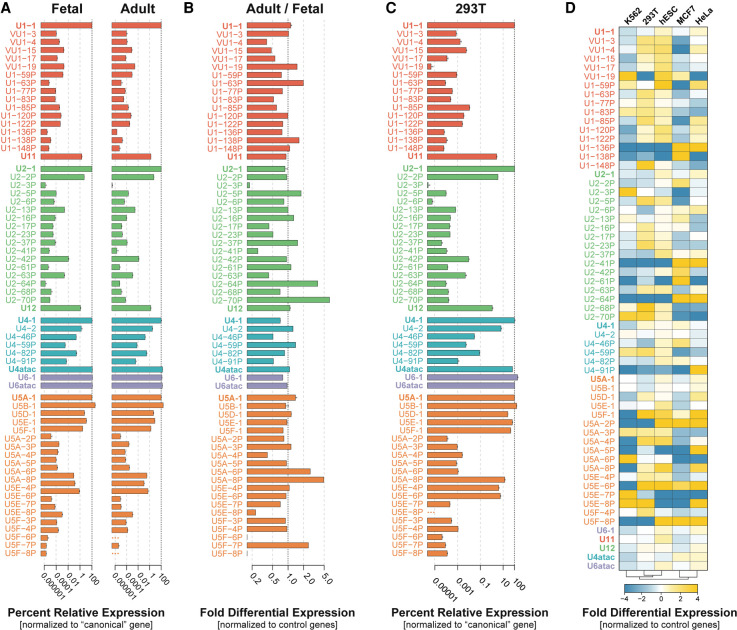
FIGURE 2.
Multiple snRNA gene variants are transcribed in human cells. (A) Expression of snRNA variants in pooled adult and fetal tissues relative to the respective canonical gene (2−ΔCt) using RT-qPCR. U6 snRNA was normalized to canonical U4 snRNA. Error bars represent the standard error of the mean (SEM) of three technical replicates. The x-axis is log10 scaled. (B) Fold differential expression of snRNA variants in pooled adult versus fetal tissues. Normalized ratios of expression between adult and fetal tissues are displayed (2−ΔΔCt). Ratios >1 indicate greater relative expression of the snRNA in adult tissues. Ratios <1 indicate greater relative expression in fetal tissues. (C) Expression of snRNA variants in 293T cells relative to the respective canonical gene (2−ΔCt) using RT-qPCR. U6 snRNA was normalized to canonical U4 snRNA. (See Supplemental Fig. 4A for expression of a subset of snRNAs relative to U1-1.) Error bars represent the standard error of the mean (SEM) of three technical replicates from human 293T cell RNA. The x-axis is log10 scaled. (D) Heat map of the fold differential expression of the snRNA genes listed at left across the human cell lines listed at top, as determined by RT-qPCR and indicated by the color scale below. Cell lines were hierarchically clustered based on their snRNA expression profiles (bottom). Three technical replicates were run for each sample.
To see if snRNA variant gene expression in a model human cell line is similar to what we observed in pooled tissues, we measured the relative expression levels of snRNA variants in human 293T cells by RT-qPCR (Fig. 2C). Most snRNA variants are expressed at levels two- to fourfold higher in 293T cells than in adult and fetal tissues, with the exception of vU1-19 and U2-2P, which are expressed at levels more than twofold lower. Most of the canonical snRNA genes, both major and minor, are expressed at comparable levels between adult and fetal tissues and 293T cells, except for U6 snRNA, which is expressed fourfold higher in 293T cells. As for the pooled tissues, expression of U1 snRNA in 293T cells is dominated by the canonical sequence, presumably due in part to gene dosage as seven U1 genes encode canonical U1 snRNA (Supplemental Fig. 4A). All U1 variants combined account for <0.1% of the cellular level of U1 snRNA in 293T cells. By comparison, the minor spliceosome U1 paralog, U11, is expressed at 1.6% of the level of canonical U1. In contrast, U2 and U4 each have a variant, U2-2P and U4-2, respectively, that is expressed at 4%–8% of the level of the canonical snRNA (Fig. 2C). All other U2 and U4 variants together were expressed at <1% of total cellular levels of each snRNA gene. All four minor snRNA genes are expressed at between 0.6% and 2.3% the level of U1, which corresponds to roughly 10,000 transcripts per cell (Fig. 2C; Supplemental Fig. 4A). Surprisingly, U4-1 is expressed at the same level as U11 and is only about twice as abundant as U4atac. This low level of U4-1 is consistent with its single gene copy, compared to ≥5 gene copies for U1-1, U2-1, and the major species of U5 (Supplemental Fig. 4A), and with the two- to threefold excess of U6 over U4 observed in HeLa and 293T cell nuclear extract (Brow and Vidaver 1995; see Supplemental Fig. 7D).
Consistent with previous findings in HeLa cells (Sontheimer and Steitz 1992), 293T cells express five dominant U5 snRNA variants (A-1, B-1, D-1, E-1, and F-1) each ranging in abundance from 7%–35% of total U5 levels. However, we identified the expression of three novel U5 variants, U5A-8P, U5E-4P, and U5E-6P, that together account for 6% of the total cellular U5 (Fig. 2C; Supplemental Fig. 4A). These variants are expressed at levels that are comparable to minor spliceosomal snRNAs and so could potentially also carry out specialized functions.
To gain insight into transcriptional regulation of snRNA variants, we examined the conservation of known promoter elements. snRNA promoters consist of distal- and proximal- sequence elements (DSE/PSE) (Dergai and Hernandez 2019), which are bound by a distinct set of transcription factors that recruit and stabilize RNAP II (Guiro and Murphy 2017). We found that the expressed variant snRNAs showed stronger sequence conservation of the PSE than the DSE (Supplemental Fig. 6A–D). This finding is in agreement with the known function of the PSE in binding the snRNA-specific SNAPc transcription factor, which recruits the general transcription initiation factors required for basal levels of RNAP II transcription (Dergai and Hernandez 2019). In contrast, the DSE acts to enhance the stability of the SNAPc complex on the PSE by associating with activator proteins POU2F1, ZNF143, and SP1 (Dergai and Hernandez 2019). Consistently, we found that variants with a more conserved DSE exhibited more robust expression in 293T cells (Supplemental Fig. 6A–D). These data are in agreement with a previous study in which variant gene promoters lacking identifiable DSEs could still associate with RNAP II, but at much lower levels, and did not show discernable association with transcriptional activators (Faresse et al. 2012).
To qualitatively assess the promoter conservation and expression level, we plotted the rank mean promoter conservation score (mean PSE and DSE rank) versus the rank expression as compared to the canonical snRNA gene (Supplemental Fig. 6E). Plotting either the mean PSE or DSE conservation score versus the rank expression did not clearly show that one was more important than the other. U4 and U5 snRNA gene promoter conservation correlates most strongly with expression level (R = 0.8–0.9). The correlation for U1 and U2 is much weaker (R = 0.4, Supplemental Fig. 6E). Thus, promoter strength likely contributes to, but is not wholly responsible for, the differing levels of variant snRNAs.
snRNA variants are differentially expressed across common human cell lines
It was previously shown that the levels of the canonical snRNAs are not constant across different human tissues (Dvinge et al. 2019). To see if the expression of snRNA variants is similarly variable, we measured their abundance and that of the canonical major and minor snRNAs in five different human cell lines: HeLa (cervical adenocarcinoma), 293T (embryonic kidney), K562 (chronic myelogenous leukemia), hESCs (human embryonic stem cells), and MCF7 (mammary gland adenocarcinoma) (Fig. 2D). This analysis revealed that many of the variants exhibit differential expression across the cell lines relative to three reference noncoding RNAs. Strikingly, numerous variant snRNA genes are differentially expressed by up to 16-fold across these cell lines, including many of the U5 snRNA variants. The canonical major snRNA genes exhibited the smallest degree of variation in expression across all cell lines (less than or equal to twofold) but, due to their high expression levels, these small fractional changes could be biologically significant (Dvinge et al. 2019). Interestingly, we find that hESCs appear to have the highest overall expression of snRNA variants relative to the other cell lines examined. This observation has previously been reported for a handful of U1 snRNA variants in hESCs compared to HeLa cells (O'Reilly et al. 2013). Our data provide further evidence that snRNA variant genes may help tailor the transcriptome in a cell-type-specific manner.
To look for potential coordinate expression of snRNAs, we next asked which snRNA variants exhibit similar changes in expression across different human cell lines by hierarchical clustering of the relative expression data (Supplemental Fig. 4B). This analysis identified three statistically significant clusters based on the similar expression of each snRNA across all cell types. The first cluster includes the most highly expressed U1-1 and U2-1 snRNAs, along with canonical U5A-1 and two U5 snRNA variants, U5B-1 and U5E-1 (P < 0.001) (Supplemental Fig. 4B, red bar). These variants exhibit similar low fold-expression changes across all of the cell lines (Fig. 2D). The second cluster of snRNA genes includes the remaining major U4 and U6 snRNAs, all minor spliceosomal snRNAs and three U5 snRNA variants (U5D-1, U5A-8P, and U5E-6P) (P < 0.001) (Supplemental Fig. 4B, orange bar). The third cluster contains two sub-clusters: snRNA genes that display the most dramatic change in expression across all cell lines (e.g., U5F-1, U5E-6P, and multiple RNVU1s) (P < 0.001) (Supplemental Fig. 4B, yellow bar), and another composed of the very low abundance or nonexpressed snRNA genes (P > 0.1) (Supplemental Fig. 4B, gray bar). The apparent coregulation of snRNA genes suggests the potential for coordinated gene expression programs.
