Abstract
Single-cell nucleic acid analysis aims at discovering the genetic differences between individual cells which is well known as the cellular heterogeneity. This technology facilitates cancer diagnosis, stem cell research, immune system analysis, and other life science applications. The conventional platforms for single-cell nucleic acid analysis more rely on manual operation or bulky devices. Recently, the emerging microfluidic technology has provided a perfect platform for single-cell nucleic acid analysis with the characteristic of accurate and automatic single-cell manipulation. In this review, we briefly summarized the procedure of single-cell nucleic acid analysis including single-cell isolation, single-cell lysis, nucleic acid amplification, and genetic analysis. And then, three representative microfluidic platforms for single-cell nucleic acid analysis are concluded as valve-, microwell-, and droplet-based platforms. Furthermore, we described the state-of-the-art integrated single-cell nucleic acid analysis systems based on the three platforms. Finally, the future development and challenges of microfluidics-based single-cell nucleic acid analysis are discussed as well.
Keywords: Single-cell, Heterogeneity, Nucleic acid analysis, Microfluidics, Manipulation
Introduction
Cell is the basic structural and functional unit in life science. The diversity of these homogenic cells exists in their populations. For a couple of decades, scientists have paid their attention on the heterogeneity among cells for better disease diagnosis and treatment (Liu et al. 2020; Nam et al. 2021; Samad and Wu 2021). Especially, the purpose of single-cell nucleic acids analysis is to discover genetic difference, for example, genetic mutations among individual cells. The small size of a single cell, and the trace content of nucleic acids in single cell cause the main difficulties in single-cell nucleic acid analysis. Traditional methods need a single-cell sorting instrument accompanied with an optical microscope to lock and relocate individual cells. The relative complicated operations need to be performed by well-trained personnel. Obviously, traditional manual operation for single-cell analysis is inefficient and prone to cause false results due to the contamination of biomaterial in the operation.
Recently, microfluidic technology has demonstrated its superiorities on accurate and automatic manipulation of single-cell with high throughput (Murphy et al. 2018). The micro-structures inside the microfluidic chip are designed in micrometers or nanometers, and this geometrical scale falls perfectly into the domain of single-cell analysis. The procedure of single-cell nucleic acid analysis with multiple-steps is allowed to be automatically implemented on microfluidic chips consisting of different functional components, for example, micro-chambers with size down to 10–9–10–18 L, micro-valves with active controllability, micro-channels for various reagent processing, etc. Microfluidic platform provides a relatively simple and straightforward method for single-cell isolation. With microfluidic chip, nucleic acid amplification can be performed more efficiently within the tiny micro-reactor because of the increased concentration. Especially, automatic single-cell nucleic acid analysis with high-throughput can be conveniently implemented on microfluidic chip with parallel and functionally-integrated flow control networks.
In this review, the major microfluidic technologies used in each step of single-cell nucleic acid analysis are described, including single-cell isolation, single-cell lysis, and nucleic acid amplification with genetic analysis. Next, the microfluidic platforms for single-cell nucleic acids analysis will be summarized into three typical groups based on their working mechanisms, e.g., valve-, microwell-, and droplet-based platforms. The details about these three typical platforms, including their characteristics, adoptability, advantages, disadvantages, system integration and applications are systematically discussed.
Single-cell nucleic acid analysis based on microfluidics
Single-cell isolation
Before microfluidic technology arising, many traditional methods have been used for single-cell separation and isolation from sample population. Including serial dilution method, fluorescence-activated cell sorting (FACS), laser capture microdissection (LCM), and immunomagnetic separation. Although serial dilution method is cost-effective and easy to handle but the inaccuracy and material loss is inevitable. Both of FACS and LCM methods are equipment-dependent, such as flow cytometry and laser generator are used for cell focusing and detection. Immunomagnetic separation needs microbeads coated with specific biochemical antigens for target cells binding as well. Compared with traditional methods, microfluidics-based single-cell isolation techniques are more suitable for single-cell nucleic acid analysis due to the integrated and closed flow network which is ideal for combined analysis. Recent years, single-cell isolation methods applied in various biochemical applications have been discussed in a couple of reviews (Alam et al. 2018; Zhang et al. 2020). In this review, the typical single-cell trapping techniques used for nucleic acid analysis are concluded as size-based trapping, hydrodynamic trapping, droplet-based trapping, and other trapping methods based on their working mechanisms.
Size-based trapping
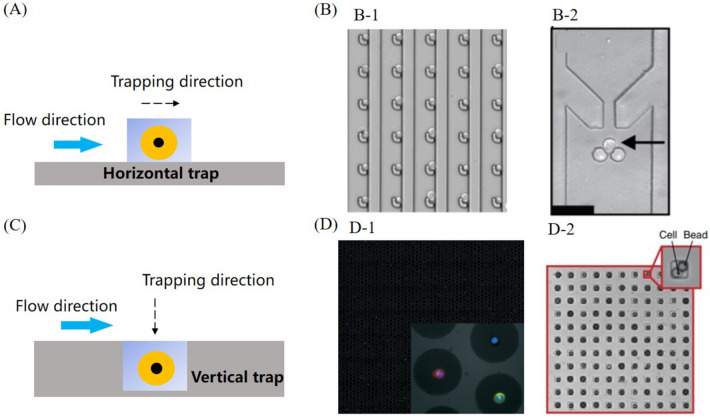
Size-based trapping is achieved by fabricating microstructures in microfluidic chips with the size close to one single cell. Furthermore, by combining cell trapping microstructures with the microchannels, cells within the flowing sample can be automatically trapped as estimated. Basically, according to the trapping direction of the microstructure, geometrical size-based microstructures can be categorized as two types, horizontal trap (White et al. 2011, 2013) and vertical trap (Gole et al. 2013; Fan et al. 2015; Li et al. 2018a). As shown in Fig. 1A and C, for horizontal trap, cell is trapped by a featured microstructure when it horizontally flows along the microchannel. On the contrary, for vertical trap, cell is trapped by microwell when it vertically falls down into the chamber. Since for the vertical trap, the process of cell trapping heavily relies on gravity, it normally costs longer time compared with horizontal trap.
Fig. 1.
Size-based single-cell trapping: A schematic of horizontal trap, B-1 horizontal trapping array for live single-cell printing (Zhang et al. 2014), B-2 horizontal trap with an open-end microstructure (VanInsberghe et al. 2018), C schematic of vertical trap, D-1 optical detection of single cells from microwells (Swennenhuis et al. 2015), D-2 microwell array used to capture and lyse single cell (Gierahn et al. 2017)
Vertical and horizontal traps are two straightforward microstructures for single cell trapping and downstream analysis. Both of them can be fabricated as a high-throughput array for multiple cell isolation when each cell is separated and limited in its own space for further optical detection and analysis (Zhang et al. 2014; Zhu et al. 2019; Zhou et al. 2020), as shown in Fig. 1B-1. For single cell-based nucleic acid analysis, it is difficult for vertical trap to integrate other flow control modules for automatic downstream analysis due to its own limitations. For horizontal trap, single cell-based nucleic acid analysis can be achieved by combining it with different modules to form an integrated flow network (VanInsberghe et al. 2018), as shown in Fig. 1B-2. In contrast, in a manual operation way, vertical trap can be used as a multi-functional chamber for cell trapping, optical detection or nucleic acid amplification, as illustrated in Fig. 1D.
Hydrodynamic trapping
In principle, hydrodynamic trapping can be regarded as an advanced version of size-based trapping since cell trapping is achieved by combining elaborate flow control with specific microstructures. Or alternatively, hydrodynamic trapping may include size-based trapping (Xu et al. 2020). Here, different from size-based trapping which purely relying on simple microstructures, hydrodynamic trapping is discussed to demonstrate how to trap single cell with more elaborate flow control as well as the cell-size microstructures. For hydrodynamic trapping, cell trapping is achieved by elaborate flow control with properly designed flow network. For example, when each single cell is trapped by a microstructure, the following cells will be automatically switched to another flow path until another single cell is trapped by another empty microstructure, and by doing this, finally all cells can be trapped independently. Furthermore, the trapped single cell can be easily released by air bubbles created by laser-based heating.
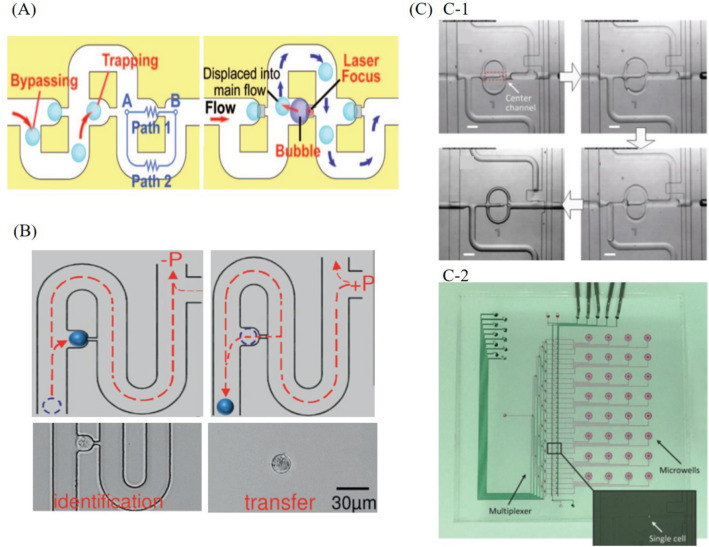
In recent years, different methods for single-cell nucleic acid analysis based on hydrodynamic trapping have been developed. Continuous hydrodynamic channel was developed by Tan and Takeuchi (2007). As shown in Fig. 2A, the main design of continuous hydrodynamic channel includes three parts, main channel for cell loading, trap site with a narrow, and a bypassing to downstream. The basic principle of this design is simple and straightforward. Cell alignment is accomplished in the main channel, and cells would be carried into the trap by the flow. As shown in Fig. 2B, Zhang et al. (2016) integrated the hydrodynamic channel into their modular single-cell pipette (mSCP) to isolate single cell from bulk volume. And they also developed a new cell-releasing mechanism instead of laser-heating by applying positive pressure from the channel outlet to push the trapped cell out of the trap. It is convenient for hydrodynamic trapping to be properly combined with downstream functional modules for integrated nucleic acid analysis based on a joint flow network. Zhang et al. upgraded hydrodynamic single-cell trapping unit (Fig. 2C-1) to a parallel processing system (Fig. 2C-2). By adding reagent and oil respectively from inlet and outlet of the hydrodynamic trapping unit, captured cells can be confined in oil droplets, and then they are transported to independent chambers for subsequent nucleic acid analysis (Zhang et al. 2019a).