Expressed snRNA variant genes exhibit sequence variation in functional domains
Sequence alignments of all the expressed snRNA variants we identified reveals interesting patterns of sequence conservation (Supplemental Fig. 5; Supplemental Table S3). In general, the 5′ half of each mature snRNA is more conserved than the 3′ half. In part, this is due to the fact that some genes are 3′-truncated, including several U2 and U4 genes. Many of the nucleotide differences that are found within snRNA variants appear in single-stranded and bulged regions of the snRNA (Supplemental Fig. 5). Key residues in sequence motifs important for RNA:protein or RNA:RNA interactions during snRNP assembly and function are substituted in a subset of snRNA variants. For example, nearly two-thirds of the U1 variants have one or more nucleotide substitutions in the 5′ splice-site recognition sequence or in sequences in Stem loops I and II that interact with the U1 snRNP proteins U1-70K and U1-A, respectively (Fig. 3A; Supplemental Fig. 5A; Scherly et al. 1989; Surowy et al. 1989;). Variants that have 5′ splice site recognition sequence deviations could recognize atypical or cryptic 5′ splice sites that may be important for alternative splicing of certain pre-mRNA substrates or contribute to splicing related disease phenotypes (Zhuang and Weiner 1986; Kyriakopoulou et al. 2006; Roca and Krainer 2009; Shuai et al. 2019; Suzuki et al. 2019).
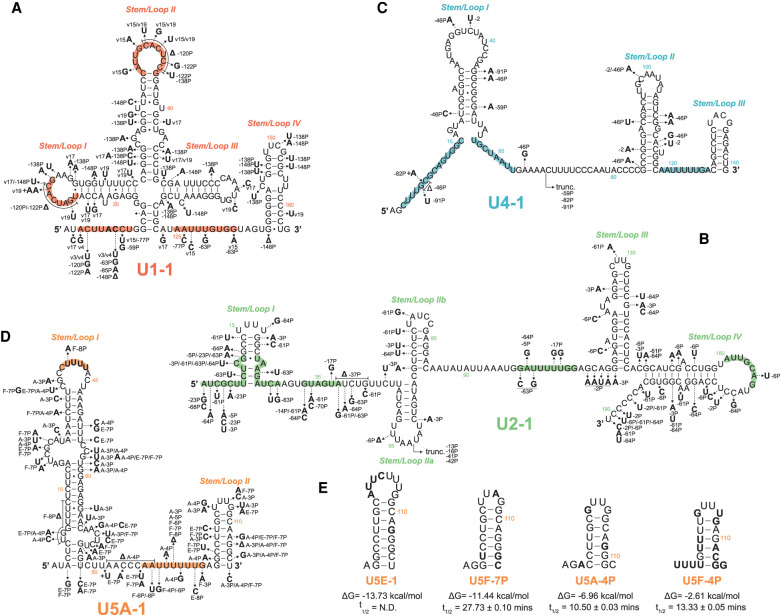
FIGURE 3.
Expressed snRNA variant genes exhibit sequence variation in RNA:RNA and RNA:protein interacting domains. Arrows show the positions of selected nucleotide variants relative to the canonical snRNA. Bold letters show the substituted base, followed by the variant snRNA(s) in which the substitution occurs. Colored regions within each structure are known functional sequences further described in Supplemental Figure 5. snRNA genes: (A) U1, (B) U2, (C) U4, (D) U5. (E) Predicted 3′ stem–loop structure of U5 snRNA variants with conserved or divergent base changes. Bold letters show the substituted base as compared to U5A. The free energy of formation (ΔG) and snRNA half-lives (t1/2) are shown below each snRNA variant structure. For reference, the free energy of formation for the 3′ stem of U5A-1 is −13.53 kcal/mol. Structures and free energy of folding were calculated using the Vienna RNAfold webserver (rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi).
The U2 snRNA variant gene family appears to be the most divergent in mature snRNA length and sequence identity (Fig. 1C). Multiple truncated U2 variants are expressed in human cells, U2-13P, -16P, -41P and -42P. These variants maintain a near canonical sequence from Stem Loop I up to Stem Loop IIa and therefore likely maintain the canonical 5′ U2 structure (Supplemental Fig. 5B). These truncated variants may, however, adopt different secondary structures downstream from Stem Loop IIa. Many U2 variants have one or more nucleotide changes in domains that function in pre-mRNA branchpoint recognition (BPRS), interactions with U6 snRNA, or stem structures required for splicing catalysis (BSL and stem IIa vs stem IIc) (Fig. 3B). These variations in snRNA sequence may impact 3′ splice site selection through binding of nonconsensus or cryptic pre-mRNA branch sites, or prevent the dynamic structural rearrangements that occur during spliceosome assembly and catalysis. Furthermore, most nucleotides in U2 that were shown to be important for binding SF3a, SF3b and U2-B″ (Dybkov et al. 2006) are unaltered in U2 variants, which may therefore assemble into variant U2 snRNPs.
There are two types of variant U4 snRNAs. One type is highly similar to U4-1 throughout the 146-nt length. For example, U4-2 and -46P have only 6 and 13 nt differences, respectively. The other, less abundant type is highly similar to U4-1 up to position 68 and then diverges completely in sequence, such as U4-59P, -82P, and -91P (Fig. 3C; Supplemental Fig. 5C). The latter type often have an oligo(A) sequence where they diverge from U4-1, suggesting they might have been created by retrotransposition of degradation products that were oligoadenylated by the TRAMP complex (LaCava et al. 2005). Only a small subset of the U4 snRNA fusion genes are expressed, and many appear to lack promoter and processing elements that are required for expression and maturation. The mechanism that apparently favors truncation at position 68 is not known, but as this corresponds to the end of the U4/U6 interaction region, it is possible that pairing with U6 blocks degradation by 3′-exonucleases (see Discussion). Since these truncated variant snRNAs may retain the ability to pair with U6 snRNA, they could potentially form aberrant U4/U6 di-snRNPs. However, the most abundant of them (U4-82P) is expressed at less than 0.1% the level of U4-1 (Fig. 2C), so the truncated variant U4 snRNAs are unlikely to have a significant dominant negative effect.
The U5 snRNA gene family has the highest percent sequence identity among its variants (Supplemental Table S1). Most of the nucleotide variation occurs within bulges of the largely paired first two-thirds or in the mostly unpaired final third of the snRNA sequence (Fig. 3D; Supplemental Fig. 5D; Sontheimer and Steitz 1992). Nearly all U5 variants maintain the canonical sequence (CUUUU) within the loop of Stem I, with U5F-8P being the only exception. In conjunction with the U5 snRNP protein PRP8, the U-rich loop is responsible for binding and aligning adjacent exons for ligation during the second step of splicing catalysis (Sontheimer and Steitz 1993; Umen and Guthrie 1995; Chiara et al. 1997; O'Keefe and Newman 1998; Maroney et al. 2000; McConnell and Steitz 2001).
In summary, while some variants retain most or all of the important functional elements of their canonical snRNA, others contain primary and secondary structure alterations that may lead to rapid turnover and/or loss of activity.
Nearly all snRNA variants are less stable than their canonical snRNA
To test if the low steady-state levels of snRNA variants are due in part to reduced stability of the RNAs, we monitored the relative levels of snRNA variants over time following treatment of cells with actinomycin D to stop transcription (a subset is shown in Fig. 4 and the full list in Supplemental Table S4). Nearly all snRNA variants displayed reduced half-lives as compared to their canonical snRNAs, whose level did not decrease over the 30-min time course. The U1 variants tested fell into two groups, a more stable group with half-lives of more than 30 min and a less stable group with half-lives of less than 30 min (Fig. 4A). RNA stability correlates only roughly with the number of nucleotide changes (Fig. 4E). The more than twofold shorter half-life of U1-148P compared to vU1-17, which both have 11 nt changes, could be due to destabilizing nucleotide changes in Stem IV in the former but not the latter (Fig. 3A). Destabilization of Stem IV could make the RNA more susceptible to degradation by 3′-exonucleases. The same explanation could apply to the more than threefold shorter half-life of U1-138P (15 changes) than vU1-19 (12 changes). While U4-46P, with 13 changes, has a half-life of 77 min, the three U4 variants that are 3′-truncated at position 68 (U4-59P, -82P, and -91P), and thus lack Stem Loops II and III and the Sm binding site, have half-lives of 17–21 min (Fig. 4C). Thus, sequence changes expected to disrupt Sm binding and/or 3′ stem–loop structures result in more rapid turnover of variant snRNAs.
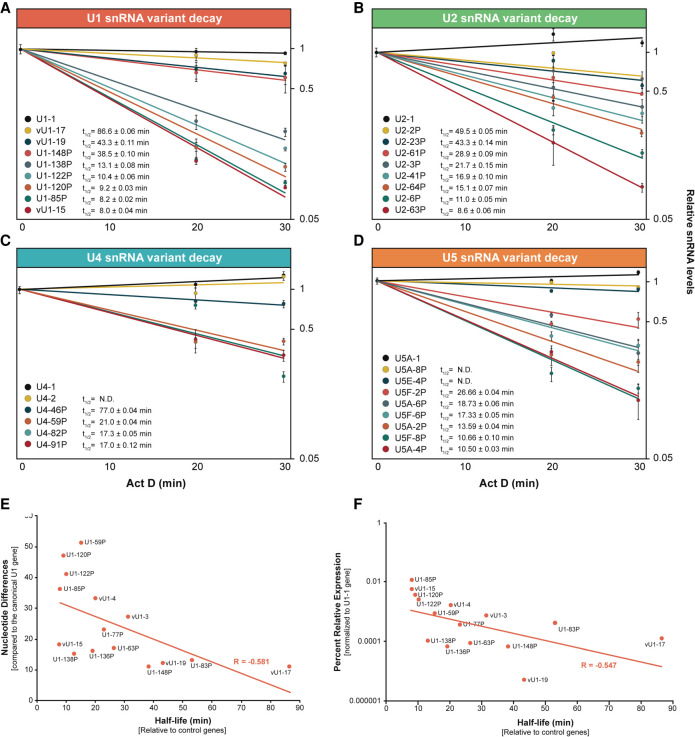
FIGURE 4.