Fig. 2.
Hydrodynamic trapping: A schematics of hydrodynamic trapping channel (Tan and Takeuchi 2007), B schematics of single-cell capturing and releasing (Zhang et al. 2016), C-1 optical micrographs showing the sequential single-cell processing in hydrodynamic channels, C-2 microfluidic chip for parallel single-cell processing based on hydrodynamic trapping (Zhang et al. 2019a)
Droplet-based trapping
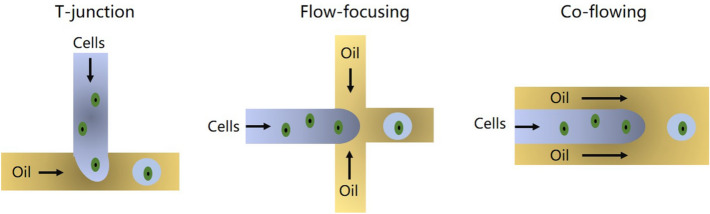
Droplet-based trapping derives from droplet-based microfluidics. For two immiscible fluids, when water containing cells is sheared by the oil with proper condition, water with a single cell will be sealed by oil to form an independent cell droplet. High-throughput cell droplets can be conveniently generated with a properly configured droplet microfluidic chip. Three typical modes for droplet-based cell trapping are as shown in Fig. 3 (Collins et al. 2015). The symmetrical geometry structure of flow-focusing or co-flowing can produce more uniform droplets than T-junction which possesses the simplest structure. These microchannels for droplet generation are commonly fabricated in PDMS substrate. Or alternatively, with a disk chip, droplet can be generated more efficiently with centrifugal force-based liquid driving (Fu et al. 2019).
Fig. 3.
Droplet-based cell trapping with different principles (Collins et al. 2015)
To get a stable droplet, fluorinated oil and surfactants are often used to modify carrier oil. Encapsulation of cells in droplets is achieved in a stochastic manner. Ideally, one cell would be confined in one droplet at the moment of interflowing between cell suspension and carrier oil. But in real situation, multiple or none occupied cases of droplet encapsulation are inevitable. To achieve efficient encapsulation in droplets, the structure and dimension of interflow channels, as well as the flow rate of reagent and oil should be properly optimized. Furthermore, for downstream biochemical reaction with single-cell nucleic acid analysis, different droplet manipulation methods, for example, merging and splitting, should be properly adopted to perform more complicated droplet-based fluidic control (Deng et al. 2013; Liu and Zhu 2020).
Electrical, optical, and magnetic trapping
In addition to the typical microfluidics-based single-cell isolation methods, other techniques are developed to capture single cell by applying external physical field forces, and they can manipulate cells with no contact in an elaborate way without complicated flow control.
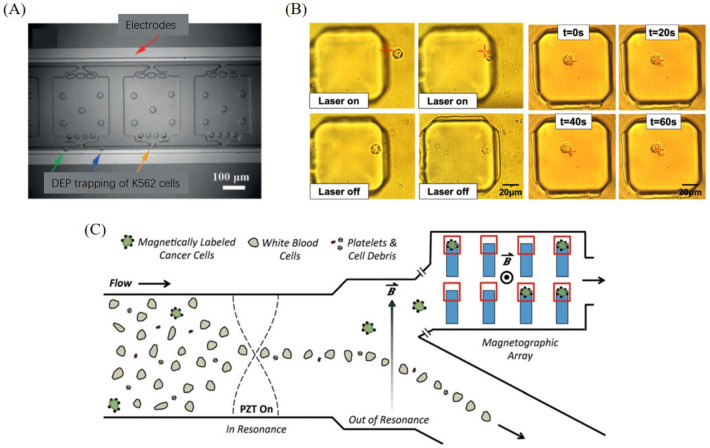
Dielectrophoretic (DEP) force is created by a nonuniform electric field applied on the micro-particle and surrounding medium. Because different cells and the reagents possess different conductivities, under a nonuniform electric field, cells will be electric polarized and moved along the direction of electric gradient until they are eventually trapped. This method is usually used for parallel single-cell manipulation (Wu et al. 2017), and biophysical properties analysis (Huang et al. 2018). Generally, DEP platforms are fabricated as planar array that has no independent reactor for nucleic acid analysis on single-cell level. To strengthen this limitation, Qin et al. (2018) designed a microfluidic chip combined electrodes and individual chambers to achieve positive DEP single-cell trapping and loop-mediated isothermal nucleic acid amplification, as shown in Fig. 4A. DEP-based cell trapping can be adopted to process multiple cells in high-throughput since the electrical field is able to cover a proper space in a microfluidic chamber.
Fig. 4.
Other cell trapping methods based on different physical fields: A DEP-based single cell trapping (Qin et al. 2018), B optical tweezer-based single cell trapping (Cong et al. 2019), C magnetic-based single cell trapping (Shields et al. 2016)
Optical tweezer can be used to trap cells by taking advantage of the differences of refractive index between cells and surrounding reagent. With this method, the target cell can be dynamically trapped in a relatively high speed, e.g., ~ 200 μm/s or more (Landry et al. 2013) which is a unique advantage among other methods. In principle, the trapping force of optical tweezer can be varied on different cells with various diameters and shapes. Normally, optical tweezer can effectively manipulate cells with a diameter from 0.5 to 10 μm. Larger or smaller cells can also be manipulated by reducing the rate of cell movement. Huang et al. developed an optoelectronic tweezers platform by implement single-wall carbon nanotube electrodes into multilayer PDMS chip. This platform can achieve selective capture of single cells with light beams (Huang et al. 2013). For increasing the range of optical trapping, Cong et al. (2019) made a focused optical beam trap by combining optical forces and convective drag forces to guide living single cancer cell into a microwell array for multidrug-resistant gene marker detection (Fig. 4B). Optical tweezer possesses the unique capacity to selectively pick up single cell with relatively controllable high-speed.
Magnetic-activated method is another popular method for single-cell trapping and manipulation, especially with the applications of target cell screening and separation. Target cells which are specifically labelled with magnetic microbeads can be separated by a properly-designed magnetic field inside the microfluidic channel (Yousuff et al. 2017). Shields et al. designed an integrated microfluidic system that can provide single circulating tumor cell (CTC) screening and isolation from blood sample. As shown in Fig. 3C, this system consists of three modules, including acoustic standing wave for cells alignment, magnetically labeled cells separation, and microwell array with micromagnets for single CTC trapping and quantification by imaging (Shields et al. 2016). Magnetic-based cell trapping is able to process multiple cells in high-throughput by duplicating the magnetic force in a magnetographic array.
For each cell trapping method, it should be properly applied to handle ideal type of samples based on its characteristics. For example, it is convenient for size-based or hydrodynamic trapping method to trap limited number of single cells from one type of cell sample or mixed different types of cells. Droplet-based cell trapping is ideal for high throughput single cell trapping and downstream analysis. Compared with size-based, hydrodynamic, or droplet-based trapping methods which can be performed with more straightforward and simpler microfluidic chips, other trapping method requires more complicated chip to provide properly-controlled physical field or force to capture cell. DEP-based cell trapping can be applied when the sample buffer is adjusted properly. Optical tweezer-based cell trapping is more suitable for one or quite few specific single cell trapping. Magnetic-based cell trapping requires that the target cell can be specifically bond to protein-labeled magnetic beads. It should be noted that, since the above summarized cell trapping methods can not handle complicated cell sample, for example, whole blood sample or mixed cell sample with lots of cell types, cell purification or isolation needs to be done before to perform cell trapping.
Single-cell lysis
The efficiency of cell lysis is critical to downstream nucleic acid amplification. A suitable lysis method should be chosen based on a couple of reasons, for example, cell type, nucleic acid stability, compatibility with downstream bioreaction, flow control in microfluidic chip, complexity of the entire system, etc. Based on different working mechanisms, major methods for single-cell lysis can be catalogued as chemical, mechanical, and electrical lysis.
Chemical lysis commonly adopts surfactants to dissolve proteins and lipids of the cell membrane that can generate pores on the membrane and let the nucleic acid released. The commonly used surfactants are proteinase K, sodium dodecyl sulfate (SDS) and Triton X-100. Chemical lysis is simple to use, in some cases it only requires a short period of heating for a better lysis performance. And chemical lysis is applicable to any microfluidic platforms for single-cell nucleic acid analysis (Wood et al. 2010; VanInsberghe et al. 2018; Li et al. 2019a). However, chemical lysis buffer may inhibit the subsequent reaction, so after cell lysis it is necessary to remove residual lysis buffer in the chamber or neutralized by other reagents. For chemical lysis, elaborate flow control is normally required to perform downstream bio-reaction, for example, purification, neutronization, or mixing before nucleic acid amplification. As a popular lysis method, chemical lysis is widely used in single-cell-based nucleic acid analysis based on its strictly verified protocol.
Mechanical lysis employed the shear force, friction force, or compressive force created by the particular microstructure fabricated inside microchip to crush cell membrane. Different from chemical lysis, mechanical lysis is harmless to target biomarkers and other intercellular components. However, to puncture cell membrane, the microstructure need to be sharp enough. The dimension of microstructures usually designed in the scale of nanometer which will extremely increase the complexity and difficulty of chip fabrication (Yun et al. 2010; Kim et al. 2012). To address this problem, Li et al. (2013) invented a bubble-induced microjets for single-cell membrane perforation instead of employing nanostructure. For mechanical lysis which relies on elaborate microstructure fabrication, the released nucleic acid templates can probably be amplified immediately without further purification step since there has no any chemicals involved. Unlike standard chemical lysis, for a specific target cells, the efficiency of mechanical lysis should be studied thoroughly before making the final choice.