Most snRNA variants are less stable than their canonical snRNA. RT-qPCR assays monitoring the relative level of snRNA variant remaining after onset of actinomycin D-mediated transcription inhibition. Half-lives (t1/2) were calculated after normalization of snRNA levels to the mean of 7SK and 5S RNA levels. (A–D) Data and values for selected U1, U2, U4, and U5 variants, respectively; see Supplemental Table S4 for all values. Error bars represent the SEM of three technical replicates from each time point. (E) Scatter plot comparing U1 variant nucleotide differences (from Supplemental Table S3) versus snRNA half-life (from Supplemental Table S4). Linear regression line and correlation coefficient (R) are indicated in red. (F) Scatter plot comparing the relative expression levels of U1 variants (log10; Fig. 2D) versus snRNA half-life (Supplemental Table S3). Linear regression line and correlation coefficient (R) are indicated in red.
Interestingly, the U5 3′ stem–loop (Stem/Loop II) varies in size and sequence across species and is nonessential in yeast (Frank et al. 1994). On the other hand, human U5 snRNAs require the 3′ stem–loop for proper expression and snRNP maturation (Hinz et al. 1996). Similarly, single point mutations in the 3′-terminal stem of U12 snRNA were found to destabilize the snRNA leading to the accumulation of truncated fragments in human cell lines (Norppa and Frilander 2021). Multiple U5 variants have a conserved Stem II, indicating a selective pressure to maintain a stable 3′-terminal stem–loop (Fig. 3E, left). However, U5 variants with less stable Stem II structures exhibit reduced half-lives (Fig. 3E, right). These data indicate that a stable 3′ stem–loop is an important determinant of human snRNA half-life.
If RNA stability is the major determinant of the steady-state level of variant snRNAs, then there should be a good correlation between the half-life and relative abundance. However, this does not seem to be the case for the U1 snRNA variants, which exhibit an inverse relationship between snRNA half-life and relative abundance in 293T cells (Fig. 4F). U1-85P and vU1-15 have the shortest half-lives, but are the most abundant of the U1 variants. Likewise, the least stable U2 variant, U2-63P, is one of the most abundant variants. However, variants such as U2-2P, U4-2, and multiple U5 snRNAs exhibit prolonged stability and higher levels of expression as compared to the majority of snRNA variant genes. Together with a recent study indicating that snRNA genes exhibit varying levels of occupancy by transcription factors and RNA polymerase II (Kosmyna et al. 2020), our data indicate that both transcription rates and snRNA stability determine variant snRNA abundances.
A subset of expressed snRNA variants is not detected in the cytoplasm
To better determine the cause of instability of variant snRNAs, we examined key steps of snRNP biogenesis, including export to the cytoplasm, binding of Sm proteins, import into the nucleus, and assembly into mature snRNPs and spliceosomes. We performed detergent-based subcellular fractionation of 293T cells to separate the nucleus from the cytoplasm (Supplemental Fig. 7A) and measured the relative levels of snRNA variants in the two fractions (Fig. 5A). We found that the canonical major and minor snRNAs other than U6/U6atac are present in the cytosol at 5% to 25% of their total cellular levels, which is generally consistent with a prior study by Pessa et al. (2008), although they detected a greater fraction of U4 and U5 in the cytosol. U6 and U6atac are present in the cytosol at 40% to 60% of their total cellular abundance, as also seen by Pessa et al. (2008). Since U6 snRNAs are transcribed by RNAP III and are capped with a γ-monomethyl phosphate, these RNAs were not thought to be targeted for nuclear export and their presence in the cytoplasm was attributed to nuclear leakage during fractionation. However, it was recently shown that yeast (S. cerevisiae) U6 snRNA is exported to the cytoplasm and imported into the nucleus along with the RNAP II-synthesized snRNAs (Becker et al. 2019). The accumulation of U6 in the mammalian cell cytoplasm may reflect a similar pathway.
FIGURE 5.
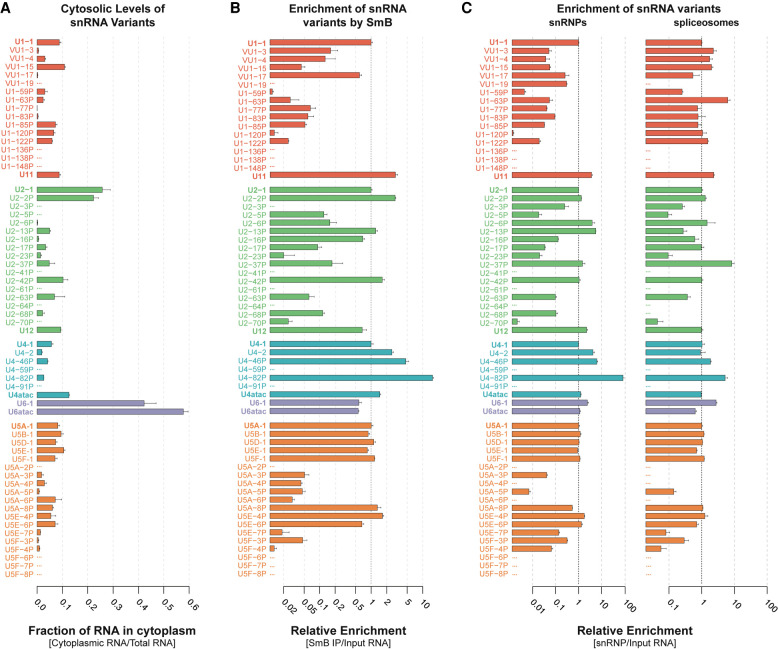
Most snRNA variants are incorporated into spliceosomes. (A) Cytosolic fraction of variant snRNAs. Nuclear and cytoplasmic fractions were normalized using cell equivalents to calculate total RNA. Error bars represent the SEM of three technical replicates from each fraction. (B) Enrichment of snRNA variants by anti-Sm (Y12 mAb) IP relative to canonical snRNAs. Fold enrichment was calculated as 2-(Ct variant - Ct canonical) for Y12 IP/2-(Ct variant - Ct canonical) for Total RNA. Error bars represent the SEM of three technical replicates. The x-axis is log2 scaled. See Supplemental Figure 7C for relative levels in Y12 mAb IP. (C) Enrichment of snRNA variants in snRNP and spliceosome fractions relative to canonical snRNAs. Fold enrichment was calculated as 2-(Ct variant - Ct canonical) for snRNP or spliceosomes /2-(Ct variant - Ct canonical) for Total RNA. Error bars represent the SEM of three technical replicates. The x-axis is log10 scaled. See Supplemental Figure 7E for relative levels in snRNPs or snRNPs or spliceosomes.
While all of the snRNA variants could be identified in the nuclear fraction, a subset were absent from the cytosol (e.g., U1-138P, U1-148P, U2-41P, U2-61P, U4-59P, and U5A-2P) (Fig. 5A). In most cases, variants that are not found in the cytoplasm tend to exhibit multiple nucleotide changes within snRNP-specific protein binding sites and are less stable (Fig. 3; Supplemental Table S4). Their absence in the cytoplasm is not due simply to low abundance, since variants of similar or lower total abundance were detected in the cytoplasm (e.g., VU1-17, U1-63P, U2-37P). These data suggest that, for a subset of variant RNAs, nuclear export is blocked, the RNA is unstable in the cytosol, or the assembled snRNP is very rapidly imported. Nuclear export of an snRNA depends on 7-methylguanosine (m7G) cap addition at the 5′ end (Hamm and Mattaj 1990), initial 3′ processing (Ohno et al. 2002; Masuyama et al. 2004), and association with snRNA-specific export factors (Matera et al. 2007). Defects in any of these initial steps may therefore impede export and preclude variant snRNAs from undergoing snRNP maturation.
Some expressed snRNA variants do not stably associate with the Sm ring
To examine the cytoplasmic fate of variant snRNAs, we investigated binding of the heteroheptameric Sm ring (Matera and Wang 2014; Gruss et al. 2017). We expected that variant snRNAs that are present in the cytoplasm and retain a single-stranded Sm binding motif (Supplemental Fig. 5E; Golembe et al. 2005) would be loaded with the Sm ring. We analyzed RNA obtained by immunoprecipitation (IP) of Sm subunits from 293T total cell lysates with the Y12 monoclonal antibody (Supplemental Fig. 7B). While all U1 snRNA variants that were detected in the cytoplasm were present in the Sm IP, they copurified with different efficiencies (Fig. 5B). Of those that co-IP with <10% efficiency relative to U1-1, some have substitutions in the Sm site (e.g., vU1-15, U1-63P, U1-77P) but others do not (e.g., U1-59P, U1-85P, U1-120P) (Fig. 3A; Supplemental Fig. 5A). However, variant U1 snRNAs in this latter group have nucleotide variations within the U1-70K protein binding site (Fig. 3A), which may explain their decreased Sm occupancy since the U1-70K protein helps recruit the SMN complex (So et al. 2016). A similar phenomenon involving other snRNP-specific proteins may account for the low Sm ring occupancy of U2-23P, U5A-6P, and U5E-7P, which also have canonical Sm sites. The fact that variant snRNAs that are not detected in the cytoplasm are also not precipitated from whole-cell lysates by anti-Sm antibody (with the exception of U2-5P and -70P) makes it unlikely that they are rapidly exiting and reentering the nucleus.
Nucleotide deviations in the Sm binding sequence also correlated with reduced relative enrichment of other snRNA variants in the Sm IP to less than ten percent of their canonical snRNA (e.g., U2-17P, U5A-4P, U5F-4P) (Figs. 3, 5). Conversely, some variants that had lower fractional cytosolic levels than their canonical snRNA were found to be more enriched than their canonical snRNA in Sm IPs (e.g., U2-42P, U4-82P). These variants may associate with snRNP biogenesis factors more readily, allowing for more rapid nuclear import, or less readily, resulting in more cytosolic degradation of unbound snRNA. Overall, nearly half of the snRNA variants tested are enriched in Sm IPs to extents comparable to their canonical snRNA despite their relatively low (<0.1%) abundance. A few variants (U2-2P, U5A-8P, U5E-4P, U5E-6P) were as abundant in Sm immunoprecipitates as the minor spliceosomal snRNAs (∼10% of canonical snRNAs, Supplemental Fig. 7C).
Surprisingly, several U4 variants are more enriched in the Y12 IP than canonical U4, especially U4-82P, which is greater than 10-fold more enriched than U4-1. This result is unexpected given that U4-82P does not have a canonical Sm binding site at the expected position. However, since the length of the U4-82P transcript is unknown, it may have an Sm binding site further downstream. Furthermore, this result implies that less than 10% of U4-1 is coprecipitated with Y12 antibody. Notably, the cryo-EM structure of the human U4/U6.U5 tri-snRNP (Charenton et al. 2019), but not the crystal structure of U4 snRNP (Leung et al. 2011), shows possible occlusion of the Y12 Sm epitopes (Hirakata et al. 1993; Brahms et al. 2000) by U4 Stem II, which could explain the poorer coprecipitation of U4-1 with Y12 antibody. The absence of a corresponding stem–loop in U4-82P may improve access to the Y12 epitopes. Another possibility is that U4-82P associates with a different protein that contains symmetric dimethylarginine residues, a key feature of the Y12 epitope, such as Coilin (Herbert et al. 2002) or another protein (Stopa et al. 2015). The ∼50% efficiency of U6 immunoprecipitation with Y12 antibody relative to U4 (Fig. 5B) is consistent with all of the U4 being paired with U6 and U6 being present in roughly twofold excess over U4.
Formation of snRNPs and spliceosomes by snRNA variants
The fact that a number of human variant snRNAs are expressed and loaded with an Sm ring raises the possibility of variant snRNA-containing spliceosomes. To determine which variant snRNAs are assembled into snRNPs and spliceosomes, we isolated RNPs by glycerol gradient fractionation of 293T nuclear extracts. Northern and western blot analysis of the gradient fractions allowed identification of peak fractions for each canonical snRNP (Supplemental Fig. 7D). Each peak fraction and the two flanking fractions were pooled to account for potential differences in sedimentation rates between variant and canonical snRNPs. To minimize contamination of spliceosomes with U4/U6.U5 tri-snRNP or prespliceosomal complexes, we used fractions from the bottom of the gradient. These fractions likely contain higher order complexes of spliceosomes, called >150S spliceosomes (Wassarman and Steitz 1993) or supraspliceosomes (Müller et al. 1998). The abundances (Supplemental Fig. 7E) and incorporation efficiencies (Fig. 5C) of variant snRNAs relative to their canonical snRNA were measured in each pool by RT-qPCR.
We found that the U1 snRNP contains <0.001% variant U1 snRNA, whereas U11 snRNP is present at about 1% the level of U1 snRNP (Supplemental Fig. 7E, left). All U1 snRNA variants tested assembled into U1 snRNP at least 10-fold less efficiently than U1-1, except for vU1-17 and vU1-19 (Fig. 5C, left). However, since vU1-19 does not co-IP with Sm or cosediment with spliceosomes (Fig. 5B,C, right), it may be in a distinct complex that cosediments with U1 snRNPs. It is likely that vU1-19 associates with U1-70K less stably due to a G28 to U mutation, as well as an AA dinucleotide insertion at position 32. Nucleotide changes at position 28 had previously been found to reduce U1-70K association with U1 (Kondo et al. 2015). In contrast to the case with U1 snRNP, several U1 snRNA variants were more strongly enriched in the spliceosomal fractions than canonical U1, strikingly so for U1-63P (Fig. 5C, right). A potential explanation for this strong enrichment is that spliceosomes containing some noncanonical U1 snRNPs are defective for activation and thus accumulate at the pre-B complex stage, prior to U1 snRNP release.
U2 snRNP exhibits higher levels of variant incorporation than U1 snRNP. In particular, about 2% of U2 snRNP contains U2-2P, which differs from U2-1 by only eight substitutions, all in the 3′ half. U2-2P snRNPs are about twofold more abundant than U12 snRNP, which in turn is about 30-fold more abundant than any other variant U2 snRNP. Similar relative abundances of canonical and variant U2 snRNAs were found in spliceosomal fractions as compared to the free U2 snRNP (Supplemental Fig. 7E). Variant U2 snRNAs that did not co-IP with Y12 were not detected in snRNPs and spliceosomes. U2-68P was detected in Sm IP and U2 snRNP but not in spliceosomes, possibly due to four substitutions in U2/U6 Helix II. In contrast, U2-37P is almost 10-fold more enriched in spliceosomes than U2-1. U2-37P has a 4-nt deletion adjacent to the branchpoint recognition sequence (Fig. 3B). Perhaps this deletion inhibits spliceosome activation but not assembly, thus causing accumulation of inactive spliceosomes.
Similar to U2 snRNP, U4 snRNP has an snRNA variant, U4-2, which is present in ∼5% of snRNPs and ∼1% of spliceosomes. U4-2 has only four substitutions compared to U4-1, one in Loop I, one in Loop II, and a compensatory base-pair change in Stem II (Fig. 3C). The relative overrepresentation of U4-2 in U4/U6 di-snRNP compared to spliceosomes might be because U4-2/U6 di-snRNP is less efficiently assembled into U4/U6.U5 tri-snRNP. In contrast, U4-46P and -82P are relatively overrepresented in spliceosomes as well as snRNPs, with U4-82P being the most enriched in snRNPs by about 10-fold (Fig. 3C). Still, there is a strong decrease in enrichment of U4-82P going from snRNPs to spliceosomes, suggesting a low efficiency of tri-snRNP addition (Supplemental Fig. 7E). A possible explanation for the overrepresentation of U4-82P in spliceosomes is that SNRNP200 (human Brr2), which binds U4 downstream from the U6 interaction region and unwinds the U4/U6 duplex during spliceosome activation, is less able to do so with the divergent sequence present in U4-82P. The resultant block to U4-82P/U6 disassembly would likely lead to spliceosome arrest. Despite their relative overrepresentation, U4-46P and U4-82P together occupy <1% of spliceosomes, due to their low abundance (Fig. 2C; Supplemental Fig. 7E).
The U5 snRNA variants are distinct in being incorporated into snRNPs and spliceosomes at much higher levels than other snRNA variants. U5A-1 was present in both U5 snRNP and spliceosomes at ∼20% of the total U5. U5B-1 occupied almost twice as many snRNPs and spliceosomes as U5A-1. Overall, canonical U5A-1 and the previously identified variants (U5B-1, D-1, E-1, F-1) comprise nearly 95% of the total U5 in both snRNPs and spliceosomes (Fig. 6). However, 5% of the total U5 levels in spliceosomes are the newly identified U5 snRNA variants U5A-8P, U5E-4P, and U5E-6P. These variants were found in the spliceosomal fractions at levels comparable to that of the minor snRNAs (1%–4%), indicating that these variants have the potential to influence splicing.
FIGURE 6.
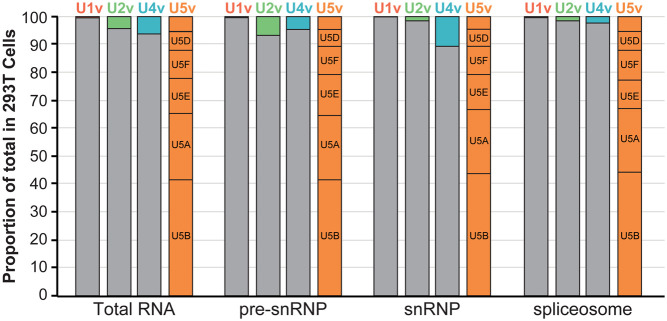
snRNA variants are expressed and incorporated into splicing complexes. Histogram of the proportion of canonical (gray) versus variant (colored) snRNAs from total RNA, Sm IPs, snRNPs, and spliceosomes isolated from 293T cells. For U5 snRNA, the five major variants are labeled, and the unlabeled box corresponds to the sum of the three minor variants. The x-axis is log10 scaled.
DISCUSSION
Pioneering work in the previous century identified the human spliceosomal snRNA genes as multigene families. Subsequent human genome sequencing identified more than 350 loci with strong sequence similarity to the RNAP II-transcribed spliceosomal snRNAs (U1, U2, U4, and U5), but the expression of these genes has not been systematically analyzed. Here we used published RNA-seq data and allele-specific RT-qPCR to identify expressed human snRNA genes across cell types and developmental stages. To distinguish between potentially functional snRNA variants and expressed pseudogenes, we examined their nuclear export, Sm ring loading, and assembly into mature snRNPs and spliceosomes. We found that at least 96% of the major spliceosomes are composed of the canonical U1, U2, and U4 snRNAs (Fig. 6). In contrast, only about 40% of spliceosomes contain the most abundant U5 snRNA, U5B (Fig. 6). Given the large fraction of spliceosomes in human cells that contain variant snRNAs, an important future goal is to determine if these variant spliceosomes carry out pre-mRNA splicing differently than canonical spliceosomes.