Applied cells in an electric field, when the transmembrane potential (TMP) surpassed the threshold of cell membrane penetration, the intracellular components will be released. If the potential is lower than the threshold, the pores on the cell membrane will close. Moreover, if the potential is too high, the cell membrane would be broken irretrievably (Nan et al. 2014). Jokilaakso et al. (2013) designed a single-cell positioning and lysis method on silicon nanowire and nanoribbon field effect transistors which needed a highly complicated chip fabrication process. The efficiency of electric lysis could be affected by different issues, for example, electric power, chip fabrication, cell type, cell state, sample buffer and et al, which limits its application in single-cell nucleic acid analysis.
Different from human cells, microbial cells are more difficult to lyse due to their diverse envelope-shaped structures (Woyke et al. 2017). Therefore, for single-cell-based nucleic acid analysis, to efficiently lyse microbial cells, a combination of different lysis methods could be a more reasonable solution than a single lysis method. For example, Liu et al. (2018) developed a combined lysis method consists of thermal, enzymatic, and chemical lysis techniques for whole genome amplification of single bacterial cell on an integrated microfluidic platform. And this combined lysis protocol achieved a 100% success for two kinds of bacterial, Gloeocapsa sp. and Sphaerocystis sp. which are extraordinarily difficult to lyse.
Single-cell nucleic acid analysis
Nucleic acid amplification for single-cell
Nucleic acid amplification is one of the critical steps in single-cell nucleic acid analysis. Unlike the analysis of cells population, the content of DNA or RNA in single-cell is negligible, normally the amount is 6–7 pg. Detection and sequencing methods cannot be accomplished from this scarce quantity of nucleic acids. Therefore, a high resolution and effective nucleic acid amplification is critical to single-cell analysis. Based on the working principle, single-cell nucleic acid amplification methods can be concluded as whole genome amplification (WGA), polymerase chain reaction (PCR), and isothermal amplification.
In single-cell nucleic acids analysis, whole genome amplifications are used for genome-wide research of DNA mutation by duplicating infinitesimal quantity of DNA. Major WGA methods include degenerate oligonucleotide-primed polymerase chain reaction (DOP-PCR), multiple displacement amplification (MDA), multiple annealing and looping-based amplification cycles (MALBAC), and linear amplification via transposon insertion (LIANTI).
By using a group of random primers, whole genome amplification could be achieved with PCR. As an initially launched tool, DOP-PCR was widely used in the early stage of whole genome analysis (Cheung and Nelson 1996). One weakness of DOP-PCR is the low genome coverage due to its exponential amplification strategy. MDA was invented in 2001 (Dean 2001), using Phi29 DNA polymerase and random primers to perform amplification. Compared with PCR, extension in MDA is achieved at a low reaction temperature in isothermal manner. MDA yields higher genome coverage than DOP-PCR, but often has the problem with sequence-dependent amplification bias. Some efforts have been made to overcome this weakness by scaling down the amplification volume, for example, with microfluidic chips (Marcy et al. 2007; Gole et al. 2013; Fu et al. 2019).
As discussed before, DOP-PCR or MDA tends to cause sequence-dependent amplification bias because it depends on the nonlinear exponential amplification mechanism. To solve this problem, Xiaoliang Sunney Xie’s team invented MALBAC (Zong et al. 2012) and LIANTI (Chen et al. 2017) in 2012 and 2017, respectively. Both of them are featured as a quasi-linear amplification method, and only the original DNA templates instead of copies from duplications will be duplicated in amplification. Therefore, accurate amplification with strictly-controlled copy number variations (CNV) can be achieved with these two methods (Yu et al. 2014).
WGA amplification method can be evaluated based on multiple factors, for example, coverage, uniformity, ADO (allelic drop out), and false positive rate (amplification preference or bias problem) as reported by other literature (Polzer and Klein 2010). As shown in Table 1, four typical WGA amplification methods are compared based on their critical performances (de Bourcy et al. 2014; Huang et al. 2015; Chen et al. 2017).
Table 1.
Comparison of critical performances of four WGA methods
| WGA methods | Coverage | Uniformity | Allelic dropout rate | False positive rate |
|---|---|---|---|---|
| DOP-PCR | Low | Medium | High | High |
| MDA | Medium | Low | Medium | Medium |
| MALBAC | Medium | High | Low | Low |
| LIANTI | High | High | Low | Low |
PCR is probably the most frequently used technique for nucleic acid amplification. Different from the genome-wide amplification mechanism of WGA, PCR is often used to amplify one or several primer-labeled fragments in the nucleic acid templets. Typically, PCR thermal cycling includes three stages: denaturation (~ 95 °C), annealing (~ 55 °C), and extension (~ 72 °C). With microfluidics-assisted PCR, the performance of mutation point detection (Gao et al. 2019; Rowlands et al. 2019) and gene-expression (Sarma et al. 2019) in single-cell level has been significantly improved with high sensitivity. Compared with WGA methods, PCR is able to more efficiently amplify and identify target genes in single cell level, which is beneficial to cancer diagnosis (Dalerba et al. 2011).
Different from PCR, isothermal amplification can be performed at a constant reaction temperature even with less time, which is quite helpful to reduce the system complexity (Asadi and Mollasalehi 2021). Loop-mediated-isothermal amplification (LAMP) and recombinase polymerase amplification (RPA) are two typical isothermal amplification methods which are widely used currently. The reaction temperature for LAMP is 60–65 °C. For RPA, this temperature is even lower, e.g., 37–39 °C. Compared with PCR, the system can be much simplified by adopting isothermal amplification into the microfluidic platform for single-cell-based nucleic acid analysis (Chung et al. 2019; Schulz et al. 2020; Gaiani et al. 2021). However, due to the longer application history, it is easier for PCR to achieve more stable and consistent analysis based on more available protocols compared with isothermal amplification.
Compared with nucleic acid amplification in large volumes (for example, μL), for single-cell-based nucleic acid amplification, one of the biggest challenges is how to achieve desired amplification efficiency which is heavily affected by the quite low quantity of original nucleic acid templates from just a single cell. In principle, amplification on microfluidic chip in the size of nanoliter will remarkably increase concentrations of genetic templates and other components, and therefore increases the probability of reaction between nucleic acid and enzyme, which will eventually improve the amplification efficiency. Therefore, compared with other platforms, it is more desirable for single-cell-based nucleic acid analysis to be performed on microfluidic chip to achieve smooth amplification with high successful probability.
Single-cell genetic analysis based on microfluidics
The emergence of next-generation sequencing (NGS) technology has made it possible for single-cell genetic analysis. The comprehensive genetic information behind a single cell can be systematically discovered by NGS. Recently, a number of review articles discussed single-cell analysis from the concepts of multi-omics (Hu et al. 2018; Leonavicius et al. 2019). Different genetic information can be respectively provided by genomics-, epigenomics-, and transcriptomics-based single cell analysis.
Both of genomics and epigenomics aim to study DNA. Genetic information of somatic cells is encoded on 46 chromosomes. For genomic-based single-cell analysis, due to cell proliferation and differentiation, related genetic variations will cause genotypic heterogeneity (Deng et al. 2019), for example, mutations, structural variations, aneuploidies, and recombination. Single-nucleotide variations (SNVs) and copy-number variations (CNVs) are two typical genomic mutations in genomic-based single-cell analysis, and both of them are quite important for precise cancer diagnosis and personnel medicine (Lim et al. 2020).
Epigenomics-based single-cell analysis is helpful for study about the epigenetic modifications, including DNA methylation, chromatin accessibility, histone modifications, chromosome conformation, and replication dynamics (Schwartzman and Tanay 2015). Epigenomics-based single-cell analysis on microfluidic platform is a valuable tool for genetic analysis. For example, chromatin immunoprecipitation (ChIP) is the most critical analysis method to study the complicated interactions between DNA and protein (O’Geen et al. 2006). Traditional ChIP analysis requires enough samples, typically 106–107 cells. Based on microfluidic chip, a novel ChIP-Seq method, which can handle even down to1000 mammalian cells, has been successfully developed by Shen et al. (2015).
Transcriptomics-based single-cell analysis is a method to discover the relationship between genotype and phenotype (Tang et al. 2011; Streets et al. 2014). Cell states and types can be identified by profiling RNA expression (Rheaume et al. 2018). And the main tool used in transcriptomics-based single-cell analysis is single-cell RNA sequencing, which including RNA extraction, RNA reverse transcription, cDNA amplification and next-generation sequencing. To avoid potential template loss, the first three steps are preferred to be performed on microfluidic platforms, especially in an enclosed chamber or a reactor, for example, in independent droplets (Ziegenhain et al. 2017). For single-cell RNA analysis, the problem with short-read length still needs to be further improved. To address this issue, Fan et al. (2020) developed a single-cell RNA sequencing technology based on the third-generation sequencing, e.g., nanopore sequencing. With this platform, 27,250 unannotated transcripts from 9338 genes were analyzed and identified.
So far, human cells are still the major targets for single-cell-based nucleic acid analysis due to its promising applications with cancer diagnosis and other genetic disease diagnosis. It has to be pointed out that, single cell analysis to other targets, for example, microbe is attracting more attention by providing significantly new insights in the fields. Usually, microbial single cell analysis is more difficult to perform because of the rigid cell wall and the extremely low amount of nucleic acid templates compared with eukaryotic cells. Recently, researchers developed different new technologies for microbial single cell analysis based on microfluidics (Liu and Walther-Antonio 2017; Tan and Toh 2020). Liu et al. (2019) achieved whole transcriptome amplification and sequencing for single Porphyromonas somerae cell based on a microfluidic platform, and a couple of interesting strategies have been properly developed to push forward this topic.