Variant snRNA incorporation into spliceosomes is influenced by the snRNP maturation pathway
Despite the expression of more than 50 snRNA variants, nuclear export of the precursor snRNA appears to be a limiting step that prevents some variants from undergoing mature snRNP formation. Multiple mechanisms likely prevent incompetent or immature snRNAs from being incorporated into snRNPs and spliceosomes (Becker et al. 2019; Lardelli and Lykke-Andersen 2020). snRNAs are processed cotranscriptionally by the Integrator complex, which generates pre-snRNA species (Matera and Wang 2014). Variant genes are potentially mis-processed and targeted for decay by the nuclear exosome. U1 variant vU1-15 has been shown to have a half-life of only 10 min, in agreement with our findings (Supplemental Table S4), and knockdown of the nuclear exosome doubles its half-life (Kawamoto et al. 2020; Lardelli and Lykke-Andersen 2020). The deadenylase TOE1 antagonizes exosomal degradation of snRNAs, but appears to act preferentially on canonical snRNAs compared to variants (Lardelli and Lykke-Andersen 2020). Thus, there is a kinetic competition between degradation and nuclear export that favors the canonical snRNAs and could explain the short half-lives of variant snRNAs that have intact Sm sites but are not detected in the cytoplasm, such as U1-138P and U2-5P. Cytoplasmic snRNA surveillance mechanisms add another quality control step, weeding out snRNAs that are defective in maturation phases including Sm protein binding (Liu and Gall 2007).
Final snRNP maturation occurs upon reimport into the nucleus, where remaining snRNP-specific proteins are loaded onto the snRNAs. Variant genes with numerous nucleotide substitutions or deletions in protein binding sites, such as U1-120P and U1-122P, may produce variant snRNPs that are inherently less stable or are incompletely assembled, as indicated by reduced Sm enrichment and half-life. Since snRNAs compete with one another for snRNP proteins, it is possible that differences in the steady state levels of snRNA variants in different human cell lines are due to differential expression of snRNP biogenesis factors. However, this remains to be determined.
Potential roles of snRNA sequence variants in splicing
Our identification of variant snRNAs in snRNPs and spliceosomes raises the possibility that these snRNAs could preferentially recognize unique splice sites, drive differential splicing kinetics, or even sequester core spliceosomal proteins into nonproductive or unstable snRNPs. All eleven U1 variants that we detected in snRNPs and spliceosomes have 1 to 3 nt changes within their 5′ splice site recognition sequence, and thus could alter 5′ splice site selection. Variants vU1-3 and vU1-4 were previously found to be incorporated into snRNP complexes (O'Reilly et al. 2013) and vU1-4 has been implicated in stem cell maintenance (Vazquez-Arango et al. 2016). Similarly, four of the U2 variants we detected in snRNPs and spliceosomes (U2-17P, -37P, -63P, and -70P) each have a single nucleotide change in the branch point recognition sequence, which could skew intron recognition. While these variant snRNPs are of very low abundance, it is conceivable that altered protein interactions could target them to specific introns.
Despite the presence of more than 90 variant U4 snRNA genes in the human genome, we were able to detect expression of only five. Surprisingly, over half of the annotated human U4 snRNA genes (51/97) are missing their 3′ half (Fig. 1C). It remains unknown what mechanism(s) generated so many truncated U4 snRNA genes. An intriguing possibility is that the endonuclease-like domain of PRP8, which is positioned near the end of the U4/U6 interaction domain in the human tri-snRNP (Agafonov et al. 2016), may have residual catalytic activity that rarely cleaves U4 adjacent to U4/U6 Stem/Loop I. Subsequent retrotransposition of these degradation products may have given rise to the large number of 3′-truncated U4 snRNA genes throughout the genome. The truncated U4 variants that are expressed in human cells have an unaltered U4/U6 Stem I, and a mostly intact U4/U6 Stem II sequence (Supplemental Fig. 5C), and thus may retain the ability to assemble into the U4/U6.U5 tri-snRNP. Indeed, we found U4-82P is associated with snRNPs and spliceosomes (Fig. 5C).
Overall, we find that most expressed snRNA variants exhibit limited sequence changes in known functional domains, allowing these variants to form snRNPs. However, variations in other regions of the snRNA variants are much more prevalent, which may promote associations with unique splicing factors and give rise to variant snRNPs with unique protein compositions.
There is no canonical U5 snRNA in humans
While the human U5 gene family is the smallest of all the major snRNA gene families (Fig. 1C), it is the only gene family to express more than one variant at greater than 1% of the total snRNA. Indeed, there is no canonical U5 snRNA. The most abundant human variant, U5B, comprises less than 50% of the U5 snRNA in the tissues and cell types examined (Fig. 6). We find that eight human U5 variants are expressed at a level of more than 1% of the total U5 snRNA. An important unanswered question is whether a subset of U5 variants is specifically enriched in the minor spliceosome. Based on the shared relative expression levels with minor spliceosomal snRNAs across human cells, the three novel U5 variants (U5A-8P, E-4P, E-6P) would be obvious candidates (Supplemental Fig. 4B). A more comprehensive study of U5 snRNA variant function is necessary to establish a causal role of variants in regulating alternative splicing events. It will also be interesting to determine if specific U5 variants are favored by the minor spliceosome.
Implications of variant spliceosomes
In addition to the core splicing proteins, human spliceosomes contain many peripheral proteins, the inclusion or exclusion of which could lead to splicing complexes with different substrate preferences. Noncanonical snRNAs may contribute to this heterogeneity by recruiting different peripheral proteins than canonical snRNAs. In addition, variant snRNAs may directly recruit specific pre-mRNAs by base-pairing, analogous to recruitment of specific mRNAs by ribosomal RNA expansion segments (Leppek et al. 2021). The minor spliceosome can be thought of as an extreme example of a variant spliceosome, with variant major spliceosomes presumably exhibiting more subtle differences in pre-mRNA substrate specificity. Differential expression of variant snRNA genes and/or use of their transcripts across cell types may direct distinct splicing programs, as has already been observed for the canonical snRNAs (Dvinge et al. 2019). Some snRNA variants may even have evolved new functions, for example, blocking splicing of certain introns by sequestering them in nonfunctional complexes.
Although transcripts of some snRNA variant genes were not detected within our cell lines, they could nevertheless function in other cell types or developmental time points. Despite the low expression of most human variant snRNA genes, knockdown of a single U1 variant leads to global pre-mRNA processing defects (O'Reilly et al. 2013), and regulation of the cellular levels of U1 variants has recently been implicated in stem cell maintenance (Vazquez-Arango et al. 2016). The role of snRNA variants in splicing regulation and disease should be an active area of further study.
MATERIALS AND METHODS
snRNA variant genes
All genes with “biotype = snRNA” were downloaded from the Ensembl database v92 (Cunningham et al. 2015) using BioMart (N = 2072). Results were filtered to include only genes annotated as belonging to the Rfam v13 families RF00003 (U1), RF00004 (U2), RF00015 (U4), RF00020 (U5), and RF00026 (U6) (N = 1840). Genes present on genomic patches (scaffold sequences) were removed, and only genes located on the 22 autosomes and the two sex chromosomes were retained (N = 1674). Only snRNA annotations that were present in both the Ensembl (v92) and RFAM (v13) database were presented to increase confidence in putative snRNA gene annotations.
RNA-seq read mapping
Fastq files containing all raw RNA-seq reads were obtained from the Sequence Read Archive, accession number SRP109305 (Marshall et al. 2017) (NCI-60) and SRP003754 (ENCODE). The ENCODE samples were sequenced using variable read lengths, and all results were therefore trimmed down to the minimal available read length (36 nt at their 5′ end), to provide more uniform results. Bowtie (Langmead et al. 2009) and RSEM (Li and Dewey 2011) were used to map all reads to the UCSC hg19 (NCBI GRCh37) human genome assembly. Reads were mapped to the gene annotation file using RSEM with the arguments –bowtie-m 100 –bowtie-chunkmbs 500 –calc-ci –output-genome- bam after modifying RSEM v1.2.4 to call Bowtie v1.0.0 with the -v 2 mapping strategy. Only alignments with zero mismatches were reported, and alignments with a mapq score below 10 were filtered out to include only uniquely mapped reads. snRNA variants that exhibited multiple regions of sequence diversity suitable for PCR primer design and expression >250 reads per kilobase million were hand-selected for primer design.
snRNA variant genomic alignments
snRNA variant genomic sequences from Ensembl were aligned using Clustal Omega, with five guide-tree/HMM iteration steps (Sievers et al. 2011). Percent sequence identity was calculated as compared to the canonical snRNA gene for each family.
Variant-specific PCR detection
For the PCR-based validation we used pooled total RNA from ten human cell lines (QPCR Human Reference Total RNA, Agilent), representing cell lines derived from tumors of the mammary gland, liver, cervix, testis, brain, skin, soft tissue, macrophages, and T and B cells. An amount of 1.0 µg of the pooled RNA was supplemented with 0.1 µg each of 293T, K562, and MCF7 total RNA. To amplify the snRNA variants, the qScript One-Step RT-qPCR kit was used (Quantabio). Reactions were scaled down from the manufacturer's protocol to a 5 µL reaction volume. RNA concentration was measured on a Nanodrop One (Thermo Scientific) machine. The amount of RNA used for each experiment is defined below. Reactions were run on a Bio-Rad CFX384 Real-Time PCR detection system following the one-step qScript cycling protocol with the annealing/amplification temperature of 61°C for all snRNA variant primer sets. A cycle threshold (Ct) cutoff of 35 was used to rule out possible false artifacts for all RT-qPCR experiments. Reactions were then run on a 12% native-PAGE, stained with Sybr Gold, gel extracted, eluted in DNA elution buffer (300 mM NaCl and 1 mM EDTA), ethanol precipitated as described in Mabin et al. (2018), and submitted to GeneWiz for Sanger sequencing to validate gene-specificity.
Real-time qPCR of snRNA variants
snRNA variants were quantitatively measured following the procedure mentioned above. The following amounts of RNA per RT-qPCR reaction were used: total cell RNA 5.0 ng, Sm RIP 1.0 ng RNA, cytoplasmic RNA 5.0 ng, gradient fractionation of snRNPs and spliceosomes 0.5–1.0 ng RNA. Data preprocessing was carried out using Bio-Rad's CFX Maestro program. Unreliable RT-qPCR values (>35) were removed from downstream analysis. Subsequent data analysis was carried out following standard ΔCt or ΔΔCt approaches as indicated in figure legends or specified below.
Human cell line maintenance
Human 293T cell lines (ATCC) were cultured at 37°C under 5% carbon dioxide in a humidified chamber in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% v/v fetal bovine serum (FBS) and 1% v/v penicillin-streptomycin.
RNA stability assays
Human 293T cells were grown to approximately 90% confluency in 12-well plates. Cells were then treated with 5 µg/mL of actinomycin D (Dot Scientific) for 30, 20, or 0 min before harvesting in TRIzol according to the manufacturer's protocols. Total RNA was analyzed by RT-qPCR (5.0 ng/sample). Relative abundance was determined by ΔCt methods using the mean of 7SK and 5S RNAs at each time point.
Subcellular fractionation
For subcellular fractionation, three 10-cm plates of cultured 293T cells were grown to approximately 90% confluency. Cells were lysed in 1 mL of RSB100 Cell Lysis Buffer (10 mM Tris-Cl pH 7.4, 100 mM NaCl, 2.5 mM MgCl2, 0.5% v/v NP-40, 0.5% v/v Triton X-100) supplemented with 40 µg/mL digitonin (Novex) and incubated on ice for 5-min. Lysates were passed through a 40-gauge needle three times and the nuclei were pelleted at 1000g for 8 min at 4°C. The supernatant was decanted and saved as cytosolic fraction. The nuclei were again resuspended in 1 mL of RSB-100 plus 40 µg/mL digitonin. Nuclear suspensions were then centrifuged at 1000g for 8 min at 4°C. The supernatant was decanted. Nuclear pellets were resuspended in RSBT (RSB-100, 0.5% v/v Triton X-100) and incubated on ice for 5 min. The nuclear lysates were centrifuged at 1000g for 8 min at 4°C. The supernatant was decanted and saved as the nuclear fraction. RNA and protein were extracted from subcellular fractions using TRIzol according to the manufacturer's protocols. RNA and protein were analyzed by RT-qPCR (5.0 ng/sample) and western blotting, respectively, the latter using antibody against GAPDH (Bethyl; A300-639A) and HP1A (Bethyl; A300-877A). Relative abundance was determined by ΔCt methods using the canonical variant, and enrichment was calculated using ΔΔCt as compared to relative total RNA (nuclear fraction + cytosolic fraction) levels. All minor snRNAs except U6atac were compared to their major snRNA counterpart. U6 and U6atac snRNAs were compared to U4-1.
Sm immunoprecipitation
Immunoprecipitation of endogenous proteins from 293T cells was carried out as described in Mabin et al. (2018). One 15-cm plate of cultured cells were lysed in 3 mL of hypotonic lysis buffer (HLB: 20 mM Tris-HCl pH 7.5, 15 mM NaCl, 10 mM EDTA, 0.5% v/v NP-40, 0.1% v/v Triton X-100, 1% v/v Sigma protease inhibitor cocktail P8340). NaCl was then increased to 225 mM and, following a 5-min incubation on ice, cell lysates were cleared for 10 min at 15,000g and 4°C. The cleared lysate (3 mL) was added to Protein A/G Dynabeads (Life Technologies) conjugated to 3 µg of mouse IgG (Santa Cruz Biotechnology; sc-2025) or anti-Sm Y12 monoclonal antibody (Invitrogen; MA5-13449) and mixed gently by inversion for 2 h at 4°C. Beads were washed in hypertonic wash buffer (20 mM Tris-HCl pH 7.5, 225 mM NaCl, 0.1% v/v NP-40) eight times and eluted in clear sample buffer (100 mM Tris-HCl pH 6.8, 4% w/v SDS, 10 mM EDTA, 100 mM DTT). RNA and protein were isolated by TRIzol following the manufacturer's protocol. The purified proteins were separated via 12% acrylamide SDS-PAGE and analyzed by western blotting using a 1:1000 dilution (1.0 µg/mL) of the anti-Sm (Y12) antibody. The RNA was analyzed by RT-qPCR (1.0 ng/sample). Relative abundance was determined by ΔCt methods using the canonical variant, and enrichment was calculated using ΔΔCt as compared to relative total RNA levels.
snRNP and spliceosome fractionation by glycerol gradients
Nuclear extract from two 15-cm plates of 293T cells were prepared as originally described by Dignam et al. (1983) and modified by Sontheimer and Steitz (1992). Nuclear extracts were sedimented in a 14 mL 10%–30% v/v glycerol gradient at 25,000 rpm at 4°C for 16 h using an SW 41 rotor, as described by Hartmuth et al. (2012). RNA or protein were extracted from each 0.5 mL fraction using TRIzol according to the manufacturer's protocols. RNA and protein were analyzed by northern and western blotting, respectively (see Supplemental Table S5 for IR fluorophore-conjugated DNA probes and primary antibodies: PRP8 [Santa Cruz Biotechnology; sc-55533], SNU114/EFTUD2 [Invitrogen; PA5-54554], PRP19 [Bethyl; A300-101A]). Image Studio Lite (Licor) was used to analyze pixel densities of both northern and western blots to determine which fractions contained each snRNP and spliceosomes. Putative U1 snRNPs corresponded to fractions 5–7, U2 snRNPs and U4/U6 snRNPs to fractions 6–8, U5 snRNPs to fractions 8–10 and spliceosomes to fractions 26–28. U4/U6.U5 tri-snRNP fractions were not tested. Spliceosomes that migrated at the bottom of the gradient were tested due to their strong enrichment of all snRNAs. RNA from each fraction was diluted 1:250, except for fractions 26–28, which were diluted 1:450. Relative abundance was determined by ΔCt methods using the canonical variant, and enrichment was calculated using ΔΔCt as compared to relative total RNA levels.
Total-RNA specimens from healthy human tissues
Total RNA from adult and fetal human tissues was obtained from Agilent Technologies. For the adult samples, breast, cerebellum, larynx, liver, lung, spleen, stomach, and trachea samples were each pooled across six donors. Both genders were represented, with median age 56 ± 16.5 yr (median ± standard deviation). For the fetal samples, all but brain, lung, and skeletal muscle were pooled across two to 17 donors, 18 to 23 wk gestational age. Equivalent weights of RNA from each adult tissue were pooled to give total adult tissue, likewise, equivalent weights of RNA from each fetal tissue were pooled to give total fetal tissue.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This paper is dedicated to the memory of Heidi Dvinge, who passed away suddenly on September 20, 2019. We thank Kathy Senn for doing the nuclear/cytoplasmic fractionation and Aaron Hoskins for constructive comments on the manuscript. This work was supported by start-up funding from the University of Wisconsin School of Medicine and Public Health, the University of Wisconsin-Madison Graduate School, and the Department of Biomolecular Chemistry. J.W.M. was supported by the National Science Foundation Graduate Research Fellowship Program under grant DGE-1747503. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. P.W.L. is a Pew Scholar in the Biomedical Sciences and is supported by P01-CA196539. D.A.B. is supported by NIH R35 GM118075.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078768.121.