Platforms and integrated systems for single-cell nucleic acid analysis
Single-cell nucleic acid analysis can be performed with traditional tools in a more manual way since each step has to be implemented on a separate device. The manual operation mode for single-cell nucleic acid analysis normally holds low efficiency and high risk of template loss or contamination. Microfluidics-based single-cell nucleic acid analysis is able to manipulate single cell in an automatic, consecutive way in an enclose chip, which is helpful to improve the critical performance including efficiency, precision, and sensitivity. Especially, single cell can be more conveniently manipulated with a cell-size functional microstructure in microfluidic chip. Based on their characteristics, three typical platforms, e.g., valve-, microwell-, and droplet-based platforms with different working mechanisms are grouped. Each platform is able to provide different solutions for different procedures relative to single cell nucleic acid analysis, e.g., single cell trapping, cell lysis and purification, and nucleic acid amplification. Representative systems based on different platforms are also summarized to demonstrate how to perform single-cell nucleic acid analysis automatically in fully integrated devices, which are quite important to commercial applications of this technology.
Valve-based platform and systems
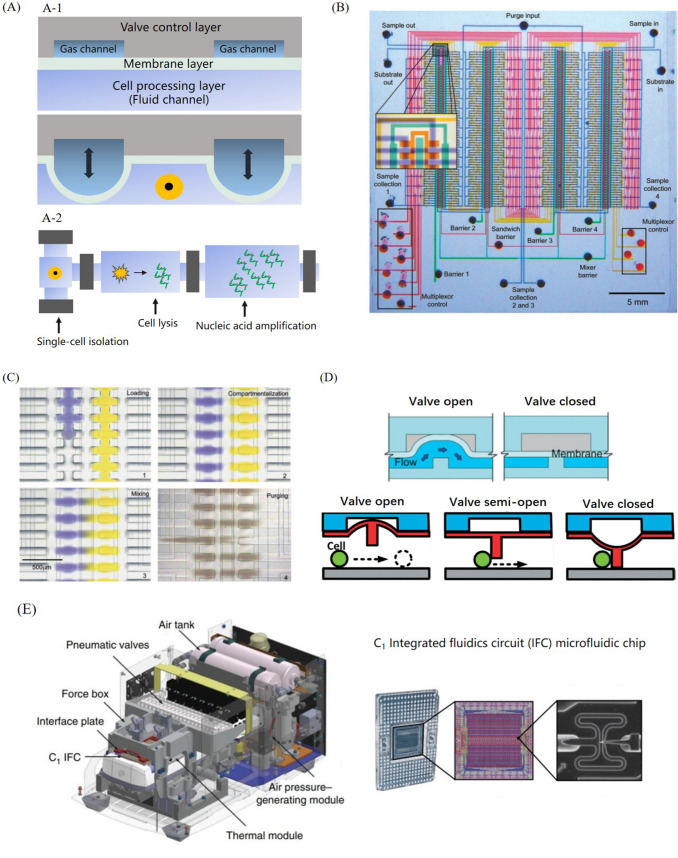
The cross-section view of an elastomeric valve in microfluidic chip was shown in Fig. 5A-1. Basic structure of a valve includes three layers, e.g., cell processing layer, membrane layer and valve control layer. This structure is normally fabricated by elastic material, such as PDMS. By changing air pressure in the gas channel of the valve control layer, the flexible membrane can be pushed down to seal the bottom cells in liquid.
Fig. 5.
Single-cell nucleic acid analysis on valve-based platform: A-1 schematic of membrane-based valve, A-2 schematic of single-cell nucleic acid analysis with valve-based microfluidic chip, B integrated chip for high throughput analysis with intensively integrated valves, reactors, and channels, C fluidic control on valve-based platform (Thorsen et al. 2002), D different working mechanisms of membrane-based valve (Li et al. 2019b; Sun et al. 2020), E integrated Fluidigm C1 system based on the concept of valve control (Pollen et al. 2014)
Originally, the multilayer elastomeric valve controlling technique was invented by the team of Stephen R. Quake in 2000 (Unger et al. 2000). Then they published a series of works boosting the development of valve-based platform applied in single-cell nucleic acid analysis. They have successively accomplished automation of nucleic acid purification, parallel picoliter RT-PCR analysis oriented to single-cell gene expression analysis, single-cell MDA efficiency analysis in 60 nL reactor, and single-cell WGA for De Novo mutation detection on valve-based platform (Hong et al. 2004; Marcus et al. 2006; Marcy et al. 2007; Wang et al. 2012; Gawad et al. 2016). With the flexible configuration of combined valves and reactors, valve-based platform is able to perform complicated bio-reaction relative to single cell nucleic acid analysis, as shown in Fig. 5A-2. High-throughput single-cell nucleic acid analysis can be achieved with a scaled up microfluidic chip integrated with a large number of valves, reactors, and channels, as shown in Fig. 5B. As shown in Fig. 5C, with valve-based platform, complicated fluid control, for example, reagent loading, mixing and driving can be conveniently achieve (Thorsen et al. 2002). For valve-based platform, elaborating mixing can be easily achieved by rapidly switching the state of the membrane between opening and closing, which can be applied to improve the efficiency of MDA amplification (Yang et al. 2014; Li et al. 2018b).
Whether the valve can completely seal the microfluidic channel is a key design point of valve-based platform. Quake’s team began to fabricate a rounded bottom of fluid channel for a perfect match between fluid channel and valve membrane (Unger et al. 2000). Most of the valve designs are following their rounded-bottom fluid channel strategy, but still there are some novel designs are proposed. A valve-base was added under the membrane for a tighter sealing. This valve-base can be fabricated on either the fluid channel bottom or the elastomeric membrane (Li et al. 2019b; Sun et al. 2020), as shown in Fig. 5D. To ensure the sealing performance, both the design concept and the fabrication technology of the membrane valve should be properly improved. The 2D layout of valve-based microfluidic chip, which including reaction units surrounded by fluid channels and gas control lines, always require elaborate design, especially when the available operation space for valving is limited. Also, the working efficiency of valve-based platform is limited by this limited planar space. Valve-based platform is more suitable to perform the complicated biochemical reaction with precise fluid control.
Based on the concept of valve-based platform, different integrated systems have been developed to perform single-cell nucleic acid analysis automatically and consecutively. For example, Zhang et al. displayed their valve-based system based on the pneumatic controlled microfluidic platform. By employing hydrodynamic flow capturing and valve controlling, this system can achieve a nearly 100% capture ratio of 32 cells for single-cell RNA sequencing (Zhang et al. 2019a). Pollen et al. (2014) introduced their Fluidigm C1 Single-Cell Auto Prep System in 2014, which is the world’s first commercial device of automatic single-cell capturing and nucleic acid amplification for downstream genetic analysis. As shown in Fig. 5E is the schematic of instrument integration, thermal module and the pneumatic valves system are two primary components reflected to the functions of valve controlling and thermal cycling of nucleic acid amplification. And on the integrated fluidics circuit (IFC) microfluidic chip, different kinds of reagents can be stored and injected into the center reaction unit for 96 single-cell isolation and RT-PCR amplification. However, due to the cell-size based isolation mechanism of IFC, C1 system required microfluidic chip renew for different cell samples (differed in the diameter of cell). And the throughput of C1 system is limited by the number of cell processing units as well. Later in 2015, the high throughout (HT) IFC was developed to simultaneously process 800 individual cells in a single run.
Microwell-based platform and systems
Microwell is usually adopted as a kind of vertical traps for single-cell isolation. Cells are loaded into microwells of their own size under the influence of gravity. The large-scale patterned microwell array has relatively high throughput with simple fabrication and low cost compared with other methods (Han et al. 2018).
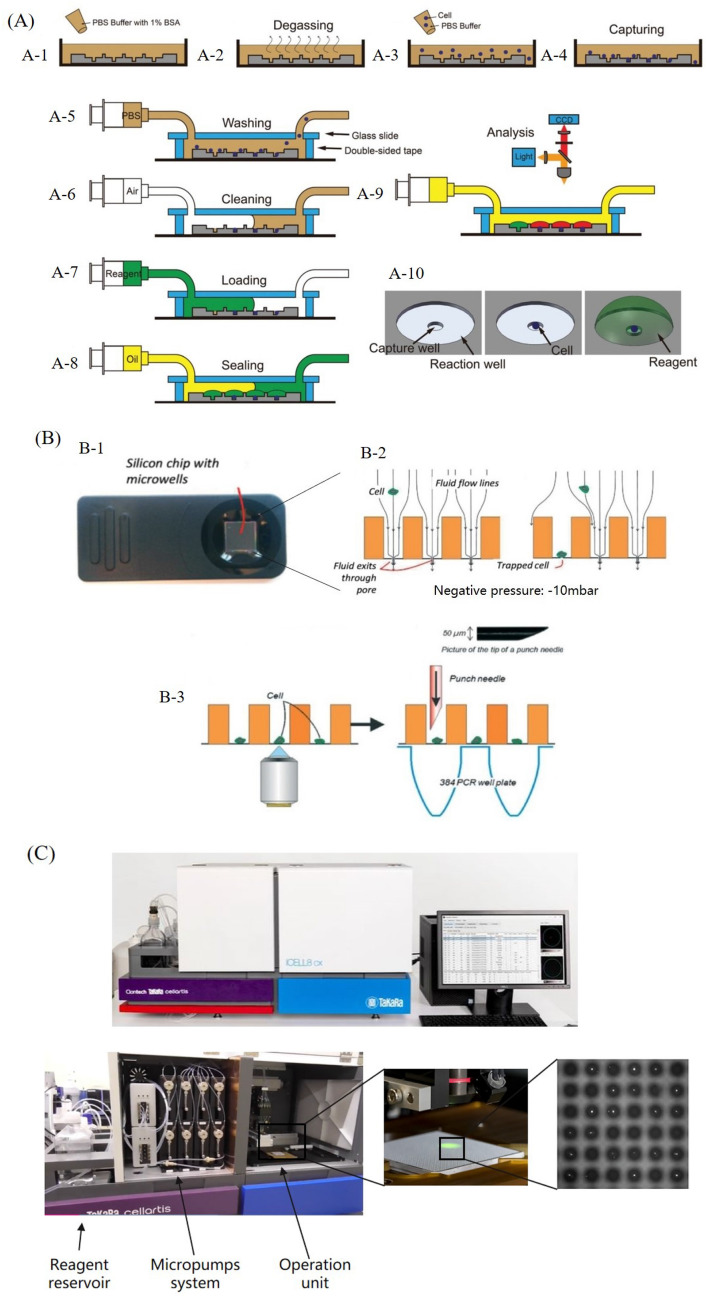
Trapping rate is the core indicator to evaluate functional performance of microwell platform, which is mainly depending on the geometry of microwell. In principle, the optimum size of the microwell to accommodate an individual cell is about 1 of depth/diameter ratio (Luan et al. 2020). Tang et al. designed a microwell chip with 200,000 wells (depth/diameter ratio: 20 μm/25 μm = 0.8) to screening metabolically active tumor cells. In this case, the fluorescent labeled single-cells were retrieved by micropipette for an off-chip sanger sequencing (Tang et al. 2017). However, for those on-chip nucleic acid analysis, the volume of cell-sized microwell is not enough for containing the reagents required by analysis protocol. So, the volume of the microwell often designed according to the total volume of all the reagents. In Gole et al.’s (2013) work, the single-cell whole genome amplification was occurred in a 12 nL microwell with depth/diameter ratio of 0.05 (20 μm/400 μm). For maintaining the single-cell capturing performance in microwells of small depth/diameter ratio, Wang et al. (2019) proposed a dual-well array chip which constructed small trapping well (20 μm in diameter) under the big reaction well (200 μm in diameter). The operation schematic of this method is shown in Fig. 6A, it should be noted that in step (A-8) oil can be used as sealing and isolation media for preventing reagent evaporation and contamination.