REFERENCES
- Agafonov DE, Kastner B, Dybkov O, Hofele RV, Liu WT, Urlaub H, Lührmann R, Stark H. 2016. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 351: 1416–1420. 10.1126/science.aad2085 [DOI] [PubMed] [Google Scholar]
- Becker D, Hirsch AG, Bender L, Lingner T, Salinas G, Krebber H. 2019. Nuclear pre-snRNA export is an essential quality assurance mechanism for functional spliceosomes. Cell Rep 27: 3199–3214. 10.1016/j.celrep.2019.05.031 [DOI] [PubMed] [Google Scholar]
- Berstein LB, Manser T, Weiner AM. 1985. Human U1 small nuclear RNA genes: extensive conservation of flanking sequences suggests cycles of gene amplification and transposition. Mol Cell Biol 5: 2159–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Lührmann R. 2000. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem 275: 17122–17129. 10.1074/jbc.M000300200 [DOI] [PubMed] [Google Scholar]
- Brow DA, Vidaver RM. 1995. An element in human U6 RNA destabilizes the U4/U6 spliceosomal RNA complex. RNA 1: 122–131. [PMC free article] [PubMed] [Google Scholar]
- Buzdin A, Ustyugova S, Gogvadze E, Vinogradova T, Lebedev Y, Sverdlov E. 2002. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of L1. Genomics 80: 402–406. 10.1006/geno.2002.6843 [DOI] [PubMed] [Google Scholar]
- Charenton C, Wilkinson ME, Nagai K. 2019. Mechanism of 5′ splice site transfer for human spliceosome activation. Science 364: 362–367. 10.1126/science.aax3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lullo DJ, Ma E, Celniker SE, Rio DC, Doudna JA. 2005. Identification and analysis of U5 snRNA variants in Drosophila. RNA 11: 1473–1477. 10.1261/rna.2141505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara MD, Palandjian L, Kramer RF, Reed R. 1997. Evidence that U5 snRNP recognizes the 3′ splice site for catalytic step II in mammals. EMBO J 16: 4746–4759. 10.1093/emboj/16.15.4746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal KB, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. 2015. Ensembl 2015. Nucleic Acids Res 43: D662–D669. 10.1093/nar/gku1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg JE, Lund E. 1988. The genes and transcription of the major small nuclear RNAs. In Structure and function of major and minor small nuclear ribonucleoprotein particles (ed Birnstiel ML), pp. 38–70. Springer, Berlin/Heidelberg. 10.1007/978-3-642-73020-7_2 [DOI] [Google Scholar]
- Denison RA, Van Arsdell SW, Bernstein LB, Weiner AM. 1981. Abundant pseudogenes for small nuclear RNAs are dispersed in the human genome. Proc Natl Acad Sci 78: 810–814. 10.1073/pnas.78.2.810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dergai O, Hernandez N. 2019. How to recruit the correct RNA polymerase? Lessons from snRNA genes. Trends Genet 35: 457–469. 10.1016/j.tig.2019.04.001 [DOI] [PubMed] [Google Scholar]
- Didychuk AL, Butcher SE, Brow DA. 2018. The life of U6 small nuclear RNA, from cradle to grave. RNA 24: 437–460. 10.1261/rna.065136.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489. 10.1093/nar/11.5.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet AJ, Droc G, Siol O, Audoux J, Gilbert N. 2015. U6 snRNA pseudogenes: markers of retrotransposition dynamics in mammals. Mol Biol Evol 32: 1815–1832. 10.1093/molbev/msv062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvinge H. 2018. Regulation of alternative mRNA splicing: old players and new perspectives. FEBS Lett 592: 2987–3006. 10.1002/1873-3468.13119 [DOI] [PubMed] [Google Scholar]
- Dvinge H, Guenthoer J, Porter PL, Bradley RK. 2019. RNA components of the spliceosome regulate tissue- and cancer-specific alternative splicing. Genome Res 29: 1591–1604. 10.1101/gr.246678.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybkov O, Will CL, Deckert J, Behzadnia N, Hartmuth K, Lührmann R. 2006. U2 snRNA-protein contacts in purified human 17S U2 snRNPs and in spliceosomal A and B complexes. Mol Cell Biol 26: 2803–2816. 10.1128/MCB.26.7.2803-2816.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeland DB, Sturtevant AP, Schuler MA. 1989. Molecular analysis of dicot and monocot small nuclear RNA populations. Plant Cell 1: 633–643. 10.1105/tpc.1.6.633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faresse NJ, Canella D, Praz V, Michaud J, Romascano D, Hernandez N. 2012. Genomic study of RNA polymerase II and III SNAPc-bound promoters reveals a gene transcribed by both enzymes and a broad use of common activators. PLoS Genet 8: e1003028. 10.1371/journal.pgen.1003028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Engelbrecht C, Chari A. 2011. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip Rev RNA 2: 718–731. 10.1002/wrna.87 [DOI] [PubMed] [Google Scholar]
- Frank DN, Roiha H, Guthrie C. 1994. Architecture of the U5 small nuclear RNA. Mol Cell Biol 14: 2180–2190. 10.1128/MCB.14.3.2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N, Lutz S, Morrish TA, Moran JV. 2005. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol 25: 7780–7795. 10.1128/MCB.25.17.7780-7795.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golembe TJ, Yong J, Dreyfuss G. 2005. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol Cell Biol 25: 10989–11004. 10.1128/MCB.25.24.10989-11004.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso AR, Gomes AQ, Barbosa-Morais NL, Caldeira S, Thorne NP, Grech G, von Lindern M, Carmo-Fonseca M. 2008. Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res 36: 4823–4832. 10.1093/nar/gkn463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss OJ, Meduri R, Schilling M, Fischer U. 2017. UsnRNP biogenesis: mechanisms and regulation. Chromosoma 126: 577–593. 10.1007/s00412-017-0637-6 [DOI] [PubMed] [Google Scholar]
- Guiro J, Murphy S. 2017. Regulation of expression of human RNA polymerase II-transcribed snRNA genes. Open Biol 7: 170073. 10.1098/rsob.170073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J, Mattaj IW. 1990. Monomethylated cap structures facilitate RNA export from the nucleus. Cell 63: 109–118. 10.1016/0092-8674(90)90292-M [DOI] [PubMed] [Google Scholar]
- Han H, Braunschweig U, Gonatopoulos-Pournatzis T, Weatheritt RJ, Hirsch CL, Ha KCH, Radovani E, Nabeel-Shah S, Sterne-Weiler T, Wang J. 2017. Multilayered control of alternative splicing regulatory networks by transcription factors. Mol Cell 65: 539–553. 10.1016/j.molcel.2017.01.011 [DOI] [PubMed] [Google Scholar]
- Hartmuth K, van Santen MA, Lührmann R. 2012. Ultracentrifugation in the analysis and purification of spliceosomes assembled in vitro. In Alternative pre-mRNA splicing: theory and protocols (ed. Stamm S, et al. ). 10.1002/9783527636778.ch13 [DOI]
- Hayashi K. 1981. Organization of sequences related to U6 RNA in the human genome. Nucleic Acids Res 9: 3379–3388. 10.1093/nar/9.14.3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. 2002. Coilin methylation regulates nuclear body formation. Dev Cell 3: 329–337. 10.1016/S1534-5807(02)00222-8 [DOI] [PubMed] [Google Scholar]
- Hernandez N. 2001. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J Biol Chem 276: 26733–26736. 10.1074/jbc.R100032200 [DOI] [PubMed] [Google Scholar]
- Hinz M, Moore MJ, Bindereif A. 1996. Domain analysis of human U5 RNA. Cap trimethylation, protein binding, and spliceosome assembly. J Biol Chem 271: 19001–19007. 10.1074/jbc.271.31.19001 [DOI] [PubMed] [Google Scholar]
- Hirakata M, Craft J, Hardin JA. 1993. Autoantigenic epitopes of the B and D polypeptides of the U1 snRNP. Analysis of domains recognized by the Y12 monoclonal anti-Sm antibody and by patient sera. J Immunol 150: 3592–3601. [PubMed] [Google Scholar]
- Ishihara T, Ariizumi Y, Shiga A, Kato T, Tan CF, Sato T, Miki Y, Yokoo M, Fujino T, Koyama A. 2013. Decreased number of gemini of coiled bodies and U12 snRNA level in amyotrophic lateral sclerosis. Hum Mol Genet 22: 4136–4147. 10.1093/hmg/ddt262 [DOI] [PubMed] [Google Scholar]
- Jia Y, Mu JC, Ackerman SL. 2012. Mutation of a U2 snRNA gene causes global disruption of alternative splicing and neurodegeneration. Cell 148: 296–308. 10.1016/j.cell.2011.11.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalvari I, Nawrocki EP, Argasinska J, Quinones-Olverar N, Finn RD, Bateman A, Petrov AI. 2018. Non-coding RNA analysis using the Rfam database. Curr Protoc Bioinformatics 62: e51. 10.1002/cpbi.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner B, Will CL, Stark H, Lührmann R. 2019. Structural insights into nuclear pre-mRNA splicing in higher eukaryotes. Cold Spring Harb Perspect Biol 11: a032417. 10.1101/cshperspect.a032417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T, Yoshimoto R, Taniguchi I, Kitabatake M, Ohno M. 2020. ISG20 and nuclear exosome promote destabilization of nascent transcripts for spliceosomal U snRNAs and U1 variants. Genes Cells 26: 18–30. 10.1111/gtc.12817 [DOI] [PubMed] [Google Scholar]
- Kondo Y, Oubridge C, van Roon A-MM, Nagai K. 2015. Crystal structure of human U1 snRNP, a small nuclear ribonucleoprotein particle, reveals the mechanism of 5′ splice site recognition. Elife 4: e04986. 10.7554/eLife.04986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmyna B, Gupta V, Query C. 2020. Transcriptional analysis supports the expression of human snRNA variants and reveals U2 snRNA homeostasis by an abundant U2 variant. bioRxiv 10.1101/2020.01.24.917260 [DOI] [Google Scholar]
- Krol A, Gallinaro H, Lazar E, Jacob M, Branlant C. 1981. The nuclear 5S RNAs from chicken, rat and man. U5 RNAs are encoded by multiple genes. Nucleic Acids Res 9: 769–787. 10.1093/nar/9.4.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakopoulou C, Larsson P, Liu L, Schuster J, Söderbom F, Kirsebom LA, Viranen A. 2006. U1-like snRNAs lacking complementarity to canonical 5′ splice sites. RNA 12: 1603–1611. 10.1261/rna.26506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724. 10.1016/j.cell.2005.04.029 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardelli RM, Lykke-Andersen J. 2020. Competition between maturation and degradation drives human snRNA 3′ end quality control. Genes Dev 34: 989–1001. 10.1101/gad.336891.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K, Byeon GW, Jujii K, Barna M. 2021. VELCRO-IP RNA-seq reveals ribosome expansion segment function in translation genome-wide. Cell Rep 34: 108629. 10.1016/j.celrep.2020.108629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKW, Nagai K, Li J. 2011. Structure of the spliceosomal U4 snRNP core domain and its implication for snRNP biogenesis. Nature 473: 536–539. 10.1038/nature09956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Gall JG. 2007. U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proc Natl Acad Sci 104: 11655–11659. 10.1073/pnas.0704977104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Matera AG. 2015. Developmental analysis of spliceosomal snRNA isoform expression. G3 5: 103–110. 10.1534/g3.114.015735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Kahan B, Dahlberg JE. 1985. Differential control of U1 small nuclear RNA expression during mouse development. Science 229: 1271–1274. 10.1126/science.2412294 [DOI] [PubMed] [Google Scholar]
- Lund E, Bostock CJ, Dahlberg JE. 1987. The transcription of Xenopus laevis embryonic U1 snRNA genes changes when oocytes mature into eggs. Genes Dev 1: 47–56. 10.1101/gad.1.1.47 [DOI] [PubMed] [Google Scholar]
- Mabin JW, Woodward JA, Patton RD, Yi Z, Jia M, Wysocki VH, Bundschuh R, Singh G. 2018. The exon junction complex undergoes a complex compositional switch that alters mRNP structure and nonsense-mediated mRNA decay activity. Cell Rep 25: 2431–2446. 10.1016/j.celrep.2018.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Romfo CM, Nilsen TW. 2000. Functional recognition of the 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol Cell 6: 317–328. 10.1016/S1097-2765(00)00032-0 [DOI] [PubMed] [Google Scholar]
- Marshall EA, Marshall EA, Sage AP, Ng KW, Martinez VD, Firmino NS, Bennewith KL, Lam WL. 2017. Small non-coding RNA transcriptome of the NCI-60 cell line panel. Sci Data 4: 170157. 10.1038/sdata.2017.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marz M, Kirsten T, Stadler PF. 2008. Evolution of spliceosomal snRNA genes in metazoan animals. J Mol Evol 67: 594–607. 10.1007/s00239-008-9149-6 [DOI] [PubMed] [Google Scholar]
- Masuyama K, Taniguchi I, Kataoka N, Ohno M. 2004. RNA length defines RNA export pathway. Genes Dev 18: 2074–2085. 10.1101/gad.1216204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Wang Z. 2014. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 15: 108–121. 10.1038/nrm3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 8: 209–220. 10.1038/nrm2124 [DOI] [PubMed] [Google Scholar]
- McConnell TS, Steitz JA. 2001. Proximity of the invariant loop of U5 snRNA to the second intron residue during pre-mRNA splicing. EMBO J 20: 3577–3586. 10.1093/emboj/20.13.3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan JB, Wang Y, Shuman S, Mills RE, Moran JV. 2019. RNA ligation precedes the retrotransposition of U6/LINE-1 chimeric RNA. Proc Natl Acad Sci 116: 20612–20622. 10.1073/pnas.1805404116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales J, Borrero M, Sumerel J, Santiago C. 1997. Identification of developmentally regulated sea urchin U5 snRNA genes. DNA Seq 7: 243–259. 10.3109/10425179709034044 [DOI] [PubMed] [Google Scholar]
- Mount SM, Gotea V, Lin CF, Hernandez K, Makalowski W. 2007. Spliceosomal small nuclear RNA genes in 11 insect genomes. RNA 13: 5–14. 10.1261/rna.259207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S, Wolpensinger B, Angenitzki M, Engel A, Sperling J, Sperling R. 1998. A supraspliceosome model for large nuclear ribonucleoprotein particles based on mass determinations by scanning transmission electron microscopy. J Mol Biol 283: 383–394. 10.1006/jmbi.1998.2078 [DOI] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol 48: 443–453. 10.1016/0022-2836(70)90057-4 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463. 10.1038/nature08909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norppa AJ, Frilander MJ. 2021. The integrity of the U12 snRNA 3′ stem-loop is necessary for its overall stability. Nucleic Acid Res 49: 2835–2847. 10.1093/nar/gkab048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Segref A, Kuersten S, Mattaj IW. 2002. Identity elements used in export of mRNAs. Mol Cell 9: 659–671. 10.1016/S1097-2765(02)00454-9 [DOI] [PubMed] [Google Scholar]
- O'Keefe RT, Newman AJ. 1998. Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J 17: 565–574. 10.1093/emboj/17.2.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly D, Dienstbier M, Cowley SA, Vazquez P, Drozdz M, Taylor S, James WS, Murphy S. 2013. Differentially expressed, variant U1 snRNAs regulate gene expression in human cells. Genome Res 23: 281–291. 10.1101/gr.142968.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett RA. 2012. New connections between splicing and human disease. Trends Genet 28: 147–154. 10.1016/j.tig.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AA, Steitz JA. 2003. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol 4: 960–970. 10.1038/nrm1259 [DOI] [PubMed] [Google Scholar]
- Pessa HKJ, Will CL, Meng X, Schneider C, Watkins NJ, Perälä N, Nymark M, Turunen JJ, Lührmann R, Frilander MJ. 2008. Minor spliceosome components are predominantly localized in the nucleus. Proc Natl Acad Sci 105: 8655–8660. 10.1073/pnas.0803646105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Krainer AR. 2009. Recognition of atypical 5′ splice sites by shifted base-pairing to U1 snRNA. Nat Struct Mol Biol 16: 176–182. 10.1038/nsmb.1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherly D, Boelens W, van Venrooij WJ, Dathan NA, Hamm J, Mattaj IW. 1989. Identification of the RNA binding segment of human U1 A protein and definition of its binding site on U1 snRNA. EMBO J 8: 4163–4170. 10.1002/j.1460-2075.1989.tb08601.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai S, Suzuki H, Diaz-Navarro A, Nadeu F, Kumar SA, Gutierrez-Fernandez A, Delgado J, Pinyol M, López-Otín C, Puente XS, et al. 2019. The U1 spliceosomal RNA is recurrently mutated in multiple cancers. Nature 574: 712–716. 10.1038/s41586-019-1651-z [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So BR, Wan L, Zhang Z, Li P, Babiash E, Duan J, Younis I, Dreyfuss G. 2016. A U1 snRNP-specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat Struct Mol Biol 23: 225–230. 10.1038/nsmb.3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer EJ, Steitz JA. 1992. Three novel functional variants of human U5 small nuclear RNA. Mol Cell Biol 12: 734–746. 10.1128/MCB.12.2.734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer EJ, Steitz JA. 1993. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262: 1989–1996. 10.1126/science.8266094 [DOI] [PubMed] [Google Scholar]
- Stopa N, Krebs JE, Shechter D. 2015. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci 72: 2041–2059. 10.1007/s00018-015-1847-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surowy CS, van Santen VL, Scheib-Wixted SM, Spritz RA. 1989. Direct, sequence-specific binding of the human U1-70K ribonucleoprotein antigen protein to loop I of U1 small nuclear RNA. Mol Cell Biol 9: 4179–4186. 10.1128/MCB.9.10.4179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Kumar SA, Shuai S, Diaz-Navarro A, Gutierrez-Fernandez A, De Antonellis P, Cavalli FMG, Juraschka K, Farooq H, Shibahara I, et al. 2019. Recurrent noncoding U1 snRNA mutations drive cryptic splicing in SHH medulloblastoma. Nature 574: 707–711. 10.1038/s41586-019-1650-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejedor JR, Papasaikas P, Valcárcel J. 2015. Genome-wide identification of Fas/CD95 alternative splicing regulators reveals links with iron homeostasis. Mol Cell 57: 23–38. 10.1016/j.molcel.2014.10.029 [DOI] [PubMed] [Google Scholar]
- Turunen JJ, Niemelä EH, Verma B, Frilander MJ. 2013. The significant other: splicing by the minor spliceosome. Wiley Interdiscip Rev RNA 4: 61–76. 10.1002/wrna.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Blencowe BJ. 2019. Alternative splicing regulatory networks: functions, mechanisms, and evolution. Mol Cell 76: 329–345. 10.1016/j.molcel.2019.09.017 [DOI] [PubMed] [Google Scholar]
- Umen JG, Guthrie C. 1995. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev 9: 855–868. 10.1101/gad.9.7.855 [DOI] [PubMed] [Google Scholar]
- Vazquez-Arango P, O'Reilly D. 2017. Variant snRNPs: new players within the spliceosome system. RNA Biol 15: 17–25. 10.1080/15476286.2017.1373238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Arango P, Vowles J, Browne C, Hartfield E, Fernandes HJR, Mandefro B, Sareen D, James W, Wade-Martins R, Cowley SA, et al. 2016. Variant U1 snRNAs are implicated in human pluripotent stem cell maintenance and neuromuscular disease. Nucleic Acids Res 44: 10960–10973. 10.1093/nar/gkw711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman DA, Steitz JA. 1993. A base-pairing interaction between U2 and U6 small nuclear RNAs occurs in >150S complexes in HeLa cell extracts: implications for the spliceosome assembly pathway. Proc Natl Acad Sci 90: 7139–7143. 10.1073/pnas.90.15.7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson ME, Charenton C, Nagai K. 2020. RNA splicing by the spliceosome. Annu Rev Biochem 89: 359–388. 10.1146/annurev-biochem-091719-064225 [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Garcia Giron C, et al. 2018. Ensemble 2018. Nucleic Acids Res 46: D754–D761. 10.1093/nar/gkx1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss D. 2008. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133: 585–600. 10.1016/j.cell.2008.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Black DL. 2013. Alternative pre-mRNA splicing in neurons: growing up and extending its reach. Trends Genet 29: 442–448. 10.1016/j.tig.2013.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM. 1986. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell 46: 827–835. 10.1016/0092-8674(86)90064-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.