Fig. 6.
Single-cell nucleic acid analysis on microwell-based platform: A integrated dual-well array chip for single-cell analysis (Wang et al. 2019), B silicon chip with microwell for single cell nucleic acid analysis (Swennenhuis et al. 2015), C TAKARA ICELL8 system based on the concept of microwell
Although microwell platform is convenient and quick for operation, but the cells settling and capturing is still time-consuming. To accelerate single-cell settling and capturing, some external field-forces and structural modification has applied. Swennenhuis et al. fabricated a 6400 microwells chip with a single pore (5 μm in diameter) on the bottom. As shown in Fig. 6B, by adding a negative pressure of − 10 mbar, single-cell would be dragged into the microwell, once the pore in the bottom was clogged by cell, the following cells were distributed to other microwells (Swennenhuis et al. 2015). Then the cells were punched into the PCR tube for single-cell whole genome amplification and sanger sequencing for DNA analysis. Morimoto et al. combined electrodes with microwell chip to attract single-cell settled in the well. After fluorescent differentiation and picking by glass capillary, single CTC genetic analysis for EGFR mutation can be achieved (Morimoto et al. 2015).
Therefore, microwell-based platform is an ideal choice for applications with simple procedure. But in practical operation, the dimensions of microwell need to be carefully designed by considering the size of target cell. In addition, except for microbeads-based nucleic acid extraction, other nucleic acid extraction methods with microwell-based platform normally possess complicated procedure which need to be done with other tools, such as glass micropipette (Gole et al. 2013). Therefore, it is more desirable for microwell-based platform to perform relatively simple single cell nucleic acid analysis. Otherwise, it is difficult to integrate elaborated fluid control with micro-well reactors.
Similarly, based on the concept of microwell-based platform, different integrated systems have been developed to perform single cell nucleic acid analysis with the assistance from a couple of outside tools. Take ICELL8 cx single-cell system for example (Fig. 6C), the system is consisting of three main components, reagent reservoir for reagents storing and fueling, micropump system for reagents quantitative transportation, and operation unit for dispenser nozzle and camera control. Moreover, the dispense workflow control, real-time monitoring, and result analysis can be integrated and functioned in Cellstudio software (Goldstein et al. 2017). Although the microwell-based system is not a fully automatic instrument, but the real-time monitoring and the linear controllable dispenser nozzle can make the single-cell nucleic acid analysis more flexible. Users can put at most 8 samples into one microwell chip, and the diameters of sample cells can range from 5 to 100 μm. Meanwhile, users also can only select single-cell containing well as the target result for analysis that easily solved the empty or multiple cells containing problem of microwell chip. Moreover, the semi-open operation manner of microwell-based system is compatible with any genetic analysis protocols and techniques. Attayek et al. (2015) designed the automated microraft (each microwell re-fabricated with an additional concave substrate) platform and imaging analysis algorithm for single-cell temporary enhanced green fluorescent protein (EGFP) expression monitoring (Attayek et al. 2017). The analysis includes following steps, non-adherent K562 cells were transfected with a CRISPR-Cas9 plasmid which contained an EGFP reporter gene, transfected cells were seeded in microraft array, automated image tracking of EGFP fluorescence, target cell release, and further gene sequencing for gene-edited clone identification.
Droplet-based platform and systems
For valve-based platform, to achieve high-throughput analysis, a quite complicated system with a large number of valves, reactors and channels has to be developed because each cell needs to analyzed with its own flow control unit. It is even more difficult for microwell-based platform to achieve high-throughput analysis because of its own limitations. In comparison, it is easier for droplet-based platform to perform high-throughput analysis since each enclosed droplet works as an independent reaction unit. When a single cell is enclosed in a droplet, it can be separately analyzed based on the operation mechanisms of droplet microfluidics. Complicated reaction in the droplet can be performed by splitting, merging, heating, or illuminating the droplet in single-cell nucleic acid analysis.
Digital droplet PCR is one of the representative applications of droplet-based platform in genetic analysis. It is used to detect the specific DNA fragment of lysate from single-cell (Aigrain et al. 2016). Zeng et al. (2010) designed a droplet generator array for a multiplex single-cell PCR detection. Microbeads coated with multiple forward primers were confined in the droplets with the target cells. After PCR, the cells can be rapidly distinguished by flow cytometry. The detection ability of this method was proved by distinguishing E. coli O157 cells from high context of K12 cells with proportion of 1/105. And another PCR-activated cell sorting (PACS) based on single-cell droplet platform was demonstrated by Lim et al. (2015). They used digital droplet PCR and Dielectrophoretic for screening microbes of rare mutation, and conducted genome sequencing on the selected droplets for advance analysis.
Moreover, droplet-based single-cell analysis platform promises a relatively accurate whole genome amplification with high amplification yield (Fu et al. 2015). This emulsion single-cell MDA technique which claimed to be the first method can simultaneously achieve CNVs detection at size of 250-kb with 50-kb resolution, and SNVs detection with error rate less than 2 × 10–5. Another droplet MDA method provided by Yohei’s team improved the genome recovery rate in WGA from 59 to 89%, it proved that the picoliter reaction volume (67 pL) of droplets can decrease amplification bias of MDA effectively (Nishikawa et al. 2015). Depending on this method, a high-throughput single-cell sequencing reads acquisition mechanism with processing rate of 21,000 single-cell/h was built.
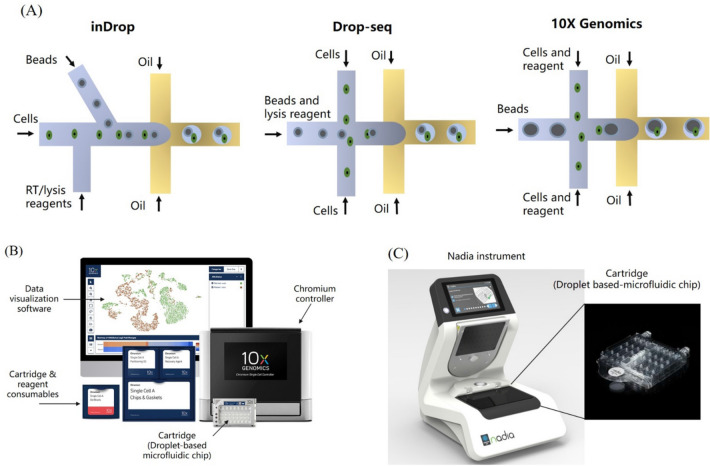
Droplet platform has proved the ability of single-cell nucleic acid amplification which is the basis of single-cell sequencing. By applying barcoded beads (Zilionis et al. 2017) for differentiating cells and unique molecular identifier (UMI) for molecular distinction, the next-generation sequencing method is perfectly implemented to droplet-based platform for single-cell genetic analysis (Lan et al. 2017; Salomon et al. 2019). For single-cell RNA sequencing on droplet platform, there are three representative methods, inDrop (Klein et al. 2015), Drop-seq (Macosko et al. 2015), and 10X Genomics Chromium (Zheng et al. 2017). The main difference of the three methods is the timing of barcode adding. As shown in Fig. 7A, Drop-Seq requires that both barcoded beads and cells are injected together into the channel with a low density that resulting a low utilization of sample cells. In inDrop method, barcoded beads are added after cell lysis which may decrease the specificity of sampling. And the 10X Genomics Chromium method combined the above two methods together leading to a better transcript capture efficiency and uniformity of sequencing library. More detailed discussion of these three methods can be found in Zhang et al. (2019b).
Fig. 7.
Single-cell nucleic acid analysis on droplet-based platform: A different droplet generation methods for single-cell RNA sequencing, B 10X Genomics Chromium system, C Dolomite Bio Nadia system
As discussed before, the two representatives of commercial droplet-based devices are Chromium system from 10X Genomics (Fig. 7B) and Nadia instrument from Dolomite Bio (Fig. 7C). The microfluidic cartridge is the core component in droplet-based instrument. The on-chip process including reagents and cell sample injection, barcoded bead and sample cell packaging in droplets, nucleic acid amplification for sequencing library preparation. In Chromium system, 100–80,000 + cells of eight samples can be prepared in 10–20 min. And 48,000 barcoded single-cell mRNA libraries can be finished by Nadia instrument. Recently, numbers of publications about single-cell sequencing and genetic analysis for cells subtypes identification have reported by utilizing these two instruments. With Chromium system, Ren et al. conducted single-cell RNA sequencing on 284 samples from 196 COVID-19 patients, to generate a comprehensive single-cell immune atlas with 1.46 million cells. This large scale of data help us to identify subtype changes of peripheral immune behind the clinical characteristics (Ren et al. 2021). Kim et al. (2020) identified gastrointestinal stromal cell populations by completing single-cell transcriptomic analysis through mechanisms of gastrointestinal stromal niches with the platform of Nadia instrument.
Other platforms and systems
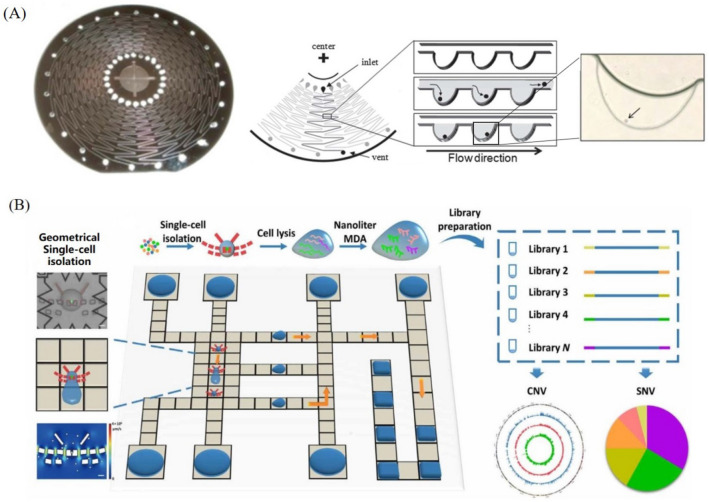
Beside the above typical platforms, there have also other platforms for single-cell nucleic acid analysis. For example, Lab-on-disk platform has been insensitively studied for automatic single-cell nucleic acid analysis. With the properly controlled centrifugal force, desirable flow control for single-cell analysis can be achieved with a properly designed disk chip. Furutani et al. (2012) designed a centrifugal platform for single-cell isolation and gene expression analysis by RT-PCR. As shown in Fig. 8A, 24 microchannels were fabricated on the disk chip, and each microchannel was designed in a curved shape from the center to the edge to impel fluid movement. 313 U-shaped microcavities were integrated on the outside of each microchannel to capture single cells. Particularly, in their work the thermal heated cell lysis occurred after the RT-PCR buffer loading, which will reduce the contamination between microcavities effectively. Moreover, centrifugal platform also can be utilized as a method to generate droplets. On the centrifugal-based droplet platform designed by Li et al. (2019c), 5μL of cell sample can be partitioned into droplets in 60 s with sample utilization more than 98%. Both of single-molecule digital droplet PCR and single-cell WGA are performed on this hybrid method to demonstrate the applicability of centrifugal platform for a rapid single-cell nucleic acid analysis.
Fig. 8.
Other platforms for single-cell nucleic acid analysis: A centrifugal disk chip for single-cell gene expression analysis (Furutani et al. 2012), B digital microfluidic chip for whole genome sequencing (Ruan et al. 2020)
Another emerging hybrid platform for single-cell nucleic acid analysis is digital microfluidics (DMF) that can manipulate droplets by electrowetting-on-dielectric (EWOD) mechanism. By controlling the droplets of cell samples and different reagents required in MDA, Ruan et al. (2020) achieved single-cell genomic detection of CNVs with minimal bin of 150-kb and SNVs with allele dropout rate of 5.2%. The schematics of DMF on-chip operation process and single-cell WGA analysis procedure was shown in Fig. 8B. The characteristics of different platforms for single-cell nucleic acid analysis are compared in Table 2.
Table 2.
Comparison of different microfluidic platforms for single-cell nucleic acid analysis
| Platforms | Advantages | Limitations |
|---|---|---|
| Valve-based |
Controllability of each step Functional integration |
Low throughput Complicated chip fabrication Complicated system |
| Microwell-based |
Straightforward operation Compatibility with other protocols |
Cross contamination Uncontrollable cell number |
| Droplet-based |
High throughput Concise system |
Elaborated flow control |
| Centrifugal-based |
Flexible flow control Parallel processing |
Limited throughput Complicated system |
| Digital microfluidics |
Controllability of each step Parallel processing |
Limited throughput Complicate system |
Conclusion and outlook
With the characteristics of high-throughput, trace-content sensitivity, miniaturization, and automation, microfluidic technology provides a competent platform for single-cell nucleic acid analysis. Analysis of genetic mutations between cells is making significant contribution to diagnosis and precision medicine for cancer and other diseases. In this review, we have summarized the major procedures of microfluidic single-cell nucleic acid analysis. The advantages and disadvantages of different microfluidic platforms have been analyzed, including valve-, microwell-, and droplet-based platforms. Moreover, the typical applications of integrated systems based on three platforms have also been discussed.
As a new technology, there are remaining unsolved problems with single-cell nucleic acid analysis based on microfluidics. Each microfluidics-based platform has its own advantages and limitations. For example, it is easy for droplet-based platform to perform high throughput analysis, but it is difficult to perform single-cell nucleic acid analysis with highly complicated procedures. In contrast, it is convenient for valve-based platform to perform single-cell nucleic acid analysis with complicated procedures because of the configurable valve-controlled flow network. However, for valve-based platform, it will be much more difficult to run high throughput (e.g., thousands of, or even more) analysis. It is easy to perform single-cell nucleic acid analysis with the microwell-based platform because of the straightforward working mechanism. However, it is difficult to perform automatic analysis with complicated procedures. Therefore, different platforms should be properly chosen and applied in different application for specific purposes.
In the future research, microfluidic technologies should be continually improved to provide more powerful tools for single-cell nucleic acid analysis. In principle, the whole process of single-cell nucleic acid analysis normally consists of a couple of steps, for example, cell trapping, cell lysis, nucleic acid purification, nucleic acid amplification and detection. One of the challenges is the contradiction between the highly integrated function and the high system complexity. In other words, when more functional modules are integrated into the microfluidic chip for a higher level of automatic analysis, the system complexity will be significantly increased. Therefore, microfluidic platforms based on new concepts need to be studied to perform fully-integrated analysis in a simpler and more reasonable way. Although droplet-based platform provides a potential solution for high-throughput analysis, the requirement for fully-integrated, highly-efficient, stable, consistent, reliable and high-throughput microfluidic platforms is a remaining challenge. In both basic research and clinical detection, normally a large number of single cells need to be analyzed to achieve accurate and comprehensive diagnosis. How to properly combine existing microfluidic platforms with the downstream detection module, for example, the sequencing module or other signal-readout module could be another challenge. Especially, based on the innovation of microfluidic technologies, to develop a fully-integrated, portable, and low-cost system to efficiently read the sequence of a single cell from the original cell sample within a limited time is a big challenge before us. Nevertheless, it is believed that more and more powerful tools based on microfluids will be invented based on the extraordinary progress which has been making in this field.
Funding
This research was funded by the National Natural Science Foundation of China (No. 81871505, 61971026), the Fundamental Research Funds for the Central Universities (XK1802-4), the National Science and Technology Major Project (2018ZX10732101-001-009), and the research fund to the top scientific and technological innovation team from Beijing University of Chemical Technology (No. buctylkjcx06).
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aigrain L, Gu Y, Quail MA. Quantitation of next generation sequencing library preparation protocol efficiencies using droplet digital PCR assays - a systematic comparison of DNA library preparation kits for Illumina sequencing. BMC Genom. 2016;17:458. doi: 10.1186/s12864-016-2757-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam MK, Koomson E, Zou H, et al. Recent advances in microfluidic technology for manipulation and analysis of biological cells (2007–2017) Anal Chim Acta. 2018;1044:29–65. doi: 10.1016/j.aca.2018.06.054. [DOI] [PubMed] [Google Scholar]
- Asadi R, Mollasalehi H. The mechanism and improvements to the isothermal amplification of nucleic acids, at a glance. Anal Biochem. 2021 doi: 10.1016/j.ab.2021.114260. [DOI] [PubMed] [Google Scholar]
- Attayek PJ, Hunsucker SA, Wang Y, et al. Array-based platform to select, release, and capture Epstein–barr virus-infected cells based on intercellular adhesion. Anal Chem. 2015;87:12281–12289. doi: 10.1021/acs.analchem.5b03579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attayek PJ, Waugh JP, Hunsucker SA, et al. Automated microraft platform to identify and collect non-adherent cells successfully gene-edited with CRISPR-Cas9. Biosens Bioelectron. 2017;91:175–182. doi: 10.1016/j.bios.2016.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Xing D, Tan L, et al. Single-cell whole-genome analyses by linear amplification via transposon insertion (LIANTI) Science. 2017;356:189–194. doi: 10.1126/science.aak9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung VG, Nelson SF. Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc Natl Acad Sci. 1996;93:14676–14679. doi: 10.1073/pnas.93.25.14676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MT, Kurabayashi K, Cai D. Single-cell RT-LAMP mRNA detection by integrated droplet sorting and merging. Lab Chip. 2019;19:2425–2434. doi: 10.1039/C9LC00161A. [DOI] [PubMed] [Google Scholar]
- Collins DJ, Neild A, deMello A, et al. The Poisson distribution and beyond: methods for microfluidic droplet production and single cell encapsulation. Lab Chip. 2015;15:3439–3459. doi: 10.1039/C5LC00614G. [DOI] [PubMed] [Google Scholar]
- Cong H, Loo F-C, Chen J, et al. Target trapping and in situ single-cell genetic marker detection with a focused optical beam. Biosens Bioelectron. 2019;133:236–242. doi: 10.1016/j.bios.2019.02.009. [DOI] [PubMed] [Google Scholar]
- Dalerba P, Kalisky T, Sahoo D, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bourcy CFA, De Vlaminck I, Kanbar JN, et al. A quantitative comparison of single-cell whole genome amplification methods. PLoS ONE. 2014;9:e105585. doi: 10.1371/journal.pone.0105585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB. Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng N-N, Wang W, Ju X-J, et al. Wetting-induced formation of controllable monodisperse multiple emulsions in microfluidics. Lab Chip. 2013;13:4047–4052. doi: 10.1039/C3LC50638J. [DOI] [PubMed] [Google Scholar]
- Deng Y, Finck A, Fan R. Single-cell omics analyses enabled by microchip technologies. Annu Rev Biomed Eng. 2019;21:365–393. doi: 10.1146/annurev-bioeng-060418-052538. [DOI] [PubMed] [Google Scholar]
- Fan HC, Fu GK, Fodor SPA. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347:1258367. doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- Fan X, Tang D, Liao Y, et al. Single-cell RNA-seq analysis of mouse preimplantation embryos by third-generation sequencing. PLOS Biol. 2020;18:e3001017. doi: 10.1371/journal.pbio.3001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Li C, Lu S, et al. Uniform and accurate single-cell sequencing based on emulsion whole-genome amplification. Proc Natl Acad Sci. 2015;112:11923–11928. doi: 10.1073/pnas.1513988112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Zhang F, Zhang X, et al. High-throughput single-cell whole-genome amplification through centrifugal emulsification and eMDA. Commun Biol. 2019;2:147. doi: 10.1038/s42003-019-0401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani S, Nagai H, Takamura Y, et al. Detection of expressed gene in isolated single cells in microchambers by a novel hot cell-direct RT-PCR method. Analyst. 2012;137:2951. doi: 10.1039/c2an15866c. [DOI] [PubMed] [Google Scholar]
- Gaiani G, Toldrà A, Andree KB, et al. Detection of Gambierdiscus and Fukuyoa single cells using recombinase polymerase amplification combined with a sandwich hybridization assay. J Appl Phycol. 2021;33:2273–2282. doi: 10.1007/s10811-021-02447-7. [DOI] [Google Scholar]
- Gao W, Zhang X, Yuan H, et al. EGFR point mutation detection of single circulating tumor cells for lung cancer using a micro-well array. Biosens Bioelectron. 2019;139:111326. doi: 10.1016/j.bios.2019.111326. [DOI] [PubMed] [Google Scholar]
- Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- Gierahn TM, Wadsworth MH, Hughes TK, et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods. 2017;14:395–398. doi: 10.1038/nmeth.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LD, Chen Y-JJ, Dunne J, et al. Massively parallel nanowell-based single-cell gene expression profiling. BMC Genom. 2017;18:519. doi: 10.1186/s12864-017-3893-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gole J, Gore A, Richards A, et al. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat Biotechnol. 2013;31:1126–1132. doi: 10.1038/nbt.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang R, Zhou Y, et al. Mapping the mouse cell atlas by Microwell-Seq. Cell. 2018;172:1091–1107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Hong JW, Studer V, Hang G, et al. A nanoliter-scale nucleic acid processor with parallel architecture. Nat Biotechnol. 2004;22:435–439. doi: 10.1038/nbt951. [DOI] [PubMed] [Google Scholar]
- Hu Y, An Q, Sheu K, et al. Single cell multi-omics technology: methodology and application. Front Cell Dev Biol. 2018;6:28. doi: 10.3389/fcell.2018.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K-W, Wu Y-C, Lee J-A, Chiou P-Y. Microfluidic integrated optoelectronic tweezers for single-cell preparation and analysis. Lab Chip. 2013;13:3721. doi: 10.1039/c3lc50607j. [DOI] [PubMed] [Google Scholar]
- Huang L, Ma F, Chapman A, et al. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genom Hum Genet. 2015;16:79–102. doi: 10.1146/annurev-genom-090413-025352. [DOI] [PubMed] [Google Scholar]
- Huang L, Zhao P, Wang W. 3D cell electrorotation and imaging for measuring multiple cellular biophysical properties. Lab Chip. 2018;18:2359–2368. doi: 10.1039/C8LC00407B. [DOI] [PubMed] [Google Scholar]
- Jokilaakso N, Salm E, Chen A, et al. Ultra-localized single cell electroporation using silicon nanowires. Lab Chip. 2013;13:336–339. doi: 10.1039/C2LC40837F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hong JW, Kim DP, et al. Nanowire-integrated microfluidic devices for facile and reagent-free mechanical cell lysis. Lab Chip. 2012;12:2914. doi: 10.1039/c2lc40154a. [DOI] [PubMed] [Google Scholar]
- Kim J-E, Fei L, Yin W-C, et al. Single cell and genetic analyses reveal conserved populations and signaling mechanisms of gastrointestinal stromal niches. Nat Commun. 2020;11:334. doi: 10.1038/s41467-019-14058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AM, Mazutis L, Akartuna I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Demaree B, Ahmed N, Abate AR. Single-cell genome sequencing at ultra-high-throughput with microfluidic droplet barcoding. Nat Biotechnol. 2017;35:640–646. doi: 10.1038/nbt.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry ZC, Giovanonni SJ, Quake SR, Blainey PC. Optofluidic cell selection from complex microbial communities for single-genome analysis. In: Methods in Enzymology. Amsterdam: Elsevier; 2013. pp. 61–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonavicius K, Nainys J, Kuciauskas D, Mazutis L. Multi-omics at single-cell resolution: comparison of experimental and data fusion approaches. Curr Opin Biotechnol. 2019;55:159–166. doi: 10.1016/j.copbio.2018.09.012. [DOI] [PubMed] [Google Scholar]
- Li ZG, Liu AQ, Klaseboer E, et al. Single cell membrane poration by bubble-induced microjets in a microfluidic chip. Lab Chip. 2013;13:1144. doi: 10.1039/c3lc41252k. [DOI] [PubMed] [Google Scholar]
- Li R, Zhou M, Li J, et al. Identifying EGFR-expressed cells and detecting EGFR multi-mutations at single-cell level by microfluidic chip. Nano-Micro Lett. 2018;10:16. doi: 10.1007/s40820-017-0168-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhou M, Yue C, et al. Multiple single cell screening and DNA MDA amplification chip for oncogenic mutation profiling. Lab Chip. 2018;18:723–734. doi: 10.1039/C7LC00924K. [DOI] [PubMed] [Google Scholar]
- Li L, Wu P, Luo Z, et al. Dean flow assisted single cell and bead encapsulation for high performance single cell expression profiling. ACS Sens. 2019;4:1299–1305. doi: 10.1021/acssensors.9b00171. [DOI] [PubMed] [Google Scholar]
- Li R, Jia F, Zhang W, et al. Device for whole genome sequencing single circulating tumor cells from whole blood. Lab Chip. 2019;19:3168–3178. doi: 10.1039/C9LC00473D. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang D, Ruan W, et al. Centrifugal-driven droplet generation method with minimal waste for single-cell whole genome amplification. Anal Chem. 2019;91:13611–13619. doi: 10.1021/acs.analchem.9b02786. [DOI] [PubMed] [Google Scholar]
- Lim SW, Tran TM, Abate AR. PCR-activated cell sorting for cultivation-free enrichment and sequencing of rare microbes. PLoS ONE. 2015;10:e0113549. doi: 10.1371/journal.pone.0113549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Lin Y, Navin N. Advancing cancer research and medicine with single-cell genomics. Cancer Cell. 2020;37:456–470. doi: 10.1016/j.ccell.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Walther-Antonio M. Microfluidics: a new tool for microbial single cell analyses in human microbiome studies. Biomicrofluidics. 2017;11:061501. doi: 10.1063/1.5002681. [DOI] [Google Scholar]
- Liu W, Zhu Y. “Development and application of analytical detection techniques for droplet-based microfluidics”-a review. Anal Chim Acta. 2020;1113:66–84. doi: 10.1016/j.aca.2020.03.011. [DOI] [PubMed] [Google Scholar]
- Liu Y, Schulze-Makuch D, de Vera J-P, et al. The development of an effective bacterial single-cell lysis method suitable for whole genome amplification in microfluidic platforms. Micromachines. 2018;9:367. doi: 10.3390/mi9080367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Jeraldo P, Jang JS, et al. Bacterial single cell whole transcriptome amplification in microfluidic platform shows putative gene expression heterogeneity. Anal Chem. 2019;91:8036–8044. doi: 10.1021/acs.analchem.8b04773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, He H, Zheng S-Y. Microfluidics in single-cell virology: technologies and applications. Trends Biotechnol. 2020 doi: 10.1016/j.tibtech.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Q, Macaraniag C, Zhou J, Papautsky I. Microfluidic systems for hydrodynamic trapping of cells and clusters. Biomicrofluidics. 2020;14:031502. doi: 10.1063/5.0002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus JS, Anderson WF, Quake SR. Parallel picoliter RT-PCR assays using microfluidics. Anal Chem. 2006;78:956–958. doi: 10.1021/ac0513865. [DOI] [PubMed] [Google Scholar]
- Marcy Y, Ishoey T, Lasken RS, et al. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3:e155. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto A, Mogami T, Watanabe M, et al. High-density dielectrophoretic microwell array for detection, capture, and single-cell analysis of rare tumor cells in peripheral blood. PLoS ONE. 2015;10:e0130418. doi: 10.1371/journal.pone.0130418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TW, Zhang Q, Naler LB, et al. Recent advances in the use of microfluidic technologies for single cell analysis. Analyst. 2018;143:60–80. doi: 10.1039/C7AN01346A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam AS, Chaligne R, Landau DA. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat Rev Genet. 2021;22:3–18. doi: 10.1038/s41576-020-0265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan L, Jiang Z, Wei X. Emerging microfluidic devices for cell lysis: a review. Lab Chip. 2014;14:1060. doi: 10.1039/c3lc51133b. [DOI] [PubMed] [Google Scholar]
- Nishikawa Y, Hosokawa M, Maruyama T, et al. Monodisperse picoliter droplets for low-bias and contamination-free reactions in single-cell whole genome amplification. PLoS ONE. 2015;10:e0138733. doi: 10.1371/journal.pone.0138733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen H, Nicolet CM, Blahnik K, et al. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 2006;41:577–580. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Shuga J, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polzer B, Klein CA. Chapter 8. Looking at the DNA of a single cell. In: Bontoux N, Potier M-C, editors. Nanoscience & nanotechnology series. Cambridge: Royal Society of Chemistry; 2010. pp. 73–80. [Google Scholar]
- Qin Y, Wu L, Schneider T, et al. A self-digitization dielectrophoretic (SD-DEP) chip for high-efficiency single-cell capture, on-demand compartmentalization, and downstream nucleic acid analysis. Angew Chem Int Ed. 2018;57:11378–11383. doi: 10.1002/anie.201807314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Wen W, Fan X, et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184:1895–1913.e19. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheaume BA, Jereen A, Bolisetty M, et al. Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat Commun. 2018;9:2759. doi: 10.1038/s41467-018-05134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands V, Rutkowski AJ, Meuser E, et al. Optimisation of robust singleplex and multiplex droplet digital PCR assays for high confidence mutation detection in circulating tumour DNA. Sci Rep. 2019;9:12620. doi: 10.1038/s41598-019-49043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q, Ruan W, Lin X, et al. Digital-WGS: automated, highly efficient whole-genome sequencing of single cells by digital microfluidics. Sci Adv. 2020;6:eabd6454. doi: 10.1126/sciadv.abd6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon R, Kaczorowski D, Valdes-Mora F, et al. Droplet-based single cell RNAseq tools: a practical guide. Lab Chip. 2019;19:1706–1727. doi: 10.1039/C8LC01239C. [DOI] [PubMed] [Google Scholar]
- Samad T, Wu SM. Single cell RNA sequencing approaches to cardiac development and congenital heart disease. Semin Cell Dev Biol. 2021 doi: 10.1016/j.semcdb.2021.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma M, Lee J, Ma S, et al. A diffusion-based microfluidic device for single-cell RNA-seq. Lab Chip. 2019;19:1247–1256. doi: 10.1039/C8LC00967H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz M, Calabrese S, Hausladen F, et al. Point-of-care testing system for digital single cell detection of MRSA directly from nasal swabs. Lab Chip. 2020;20:2549–2561. doi: 10.1039/D0LC00294A. [DOI] [PubMed] [Google Scholar]
- Schwartzman O, Tanay A. Single-cell epigenomics: techniques and emerging applications. Nat Rev Genet. 2015;16:716–726. doi: 10.1038/nrg3980. [DOI] [PubMed] [Google Scholar]
- Shen J, Jiang D, Fu Y, et al. H3K4me3 epigenomic landscape derived from ChIP-Seq of 1 000 mouse early embryonic cells. Cell Res. 2015;25:143–147. doi: 10.1038/cr.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields CW, IV, Wang JL, Ohiri KA, et al. Magnetic separation of acoustically focused cancer cells from blood for magnetographic templating and analysis. Lab Chip. 2016;16:3833–3844. doi: 10.1039/C6LC00719H. [DOI] [PubMed] [Google Scholar]
- Streets AM, Zhang X, Cao C, et al. Microfluidic single-cell whole-transcriptome sequencing. Proc Natl Acad Sci. 2014;111:7048–7053. doi: 10.1073/pnas.1402030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Tayagui A, Garrill A, Nock V. Microfluidic platform for integrated compartmentalization of single zoospores, germination and measurement of protrusive force generated by germ tubes. Lab Chip. 2020;20:4141–4151. doi: 10.1039/D0LC00752H. [DOI] [PubMed] [Google Scholar]
- Swennenhuis JF, Tibbe AGJ, Stevens M, et al. Self-seeding microwell chip for the isolation and characterization of single cells. Lab Chip. 2015;15:3039–3046. doi: 10.1039/C5LC00304K. [DOI] [PubMed] [Google Scholar]
- Tan W-H, Takeuchi S. A trap-and-release integrated microfluidic system for dynamic microarray applications. Proc Natl Acad Sci. 2007;104:1146–1151. doi: 10.1073/pnas.0606625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H-Y, Toh Y-C. What can microfluidics do for human microbiome research? Biomicrofluidics. 2020;14:051303. doi: 10.1063/5.0012185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Lao K, Surani MA. Development and applications of single-cell transcriptome analysis. Nat Methods. 2011;8:S6–S11. doi: 10.1038/nmeth.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Wang Z, Li Z, et al. High-throughput screening of rare metabolically active tumor cells in pleural effusion and peripheral blood of lung cancer patients. Proc Natl Acad Sci. 2017;114:2544–2549. doi: 10.1073/pnas.1612229114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen T, Maerkl SJ, Quake SR. Microfluidic large-scale integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- Unger MA, Chou H-P, Thorsen T, et al. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- VanInsberghe M, Zahn H, White AK, et al. Highly multiplexed single-cell quantitative PCR. PLoS ONE. 2018;13:e0191601. doi: 10.1371/journal.pone.0191601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Ren L, Liu W, et al. Fluorescence quantification of intracellular materials at the single-cell level by an integrated dual-well array microfluidic device. Analyst. 2019;144:2811–2819. doi: 10.1039/C9AN00153K. [DOI] [PubMed] [Google Scholar]
- White AK, VanInsberghe M, Petriv OI, et al. High-throughput microfluidic single-cell RT-qPCR. Proc Natl Acad Sci. 2011;108:13999–14004. doi: 10.1073/pnas.1019446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AK, Heyries KA, Doolin C, et al. High-throughput microfluidic single-cell digital polymerase chain reaction. Anal Chem. 2013;85:7182–7190. doi: 10.1021/ac400896j. [DOI] [PubMed] [Google Scholar]
- Wood DK, Weingeist DM, Bhatia SN, Engelward BP. Single cell trapping and DNA damage analysis using microwell arrays. Proc Natl Acad Sci. 2010;107:10008–10013. doi: 10.1073/pnas.1004056107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woyke T, Doud DFR, Schulz F. The trajectory of microbial single-cell sequencing. Nat Methods. 2017;14:1045–1054. doi: 10.1038/nmeth.4469. [DOI] [PubMed] [Google Scholar]
- Wu C, Chen R, Liu Y, et al. A planar dielectrophoresis-based chip for high-throughput cell pairing. Lab Chip. 2017;17:4008–4014. doi: 10.1039/C7LC01082F. [DOI] [PubMed] [Google Scholar]
- Xu X, Wang J, Wu L, et al. Microfluidic single-cell omics analysis. Small. 2020;16:1903905. doi: 10.1002/smll.201903905. [DOI] [PubMed] [Google Scholar]
- Yang Y, Swennenhuis JF, Rho HS, et al. Parallel single cancer cell whole genome amplification using button-valve assisted mixing in nanoliter chambers. PLoS ONE. 2014;9:e107958. doi: 10.1371/journal.pone.0107958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousuff C, Ho E, Hussain KI, Hamid N. Microfluidic platform for cell isolation and manipulation based on cell properties. Micromachines. 2017;8:15. doi: 10.3390/mi8010015. [DOI] [Google Scholar]
- Yu Z, Lu S, Huang Y. Microfluidic Whole genome amplification device for single cell sequencing. Anal Chem. 2014;86:9386–9390. doi: 10.1021/ac5032176. [DOI] [PubMed] [Google Scholar]
- Yun S-S, Yoon SY, Song M-K, et al. Handheld mechanical cell lysis chip with ultra-sharp silicon nano-blade arrays for rapid intracellular protein extraction. Lab Chip. 2010;10:1442. doi: 10.1039/b925244d. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Novak R, Shuga J, et al. High-performance single cell genetic analysis using microfluidic emulsion generator arrays. Anal Chem. 2010;82:3183–3190. doi: 10.1021/ac902683t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Chou C-K, Xia X, et al. Block-cell-printing for live single-cell printing. Proc Natl Acad Sci. 2014;111:2948–2953. doi: 10.1073/pnas.1313661111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Gao M, Chong Z, et al. Single-cell isolation by a modular single-cell pipette for RNA-sequencing. Lab Chip. 2016;16:4742–4748. doi: 10.1039/C6LC01241H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Xu H, Huang Y, et al. Improving single-cell transcriptome sequencing efficiency with a microfluidic phase-switch device. Analyst. 2019;144:7185–7191. doi: 10.1039/C9AN00823C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li T, Liu F, et al. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-Seq systems. Mol Cell. 2019;73:130–142.e5. doi: 10.1016/j.molcel.2018.10.020. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wei X, Wei Y, et al. The up-to-date strategies for the isolation and manipulation of single cells. Talanta. 2020;218:121147. doi: 10.1016/j.talanta.2020.121147. [DOI] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Shao N, Bessa de Castro R, et al. Evaluation of single-cell cytokine secretion and cell-cell interactions with a hierarchical loading microwell chip. Cell Rep. 2020;31:107574. doi: 10.1016/j.celrep.2020.107574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wang Y, Chen P, et al. Highly efficient microfluidic device for cell trapping and pairing towards cell-cell communication analysis. Sens Actuators B Chem. 2019;283:685–692. doi: 10.1016/j.snb.2018.12.078. [DOI] [Google Scholar]
- Ziegenhain C, Vieth B, Parekh S, et al. Comparative analysis of single-cell RNA sequencing methods. Mol Cell. 2017;65:631–643.e4. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- Zilionis R, Nainys J, Veres A, et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc. 2017;12:44–73. doi: 10.1038/nprot.2016.154. [DOI] [PubMed] [Google Scholar]
- Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]